SUMMARY
Herpes Zoster (shingles) causes significant morbidity in immune compromised hosts and older adults. While a vaccine is available for prevention of shingles, its efficacy declines with age. To help to understand the mechanisms driving vaccinal responses, we constructed a multiscale, multifactorial response network (MMRN) of immunity in healthy young and older adults immunized with the live attenuated shingles vaccine Zostavax®. Vaccination induces robust antigen-specific antibody, plasmablasts and CD4+ T cells, yet limited CD8+ T cell and antiviral responses. The MMRN reveals striking associations between orthogonal datasets such as transcriptomic and metabolomics signatures, cell populations and cytokine levels, and identifies immune and metabolic correlates of vaccine immunity. Networks associated with inositol phosphate, glycerophospholipids and sterol metabolism are tightly coupled with immunity. Critically, the sterol regulatory binding protein 1 and its targets are key integrators of antibody and T follicular cell responses. Our approach is broadly applicable to study human immunity, and can help to identify predictors of efficacy as well as mechanisms controlling immunity to vaccination.
Keywords: Herpes zoster vaccine, Zostavax®, immune response, shingles, transcriptomics, metabolomics, multiscale, systems biology
INTRODUCTION
Recent advances in high-throughput technologies have enabled the collection of detailed data of immune response in humans, especially via controlled studies of vaccination (Davis, 2008; Kidd et al., 2014; Mooney et al., 2013; Pulendran and Ahmed, 2011; Pulendran et al., 2010; Zak and Aderem, 2015). Blood transcriptomics, albeit complex, has been very informative about early events of the immune response. For example, the transcriptomic signatures after vaccination have been used to predict vaccine-induced antibody and T cell responses in multiple studies, and different classes of vaccines induced distinct gene expression programs (Franco et al., 2013; Furman et al., 2013; Gaucher et al., 2008; Kazmin et al., 2017; Li et al., 2014; Nakaya et al., 2016; Nakaya et al., 2015; Nakaya et al., 2011; Qi et al., 2016; Querec et al., 2009; Sobolev et al., 2016; Tsang et al., 2014; Zak et al., 2012). These studies contribute to the emerging field of systems vaccinology, and open up new ways to understand the molecular mechanisms of vaccine induced immunity, and to facilitate the development of new vaccines (Pulendran et al., 2010).
Herpes zoster (HZ) vaccination provides a unique case to investigate the stimulation of immune memory. HZ (shingles) is the clinical manifestation of varicella zoster virus (VZV) reactivation. After acute infection, VZV remains dormant in sensory neurons. Viral reactivations in immune compromised hosts and older adults may result in HZ accompanied by significant morbidity. Zostavax® is licensed as a vaccine for the prevention of HZ in individuals 50 years of age and older. Zostavax® is a live attenuated virus vaccine with the same Oka/Merck vaccine strain, but 14 times the dose, used in the varicella (chickenpox) vaccine. Zostavax® was shown to confer ~50% overall reduction in HZ, and ~66% reduction in HZ-associated pain (Oxman et al., 2005). Interestingly, the efficacy against HZ was 63.9% in subjects who were 60–69 years old, but only 37.6% in subjects older than 70 years, highlighting the age related decline of vaccine efficacy in the elderly (Oxman et al., 2005). This varying efficacy, whilst far from optimal in generating robust antiviral immunity, nevertheless provides an opportunity to understand how individuals respond differently to the same vaccine.
In this study, we measured the magnitude and quality of innate and adaptive immune response to Zostavax® vaccination for up to 180 days. Complementing these assays with transcriptomics and metabolomics, our integrative computational analysis reveals a highly interconnected immune network of gene and metabolic pathways that correlate with the later adaptive response. The findings are of considerable interest for the understanding of human immunology, in the context of immunity to a live virus vaccine.
RESULTS
Study design for integrative profiling of immunity to Zostavax® vaccination in humans
A longitudinal study of Zostavax® vaccination was conducted in healthy adults between August 2011 and November 2012. Seventy-seven participants were enrolled in Atlanta, Georgia and Denver, Colorado, USA. The cohort contained two age groups: 33 young adults between the age of 25 to 40 years, and 44 so called elderly subjects between the age of 60 to 79 years (Table S1). The subjects’ blood samples were collected at baseline prior to vaccination and at days 1, 3, 7, 14, 28, 90 and 180 post-vaccination (Figure 1A). Throughout the course of study, we measured VZV specific antibody titers, antigen-specific B and T cell populations, and various cell populations involved in innate immunity, including dendritic cells and monocyte subsets. Cytokines were assayed by a 27-plex Luminex platform. VZV DNA in the whole blood was measured by polymerase chain reactions (PCR). Transcriptomics of PBMCs and high-resolution metabolomics of plasma were obtained at early time points. As a result, each subject was profiled by multiple technologies in a time series (illustrated in left panel, Figure 1A). This rich collection of immune profiles, including high-dimensional data from transcriptomics and metabolomics, provided a unique opportunity to construct an integrated view of the immune response to Zostavax® in humans. This also poses major challenges in computational analysis concerning approaches to integrate data across different technologies, and to dealing with the redundancy in omics data. In the current paper, we will first present each data type separately. Then, the integrative analysis is presented in a framework of “multiscale, multifactorial response network” (MMRN). We will use MMRN to interrogate the mechanisms of Zostavax® induced immune response, and to interpret predictive models of adaptive response.
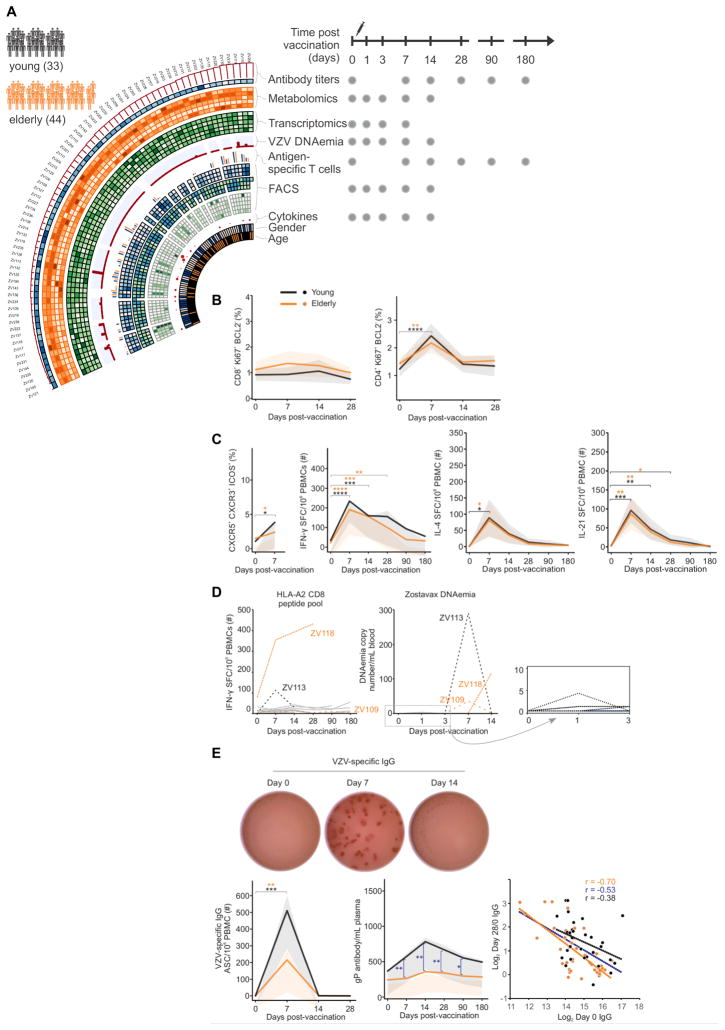
Figure 1. Zostavax® study overview, cellular and antibody responses.
A) Study overview. 77 subjects as either young adults (25–40 yo, n=33) or elderly (60–79 yo, n=44). Different samples were taken at baseline (day 0), and 1, 3, 7, 14, 28, 90 and 180 days. Please refer to STAR methods (supplemental) for details.
B) CD8+ and CD4+ T cell activation as measured by Ki67+Bcl2- in young adults (black, n=13 for CD8+, n=15 for CD4+) and the elderly (orange, n=19 for CD8+, n=21 for CD4+). Mean values in solid lines, and shades for 90% confidence interval.
C) Increase in VZV-specific Tfh-like cells (as measured by CXCR5+ICOS+CXCR3+) in young adults (n=16) and the elderly (n=20). Antigen-specific IFN-γ, IL-4 or IL-21 production by CD4+ T cells as measured by ELISPOT in young adults (n=17) and the elderly (n=22). SFC = Spot forming cells.
D) Antigen-specific CD8+ T cell responses and Zostavax DNAemia in a subset of HLA-A2+ individuals (n=13). PBMCs stimulated with previously published VZV-HLA-A2 restricted peptides overnight and IFN-γ measured via ELISPOT. Viral DNA copies in blood measured by PCR
E) Representative VZV-specific IgG ELISPOT. Number of Zostavax®-specific plasmablasts in young adults (n=5) and the elderly (n=8). Kinetic analysis of Zostavax® glycoprotein-specific IgG antibodies as measured by ELISA in young adults (n=16) and the elderly (n=21). Inverse correlation between VZV-specific IgG at baseline and day 28 response is shown for young (black), elderly (organge) and the total cohort (blue). Comparison between the two groups was done using unpaired t-test. Comparison of response between different time points was done using paired t-test. *p<0.05, ** p<0.01, ***p<0.005, **** p<0.0001.
Zostavax®-induced significant CD4+ T cell and antibody responses
Since Zostavax® comprises a live-attenuated virus, we were surprised to observe a weak CD8+ T cell response in peripheral blood, as measured by the downregulation of the pro-survival molecule (Bcl2) and upregulation of Ki67 (expressed in proliferating cells, (Miller et al., 2008) (Figure 1B), unlike the robust CD8+ T cell responses seen with other live vaccines such as yellow fever and smallpox (Akondy et al., 2009; Miller et al., 2008). In contrast significant CD4+ T cell responses were detected in the majority of subjects (Figure 1B), consistent with a recent report (Qi et al., 2016). To understand this CD4+ T cell response in more detail, we assessed the phenotype of the cells, including blood follicular T cell (Tfh)-like CD4+ T cells. Activated Tfh cells are important in providing B cell help and express the chemokine receptor CXCR5, ICOS (a co-stimulatory molecule expressed on activated T cells), PD-1 and secretion of IL-4 and IL-21 (Crotty, 2011; Yusuf et al., 2010). Blood Tfh-like cells that expressed CXCR5 and CXCR3 (a chemokine receptor on Th1 CD4+ T cells (Groom and Luster, 2011)) and ICOS, measured seven days after influenza vaccination correlated with the antibody response (Bentebibel et al., 2013). Figure 1C shows an increase in CD4+CXCR5+CXCR3+ICOS+ T cells after Zostavax® in both the young and elderly adults. Significant increase in CD4+ T cells that are positive for IFN-γ, IL-4 and IL-21 were also detected, indicating a mixed Th1/Th2/Th21 profile (Figure 1C). We then examined the phenotype of VZV-specific CD4+ T cells by gating on Ki67+/Bcl2− CD4+ T cells. These cells express T-bet, CXCR3, PD-1 and some ICOS whereas they express less RORγT and CCR7 (Figure S1A). Finally, we assessed if CD4+ or CD8+ T cells were producing IFN-γ after vaccination. Figure S1B shows that depletion of CD4+ T cells, but not of CD8+ T cells, abolished IFN-γ production in the ELISPOT assay.
To further assess the antigen specific CD8+ T cell response, we stimulated PBMCs with known VZV-specific HLA-A2 restricted peptides in a subset of HLA-A2+ individuals (Chiu et al., 2014; van der Heiden et al., 2009). Despite detectable VZV-specific CD8+ T cell responses at baseline, the CD8+ T cells did not respond to the vaccine peptides with the exception of two individuals (ZV113 and ZV118, Figure 1D). Interestingly, when we examined VZV DNA in the blood, although residual amount of viral DNA was seen in some subjects at day 1 (Table S2), only three showed positive viral replication in the blood, including both the CD8+ responders (Figure 1D). The subject that had prolonged DNAemia at day 14 (ZV118) also showed the highest CD8+ response to the vaccine peptides (Figure 1D). Overall, these results demonstrate that Zostavax® induces a a mixed Th1/Tfh-like profile (Th2/Th21) profile. The CD8+ T cell response was limited, but clear in the few participants with positive replication of VZV.
