ABSTRACT
The heterotrimeric pre-mRNA retention and splicing (RES) complex, consisting of Bud13p, Snu17p and Pml1p, promotes splicing and nuclear retention of a subset of intron-containing pre-mRNAs. Yeast cells deleted for individual RES genes show growth defects that are exacerbated at elevated temperatures. Although the growth phenotypes correlate to the splicing defects in the individual mutants, the underlying mechanism is unknown. Here, we show that the temperature sensitive (Ts) growth phenotype of bud13Δ and snu17Δ cells is a consequence of inefficient splicing of MED20 pre-mRNA, which codes for a subunit of the Mediator complex; a co-regulator of RNA polymerase II transcription. The MED20 pre-mRNA splicing defect is less pronounced in pml1Δ cells, explaining why they grow better than the other 2 RES mutants at elevated temperatures. Inactivation of the cytoplasmic nonsense-mediated mRNA decay (NMD) pathway in the RES mutants leads to accumulation of MED20 pre-mRNA, indicating that inefficient nuclear retention contributes to the growth defect. Further, the Ts phenotype of bud13Δ and snu17Δ cells is partially suppressed by the inactivation of NMD, showing that the growth defects are augmented by the presence of a functional NMD pathway. Collectively, our results demonstrate an important role of the RES complex in maintaining the Med20p levels.
KEYWORDS: Med20, mediator, NMD, RES complex, splicing
Introduction
The removal of introns from RNA polymerase II-transcribed pre-mRNAs is catalyzed by the spliceosome, a large ribonucleoprotein complex assembled from 5 small nuclear ribonucleoprotein particles (snRNPs) and numerous protein factors.1 Spliceosomes are assembled through a stepwise recruitment of snRNP and non-snRNP factors to pre-mRNA where the introns are defined by short conserved sequences, including the 5′ splice site, branch point, and 3′ splice site.1 During assembly and the subsequent 2 catalytic steps of splicing, the spliceosome undergoes multiple rearrangements of its structure and composition, which involves both recruitment and release of individual factors.1
The Saccharomyces cerevisiae pre-mRNA retention and splicing (RES) complex is a heterotrimeric non-snRNP complex that associates with spliceosomes before the first catalytic step.2 The RES complex is composed of Bud13p, Snu17p and Pml1p of which Snu17p is the core subunit that binds the other 2 factors.2-5 RES factors are found in stoichiometric amounts in activated spliceosomes where they interact with intronic pre-mRNA sequences,6,7 as well as with the U2 snRNP.8-11 Homologues to the RES subunits are found in humans and these proteins also associate with spliceosomes before the first catalytic step.12,13 In yeast, the RES complex is not essential for splicing as strains deleted for BUD13, SNU17, or PML1 are viable.2 Although genome-wide analyses of bud13Δ and snu17Δ strains revealed that they show increased accumulation of many intron-containing pre-mRNAs14,15 direct tests have suggested that the RES complex preferentially promotes splicing of a subset of pre-mRNAs. The RES complex was originally proposed to enhance splicing of transcripts in which the 5′ splice site does not conform to the consensus sequence,2,16,17 but more recent studies have shown that RES-regulated transcripts also include transcripts with consensus 5′ splice sites.18-20 In addition to influencing splicing, the inactivation of any RES subunit leads to export of unspliced pre-mRNAs to the cytoplasm.2 This finding in combination with the observation that the absence of Pml1p can, under some circumstances, induce pre-mRNA leakage without any apparent defect in splicing indicates that the RES complex may also have an important role in the nuclear retention of unspliced pre-mRNAs.2
The pre-mRNAs that enter the cytoplasm are typically degraded by the nonsense-mediated mRNA decay (NMD) pathway, as the presence of an intron often leads to the generation or inclusion of a premature translation termination codon.21-24 The destabilization of transcripts containing premature translation termination codons requires their translation and a set of trans-acting factors, including the Upf1, Upf2, and Upf3 proteins.25 By eliminating nonsense-containing transcripts, the NMD pathway ensures that protein-encoding transcripts encompass proper open reading frames (ORFs).25
Although the RES complex is dispensable for the growth of yeast cells, the inactivation of individual RES genes induces several different phenotypes. All 3 RES mutants show growth defects that are exacerbated at elevated temperatures but the cause of these phenotypes are not known.2 The individual RES mutants show somewhat different growth phenotypes, i.e., cells deleted for PML1 grow significantly better than cells deleted for BUD13 or SNU17 and snu17Δ mutants grow slightly better than bud13Δ cells.2 As the splicing defects are more pronounced in bud13Δ and snu17Δ than in pml1Δ cells, the growth phenotypes of the individual mutants correlate to the importance of the respective factor in splicing.2 In addition to the growth phenotypes, diploid cells deleted for BUD13 or SNU17 show a haploid-like bud-site selection pattern and an increased ability to mate as MATα cells.,19,26 These phenotypes are caused by inefficient splicing of MATa1 pre-mRNA and the consequent effect of reduced MATa1p levels on the ability to turn off haploid-specific genes and allow diploid gene expression.18,19 Another phenotype associated with the absence of Bud13p or Snu17p is a severely reduced level of the modified nucleoside N4-acetylcytidine (ac4C) in tRNA.20 At elevated temperatures, a reduction in ac4C levels is also observed in cells lacking Pml1p. This tRNA modification defect is caused by inefficient splicing of TAN1 pre-mRNA20 which encodes a tRNA binding protein required for formation of ac4C.27 The absence of individual RES factors also leads to degradation of TAN1 pre-mRNA by the cytoplasmic nonsense-mediated mRNA decay (NMD) pathway, indicating that poor nuclear retention may contribute to the tRNA modification defect.20
The notion that the RES complex enhances splicing of a distinct subset of pre-mRNAs suggests that the growth defects of RES mutants may be a consequence of inefficient processing of a specific transcript. To investigate this hypothesis, we screened for genes that in high dosage suppress the temperature sensitive (Ts) phenotype of bud13Δ cells. Here, we provide a detailed analysis of one dosage suppressor and show that the Ts phenotype of bud13Δ and snu17Δ mutants is largely a consequence of inefficient MED20 pre-mRNA processing.
