Abstract
Background
Clostridium difficile infection (CDI) is a common nosocomial diarrheal illness increasingly associated with mortality in United States. The underlying factors and mechanisms behind the recent increases in morbidity from CDI have not been fully elucidated. Murine models suggest a mucosal barrier breakdown leads to bacterial translocation and subsequent bloodstream infection (BSI). This study tests the hypothesis that CDI is associated with subsequent BSI in humans.
Methods
We conducted a retrospective cohort study on 1132 inpatients hospitalized >72 hours with available stool test results for toxigenic C. difficile. The primary outcome was BSI following CDI. Secondary outcomes included 30-day mortality, colectomy, readmission, and ICU admission. Unadjusted and adjusted logistic regression models were developed.
Results
CDI occurred in 570 of 1132 patients (50.4%). BSI occurred in 86 (7.6%) patients. Enterococcus (14%) and Klebsiella (14%) species were the most common organisms. Patients with BSI had higher comorbidity scores and were more likely to be male, on immunosuppression, critically ill, and have a central venous catheter in place. Of the patients with BSI, 36 (42%) had CDI. CDI was not associated with subsequent BSI (OR 0.69; 95% CI 0.44–1.08; P =.103) in unadjusted analysis. In multivariable modeling, CDI appeared protective against subsequent BSI (OR 0.57; 95% CI 0.34–0.96; P = .036). Interaction modeling suggests a complicated relationship among CDI, BSI, antibiotic exposure, and central venous catheter use.
Conclusions
In this cohort of inpatients that underwent testing for CDI, CDI was not a risk factor for developing subsequent BSI.
Keywords: Clostridium difficile infection, colitis, bloodstream infection, central line
Introduction
Clostridium difficile infection (CDI) causes a colitis that has become the leading source of nosocomial diarrhea worldwide [1]. In the United States, it is estimated to cause 453,000 infections per year, 107,600 of which are hospital-onset [2]. The clinical presentation varies from mild diarrhea to debilitating disease with high fever, severe abdominal pain, and a paralytic ileus (megacolon) or perforation [3]. The burden of disease, despite best efforts, is not decreasing—CDI mortality has dramatically increased in the past decade, and is now the leading cause of gastroenteritis-associated death in the United States [4, 5]. The cause of mortality in CDI is likely multifactorial, but as discussed below evidence from murine models posits a role for bacterial translocation in the pathogenesis of complicated CDI.
It has been established that C. difficile cytopathic toxins cause mucosal injury and a pseudomembranous colitis [6]. These toxins, identified as C. difficile toxins A and B (TcdA and TcdB), inactivate members of mucosal Rho GTP-ases, and this is proposed to result in colonocyte death, neutrophilic colitis, and loss of intestinal barrier function [7]. Recent murine models suggest that loss of barrier function during CDI predisposes to bacterial translocation and systemic dissemination, with ensuing septic shock and death [8, 9]. This dissemination is proposed to be a primary cause of mortality; mice treated with antibiotics targeted at gut bacteria had significantly improved survival [9].
In humans, there are only two studies in the literature thus far examining if CDI and BSI are related. CDI was determined to be a risk factor for VRE bacteremia in a small cohort (N=59) of VRE-colonized acute leukemia patients [10]. A more recent retrospective analysis of CDI+ patients found no difference in the incidence of non-Staphylococcal BSI relative to CDI, but BSI occurring after CDI was more likely to be without an obvious source, suggesting colonic translocation [11]. In addition to the above studies regarding gut bacterial translocation, C. difficile bacteremia itself is a rare clinical entity [12].
In non-CDI colitis, there is some evidence that translocation can be clinically significant. Inflammatory bowel disease (IBD) was found to be a risk factor for anaerobic bacteremia in a recent large population surveillance study [13]. Also, antibiotic induced diarrhea was associated with Klebsiella oxytoca bacteremia, suggesting translocation [14]. Additionally, there are reports of BSI attributed to colonic infections with CMV, Strongyloides stercoralis, and Entamoeba histolytica [15–18].
Based on the available evidence, it is not yet established whether CDI is a risk factor for subsequent BSI in humans. As CDI is becoming a more frequent and virulent nosocomial pathogen, determining if it predisposes to potentially lethal BSI could inform clinical decision-making. The objective of this study was to determine if CDI is associated with BSI in humans.
Methods
Study design and population
This study was approved by the University of Michigan Institutional Review Board. We conducted a retrospective cohort study on inpatients at the University of Michigan Hospital who were tested for possible healthcare associated C. difficile infection (>48 hours after admission) between October 2010 and January 2013. All laboratory testing of inpatients was performed at the discretion of the inpatient care team, which ordered C. difficile testing per institutional guidelines that mirror national guidelines recommending testing only symptomatic patients with suspected CDI [19, 20]. Patients hospitalized <72 hours, those with a prior positive and redundant blood culture or a positive CDI within the prior 8 weeks were excluded.