Finally, we asssed the antibody response induced by Zostavax®. We detected a transient peak of antigen-specific plasmablast cells 7 days after Zostavax® administration (Figure 1E), consistent with the plasmablast responses observed to other vaccines such as influenza (Wrammert et al., 2008). The magnitude of this cellular response was higher in the younger group (Figure 1E), consistent with previous studies with influenza vaccine (Sasaki et al., 2011). The measurement of VZV glycoprotein-specific IgG over the course of the study showed significant increases induced by Zostavax® in both the young and elderly adults (Figure 1E). All subjects had VZV antibodies prior to vaccination (as per study inclusion criteria). The IgG antibody peaked between two and four weeks after vaccination, and dropped by 180 days after vaccination. Direct comparison between the two groups indicates that the younger group had a greater increase of VZV-specific antibody after vaccination (Figure 1E). We also assayed the IgG response against total VZV infected cell lysate and the results were consistent with those using VZV glycoproteins (Figure S1C–D). Additionally, we measured plasma VZV-specific IgA, which was significantly increased in both groups. However, there was not a significant difference between the young and elderly groups (Figure S1E). Taken together, these results show that Zostavax® induces a robust plasmablast and antibody response that is greater in the younger adults compared to the elderly. The increase of VZV-specific IgG after vaccination showed an inverse correlation to the baseline IgG (Figure 1E). Such inverse correlation to baseline titer was also reported in influenza vaccine studies (Nakaya et al., 2015; Nakaya et al., 2011; Tsang et al., 2014).
Transcriptomic signatures of Zostavax® vaccination
We assessed the gene expression changes in PBMCsinduced by Zostavax® vaccination. Besides the interferon induced antiviral genes, such as MX1 and IFI44L, a phosphoserine phosphatase, PSPH, was highly upregulated at day 1/0 and day 3/0 (Figure S2A). At day 7, immunoglobulin genes were highly upregulated, as well as TNFRSF17 and MZB1, two genes extensively reported for their roles in antibody production (Li et al., 2014; Nakaya et al., 2011; Querec et al., 2009) (Figure S2A). We have previously reported a set of blood transcription modules (BTMs) as a tool for transcriptomic analysis (Li et al., 2014). In this study, the activity of each BTM module was taken as the mean value of its member genes. BTM analysis of the response to Zostavax® revealed signatures seen with several other vaccines, including the seasonal influenza vaccines and the meningococcal vaccine such as a strong plasmablast signature at day 7 (Figure S2B), consistent with the robust plasmablast response at this time point (Figure 1E).
Surprisingly however, several BTMs related to innate immunity including those relating to antigen presentation, dendritic cells, antiviral innate immune sensing including cytosolic DNA sensing, RIG-I receptor signaling and type I IFN responses were weakly induced by Zostavax® relative to YF-17D, which is another live viral vector (Figure S2B). This suggests that Zostavax® is a weak stimulator of innate responses and DC activation. We tested this by stimulating human PBMCs with Zostavax® in vitro. We could detect VZV glycoprotein in a substantial proportion of monocytes, myeloid DC (mDC) and plasmacytoid DC (pDC) by intracellular FACS staining, suggesting uptake of virus by these cells (Figure S3A). However, there did not appear to be activation of these cell types in response to stimulation of PBMCs with Zostavax®, as evidenced by the failure to upregulate CD80 (Figure S3A), or release of proinflammatory cytokines in the supernatants (Figure S3B). Intracellular FACS staining of cytokines in these cultures revealed that Zostavax® failed to induce IFNα in pDCs (Figure S3C) or IL-6 in monocytes or mDCs (Figure S3D). However, Zostavax® induced several chemokines, such as CXCL8, CCL2, CXCL9 and CXCL10 in PBMCs (Figure S3E). Thus, although Zostavax® contains live VZV which infects monocytes and DCs, it fails to induce significant innate activation of such cells.
Metabolomic signatures of Zostavax® vaccination
Using untargeted high-resolution metabolomics, thousands of metabolite peaks were detected in vaccinee plasma samples (Figure 2A, B). Matched by accurate mass-to-charge ratio (m/z) and retention time in liquid chromatography (Figure 2A), a small subset of metabolite peaks were also found in the Zostavax® vaccine itself (Figure 2B). Since metabolomics is a rapidly evolving field and the primary goal of this study is to quantitatively model immune response, we decided to filter the metabolite peaks by only keeping the peaks that were detected in over 90% of all samples. This resulted in 4,048 peaks from the negative ionization and 2,216 peaks in the positive ionization. Compared to the baseline, both negative and positive ionization data indicated the largest numbers of differential metabolites at day 1 and day 14 (under negative ionization, 429 and 425 peaks at FDR < 0.1, respectively). Few overlap peaks were found between the day 1/0 significant metabolite peaks and chemicals in the vaccine (data not shown), indicating that the human metabolomic changes were not simply due to carryover from chemicals in the vaccine. We used the mummichog software, a pathway tool designed for untargeted metabolomics data (Li et al., 2013), to evaluate the day 1/0 significant metabolic pathways shown in Figure 2C. Strikingly, TCA cycle (and overlapping glycolysis and gluconeogenesis) and propanoate metabolism was also identified in the day 3/0 transcriptomics data (Table S3). Ascorbate and aldarate metabolism and tryptophan metabolism were also upregulated in transcriptomics but with marginal significance (Table S3). This concordance between the vaccine-induced metabolite pathways at day 1 and transcriptional signatures in PBMCs 2 days later at day 3, is intriguing. We thus sought to test the association between metabolomics and transcriptomics systematically.
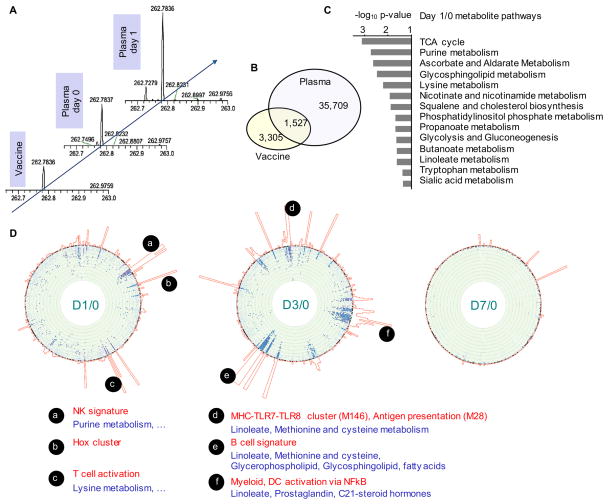
Figure 2. Plasma metabolomics and association with PBMC transcriptomics after Zostavax® vaccination.
A) Illustration of chromatographic peaks identified in high-resolution metabolomics. The peaks across individuals and time points were aligned based on accurate mass-to-charge ratio (m/z) and LC retention time (not shown).
B) Common peaks detected in both Zostavax® vaccine and vaccinee plasma samples (across days 0, 1, 3, 7, 14). Data shown for negative ionization.
C) Significantly enriched pathways for Day 1 vs Day 0 (both negative and positive ionization), by the mummichog software (Li et al., 2013).
D) Metabolome-wide association with transcriptomics. The “chromosome” bands on the circular edge are 346 BTM gene modules in fixed order. Towards the center of the circle are scatter plots of −log10 p-values of metabolites that are significantly associated with the respective gene module (p < 0.01). Each dot represents a metabolite peak, blue from positive ionization, purple from negative ionization. Only metabolite peaks with FDR < 0.1 are shown on the foreground. The joined histograms outside of the BTM bands indicate the number of significant metabolites for each gene module.
Integration of metabolomics, transcriptomics and flow cytometry
To integrate metabolomics and transciptomics data, we used a method similar to genome-wide association study, and constructed a regression model that accounted for age, gender and race, to test the association of the relative change in each metabolite feature with that of each BTM activity in response to vaccination. Mummichog pathway analysis was then performed on the significantly associated metabolite peaks per BTM. The results for all gene modules are combined into circular plots, whereas BTM modules and metabolic pathways are highlighted for the most significant associations (Figure 2D). At day 1 after vaccination, both NK signature and T cell modules were associated with a number of metabolites (Figure 2D). Based on their regression coefficients, these were negative association, and the corresponding pathways, including purine metabolism and lysine metabolism (Figure 2D), largely overlapping with the day 1/0 significant pathway in Figure 2C. At day 3/0, strong association to transcriptomics was seen in many metabolic pathways, including linoleate metabolism, methionine and cysteine metabolism, glycerophospholipid metabolism, glycosphingolipid metabolism. (Figure 2D). They were most strongly associated with gene expression for MHC-TLR7-TLR8 cluster, antigen presentation and myeloid, DC activation via NF-κB, and with B cell signatures. By day 7/0, no significant association was observed between transcriptomics and metabolomics (Figure 2D).
Besides significant correlation with metabolomics, further analysis of the transcriptomics data also revealed many correlations with cell populations, manifesting a high degree of redundancy (Figure S4A). For example, in Figure S4B, the gene module for “plasma cells and immunoglobulins” was correlated with antibody secreting cells at day 7/0, and the “T cell activation and signaling” gene module was correlated with total CD3+ T cells at day 3/0. Even though transcriptomics was measured on PBMCs, the gene module of “recruitment of neutrophils” did correlate with the number of neutrophils measured by flow cytometry in whole blood at day 7/0 (Figure S4B). Across time points, early gene expression for calcium signaling and steroid metabolism is correlated with later CD4+ T cell activation (Figure S4C). Moreover, there were striking correlations between the most variable gene modules and cell populations on a temporal course, as shown in Figure S4D. For example at baseline, there was a correlation between transcriptional signatures of the KIR clusters and the frequencies of NK cells. Furthermore, at baseline the transcriptional signatures of cytosolic DNA sensing, chemokines and inflammation were positively correlated with nuclear receptor 4 family, with the number of neutrophils, but negatively correlated with day 1/0 genes of myeloid and dendritic cells activation via NF-κb. In addition, day 3/0 B cell signature genes are correlated with the number of day 7/0 CXCR3+ Tfh like cells (Figure S4D). The data in Figure S4D also suggest that major cell populations were stable for each study subject, which was confirmed by plotting the correlations between time points (Figure S4E), consistent with previous findings (Tsang et al., 2014). The majority of cytokines and metabolite clusters also exhibited high stability within each subject, while gene modules showed a more mixed range (Figure S4E). Therefore, many individual characteristics, including metabolic phenotypes, persisted through the study.
A Multiscale, Multifactorial Response Network (MMRN)
Effective integration of multiple data types requires dimension reduction and needs to account for their different variance structures. We constructed a multiscale, multifactorial response network (MMRN) as illustrated in Figure S5. Gene expression was reduced to BTMs, and metabolite peaks were grouped by unsupervised clustering (Ben-Dor et al., 1999). We computed the correlation network of all BTMs, using Pearson correlation with Bonferroni correction (p < 0.001). The correlation network was partitioned into sub-graphs that are closely correlated, using an established topological algorithm (Kalinka and Tomancak, 2011). Each resulting subgraph, grouping multiple BTM modules of similar profiles, was treated as a gene network. Similarly, the same network partition method was applied to metabolite clusters, cytokines and cell populations (Figure S5). Of note, these resulting networks are not meant to infer regulatory relationship, but serve as units of dimension reduction.
To deal with the different variance structure across data types, we used partial least square (PLS) regression. PLS finds the principal components with the best covariance between two matrices. The statistical significance of such association can be thoroughly assessed by permutation. This approach has been successfully applied to the modeling of signaling networks that were measured by multiple technologies (Janes et al., 2005). In this study, data matrices from the four types of networks (genes, metabolites, cytokines and cells) were used as input to PLS regression, and the significantly associated networks were connected to a multiscale, multifactorial response network (MMRN, Figure 3A). The MMRN was a dense network of 608 nodes and 3476 connections (at p< 0.01; 261 nodes and 771 connections at p < 0.001; Data S1). It should also be noted that each node in MMRN is a network of its own right. This hierarchical structure allows an overview of the super-network and multiple zoom levels for additional details.