Results
Increased dosage of the MED20 gene suppresses the Ts phenotype of bud13Δ and snu17Δ cells
To investigate the cause of the growth phenotypes of RES mutants, we screened for genes that in high dosage suppress the growth defect of bud13Δ cells at 37°C. Consistent with the notion that the growth defect may be a consequence of inefficient splicing of a specific transcript, we found that high-copy plasmids harboring the intron-containing MED20 (SRB2) gene partially suppressed the Ts phenotype (Fig. 1A). The MED20 gene harbors a 101 nt intron at the 5′ part of the ORF and codes for a nonessential subunit of the Mediator complex; an important co-regulator of RNA polymerase II transcription.28-30 Cells deleted for MED20 show growth defects that are similar to those observed for bud13Δ cells, i.e., they grow slowly and this phenotype is exacerbated at elevated temperatures29 (Fig. S1).
Figure 1.
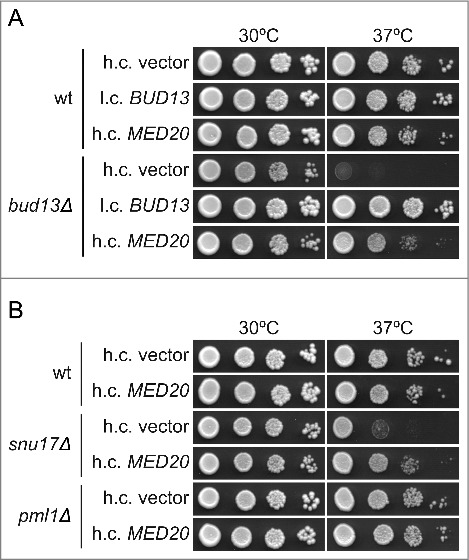
Increased dosage of the MED20 gene suppresses the temperature sensitive growth defect of bud13Δ and snu17Δ mutants. (A) Growth of the wild-type (UMY2219) and bud13Δ (MJY546) strains carrying the indicated high-copy (h.c.) or low-copy (l.c.) URA3 plasmids. The strains were grown over-night in liquid SC-ura medium, serially diluted, spotted on SC-ura plates, and incubated at 30°C or 37°C for 2 d. (B) Growth of the wild-type (UMY2219), snu17Δ (MJY548), and pml1Δ (MJY535) strains carrying the indicated plasmids. The strains were grown over-night in liquid SC-ura medium, serially diluted, spotted on SC-ura plates, and incubated at 30°C or 37°C for 2 d.
Individual RES mutants show somewhat different growth characteristics, i.e., cells lacking Pml1p grow considerably better than those lacking Bud13p or Snu17p and snu17Δ mutants grow slightly better than bud13Δ cells.2 To assess if the suppression induced by increased MED20 dosage is relevant for RES-deficient cells in general, we introduced the high-copy MED20 plasmid into cells deleted for SNU17 or PML1. Increased dosage of MED20 partially suppressed the Ts phenotype of snu17Δ cells whereas no apparent suppression was observed for the pml1Δ mutant (Fig. 1B). Thus, the 2 RES mutants with the strongest growth defects are suppressed by increased MED20 dosage.
Inefficient MED20 pre-mRNA splicing accounts for the Ts phenotype of bud13Δ and snu17Δ cells
To investigate if the RES complex is important for splicing of MED20 pre-mRNA, we used northern blotting to analyze MED20 transcripts in wild-type, bud13Δ, snu17Δ, and pml1Δ cells grown at 30°C or 37°C. The blots were also probed for the intron-less PGK1 mRNA and 18S rRNA, which served as a loading control. The analyses revealed a decreased abundance of spliced and increased abundance of unspliced MED20 transcripts in all 3 mutants (Fig. 2A). As expected, the splicing defect was more pronounced in the bud13Δ and snu17Δ mutants than in the pml1Δ strain at both 30°C and 37°C (Fig. 2A and B). The MED20 mRNA level in the individual RES mutants was comparable at 30°C and 37°C, indicating that the growth temperature does not influence the efficiency of MED20 pre-mRNA splicing.
Figure 2.