Microbiology
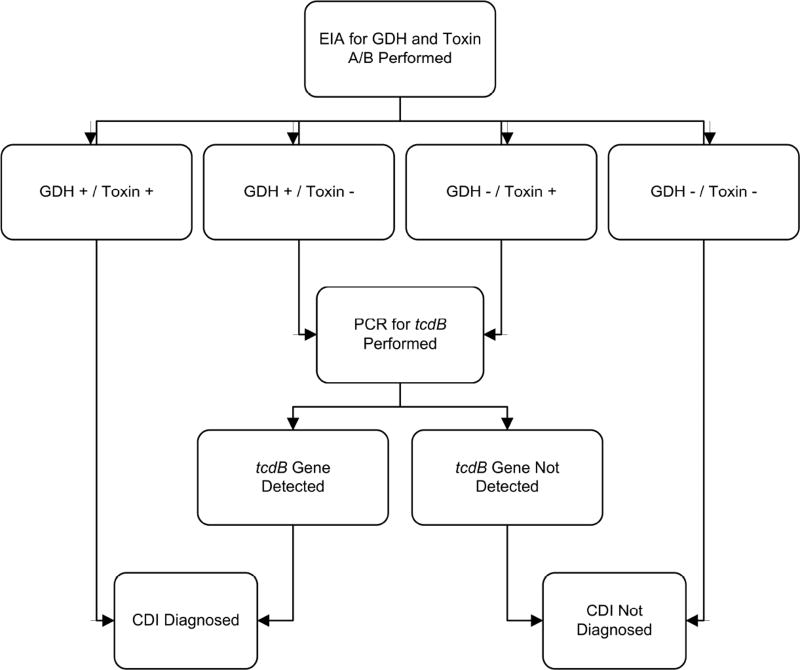
Testing was performed on stools in the clinical microbiology laboratory via an algorithm (Figure 1) using the C. DIFF QUIK CHEK COMPLETE® test for C. difficile glutamate dehydrogenase (GDH) and toxins A or B (Techlab, Inc., Blacksburg, VA) by EIA. All GDH+/toxin− stool tests were subjected to analysis for the tcdB gene by real-time PCR using the GeneOhm™ Cdiff Assay (BD, Franklin Lakes, NJ) run on a Cepheid SmartCycler® System (Cepheid, Sunnyvale, CA). Confirmation of all positive C. difficile tests was attempted by anaerobic culture on taurocholate-cycloserine-cefoxitin-fructose agar at 37°C. Attempts were made to ribotype samples using high-throughput, fluorescent PCR-ribotyping as described elsewhere [21, 22]. Blood culture collection and detection of positives was performed using the aerobic and anaerobic BacT/Alert system (BioMerieux, Durham, NC) and handled per the clinical microbiology laboratory protocol [23]. Briefly, all culture bottles are read by the automated system and positives reported to the laboratory staff every 15 minutes, followed by an initial Gram stain and subculturing / identification as appropriate.
Figure 1. Testing algorithm for Clostridium difficile infection.
This flow diagram illustrates this University of Michigan diagnostic testing algorithm for detecting toxigenic Clostridium difficile in stool [38]. Abbreviations: CDI, Clostridium difficile infection; EIA, enzyme immunoassay; GDH, glutamate dehydrogenase; PCR, polymerase chain reaction.
Data Extraction
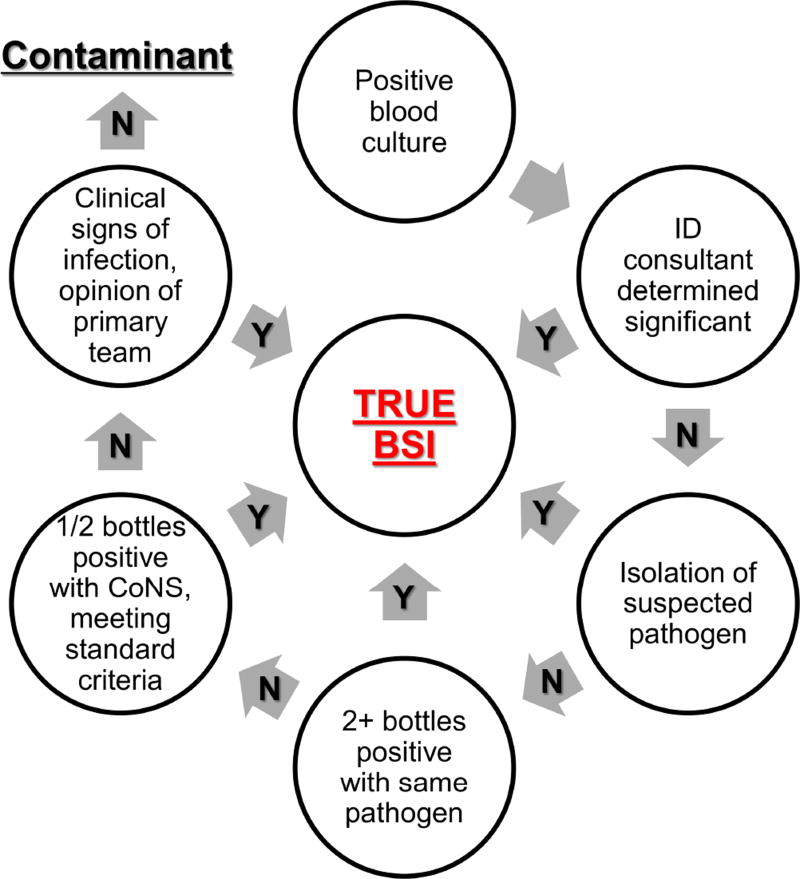
The electronic medical record of each subject with a positive blood culture subsequent to CDI onset was reviewed by a physician to determine if it represented a true BSI, using the algorithm outlined in Figure 2, and not a contaminant. If the blood culture was determined to be a contaminant, this patient was treated as a non-BSI case in our analysis. Concurrent presence/absence of a central venous catheter and the type was also obtained. Other variables were also extracted: demographics, medications, laboratory results including C. difficile test results, and 30 day outcomes including mortality, colectomy, ICU admission, and readmission.
