Abstract
Background and Objectives
Optical imaging to guide cancer resections is rapidly transitioning into the operating room. However, the sensitivity of this technique to detect subclinical disease is yet characterized. The purpose of this study was to determine the minimum range of cancer cells that can be detected by antibody-based fluorescence imaging.
Methods
2LMP (breast), COLO-205 (colon), MiaPaca-2 (pancreas), and SCC-1 (head and neck) cells incubated in vitro with cetuximab-IRDye800CW (dose range 8.6μM to 86nM) were implanted subcutaneously in mice (n=3 mice, 5 tumors/mouse). Following incubation with 8.6×10−2μM of cetuximab-IRDye800CW in vitro, serial dilutions of each cell type (1×103–1×106) were implanted subcutaneously (n=3, 5 tumors/mouse). Tumors were imaged with Pearl Impulse and Xenogen IVIS 100 imaging systems. Scatchard analysis was performed to determine receptor density and kinetics for each cell line.
Results
Under conditions of minimal cetuximab-IRDye800CW exposure to low cellular quantity, closed-field fluorescence imaging theoretically detected a minimum of 4.2×104–9.5×104 2LMP cells, 1.9×105–4.5×105 MiaPaca-2 cells, and 2.4×104–6.7×104 SCC-1 cells; COLO-205 cells could not be identified. Higher EGFR-mediated uptake of cetuximab correlated with sensitivity of detection.
Conclusion
This study supports the clinical utility of cetuximab-IRDye800CW to sensitively localize subclinical disease in the surgical setting.
Keywords: Clinical, optical imaging, antibody, fluorescence, near-infrared
INTRODUCTION
Surgical resection with negative margins remains the cornerstone of treatment for many tumor types. Currently, this involves intraoperative analysis of frozen tissue sections to assess margin negativity. However, the recent union between optical imaging and surgical oncology has the potential to permit more accurate identification of tumor margins using real-time intraoperative imaging techniques.[1] This has demonstrated significant potential in surgical oncology by reducing the need for frozen tissue histopathology, which is costly, onerous, and subject to sampling error.[2,3]
Optical imaging technologies often utilize fluorescent light in the near-infrared (NIR) range (700–900nm), which provides favorable depth of tissue penetration and minimizes tissue autofluorescence by reducing background scattering.[4] Regardless of the specific modality used, an important consideration in optical imaging is tumor specificity of the optical probe, which can improve resolution of the tumor for in vivo imaging. For example, fluorescently labeled monoclonal antibodies that target ligands overexpressed in tumors are ideal probes due to their high affinity and specificity for the target antigen, and they can be administered systemically.
However, the ability of these optical probes to identify small nests of subclinical disease, not otherwise visible with the naked eye, is not well characterized. Traditionally, a 1cm3 tumor (1×109 cells) is generally considered to be the amount of tumor that is clinically detectable[5], but the true limit of sensitivity of fluorescent optical probes is not well defined. Therefore, the primary objective of this study is to quantify this tumor detection threshold by utilizing cetuximab, a monoclonal antibody to epidermal growth factor receptor (EGFR), conjugated to the NIR fluorescent dye molecule IRDye800CW (IRDye800CW-N-hydroxysuccinimide ester, LI-COR Biosciences, Lincoln, Nebraska). IRDye800CW is a NIR probe with a broad absorption (778nm) and emission (794nm) spectrum, and has been shown to be nontoxic in humans.[6–8] EGFR is overexpressed in head and neck squamous cell carcinoma (HNSCC), with over 90% of tumors overexpressing the receptor.[9–11] In fact, several cancer types are known to over express EGFR, including breast, pancreas, and colon; thus, it serves as a versatile and reliable probe target.[12–16] Therefore, in this study, multiple cancer types were tested to demonstrate the adaptability and application of this probe to detect subclinical disease in various patient populations.
MATERIALS AND METHODS
Reagents
Anti-EGFR antibody, cetuximab, (Erbitux, ImClone, New York, NY; 152 kDa) was supplied at 2mg/mL. The fluorescent probe, IRDye800CW, (IRDye800CW-N-hydroxysuccinimide ester; LI-COR Biosciences, Lincoln, Nebraska) was supplied as a GMP-compliant reagent. The University of Alabama at Birmingham Vector Production Facility performed the conjugation reaction to prepare the cetuximab-IRDye800CW. Antibodies were incubated with IRDye800CW in 1.00M potassium phosphate buffer (pH 9.0) for 2-hours at room temperature. Desalting spin columns (Pierce Biotechnology, Rockford, IL) removed the unconjugated dye. A final dye:protein ratio of 1.5–2.0 was determined using spectrophotometry.
To produce lower concentrations of cetuximab-IRDye800CW, stock agent (2mg/mL) was diluted in 1X PBS (21-040-CV, Mediatech, Inc., Manassas, VA).
