3D printing of bacteria-laden hydrogels enables the digital fabrication of complex functional materials.
Abstract
Despite recent advances to control the spatial composition and dynamic functionalities of bacteria embedded in materials, bacterial localization into complex three-dimensional (3D) geometries remains a major challenge. We demonstrate a 3D printing approach to create bacteria-derived functional materials by combining the natural diverse metabolism of bacteria with the shape design freedom of additive manufacturing. To achieve this, we embedded bacteria in a biocompatible and functionalized 3D printing ink and printed two types of “living materials” capable of degrading pollutants and of producing medically relevant bacterial cellulose. With this versatile bacteria-printing platform, complex materials displaying spatially specific compositions, geometry, and properties not accessed by standard technologies can be assembled from bottom up for new biotechnological and biomedical applications.
INTRODUCTION
Bacteria are able to thrive in virtually any ecological niche because of their adaptive and diverse metabolic activity (1). Owing to such a metabolic diversity, richer than in any other types of organisms, bacteria create, for example, physical matter in the form of biofilms that warrant survival even in hostile environments (2, 3). Biofilms adapt their mechanical properties under stress to match conditions imposed by the surrounding environment with a great diversity of biopolymers (4–6). During growth in biofilms, bacteria can also form and degrade a plethora of compounds, which are often used to synthesize chemicals, biopolymers, enzymes, and proteins relevant for the food, medical, and chemical industries (7). Moreover, bacteria are able to form calcium carbonate (8), magnetites (9), and biopolymers (10), which could lead to a new generation of biomineralized composites (11), biodegradable plastics (12), and functional materials for biomedical applications (13). This provides these microorganisms with a wide spectrum of functional properties that hold potential for a variety of novel applications. Despite these remarkable features, the use of the programmable biochemical machinery of bacteria to create “living materials” with controlled three-dimensional (3D) shape, microstructure, and dynamic metabolic response remains largely unexplored. This is due to the lack of manufacturing tools that enable the immobilization of bacteria in a biocompatible medium that can be further processed into functional materials with well-defined 3D geometry and site-specific cellular and chemical composition.
Several strategies to immobilize bacteria while maintaining their metabolic activity have been used in biotechnological applications. Typically, microorganisms are immobilized through specific processes, such as adsorption on surfaces, cell cross-linking, encapsulation, and entrapment, to increase the production yield and to provide safety for the bacteria from toxic substances (14). In contrast to immobilization on hard surfaces, embedding bacteria in hydrogels provides an ideal living environment with a high water content to allow nutrients to flow in and waste products to diffuse out of the gel. Naturally, bacteria produce their own hydrogel in the form of protective biofilms with very diverse mechanical properties. For example, biofilms made by Bacillus subtilis are formed at the water-air interface through amyloid fibers that provide biofilm cohesion and relatively strong mechanical properties (15, 16). Other bacteria, such as Acetobacter xylinum, also referred to as Gluconacetobacter xylinus, are able to secrete nanocellulose hydrogels directly at the water-air interface with astonishing mechanical strength (17, 18). For example, this biofilm formation process is exploited by the medical industry for the production of artificial skin, medical patches, and food products (19–22). In particular, bacterial cellulose has been found to be extremely useful in the medical industry because of its cell biocompatibility, making it ideal for tissue engineering (13, 18). So far, bacterial cellulose has been manufactured in situ in the form of surface-patterned implants (13), ear transplants (23), and potential blood vessels (18, 24) by diffusing oxygen through nanopatterned and 3D-shaped silicone molds. In these examples, biofilms have been mainly formed in a nonimmobilized state at a variety of surfaces and interfaces by depositing a layer of bacteria initially suspended in a fluid culture medium on the desired substrate. Immobilizing bacteria in a viscoelastic matrix that could be free-formed into intricate geometries and combined with different microorganisms to achieve spatial cellular and chemical composition control would enable the fabrication of materials with thus far inaccessible dynamic functionalities.
For instance, tackling the geometrical constraints and the lack of spatial control of current biofilm growth approaches could lead to novel design possibilities in microbial fuel cells (25) and biosensors (26) by providing mechanical stability to entrapped cells in biotechnological applications (14). This shape and structural and compositional control can potentially enable the implementation of enticing dynamic properties to otherwise static functional materials. Site-specific control over bacteria distribution has been recently shown to be a very interesting technique to study bacteria dynamics in biofilms (27, 28). In this example, 3D printing technology was used to localize bacteria into different compartments, providing an ideal environment to study bacterial communication in biofilms, known as quorum sensing (29). However, direct printing of multiple bacteria species has been so far limited to millimeter-scale structures (30). The incorporation of bacteria in larger synthetic structures has been achieved by multimaterial printing of support structures containing fluidic channels that are later infiltrated with a suspension of the living organisms. In contrast to this simple bacterial impregnation (31), 3D multimaterial printing of bacteria should enable the homogeneous incorporation of bacteria throughout the printed hydrogel in a one-step approach. Moreover, multiple bacterial strains can potentially be localized at specific sites within a complex architecture to study quorum sensing, bacterial growth, and migration. Embedded bacteria are also more resilient to external adverse effects, such as toxic components, compared to bacteria that live on surfaces only. Despite all these envisioned advantages, the direct processing of immobilized bacteria into self-supporting structures with complex 3D geometries, site-specific cellular composition, and dynamic functionalities has not yet been shown.
Here, we report on a 3D printing platform that enables the digital fabrication of free-standing cell-laden hydrogels with full control over the spatial distribution and concentration of cells or microbes in complex and self-supporting 3D architectures. To this end, we develop and study a functional living ink, called “Flink,” that is a biocompatible immobilization medium that exhibits the viscoelastic properties required for 3D printing of various cells through multimaterial direct ink writing (DIW). The freedom of shape and material composition provided by this printing technique is combined with the metabolic response of microorganisms to enable the digital fabrication of bacteria-derived living materials featuring unprecedented functionalities, such as adaptive behavior, pollutant degradation, and structure formation in the form of cellulose reinforcement. As a novel additive manufacturing approach, Flink 3D printing opens the possibility to combine different organisms and chemistries in a single process, allowing for the digital shaping of living materials into new geometries and adaptive functional architectures (32). The potential of this approach is demonstrated by printing a biocompatible and rheologically optimized bioink into 3D cellular structures for bioremediation and complex-shaped synthetic skin scaffolds for biomedical applications.
RESULTS AND DISCUSSION
The 3D printing technology developed for the digital manufacturing of living materials using functional bacteria-laden inks is shown in Fig. 1. Owing to their diverse metabolism, bacteria that are capable of degrading toxins, synthesizing vitamins, forming cellulose, and performing photosynthesis can be loaded in the ink. The possibility to implement this inherent diverse metabolic activity in a 3D-printed living material is harnessed by loading the desired bacteria in a hydrogel ink that can be extruded in the form of filaments while providing the environment to keep cells alive and functional. Growth and metabolic activity in the printed structure is possible by embedding the bacteria in a hydrogel composed of biocompatible hyaluronic acid (HA), κ-carrageenan (κ-CA), and fumed silica (FS). By including bacteria and their metabolic activity in the hydrogel, the functional living ink Flink is formed and 3D-printed into scaffolds with functionalities inaccessible by nonliving materials.