Figure 2. Algorithm for determining BSI significance.
This figure illustrates the decision pathway used to determine if a positive blood culture is a true subsequent BSI or a contaminant. Definitions of “suspected pathogen” and “standard criteria” are found in the methods section. Abbreviations: ID, infectious disease; CDI, Clostridium difficile infection; BSI, Bloodstream infection; CoNS, Coagulase negative Staphylococcus.
Study Definitions
In our study, subsequent BSI was defined as positive blood culture results obtained <48 hours prior to, or any time after, CDI stool assay collection that was determined to be a true (non-contaminant) BSI using the Figure 2 algorithm. The <48 hours prior criterion for BSI was selected as there is often a delay between CDI symptom onset and test result availability. In this algorithm, suspected pathogen was defined as a bacterial species documented to be overwhelmingly pathogenic (>88% clinical significance) [24]. Standard criteria (to determine positivity of Coagulase-negative Staphylococcus) was defined using a previously determined algorithm (sensitivity 62%, specificity 91%) combining white blood cell count, presence or absence of fever and/or hypotension [25]. Charlson comorbidity index (weighted and non-weighted) are previously validated comorbidity indices based off ICD-9 codes, shown to predict patient mortality [26, 27], and these were calculated. Ventilator status was defined by evidence in the electronic medical record of endotracheal intubation during the index hospitalization.
Immunosuppression was defined as prednisone use of ≥5 mg per day or any steroid-sparing agent. Prior fluoroquinolone use and concurrent non-CDI antibiotic use are in relation to CDI stool assay testing, during the index hospitalization. Prior CDI was defined as a positive CDI test >8 weeks prior to the index CDI stool assay. Modified SIRS criteria was positive if any two of the following criteria were present: leukocytosis >12,000 cells/mm3 or leukopenia <4,000 cells/mm3 ; tachypnea (respiratory rate >20); fever (>38° C) or hypothermia (<36° C). The primary outcome was presence of subsequent bloodstream infection. Secondary outcomes included 30-day mortality, colectomy, readmission, and ICU admission.
Statistical Analysis
After data cleaning, descriptive statistics were prepared using proportions for categorical variables and measures of central tendency/spread for continuous variables. For all analytical statistics, P values <.05 were considered statistically significant. To analyze relationships between predictor variables and the primary/secondary outcomes, simple, unconditional logistic regression was employed. Multiple logistic regression was employed for adjusted analysis. Multivariable models were constructed through stepwise addition, incorporating variables significant on unadjusted analysis. To assess for possible confounding or collinearity, the point estimates for coefficients and the significance testing results were scrutinized as each new variable was added to the model. Likelihood ratio testing was used to exclude variables from the models with P >.05. As noted below, given the complex relationship between two predictors and the primary outcome, interaction terms from the final model were examined but only included in the final model if significant.
Results
Population characteristics and outcomes
A summary of study population demographics and outcomes is shown in Table 1. There were 1132 patients eligible for inclusion in the study, of which 86 (7.6%) were determined to have a true subsequent (post-CDI) BSI. Average age was 55 years, gender was evenly split, and the predominant race was white (80.5%). Half (50.4%) of the sample population’s CDI assays returned positive, and nearly half (49.0%) had a central line in place. In unadjusted analysis, those with BSI were more likely to be male, immunosuppressed, medically comorbid (per Charlson-Deyo score), and have a central venous catheter. They had an average hospital stay 18.6 days longer than those without BSI (P <.001). Regarding the primary result of this study, there was no association between CDI and subsequent BSI (OR 0.69, CI 0.44–1.08, P =.103). Regarding secondary outcomes, death and ICU admission within 30 days were both positively associated with BSI. To fully evaluate for confounders, we also examined the relationship of patient characteristics and CDI, summarized in Table 2. The only significant association was an increase in CDI in those with a history of CDI (>8 weeks prior).
Table 1.
Selected characteristics1 and outcomes of the study population.
