Abstract
Background
Alcohol increases intestinal permeability to pro-inflammatory microbial products that promote liver disease, even after a period of sobriety. We sought to test the hypothesis that alcohol affects intestinal stem cells using an in vivo model and ex vivo organoids generated from jejunum and colon from mice fed chronic alcohol.
Methods
Mice were fed a control or an alcohol diet. Intestinal permeability, liver steatosis/inflammation, and stool short chain fatty acids (SCFA) were measured. Jejunum and colon organoids and tissue were stained for stem cell, cell lineage, and apical junction markers with assessment of mRNA by PCR and RNA-seq. ChIP-PCR analysis was carried out for Notch1 using an antibody specific for acetylated histone 3.
Results
Alcohol-fed mice exhibited colonic (but not small intestinal) hyperpermeability, steatohepatitis and decreased butyrate/total SCFA ratio in stool. Stem cell, cell lineage, and apical junction marker staining in tissue or organoids from jejunum tissue were not impacted by alcohol. Only chromogranin A (Chga) was increased in jejunum organoids by qPCR. However, colon tissue and organoid staining exhibited an alcohol-induced significant decrease in cytokeratin 20+ (Krt20) absorptive lineage enterocytes, a decrease in occludin and E-cadherin apical junction proteins, an increase in Chga, and an increase in the Lgr5 stem cell marker. qPCR revealed an alcohol-induced decrease in colon organoid and tissue Notch1, Hes1, and Krt20 and increased Chga, supporting an alteration in stem cell fate due to decreased Notch1 expression. Colon tissue ChIP-PCR revealed alcohol feeding suppressed Notch1 mRNA expression (via deacetylation of histone H3) and decreased Notch1 tissue staining.
Conclusions
Data support a model for alcohol-induced colonic hyperpermeability via epigenetic effects on Notch1, and thus Hes1, suppression through a mechanism involving histone H3 deacetylation at the Notch1 locus. This decreased enterocyte and increased enteroendocrine cell colonic stem cell fate and decreased apical junctional proteins leading to hyperpermeability.
Keywords: Alcohol, organoids, intestinal permeability, Notch1, stem cells
INTRODUCTION
Chronic alcohol abuse results in end organ damage such as alcoholic liver disease (ALD).(Wang et al., 2010) ALD is currently the second leading cause of liver transplantation. Data from our laboratory and others have shown that alcohol-induced increased intestinal epithelial permeability (leaky gut) is an essential component of alcohol-induced organ damage and ALD.(Wang et al., 2010, Keshavarzian et al., 1999, Keshavarzian et al., 2009) Prior studies have shown that the effect of alcohol on the gut barrier could vary among different sites of the intestine.(Elamin et al., 2013) However, the mechanisms contributing to alcohol-induced intestinal hyperpermeability and its differential effects on large and small intestine have not been established. Intestinal epithelial cells bind to one another through apical junctional complex (AJC) proteins that include tight junction, adherens junction and desmosome junctional proteins(Laukoetter et al., 2006) to form the major component of the gastrointestinal barrier. The integrity of this barrier relies on normal and mature epithelial cells. Mature epithelial cells in the small and large intestine show rapid turnover rates, and die within 3–5 days after formation.(Barker, 2014) Therefore, maintenance of continuous epithelial cell differentiation and the presence of mature epithelial cells are critically important for the integrity of the barrier. Several studies have revealed the important role of intestinal crypt stem cells in the continual replenishment of gastrointestinal epithelium and barrier by supplying cells that differentiate into different cell lineages.(Barker, 2014)
Two key cell lineage pathways are derived from intestinal stem cells.(Barker, 2014, Clevers, 2013) The secretory cell lineage pathway results in Paneth, enteroendocrine, and goblet cells in the small intestine and enteroendocrine and goblet cells in the colon. The absorptive cell lineage pathway results in differentiation into enterocytes (about 80% of epithelial cells) in both the small intestine and colon. Stem cell Notch1 signaling is the critical determinant that regulates which cell fate pathway is followed: secretory or absorptive.(Barker, 2014, Noah and Shroyer, 2013). Therefore, effects on the stem cells, such as alcohol-induced suppression of Notch1 signaling, could potentially dysregulate epithelial cell differentiation, disrupting the mucosal barrier function as recently shown for decreased intestinal Notch1 signaling in diabetic mice.(Min et al., 2014)