Cell Lines and Tissue Culture
Both luciferase positive (Luc+) and negative (Luc−) breast cancer (2LMP), colon cancer (COLO-205), pancreatic cancer (MiaPaca-2), and head and neck cancer (SCC-1) cell lines were maintained in DMEM containing 10% FBS and incubated at 37°C in 5% CO2. COLO-205, MiaPaca-2, and SCC-1 cell lines were acquired from American Type Culture Collection (ATCC, Manassas, VA). 2LMP cells (a 2× lung metastatic pooled subclone of MDA-MB-23) were provided as a kind gift from Dr. Donald Buchsbaum (UAB, Birmingham, AL). Luciferase expression was stably induced using the ViraPort retroviral vector (Stratagene, La Jolla, CA). Cell counts were performed using a BD Accuri C6 Flow Cytometer.
Animal Models
Female nude athymic mice, aged 4–6 weeks (Charles River Laboratories, Hartford, Connecticut), were used for the studies. All experiments and euthanasia procedures were performed in accordance with the University of Alabama at Birmingham Institutional Animal Care and Use Committee (IACUC) guidelines. For cetuximab-IRDye800CW and cell tittering experiments, each tumor type (n=4 cell lines) was injected subcutaneously to form flank xenograft tumors (n=5) containing 1×106 tumor cells in each bolus inoculation (n=3 mice per tumor type). Additional assessments of in vitro cetuximab-IRDye800CW incubation time effects were conducted (24h vs. 72h) utilizing 2LMP (n=3 mice per group) for both cetuximab-IRDye800CW (n=4 tumor foci per animal) and cell titers (n=5 tumor foci per animal).
Imaging Devices
The Pearl Impulse (LI-COR Biosciences, Lincoln, Nebraska) is a NIR closed-field imaging system with a cooled charge-coupled camera that is optimized for IRDye800CW spectrum. A Xenogen IVIS 100 bioluminescence-imaging (BLI) device (PerkinElmer, Waltham, MA) was used to localize and quantify the bioluminescent signal from implanted Luc+ cells in the tumor boluses.
Titer of Cetuximab-IRDye800CW Probe Concentration
To assess the minimum cetuximab-IRDye800CW availability needed to fluorescently localize disease, serially diluted amounts of cetuximab-IRDye800CW ((A) 8.6μM, (B) 8.6×10−1 μM, (C) 8.6×10−2 μM, (D) 8.6×10−3 μM, (E) 8.6×10−4 μM) were each incubated in vitro to 1×105 (Luc+) cancer cells of the four tumor types. Tested concentrations represent a range based on the 25mg/m2 dose, equivalent to 7.1μM, used for clinical application.[6] Cells were allowed to incubate for 1-hour after fluorescent probe addition. Post incubation, non-trypsinized cells were washed thrice in the absence of trypsin. For each concentration group, the fluorescently labeled 1×105 Luc+ cells were added to 9×105 Luc− unlabeled cells to reach a total tumor bolus of 1×106 cells (see Figure 1A). Each tumor bolus was injected subcutaneously along the flank, with the highest concentration of probe at the caudal end, moving cranially. After a 24-hour tumor incubation period, mice received an intraperitoneal dose of D-Luciferin (2.5mg/mouse, Perkin Elmer, Waltham, MA). Mice were then anesthetized and euthanized, and a skin flap was surgically created to visualize the tumors on the subcutaneous surface. Tumors were imaged with BLI and FLI modalities.
Figure 1.
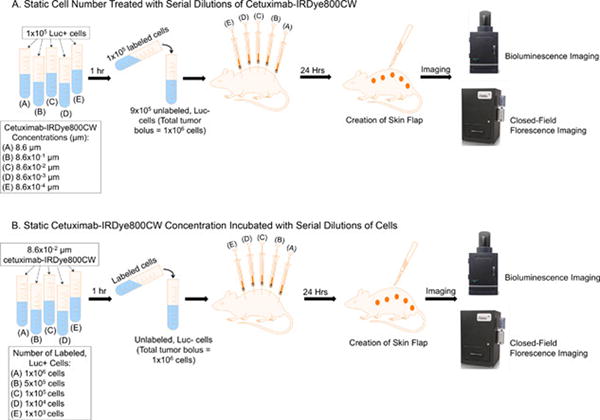
(A) Experimental workflow used to identify the lowest cetuximab-IRDye800CW dose to produce positive fluorescence contrast between tumor and normal tissue. (B) Experimental workflow used to determine minimum number of cetuximab-IRDye800CW labeled cells needed for positive disease localization.
Titer of Cetuximab-IRDye800CW-labeled Tumor Cells
To further understand fluorescence detection limitations, the minimal cetuximab-IRDye800CW concentration needed to localize disease was used to identify the least number of cetuximab-IRDye800CW exposed cells to produce fluorescence contrast for accurate disease delineation. A constant concentration of cetuximab-IRDye800CW (8.6×10−2μM) was incubated with serially decreasing amounts of Luc+ tumor cells from each tumor type ((A) 1×106, (B) 5×105, (C) 1×105, (D) 1×104, (E) 1×103). The cetuximab-IRDye800CW in vitro incubation was performed for 1-hour; non-trypsinized cells were subsequently washed three times with 1X PBS. After which (A)–(E) Luc+ cells were combined with unlabeled Luc− cells to create a total tumor bolus of 1×106 cells (see Figure 1B). Boluses were injected subcutaneously along the flank, with the highest number of Luc+ cells being at the caudal end, moving cranially. After a 24-hour incubation period, mice were anesthetized and euthanized, and a skin flap was surgically created as described previously. Mice were imaged with BLI and FLI modalities.