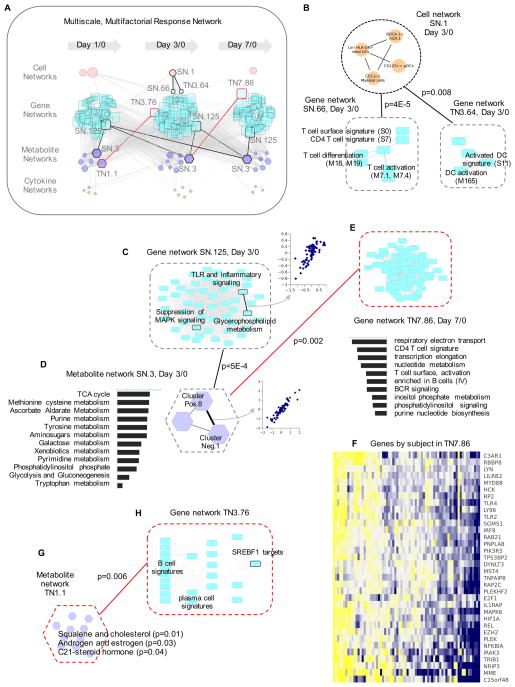
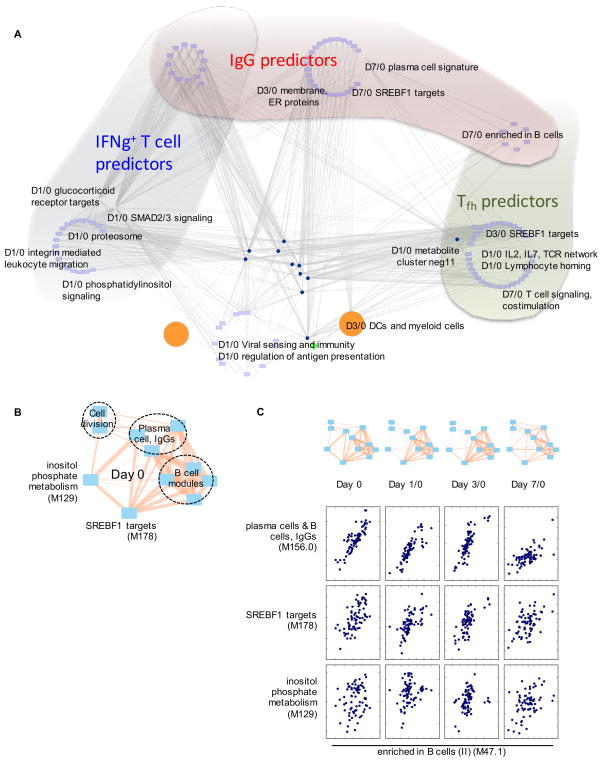
Figure 3. Multiscale, multifactorial response network (MMRN) of Zostavax® vaccination.
A) MMRN consists of correlation networks using data from metabolomics, transcriptomics, cytokines and cell frequencies. Each node is a child network of one data type. The links between nodes were established by significant association using partial least square regression and permutation test. In the highlighted examples, black lines are for stable networks, red lines transient networks.
B–H) are examples taken from the MMRN, where black dashed outlines indicate stable networks, red transient networks. B) Connections between a cell network and two gene networks.
C) Gene network SN.125, where the callout scatter plot shows the correlation between two gene modules, TLR and inflammatory signaling and glycerophospholipid metabolism.
D) Metabolite network SN.3. The callout scatter plot on the right shows the correlation between two metabolite clusters measured by positive ionization (Pos.8) and negative ionization (Neg.1). The histogram on the left shows the top pathways in this metabolite network, ranked by −log10 p-value.
E) Gene network TN7.86, a day 7/0 transient network connected with day 3/0 metabolite network SN.3. The histogram under the network shows its top gene modules, ranked by the number of connections.
F) Selected genes from the gene network TN7.86.
A few examples are highlighted from the MMRN (Figure 3A). For example, the cell network SN.1 consisting of cell populations, including BDCA1+ myeloid DCs and CD11c+ myeloid cells, is significantly associated with a gene network on T cell activation and differentiation (SN.66) and a gene network on DC activation (TN3.64) at day 3/0 (Figure 3B). As another example, the largest stable gene network (SN.125) and the largest stable metabolite network (SN.3) were connected across all time points (Figure 3A). The network SN.125 contains many gene modules on TLR and inflammatory signaling, which also showed a tight correlation with the gene module on glycerophospholipid metabolism (Figure 3C). The metabolite network SN.3 consisted of three clusters (Figure 3D). Using mummichog, the metabolic pathways in this network were enriched for TCA cycle, nucleotide metabolic pathways, and phosphatidylinositol metabolism (Figure 3D, left). The metabolite network SN.3 at day 3 is also associated with the largest transient gene network at day 7, TN7.86, which is shown in Figure 3E. Interestingly, this gene network contains the same metabolic pathways (Figure 3D) that were seen in the metabolite network SN.3 (Figure 3D). A subset of genes in the TN7.86 gene network is shown in Figure 3F, illustrating how our network nodes represent concerted yet redundant variables. Not surprisingly, these genes also indicate close correlation between TLR2/4 signaling and NF-κB pathways. As a third example, largest transient metabolite network appeared at day 1/0 (TN1.1, Figure 3G), which contains multiple pathways associated with steroid hormones (Figure 3G). Of note, the day 3/0 gene network associated with TN1.1 (TN3.76) consisted of B cell and plasma cell genes and targets of the sterol regulatory element binding (SREBF1) transcription factor (Figure 3H).
Thus the MMRN provides a means to contextualize orthogonal variables, and construct networks which integrate multiple data types. Here we query an outcome on MMRN by combining two methods: a) the GSA algorithm (Efron and Tibshirani, 2007), where each node network is treated as a gene set and its significance is estimated by permutation; b) context lookup, where a node network is selected because it contains individual features that are significantly associated with the outcome. Node networks selected by these two methods are then connected via edges in MMRN, and a parsimonious network is presented as the query result. For example, the upregulation of PSPH gene (Figure S2A) is an interesting and unique observation in this study, as it’s not seen in other vaccine studies. The PSPH gene is not even in any BTM module. By querying MMRN, the day 3 upregulation of PSPH is found to be associated with a number of node networks, most significantly a baseline network of inositol phosphate metabolism, transmembrane transport and ubiquitination (Figure S6A). The expression of PSPH at day 3 relative to day 0 was inversely correlated with day 1/0 G-protein coupled receptors (Figure S6B), viral sensing genes and NK signature (not shown), and IL-2 and MIP-1β at day 1/0, and IL-17 at day 3/0 (Figure S6B). Taken together, these data suggest that PSPH is extensively connected with inositol phosphate metabolism. Given the activity of a phosphatase to revert phosphorylation, the upregulation of PSPH may counteract the kinases in NF-κB or MAPK signaling that resulted from VZV infection.
MMRN reveals networks associated with age, sex and viral load
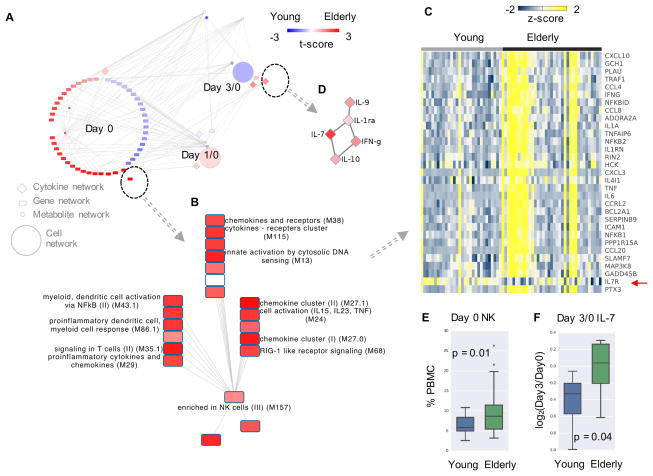
An age related network was obtained by querying the MMRN using the above method (Figure 4A). It was striking that the majority of age related networks were from day 0 prior to vaccination. The most significant gene network consisted of gene modules related to inflammation, cytosolic DNA sensing and NK cells, which were significantly higher in the elderly group (Figure 4B), consistent with our recent results(Nakaya et al., 2015). In fact, this difference was driven by a subset of elderly vaccinees (Figure 4C), indicating biological heterogeneity amongst individuals within a chronological category. Among the few different networks at day 3/0 is an IL-7 cytokine network (Figure 4D). Interestingly, there was lower expression level of IL7R gene in the elderly group (Figure 4C, by red arrow), suggesting a potential compensatory mechanism. Furthermore, difference in NK cells measured by flow cytometry (Figure 4E) was consistent with the transcriptomics data, and Luminex data supported the higher level of IL-7 in the elderly group (Figure 4F). In stark contrast to the age network, the sex related network shows the majority of activities that were higher in females at day 3/0 (Figure S6C), with many T cell related gene modules (Figure S6D). Such differences may account for slightly higher IgG response in females by day 28/0 (Figure S6D).
Figure 4. Age difference explained by MMRN.
A) The subnetwork from MMRN related to age groups, colored by t-score in comparison between the young and elderly groups. Red indicates higher value in elderly. The most significant gene network is shown in B) and C), cytokine network in D). In C), each column represents a subject at day 0, colored by z-scores of gene expression values. IL7R is marked by a red arrow.
E) Baseline NK cell frequencies measured by flow cytometry.
F) Day 3/0 IL-7 levels measured by Luminex.
The presence of VZV DNA in the blood of vaccinees is compounded by temporal differences (Table S2), and could be due to viral replication or DNA carryover from the vaccine injection. If day 1 viral DNA is deemed as carryover from vaccine injection, eight individuals showed clear viral replication (PCR+). The comparison between these 8 PCR+ subjects and 27 PCR− individuals revealed significant differences at all time points (Figure S6E). The PCR+ group had higher baseline levels of plasma cells signature genes and BCR signaling, although these signatures were unlikely to have derived from VZV specific plasmablasts; they also had higher gene expression for heme biosynthesis and inositol phosphate metabolism (Figure S6E). By day 7/0, the PCR+ group had higher activity in cytoskeleton remodeling (enriched for SRF targets), cell adhesion, platelet activation and G protein mediated calcium signaling (Figure S6E), possibly indicative of a response to viral replication. The PCR+ group also had lower activity in plasma cells signature genes at day 7/0 (Figure S6F), indicating a compromised antibody response. Interestingly, the day 7/0 cytokine RANTES is significantly higher in the PCR− group (Figure S6F).
Predictors of the adaptive responses form a redundant and dynamic network
An important goal of systems vaccinology is to identify predictors of the adaptive response to the vaccination. The antibody response induced by Zostavax® was strongly influenced by the baseline antibody level (Figure 1E). Age, sex and study site also had small but significant contributions to the IgG response (Figure S7A). We used a multivariate regression model, accounting for gender, age, study site and baseline VZV IgG, to test independent contribution from each variable of gene modules, metabolite clusters, cytokines or cell populations. All significant predictors of day 28/0 IgG response by this model (p < 0.05) are listed in Figure S7B. These include the gene modules for plasma cells, immunoglobulin and B cells at day 7/0; a module on membrane and ER proteins at day 3/0, and small GTPase mediated signaling as a negative predictor at day 1/0. Among the cell populations, day 7 IFN-γ+ T cells are the most significant predictor of IgG response (Figure S7B). Similarly, the significant predictors of T cell responses at day 7/0 from multivariate regression are listed in Table S5.