Notch receptors are type one transmembrane receptors that are activated by ligands located on adjacent cells. Upon activation, the Notch receptor is first cleaved by an ADAM protease and subsequently by a gamma secretase to yield an NICD intracellular domain. The NICD translocates to the nucleus and pairs with CBF-1/Rbpj in humans/mice to activate transcription of Notch target genes. There are four Notch receptors (Notch 1–4) in mammals and five Notch ligands (Dll1,3,4 and Jagged 1–2).(Noah and Shroyer, 2013) Notch1 is by far the predominant Notch receptor in the mammalian intestine and is expressed by intestinal stem cells.(Fre et al., 2011) Notch signaling regulates cell differentiation, proliferation and especially cell fate and is conserved from drosophila to mammals. In the mammalian intestine, Notch1 is the principal regulator of intestinal stem cell fate with increased Notch1 signaling promoting absorptive lineage enterocytes and decreased Notch1 signaling resulting in secretory lineage enteroendocrine, goblet, or Paneth cells (small intestine).(Noah and Shroyer, 2013)
Our group has shown that alcohol effects on intestinal epithelial cells and barrier function persists beyond the 3–5 days which is the turnover rate of the mature epithelial layer in vivo.(Barker, 2014, Wood et al., 2013) Also, alcoholics exhibit intestinal hyperpermeability for weeks after cessation of drinking alcohol.(Bjarnason et al., 1984) Based on this information, we hypothesized that alcohol disrupts intestinal barrier function, at least in part, by influencing stem cell function. To test our hypothesis, we first used an in vivo model to assess gut leakiness and collect tissue from control and alcohol-fed mice. This tissue was used to generate ex vivo intestinal epithelial organoids to examine alcohol-induced changes to the stem cells of small (jejunum) and large (colon) intestinal epithelium. Organoids grow from intestinal stem cells into 3D mini-gut epithelial crypt structures that recapitulate many aspects of intestinal epithelial biology in vivo.(Leushacke and Barker, 2014, Sato and Clevers, 2013) In the organoid culture system, Lgr5+ intestinal stem cells (ISC) continue to differentiate to form the major cell types of the intestinal epithelium, providing an appropriate ex vivo model to study in vivo effects on intestinal stem cells.(Leushacke and Barker, 2014) In this study, our permeability data show that chronic alcohol feeding promoted colonic but not small intestinal hyperpermeability in mice. Using the organoid model we show that chronic alcohol feeding disrupts AJC proteins in the colonic (but not jejunum) organoids and this disruption was associated with changes in stem cell fate possibly via an epigenetic suppression of Notch1 expression via deacetylation of Histone H3 at the Notch1 locus.
MATERIALS AND METHODS
Mice and Chronic Alcohol Feeding Model
All animal studies in these experiments were approved by the Rush University Institutional Animal Care and Use Committee (IACUC). All experiments used 6–8 week old male C57BL/6J mice; hereafter BL/6 mice; from Jackson Labs (Bar Harbor, ME). Mice were maintained on a standard 12h light/12h dark (6am–6pm) schedule under SPF conditions in isolator cages with 3–5 mice/cage. Mice were fed a standard chow diet with either water or 15% alcohol v/v for a total of 8 weeks with a 2 week alcohol ramp period. The 2 week ramp period was as follows: 4d at 3% ethanol, 5d at 6% ethanol, then 5d 10% ethanol then 8 weeks at 15% ethanol. Consumption of alcohol was measured daily, with average consumption of 5ml of 15% alcohol/day equaling about 24g/kg/day ethanol consumption during the 8 week treatment period. After 10 weeks, mice were sacrificed by decapitation and trunk blood was collected. Liver was collected and fixed overnight in 10% buffered formalin for staining or flash frozen for MPO analysis. For organoid culture, intestines were immediately placed in cold PBS and processed as described below. Additional sections of jejunum and colon were placed in RNAlater and stored at −80°C for RNA analysis (qPCR or RNA-Seq).