In Vitro Binding Assay
Scatchard plot assays were performed on each cell line as previously described to determine the density of EGFR receptor internalization for cetuximab.[17] COLO-205, 2LMP, MiaPaca-2, and SCC-1 cells were washed with 1X PBS (pH 7.4) at room temperature and plated in separate 24-well plates (5.0×105 cells/well, 3 plates/cell line) and incubated overnight at 37°C in complete growth medium. An internalization medium (pH 7.4) was prepared using DMEM with 30mM Hepes, 2mM L-glutamine, 1mM Na pyruvate, and 1% BSA. The cetuximab antibody was labeled with Tc-99m using SnCl2/tricine as the transfer ligand [18] and serially diluted by two-fold in internalization media.
Incubation Time and Florescence Detection
To further understand the effect of incubation time on fluorescence detection for serial titers of both cetuximab-IRDye800CW and tumor cells, experiments described previously and outlined in Figure 1A/B were repeated. However, the 2LMP cells were incubated in vitro with the fluorescent probe for 24 or 72-hours. Cetuximab-IRDye800CW concentrations of 8.6×10−1 μM, 8.6×10−2 μM, 8.6×10−3 μM, and 8.6×10−4 μM were tested. Post incubation, non-trypsinized cells were washed thrice with 1X PBS. Again, a skin flap was created at 24-hours post-tumor implantation and mice were imaged with BLI and FLI devices.
Data Analysis
BLI images were used to quantify the bioluminescent signal within a hand drawn region-of-interest (ROI) based on brightfield imaging using onboard system software to quantify the signal emitted from each tumor foci. The bioluminescent signal was normalized to total counts/second exposure (CpS). CpS values from tumor foci with the same cell type and injected amount of Luc+ cells were averaged together. A corresponding correction factor (CF) was determined by dividing averaged CpS by individual CpS values. The CF was multiplied by total fluorescent counts (TFC) from individual tumor foci measured on FLI within a hand drawn ROI using ImageStudio (LI-COR Biosciences) software. CF adjusted FLI signals loaded with the same cell type/amount and cetuximab-IRDye800CW concentration were averaged together. Group tumor TFCs were divided by background TFCs measured in a tumor-free mouse 24-hours after systemic injection of cetuximab-IRDye800CW (200μg), allowing for tumor-to-background ratio (TBR) calculation, where TBR>1 was considered detectable.
Calculation of Theoretical Minimum Number of Cells Detectable
The theoretical minimum number of cells detectable was determined by interpolation to find the cell count when TBR=1, the threshold of positive fluorescence contrast.[19] A theoretical detection range was calculated for each cell line. The interpolation was drawn between loaded cell count at a TBR>1±SD to the loaded cell count at TBR<1±SD. That is, the assessed linear regression was determined at cell count=TBR>1+SD to cell count=TBR<1+SD, and cell count=TBR>1–SD to cell count=TBR<1–SD.
Statistics
Calculated TBRs were assessed with student T-tests between cohorts to determine statistical significance. An exponential regression was performed to evaluate the trend between internalized Tc-99m-cetuximab percentage observed in vitro and the theoretical minimum cell number detected with in vivo imagining. Statistical significance between percent-internalized Tc-99m-cetuximab/cell line was assessed with a one-way ANOVA run in Prism 5 (GraphPad Software, Inc., La Jolla, CA).
RESULTS
Lowest cetuximab-IRDye800CW concentration required for detection
Figure 1A illustrates the workflow for identifying the lowest cetuximab-IRDye800CW titer to accurately localize disease. Results of this experiment are demonstrated both qualitatively and quantitatively in Figure 2. BLI images in Figure 2A identified all 5 foci of implanted disease on the skin flap in each cell line. Disease was also appreciably localized with FLI, with a trend of increasing florescence intensity in a caudal direction with increasing cetuximab concentration. Evaluation of TBR values at each cetuximab-IRDye800CW concentration per cell line revealed, in Figure 2B, TBR values>1 for 2LMP at 8.6μM (TBR: 3.3) and 8.6×10−1μM (TBR: 1.8); COLO-205 at 8.6μM (TBR: 4.6); MiaPaca-2 at 8.6μM (TBR: 7.7); and SCC-1 at 8.6μM (TBR: 16.7), 8.6×10−1μM (TBR: 3.6), and 8.6×10−2μM (TBR: 1.8). As shown, 8.6×10−2μM was the lowest dose capable of localizing disease.
Figure 2.