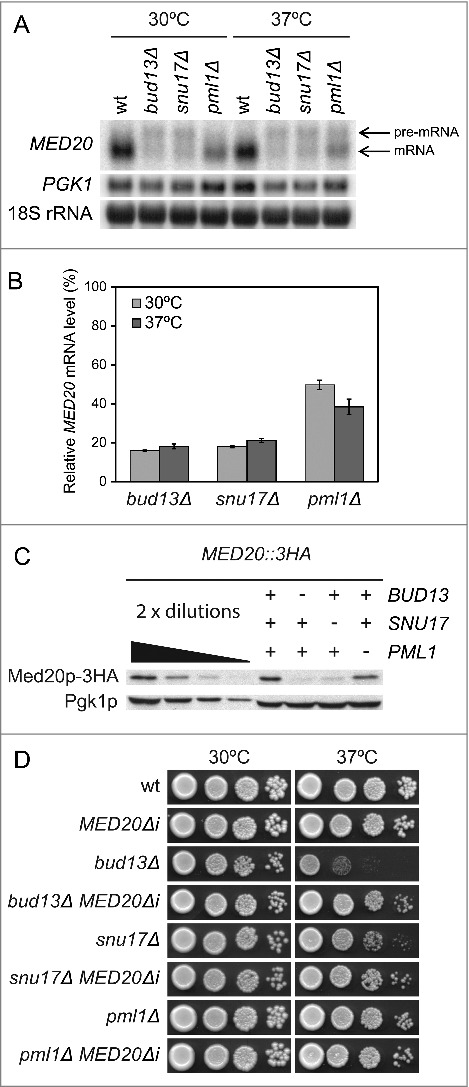
The RES complex promotes splicing of MED20 pre-mRNA. (A) Northern analysis of total RNA isolated from wild-type (UMY2219), bud13Δ (MJY546), snu17Δ (MJY548), and pml1Δ (MJY535) cells grown in YEPD medium at 30°C or 37°C. The blot was probed for MED20 and PGK1 transcripts using randomly labeled DNA fragments. 18S rRNA was detected using an oligonucleotide probe. (B) Levels of spliced MED20 mRNA in RES mutants. The MED20 mRNA signal was normalized to the corresponding 18S rRNA signal and the value for each strain expressed relative to that for the wild-type at the same temperature, which was set to 100%. The MED20 mRNA levels represent the average from the experiment shown in Fig. 2A and 2 additional independent experiments. The standard deviation is indicated. (C) Western analysis of the indicated strains (MJY775, MJY779, MJY800, and MJY802) grown in YEPD medium at 30°C. Monoclonal antibodies against HA or Pgk1p were used to detect the indicated proteins. Control experiments showed that the sequence for the 3HA-tag did not influence the MED20 pre-mRNA splicing efficiency in either wild-type or RES-deficient cells (data not shown). (D) Effects of the MED20Δi allele on growth of bud13Δ, snu17Δ, and pml1Δ cells. The wild-type (UMY2219), MED20Δi (MJY736), bud13Δ (MJY546), bud13Δ MED20Δi (MJY738), snu17Δ (MJY548), snu17Δ MED20Δi (MJY809), pml1Δ (MJY535), and pml1Δ MED20Δi (MJY808) strains were grown over-night in YEPD medium, serially diluted, spotted on YEPD plates, and incubated at 30°C or 37°C for 2 d.
To assess if the reduction in spliced MED20 mRNA in the RES mutants correlates with reduced Med20 protein levels, we performed western blot analyses of strains in which the DNA sequence for 3 tandem influenza virus hemagglutinin epitopes (3HA) was fused to the endogenous MED20 ORF. In good agreement with the mRNA levels, the abundance of Med20 protein was reduced in all 3 RES mutants and the Med20p levels were considerably lower in bud13Δ and snu17Δ than in pml1Δ cells (Fig. 2C). Thus, our results suggested a model where the Ts growth phenotypes of bud13Δ and snu17Δ cells are caused by inefficient MED20 pre-mRNA splicing and the consequent reduction in Med20p levels.
If inefficient MED20 pre-mRNA splicing contributes to the growth defect of bud13Δ and snu17Δ cells, then the growth defect should be suppressed by expressing an allele of MED20 that lacks the intron. Because pml1Δ cells show only a modest growth defect, which is not suppressed by increased MED20 dosage, the removal of the intron should have no or small effects. To test these predictions, we removed the sequence for the intron in the endogenous MED20 gene and combined this MED20Δi allele with a bud13Δ, snu17Δ, or pml1Δ mutation. Northern blot analyses revealed that the introduction of the MED20Δi allele into the RES mutants generated MED20 mRNA levels that were comparable to those normally observed in the wild-type (Fig. S2). As predicted, the growth defects of bud13Δ and snu17Δ cells at 37°C were largely suppressed by the MED20Δi allele whereas no effect was observed in pml1Δ cells (Fig. 2D and Table S1). The MED20Δi allele did, however, not counteract the growth defect of bud13Δ and snu17Δ cells at 30°C (Fig. 2D and Table S1), indicating that inefficient splicing of another transcript(s) determines the growth behavior at this temperature. We conclude that inefficient MED20 pre-mRNA splicing is the primary cause for the Ts phenotype of bud13Δ and snu17Δ cells.
Inactivation of NMD in the RES mutants leads to accumulation of MED20 pre-mRNA
In addition to its role in pre-mRNA splicing, the RES complex is thought to promote nuclear retention of unspliced pre-mRNAs.2 Consistent with this notion, we previously showed that the absence of individual RES factors leads to degradation of TAN1 pre-mRNA by the cytoplasmic NMD pathway.20 The low total abundance of MED20 transcripts in the RES mutants suggested that the MED20 pre-mRNA may also be exported to the cytoplasm and degraded by NMD. To assess this possibility, we inactivated NMD by deleting the UPF1 gene31 and combined this upf1Δ allele with a bud13Δ, snu17Δ, or pml1Δ mutation. Unexpectedly, the Ts phenotype of bud13Δ and snu17Δ cells was partially suppressed by the inactivation of NMD whereas no apparent suppression was observed in the pml1Δ mutant (Fig. 3A and Table S1). Northern blotting was used to analyze the effect of NMD on the abundance of MED20 transcripts in RES-deficient and RES-proficient cells. As controls, the blots were probed for the intron-containing TAN1, GOT1 and CYH2 transcripts. The latter 2 transcripts were selected because the abundance of GOT1 pre-mRNA is largely unaffected by RES- and/or NMD-inactivation whereas the CYH2 pre-mRNA is a well-characterized NMD substrate whose abundance is not influenced by the inactivation of RES factors..20,21 The analyses revealed that the MED20 pre-mRNA accumulates in NMD-deficient bud13Δ, snu17Δ and pml1Δ cells (Fig. 3B). Even though these findings suggest that RES inactivation leads to degradation of MED20 pre-mRNA by the NMD pathway, the low abundance of MED20 pre-mRNA in the bud13Δ, snu17Δ and pml1Δ single mutants prevented us from confirming that NMD directly influences its half-life.