| BSI | Non-BSI | Total | ||||
|---|---|---|---|---|---|---|
| Characteristic | N (%) or mean ±SD |
N (%) or mean ±SD |
N | OR (95% CI) | P | |
| Gender | 1.74 (1.11–2.72) | .016 | ||||
| Male | 52 (60.5) | 490(46.8) | 542 | |||
| Female | 34 (39.5) | 556 (53.2) | 590 | |||
|
| ||||||
| Age (years) | 56.5 ±14.9 | 55.1 ±20.3 | 1.00 (0.99–1.02) | .529 | ||
|
| ||||||
| Race2 | 0.74 (0.44–1.23) | .244 | ||||
| White | 65 (75.6) | 845 (80.8) | 910 | |||
| Non-white | 21 (24.4) | 201 (19.2) | 222 | |||
|
| ||||||
| Length of Stay (days) | 35.5 ±31.7 | 16.9 ±15.9 | 1.03 (1.02–1.04) | <.001 | ||
|
| ||||||
| Charlson-Deyo Score3 | 3.2 ±2.9 | 2.3 ±2.5 | 1.12 (1.04–1.21) | .004 | ||
|
| ||||||
| Ventilator status | 2.63 (1.68–4.11) | <.001 | ||||
| Yes | 40 (46.5) | 260 (24.9) | 300 | |||
| No | 46 (53.4) | 786 (75.1) | 832 | |||
|
| ||||||
| Immunosuppression | 1.94 (1.22–3.07) | .005 | ||||
| Yes | 56 (65.1) | 513 (49.0) | 569 | |||
| No | 30 (34.9) | 533 (51.0) | 563 | |||
|
| ||||||
| Non-CDI antibiotics | 3.90 (1.86–8.18) | <.001 | ||||
| Yes | 78 (90.7) | 747 (71.4) | 825 | |||
| No | 8 (9.3) | 299 (28.6) | 307 | |||
|
| ||||||
| Prior to CDI FQ use | 1.59 (1.02–2.47) | .041 | ||||
| Yes | 39 (45.3) | 359 (34.3) | 398 | |||
| No | 47 (54.7) | 687 (65.7) | 734 | |||
|
| ||||||
| Prior CDI | 1.16 (0.54–2.48) | .703 | ||||
| Yes | 8 (9.3) | 85 (8.1) | 93 | |||
| No | 78 (90.7) | 961 (91.9) | 1039 | |||
|
| ||||||
| CDI | 0.69 (0.44–1.08) | .103 | ||||
| Yes | 36 (41.9) | 534 (51.1) | 570 | |||
| No | 50 (58.1) | 512 (48.9) | 562 | |||
|
| ||||||
| CDI Ribotype4 | 2.16 (1.01–4.22) | .033 | ||||
| 027 | 10 (31.3) | 60 (13.3) | 70 | |||
| 014–020 | 2 (6.2) | 75 (16.6) | 77 | |||
| 053–163 | 2 (6.2) | 32 (7.1) | 34 | |||
| Other | 18 (56.3) | 284 (63.0) | 302 | |||
|
| ||||||
| Central line | 3.66 (2.19–6.13) | <.001 | ||||
| Yes | 65 (75.6) | 490 (47.0) | 555 | |||
| No | 20 (24.4) | 552 (53.0) | 572 | |||
|
| ||||||
| Modified SIRS5 | 2.03 (1.17–3.53) | .012 | ||||
| Yes | 51 (59.3) | 497 (47.5) | 548 | |||
| No | 18 (20.9) | 356 (34.1) | 374 | |||
|
| ||||||
| Outcomes (30 day) | N (%) | N (%) | N | OR (95% CI) | P | |
|
| ||||||
| Death | 3.48 (1.96–6.15) | <.001 | ||||
| Yes | 18 (20.9) | 74 (7.1) | 92 | |||
| No | 68 (79.1) | 972 (92.9) | 1040 | |||
|
| ||||||
| Colectomy | N/A | |||||
| Yes | 0 (0.0) | 7 (0.7) | 7 | |||
| No | 86 (100.0) | 1039 (99.3) | 1125 | |||
|
| ||||||
| Re-admission | 0.56 (0.31–1.03) | .064 | ||||
| Yes | 13 (15.1) | 251 (24.0) | 264 | |||
| No | 73 (84.9) | 795 (76.0) | 868 | |||
|
| ||||||
| ICU | 3.38 (2.09–5.47) | <.001 | ||||
| Yes | 29 (33.7) | 137 (13.1) | 166 | |||
| No | 57 (66.3) | 909 (86.9) | 966 | |||
|
| ||||||
| Total | 86 (7.6) | 1046 (92.4) | 1132 | |||
Abbreviations: BSI, bloodstream infection; SD, standard deviation; OR, odds ratio; CI, confidence interval; CDI, Clostridium difficile infection; SIRS, systemic inflammatory response syndrome; N/A, not available; ICU, intensive care unit; FQ, fluoroquinolone.
The following were studied but not reported in this table: ethnicity, unweighted Charlson comorbidity index, and type of central venous catheter. All definitions of characteristics can be found in the methods section.
Non-white included Black/African American, Asian, American Indian, Alaska native, Other, and Unknown.
Weighted Charlson comorbidity index. 40 patients (3.5%) had insufficient data.
OR, CI and P were calculated using ribotype 027 vs. all other ribotypes.
210 patients (18.6%) had missing data precluding the ability to calculate modified SIRS.
Table 2.
Unadjusted analysis of selected characteristics in relation to CDI.
| Variable1 | Odds Ratio | (95% CI) | P |
|---|---|---|---|
| Gender | 1.11 | (0.88–1.40) | .399 |
| Age | 1.00 | (0.99–1.00) | .429 |
| Caucasian race | 0.89 | (0.66–1.19) | .435 |
| Charlson comorbidity score (w) | 1.03 | (0.99–1.08) | .162 |
| Ventilator status | 1.24 | (0.95–1.62) | .108 |
| Concurrent non-CDI antibiotics | 0.79 | (0.60–1.02) | .073 |
| Immunosuppressed | 0.85 | (0.67–1.07) | .171 |
| Prior-to-CDI Fluoroquinolones | 0.84 | (0.66–1.07) | .156 |
| Prior CDI (>8 weeks prior) | 2.45 | (1.55–3.89) | <.001 |
| Central venous catheter | 1.10 | (0.87–1.39) | .420 |
| Modified SIRS | 1.27 | (0.97–1.65) | .078 |
Abbreviations: CI, confidence interval; CDI, Clostridium difficile infection; w, weighted; SIRS, systemic inflammatory response syndrome.
Definitions of select variables found in the methods section.