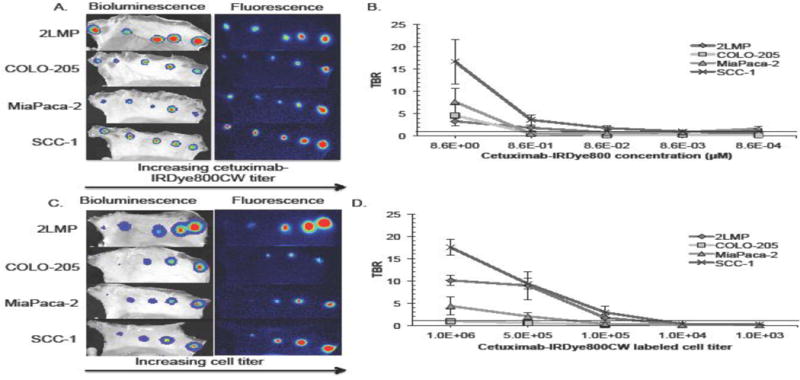
Results from the experiment described in Figure 1A, performed to identify the lowest cetuximab-IRDye800CW dose that provides positive contrast. (A) Visual representation first of disease verification at all 5 implantation sites for all cell lines on bioluminescence imaging and then the respective fluorescence signals detected on fluorescence imaging. (B) Graphically displays the tumor-to-background mean fluorescence counts for each cell line at all 5 tested cetuximab-IRDye800CW concentrations. Results from the experiment used to identify the minimum amount of cells fluorescently detected using cetuximab-IRDye800CW as depicted in Figure 1B. (C) Representative bioluminescence and fluorescence acquisitions are shown. (D) Graphical representation of tumor-to-background mean fluorescence signal recorded on fluorescence imaging for each cell line at each labeled cell concentration. (B and D) Threshold lines drawn at TBR=1 represent the theoretical disease detection limit.
Minimum number of tumor cells detectable
Next, disease quantity exposed to cetuximab-IRDye800CW was varied to determine the lowest fluorescently active cell titer required for disease detection (Figure 1B). BLI was used as the gold standard for tumor localization and normalization of tumor fluorescence signal. In Figure 2C, FLI imaging of cetuximab-IRDye800CW (8.6×10−2μM dose) in 2LMP, COLO-205, MiaPaca-2, and SCC-1 followed a trend of increasing fluorescence as the total number of labeled cells increased in a caudal direction. As before, TBR>1 was used as the lower tumor threshold detection limit. Cell titers positively identified were as follows (Figure 2D): SCC-1 at 1.0×106 (TBR: 17.5), 5.0×105 (TBR: 9.3), and 1.0×105 (TBR: 2.9), which were significantly greater than 2LMP at 1.0×106 (TBR: 10.1; p<0.01), 5.0×105 (TBR: 8.9), and 1.0×105 (TBR: 1.7); and MiaPaca-2 at 1.0×106 (TBR: 4.3; p<0.004) and 5.0×105 (TBR: 2.0; p<0.01). COLO-205 was not localized with a TBR>1 at any tested cell number.
Correlating EGFR expression to theoretical florescence detection limit
To account for differences in EGFR expression among tested cell lines, a Scatchard assay was performed to measure both surface-bound and internalized conjugate after specific binding of Tc-99m-cetuximab to EGFR. Values listed (Figure 3A) represent the percentage of internalized Tc-99m-cetuximab associated with the cells. This percentage serves as a surrogate of EGFR internalization kinetics as the value reflects the Tc-99m-cetuximab association rate, since radioactive quantification was performed at 1-hour for all cell lines. The internalized percentages are as follows: SCC-1 96±4% (1.2×105/1.3×105), 2LMP 70±6% (1.2×105/1.8×105), COLO-205 50±4% (6.0×104/1.2×104), and MiaPaca-2 20±6% (5.3×104/2.5×105) (p<0.0001) (internalized Tc-99m-cetuximab/total associated Tc-99m-cetuximab). Figure 3B presents results from a regression analysis developed to interpolate the theoretical minimum cell number that could be detected with cetuximab-IRDye800CW fluorescence probe technology for each cell line. This value was obtained by calculating the theoretical cell number where TBR=1. Using this method, the minimum SCC-1 cell number range required for detection was 2.4×104–6.7×104 cells, which was significantly less (p<0.05) than MiaPaca-2 (1.9×105–4.5×105 cells). The theoretical detection limit of 2LMP was 4.2×104–9.5×104 cells. The lowest number of COLO-205 required for detection was out of range, and could not be calculated with FLI. An inverse correlation between the percentage of internalized Tc-99m-cetuximab and the theoretical minimal number of cells detectable with cetuximab-IRDye800CW was observed during the exponential regression (R2 = 0.98).
Figure 3.
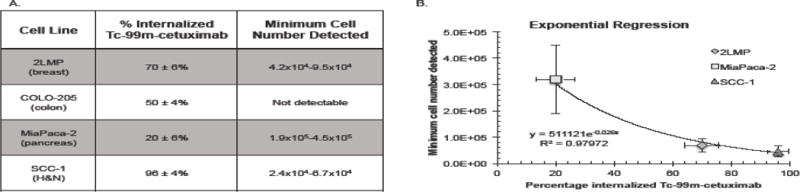
(A) Percentage of internalized Tc-99m-cetxumab as determined by Scatchard assay and the theoretical minimum cell number detected per cell line. (B) An exponential regression is shown demonstrating the relationship between percentages internalized Tc-99m-cetuximab and the theoretical minimum cell number detected per cell line.