Liver Steatosis and Inflammation
Slides cut from formalin fixed and paraffin embedded liver tissue were analyzed after H&E staining as we have previously described.(Summa et al., 2013) Briefly, slides were scored by a blinded liver pathologist (SS) using a 0–3 point scale. For liver steatosis: severity was scored as percent hepatocyte involvement (0 = <5%, 1 = 5–33%, 2 = 34–66%, 3 = >67%), corresponding to the fraction of lipid-containing hepatocytes. For inflammation: severity was scored 0–3 based on the number of inflammatory foci per 200x field (0 = no foci, 1 = 1 focus, 2 = 2–4 foci, 3 = >4 foci). These two scores were combined to yield an average composite score (n = 8 mice/group). As an alternative measure of liver inflammation we measured myeloperoxidase (MPO) in liver tissue by ELISA (Sigma, St. Louis, MO) (n = 4 mice/group).
Intestinal Permeability
In vivo intestinal permeability was assessed at the end of the 8 week alcohol treatment period as we have previously described.(Shaikh et al., 2015, Summa et al., 2013) Briefly, mice were fasted for eight hours prior to the test performed at time ZT0 (lights on). A 200ul solution was orally gavaged containing lactulose (3.2 mg), sucrose (0.45 mg), sucralose (0.45 mg) and mannitol (0.9 mg), after which 2ml of 0.9% saline was administered subcutaneously to promote urine output. Mice were placed in metabolic cages and urine was collected for five hours and the total volume recorded. Intestinal permeability was calculated by measuring urinary sugar concentration with gas chromatography and expressed as percent excretion of oral dose of sugar (n=4 mice/group).(Shaikh et al., 2015)
SCFA Measurement for Stool Samples
The levels of SCFA in stool were measured according to our modified protocol of Rose et al.(Rose et al., 2010) using gas chromatography and an internal standard as detailed in Supplementary Information (n = 7 mice/group).
Mouse Intestinal Organoid Preparation and Culture
Mouse intestinal organoid cultures were prepared from for both jejunum and colon using the methods described by Sato et al. (Jung et al., 2011, Sato et al., 2011, Sato et al., 2009) in which intact intestinal crypts are isolated and grown with crypt niche growth factors and Matrigel in 24-well plates as described in detail in Supplementary Materials and Methods and Supplementary Table 1.
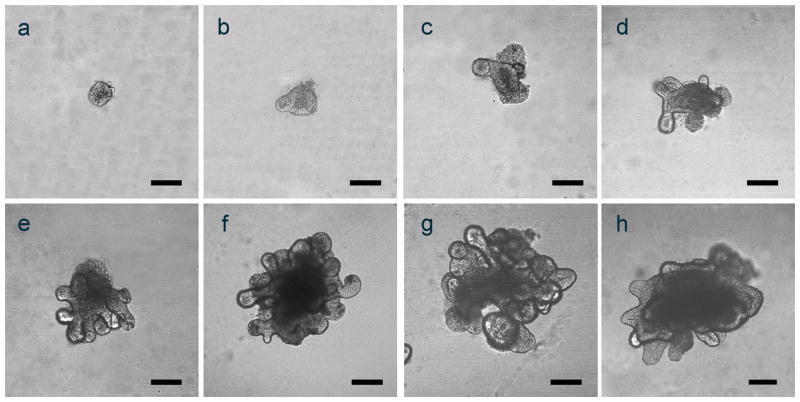
Crypts were isolated from small and large intestine of alcohol and control fed mice and used to grow organoids in culture.(Sato et al., 2011) In order to verify/document that our organoid cultures were similar to those described by others, we took images of jejunum and colon organoid cultures on days 0–7 for the jejunum (Figure 1a–g) and on day 21 for the colon (Figure 1h). Based on these results, jejunum organoids at day 7 and colon organoids at day 21 were used for all analyses. The time difference (i.e., day 7 vs. day 21) is due to the slower growth of the colon organoids compared to the growth of jejunum organoids to attain similar size.
Figure 1. Representative images of small intestinal (jejunum) and colonic organoids.
Confocal microscopic images were taken of representative small intestinal (days 0–7; a–g) and colonic organoids (day 21, h) growing in 24 well plates as described in Methods. Scale bars in all panels (a–h) represent 100μm.