Microbiology of bloodstream isolates and C. difficile
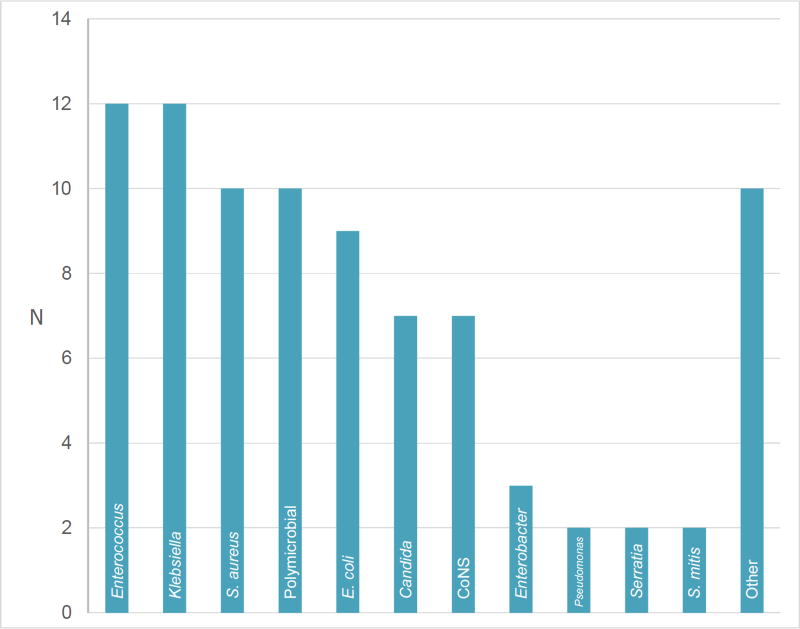
The most common BSI genera isolated were Enterococcus (14%) and Klebsiella (14%). S. aureus (11.6%) and polymicrobial (11.6%) culture results were the second most common. The microbiology of the isolates is depicted in Figure 3. The most common C. difficile ribotypes were ribotype 027 and 014–020, and ribotype 027 infection was associated with bacteremia (OR 2.16, CI 1.01–4.22, P = .033) (Table 1).
Figure 3. Microbiology of subsequent BSI.
The graph shows the number of each species of BSI detected. If category is a genus, this included multiple species. Abbreviations: CoNS, Coagulase negative Staphylococci.
Multivariable model
To account for confounding between variables, multivariable modeling was done with the strongest associations in the unadjusted analysis, though concomitant use of non-CDI antibiotics was not eligible for inclusion in the model due to collinearity with the primary outcome of BSI (patients with BSI are often treated with intravenous [IV] antimicrobials). Notably, Ribotype 027, though associated in unadjusted analysis, did not enter the final model as it was not significant and forcing it into the model reduced the model’s performance. Immunosuppression also failed the likelihood ratio test and was excluded from final modeling. The most parsimonious model is summarized in Table 3. In this model, an unexpected negative association between CDI and BSI became significant (OR 0.57, 95% CI 0.34–0.96, P = .036). However, we suspected that this protective effect of CDI was likely due to a complex relationship between concomitant IV antibiotic use, presence of a central venous catheter, and the increased subsequent likelihood of both CDI and a BSI. When this was modeled with an interaction term accounting for a putative interaction between CDI and central venous catheter presence, the association reversed and no association was present between CDI and BSI (OR 1.15, 95% CI 0.44–3.00, P = .774) but the interaction term did not achieve significance (P = .085). Inclusion of concomitant antibiotic use further increased the significance of the interaction term (P = .058), suggesting a complex relationship between these three variables. A putative causal pathway is IV antimicrobial use, possibly for an earlier BSI that is not related to CDI, predisposes to CDI but makes subsequent BSI less likely since it is already being treated. Properly teasing apart these relationships was not possible in our study, as the models constructed with other appropriate interaction terms to test them failed to converge (data not shown).
Table 3.
Multivariable model for BSI.
| Variable | Odds Ratio | (95% CI) | P |
|---|---|---|---|
| Male gender | 1.75 | (1.03–2.98) | .037 |
| Charlson comorbidity score (w) | 1.15 | (1.05–1.26) | .002 |
| Modified SIRS | 1.88 | (1.03–3.41) | .039 |
| Central venous catheter | 2.64 | (1.48–4.71) | .001 |
| CDI | 0.57 | (0.34–0.96) | .036 |
Abbreviations: CDI, Clostridium difficile infection; BSI, bloodstream infection; w, weighted; SIRS, systemic inflammatory response syndrome.
Discussion
This study examined if infection with Clostridium difficile predisposes patients to subsequent bloodstream infection, with results showing no evidence to support this hypothesis. Ultimately, this lack of association remained true using both unadjusted analysis and multivariable modeling controlling for gender, comorbidities, modified SIRS criteria and presence of a central venous catheter. It was important to analyze this hypothesis, as CDI is becoming a more prevalent infection with striking recent increases in morbidity and mortality [5]. The mechanisms by which mortality is increasing needs to be fully understood in order to appropriately combat this virulent organism. Murine models suggest a potential role for bacterial translocation and bloodstream dissemination [8, 9, 28], but our results do not support this finding in humans. Overall strengths of our study include the size of our sample, inclusion of several possible confounders, careful modeling that evaluated for interactions, and concordance of results with clinical experience.
A probable explanation for the lack of association is that our patients had relatively intact mucosal immunity at the time of infection. In the murine models with bacterial dissemination, there was genetic knockout of key mediators (Nod-1, IL1-β, IL-22) in neutrophil recruitment and innate immunity essential to maintain barrier function in the setting of mucosal damage [8, 9, 28]. However, our human population was likely able to protect against translocation and prevent significantly increased dissemination. This ability to induce a physiologic inflammatory response to CDI actually seemed to be protective against BSI after adjusting for other variables using the most parsimonious modeling. At a barrier level, a strong immune response to a slightly damaged mucosal barrier could be protective, as intestinal epithelial cells are remarkably sensitive to the inflammatory milieu [29]. At a broader level, systemic immune activation in response to CDI could be protective as immune mediators have been shown to be paramount to clearance of bloodstream pathogens [30, 31].