Effect of incubation time on disease detection
To determine the effect of prolonged probe exposure time on fluorescence localization, tumor cells were incubated for 24 or 72-hours prior to imaging. First, the lowest cetuximab-IRDye800CW concentration to localize disease was re-evaluated. 2LMP cells incubated for 24-hours were localized at a cetuximab-IRDye800CW concentration of 8.6×10−1μM (TBR: 2.3), 8.6×10−2μM (TBR: 1.8), 8.6×10−3μM (TBR: 1.8), and 8.6×10−4μM (TBR: 1.5) while 2LMP at 72-hours was detected at 8.6×10−1μM (TBR: 2.6) and 8.6×10−2 μM (TBR: 1.7) (Figure 4A). The 8.6×10−2 μM cetuximab-IRDye800CW dose was the minimum dose to sufficiently localize disease (Figure 4B). Next, the determination of minimum numbers of cells required for detection was re-examined (Figure 4C). As shown (Figure 4D), 2LMP incubated for 24-hours positively identified disease at 1.0×106 (TBR: 21.9; p<0.001), 5.0×105 (TBR: 6.5), and 1.0×105 (TBR: 2.9; p<0.05) cells. After 72-hours incubation, disease was identified in 2LMP at 1.0×106 (TBR: 17.1), 5.0×105 (TBR: 8.1), and 1.0×105 (TBR: 1.2) cells. Additionally, the theoretical minimum number of cells detectable was determined by interpolating fluorescence values to find where TBR=1. Using 2LMP cells, FLI was able to positively detect disease in as few as 2.5×104–7.0×104 cells if incubated for 24-hours, which was less than both the 7.8×104–8.8×104 cells at 72-hours and 4.2×104–9.5×104 cells at 1-hour.
Figure 4.
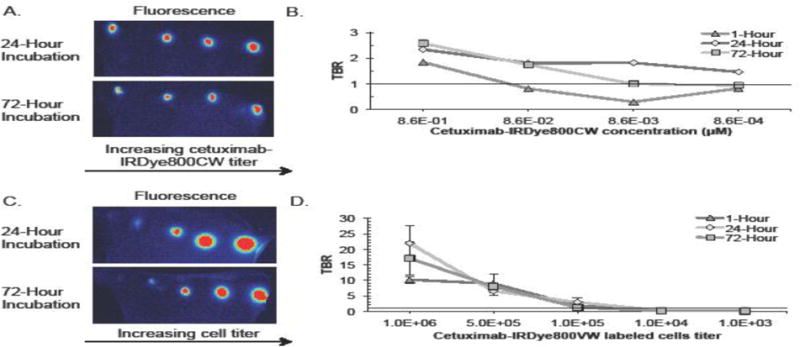
(A) Representative images of tittered cetuximab-IRDye800CW concentrations in tumors containing cells incubated at either 24 or 72-hours. (B) Tumor-to-background ratios of tumors following 1, 24, and 72-hour incubation times are shown for each concentration tested. (C) Representative images of tittered cetuximab-IRDye800CW (8.6×10−2 μM) incubated cell numbers in tumors containing cells incubated at either 24 or 72-hours. (D) Tumor-to-background ratios of tumors following 1, 24, and 72-hour incubation times are shown for each cell number tested.
DISCUSSION
Fluorescently labeled antibodies have the potential to enhance intra-operative detection of subclinical disease. However, the full potential to improve clinical outcomes has yet to be realized. Results from a recent clinical trial of cetuximab-IRDye800CW demonstrated sufficient specificity to fluorescently localize disease in HNSCC patients.[6] Using this antibody-based method, fluorescence contrast delivers a more sensitive and specific approach when compared to a surgical and pathological evaluation.[20] When compared to standard histopathological evaluation, florescence imaging has the potential to permit evaluation of the entire wound bed, which could greatly reduce sampling error. Accurate localization is further supported at the cellular level as histologic assessment of florescence sequestration demonstrates a statistically significant (p<0.001) delineation between cancerous/non-cancerous tissues at 1mm from the tumor border.[21]
This report defines the detection limits of cetuximab-IRDye800CW for florescence-guided surgery to accurately identify disease. First, the minimal cetuximab-IRDye800CW dose to localize diseased was determined using a TBR value greater than 1. While high TBR values increase ease of disease localization, a TBR>1 is the disease detection threshold as mathematically defined: tumor fluorescence signal>background fluorescence signal.[19] This value is justified by utilizing modern imaging devices with high dynamic range, where tumors with TBR>1 can be successfully delineated by adjusting the upper and lower window limits using a spectral color look up table. Furthermore, images were interpreted in context of TBR values as it functionally mitigates regional non-specific probe accumulation and tissue auto-fluorescence by including resultant fluorescence counts. Selecting a 8.6×10−2μM conjugate concentration, representative of 1.2% of total dose administered in patients during a recent clinical trial [6], we simulate, in vitro, the result of systemic drug elimination in vivo secondary to native clearance pathways that reduces final delivered drug quantity in a controlled manner. We then evaluate the minimal cellular concentration requiring cetuximab-IRDye800CW incubation to yield accurate fluorescence disease detection. By only exposing fractions of the implanted total tumor bolus to cetuximab-IRDye800CW, in vivo tumor heterogeneity is simulated. This heterogeneity, secondary to continual, sporadic mutations among malignant cells, is responsible for intratumoral areas responding differently to a particular drug.[22]
Interpolation of the data for each cell line at the labeled cell concentration where TBR>1 reveals the theoretical minimum number of cells detectable using cetuximab-IRDye800CW. Interestingly, efficacy was cell line-specific with as few as 2.4×104–6.7×104 cells detectable for SCC-1, which was significantly less (p<0.05) than the theoretical 1.9×105–4.5×105 cells in MiaPaca-2. These data show markedly improved detection when compared to CT and MRI, which are limited to an anatomical resolution of 1×109 cells, approximately 1mm3.[5] Collectively, the use of cetuximab-IRDye800CW technology has the ability to localize disease on a scale of 2.2×104, 1.5×104, and 3.1×103 times smaller than contemporary methods for SCC-1, 2LMP, and MiaPaca-2 tumors, respectively. Moreover, this technology is cancer-specific as evidenced by setting a detection threshold of TBR>1. This represents a novel advantage over standard CT and MRI, which are not yet functionally capable of discriminating between benign versus cancerous growths. While PET scans are often utilized to fill the gaps in currently available imaging modalities, they also fall short in their specificity as metabolic activity is a relatively non-specific marker of malignancy and resolutions are poor. As such, antibody-targeted fluorescent probes provide a modality that is both more sensitive and specific for tumor localization that can be used in real-time to enhance the surgeon’s range of cancer detection.