Fig. 1. Schematics of the 3D bacteria-printing platform for the creation of functional living materials.
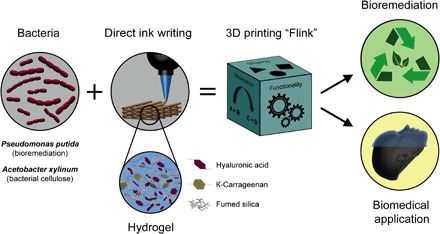
Multifunctional bacteria are embedded in a bioink consisting of biocompatible HA, κ-CA, and FS in bacterial medium. 3D printing of bacteria-containing hydrogels enables the creation of structures in arbitrary shape and added functionality due to the manifold products of bacterial metabolism. The inclusion of specific bacterial strains leads to a living and responsive hydrogel, a novel class of material named Flink. For example, the inclusion of P. putida and A. xylinum yields 3D-printed materials capable of degrading environmental pollutants and forming bacterial cellulose in situ for biomedical applications, respectively.
Two examples of living materials are presented here to demonstrate the possible functionalities arising from the metabolic activity and growth of bacteria embedded within 3D-printed structures. As a first example, the phenol degradation capability of Pseudomonas putida immobilized in a 3D-printed lattice is demonstrated for bioremediation applications. A second example shows the growth of A. xylinum in a complex-shaped 3D-printed architecture that enables the in situ formation of bacterial cellulose relevant for biomedical applications. The presented multimaterial additive manufacturing process allows for the creation of versatile and functional 3D structures with spatial control over the hydrogel composition and localization of different bacterial strains. In contrast to previous works on the formation of cellulose by immersion of a 3D-printed substrate into a bacteria culture medium (23, 33), we 3D-print functional bulk structures using inks that are already preloaded with bacteria. This eliminates the need for support material and opens the possibility to create complex structures of truly 3D shapes with spatially defined bacteria type and concentration. This is achieved by designing inks with rheological behavior that are adequate for extrusion-based printing and retain bacterial survival and metabolic activity after the manufacturing process.
To benefit from the inherent freedom of shape provided by 3D printing (34, 35), the rheological properties of the hydrogel ink have to be designed to ensure the additive deposition of distortion-free and accurate 3D structures. A viscoelastic and shear-thinning bioink with structure recovery was developed to fulfill these rheological requirements while ensuring a high survival rate of bacteria. This was achieved by using κ-CA, HA, and FS as nontoxic constituents. As opposed to synthetic polymers, κ-CA and HA are natural and widely available biopolymers that can be solubilized in water-based media such as Lysogeny broth (LB) and other typical bacterial media to form a versatile ink.
In addition to significantly increasing the viscosity of the solutions at minor concentrations, the natural hydrogel formers κ-CA and HA also retain sufficient water to create an environment that is biocompatible and favorable for the growth of bacteria. The viscous behavior achieved with HA and FS combined with the strong elasticity arising from κ-CA and FS makes these three constituents suitable ingredients to tune the rheological behavior of bioinks. To understand the role of these three individual constituents on the rheological behavior of the bioink, we first investigate the flow response and viscoelastic properties of each hydrogel constituent alone (κ-CA, HA, and FS). All constituents show a shear-thinning behavior with variable viscosities across a wide range of shear rate (Fig. 2A, left). Moreover, no hysteresis was observed for increasing and decreasing shear rates, indicating fast recovery of the single-constituent hydrogels within a few seconds. Oscillatory amplitude sweeps show that solutions with 1 weight % (wt %) κ-CA are predominantly elastic at low strains and viscous above a critical shear strain (εy), confirming the formation of a network below εy. A similar but much less pronounced formation of a network is observed for 1 wt % FS at low shear strains. In contrast, a solution of 1 wt % HA exhibits predominantly viscous behavior (Fig. 2A, right). Combining the individual constituents at equal weight fractions (1 wt % each) leads to a synergistic effect shown by the higher viscosity (Fig. 2B, left) of the Flink compared to the viscosities of each constituent alone at the same individual concentrations (Fig. 2A, left). Altering one of the constituents away from the 1:1:1 ratio was experimentally found to impair the printability of the ink by an increased G′ at higher κ-CA and FS concentrations and an increased G″ at higher HA concentrations. Therefore, the three constituents were combined at a fixed weight fraction of 1 wt % each to form a viscoelastic ink that is shear-thinning over the entire applied shear rate regime (Fig. 2B). The rheological properties and extrudability of this final ink is not altered by the presence of bacteria. Oscillatory frequency sweeps also show that this ink is predominantly elastic over the frequency range of 0.1 to 100 rad/s (fig. S2). Increasing the concentrations of individual constituents from 1 to 2 and 3 wt % at a fixed 1:1:1 ratio leads to higher overall viscosity and elasticity while keeping the shear-thinning response. We name these additional compositions according to the total weight fraction of the added constituents. For example, the 3 wt % Flink contains 1 wt % κ-CA, 1 wt % HA, and 1 wt % FS, whereas a 9 wt % Flink contains 3 wt % of each constituent.
Fig. 2. Rheology and shape retention properties of bacteria-laden Flink used for 3D printing.
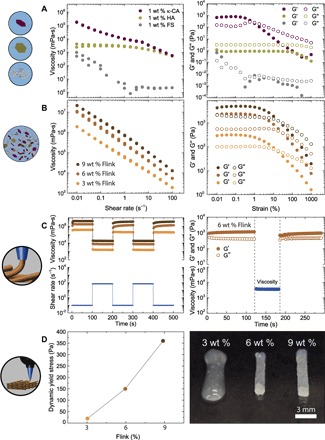
(A and B) Rheological properties of the individual components (1 wt % κ-CA, 1 wt % HA, and 1 wt % FS) in LB bacterial medium (A) and in combination as Flink at concentrations of 1, 2, and 3 wt % for each individual constituent (hereafter called 3, 6, and 9 wt % Flinks, respectively) (B). The steady-state flow behavior of the inks is measured by viscosity curves at increasing and decreasing shear rates. The elastic (G′) and viscous (G″) moduli are measured by oscillatory amplitude sweeps (strain, 0.01 to 1000%; angular frequency, 1 rad/s). (C) Left: To simulate printing conditions, alternating high (70 s−1) and low (0.1 s−1) shear rates in steady-state rotation mode were applied to 3, 6, and 9 wt % Flinks. Right: Instantaneous recovery of the viscoelastic network of 6 wt % Flink is shown by a sudden shear process in a steady state (70 s−1), followed by an oscillatory time sweep. (D) Left: Dynamic yield stresses of different Flink concentrations measured in strain-controlled measurements. Right: The effect of yield stress and elasticity on structure retention in printed filaments for different Flinks.