Organoid and Tissue Immunofluorescent and Immunoperoxidase Staining
The organoid immunofluorescent staining was performed on 8 well chamber slides (Nunc, Rochester, NY) as fully described in the Supplementary Materials and Methods. Briefly, the organoids were fixed with warm 4% PFA on chamber slides for one hour at RT and then permeabilized with 1% Triton-X100. The samples were blocked with a solution of 3% goat serum/1% BSA/0.2% Triton X-100 for 1 hour. Primary antibody staining for the organoids was done overnight. Organoids were washed and incubated for 2 hours with the appropriate secondary antibody conjugated to Alexa Flour 488 (green). Organoids were then washed and double stained for DAPI and mounted using Flouromount aqueous mounting medium (Sigma Aldrich). Organoids were imaged using a Zeiss LSM 700 confocal microscope (Zeiss, Oberkochen, Germany). The mouse colon tissue immunofluorescent and immunoperoxidase staining was performed on paraffin-embedded formalin-fixed proximal colon tissue samples and is fully described in Supplementary Materials and Methods. Specific antibodies used for staining are detailed in Supplementary Table 2. All organoid and tissue staining data is from n = 4 mice/group with 7–10 images of stained organoids (or tissue) to determine relative expression of each marker and to pick the representative images. Staining was evaluated by two observers blinded to treatment group for all staining data.
qPCR mRNA Analysis
Cultured intestinal organoids and tissue samples from jejunum and colon were processed for RNA isolation according to the manufactures protocol (RNeasy, Qiagen, Hilden, Germany). For real time PCR, cDNA was prepared using the high capacity cDNA reverse transcription kit from the manufacturer (Applied Biosystems, Foster City, CA). The real time PCR was performed on an Applied Biosystems 7500 apparatus using primers (IDT, Coralville, IA) and Fast Sybr green (Applied Biosystems). The quantitative analysis was calculated from the ΔΔCt values normalized against the β actin values used as a housekeeping gene.(Forsyth et al., 2010) Specific primer sequences are detailed in Supplemental Table 3. All qPCR data are means ± SEM from n = 4 mice/group.
RNA Extraction and Library Preparation for RNA-Seq
This is fully described in Supplementary Materials and Methods.
RNA-seq QC and Quantification and RNA-Seq Expression Analysis
Complete RNA seq data counts and reads can be found at Gene Expression Omnibus (GEO): GSE99291. This is fully described in Supplementary Materials and Methods.
ChIP-PCR Analysis
Tissue samples from colon were processed for DNA chromatin shearing according to the manufactures protocol (truChIP™ Tissue Chromatin Shearing Kit, Covaris, Woburn MA, USA). Subsequently, immunoprecipitation and qPCR protocol was performed as described in Supplementary Materials and Methods. Specific PCR primer sequences are detailed in Supplementary Table 3. All ChIP-PCR data are means ± SEM from n = 4 mice/group.
Statistical Analysis
Statistical evaluations of data were performed using GraphPad Prism software (GraphPad Software, Inc, San Diego, CA). The intestinal permeability, MPO, SCFA, and qPCR data are represented as means ± SEM. A t-test was used for analysis. Data was considered statistically significant at p<0.05. The liver histology data was analyzed via non-parametric analysis with a significance value of p<0.05.