When comparing EGFR kinetics, a strong inverse relationship (R2=0.98) was found between the internalized Tc-99m-cetuximab percentage and the theoretical minimum detectable cells. This suggests that cetuximab-IRDye800CW induced contrast is more robust in cancers with greater EGFR turnover. These results support findings from preliminary work demonstrating florescence intensity is directly related to EGFR expression density since accumulation is dependent on EGFR uptake.[23] Therefore, we speculate that this modality may have a greater effect on improving clinical outcomes in diseases such as HNSCC, which has characteristically high EGFR activity. Additionally, this study serves as a proof of concept that demonstrates fluorescently labeled antibodies against ligands upregulated in cancerous tissue can be effectively applied to other tumor types.
Lastly, the effect of incubation time on both the minimum cetuximab-IRDye800CW concentration and the minimum number of cells exposed to the conjugate capable of accurately identifying disease was determined. Assessment was conducted with 2LMP as the cell line’s EGFR concentration was approximately equal to the mean of all 4 EGFR positive cell lines tested. During the experiment, an incubation time of 24-hours significantly decreased the theoretical minimal cells detected threshold to 2.5×104–7.0×104 vs. 7.8×104–8.8×104 cells at 72-hours and 4.2×104–9.5×104 cells at 1-hour. While the early phase clinical trial had an incubation time of 72-hours[6], these data suggest the need for further in-human study at 24-hours as enhanced optical contrast may further improve complete resection rates.
The murine model utilized was developed with several aspects in mind to best reveal the cetuximab-IRDye800CW detection threshold in a clinically significant manner. First, background fluorescence values were obtained from a naïve mouse 1, 24, or 72-hours after systemic administration of cetuximab-IRDye800CW. Background circulation time was equal to probe-cell incubation time to account for systemic, non-specific uptake/fluorescent signals and probe washout for each experiment.[19,24] Probe-cell tittering required in vitro incubation to ensure accurate dose and cellular count exposures. The 10% total tumor labeling was chosen as a measurable value to simulate reduced tumor delivery due to poor vascular extravasation and tumor heterogeneity. Moreover, cetuximab efficacy studies reveal a 10% tumor response rate to monotherapy.[25] Next, tumoral implantation was subcutaneous for skin flap evaluation that would simulate a post-resection wound bed with residual islands of different disease types. Thus, results highlight the detection abilities of disease at the resection plane and may not reflect capabilities of deeper situated disease, to and from which photons have an increased probability of scatter/absorption events.[1] IRDye800CW is a low energy photon emission source heavily influenced by surrounding tissue that attenuates the final signal.[1] Murine tissue confounders affecting final energy signals are similar to those of human tissues, providing translatable results. Utilization of a higher energy source emitter such as radioactivity may further decrease the disease detection threshold. However, IRDye800CW requires immediate sensitivity assessment due to its quick clinical integration.[6,26]
Employing a cetuximab-IRDye800CW dose that is 1.2% of a clinical dose is presumably sub-saturating. As a result, probe accumulation is decreased, which consequentially reduces detection capabilities. The small probe dose was used intentionally to simulate reduced tumoral uptake secondary to poor tumor hemodynamics and hemokinetics.[27] This is especially needed in making suppositions about detection of micro disease foci as smaller islands, less than 1mm, which may have yet increased angiogenesis and thus survive solely from host vasculature. [28–30] Therefore, a tumoral milieu promoting cetuximab-IRDye800CW delivery, like an angiogenic-active environment, may further enhance disease detection.