To more closely simulate the time scales and mechanical forces acting on the ink during printing, high (70 s−1) and low (0.1 s−1) shear rate cycles were applied to formulations with different constituent concentrations (Fig. 2C). All the presented inks show an immediate recovery in viscosity even after multiple cycles. This elastic recovery after deposition was quantified by performing additional experiments, in which an oscillatory time sweep at constant amplitude and frequency was interrupted by a steady-state shear step of 70 s−1, followed by a second oscillatory measurement. The high rotational shear applied reproduces the stresses experienced by the ink during extrusion, whereas the oscillatory measurements are used to quantify the storage modulus of the ink before and after shearing. The elastic modulus recovers almost fully within 1 s for all ink designs. The viscoelastic properties of the hydrogel are regained quickly enough to avoid flow-induced distortion after 3D printing if the Flink concentration is equal or higher than 6 wt % (Fig. 2D, right).
After extrusion, the storage modulus of the ink has to ensure minimum gravity-induced distortion of the deposited filaments. This is particularly relevant for grid structures exhibiting free-spanning filaments (36). Flinks with total constituent concentrations above 4.5 wt % display recovered elastic modulus that is high enough to prevent excessive deflection of free-spanning filaments in typical grid-type printed structures (table S1). Using simple beam theory and typical grid dimensions, we estimate that the recovered storage modulus in the order of 1 kPa obtained here allows a supported filament to span over a distance larger than three times its own diameter (see section S1) (32, 36, 37).
Besides a high-storage modulus and quick elasticity recovery, an adequate yield stress after extrusion is critical to prevent distortion of the print due to capillary forces. As expected, Flinks with increasing constituent concentrations also show higher yield stresses (Fig. 2D). The oscillatory measurements reveal that Flinks with constituent concentrations above 4.5 wt % exhibit yield stresses above 100 Pa. This yield stress level is higher than the capillary forces expected to arise from the curved surface of filaments typically produced through DIW (diameter, 0.58 mm; surface tension, 0.009 N/m) (32). By contrast, the 3 wt % Flink displays an elastic modulus under rest of 300 Pa and a yield stress of 20 Pa (Fig. 2B), which are not high enough to enable structure retention after deposition (Fig. 2D, right). Because they provide fast recovered elastic moduli of approximately 1 kPa and yield stress above 100 Pa, Flinks with 4.5 wt % of constituents or higher were chosen as 3D printing inks for the inclusion of bacteria.
Using multimaterial DIW (32) of Flinks, bacteria can be incorporated and grown in specific regions of printed structures with high accuracy and freedom of shape (Fig. 3, A to C). A grid comprising two bacterial strains, B. subtilis (stained with Nile Red) and P. putida [stained with 4′,6-diamidino-2-phenylindole (DAPI)], was printed with both strains spatially segregated into different struts of the structure, as shown in Fig. 3D. Full control of bacteria local concentration is possible by multimaterial DIW because each of the cartridges can be loaded with different bacterial strains at various concentrations and eventually extruded at any point in the object. This illustrates the versatility of the method, which allows combining multiple types of bacteria with various nutrient requirements, metabolic activities, and functionalities within the same structure.
Fig. 3. 3D printing accuracy, swelling, and mechanical properties after cross-linking and bacterial growth using a 4.5 wt % Flink and Flink-GMHA.
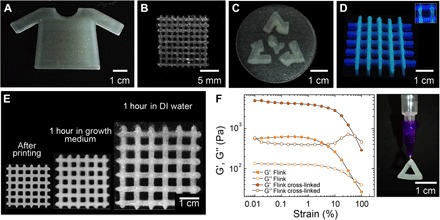
(A to C) Various shapes of Flink hydrogels containing localized bacteria with high precision in 3D. (D) Multimaterial 3D printing with spatial segregation of two bacterial strains. P. putida is labeled with DAPI (blue) and localized in the horizontal lines, whereas B. subtilis is colored with Nile Red (green) and embedded in the vertical lines. (E) Flink (6 wt %; without bacteria) was modified with GMHA to form a UV–cross-linked water-insoluble hydrogel after UV light exposure (365 nm for 60 s at 90 mW). The hydrogels are shown after printing, after 1 hour in bacterial medium and after 1hour in DI water, respectively. (F) Amplitude oscillatory sweeps of the modified Flink-GMHA (4.5 wt %) before and after cross-linking. Strains in the range from 0.01 to 100% at an angular frequency of 1 rads−1 were applied in these measurements. All scaffolds are self-sustaining and resilient.
In addition to spatial control, the survivability and proliferation of bacteria within the printed structures are crucial to obtain functional living materials through this process. To determine bacterial survivability and proliferation in the ink, a toxicity assay for Gram-positive (B. subtilis) and Gram-negative (P. putida) bacteria revealed all ink constituents to be fully biocompatible. Further, the presence of radicals during ultraviolet (UV) exposure (365 nm for 60 s at 90 mW) is not hazardous to the bacteria (100% viability; fig. S1). Thus, the presented Flinks provide maximal bacteria survivability combined with freeform printing at high accuracy and small length scales (Fig. 3, A to C).
Resilient self-supporting structures are obtained after printing by replacing HA with a chemically modified glycidyl methacrylate HA (GMHA). The exchange of HA with GMHA (Flink-GMHA) does not significantly alter the viscosity (fig. S3), and it allows the hydrogel to be UV–cross-linked (Fig. 3F) at low exposure dose and innocuous wavelengths (365 nm for 60 s at 90 mW) to form a water-insoluble hydrogel (Fig. 3E). A strong hydrogel is formed with sufficient mechanical strength to be handled and swollen in various media, thereby providing the high mechanical stability needed to form viable scaffolds. Several other design strategies can be envisioned to impart additional functionalities to the hydrogel (38–40).
When immersed in bacterial medium, the cross-linked hydrogel swells to 1.5 times its size, whereas in water, the grid doubles its original size. The swelling of the hydrogel is affected by the difference in ion concentration present in the hydrogel and the surrounding aqueous medium. Because Flinks are formulated with bacterial broth containing high salt concentrations, osmotic pressure leads to an uptake of the surrounding liquid, when the printed object is placed in Milli-Q water, causing the hydrogel to swell. In contrast, there is only a small exchange of ions when immersing the structure in bacterial media.
Amplitude sweeps on the chemically modified Flink-GMHA reveal an increase in the elastic (G′) and viscous (G″) moduli of the Flink-GMHA after cross-linking (Fig. 3F). The increase of G″ at higher strains in the form of a weak strain overshoot suggests that cross-linking generates an interconnected network (41).
The immobilization of bacteria in these complex-shaped hydrogels allows us to harness the nearly ad infinitum richness of bacteria diversity and their metabolic products. By partially mimicking biofilms that provide a natural habitat of bacteria, hydrogels allow for the control of bacterial metabolisms in predesigned environments. As demonstrated in biotechnological applications, this increases the product yield, protects the bacteria from environmental influences, and permits growth. Continuous release of bacteria and high metabolic activity are secured through immobilization in hydrogels. In addition, structures containing immobilized bacteria can easily be separated from the surrounding medium and be reused for many cycles.