Figure 3.
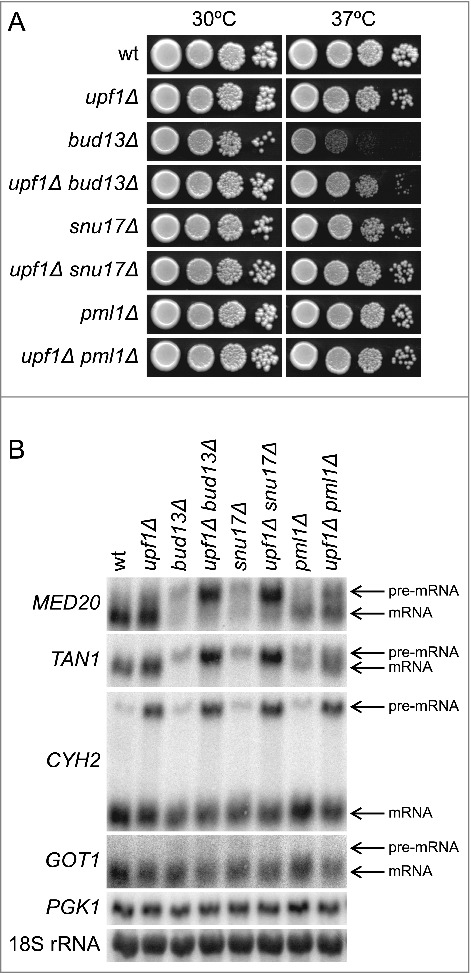
The inactivation of NMD in RES mutants causes accumulation of MED20 pre-mRNA. (A) Effects of the upf1Δ allele on growth of bud13Δ, snu17Δ, and pml1Δ cells. The wild-type (UMY2219), upf1Δ (MJY537), bud13Δ (MJY546), bud13Δ upf1Δ (MJY555), snu17Δ (MJY548), snu17Δ upf1Δ (MJY557), pml1Δ (MJY535), and pml1Δ upf1Δ (MJY559) strains were grown over-night in YEPD medium, serially diluted, spotted on YEPD plates, and incubated at 30°C or 37°C for 2 d. (B) Northern analysis of total RNA isolated from wild-type (UMY2219), upf1Δ (MJY537), bud13Δ (MJY546), bud13Δ upf1Δ (MJY555), snu17Δ (MJY548), snu17Δ upf1Δ (MJY557), pml1Δ (MJY535), and pml1Δ upf1Δ (MJY559) cells grown in YEPD medium at 30°C. The blot was probed for MED20, TAN1, CYH2, GOT1, and PGK1 transcripts using randomly labeled DNA fragments. 18S rRNA was detected using an oligonucleotide probe.
The first exon and the intron in MED20 are required for RES dependency
The RES complex was originally thought to be particularly important for splicing of transcripts that harbor introns with non-canonical 5′ splice sites.2,16,17 However, more recent studies have shown that the RES-regulated transcripts also include transcripts with consensus 5′ splice sites.18-20 In fact, we previously showed that the sequences between the 5′ splice site and branch point in TAN1 pre-mRNA are necessary and sufficient to induce RES dependent splicing.20 Although, the intron in MED20 pre-mRNA encompasses canonical 5′ and 3′ splice sites, the branch site sequence differs at one position from the consensus. To determine if the non-canonical branch site is causing the RES dependency, we changed the branch site sequence in the endogenous MED20 gene to the consensus. Analyses of wild-type, upf1Δ, bud13Δ, and upf1Δ bud13Δ cells harboring this allele (MED20-BP*) revealed that the transcript was still largely RES dependent (Fig. 4A). This finding suggested that some other feature(s) of MED20 pre-mRNA induces the RES dependency. To assess if the intron is required, we replaced the endogenous MED20 intron with the intron from the GOT1 gene. The MED20 pre-mRNA encompassing the GOT1 intron was spliced more efficiently than both wild-type and branch site-altered MED20 transcripts in bud13Δ and upf1Δ bud13Δ cells (Fig. 4A). Thus, the MED20 intron encompasses a cis-acting feature that promotes RES dependency.
Figure 4.
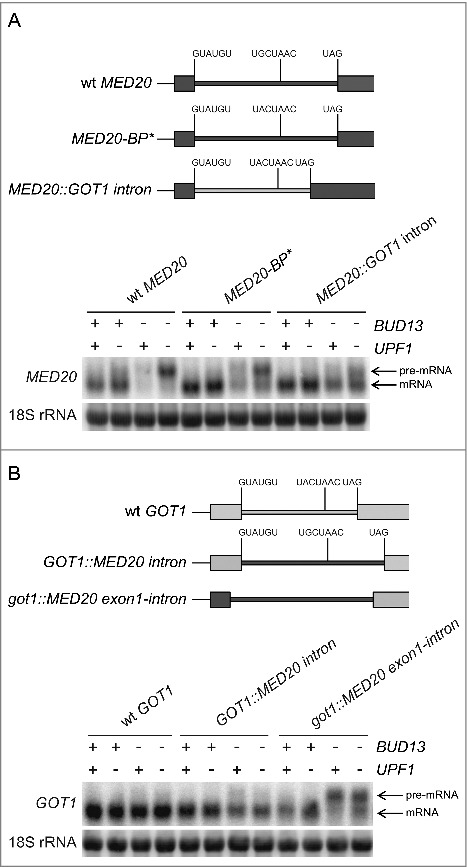
RES-controlled splicing of MED20 pre-mRNA requires both exonic and intronic elements. (A) Northern analysis of total RNA isolated from the indicated strains (UMY2219, MJY683, MJY546, MJY754, MJY817, MJY819, MJY821, MJY823 MJY735, MJY744, MJY746, and MJY748) grown in YEPD medium at 30°C. A schematic representation of the different MED20 alleles is shown above the blot. The blot was probed for MED20 transcripts using randomly labeled DNA fragments and for 18S rRNA using an oligonucleotide probe. (B) Northern analysis of total RNA isolated from the indicated strains (UMY2219, MJY683, MJY546, MJY754, MJY759, MJY761, MJY763, MJY765, MJY933, MJY934, MJY935, and MJY936) grown in YEPD medium at 30°C. A schematic representation of the different GOT1 alleles is shown above the blot. The blot was probed for GOT1 transcripts using randomly labeled DNA fragments and for 18S rRNA using an oligonucleotide probe.