RESULTS
Alcohol Fed Mice Have Increased Colonic Permeability and Liver Pathology
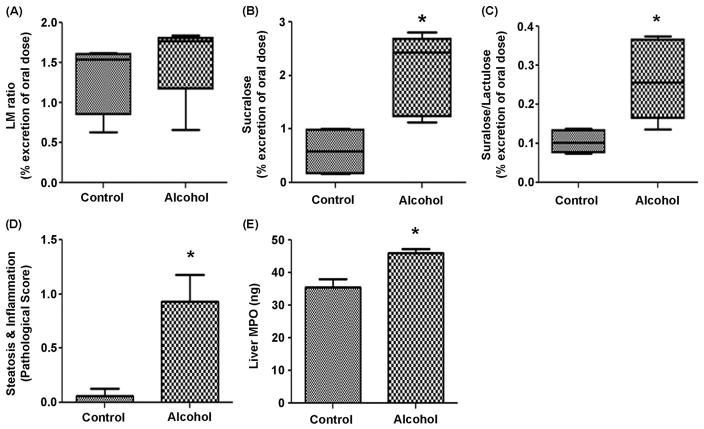
Following a two week gradual increase in alcohol concentration, 15% v/v alcohol in the drinking water was administered to BL/6 mice for eight weeks. One week before sacrifice, intestinal permeability was assessed following a sugar gavage and analysis of urinary sugar content as described in the Methods. The urinary lactulose (control 4.96±1.52 vs. alcohol 7.8±0.44) and urinary mannitol (control 3.754±0.8 vs. alcohol 4.409±0.32) were not significantly different in the control vs. alcohol-fed groups. The ratio of urinary lactulose to mannitol (L/M ratio), primarily a measure of small intestinal permeability,(Shaikh et al., 2015) was also not different between control (1.3±0.5) and alcohol-fed (1.5±0.5) mice (Figure 2A). However, a significant difference in colonic permeability was observed. Urinary sucralose (a measure of whole gut permeability)(Shaikh et al., 2015) was significantly elevated in alcohol-fed (2.1±0.5) vs. control-fed mice (0.8±0.5; p<0.05) (Figure 2B). The urinary sucralose/lactulose (S/L) ratio, another measure of colonic permeability,(Shaikh et al., 2015) was also significantly elevated in alcohol-fed (0.26±0.5) vs. control-fed mice (0.1±0.2; p<0.05) (Figure 2C). Increased intestinal permeability to microbial contents such as LPS is required for alcohol-induced steatohepatitis in rodents.(Wang et al., 2010, Keshavarzian et al., 2009) Indeed, alcohol-fed mice (0.97±0.3) exhibited significantly greater combined steatosis-inflammation scores compared to control-fed mice (0.1±0.1; p<0.05) (Figure 2D). We performed an ELISA for myeloperoxidase (MPO) content of liver tissue (MPO/ng tissue). Liver MPO content (marker of hepatic PMN infiltration) in alcohol-fed mice (47.2±2.3) was also significantly higher than in control-fed mice (36.2±4.2; p<0.05) (Figure 2E).
Figure 2. Alcohol-fed mice demonstrate intestinal hyperpermeability and liver pathology.
C57BL/6 mice were fed chow or chow + alcohol in water for a 2 week ramp period followed by 8 weeks of 15% alcohol. After week 7 of 15% alcohol, fasted mice were gavaged with a solution containing sucrose, mannitol, lactulose, and sucralose and placed in a metabolic chamber for 5h for urine collection. Urine was analyzed for sugar content by gas chromatography (Methods). Data are expressed as mean urinary % excretion of the original oral dose ± SEM (n = 4 mice/group). (A.) Lactulose/Mannitol (L/M) ratio; (B.) Sucralose; (C.) Sucralose/Lactulose ratio. p=0.11 for L/M ratio; *p<0.016 for sucralose; *p<0.032 for sucralose/lactulose ratio (t-tests). (D.) Liver samples were collected, formalin fixed, paraffin embedded, and slides stained with H&E for evaluation of combined steatosis and inflammation score (0–6) (n = 8 mice/group) (Methods). Data are means of this score ± SEM p*<.05 (Mann-Whitney). (E.) A portion of the liver was flash frozen and later analyzed for MPO by ELISA. Data are means ± SEM of ng MPO/gm liver tissue with *p<.05 (t test)(n = 4 mice/group).
Taken together, these data demonstrate alcohol-induced colonic hyperpermeability and liver steatohepatitis pathology in our mice model of alcoholic liver disease.