To evince this concept in hydrogels with 3D programmable geometries, a bacterial strain of P. putida that is capable of phenol degradation was immobilized in a 3D-printed Flink-GMHA grid (DSM 50222) (42). The resulting functional living structure converts environmentally hazardous phenol into biomass, taking advantage of the high surface area of the grid architecture to maximize contact between immobilized cells and the liquid medium. To demonstrate these unique features, the grid was photo–cross-linked after printing and incubated in a minimal salts medium (MM) with phenol as the only carbon source. The degradation of phenol was followed using UV-visible adsorption spectroscopy over time, and the concentration of bacteria released from the grid was quantified by measuring the optical density (OD) of the liquid medium (43, 44). The observed decrease in phenol concentration confirms the degradation of this chemical by the incubated bacteria (Fig. 4A). The degradation time window decreases from 130 to 24 hours in a second incubation period in which the same initial concentration of phenol was used after extensive rinsing of the grid in deionized (DI) water. For both incubations, the phenol concentration reduction is accompanied by an increase in the concentration of bacteria in the liquid medium, as indicated by the increase in OD. This reveals that part of the immobilized bacteria is released into the medium and grows at the expense of phenol. Bacterial growth ceased after the initial phenol was fully converted into biomass.
Fig. 4. 3D-printed bacteria-functionalized structures with complex shapes for bioremediation and biomedical applications.
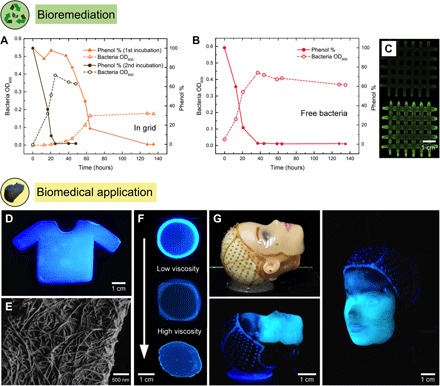
(A) A photo–cross-linked grid structure printed using a 4.5 wt % Flink-GMHA loaded with P. putida, a known phenol degrader, was incubated in an MM with phenol as the only carbon source. Phenol concentration and bacterial optical density (OD600) are shown as a function of time, as indicators of phenol degradation and bacterial growth. As a control, an equivalent bacteria concentration was incubated as a free-floating culture. (B) The preincubated grid was inoculated for a second time in a phenol containing MM. This time, phenol degradation happened as fast as with the free-floating control due to the higher bacterial concentration now present inside the grid. (C) Staining of bacterial DNA within the grid by ethidium bromide before (top) and after (bottom) incubation in the phenol-containing medium (343 nm). (D) In situ formation of bacterial cellulose by A. xylinum is used to generate a 3D-printed scaffold with a 4.5 wt % Flink in the shape of a T-shirt. Bacterial cellulose is visualized with a specific fluorescent dye at 365 nm. (E) Bacterial cellulose nanofibril network under SEM printed with a 3 wt % Flink. (F) Growth of bacterial cellulose depends on oxygen availability and the viscosity of the Flink. A dense cellulose network is only present in regions of high oxygen levels and low to medium viscosities. Images show, from top to bottom, circular prints using 3, 6, and 9 wt % Flinks. (G) A doll face was scanned, and a 4.5 wt % Flink containing A. xylinum was deposited onto the face using a custom-built 3D printer. In situ cellulose growth leads to the formation of a cellulose-reinforced hydrogel that, after removal of all biological residues, can serve as a skin transplant.
The relative contributions of immobilized and free bacteria to the total phenol degradation process were quantified by carrying out a control experiment, in which the same total concentration of bacteria used in the first incubation of the immobilized system was exposed to an equivalent level of phenol. In this control experiment, the initial phenol was entirely degraded into biomass after 40 hours. Because the OD measured in the grid after the first incubation is 50% of the value observed for the liquid with free bacteria, we conclude that phenol degradation in the printed structures is caused not only by bacteria released from the grid but also by those immobilized inside and on the surface of the scaffold. The control experiment also reveals that two incubations of the grid are enough to reach the fast degradation kinetics observed for the liquid containing free bacteria (Fig. 4B). Despite this additional incubation step needed to reach equivalent degradation kinetics, the bacteria-laden 3D-printed grid exhibits the intrinsic advantages of immobilized systems combined with the freedom in geometrical design enabled by printing. Ethidium bromide staining of the grid before and after incubation confirmed the presence of bacterial DNA throughout the entire grid (Fig. 4C and fig. S4), underlining the assertion that growth took place within the gel. Hence, bacterial growth and metabolic activity were secured through immobilization in the predesigned environment of the printed structure.
Another example of added functionality through bacterial incorporation is demonstrated by printing Flink loaded with the bacterium A. xylinum, which is capable of producing cellulose when exposed to oxygen in a culture medium. In situ cellulose formation by A. xylinum leads to mechanically robust complex-shaped 3D-printed structures with potential biomedical applications. This is demonstrated by embedding A. xylinum in the hydrogel containing κ-CA, HA, and FS, followed by incubation of the printed structure for 4 to 7 days. To promote biofilm formation and, therefore, bacterial cellulose growth, the inks in this case were deliberately not cross-linked. After 3D printing and cellulose growth, most of the ink constituents are washed out, leaving only a network of nanofibrillated bacterial cellulose (see fig. S5). To use these constructs for biomedical applications, the printed structures can be washed in water/ethanol mixtures. In addition, bacteria can also be removed by using standard protocols, as for example, immersion in a 1 M NaOH solution at 80°C (13).
The formation of bacterial cellulose in the 3D-printed scaffold was confirmed by scanning electron microscopy (SEM) imaging and by staining the grown cellulose with Fluorescent Brightener 28, a cellulose-specific fluorescent dye (Fig. 4, D and E). Bacterial cellulose formation depends on oxygen availability and the viscosity of the hydrogel ink. To analyze the growth of A. xylinum as a function of oxygen availability and ink viscosity, circular prints of 3, 4.5, and 6 wt % Flinks were covered with two glass slides such that air was only accessible from the sides (Fig. 4F). Because the proliferation of A. xylinum and therewith the production of cellulose requires high oxygen levels (17), cellulose was solely formed at the rim of the printed structure. Thus, the availability of oxygen controls the growth depth of bacteria inside the 3D-printed structure. For the model experiments with circular print geometries, we observe the growth depth to vary between 0.5 and 2 mm, depending on the weight fraction of constituents and, thus, the viscosity of the ink. This self-limited depth makes the bacteria-driven cellulose formation process convenient for the preparation of films and coatings. For example, coating an implant with bacterial cellulose has been shown to be very effective to avoid organ rejection (13). Alternatively, the manufacturing of bulk parts can be envisaged if the immobilization of A. xylinum is combined, for example, with the possible incorporation of oxygen-producing cyanobacteria in the hydrogel (45).