To determine if the MED20 intron is sufficient to trigger RES dependency, we replaced the intron in the endogenous GOT1 gene with the intron from MED20. Although the abundance of the spliced GOT1 transcript decreased in bud13Δ and upf1Δ bud13Δ cells, no accumulation of the hybrid pre-mRNA was observed (Fig. 4B). This finding indicated that MED20 may also encompass a cis-acting element in an exon. To investigate the validity of this hypothesis, we replaced the first exon as well as the intron in GOT1 with the corresponding sequences of MED20. The analyses revealed that the hybrid pre-mRNA accumulated in both bud13Δ and upf1Δ bud13Δ cells, indicating the presence of a cis-acting element in exon 1 of MED20 (Fig. 4B). The lack of an effect of the upf1Δ allele on MED20-GOT1 pre-mRNA abundance is due to the absence of a premature stop codon in the transcript, i.e., the pre-mRNA encompasses an ORF that begins at the MED20 start codon and ends at the normal stop codon in GOT1. Collectively, these results indicate that the RES-dependent splicing of MED20 pre-mRNA requires both exonic and intronic features.
Discussion
The RES complex associates with spliceosomes and is thought to promote splicing of a subset of intron-containing pre-mRNAs. It has however been unclear if the growth defects of RES mutants are caused by inefficient splicing of specific transcripts or if they reflect a more general splicing defect. In this study, we find that the Ts phenotype of bud13Δ and snu17Δ cells is largely attributable to inefficient splicing of MED20 pre-mRNA, which codes for a subunit of the Mediator complex. We also find that the MED20 splicing defect is less pronounced in cells deleted for PML1, explaining why pml1Δ cells grow better than the other 2 RES mutants at 37°C. Thus, our findings suggest that the Ts phenotype of bud13Δ and snu17Δ mutants is caused by decreased Med20p levels and the consequent effects on the functionality of the Mediator complex.
Many factors that elicit their function at an early step of splicing also promote nuclear retention of unspliced pre-mRNAs. The effect on retention is usually not separable from that on splicing, but some factors, including Pml1p, have been suggested to promote nuclear retention without any apparent role in splicing.2,23,32,33 Our finding that the inactivation of NMD in RES mutants leads to an accumulation of MED20 pre-mRNA is consistent with the notion that the RES factors promote nuclear retention of unspliced pre-mRNAs.2 However, the observation that MED20 pre-mRNA accumulates in the pml1Δ single mutant makes it difficult to differentiate between effects on pre-mRNA retention and splicing.
Although the stabilization of unspliced pre-mRNAs in the cytoplasm is expected to be detrimental for growth, the inactivation of NMD in individual splicing factor mutants can lead to either enhanced or suppressed growth defects.23,34 The mechanism by which the inactivation of NMD suppresses the growth defects of specific splicing factor mutants remains unknown.34 The inactivation of NMD in the RES mutants causes a slight increase in the levels of spliced MED20 mRNA (Fig. 3B), providing possible explanation to the finding that the Ts phenotype of bud13Δ and snu17Δ cells is partially suppressed by introduction of a upf1Δ allele. However, the upf1Δ allele also counteracts the growth defect of bud13Δ cells at 30°C, indicating that altered expression of other gene products contributes to the suppression.
In addition to MED20, 4 endogenous transcripts, AMA1, MER2, MATa1 and TAN1 are known to require Bud13p and Snu17p for efficient splicing16-20 The AMA1 and MER2 pre-mRNAs are under control of the meiotic splicing regulator Mer1 and they both encompass an intron with a suboptimal 5´ splice site. In contrast, MATa1, TAN1 and MED20 contain introns with consensus 5´ splice sites, suggesting that the 5´ splice site may not be the defining feature for RES dependency. Accordingly, we previously showed that the sequence between the 5´ splice site and branch point is necessary and sufficient to induce RES-dependent splicing of TAN1 pre-mRNA.20 The cis-acting elements that induce RES dependency of MED20 pre-mRNA are present in both the first exon and intron, suggesting that the defining element(s) may be different from that in TAN1. In the activated spliceosome, the RES complex interacts with intronic sequences down-stream of the branch site,6,7,11 indicating that the cis-acting elements that trigger RES dependency may either represent sites where the RES complex initially interacts or they may indirectly influence the requirement for the RES factors. As the mechanism by which the RES complex preferentially enhances splicing of the target transcripts remains unclear, additional studies are needed to define the specificity and regulatory roles of the RES complex.
Materials and methods
Strains, media, and genetic procedures
Yeast strains used in this study are listed in Table S2. Yeast transformations, media, and genetic procedures have been described.35 Gene disruptions were confirmed by PCR using primers that annealed outside of sequences present in the transformed DNA fragment. Integrations of tags and sequences for altered introns were confirmed by DNA sequencing of the relevant PCR products.