A limitation of this study is the device type used to evaluate disease detection thresholds. First, Pearl, the closed-field fluorescence device, is optimized to image at 800nm, the excitation wavelength of the IRDye800CW molecule, thereby enhancing the sensitivity and accuracy of the presented theoretical suppositions. While an open-field, NIR device is required for intraoperative translatability, clinical devices are being rapidly developed and improved. LUNA (Novadaq, Canada), a contemporary open-field device, though inferior in sensitivity to open-field counterparts, yields similar detection capabilities.[31] Nonetheless, Pearl does represent an effective prototype for the utility of both current and future NIR devices. However, Pearl could be rapidly integrated into patient care as a post-resection, specimen-imaging tool. While Pearl may not function to provide real-time resection guidance, it may improve disease eradication by assessing disease margins more quickly than histopathologic methods. A limitation of fluorescence detection as a whole includes local, non-specific fluorescence due to increased photon scattering events near disease that may limit precise localization. This results in a fluorescent halo illuminating non-diseased tissues. While the perfect probe will produce disease specific fluorescence, the minimal, collaterally resected tissue with non-ideal agents is much less than that resected when disease is evaluated on palpation and visual aspects alone. As a result, decreased excision of non-diseased tissue may improve post-resection functionality.[1]
In conclusion, we showed cetuximab-IRDye800CW has the potential to detect subclinical islands of disease in the surgical setting. Further study is required for each tumor type to realize the specific, beneficial gain this technology offers compared to contemporary resection[1]. Results from the current study support the use of this modality to augment complete resection rates and decrease morbidity and mortality in EGFR-positive disease.
Synopsis.
Fluorescence-guided surgery offers promise in the field of surgical oncology by significantly improving morbidity and mortality outcomes by augmenting disease excision. However, disease detection sensitivity and the affects of receptor expression have yet been evaluated. Our results reveal the robustness of fluorescence-guided surgery to localize subclinical nests of disease.
Acknowledgments
Funding Support: This work was supported by grants from the National Institute of Health (R21CA179171 and T32CA091078). Institutional device loans by LICOR Biosciences.
Footnotes
Conflicts of Interest: None of the authors have any conflicts of interest to declare.
References
- 1.Keereweer S, Van Driel PB, Snoeks TJ, et al. Optical image-guided cancer surgery: challenges and limitations. Clin Cancer Res. 2013;19:3745–3754. doi: 10.1158/1078-0432.CCR-12-3598. [DOI] [PubMed] [Google Scholar]
- 2.Twaalfhoven FC, Peters AA, Trimbos JB, et al. The accuracy of frozen section diagnosis of ovarian tumors. Gynecol Oncol. 1991;41:189–192. doi: 10.1016/0090-8258(91)90306-p. [DOI] [PubMed] [Google Scholar]
- 3.Rosenthal EL, Warram JM, de Boer E, et al. Successful Translation of Fluorescence Navigation During Oncologic Surgery: A Consensus Report. J Nucl Med. 2016;57:144–150. doi: 10.2967/jnumed.115.158915. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Hadjipanayis CG, Jiang H, Roberts DW, Yang L. Current and future clinical applications for optical imaging of cancer: from intraoperative surgical guidance to cancer screening. Semin Oncol. 2011;38:109–118. doi: 10.1053/j.seminoncol.2010.11.008. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Del Monte U. Does the cell number 10(9) still really fit one gram of tumor tissue? Cell Cycle. 2009;8:505–506. doi: 10.4161/cc.8.3.7608. [DOI] [PubMed] [Google Scholar]
- 6.Rosenthal EL, Warram JM, de Boer E, et al. Safety and Tumor Specificity of Cetuximab-IRDye800 for Surgical Navigation in Head and Neck Cancer. Clin Cancer Res. 2015;21:3658–3666. doi: 10.1158/1078-0432.CCR-14-3284. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Zinn KR, Korb M, Samuel S, et al. IND-directed safety and biodistribution study of intravenously injected cetuximab-IRDye800 in cynomolgus macaques. Mol Imaging Biol. 2015;17:49–57. doi: 10.1007/s11307-014-0773-9. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Marshall MV, Draney D, Sevick-Muraca EM, Olive DM. Single-dose intravenous toxicity study of IRDye 800CW in Sprague-Dawley rats. Mol Imaging Biol. 2010;12:583–594. doi: 10.1007/s11307-010-0317-x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Ang KK, Berkey BA, Tu X, et al. Impact of epidermal growth factor receptor expression on survival and pattern of relapse in patients with advanced head and neck carcinoma. Cancer Res. 2002;62:7350–7356. [PubMed] [Google Scholar]
- 10.Grandis JR, Tweardy DJ. Elevated levels of transforming growth factor alpha and epidermal growth factor receptor messenger RNA are early markers of carcinogenesis in head and neck cancer. Cancer Res. 1993;53:3579–3584. [PubMed] [Google Scholar]