When oxygen is made available only at the periphery of the printed hydrogel, the growth depth was found to decrease for inks with increasing viscosity. At low viscosities (3 wt % Flink; 200 Pa∙s), cellulose formation is high, visible by the bright ring of densely nanofibrillated cellulose network (Fig. 4F). When incorporated at this low concentration, the ink constituents can be washed out, leaving a stable interconnected bacterial cellulose network (fig. S5A). With progressively higher viscosities (6 wt % Flink; 2000 Pa∙s), less bacterial cellulose growth was observed at the rim of the print. This suggests that the immobilization of bacteria inside a highly viscous gel reduces the formation of cellulose microfibrils by restricting cell locomotion in the culture medium (46). At the intermediate constituent concentrations of the 4.5 wt % Flink, a highly interconnected bacterial cellulose network was formed while maintaining a high enough elasticity and yield stress to ensure printing accuracy (Fig. 3). In this case, the ink constituents can be dissolved in water, leaving a bacterial cellulose scaffold (fig. S5, B to E). Above 6 wt % Flink, the bacterial cellulose formation is not high enough to remain cohesive after submersion in water.
To illustrate the full potential of Flinks in creating bacteria-derived functional materials with intricate geometries, we used a custom-built four-axis 3D printer capable of scanning and printing on arbitrary, nonplanar surfaces to deposit a hydrogel loaded with A. xylinum onto a substrate representing a human face (Fig. 4G). As in previous examples, the rheology of the ink was designed to ensure fluidity in the nozzle combined with quick elasticity recover to enable the deposition of distortion-free nondripping filaments. This was possible using the 4.5 wt % Flink. Bacterial cellulose was formed in situ after incubating the face under optimal growth conditions (at 30°C at high humidity for 4 days), as confirmed by direct visualization using a cellulose-specific fluorescent dye. In the region of the forehead, less cellulose formation was visible due to a thinner hydrogel thickness compared to the thickness around the eyes, nose, and mouth. Given the emerging importance of bacterial cellulose as skin replacements and as tissue envelops in organ transplantation (13, 18, 22), the possibility to form bacterial cellulose in any desired 3D shape allows us to apply the decellularized cellulose onto the site of interest without risking the detachment of the skin graft upon movement. The in situ formation of reinforcing cellulose fibers within the hydrogel is particularly attractive for regions under mechanical tension, such as the elbow and knee, or when administered as a pouch onto organs to prevent fibrosis after surgical implants and transplantations. Cellulose films grown in complex geometries precisely match the topography of the site of interest, preventing the formation of wrinkles and entrapments of contaminants that could impair the healing process. We envision that long-term medical applications will benefit from the presented multimaterial 3D printing process by locally deploying bacteria where needed.
CONCLUSIONS
We have developed a 3D printing platform that enables additive manufacturing of complex 3D living architectures of bacteria-laden hydrogels with full localization and concentration control of bacteria. With the freedom of shape provided by this printing technique and the inherent diverse metabolic activity of bacteria, living bacteria-derived materials with unprecedented functionalities can be created, adding a new dimension to 3D printing. This was achieved by developing a biocompatible hydrogel with optimized rheological properties that allows for the immobilization of bacteria into 3D-printed architectures at a high accuracy. The potential of 3D-printed bacterial structures for biotechnological applications was demonstrated through the immobilization of the two model bacterial strains P. putida and A. xylinum in the hydrogel ink. P. putida, a known phenol degrader, was immobilized and allowed to degrade phenol into biomass, showing the potential of the 3D bacteria printing platform for biotechnological applications. Immobilization of A. xylinum in a predesigned 3D matrix enabled the in situ formation of bacterial cellulose scaffolds on nonplanar surfaces, relevant for personalized biomedical applications. Apart from biomedical and biotechnological applications, we envision this versatile bacteria-printing platform to be used for the additive manufacturing of a new generation of biologically generated functional materials.
MATERIALS AND METHODS
Flink formulations and preparation
The following chemicals were used as received if not otherwise mentioned: sodium hyaluronate (HA; BulkSupplements), κ-CA (Acros Organics), fumed silica WDK V15 (FS; Wacker Chemie), ethanol, triethylamine, tetrabutylammonium bromide, glycidyl methacrylate (GM), 2-hydroxy-4′-(2-hydroxyethoxy)-2-methylpropiophenone (Irgacure 2959), and hexamethyldisilazane (HDMS; Sigma-Aldrich). All Flink formulations contained the same weight ratio of 1:1:1 of HA, κ-CA, and FS or a multiple thereof. For a typical Flink, the bacterial broth (containing no bacteria) was first heated to 80°C. κ-CA and HA were added simultaneously in concentrations of χ weight % each and dissolved using a dynamic mixer (2000 rpm for 5 min). Once all components were dissolved, the resulting gel was cooled to room temperature under rigorous stirring, forming a microgel. Further cooling to 4°C was performed in an ice bath. To prevent the formation of larger gel aggregates, mixing was continued at a high shear rate (2000 rpm) until a soft gel was formed. Thereafter, χ weight % FS was added and mixed until a viscoelastic gel was reached.
For the bacteria-laden Flinks, 500 μl of a concentrated fully grown bacteria stem solution was added to 10 ml of Flink and gently stirred to inoculate the Flink. Colony-forming units (CFU) were used to quantify the amount of viable bacteria used: a maximum of 107 CFU/ml for A. xylinum and 109 CFU/ml for B. subtilis.
HA functionalization and preparation of light-curable Flink
HA was functionalized with GM by following a procedure published by Leach and Schmidt (47). In short, 1 g of HA was dissolved in DI H2O. Triethylamine (2.2 ml), GM (2.2 ml), and tetrabutylammonium bromide (2.2 g) were added separately and thoroughly mixed before the next component was added. Once all components were dissolved, the solution was incubated at 60°C for 1 hour, resulting in GM-functionalized HA, abbreviated as GMHA. The product was extracted through precipitation in cold ethanol (0°C) before lyophilization.
The ink formulation procedure described above was also used for the preparation of light-curable Flinks by solely replacing HA with GMHA. In addition, 1 wt % Irgacure 2959 was added to the final gel as a water-soluble photoinitiator.
3D printing of Flinks for bioremediation
3D objects printed for bioremediation and multimaterial demonstrations were fabricated using the following workflow: BioCAD 1.1 (regenHU Ltd.) was used to make computer-aided design drawings of the objects. The resulting G-code was then transferred to a commercially available 3D printer (3D Discovery, regenHU Ltd.). The Flinks were transferred to 10-cm3 amber printing cartridges and subsequently degassed in a planetary mixer for 5 min at 2200 rpm (ARE-250, Thinky). Every Flink formulation was sequentially deposited by a separate pressure-driven printhead. Printing pressures of less than 0.3 MPa are required for extrusion. These low pressures do not adversely affect bacterial viability because pressures higher than 100 MPa are required to kill bacteria (48). Moreover, the viscoelastic nature of the inks leads to shear stresses predominantly at the inner wall of the orifice and not in the central bulk of the extrudate. This protects bacteria from shear stresses occurring during printing.