Even though our modeling suggests CDI is protective against BSI, there is likely a contribution from complex interactions both measured and unmeasured. To exclude any simple confounders from our analysis, we performed unadjusted analysis of patient characteristics to CDI (Table 2). This did not show any association other than prior CDI, which was not related to BSI in our data, therefore any confounding is likely to be complex through multiple steps. For example, although having a central line was clearly a BSI risk factor in our analysis (consistent with the preponderance of prior research [32, 33]), interaction modeling suggested a trend (P =.085) towards a protective effect of having a central catheter during CDI on subsequent BSI. A possible explanation is the early administration of broad spectrum IV antibiotics in patients who have a central line (more likely admitted to an ICU) with an undifferentiated diagnosis, who end up developing CDI as a result. However, the lack of significance precluded this interaction from being included in the final models. Overall, we suspect very complex multifactorial confounders are responsible for the protective effect observed and deserve further research.
When comparing our results to previous human research, our results are discordant from the small study of acute leukemia patients where CDI was a risk factor for VRE [10]. This is likely due to our larger sampling size, more varied (including patients with and without cancer) patient population, and our inclusion of various types of non-VRE organisms in our analysis. We believe this makes our findings more applicable to a general inpatient population. Our results are overall consistent with more recent findings, based off a much larger sample population, that there is no difference in frequency of BSI relative to CDI [11].
The secondary outcomes related to BSI were as expected; patients with BSI had higher levels of 30 day mortality and ICU admission. This concordance with well-established outcomes data on sepsis and septic shock [34] supports our sample population being a representative cohort, the accuracy in our data collection, and the use of sound statistical techniques.
Our study has multiple potential limitations. First, it is a retrospective analysis using electronic medical records. As our primary outcome is based off correct determination of true BSI vs. contaminant, we developed a rigorous algorithm (Figure 2) using available literature to minimize incorrect classification of BSI. The other major variable is CDI diagnostic testing, but overall sensitivity of our diagnostic algorithm (Figure 1) was >90% even in Cary-Blair media, as demonstrated in a study conducted by our clinical laboratory [35], though specificity was likely not an issue since we confirmed all positive tests via anaerobic culture. An additional possible limitation is our attempt to encompass all subsequent BSI and we included blood culture media collected up to 48 hours prior to CDI assay collection. This is to account for the undiagnosed, but clinically present CDI that may have preceded BSI. Antibiotics given during this time are unlikely to be confounders as previous studies have shown the colonic microbiome can take days to change after antibiotic exposure [36]. Our study included all age groups, however we feel the results are more generalizable to adults as our pediatric population made up only 5.3% of the sample and there were no pediatric cases of BSI in our population. Finally, including S. aureus in our analysis could be considered a limitation, as this bacteria is unlikely of colonic origin [37]. However, this study was also interested if CDI increases BSI through mechanisms other than translocation, including modulating inflammatory response. Furthermore, the amount of S. aureus BSI was relatively small (10%) in our cohort.
Conclusion
In summary, this large retrospective analysis of inpatients tested for healthcare associated CDI shows no increased risk of BSI following CDI. As CDI is becoming a more prevalent, morbid disease, it is important to determine all of the CDI-related complications and mechanisms leading to mortality. Further studies will be required to tease apart the relationship between CDI, concomitant antibiotic use, and subsequent BSI.
Key Points.
Clostridium difficile infection is not associated with subsequent bloodstream infection. It is associated with male gender, immunosuppression, comorbid disease burden, and systemic inflammatory response syndrome.
HIGHLIGHTS.
C. difficile infection is not a risk factor for subsequent bloodstream infection
Bloodstream infections were most often from Klebsiella and Enterococcus species
Central venous catheter presence was a major risk factor for bloodstream infection
Acknowledgments
Funding
This work was supported by grants from the Claude D. Pepper Older Americans Independence Center [grant number AG- 024824], the Michigan Institute for Clinical and Health Research [grant number 2UL1TR000433], and the National Institutes of Health, National Institute of Allergy and Infectious Diseases [grant numbers U19-AI090871 and R21-AI120599]. The funders had no role in study design, data collection and analysis, decision to publish, or preparation of the manuscript.
We would like to thank the University of Michigan Health System’s Medical Center Information Technology team for database support. Informatics assistance with data abstraction and the EMERSE tool was provided by the University of Michigan’s Cancer Center’s (UMCC) Biomedical Informatics Core with partial support from the National Institutes of Health through the UMCC Support Grant (CA46952).
Footnotes
Publisher's Disclaimer: This is a PDF file of an unedited manuscript that has been accepted for publication. As a service to our customers we are providing this early version of the manuscript. The manuscript will undergo copyediting, typesetting, and review of the resulting proof before it is published in its final citable form. Please note that during the production process errors may be discovered which could affect the content, and all legal disclaimers that apply to the journal pertain.
Conflicts of Interest
All authors: no reported conflicts.