Collinearity is an inherent limit of such regression models. Indeed, the majority of these predictors of antibody response and of T cell responses were highly connected or even overlapping in the MMRN (Figure 5A). For example, multiple B cell related networks are both predictors of the IgG response and the Tfh like cell response (Figure 5A). The putative SREBF1 (sterol regulatory element binding transcription factor) target network at day 1/0 and day 3/0 was predictive of Tfh response, and the same network at day 7/0 was predictive of IgG response (Table S5, Figure 5A). Many features of similar correlations to the adaptive responses were captured by MMRN subnetworks, with examples shown in Figure S7C–E. The dynamics of the predictor networks is further illustrated in Figure 5B–C: the plasma and B cell modules were correlated with each other, and with SREBF1 targets and inositol phosphate metabolism at all time points, but the correlation pattern and strength differed. Taken together, the MMRN elevates the statistical prediction beyond linear models by network connections that accommodate indirect steps and temporal developments.
Figure 5. Predictors of adaptive immune response are redundant and dynamic.
A) The significant predictors of IgG (Figure S7B), IFNg+ T cells and Tfh like cells (Table S5) were identified using multivariate regression, then queried against the MMRN. The query results are extensively connected in MMRN, whereas different groups of predictors are placed in different shades. The nodes at the bottom center were not part of the predictors, but included due to their network connections.
B–C) The top antibody predictors are correlated with each other, while the correlation patterns change over time. Inositol phosphate metabolism is a predictor of T cell response, included here as reference. The edge width in the networks is proportional to Pearson correlation coefficient (0.30 to 0.99).
Sterol metabolism integrates humoral and cellular responses
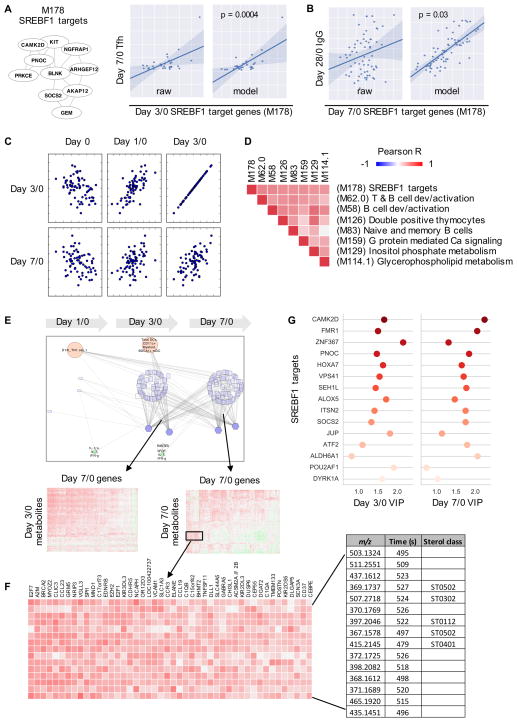
The predictive power of SREBF1 target genes on adaptive responses was brought by a gene module that we identified in a previous study (Li et al., 2014) (Figure 6A). This module contains 6 genes that were putatively identified as SREBF1 targets at the time, but all 10 member genes are now verified SREBF1 targets according to the TRANSFAC database (gene-regulation.com). SREBF1 (also known as SREBP1) is a transcription factor that modulates cellular cholesterol homeostasis, and emerging evidence suggests an important role for it in the immune system (Spann and Glass, 2013; York et al., 2015). In this study, we found that day 3/0 SREBF1 targets was predictive of Tfh response (Figure 6A), and day 7/0 SREBF1 targets was predictive of antibody response (Figure 6B). Post-vaccination, the expression of this gene module showed good correlation between time points, indicating that it captured a stable metabolic characteristic of the individuals; the induced response was inversely correlated to the baseline level (Figure 6C). Furthermore, the activity of these genes was well correlated with several modules on T and B cells, inositol phosphate metabolism and glycerophospholipid metabolism (Figure 6D).
Figure 6. Sterol metabolism integrates cellular and humoral responses.
A–B) The BTM module M178 contains 10 SREBF1 target genes. The expression level of this module at day 3/0 is predictive of the increase of Tfh-like cells, at day 7/0 predictive of IgG response. Each dot represents one subject. Model shows predicted values using a regression model accounting for age, sex and study site (and baseline IgG for the last panel).
C) Scatter plots showing the expression level of M178 at different time points on both axes.
D) Correlation of M178 with other gene modules at day 7/0. The orders on X and Y-axes are symmetrical.
E–F) The MMRN subnetwork associated with M178. Gene and metabolite correlations are further shown in callout heatmaps and in F). A subset of the metabolites in F) are matched to sterol classes as designated in the LIPID MAPS database.
G) Top SREBF1 target genes included in the network in E), indicated by their variable importance by projection (VIP) at days 3/0 and 7/0.
The MMRN subnetwork queried by SREBF1 activity revealed broad associations between genes and metabolites (Figure 6E). The examples in day 7/0 (Figure 6F) include chemokine signaling (CCL23, CCL19, CCR3), TNF/MAPK signaling (TNFSF11, VACM1, DUSP6), complement genes (C1QA, C1QB), killer cell immunoglobulin like receptors (KIR3DL3, KIR2DL3, KIR2DS5), and lipid metabolic enzymes (ACSM2A, ACSM2B, DGAT2). The corresponding metabolite features were matched to several sterol classes in the LIPID MAPS database (lipidmaps.org), while many the peaks showed closely related chromatographic time and correlated intensity, suggesting that they belong to the same lipid category (Figure 6F). Interestingly and reassuringly, the queried network returned many additional SREBF1 target genes, totaling 133 known target genes. The top genes, ranked by importance in PLS regression, include CAMK2D, a kinase downstream of calcium signaling, SOCS2, suppressor of cytokine signaling, POU2AF1, an essential regulator of B cell response, and ALOX5, an enzyme for synthesizing leukotrienes (Figure 6G). Taken together, SREBF1 activity integrated a broad array of both intracellular programs and extracellular signals from cytokines and metabolites.
Inositol phosphate metabolism provides deep immune phenotyping
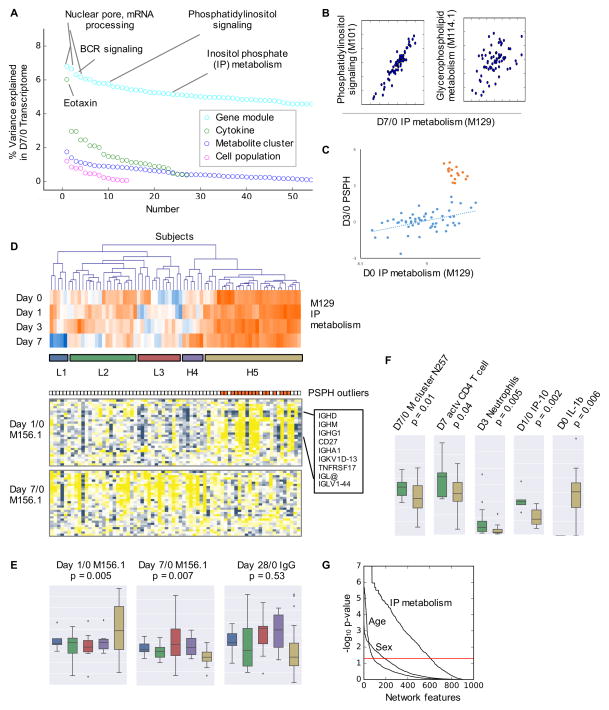
A gene module of inositol phosphate metabolism (M129, day 1/0) is a significant predictor of T cell response (Table S5). Metabolites related to this pathway were significantly changed 1 day after vaccination (Figure 2C), and were extensively associated with gene signatures (Figure 3). Indeed, when we examined the contributing factors to the day 7/0 transcriptome by variance analysis (Nath et al., 2012), BCR signaling, phosphatidylinositol signaling and inositol phosphate metabolism were on the top of the list, besides house-keeping genes such as mRNA processing (Figure 7A). Inositol phosphate (IP) metabolism is known to overlap with phosphatidylinositol signaling and glycerophospholipid metabolism (Figure 7B), and these pathways account for many messenger metabolites essential to immune signaling, including phosphoinositide, diacylglycerol and arachidonic acids (Fritsche, 2006; Okkenhaug, 2013; Zhong et al., 2008).
Figure 7. Immune phenotyping by inositol phosphate metabolism.
A) Percent of variance in the day 7/0 transcriptome explained by each data types, using weighted average of the variance captured by the first five principal components (Nath et al., 2012).
B) Scatter plots of day 7/0 expression of IP metabolism (M129) in relation to day 7/0 phosphatidylinositol signaling and day 7/0 glycerophospholipid metabolism. Each dot represents one subject.
C) The expression of PSPH gene at day 3/0, correlated with baseline inositol phosphate metabolism, reveals an outlier group in the cohort.
D) Hierarchical clustering of subjects based on their gene expression of IP metabolism (M129). The clusters are color coded in the same way as for E–F). The expression of plasma cell signature genes (M156.1) is shown in the two heat maps on the bottom. Each column corresponds to a subject, matched in all three heat maps. The outliers in C are colored in orange, shown to the top of gene heat maps.
E–F) Boxplots for the clusters in D). The p-values are given by Student’s t-test comparing clusters L2 (low) and H5 (high in IP metabolism).
G) Association of all nodes (including unconnected nodes) in the full MMRN network with sex, age (young vs elderly), and IP groups (low vs high). The p-values on Y-axis are based on the GSA query method.
As discussed previously the most upregulated gene PSPH was associated with IP metabolism (Figure S6A). However, a closer examination revealed a dichotomous pattern: for the majority of subjects, the expression of PSPH gene at day 3/0 displayed a linear correlation with the baseline gene module on IP metabolism (blue dots in Figure 7C), but a subset of subjects had a hyper-upregulation of PSPH over 23 folds (orange dots in Figure 7C). Combining data from all time points, groups of subjects of low or high IP metabolism emerged from unsupervised clustering (Figure 7D). Interestingly, all the subjects with hyper-PSPH activity fall into the H5 cluster of high IP metabolism (Figure 7D). This pattern in IP metabolism penetrated into the expression of plasma cell signature genes (M156.1), whereas the group of high IP metabolism showed high expression of immunoglobulins at day 1/0, but low expression at day 7/0 (Figure 7D–E). The Zostavax® specific IgG at day 28/0 followed a similar pattern as day 7/0 M156.1, but the difference between subject groups did not reach statistical significance (Figure 7E). Similarly, the group of high IP metabolism had significantly lower number of activated CD4+ T cells (Figure 7F). Lower level of plasma metabolite cluster Neg257, which is tentatively matched to glycerophospholipids, was also seen in this group, together with differences in neutrophils and cytokines (Figure 7F).
These data suggest that high activity in IP metabolism was detrimental to both T cell and B cell responses. More importantly, IP metabolism manifested a fundamental phenotype in the immune response to Zostavax®. When we examined the association of all nodes in the full MMRN, while 9% was significant for age and 21% for sex, 67% (608/909) of MMRN was significantly associated with IP metabolism (Figure 7G).
DISCUSSION
This study is to our knowledge is the first to combine orthogonal datasets, including PBMC transcriptomics, flow cytometry of blood cell populations and plasma cytokine analysis and metabolomics, to identify molecular networks induced by vaccination. A striking result to emerge from this study is that Zostavax®, which is a live attenuated viral vaccine, induced negligible effector CD8+ T cell response, and a weak type I IFN antiviral response compared to that observed with other acute viral infections such as yellow fever or dengue (Gaucher et al., 2008; Kwissa et al., 2014; Querec et al., 2009). In contrast, there were substantial CD4+ T cell and plasmablast responses. Two factors may have accounted for the weak CD8+ T cell response. First, preexisting antibody against VZV, generated by previous exposure VZV due to infection or vaccination against chickenpox, could attenuate viral replication and the ensuing CD8+ T cell response. Consistent with this, persistent viral DNA was only detected in in a small number of individuals that mostly exhibited CD8+ response. In addition, VZV may have an intrinsic impairment in its ability to stimulate pro-inflammatory cytokines and type I IFN from mDCs and pDCs respectively. Consistent with this, in vitro infection of mDCs, pDCs or monocytes by VZV also failed to induce pro-inflammatory cytokines or IFNα, despite the fact that a substantial fraction of these cells contained viral components (Figure S3). The evasion of VZV from the immune system by latency and attenuation of mDC responses (Abendroth et al., 2010) may also account for our observations.