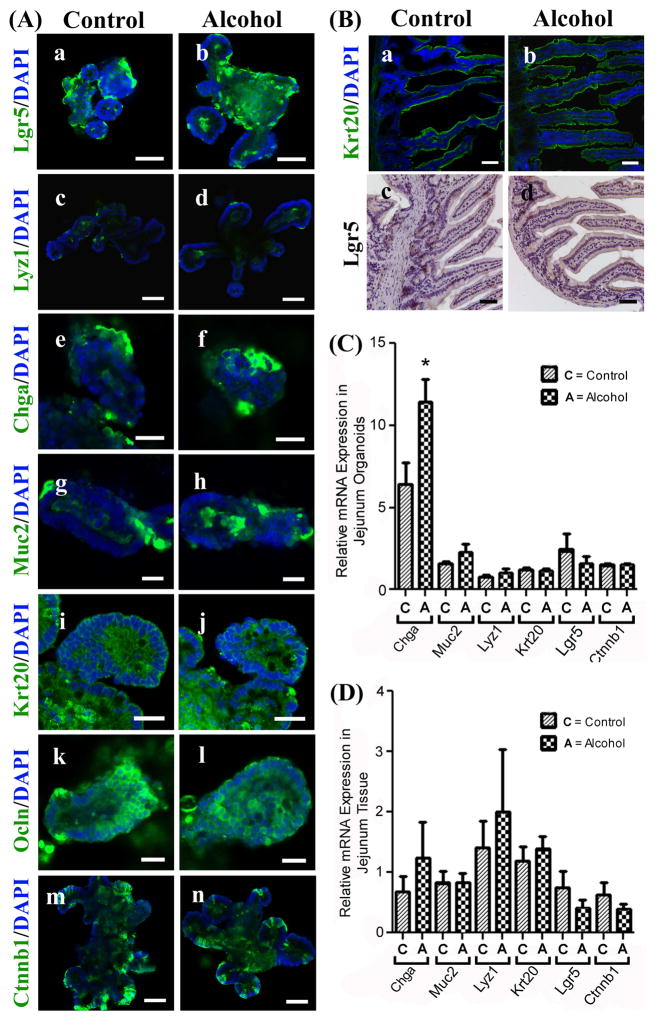
Jejunum organoids and tissue are largely unaffected by chronic alcohol feeding
Small intestine organoids recapitulate the development of the small intestinal cell types with leucine rich repeat containing G protein coupled receptor 5 (Lgr5+) crypt base columnar (CBC) stem cells differentiating to form the four major cell types in the small intestine. Cell fate markers include secretory cell lineage Paneth cells (lysozyme, Lyz1), enteroendocrine cells (chromogranin A, Chga), and goblet cells (mucin 2, Muc2) as well as absorptive cell lineage enterocytes (cytokeratin 20, Krt20). Organoids generated from jejunum tissue of control and alcohol-fed mice showed no differences in Lgr5 staining (stem cell marker) or any other cell fate markers including Chga (see PCR data below) (Figure 3Aa–j). Likewise, no staining differences were observed between alcohol-fed and control groups for the apical junction proteins occludin (tight junction protein) or E-cadherin (adherens junction protein; data not shown), or β-catenin (Ctnnb1) a key target transcription factor of the Wnt pathway (Figure 3Ak–n). Staining analysis of jejunum tissue revealed no alcohol-induced effects on staining for Krt20 or Lgr5 (Figure 3B).
Figure 3. Jejunum organoids and tissue are largely unaffected by chronic alcohol feeding in mice.
(A.) Organoids grown from small intestinal jejunum tissue from control (Control) or alcohol-fed (Alcohol) mice were stained for cell lineage and other key markers as described in Methods; (a/b) Stem cell marker: leucine rich repeat containing G protein coupled receptor 5 (Lgr5); (c/d) Paneth cells: lysozyme 1 (Lyz1); (e/f) Enteroendocrine cells: chromogranin A (Chga); (g/h) Goblet cells: mucin 2 (Muc2); (i/j) Enterocytes: cytokeratin 20 (Krt20); (k/l) Tight junction marker occludin (Ocln); or (m/n) beta-catenin (Ctnnb1). (B.) Whole intestinal tissue from jejunum of control (a/c) or alcohol fed (b/d) mice were stained for cytokeratin 20 (Krt20)(green) or Lgr5 (DAB staining, brown) as described in Methods. Scale bars in A (a, b, c, d, m, n) represent 50μm; A (e, f, g, h, i, j, k, l) 20μm. Scale bars in Panel B (a, b, c, d) represent 50μm. Jejunum organoids (C.) or Jejunum tissue (D.) were analyzed by qPCR as described in Methods for Chga, Muc2, Lyz1, Krt20, Lgr5, and Ctnnb1 expression.*p<.05 (t-test).
qPCR analysis of mRNA expression for lineage-specific marker genes as well as Lgr5 and beta catenin (i.e., Chga, Muc2, Lyz1, Krt20, Lgr5, Ctnnb1) revealed that in jejunum organoids there were no significant alcohol-induced effects with the exception of Chga which was significantly increased (p<0.05) in alcohol-fed mice (Figure 3C). qPCR analysis of jejunum tissue showed that there were no significant effects of alcohol on cell lineage marker mRNA expression (Figure 3D).