References
- 1.Hurley BW, Nguyen CC. The spectrum of pseudomembranous enterocolitis and antibiotic-associated diarrhea. Arch Intern Med. 2002;162(19):2177. doi: 10.1001/archinte.162.19.2177. [DOI] [PubMed] [Google Scholar]
- 2.Lessa FC, Mu Y, Bamberg WM, et al. Burden of Clostridium difficile infection in the United States. N Engl J Med. 2015;372(9):825–34. doi: 10.1056/NEJMoa1408913. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Eleftherios Mylonakis METR, MD, Calderwood Stephen B., MD Clostridium difficile–Associated Diarrhea - A review. Archives of Internal Medicine. 2001;161:525–33. doi: 10.1001/archinte.161.4.525. [DOI] [PubMed] [Google Scholar]
- 4.Hall AJ, Curns AT, McDonald LC, Parashar UD, Lopman BA. The roles of Clostridium difficile and norovirus among gastroenteritis-associated deaths in the United States, 1999–2007. Clin Infect Dis. 2012;55(2):216–23. doi: 10.1093/cid/cis386. [DOI] [PubMed] [Google Scholar]
- 5.Lessa FC, Gould CV, McDonald LC. Current Status of Clostridium difficile Infection Epidemiology. Clin Infect Dis. 2012;55(suppl 2):S65–S70. doi: 10.1093/cid/cis319. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Bartlett JG, Moon N, Chang TW, Taylor N, Onderdonk AB. Role of Clostridium difficile in antibiotic-associated pseudomembranous colitis. Gastroenterology. 1978;75(5):778–82. [PubMed] [Google Scholar]
- 7.Leffler DA, Lamont JT. Clostridium difficile infection. N Engl J Med. 2015;372(16):1539–48. doi: 10.1056/NEJMra1403772. [DOI] [PubMed] [Google Scholar]
- 8.Hasegawa M, Kamada N, Jiao Y, Liu MZ, Nunez G, Inohara N. Protective role of commensals against Clostridium difficile infection via an IL-1beta-mediated positive-feedback loop. J Immunol. 2012;189(6):3085–91. doi: 10.4049/jimmunol.1200821. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Hasegawa M, Yada S, Liu MZ, et al. Interleukin-22 regulates the complement system to promote resistance against pathobionts after pathogen-induced intestinal damage. Immunity. 2014;41(4):620–32. doi: 10.1016/j.immuni.2014.09.010. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Mary-Claire Roghmann RJM, Jr, Brewrink Jeanine, Cross Alan S, Morris J Glenn., Jr Clostridium difficile Infection Is a Risk Factor for Bacteremia Due to Vancomycin-Resistant Enterococci (VRE) in VRE-Colonized Patients with Acute Leukemia. Clinical Infectious Diseases. 1997;(25):1056–9. doi: 10.1086/516112. [DOI] [PubMed] [Google Scholar]
- 11.Thomas JA, Newman KC, Doshi S, Logan N, Musher DM. Bacteraemia from an unrecognized source (occult bacteraemia) occurring during Clostridium difficile infection. Scand J Infect Dis. 2011;43(4):269–74. doi: 10.3109/00365548.2010.546366. [DOI] [PubMed] [Google Scholar]
- 12.Libby DB, Bearman G. Bacteremia due to Clostridium difficile--review of the literature. Int J Infect Dis. 2009;13(5):e305–9. doi: 10.1016/j.ijid.2009.01.014. [DOI] [PubMed] [Google Scholar]
- 13.Ngo JT, Parkins MD, Gregson DB, et al. Population-based assessment of the incidence, risk factors, and outcomes of anaerobic bloodstream infections. Infection. 2013;41(1):41–8. doi: 10.1007/s15010-012-0389-4. [DOI] [PubMed] [Google Scholar]
- 14.Cox ER, Nayak SU, Kuruppu JC. Klebsiella oxytoca bacteremia-causal relationship to symptomatic colitis? Int J Infect Dis. 2013;17(6):e472–3. doi: 10.1016/j.ijid.2013.02.003. [DOI] [PubMed] [Google Scholar]
- 15.João Santos-Antunes FM, Macedo Guilherme. Listeria monocytogenes bacteremia and CMV colitis in a patient with Ulcerative Colitis. Journal of Crohn's and Colitis. 2004;8(3):254–5. doi: 10.1016/j.crohns.2013.10.003. [DOI] [PubMed] [Google Scholar]
- 16.Verstreken I, Laleman W, Wauters G, Verhaegen J. Desulfovibrio desulfuricans bacteremia in an immunocompromised host with a liver graft and ulcerative colitis. J Clin Microbiol. 2012;50(1):199–201. doi: 10.1128/JCM.00987-11. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Linder JD, Monkemuller KE, Lazenby AJ, Wilcox CM. Streptococcus bovis bacteremia associated with Strongyloides stercoralis colitis. Gastrointest Endosc. 2000;52(6):796–8. doi: 10.1067/mge.2000.109717. [DOI] [PubMed] [Google Scholar]
- 18.Ishioka H, Hayakawa K, Shinozaki S, Umezawa M, Hatakeyama S Group G. Streptococcal Bacteremia and Vertebral Osteomyelitis in a Homosexual Man with Amebic Colitis. Internal Medicine. 2008;47(4):317–20. doi: 10.2169/internalmedicine.47.0516. [DOI] [PubMed] [Google Scholar]
- 19.Cohen SH, Gerding DN, Johnson S, et al. Clinical practice guidelines for Clostridium difficile infection in adults: 2010 update by the society for healthcare epidemiology of America (SHEA) and the infectious diseases society of America (IDSA) Infect Control Hosp Epidemiol. 2010;31(5):431–55. doi: 10.1086/651706. [DOI] [PubMed] [Google Scholar]
- 20.Surawicz CM, Brandt LJ, Binion DG, et al. Guidelines for Diagnosis, Treatment, and Prevention of Clostridium difficile Infections. Am J Gastroenterol. 2013;108(4):478–98. doi: 10.1038/ajg.2013.4. [DOI] [PubMed] [Google Scholar]
- 21.Walk ST, Micic D, Jain R, et al. Clostridium difficile Ribotype Does Not Predict Severe Infection. Clin Infect Dis. 2012;55(12):1661–8. doi: 10.1093/cid/cis786. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Martinson JNV, Broadaway S, Lohman E, et al. Evaluation of portability and cost of a fluorescent PCR ribotyping protocol for Clostridium difficile epidemiology. Journal of Clinical Microbiology. 2015 doi: 10.1128/JCM.03591-14. published online January 28, 2015. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.University of Michigan Pathology Handbook: Blood Culture, Bacteria and Yeast. Available at: https://www.pathology.med.umich.edu/handbook/details.php?testID=175.