Analysis of orthogonal datasets revealed highly significant correlations between distinct signatures identified using transcriptomics, metabolomics, flow cytometry and cytokine profiling. Surprisingly, there was striking biological convergence between the pathways identified by metabolomics and transcriptomics. For example, pathways relating to TCA cycle (and overlapping glycolysis and gluconeogenesis) and propanoate metabolism identified by metabolomics of plasma at day 1 were also identified in the day 3/0 transcriptomics data. This surprising antecedence of the metabolic signature relative to the corresponding transcriptomic signature might suggest that blood cells could potentially regulate their transcriptional program in response to metabolic cues. A second insight provided by the integrated analysis of MMRN network was the significant differences between the young and older adults, including baseline NK cells and expression of inflammatory genes. Interestingly, the same gene network, consisting of heme biosynthesis, BCR signaling and inositol phosphate metabolism, was highly expressed in viremic subjects (Figure S6E) and was also strongly associated with poor IgG response (Figure S7C). This baseline metabolic phenotype of individuals appeared to have a large effect on their immune response to vaccination, where inositol phosphate metabolism was a dominating variable in MMRN, which also accounted for the hyper response of PSPH gene. It has been long known that phosphatases are required for the NF-κB/MAPK pathways downstream to TLR signaling (Germain et al., 2011), but which phosphatases in which step are often unknown. Thus PSPH emerged as a major candidate from this study.
In addition, MMRN revealed a striking association of SREBF1 target genes and Tfh cell and antibody responses. SREBF1 is a transcription factor that plays a central role in the modulation of cellular cholesterol homeostasis (Spann and Glass, 2013; York et al., 2015), and emerging evidences show a connection between homeostatic cholesterol sensing and intracellular sterol flux and immunity, such as innate responses and the integrated stress response in macrophages (Im et al., 2011; Shibata et al., 2013), type I IFN signaling and CD4 T cell activation (Bensinger et al., 2008; York et al., 2015). A recent population study of 712 subjects (Bartel et al., 2015) identified the SREBF genes among the most significantly associated with blood metabolites. Our MMRN analysis revealed a large number of genes and metabolites that were integrated via SREBP1 activities, key to the development of B and T cells. The pathways of linoleate and glycerophospholipid metabolism appeared to be a major mediator for antigen presenting cells, highly correlated with other innate pathways at day 3 (Figure 3C), then with T and B cells at day 7 (Figure 6D, S4D). Further involvement of nuclear receptors was also supported by the data on glucocorticoid receptor targets. These data also suggest a potential link between the immune system and the endocrine system, as steroid and xenobiotic receptors and NF-κB signaling suppress each other (Zhou et al., 2006). Should the cytochrome P450 enzymes be perturbed by vaccination (possibly via the lipids in vaccine formula), further questions can be raised on potential interactions with many pharmaceutical drugs, as they are the same enzymes in steroid hormone pathways (Morgan, 2009).
In summary, we demonstrate the utility in analyzing orthogonal data types in an integrated manner to create an MMRN, which provides a powerful tool to contextualize immune responses to vaccination, and investigate their dependency on extraneous variables such as age, sex and viral load. MMRN has revaled new biological insights about the immune response, and the network properties and the importance of metabolic phenotypes will facilitate future biomarker studies and vaccine development.
STAR METHODS
CONTACT FOR REAGENT AND RESOURCE SHARING
Further information and requests for resources and reagents should be directed to and will be fulfilled by the lead contact, Bali Pulendran (bpulend@emory.edu).
EXPERIMENTAL MODEL AND SUBJECT DETAILS
A total of 77 subjects, screened to have a positive VZV antibody titer, were vaccinated with Zostavax®. Written informed consent was obtained from each subject and protocols were approved by Institutional Review Boards of Emory University and the University of Colorado School of Medicine.
METHOD DETAILS
PBMC and plasma collection
PBMCs were isolated using BD Vacutainer CPT tubes, washed and resuspended in complete FBS media (RPMI 1640 plus 10% FBS, 50 U/ml penicillin, 50 μg/ml streptomycin, 2 mM L-glutamine and 50 μM 2-mercaptoethanol) for either immediate use or frozen in FBS with 10% dimethyl sulfoxide for subsequent analysis. Plasma samples were collected and stored at −20°C.
Flow cytometry
Flow cytometry analysis was done on either heparinized whole blood or cryopreserved PBMCs. For whole blood assays, cells were stained with anti-CD4 APC-H7, anti-CD8 V500 and anti-CD3 PerCP for 20 min at room temperature followed by erythrocyte lysis using BD FACS lysing solution (BD Biosciences). Cells were then fixed and permeabilized with BD Cytofix/Cytoperm for 30 min on ice and then anti-Ki67 FITC and anti-Bcl6 PE added for 30 min on ice in BD permeabilization buffer. To measure blood follicular T helper like cells, cryopreserved cells were resuscitated and stained with anti-CD45RA FITC, anti-PD-1 PE, anti-ICOS PerCP, anti-CXCR5 Alexa 647, anti-CD4 APC-H7, anti-CXCR3 PE-Cy7, anti-CD3 Alexa 700 and live/dead aqua (Invitrogen). All antibodies except anti-PD-1 PE (Biolegend) and anti-CD3 Alexa 700 (Invitrogen) were purchased from BD. To determine the frequency of innate cell populations and their activation status, flow cytometry analysis was done on cryopreserved PBMCs. Following live/dead staining (Alexa Fluor 430, Life Technologies), surface staining of cells was done with an appropriate antibody cocktail. All samples were run on an LSR II (BD Biosciences) and analyzed via FlowJo software. Each cell population was quantified as percentage of total PBMC.
Zostavax®-specific IFN-γ, IL-4 and IL-21 ELISPOT
Ninety-six well filter plates (Millipore–S2EM004M99 for IFN-γ; MAIPS4510 for IL-4 and IL-21) were coated with either purified anti-human IFN-γ (BD Biosciences; 1:200), anti-human IL-4 (BD Biosciences; 1:200) or anti-human IL-21 (MABTECH; 1:50) in PBS overnight at 4°C. Plates were then washed once and blocked with complete FBS media for 2 hr at 37°C. Cryopreserved cells (3–5×105/well) were resuspended in RPMI 1640 + 10% human AB serum (Sigma Aldrich), penicillin, streptomycin, L-glutamine and 2-mercaptoethanol and added to each plate, stimulated with either media alone, 0.25 μg of VZV cell lysate (Meridian Life Sciences - Strain Ellen grade II detergent free) or VZV HLA-A2-restricted peptide pool (ILIEGIFFV (Ribonucleotide reductase subunit 2 (Chiu et al., 2014)), DLLYFVCLGV (ORF34; previously identified in Ahmed laboratory), ALWALPHAA (IE62 (van der Heiden et al., 2009)), SLPRSRTPI (IE62 (Frey et al., 2003)) and SAPLPSNRV (IE62 (Frey et al., 2003))). The cells were then incubated overnight at 37°C. The following day, plates were washed with H2O for 5 min × 2, four times with PBS-0.05% Tween (PBS-T) and then biotin anti-human IFN-γ (BD Biosciences; 1:250), anti-human IL-4 (BD Biosciences; 1:250) or anti-human IL-21 (MABTECH; 1:500) added in PBS + 10% FBS for 2 hr at room temperature. Plates were washed with PBS-T and streptavidin-HRP (Jackson Immunoresearch) added at 1:1000 in PBS + 10% FCS for 1 hr at room temperature. Plates were washed four times with PBS-T, four times with PBS and developed using AEC substrate (3 amino-9 ethyl-carbazole; Sigma Aldrich). Developed plates were dried, scanned and analyzed using an automated ELISPOT counter (Cellular Technologies, Ltd).
Zostavax®-specific and total IgG B cell ELISPOT
Ninety-six well filter plates (Millipore–MSHAN4B50) were coated with either VZV cell lysate (Meridian Life Sciences) at 5 μg/well or Affinipure donkey anti-human IgG (H+L; Jackson Immunoresearch) at 100 ng/well in PBS overnight at 4°C. The plates were washed with PBS four times and complete FBS media added for 2 hr at 37°C. Freshly prepared PBMCs were resuspended in complete FBS media at 1×107/ml and added to each well, followed by threefold dilution and then incubated overnight at 37°C. The next day, the plates were washed four times with PBS, four times with PBS-T and incubated for 1.5 hr at room temperature with Biotin-SP-Affinipure F(ab′)2 donkey anti-human IgG (Jackson Immunoresearch) at 100 ng/well in PBS + 10% FCS + 0.05% Tween 20 (PBS-T-FBS). Plates were washed four times with PBS-T and incubated for 1.5 hr with HRP-avidin D (Vector laboratories) at 1:1000 in PBS-T-FBS. Plates were washed four times with PBS-T, four times with PBS and then developed using AEC substrate (3 amino-9 ethyl-carbazole; Sigma Aldrich). Developed plates were dried, scanned and analyzed using an automated ELISPOT counter (Cellular Technologies, Ltd).
Zostavax®-specific IgG/IgA ELISA
Maxisorp plates (Nalgene-Nunc) were coated with either 0.5 μg/well VZV cell lysate (Meridian Life Sciences) or VZV glycoprotein-only protein (Merck) diluted in PBS and incubated overnight at 4°C. Plates were washed with PBS+0.1% Tween 20 and blocked with PBS+10%FBS+0.05% Tween 20 (blocking buffer) for 1.5 hr at room temperature. Plasma samples were serially diluted in blocking buffer and incubated for 1.5 hr at room temperature. Plates were washed four times with PBS+0.1% Tween 20 and goat anti-mouse IgG or IgA HRP added at 1:5000 in blocking buffer for 1.5 hr at room temperature. Plates were washed four times with PBS+0.1% FBS, four times with PBS and developed using the OPD peroxidase substrate (Sigma Aldrich), stopped by addition of 1M HCL and read at 415 nm on a microplate reader. The antibody titer was measured at the EC50.
Viral DNA detection
The DNA was extracted with the QuickGene DNA Whole Blood kit per manufacturer’s recommendations (Autogen, Cat. No DB-S). Typical yield was 4–8 μg of DNA from 200 μL blood. PCR was carried out using 200 ng of extracted DNA in 5 μL of water and 15 μL Master Mix containing 50 mM Tris-Cl (pH8.3), 10 mM KCl, 5 mM (NH4)2SO4, 2 mM MgCl2, 200 mM of each deoxynucleoside triphosphate, 0.5 μM of each primer, and 1.25 U of FasStart Taq DNA polymerase (Roche). VZV genome open reading frame 63 (ORF63) was detected with a downstream primer sequence (VZV63-f) of 5′-CGAGATTCACGAAGATTGCG-3′ and an upstream primer sequence (VZV63-r) of 5′-CAATTACATCCGATGGCGTAG-3′. Amplification with these primers yielded a DNA fragment of 292 nucleotides. In order to increase the sensitivity to detect VZV DNA ORF63, and to verify the PCR products having the same DNA sequence of VZV ORF63, nested PCR analysis was carried out on 1/40 of the volume of PCR product that did not initially contain detectable VZV DNA. This utilized a second pair of VZV ORF63 primers. The downstream nested primer (V63-315d) sequence was 5′-GTGCTGGGAGGAATTGTTACAG-3′, and the upstream primer (V63-500u) sequence was 5′-TCGTCGCTATCGTCTTCACCAC-3′. Amplification with these primers yielded a DNA fragment of 186 nucleotides. Amplification was performed with a RotorGene (RG-3000) real-time PCR System or the Bio-Rad CFX96 real-time PCR system, depending on availability. For the latter we used Bio-Rad’s STBR Green Supermix (catalog number 172–5260) as the master mix. The primers and thermal profile were the same for both systems and they provided similar sensitivity in reconstruction experiments. The initial DNA polymerase activation step was 95°C for 6 minutes, followed by 50 cycles of 95 C for 10 seconds, 65 C for 20 seconds, and 72 C for 30 seconds. The nested PCR initial DNA polymerase activation step was 95 C for 6 minutes, followed by 50 cycles of 95°C for 10 seconds, 66 C for 20 seconds, and 72 C for 20 seconds. Melting curve analysis from either type of amplification was carried out from 60 C to 95 C. Each sample in either type of amplification was tested in triplicate. The type of VZV DNA detected was identified on random positive samples by partial sequencing of amplicons to include codon 958 of ORF62, which distinguishes vaccine strain VZV (V-Oka) from wild type varicella (Gomi et al., 2002). Sensitivity of the PCR assay was determined by adding cell-associated VZV from 500 to 16000 VZV DNA copies to 1 mL of repeatedly PCR-negative control blood. The DNA was extracted from the reconstruction samples and tested for VZV DNA as described above.