The results generated in jejunum whole tissue generally match the data for organoids prepared from the jejunum tissue, and are overall consistent with the observed lack of alcohol effect on small intestinal stem cells and small intestinal permeability in our model.
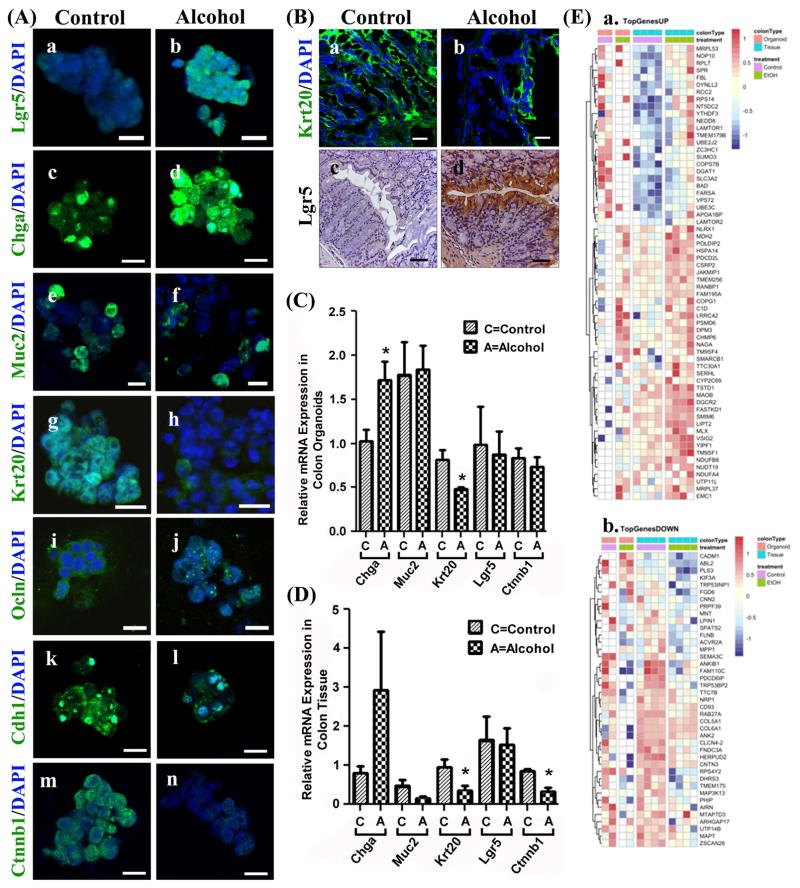
Colonic organoids and tissue demonstrate alcohol-induced effects on stem cells, cell lineage markers and apical junctional proteins
In colonic organoids Lgr5+ stem cells differentiate into three major cell types including secretory cell lineage enteroendocrine cells (ChgA+), goblet cells (Muc2+) and absorptive lineage enterocytes (Krt20+). Staining analysis revealed striking alcohol-induced changes in the colon organoids stem cell and cell fate markers including an increase in Lgr5 (stem cell marker), increase in Chga (secretory lineage, enteroendocrine cell), no change in Muc2 (goblet cell), and markedly diminished staining for Krt20 (absorptive lineage, enterocytes). We also found decreased levels of the apical junction proteins Ocln (occludin tight junction protein), and Cdh1 (adherens junction protein E-cadherin), as well as decreased Ctnnb1 (β-catenin) (Figure 4Aa–n). Staining of colon tissue revealed markedly lower Krt20 and elevated Lgr5 staining (Figure 4B) in alcohol-fed mice compared to control mice. Colon tissue was not stained for apical junction markers Ocln and Cdh1 or for Chga and Ctnnb.