- 24.Pien BC, Sundaram P, Raoof N, et al. The clinical and prognostic importance of positive blood cultures in adults. Am J Med. 2010;123(9):819–28. doi: 10.1016/j.amjmed.2010.03.021. [DOI] [PubMed] [Google Scholar]
- 25.Beekmann SE, Diekema DJ, Doern GV. Determining the clinical significance of coagulase-negative staphylococci isolated from blood cultures. Infect Control Hosp Epidemiol. 2005;26(6):559–66. doi: 10.1086/502584. [DOI] [PubMed] [Google Scholar]
- 26.Needham DM, Scales DC, Laupacis A, Pronovost PJ. A systematic review of the Charlson comorbidity index using Canadian administrative databases: a perspective on risk adjustment in critical care research. J Crit Care. 2005;20(1):12–9. doi: 10.1016/j.jcrc.2004.09.007. [DOI] [PubMed] [Google Scholar]
- 27.Sharabiani MT, Aylin P, Bottle A. Systematic review of comorbidity indices for administrative data. Med Care. 2012;50(12):1109–18. doi: 10.1097/MLR.0b013e31825f64d0. [DOI] [PubMed] [Google Scholar]
- 28.Hasegawa M, Yamazaki T, Kamada N, et al. Nucleotide-binding oligomerization domain 1 mediates recognition of Clostridium difficile and induces neutrophil recruitment and protection against the pathogen. J Immunol. 2011;186(8):4872–80. doi: 10.4049/jimmunol.1003761. [DOI] [PubMed] [Google Scholar]
- 29.Onyiah JC, Colgan SP. Cytokine responses and epithelial function in the intestinal mucosa. Cell Mol Life Sci. 2016;73(22):4203–12. doi: 10.1007/s00018-016-2289-8. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Verschoor A, Neuenhahn M, Navarini AA, et al. A platelet-mediated system for shuttling blood-borne bacteria to CD8alpha+ dendritic cells depends on glycoprotein GPIb and complement C3. Nat Immunol. 2011;12(12):1194–201. doi: 10.1038/ni.2140. [DOI] [PubMed] [Google Scholar]
- 31.Broadley SP, Plaumann A, Coletti R, et al. Dual-Track Clearance of Circulating Bacteria Balances Rapid Restoration of Blood Sterility with Induction of Adaptive Immunity. Cell Host Microbe. 2016;20(1):36–48. doi: 10.1016/j.chom.2016.05.023. [DOI] [PubMed] [Google Scholar]
- 32.Safdar N, Maki DG. The pathogenesis of catheter-related bloodstream infection with noncuffed short-term central venous catheters. Intensive Care Med. 2004;30(1):62–7. doi: 10.1007/s00134-003-2045-z. [DOI] [PubMed] [Google Scholar]
- 33.Pronovost P, Needham D, Berenholtz S, et al. An Intervention to Decrease Catheter-Related Bloodstream Infections in the ICU. New England Journal of Medicine. 2006;355(26):2725–32. doi: 10.1056/NEJMoa061115. [DOI] [PubMed] [Google Scholar]
- 34.Angus DC, van der Poll T. Severe sepsis and septic shock. N Engl J Med. 2013;369(9):840–51. doi: 10.1056/NEJMra1208623. [DOI] [PubMed] [Google Scholar]
- 35.Brown NA, Lebar WD, Young CL, Hankerd RE, Newton DW. Diagnosis of Clostridium difficile infection: comparison of four methods on specimens collected in Cary-Blair transport medium and tcdB PCR on fresh versus frozen samples. Infect Dis Rep. 2011;3(1):e5. doi: 10.4081/idr.2011.e5. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Young VB, Schmidt TM. Antibiotic-Associated Diarrhea Accompanied by Large-Scale Alterations in the Composition of the Fecal Microbiota. Journal of Clinical Microbiology. 2004;42(3):1203–6. doi: 10.1128/JCM.42.3.1203-1206.2004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Hold GL, Pryde SE, Russell VJ, Furrie E, Flint HJ. Assessment of microbial diversity in human colonic samples by 16S rDNA sequence analysis. FEMS Microbiol Ecol. 39:33–9. doi: 10.1111/j.1574-6941.2002.tb00904.x. [DOI] [PubMed] [Google Scholar]
- 38.Rao K, Erb-Downward JR, Walk ST, et al. The systemic inflammatory response to Clostridium difficile infection. PLoS One. 2014;9(3):e92578. doi: 10.1371/journal.pone.0092578. [DOI] [PMC free article] [PubMed] [Google Scholar]