A strain expressing a C-terminal 3HA-tagged Med20 protein was constructed by transforming the diploid strain UMY2366 with a 3HA-KanMX6 fragment amplified from pFA6a-3HA-KanMX6.36 The oligonucleotides used were: 5′- ATGTGATTTGGCGTACCAGTATGTTCGTGCTCTGGAGCTGCGGATCCCCGGGTTAATTAA-3′ and 5′- CAAACAGAAAAAAAAAAAAAGAAAAAGCATTCGTAAGAACGAATTCGAGCTCGTTTAAAC-3′. The diploid strain was allowed to sporulate and a haploid MED20::3HA-kanMX6 strain obtained from a tetrad. Strains MJY775, MJY779, MJY800, and MJY802 were obtained from subsequent crosses with the appropriate strains.
To construct strains expressing MED20 transcripts in which the intron was removed, modified, or replaced, we first replaced the intron in one copy of MED20 gene in the diploid strain UMY2366 with a URA3 marker. The URA3 marker was PCR-amplified from pRS40637 using the oligonucleotides 5′- GCTCGAAAGCACAGTAGCAATCCATCATGGGAAAATCAGCAGATTGTACTGAGAGTGCAC-3′ and 5′- CGTTAGTGTAGCGGGAGTGGCTCTTTCCACGAATATAACGCTGTGCGGTATTTCACACCG-3′. The strain was allowed to sporulate and a haploid med20::URA3 strain (MJY724) obtained from a tetrad. The haploid strain was transformed with DNA fragments containing the relevant sequence alterations and homologies to the exons of MED20. Following selection on 5-fluoroorotic acid (5-FOA)-containing plates, individual clones were screened for integration of the relevant DNA fragment. This approach combined with subsequent crosses with the appropriate strains generated MJY736, MJY738, MJY809, MJY808, MJY817, MJY819, MJY821, MJY823, MJY735, MJY744, MJY746, and MJY748. A similar approach was used to construct strains expressing hybrid GOT1 transcripts. For these strains, MJY574 was transformed with the relevant DNA fragments. The generated strains were crossed to MJY696 and MJY759, MJY761, MJY763, MJY765, MJY933, MJY934, MJY935, MJY936 obtained from tetrads.
High copy suppressor screen
To identify genes that in high dosage suppress the Ts phenotype of bud13Δ cells, we transformed strain MJY546 with a YEp24-based high copy genomic library38 and selected for growth on SC-ura plates at 37°C. Plasmids from transformants that showed improved growth at 37°C were isolated and retransformed and those that stayed positive were partially sequenced and subjected to homology searches against the S. cerevisiae genome. The gene responsible for the suppression was identified by subclonings and retransformations.
Plasmid constructions
A high copy plasmid harboring the MED20 gene (pMJ48) was constructed by cloning an EcoRI/KpnI DNA fragment from a suppressing YEp24 library plasmid into the corresponding sites of pRS426.39 Plasmid pRS316-BUD13 (pMJ51) has been described.20
RNA and protein methods
RNA isolations and RNA gel blot analyses were performed as described .20,40,41 For western blotting experiments, ∼18 OD600 units of exponentially growing cells were harvested, washed once with buffer B (50 mM Tris-HCl at pH 7.5, 150 mM NaCl, 5% glycerol, 1 mM DTT), and resuspended in 250 µl buffer B containing 1 х protease inhibitor cocktail (Roche Applied Science). Samples were transferred to 2-ml tubes containing ∼0.15 g of glass beads (0.5 mm diameter, Biospec) and vortexed 6 times for 30 s with one minute on ice between steps. Following centrifugation, 15 µg of total protein was resolved by 10% SDS-PAGE and transferred to Immobilon-P membrane (Millipore). Proteins were detected as described previously.20,42
Supplementary Material
Disclosure of potential conflicts of interest
No potential conflicts of interest were disclosed.
Acknowledgments
We thank Tracy Nissan, Vasili Hauryliuk, Anders Byström and Fu Xu for helpful comments on the manuscript.
Funding
This work was supported by Carl Tryggers Foundation (CTS13:206 to MJ); Åke Wibergs Foundation (M14–0207 to MJ); and Insamlingsstiftelsen Umeå universitet (to MJ).