Multiplex cytokine assays
Plasma samples from CPTs were stored at −80°C prior to cytokine analysis. Luminex assays were performed with the Beadlyte Human 10-Plex Multi-Cytokine Detection System (Upstate). Plasma samples were run in duplicate following the manufacturer’s protocol on a Bio-Plex Luminex-100 station (Bio-Rad). Data were log2 transformed. Only cytokines above limit of detection were included in further analysis.
In vitro stimulation of PBMCs with Zostavax®
PBMCs isolated from healthy donors were cultured in RPMI and stimulated with Zostavax® (Zos at multiplicity of infection; 0.00028 or 0.0011 or 0.0045) or TLR9 ligand CpG-A (8 μg/ml, invivogen) or R848 (1 μg/ml, invivogen) for 20 hours. Following live/dead staining using zombie fixable viability kit (Biolegend), surface staining of cells was done to identify different innate cell populations and also for the expression of activation marker, CD80 and HLA-DR. For intracellular staining, cells were fixed and permeabilized (BD Fix/Perm kits) and stained for VZV-gI to detect the replication of VZV and that of IL-6 in CD14+ monocytes and CD1c+ myeloid DCs (mDCs), and IFN-α in CD123hi plasmacytoid DCs (pDCs) to analyze the cytokine production. Multi-color flow cytometry acquisition was performed on a BD LSR II and data was analyzed using FlowJo (Treestar). Cell-free culture supernatants collected were analyzed for cytokines and chemokines, including IL-6, TNF, CXCL8, CCL2, CXCL9 and CXCL10 using Cytokine Bead Array (CBA) kits (BD Biosciences, San Diego, US)
PBMC Transcriptomics analysis
The transcriptomics analysis was performed as previously described (Li et al., 2014; Nakaya et al., 2011). Total RNA from PBMCs (~1.5 × 106 cells) was isolated using Trizol® (Invitrogen, Life Technologies Corporation) according to the manufacturer’s instructions. Samples, after quality control, were hybridized on Human U133 Plus 2.0 Arrays (Affymetrix).
Plasma Metabolomics analysis
Metabolomics analysis was performed similarly as previously described (Li et al., 2013). Briefly, metabolites were extracted by the addition of 130 μL of acetonitrile to 65 μL of plasma, followed by mixing, and 30 minutes of incubation on ice and centrifugation (13,400 × rpm at 4°C) for 10 minutes. The supernatant was transferred into autosampler vials for LC-MS analysis. Mass spectral data were collected with a 10-minute reversed phase gradient on a Thermo Q Exactive mass spectrometer (Thermo Fisher, San Diego, CA) set to record from mass-to-charge ratio (m/z) 85 to 2000. Three technical replicates were run for each sample using reversed phase chromatography (Higgins Analytical, Targa C18 5 μM, 100 × 2.1 mm) using both positive and negative electrospray ionization. Stable isotope standards ([13C6]-D-glucose, [13C5]-L-glutamic acid, [15N]-L-tyrosine, [trimethyl-13C3]-caffeine, [3,3-13C2]-cystine, [15N,13C5]-L-methionine, Cambridge Isotope Laboratories (>98% isotopic purity)) were added to each sample prior to metabolite extraction, and pooled reference plasma samples were analyzed with each batch for quality control. Metabolite confirmation was performed with tandem MS-MS experiments on Thermo QE HF, Orbitrap Fusion, or Finnigan LTQ FT Ultra mass spectrometers. To identify vaccine-derived metabolites, the ZostavaxR vaccine was resuspended according to the manufacturer’s instructions, and analyzed with the same metabolomics protocol as for plasma.
QUANTIFICATION AND STATISTICAL ANALYSIS
Transcriptomics data processing and pathway/module analysis
The transcriptomics data were processed using RMA algorithm in BioConductor (Irizarry et al., 2003). Enrichment tests of KEGG pathways and transcription factor targets were performed using GSEA (Subramanian et al., 2005). For BTM analysis, the transcriptomics data were summarized into module level score by taking the mean value of member genes (Li et al., 2014). Paired t-test was used to compare genes or modules between different time points.
Metabolomics and pathway analysis
Metabolite peaks were extracted using the metabolomics software package apLCMS (Yu et al., 2013). Only peaks that were present in > 90% samples were used for further analysis. Pearson correlation coefficient R between the three technical replicates was used as an indicator of analytical quality. For data from positive ionization, 94% of all injections produced R > 0.90. Eight replicates with R < 0.76 were removed from analysis. For each sample, data were log2 transformed and averaged over replicates. Twenty-three metabolites were confirmed by matching to chemical standards, including linoleate, methionine, cystine, 2-aminobutyrate, hypoxanthine, carnitine, o-acetylcarnitine, myo-inositol and glycerophosphocholine. Data from negative ionization were processed similarly. A total of 2216 peaks and 4048 peaks were used for positive and negative analysis. Using accurate m/z search under 10 ppm, 838 and 1206 metabolites were matched in mummichog, which include metabolites from KEGG and other databases, from positive ionization and negative ionization respectively. Mummichog further tests pathway enrichment patterns using permutations, and computes the probability for each pathway (Li et al., 2013).
Data visualization using Circos
Figure 1A and Figure 2D were generated using Circos 0.66 (http://circos.ca). In Figure 1A: each column represents a subject, with each track as one type of data. In all tracks, higher bar or darker color corresponding to greater value, except in Gender, males are colored blue and females colored in red; missing data in white. The number of measured subjects varies by data types. N=72 for transcriptomics and metabolomics; N=70 for anti-VZV IgG; N=62 for flow cytometry; N=67 for VZV DNAemia; N=20 for cytokines. The antigen-specific T cell assays were mostly performed in Emory subset of subjects, and sample size is reported in individual figures.
Cytokines, examples shown for IL-12 (p70) levels on day 1.
Innate cells, examples from top to bottom: day1 CD16+ Neutrophils, D1 BDCA-1+ mDC1, day1 CD11c+ Myeloid cells, day1 Lin-HLA-DR+ total DCs, day3 CD56+ NK cells.
B cells, examples of CD19+ B-cell counts for days 1, 3, and 7.
T cells, examples of CD3+ T-cell counts for days 1, 3, and 7.
Specific T cells, antigen specific CD4+ T cell counts for day 7 vs. day 0.
VZV DNAemia, histogram of copy numbers for days 3, 7, and 14.
Transcriptomics, example gene modules shown from top to bottom: Day3/0_IL2 IL7 TCR network (M65), Day3/0_B cell development (M9), Day3/0_T cell differentiation (M14), Day7/0_plasma cells immunoglobulins (M156.1), Day7/0_enriched in B cells (III) (M47.2), and Day3/0_enriched in T cells (II) (M223).
Metabolomics, example cluster shown from top to bottom: Day3/0_Clus_neg_25, Day3/0_Clus_neg_5, Day3/0_Clus_neg_195, Day7/0_Clus_neg_257, Day14/0_Clus_neg_10, and Day14/0_Clus_neg_1320.
IgG, antigen specific IgG increase at day 30 (bars), and baseline at day 0.
Linear regression models in MWAS and predictors
All transcriptomics and metabolomics data were log2 transformed. For correlation and regression analyses, difference to baseline samples was used. All regression models were run in R. For MWAS, each metabolite peak was regressed against a BTM score, taking into account age, sex and study site, using the lm function and Storey FDR (Storey and Tibshirani, 2003). For predictors of adaptive response, including IgG, IFN-γ+ T cells and Tfh-like cells, R function glm and epicalc library were used, accounting for age, sex and study site.
Construction and query of MMRN
As illustrated in Figure S5, for dimension reduction, gene expression was reduced to BTM activities, and metabolite peaks were grouped by unsupervised clustering (Ben-Dor et al., 1999). We computed the correlation network of all BTMs, using Pearson correlation with Bonferroni correction (p < 0.001). The metabolite peaks from metabolomics were first clustered by an unsupervised algorithm (Ben-Dor et al., 1999). The resulting clusters often grouped peaks that belong to the same metabolite (isotopes or adducts) or pathway, as evidenced by matched retention time and mass defect. The mean value was then taken for each cluster, used for computing all pair-wise Pearson correlation, and constructed into a correlation network of metabolite clusters (Bonferroni p < 0.001). These correlation networks, as undirected and unweighted, were partitioned into subnetworks by linkcomm algorithm, which optimizes for partition density (Kalinka and Tomancak, 2011). Since the linkcomm algorithm deals with edge clusters not node clusters, it allows redundancy in the results, i.e., the same node can belong to multiple subnetworks. The resulting metabolite networks were used as input for MMRN construction. The same procedure was performed for BTM networks, cytokine networks and cell networks. Of note, these resulting networks are not meant to infer regulatory relationship, but serve as units of dimension reduction since their members manifested similar patterns in this dataset. These networks were also distinguished into two categories, whereas “stable networks” were derived from correlations that were observed at all time points, while “transient networks” were derived from correlations that were observed at a particular time point. “Stable” and “transient” are referring to network topology, as both types of networks are dynamic and their quantitative levels vary over time, but the stable networks have conserved members and structures.
Partial least square (PLS) regression was used to assess the association across different data types. PLS finds the principal components with the best covariance between two matrices (Janes et al., 2005). In this study, data matrices from the four types of networks (genes, metabolites, cytokines and cells) were used as input to PLS regression. The p-values of association is computed on 1 million permutations for each pair of data types, and the significantly associated networks were connected to a multiscale, multifactorial response network (MMRN). It should also be noted that each node in MMRN is a network of its own right. This hierarchical structure allows an overview of the super-network and multiple zoom levels for additional details. A query program was written for MMRN, to extract a parsimonious subnetwork which connects the query nodes. To use GSA algorithm on member networks, t-score or Pearson correlation coefficient was used as primary score, and the significance is based permutation (Efron and Tibshirani, 2007). VIP of PLS regression was computed according to (Mehmood, 2012). The networks were visualized using Cytoscape 3 (cytoscape.org). The MMRN in Cytoscape format, with all source data, is provided in Data S1.
DATA AND SOFTWARE AVAILABILITY
The GEO accession number for the microarray data reported in this paper is GSE79396.