- 11.Kalyankrishna S, Grandis JR. Epidermal growth factor receptor biology in head and neck cancer. Journal of clinical oncology : official journal of the American Society of Clinical Oncology. 2006;24:2666–2672. doi: 10.1200/JCO.2005.04.8306. [DOI] [PubMed] [Google Scholar]
- 12.Salomon DS, Brandt R, Ciardiello F, Normanno N. Epidermal growth factor-related peptides and their receptors in human malignancies. Crit Rev Oncol Hematol. 1995;19:183–232. doi: 10.1016/1040-8428(94)00144-i. [DOI] [PubMed] [Google Scholar]
- 13.Sasada T, Azuma K, Ohtake J, Fujimoto Y. Immune Responses to Epidermal Growth Factor Receptor (EGFR) and Their Application for Cancer Treatment. Front Pharmacol. 2016;7:405. doi: 10.3389/fphar.2016.00405. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Davis NM, Sokolosky M, Stadelman K, et al. Deregulation of the EGFR/PI3K/PTEN/Akt/mTORC1 pathway in breast cancer: possibilities for therapeutic intervention. Oncotarget. 2014;5:4603–4650. doi: 10.18632/oncotarget.2209. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Nicholson RI, Gee JM, Harper ME. EGFR and cancer prognosis. Eur J Cancer. 2001;37(Suppl 4):S9–15. doi: 10.1016/s0959-8049(01)00231-3. [DOI] [PubMed] [Google Scholar]
- 16.Troiani T, Martinelli E, Capasso A, et al. Targeting EGFR in pancreatic cancer treatment. Curr Drug Targets. 2012;13:802–810. doi: 10.2174/138945012800564158. [DOI] [PubMed] [Google Scholar]
- 17.Kim H, Zhai G, Liu Z, et al. Extracelluar matrix metalloproteinase as a novel target for pancreatic cancer therapy. Anticancer Drugs. 2011;22:864–874. doi: 10.1097/CAD.0b013e328349311e. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Larsen SK, Solomon HF, Caldwell G, Abrams MJ. [99mTc]tricine: a useful precursor complex for the radiolabeling of hydrazinonicotinate protein conjugates. Bioconjugate chemistry. 1995;6:635–638. doi: 10.1021/bc00035a020. [DOI] [PubMed] [Google Scholar]
- 19.Frangioni JV. New technologies for human cancer imaging. J Clin Oncol. 2008;26:4012–4021. doi: 10.1200/JCO.2007.14.3065. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Warram JM, de Boer E, van Dam GM, et al. Fluorescence imaging to localize head and neck squamous cell carcinoma for enhanced pathological assessment. J Pathol Clin Res. 2016;2:104–112. doi: 10.1002/cjp2.40. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.de Boer E, Warram JM, Tucker MD, et al. In Vivo Fluorescence Immunohistochemistry: Localization of Fluorescently Labeled Cetuximab in Squamous Cell Carcinomas. Sci Rep. 2015;5:10169. doi: 10.1038/srep10169. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Calderwood SK. Tumor heterogeneity, clonal evolution, and therapy resistance: an opportunity for multitargeting therapy. Discov Med. 2013;15:188–194. [PMC free article] [PubMed] [Google Scholar]
- 23.Sorace AG, Korb M, Warram JM, et al. Ultrasound-stimulated drug delivery for treatment of residual disease after incomplete resection of head and neck cancer. Ultrasound Med Biol. 2014;40:755–764. doi: 10.1016/j.ultrasmedbio.2013.11.002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Gioux S, Choi HS, Frangioni JV. Image-guided surgery using invisible near-infrared light: fundamentals of clinical translation. Mol Imaging. 2010;9:237–255. [PMC free article] [PubMed] [Google Scholar]
- 25.Cunningham D, Humblet Y, Siena S, et al. Cetuximab monotherapy and cetuximab plus irinotecan in irinotecan-refractory metastatic colorectal cancer. N Engl J Med. 2004;351:337–345. doi: 10.1056/NEJMoa033025. [DOI] [PubMed] [Google Scholar]
- 26.Tipirneni KE, Warram JM, Moore LS, et al. Oncologic Procedures Amenable to Fluorescence-guided Surgery. Ann Surg. 2016 doi: 10.1097/SLA.0000000000002127. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Thurber GM, Weissleder R. Quantitating antibody uptake in vivo: conditional dependence on antigen expression levels. Mol Imaging Biol. 2011;13:623–632. doi: 10.1007/s11307-010-0397-7. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Groebe K, Mueller-Klieser W. On the relation between size of necrosis and diameter of tumor spheroids. Int J Radiat Oncol Biol Phys. 1996;34:395–401. doi: 10.1016/0360-3016(95)02065-9. [DOI] [PubMed] [Google Scholar]
- 29.Naumov GN, Akslen LA, Folkman J. Role of angiogenesis in human tumor dormancy: animal models of the angiogenic switch. Cell Cycle. 2006;5:1779–1787. doi: 10.4161/cc.5.16.3018. [DOI] [PubMed] [Google Scholar]
- 30.Niu G, Li Z, Xie J, et al. PET of EGFR antibody distribution in head and neck squamous cell carcinoma models. J Nucl Med. 2009;50:1116–1123. doi: 10.2967/jnumed.109.061820. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Moore LS, Rosenthal EL, Chung TK, et al. Characterizing the Utility and Limitations of Repurposing an Open-Field Optical Imaging Device for Fluorescence-Guided Surgery in Head and Neck Cancer Patients. J Nucl Med. 2017;58:246–251. doi: 10.2967/jnumed.115.171413. [DOI] [PMC free article] [PubMed] [Google Scholar]