References
- 1.Will CL, Lührmann R. Spliceosome structure and function. Cold Spring Harb Perspect Biol 2011; 3:pii: a003707; PMID:21441581; https://doi.org/ 10.1101/cshperspect.a003707 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Dziembowski A, Ventura AP, Rutz B, Caspary F, Faux C, Halgand F, Laprevote O, Seraphin B. Proteomic analysis identifies a new complex required for nuclear pre-mRNA retention and splicing. EMBO J 2004; 23:4847-56; PMID:15565172; https://doi.org/ 10.1038/sj.emboj.7600482 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Brooks MA, Dziembowski A, Quevillon-Cheruel S, Henriot V, Faux C, van Tilbeurgh H, Seraphin B. Structure of the yeast Pml1 splicing factor and its integration into the RES complex. Nucleic Acids Res 2009; 37:129-43; PMID:19033360; https://doi.org/ 10.1093/nar/gkn894 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Trowitzsch S, Weber G, Lührmann R, Wahl MC. An unusual RNA recognition motif acts as a scaffold for multiple proteins in the pre-mRNA retention and splicing complex. J Biol Chem 2008; 283:32317-27; PMID:18809678; https://doi.org/ 10.1074/jbc.M804977200 [DOI] [PubMed] [Google Scholar]
- 5.Trowitzsch S, Weber G, Lührmann R, Wahl MC. Crystal structure of the Pml1p subunit of the yeast precursor mRNA retention and splicing complex. J Mol Biol 2009; 385:531-41; PMID:19010333; https://doi.org/ 10.1016/j.jmb.2008.10.087 [DOI] [PubMed] [Google Scholar]
- 6.Schneider C, Agafonov DE, Schmitzova J, Hartmuth K, Fabrizio P, Lührmann R. Dynamic Contacts of U2, RES, Cwc25, Prp8 and Prp45 Proteins with the Pre-mRNA Branch-Site and 3′ Splice Site during Catalytic Activation and Step 1 Catalysis in Yeast Spliceosomes. PLoS Genet 2015; 11:e1005539; PMID:26393790; https://doi.org/ 10.1371/journal.pgen.1005539 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Wysoczanski P, Schneider C, Xiang S, Munari F, Trowitzsch S, Wahl MC, Lührmann R, Becker S, Zweckstetter M. Cooperative structure of the heterotrimeric pre-mRNA retention and splicing complex. Nat Struct Mol Biol 2014; 21:911-8; PMID:25218446; https://doi.org/ 10.1038/nsmb.2889 [DOI] [PubMed] [Google Scholar]
- 8.Gottschalk A, Bartels C, Neubauer G, Lührmann R, Fabrizio P. A novel yeast U2 snRNP protein, Snu17p, is required for the first catalytic step of splicing and for progression of spliceosome assembly. Mol Cell Biol 2001; 21:3037-46; PMID:11287609; https://doi.org/ 10.1128/MCB.21.9.3037-3046.2001 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Wang Q, He J, Lynn B, Rymond BC. Interactions of the yeast SF3b splicing factor. Mol Cell Biol 2005; 25:10745-54; PMID:16314500; https://doi.org/ 10.1128/MCB.25.24.10745-10754.2005 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Fabrizio P, Dannenberg J, Dube P, Kastner B, Stark H, Urlaub H, Lührmann R. The evolutionarily conserved core design of the catalytic activation step of the yeast spliceosome. Mol Cell 2009; 36:593-608; PMID:19941820; https://doi.org/ 10.1016/j.molcel.2009.09.040 [DOI] [PubMed] [Google Scholar]
- 11.Yan C, Wan R, Bai R, Huang G, Shi Y. Structure of a yeast activated spliceosome at 3.5 A resolution. Science 2016; 353:904-11; PMID:27445306; https://doi.org/ 10.1126/science.aag0291 [DOI] [PubMed] [Google Scholar]
- 12.Bessonov S, Anokhina M, Will CL, Urlaub H, Lührmann R. Isolation of an active step I spliceosome and composition of its RNP core. Nature 2008; 452:846-50; PMID:18322460; https://doi.org/ 10.1038/nature06842 [DOI] [PubMed] [Google Scholar]
- 13.Deckert J, Hartmuth K, Boehringer D, Behzadnia N, Will CL, Kastner B, Stark H, Urlaub H, Lührmann R. Protein composition and electron microscopy structure of affinity-purified human spliceosomal B complexes isolated under physiological conditions. Mol Cell Biol 2006; 26:5528-43; PMID:16809785; https://doi.org/ 10.1128/MCB.00582-06 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Clark TA, Sugnet CW, Ares M Jr.. Genomewide analysis of mRNA processing in yeast using splicing-specific microarrays. Science 2002; 296:907-10; PMID:11988574; https://doi.org/ 10.1126/science.1069415 [DOI] [PubMed] [Google Scholar]
- 15.Khanna M, Van Bakel H, Tang X, Calarco JA, Babak T, Guo G, Emili A, Greenblatt JF, Hughes TR, Krogan NJ, et al.. A systematic characterization of Cwc21, the yeast ortholog of the human spliceosomal protein SRm300. RNA 2009; 15:2174-85; PMID:19789211; https://doi.org/ 10.1261/rna.1790509 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Scherrer FW Jr, Spingola M. A subset of Mer1p-dependent introns requires Bud13p for splicing activation and nuclear retention. RNA 2006; 12:1361-72; PMID:16738408; https://doi.org/ 10.1261/rna.2276806 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Spingola M, Armisen J, Ares M Jr.. Mer1p is a modular splicing factor whose function depends on the conserved U2 snRNP protein Snu17p. Nucleic Acids Res 2004; 32:1242-50; PMID:14973223; https://doi.org/ 10.1093/nar/gkh281 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Tuo S, Nakashima K, Pringle JR. Apparent defect in yeast bud-site selection due to a specific failure to splice the pre-mRNA of a regulator of cell-type-specific transcription. PLoS One 2012; 7:e47621; PMID:23118884; https://doi.org/ 10.1371/journal.pone.0047621 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Schmidlin T, Kaeberlein M, Kudlow BA, MacKay V, Lockshon D, Kennedy BK. Single-gene deletions that restore mating competence to diploid yeast. FEMS Yeast Res 2008; 8:276-86; PMID:17995956; https://doi.org/ 10.1111/j.1567-1364.2007.00322.x [DOI] [PubMed] [Google Scholar]