Supplementary Material
KEY RESOURCES TABLE.
| REAGENT or RESOURCE | SOURCE | IDENTIFIER |
|---|---|---|
| Antibodies | ||
| Anti-human Ki-67 Alexa 488 (clone B56) | BD | 561165 |
| Anti-human Bcl2 PE (clone BCL2/100) | BD | 340651 |
| Anti-human CD3 PerCP (clone SK7) | BD | 347344 |
| Anti-human CD4 APC-H7 (clone RPA-T4) | BD | 560158 |
| Anti-human CD8 V500 (clone RPA-T8) | BD | 560774 |
| Anti-human ICOS Alexa 488 (clone DX29) | BD | 564549 |
| Anti-human CXCR5 Alexa 647 (clone RF8B2) | BD | 558113 |
| Anti-human CD45RA PE-Cy5 (clone HI100) | BD | 555490 |
| Anti-human CXCR3 PE-Cy7 (clone 1C6/CXCR3) | BD | 560831 |
| Anti-human CCR7 PE-CF594 (clone 150503) | BD | 562381 |
| Anti-human CD3 Alexa700 (clone UCHT1) | Invitrogen | CD0329 |
| Anti-human Ki-67 V450 (clone B56) | BD | 561281 |
| Anti-human PD-1 BV605 (clone EH12.2H7) | Biolegend | 329924 |
| Anti-human T-bet V450 (O4-46) | BD | 561312 |
| Anti-human RORgT PE (Q21-559) | BD | 563081 |
| Anti-human IFNg ELISPOT antibody pair | BD | 551873 |
| Anti-human IL-4 ELISPOT antibody pair | BD | 551885 |
| Anti-human IL-21 ELISPOT antibody pair | Mabtech | 3540-2H |
| Avidin-HRP | Vector Laboratories | A-2014 |
| B cell antibody ELISPOT plate | Millipore | MSHAN4B50 |
| Affinipure donkey anti-human IgG (H+L) | Jackson Immunoresearch | 709-005-149 |
| Biotin-SP-Affinipure F(ab′)2 donkey anti-human IgG | Jackson Immunoresearch | 709-066-149 |
| Goat anti-human IgG HRP | Southern biotech | 2010-05 |
| Goat anti-human IgA HRP | Southern biotech | 2010-05 |
| Anti-human CD4 microbeads | Miltenyi Biotec | 130-045-101 |
| Anti-human CD8 microbeads | Miltenyi Biotec | 130-045-201 |
| Mouse monoclonal anti-Lineage-FITC (clone multiple) | BD | 340546 |
| Mouse monoclonal anti-CD69-FITC (Clone FN50) | BD | 557049 |
| Mouse monoclonal IgG1 Isotype–FITC (clone MOPC-21) | BD | 555909 |
| Mouse monoclonal anti-CD86-PE (clone 2331) | BD | 555658 |
| Mouse monoclonal anti-IgG1-PE (clone MOPC-21) | BD | 555749 |
| Mouse monoclonal anti-CCR7-PE (clone 150503) | R&D Systems | FAB197P |
| Mouse monoclonal anti-IgG2a-PE (clone 201102) | R&D Systems | IC003P |
| Mouse monoclonal anti-HLA-DR-PE Texas-Red (clone TU36) | Invitrogen | MHLDR17 |
| Mouse monoclonal anti-BDCA1-PerCpCy5.5 (clone L161) | BioLegend | 331514 |
| Mouse monoclonal anti-CD56-PerCpCy5.5 (clone HCD56) | BioLegend | 318322 |
| Mouse monoclonal anti-CD123-PECy7 (clone 6H6) | BioLegend | 306010 |
| Mouse monoclonal anti-CD3-PECy7 (clone SP34-2) | BD | 557749 |
| Mouse monoclonal anti-CD86-APC (clone 2331/FUN-1) | BD | 555660 |
| Mouse monoclonal anti-CD19-APC (clone SJ25C1) | eBioscience | 17-0198-42 |
| Mouse monoclonal anti-IgG1 APC (clone MOPC-21) | BD | 555751 |
| Mouse monoclonal anti-HLA-DR-APC-Cy7 (clone L243) | BD | 335814 |
| Mouse monoclonal anti-CD4-APC-Cy7 (clone RPA-T4) | BD | 557871 |
| Mouse monoclonal anti-CD11c-AF700 (clone B-ly6) | BD | 561352 |
| Mouse monoclonal anti-CD16 AF700 (clone 3G8) | BD | 557920 |
| Mouse monoclonal anti-CD11b-Pacific blue (clone ICR44) | BioLegend | 301315 |
| Mouse monoclonal anti-IgG1-Pacific blue (clone MOPC-21) | BioLegend | 400151 |
| Mouse monoclonal anti-CCR5-V450 (clone 2D7/CCR5) | BD | 562121 |
| Mouse monoclonal anti-IgG2a-V450 (clone G155-178) | BD | 560550 |
| Aqua live/dead (Alexa 430) | Invitrogen | L34957 |
| Mouse monoclonal anti-CD14-Qdot605 (clone TUK4) | Invitrogen | Q10013 |
| Mouse monoclonal anti-CD80-FITC (clone L307.4) | BD | 557226 |
| Mouse monoclonal anti-CD1c-PE (clone AD5 8E7) | Miltenyi Biotech | 130-098-007 |
| Mouse monoclonal anti-CD3-PE-CF594 (clone UCHT1) | BD Horizon | 562280 |
| Mouse monoclonal anti-CD19-PE-CF594 (clone HIB19) | BD Horizon | 562294 |
| Mouse monoclonal anti-CD40-PerCP-CY5.5 (clone 5C3) | Biolegend | 334316 |
| Mouse monoclonal anti-CD86-BV421 (clone IT2.2) | Biolegend | 305426 |
| Mouse monoclonal anti-CD16-BV650 (clone 3G8) | Biolegend | 302041 |
| Mouse monoclonal anti-CD11c-BV711 (clone B-ly6) | BD Horizon | 563130 |
| Mouse monoclonal anti-anti-VZV-gE (clone MAB8612) | Millipore | MAB8612 |
| Mouse monoclonal anti-CD69-AF700 (clone FN50) | BD | 560739 |
| Mouse monoclonal anti-IL-1beta-FITC (clone AS10) | BD | 340515 |
| Mouse monoclonal anti-IL-8-PerCP-Cy5.5 (clone BH0814) | Biolegend | 514605 |
| Mouse monoclonal anti-IFN-alpha2b-V450 (clone 7N4-1) | BD | 561382 |
| Mouse monoclonal anti-IL-6-APC (clone MQ2-13A5) | Biolegend | 501112 |
| Mouse monoclonal anti-TNF-alpha-AF700 (Clone MAb11) | eBioscience | 56-7349 |
| Human Inflammatory Cytokine Kit-CBA | BD | 551811 |
| Human Chemokine Kit-CBA | BD | 552990 |
| Bacterial and Virus Strains | ||
| VZV infected cell lysate (strain Ellen; detergent free | Meridian Life Sciences | 7740 |
| VZV glycoprotein only protein | Merck | |
| Chemicals, Peptides, and Recombinant Proteins | ||
| Live/dead aqua | Invitrogen | L34966 |
| ILIEGIFFV | Mimitopes | |
| DLLYFVCLGV | Mimitopes | |
| ALWALPHAA | Mimitopes | |
| SLPRSRTPI | Mimitopes | |
| SAPLPSNRV | Mimitopes | |
| 3 amino-9 ethyl-carbazole | Sigma-Aldrich | A5754 |
| OPD peroxidase substrate | Sigma-Aldrich | P5412 |
| SYBR Green supermix | Bio-Rad | 172-5260 |
| TLR9 ligand ODN 2216 (Class A CpG) | Invivogen | tlrl-2216 |
| TLR7/8 ligand Imidazoquinoline compound | Invivogen | tlrl-r848 |
| TLR4 ligand LPS-EK Ultrapure | Invivogen | tlrl-peklps |
| Critical Commercial Assays | ||
| Quickgene DNA whole blood kit | Autogen | DB-S |
| Deposited Data | ||
| Microarray data | This paper | GSE79396 |
| Oligonucleotides | ||
| VZV 63For CGAGATTCACGAAGATTGCG | Invitrogen | |
| VZV 63Rev CAATTACATCCGATGGCGTAG | Invitrogen | |
| VZV-315d GTGCTGGGAGGAATTGTTACAG | Invitrogen | |
| V63-500u TCGTCGCTATCGTCTTCACCAC | Invitrogen | |
| Software and Algorithms | ||
| FlowJo 9.9.3 | FlowJo | |
| Prism 6 | Graphpad | |
| Bioconductor 2.13 | http://bioconductor.org | |
| Circos 0.66 | http://circos.ca | |
| apLCMS 6 | Yu et al, 2013 | http://web1.sph.emory.edu/apLCMS/ |
| Mummichog 0.10.3 | Li et al, 2013 | http://mummichog.org |
| Other | ||
| IFNg ELISPOT plate | Millipore | S2EM004M99 |
| IL-2/IL-21 ELISPOT plate | Millipore | MAIPS4510 |
| DNA microarray data from the PBMC samples before and after VZV vaccination | This paper | https://www.ncbi.nlm.nih.gov/geo/query/acc.cgi?acc=GSE79396 |
Acknowledgments
We are grateful to Mary Bower, Ildefonzo Tellez, Srilatha Edugupanti, David Rimland and the Hope Clinic Staff who assisted with the clinical work. We also thank Emory Immunology/Flow Cytometry Core for technical assistance. This work was supported by NIH grants U19AI090023 (B.P.), U19AI057266 (R.A. and B.P.).
Footnotes
AUTHOR CONTRIBUTIONS
S.L. led the bioinformatics and metabolomics analyses, and performed analysis and integration of transcriptomics, metabolomics, MMRN and predictive modeling. N.L.S. led the collection and analysis of adaptive immune data. The clinical visits and sample collection were performed by N.R., A.M., J.W. and J.C. T.Y. contributed to metabolomics data processing and MWAS. S.B. and T.H. contributed to transcriptomics data analysis and data management. M.S.M performed in vitro analysis of IFN response, and flow cytometry analysis with S.D. M.M., C.C. and A. Wieland contributed to antibody, B cell and T cel analyses. S.D. and K.V. analyzed gP specific IgG response. K.L., V.T. and D.P.J. performed the mass spectrometry analyses. A.S. contributed to antigen specific T cell analysis. H.I.N. led the microarray sample analysis. H.I.N. and S.S. participated in and supervised the overall data analysis. B.P., R.A., A.W., M.L. and M.J.M. conceived the study and designed the experiments. S.L., N.L.S. and B.P. wrote the paper.
Publisher's Disclaimer: This is a PDF file of an unedited manuscript that has been accepted for publication. As a service to our customers we are providing this early version of the manuscript. The manuscript will undergo copyediting, typesetting, and review of the resulting proof before it is published in its final citable form. Please note that during the production process errors may be discovered which could affect the content, and all legal disclaimers that apply to the journal pertain.