The light-curable Flinks were cross-linked in a layer-by-layer fashion using an OmniCure S1000 (Lumen Dynamics). Each layer was illuminated for 60 s.
3D printing of Flinks for biomedical applications
A custom-built four-axis 3D printer was used to scan and print onto nonplanar surfaces. First, a doll face was scanned using a high-accuracy laser displacement sensor (LK-H150, Keyence Corporation). Grasshopper (Rhinoceros, Robert McNeel & Associates) was used to generate a nonplanar print path on the previously scanned surface (detailed information in section S2).
Flink containing A. xylinum was extruded with a volumetric-controlled eco-PEN 300 (ViscoTec). The prints were incubated in a sealed container at saturated humidity and at 28°C for 5 days. After incubation, the bacterial cellulose scaffolds were strong enough to be removed from the substrate by hand and were then placed in distilled H2O to prevent them from drying. For fluorescent labeling of the bacterial cellulose, 0.5 wt % of Fluorescent Brightener 28 (Sigma-Aldrich) was added to the solution. To activate the dye, a few drops of 1 M NaOH were added to achieve a slightly alkaline solution (pH 7.5 to 8). After 5 min, the labeled cellulose scaffold was rinsed extensively with distilled H2O to wash away excess dye. A handheld UV lamp (365 nm; UVP LLC) was used to induce fluorescence of the labeled bacterial cellulose.
To prepare the samples for SEM, the substrates were immersed into increasing concentrations of ethanol (30, 50, 70, 90, and 95%) for 10 min each. Finally, they were rinsed twice in 100% ethanol for 5 min each. Ethanol dehydration was followed by a gradual replacement with HDMS that was left to evaporate in a fume hood overnight (13).
Rheology
A full rheological characterization was performed using couette, plate-plate, and cone-plate geometries mounted on strain- and stress-controlled rheometers (MCR501 and MCR702, Anton Paar). In a precharacterization step, the single components (κ-CA, HA, and FS) were measured at a concentration of 1 wt % in water. The three components combined formed the Flink at 4.5, 6, and 9 wt % with equal amounts of each constituent (multiple of 1:1:1 ratio of κ-CA/HA/FS). Flow curves were obtained in strain rate–controlled measurements at shear rates from 0.01 to 100 s−1 in time-controlled measurements. Oscillatory amplitude sweeps were carried out at a frequency of 1 rad/s, with deformations ranging from 0.01 to 100%. The ink with modified HA was also measured under the conditions described above. For the Flinks and single components, a CC17/CC27 cocylindrical couette geometry was used.
To study the recovery behavior of the ink after printing, the shear forces exerted during extrusion were simulated by shearing the ink at a shear rate of 70 s−1 for 100 s, followed by the measurement of the viscosity over time at a shear rate of 0.1 s−1. In addition, the 4.5 wt % Flink was measured in an oscillatory time sweep, followed by a shear period of 1 min at 70 s−1 and finally by a time sweep. Cast hydrogel circles with a diameter of 26 mm were cross-linked in situ while measuring with a plate-plate geometry in oscillatory mode.
Strains and culture media
P. putida DSM50222 and Kt2442 were grown at 30°C in 0.5× concentrated brain-heart infusion (1/2 BHI) medium (BioLife) or in MM containing Na2HPO4·12H2O (5.4 g/liter), KH2PO4 (1.4 g/liter), (NH4)2SO4 (0.5 g/liter), MgSO4·7H2O (0.2 g/liter), and trace mineral supplement [5 ml/liter; MD-TMS, American Type Culture Collection (ATCC)]. B. subtilis DSM675 was grown at 30°C in LB medium. A. xylinum ATCC-700178 (also known as G. xylinus) was provided by A. Ferrari from the Laboratory of Thermodynamics of Emerging Technologies at ETH Zürich and grown in yeast extract (5 g/liter; BioLife), mannitol (25 g/liter; Sigma-Aldrich), and peptone (3 g/liter) in water.
Phenol degradation assay
To adapt bacteria to growth in the presence of phenol, cultures of P. putida DSM50222 were incubated for 12 hours at 30°C in 1/2 BHI medium supplemented with phenol (400 mg/liter). Cultures were pelleted by centrifugation and resuspended in MM for ink preparation or direct inoculation. Cells (1 × 108) were either printed as a grid before incubation or directly incubated at 30°C in 25 ml of MM containing phenol (400 mg/liter). Cell growth (OD600) and phenol degradation were monitored over 135 hours. Phenol concentrations were determined essentially as described by Yang and Humphrey (43). Briefly, cells were removed by centrifugation, and 25 liters of cell-free sample was mixed with 250 liters of 1% Fe(CN)6 and 1250 liters of 1% 4-aminoantipyrine, both dissolved in 0.1 M glycine buffer (pH 9.7). Absorbance of the red product was measured at 505 nm using a spectrophotometer and compared to a standard curve (43, 44).
Toxicity assay
Overnight cultures of B. subtilis and P. putida were incubated with ink components for 1 hour and/or exposed to UV light (365 nm for 60 s at 90 mW), as indicated. Serial dilutions were subsequently plated on LB and 1/2 BHI agar, respectively. After 12 hours of growth, bacterial colonies were quantified.
Supplementary Material
Acknowledgments
We would like to thank Y. López for media preparation and the Laboratory of Food Biotechnology for allowing us to use their facilities. We thank P. Fischer and the Laboratory of Food Process Engineering for their support and for providing access to their rheometer. Funding: M.S., P.A.R., F.C., and A.R.S. acknowledge the financial support from the Swiss National Science Foundation (Consolidator Grant no. BSCGI0_157696). P.A.R. and A.R.S. also acknowledge the financial support of the Swiss National Center of Competence in Research (NCCR) for Bio-Inspired Materials. Author contributions: A.R.S., P.A.R., S.K., and M.S. conceived and designed the experiments. M.S., S.K., and P.A.R. performed the experiments. P.A.R. and M.S. analyzed the data. F.C. and S.K. provided the materials and analysis tools. A.R.S., M.S., and P.A.R. wrote the manuscript, with contributions from all the authors. Competing interests: A.R.S., P.A.R., and M.S. are inventors on a patent application related to this work (European patent application no. 7156787.8, filed 17 February 2017). All other authors declare that they have no competing interests. Data and materials availability: All data needed to evaluate the conclusions in the paper are present in the paper and/or the Supplementary Materials. Additional data related to this paper may be requested from the authors.