- 20.Zhou Y, Chen C, Johansson MJO. The pre-mRNA retention and splicing complex controls tRNA maturation by promoting TAN1 expression. Nucleic Acids Res 2013; 41:5669-78; PMID:23605039; https://doi.org/ 10.1093/nar/gkt269 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.He F, Peltz SW, Donahue JL, Rosbash M, Jacobson A. Stabilization and ribosome association of unspliced pre-mRNAs in a yeast upf1- mutant. Proc Natl Acad Sci USA 1993; 90:7034-8; PMID:834621314690598 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.He F, Li X, Spatrick P, Casillo R, Dong S, Jacobson A. Genome-wide analysis of mRNAs regulated by the nonsense-mediated and 5′ to 3′ mRNA decay pathways in yeast. Mol Cell 2003; 12:1439-52; PMID:14690598; https://doi.org/ 10.1016/S1097-2765(03)00446-5 [DOI] [PubMed] [Google Scholar]
- 23.Rutz B, Seraphin B. A dual role for BBP/ScSF1 in nuclear pre-mRNA retention and splicing. EMBO J 2000; 19:1873-86; PMID:10775271; https://doi.org/ 10.1093/emboj/19.8.1873 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Sayani S, Janis M, Lee CY, Toesca I, Chanfreau GF. Widespread impact of nonsense-mediated mRNA decay on the yeast intronome. Mol Cell 2008; 31:360-70; PMID:18691968; https://doi.org/ 10.1016/j.molcel.2008.07.005 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Kervestin S, Jacobson A. NMD: a multifaceted response to premature translational termination. Nat Rev Mol Cell Biol 2012; 13:700-12; PMID:23072888; https://doi.org/ 10.1038/nrm3454 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Ni L, Snyder M. A genomic study of the bipolar bud site selection pattern in Saccharomyces cerevisiae. Mol Biol Cell 2001; 12:2147-70; PMID:11452010; https://doi.org/ 10.1091/mbc.12.7.2147 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Johansson MJO, Byström AS. The Saccharomyces cerevisiae TAN1 gene is required for N4-acetylcytidine formation in tRNA. RNA 2004; 10:712-9; PMID:15037780; https://doi.org/ 10.1261/rna.5198204 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Kim YJ, Björklund S, Li Y, Sayre MH, Kornberg RD. A multiprotein mediator of transcriptional activation and its interaction with the C-terminal repeat domain of RNA polymerase II. Cell 1994; 77:599-608; PMID:8187178; https://doi.org/ 10.1016/0092-8674(94)90221-6 [DOI] [PubMed] [Google Scholar]
- 29.Nonet ML, Young RA. Intragenic and extragenic suppressors of mutations in the heptapeptide repeat domain of Saccharomyces cerevisiae RNA polymerase II. Genetics 1989; 123:715-24; PMID:2693207 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Allen BL, Taatjes DJ. The Mediator complex: a central integrator of transcription. Nat Rev Mol Cell Biol 2015; 16:155-66; PMID:25693131; https://doi.org/ 10.1038/nrm3951 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Leeds P, Peltz SW, Jacobson A, Culbertson MR. The product of the yeast UPF1 gene is required for rapid turnover of mRNAs containing a premature translational termination codon. Genes Dev 1991; 5:2303-14; PMID:1748286; https://doi.org/ 10.1101/gad.5.12a.2303 [DOI] [PubMed] [Google Scholar]
- 32.Galy V, Gadal O, Fromont-Racine M, Romano A, Jacquier A, Nehrbass U. Nuclear retention of unspliced mRNAs in yeast is mediated by perinuclear Mlp1. Cell 2004; 116:63-73; PMID:14718167; https://doi.org/ 10.1016/S0092-8674(03)01026-2 [DOI] [PubMed] [Google Scholar]
- 33.Palancade B, Zuccolo M, Loeillet S, Nicolas A, Doye V. Pml39, a novel protein of the nuclear periphery required for nuclear retention of improper messenger ribonucleoparticles. Mol Biol Cell 2005; 16:5258-68; PMID:16162818; https://doi.org/ 10.1091/mbc.E05-06-0527 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Kawashima T, Pellegrini M, Chanfreau GF. Nonsense-mediated mRNA decay mutes the splicing defects of spliceosome component mutations. RNA 2009; 15:2236-47; PMID:19850912; https://doi.org/ 10.1261/rna.1736809 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Amberg DC, Burke DJ, Strathern JN. Methods in Yeast Genetics Cold Spring Harbor. N Y: Cold Spring Harbor Laboratory Press, 2005 [Google Scholar]
- 36.Longtine MS, McKenzie A 3rd, Demarini DJ, Shah NG, Wach A, Brachat A, Philippsen P, Pringle JR. Additional modules for versatile and economical PCR-based gene deletion and modification in Saccharomyces cerevisiae. Yeast 1998; 14:953-61; PMID:9717241; https://doi.org/ [DOI] [PubMed] [Google Scholar]
- 37.Sikorski RS, Hieter P. A system of shuttle vectors and yeast host strains designed for efficient manipulation of DNA in Saccharomyces cerevisiae. Genetics 1989; 122:19-27; PMID:2659436 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Carlson M, Botstein D. Two differentially regulated mRNAs with different 5′ ends encode secreted with intracellular forms of yeast invertase. Cell 1982; 28:145-54; PMID:7039847; https://doi.org/ 10.1016/0092-8674(82)90384-1 [DOI] [PubMed] [Google Scholar]
- 39.Christianson TW, Sikorski RS, Dante M, Shero JH, Hieter P. Multifunctional yeast high-copy-number shuttle vectors. Gene 1992; 110:119-22; PMID:1544568; https://doi.org/ 10.1016/0378-1119(92)90454-W [DOI] [PubMed] [Google Scholar]
- 40.He F, Amrani N, Johansson MJO, Jacobson A. Chapter 6. Qualitative and quantitative assessment of the activity of the yeast nonsense-mediated mRNA decay pathway. Methods Enzymol 2008; 449:127-47; PMID:19215756; http:/dx.doi.org/ 10.1016/S0076-6879(08)02406-3 [DOI] [PubMed] [Google Scholar]
- 41.Johansson MJO. Determining if an mRNA is a substrate of nonsense-mediated mRNA decay in Saccharomyces cerevisiae. Methods Mol Biol 2017; 1507:169-77; PMID:27832540; http:/dx.doi.org/ 10.1007/978-1-4939-6518-2_13 [DOI] [PubMed] [Google Scholar]
- 42.Johansson MJO, Jacobson A. Nonsense-mediated mRNA decay maintains translational fidelity by limiting magnesium uptake. Genes Dev 2010; 24:1491-5; PMID:20634315; https://doi.org/ 10.1101/gad.1930710 [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.