References
- Abendroth A, Kinchington PR, Slobedman B. Varicella zoster virus immune evasion strategies. Curr Top Microbiol Immunol. 2010;342:155–171. doi: 10.1007/82_2010_41. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Akondy RS, Monson ND, Miller JD, Edupuganti S, Teuwen D, Wu H, Quyyumi F, Garg S, Altman JD, Del Rio C, et al. The yellow fever virus vaccine induces a broad and polyfunctional human memory CD8+ T cell response. Journal of immunology. 2009;183:7919–7930. doi: 10.4049/jimmunol.0803903. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bartel J, Krumsiek J, Schramm K, Adamski J, Gieger C, Herder C, Carstensen M, Peters A, Rathmann W, Roden M, et al. The Human Blood Metabolome-Transcriptome Interface. PLoS genetics. 2015;11:e1005274. doi: 10.1371/journal.pgen.1005274. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ben-Dor A, Shamir R, Yakhini Z. Clustering gene expression patterns. Journal of computational biology : a journal of computational molecular cell biology. 1999;6:281–297. doi: 10.1089/106652799318274. [DOI] [PubMed] [Google Scholar]
- Bensinger SJ, Bradley MN, Joseph SB, Zelcer N, Janssen EM, Hausner MA, Shih R, Parks JS, Edwards PA, Jamieson BD, et al. LXR signaling couples sterol metabolism to proliferation in the acquired immune response. Cell. 2008;134:97–111. doi: 10.1016/j.cell.2008.04.052. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bentebibel SE, Lopez S, Obermoser G, Schmitt N, Mueller C, Harrod C, Flano E, Mejias A, Albrecht RA, Blankenship D, et al. Induction of ICOS+CXCR3+CXCR5+ TH cells correlates with antibody responses to influenza vaccination. Science translational medicine. 2013;5:176ra132. doi: 10.1126/scitranslmed.3005191. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chiu C, McCausland M, Sidney J, Duh FM, Rouphael N, Mehta A, Mulligan M, Carrington M, Wieland A, Sullivan NL, et al. Broadly reactive human CD8 T cells that recognize an epitope conserved between VZV, HSV and EBV. PLoS pathogens. 2014;10:e1004008. doi: 10.1371/journal.ppat.1004008. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Crotty S. Follicular helper CD4 T cells (TFH) Annual review of immunology. 2011;29:621–663. doi: 10.1146/annurev-immunol-031210-101400. [DOI] [PubMed] [Google Scholar]
- Davis MM. A prescription for human immunology. Immunity. 2008;29:835–838. doi: 10.1016/j.immuni.2008.12.003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Efron B, Tibshirani R. On testing the significance of sets of genes. The annals of applied statistics. 2007:107–129. [Google Scholar]
- Franco LM, Bucasas KL, Wells JM, Nino D, Wang X, Zapata GE, Arden N, Renwick A, Yu P, Quarles JM, et al. Integrative genomic analysis of the human immune response to influenza vaccination. eLife. 2013;2:e00299. doi: 10.7554/eLife.00299. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Fritsche K. Fatty acids as modulators of the immune response. Annual review of nutrition. 2006;26:45–73. doi: 10.1146/annurev.nutr.25.050304.092610. [DOI] [PubMed] [Google Scholar]
- Furman D, Jojic V, Kidd B, Shen-Orr S, Price J, Jarrell J, Tse T, Huang H, Lund P, Maecker HT, et al. Apoptosis and other immune biomarkers predict influenza vaccine responsiveness. Molecular systems biology. 2013;9:659. doi: 10.1038/msb.2013.15. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gaucher D, Therrien R, Kettaf N, Angermann BR, Boucher G, Filali-Mouhim A, Moser JM, Mehta RS, Drake DR, 3rd, Castro E, et al. Yellow fever vaccine induces integrated multilineage and polyfunctional immune responses. The Journal of experimental medicine. 2008;205:3119–3131. doi: 10.1084/jem.20082292. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Germain RN, Meier-Schellersheim M, Nita-Lazar A, Fraser ID. Systems biology in immunology: a computational modeling perspective. Annual review of immunology. 2011;29:527–585. doi: 10.1146/annurev-immunol-030409-101317. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Groom JR, Luster AD. CXCR3 in T cell function. Experimental cell research. 2011;317:620–631. doi: 10.1016/j.yexcr.2010.12.017. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Im SS, Yousef L, Blaschitz C, Liu JZ, Edwards RA, Young SG, Raffatellu M, Osborne TF. Linking lipid metabolism to the innate immune response in macrophages through sterol regulatory element binding protein-1a. Cell metabolism. 2011;13:540–549. doi: 10.1016/j.cmet.2011.04.001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Janes KA, Albeck JG, Gaudet S, Sorger PK, Lauffenburger DA, Yaffe MB. A systems model of signaling identifies a molecular basis set for cytokine-induced apoptosis. Science. 2005;310:1646–1653. doi: 10.1126/science.1116598. [DOI] [PubMed] [Google Scholar]
- Kalinka AT, Tomancak P. linkcomm: an R package for the generation, visualization, and analysis of link communities in networks of arbitrary size and type. Bioinformatics. 2011;27:2011–2012. doi: 10.1093/bioinformatics/btr311. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kazmin D, Nakaya HI, Lee EK, Johnson MJ, Most Rvd, Berg RAvd, Ballou WR, Jongert E, Wille-Reece U, Ockenhouse C, et al. Systems analysis of protective immune responses to RTS,S malaria vaccination in humans. PNAS. 2017 doi: 10.1073/pnas.1621489114. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kidd BA, Peters LA, Schadt EE, Dudley JT. Unifying immunology with informatics and multiscale biology. Nature immunology. 2014;15:118–127. doi: 10.1038/ni.2787. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kwissa M, Nakaya HI, Onlamoon N, Wrammert J, Villinger F, Perng GC, Yoksan S, Pattanapanyasat K, Chokephaibulkit K, Ahmed R, et al. Dengue virus infection induces expansion of a CD14(+)CD16(+) monocyte population that stimulates plasmablast differentiation. Cell host & microbe. 2014;16:115–127. doi: 10.1016/j.chom.2014.06.001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Li S, Park Y, Duraisingham S, Strobel FH, Khan N, Soltow QA, Jones DP, Pulendran B. Predicting network activity from high throughput metabolomics. PLoS computational biology. 2013;9:e1003123. doi: 10.1371/journal.pcbi.1003123. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Li S, Rouphael N, Duraisingham S, Romero-Steiner S, Presnell S, Davis C, Schmidt DS, Johnson SE, Milton A, Rajam G, et al. Molecular signatures of antibody responses derived from a systems biology study of five human vaccines. Nature immunology. 2014;15:195–204. doi: 10.1038/ni.2789. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Miller JD, van der Most RG, Akondy RS, Glidewell JT, Albott S, Masopust D, Murali-Krishna K, Mahar PL, Edupuganti S, Lalor S, et al. Human effector and memory CD8+ T cell responses to smallpox and yellow fever vaccines. Immunity. 2008;28:710–722. doi: 10.1016/j.immuni.2008.02.020. [DOI] [PubMed] [Google Scholar]
- Mooney M, McWeeney S, Canderan G, Sekaly RP. A systems framework for vaccine design. Current opinion in immunology. 2013;25:551–555. doi: 10.1016/j.coi.2013.09.014. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Morgan ET. Impact of infectious and inflammatory disease on cytochrome P450-mediated drug metabolism and pharmacokinetics. Clinical pharmacology and therapeutics. 2009;85:434–438. doi: 10.1038/clpt.2008.302. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Nakaya HI, Clutterbuck E, Kazmin D, Wang L, Cortese M, Bosinger SE, Patel NB, Zak DE, Aderem A, Dong T, et al. Systems biology of immunity to MF59-adjuvanted versus nonadjuvanted trivalent seasonal influenza vaccines in early childhood. Proceedings of the National Academy of Sciences of the United States of America. 2016;113:1853–1858. doi: 10.1073/pnas.1519690113. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Nakaya HI, Hagan T, Duraisingham SS, Lee EK, Kwissa M, Rouphael N, Frasca D, Gersten M, Mehta AK, Gaujoux R, et al. Systems Analysis of Immunity to Influenza Vaccination across Multiple Years and in Diverse Populations Reveals Shared Molecular Signatures. Immunity. 2015;43:1186–1198. doi: 10.1016/j.immuni.2015.11.012. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Nakaya HI, Wrammert J, Lee EK, Racioppi L, Marie-Kunze S, Haining WN, Means AR, Kasturi SP, Khan N, Li GM. Systems biology of vaccination for seasonal influenza in humans. Nature immunology. 2011;12:786–795. doi: 10.1038/ni.2067. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Nath AP, Arafat D, Gibson G. Using blood informative transcripts in geographical genomics: impact of lifestyle on gene expression in fijians. Front Genet. 2012;3:243. doi: 10.3389/fgene.2012.00243. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Okkenhaug K. Signaling by the phosphoinositide 3-kinase family in immune cells. Annual review of immunology. 2013;31:675–704. doi: 10.1146/annurev-immunol-032712-095946. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Oxman M, Levin M, Johnson G, Schmader K, Straus S, Gelb L, Arbeit R, Simberkoff M, Gershon A, Davis L. A vaccine to prevent herpes zoster and postherpetic neuralgia in older adults. New England Journal of Medicine. 2005;352:2271–2284. doi: 10.1056/NEJMoa051016. [DOI] [PubMed] [Google Scholar]
- Pulendran B, Ahmed R. Immunological mechanisms of vaccination. Nature immunology. 2011;12:509–517. doi: 10.1038/ni.2039. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Pulendran B, Li S, Nakaya HI. Systems vaccinology. Immunity. 2010;33:516–529. doi: 10.1016/j.immuni.2010.10.006. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Qi Q, Cavanagh MM, Le Saux S, Wagar LE, Mackey S, Hu J, Maecker H, Swan GE, Davis MM, Dekker CL, et al. Defective T Memory Cell Differentiation after Varicella Zoster Vaccination in Older Individuals. PLoS pathogens. 2016;12:e1005892. doi: 10.1371/journal.ppat.1005892. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Querec TD, Akondy RS, Lee EK, Cao W, Nakaya HI, Teuwen D, Pirani A, Gernert K, Deng J, Marzolf B. Systems biology approach predicts immunogenicity of the yellow fever vaccine in humans. Nature immunology. 2009;10:116–125. doi: 10.1038/ni.1688. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sasaki S, Sullivan M, Narvaez CF, Holmes TH, Furman D, Zheng NY, Nishtala M, Wrammert J, Smith K, James JA. Limited efficacy of inactivated influenza vaccine in elderly individuals is associated with decreased production of vaccine-specific antibodies. The Journal of clinical investigation. 2011;121:3109–3119. doi: 10.1172/JCI57834. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Shibata N, Carlin AF, Spann NJ, Saijo K, Morello CS, McDonald JG, Romanoski CE, Maurya MR, Kaikkonen MU, Lam MT, et al. 25-Hydroxycholesterol activates the integrated stress response to reprogram transcription and translation in macrophages. J Biol Chem. 2013;288:35812–35823. doi: 10.1074/jbc.M113.519637. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sobolev O, Binda E, O’Farrell S, Lorenc A, Pradines J, Huang Y, Duffner J, Schulz R, Cason J, Zambon M, et al. Adjuvanted influenza-H1N1 vaccination reveals lymphoid signatures of age-dependent early responses and of clinical adverse events. Nature immunology. 2016;17:204–213. doi: 10.1038/ni.3328. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Spann NJ, Glass CK. Sterols and oxysterols in immune cell function. Nature immunology. 2013;14:893–900. doi: 10.1038/ni.2681. [DOI] [PubMed] [Google Scholar]
- Tsang JS, Schwartzberg PL, Kotliarov Y, Biancotto A, Xie Z, Germain RN, Wang E, Olnes MJ, Narayanan M, Golding H. Global analyses of human immune variation reveal baseline predictors of postvaccination responses. Cell. 2014;157:499–513. doi: 10.1016/j.cell.2014.03.031. [DOI] [PMC free article] [PubMed] [Google Scholar]
- van der Heiden PL, de Boer R, van der Steen DM, Kester MG, van der Hoorn MW, Haarman WM, Barnby-Porritt HE, Fry JW, Napper C, Marijt EW. Identification of varicella-zoster virus-specific CD8 T cells in patients after T-cell-depleted allogeneic stem cell transplantation. Journal of virology. 2009;83:7361–7364. doi: 10.1128/JVI.02662-08. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wrammert J, Smith K, Miller J, Langley WA, Kokko K, Larsen C, Zheng NY, Mays I, Garman L, Helms C. Rapid cloning of high-affinity human monoclonal antibodies against influenza virus. Nature. 2008;453:667–671. doi: 10.1038/nature06890. [DOI] [PMC free article] [PubMed] [Google Scholar]
- York AG, Williams KJ, Argus JP, Zhou QD, Brar G, Vergnes L, Gray EE, Zhen A, Wu NC, Yamada DH, et al. Limiting Cholesterol Biosynthetic Flux Spontaneously Engages Type I IFN Signaling. Cell. 2015;163:1716–1729. doi: 10.1016/j.cell.2015.11.045. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yusuf I, Kageyama R, Monticelli L, Johnston RJ, DiToro D, Hansen K, Barnett B, Crotty S. Germinal center T follicular helper cell IL-4 production is dependent on signaling lymphocytic activation molecule receptor (CD150) The Journal of Immunology. 2010;185:190–202. doi: 10.4049/jimmunol.0903505. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zak DE, Aderem A. Systems integration of innate and adaptive immunity. Vaccine. 2015;33:5241–5248. doi: 10.1016/j.vaccine.2015.05.098. [DOI] [PubMed] [Google Scholar]
- Zak DE, Andersen-Nissen E, Peterson ER, Sato A, Hamilton MK, Borgerding J, Krishnamurty AT, Chang JT, Adams DJ, Hensley TR, et al. Merck Ad5/HIV induces broad innate immune activation that predicts CD8(+) T-cell responses but is attenuated by preexisting Ad5 immunity. Proceedings of the National Academy of Sciences of the United States of America. 2012;109:E3503–3512. doi: 10.1073/pnas.1208972109. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhong XP, Guo R, Zhou H, Liu C, Wan CK. Diacylglycerol kinases in immune cell function and self-tolerance. Immunol Rev. 2008;224:249–264. doi: 10.1111/j.1600-065X.2008.00647.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhou C, Tabb MM, Nelson EL, Grün F, Verma S, Sadatrafiei A, Lin M, Mallick S, Forman BM, Thummel KE. Mutual repression between steroid and xenobiotic receptor and NF-κB signaling pathways links xenobiotic metabolism and inflammation. The Journal of clinical investigation. 2006;116:2280–2289. doi: 10.1172/JCI26283. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.