SUPPLEMENTARY MATERIALS
Supplementary material for this article is available at http://advances.sciencemag.org/cgi/content/full/3/12/eaao6804/DC1
section S1. Free-spanning length calculation
section S2. Printer software and hardware
fig. S1. Toxicity assay for P. putida and B. subtilis.
fig. S2. Frequency sweep of a 3 wt % Flink from 0.1 to 100 rad/s at a strain of 1%.
fig. S3. Flow curve of 3 wt % Flink-GMHA and 3 wt % Flink.
fig. S4. A printed grid structure using a 4.5 wt % Flink-GMHA loaded with P. putida, a known phenol degrader, was incubated in an MM with phenol as the only carbon source (left).
fig. S5. Examples of 3D printed shapes after bacterial cellulose growth and removal of the Flink.
table S1. Data obtained from the oscillatory amplitude sweep measurements for 3, 4.5, 6 and 9 wt % Flinks.
REFERENCES AND NOTES
- 1.Prosser J. I., Bohannan B. J. M., Curtis T. P., Ellis R. J., Firestone M. K., Freckleton R. P., Green J. L., Green L. E., Killham K., Lennon J. J., Mark Osborn A., Solan M., van der Gast C. J., Young J. P. W., The role of ecological theory in microbial ecology. Nat. Rev. Microbiol. 5, 384–392 (2007). [DOI] [PubMed] [Google Scholar]
- 2.Hall-Stoodley L., Costerton J. W., Stoodley P., Bacterial biofilms: From the natural environment to infectious diseases. Nat. Rev. Microbiol. 2, 95–108 (2004). [DOI] [PubMed] [Google Scholar]
- 3.Costerton J. W., Lewandowski Z., Caldwell D. E., Korber D. R., Lappin-Scott H. M., Microbial biofilms. Annu. Rev. Microbiol. 49, 711–745 (1995). [DOI] [PubMed] [Google Scholar]
- 4.Flemming H.-C., Wingender J., The biofilm matrix. Nat. Rev. Microbiol. 8, 623–633 (2010). [DOI] [PubMed] [Google Scholar]
- 5.Flemming H.-C., Wingender J., Szewzyk U., Steinberg P., Rice S. A., Kjelleberg S., Biofilms: An emergent form of bacterial life. Nat. Rev. Microbiol. 14, 563–575 (2016). [DOI] [PubMed] [Google Scholar]
- 6.Branda S. S., Vik Å., Friedman L., Kolter R., Biofilms: The matrix revisited. Trends Microbiol. 13, 20–26 (2005). [DOI] [PubMed] [Google Scholar]
- 7.Demain A. L., Microbial biotechnology. Trends Biotechnol. 18, 26–31 (2000). [DOI] [PubMed] [Google Scholar]
- 8.Douglas S., Beveridge T. J., Mineral formation by bacteria in natural microbial communities. FEMS Microbiol. Ecol. 26, 79–88 (1998). [Google Scholar]
- 9.Bazylinski D. A., Frankel R. B., Magnetosome formation in prokaryotes. Nat. Rev. Microbiol. 2, 217–230 (2004). [DOI] [PubMed] [Google Scholar]
- 10.Rehm B. H. A., Bacterial polymers: Biosynthesis, modifications and applications. Nat. Rev. Microbiol. 8, 578–592 (2010). [DOI] [PubMed] [Google Scholar]
- 11.Klaus-Joerger T., Joerger R., Olsson E., Granqvist C.-G., Bacteria as workers in the living factory: Metal-accumulating bacteria and their potential for materials science. Trends Biotechnol. 19, 15–20 (2001). [DOI] [PubMed] [Google Scholar]
- 12.Bhuwal A. K., Singh G., Aggarwal N. K., Goyal V., Yadav A., Isolation and screening of polyhydroxyalkanoates producing bacteria from pulp, paper, and cardboard industry wastes. Int. J. Biomater. 2013, 752821 (2013). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Bottan S., Robotti F., Jayathissa P., Hegglin A., Bahamonde N., Heredia-Guerrero J. A., Bayer I. S., Scarpellini A., Merker H., Lindenblatt N., Poulikakos D., Ferrari A., Surface-structured bacterial cellulose with guided assembly-based biolithography (GAB). ACS Nano 9, 206–219 (2015). [DOI] [PubMed] [Google Scholar]
- 14.Cassidy M. B., Lee H., Trevors J. T., Environmental applications of immobilized microbial cells: A review. J. Ind. Microbiol. 16, 79–101 (1996). [Google Scholar]
- 15.Rühs P. A., Böni L., Fuller G. G., Inglis R. F., Fischer P., In-situ quantification of the interfacial rheological response of bacterial biofilms to environmental stimuli. PLOS ONE 8, e78524 (2013). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Vlamakis H., Chai Y., Beauregard P., Losick R., Kolter R., Sticking together: Building a biofilm the Bacillus subtilis way. Nat. Rev. Microbiol. 11, 157–168 (2013). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Brown R. M. Jr, Willison J. H., Richardson C. L., Cellulose biosynthesis in Acetobacter xylinum: Visualization of the site of synthesis and direct measurement of the in vivo process. Proc. Natl. Acad. Sci. U.S.A. 73, 4565–4569 (1976). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Bäckdahl H., Helenius G., Bodin A., Nannmark U., Johansson B. R., Risberg B., Gatenholm P., Mechanical properties of bacterial cellulose and interactions with smooth muscle cells. Biomaterials 27, 2141–2149 (2006). [DOI] [PubMed] [Google Scholar]
- 19.Chin N. L., Man H. C., Talib R. A., 2nd international conference on agricultural and food engineering (CAFEi 2014): New trends forward. Agric. Agric. Sci. Procedia 2, 1–414 (2014). [Google Scholar]
- 20.Shi Z., Zhang Y., Phillips G. O., Yang G., Utilization of bacterial cellulose in food. Food Hydrocoll. 35, 539–545 (2014). [Google Scholar]
- 21.Sulaeva I., Henniges U., Rosenau T., Potthast A., Bacterial cellulose as a material for wound treatment: Properties and modifications. A review. Biotechnol. Adv. 33, 1547–1571 (2015). [DOI] [PubMed] [Google Scholar]
- 22.Czaja W. K., Young D. J., Kawecki M., Brown R. M. Jr, The future prospects of microbial cellulose in biomedical applications. Biomacromolecules 8, 1–12 (2007). [DOI] [PubMed] [Google Scholar]
- 23.Nimeskern L., Ávila H. M., Sundberg J., Gatenholm P., Müller R., Stok K. S., Mechanical evaluation of bacterial nanocellulose as an implant material for ear cartilage replacement. J. Mech. Behav. Biomed. Mater. 22, 12–21 (2013). [DOI] [PubMed] [Google Scholar]
- 24.Kowalska-Ludwicka K., Cala J., Grobelski B., Sygut D., Jesionek-Kupnicka D., Kolodziejczyk M., Bielecki S., Pasieka Z., Modified bacterial cellulose tubes for regeneration of damaged peripheral nerves. Arch. Med. Sci. 9, 527–534 (2013). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Logan B. E., Hamelers B., Rozendal R., Schröder U., Keller J., Freguia S., Aelterman P., Verstraete W., Rabaey K., Microbial fuel cells: Methodology and technology. Environ. Sci. Technol. 40, 5181–5192 (2006). [DOI] [PubMed] [Google Scholar]
- 26.Su L., Jia W., Hou C., Lei Y., Microbial biosensors: A review. Biosens. Bioelectron. 26, 1788–1799 (2011). [DOI] [PubMed] [Google Scholar]
- 27.Connell J. L., Kim J., Shear J. B., Bard A. J., Whiteley M., Real-time monitoring of quorum sensing in 3D-printed bacterial aggregates using scanning electrochemical microscopy. Proc. Natl. Acad. Sci. U.S.A. 111, 18255–18260 (2014). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Connell J. L., Ritschdorff E. T., Whiteley M., Shear J. B., 3D printing of microscopic bacterial communities. Proc. Natl. Acad. Sci. U.S.A. 110, 18380–18385 (2013). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Bodelón G., Montes-García V., López-Puente V., Hill E. H., Hamon C., Sanz-Ortiz M. N., Rodal-Cedeira S., Costas C., Celiksoy S., Pérez-Juste I., Scarabelli L., La Porta A., Pérez-Juste J., Pastoriza-Santos I., Liz-Marzán L. M., Detection and imaging of quorum sensing in Pseudomonas aeruginosa biofilm communities by surface-enhanced resonance Raman scattering. Nat. Mater. 15, 1203–1211 (2016). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Lehner B. A. E., Schmieden D. T., Meyer A. S., A straightforward approach for 3D bacterial printing. ACS Synth. Biol. 6, 1124–1130 (2017). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Bader C., Patrick W. G., Kolb D., Hays S. G., Keating S., Sharma S., Dikovsky D., Belocon B., Weaver J. C., Silver P. A., Oxman N., Grown, printed, and biologically augmented: An additively manufactured microfluidic wearable, functionally templated for synthetic microbes. 3D Print. Addit. Manuf. 3, 79–89 (2016). [Google Scholar]
- 32.Kokkinis D., Schaffner M., Studart A. R., Multimaterial magnetically assisted 3D printing of composite materials. Nat. Commun. 6, 8643 (2015). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.P. Gatenholm, H. Backdahl, T. J. Tzavaras, R. V Davalos, M. B. Sano, “Three-dimensional bioprinting of biosynthetic cellulose (BC) implants and scaffolds for tissue engineering,” U.S. Patent 8,691,974 (2014).
- 34.Fu Q., Saiz E., Tomsia A. P., Direct ink writing of highly porous and strong glass scaffolds for load-bearing bone defects repair and regeneration. Acta Biomater. 7, 3547–3554 (2011). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Lewis J. A., Direct ink writing of 3D functional materials. Adv. Funct. Mater. 16, 2193–2204 (2006). [Google Scholar]
- 36.Smay J. E., Cesarano J. III, Lewis J. A., Colloidal inks for directed assembly of 3-D periodic structures. Langmuir 18, 5429–5437 (2002). [Google Scholar]
- 37.Minas C., Carnelli D., Tervoort E., Studart A. R., 3D printing of emulsions and foams into hierarchical porous ceramics. Adv. Mater. 28, 9993–9999 (2016). [DOI] [PubMed] [Google Scholar]
- 38.Rose S., Prevoteau A., Elzière P., Hourdet D., Marcellan A., Leibler L., Nanoparticle solutions as adhesives for gels and biological tissues. Nature 505, 382–385 (2014). [DOI] [PubMed] [Google Scholar]
- 39.Jungst T., Smolan W., Schacht K., Scheibel T., Groll J., Strategies and molecular design criteria for 3D printable hydrogels. Chem. Rev. 116, 1496–1539 (2016). [DOI] [PubMed] [Google Scholar]
- 40.Lee Y., Chung H. J., Yeo S., Ahn C.-H., Lee H., Messersmith P. B., Park T. G., Thermo-sensitive, injectable, and tissue adhesive sol–gel transition hyaluronic acid/pluronic composite hydrogels prepared from bio-inspired catechol-thiol reaction. Soft Matter 6, 977–983 (2010). [Google Scholar]
- 41.Hyun K., Wilhelm M., Klein C. O., Cho K. S., Nam J. G., Ahn K. H., Lee S. J., Ewoldt R. H., McKinley G. H., A review of nonlinear oscillatory shear tests: Analysis and application of large amplitude oscillatory shear (LAOS). Prog. Polym. Sci. 36, 1697–1753 (2011). [Google Scholar]
- 42.Allsop P. J., Chisti Y., Moo-Young M., Sullivan G. R., Dynamics of phenol degradation by Pseudomonas putida. Biotechnol. Bioeng. 41, 572–580 (1993). [DOI] [PubMed] [Google Scholar]
- 43.Yang R. D., Humphrey A. E., Dynamic and steady state studies of phenol biodegradation in pure and mixed cultures. Biotechnol. Bioeng. 17, 1211–1235 (1975). [DOI] [PubMed] [Google Scholar]
- 44.Hasan S. A., Jabeen S., Degradation kinetics and pathway of phenol by Pseudomonas and Bacillus species. Biotechnol. Biotechnol. Equip. 29, 45–53 (2015). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Das A. A. K., Bovill J., Ayesh M., Stoyanov S. D., Paunov V. N., Fabrication of living soft matter by symbiotic growth of unicellular microorganisms. J. Mater. Chem. B 4, 3685–3694 (2016). [DOI] [PubMed] [Google Scholar]
- 46.Tittsler R. P., Sandholzer L. A., The use of semi-solid agar for the detection of bacterial motility. J. Bacteriol. 31, 575–580 (1936). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.Leach J. B., Schmidt C. E., Characterization of protein release from photocrosslinkable hyaluronic acid-polyethylene glycol hydrogel tissue engineering scaffolds. Biomaterials 26, 125–135 (2005). [DOI] [PubMed] [Google Scholar]
- 48.Bertucco A., Spilimbergo S., Treating micro-organisms with high pressure. Ind. Chem. Libr. 9, 626–640 (2001). [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
Supplementary material for this article is available at http://advances.sciencemag.org/cgi/content/full/3/12/eaao6804/DC1
section S1. Free-spanning length calculation
section S2. Printer software and hardware
fig. S1. Toxicity assay for P. putida and B. subtilis.
fig. S2. Frequency sweep of a 3 wt % Flink from 0.1 to 100 rad/s at a strain of 1%.
fig. S3. Flow curve of 3 wt % Flink-GMHA and 3 wt % Flink.
fig. S4. A printed grid structure using a 4.5 wt % Flink-GMHA loaded with P. putida, a known phenol degrader, was incubated in an MM with phenol as the only carbon source (left).
fig. S5. Examples of 3D printed shapes after bacterial cellulose growth and removal of the Flink.
table S1. Data obtained from the oscillatory amplitude sweep measurements for 3, 4.5, 6 and 9 wt % Flinks.