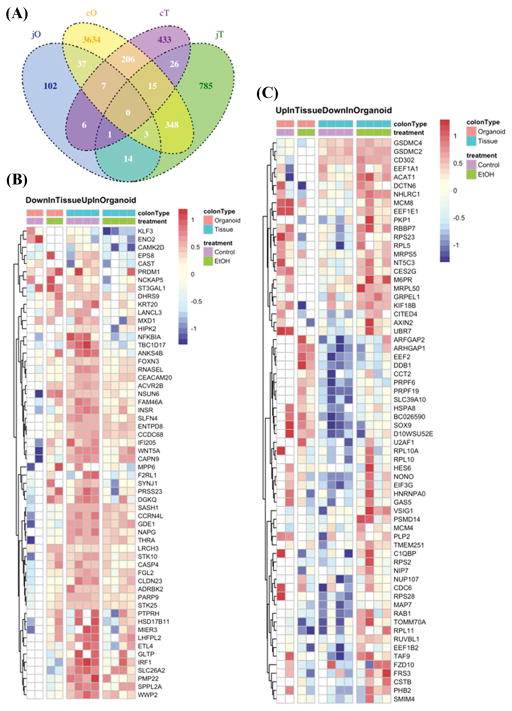
Figure 4. Colonic organoids and tissue from alcohol fed mice exhibit differences in cell lineage markers, apical junction proteins, stem cell markers and RNA-seq.
(A.) Organoids grown from colonic intestinal tissue from control (Control) or alcohol-fed (Alcohol) mice were stained for cell lineage and other key markers (Methods): (a/b) Lgr5; (c/d) Enteroendocrine cells: chromogranin A (Chga); (e/f) Goblet cells: mucin 2 (Muc2); (g/h) Colonocytes/enterocytes: cytokeratin 20 (Krt20); (i/j) tight junction protein occludin (Ocln); (k/l) E-cadherin (Cdh1); and (m/n) β-catenin (Ctnnb1). (B.) Whole intestinal tissue from colons of Control (left) or Alcohol fed (right) mice were stained for (a/b) cytokeratin 20 (Krt20)(green) or (c/d) Lgr5 (DAB staining, brown) as described in Methods. Tissue from colon organoids (C.) or colon tissue (D.) were analyzed by qPCR (Methods) for gene expression including Chga, Muc2, Krt20, Lgr5, and Ctnnb1(*p<.05)(t-test) (E.) RNA-seq analysis data from colon tissue and organoids from control and alcohol fed mice was used to construct heat maps to depict differential gene expression with alcohol feeding (Methods). (a.) TopGenesUP (43 genes) and (b.)TopGenesDOWN (33 genes): Heat maps of genes from colon samples that are common between organoids and colon tissue that are upregulated (TopgenesUP) or downregulated (TopGenesDOWN) in alcohol fed mice. LogCPM data was z-scored transformed and plotted using the Bioconductor pheatmap.(Kolde, 2015) Genes were clustered by complete linkage method using Euclidean distance. The analysis was performed for the contrast of alcohol fed vs. control, and so the red color indicates increased expression in alcohol fed mice, while blue indicates increased expression in control mice. The first colored row at the top of the plot indicate the type of colon sample: orange is organoid cells, blue is tissue. The second colored band indicates the treatment: green are alcohol fed samples while purple are controls. Scale bars in A (a–n) represent 10μm and in D (a, b) represent 20μm and D (c, d) represent 50μm.
qPCR analysis revealed that colon organoids generated from alcohol-fed mice had significantly higher Chga and lower Krt20 expression (both p<0.05) compared to control-fed mice organoids, matching the results obtained from staining (Figure 4C). However, there was no increase in colon organoid Lgr5 or decrease in Ctnnb mRNA which did not match the staining data. Colon tissue assessed by qPCR showed alcohol feeding increased Chga and decreased Krt20 and Ctnnb mRNA expression in colon tissue (all p<0.05) but no mRNA increase in Lgr5 (Fig. 4D).
Thus, the Krt20 (decreased) and Lgr5 (increased) results for colon tissue staining match the staining data for these markers in colon organoids. However in both organoids and colon tissue there was no increase in Lgr5 mRNA. Taken together, the colon organoid and colon tissue staining, and PCR results support decreased colonic absorptive pathway (Krt20+ enterocytes) differentiation and increased differentiation of Chga+ secretory enteroendocrine cells.
Next, we used RNA-seq analysis to examine global gene expression similarities and differences between colon tissue and colon organoids from control and alcohol-fed mice. The data were analyzed for differential expression with a nominal p<0.05. Comparison between colon organoids (cO) and colon tissue (cT) revealed that 206 genes were commonly affected by alcohol (Figure 5A). Of these genes, 43 were significantly increased due to alcohol feeding (Figure 4Ea; TopGenesUP) including genes involved in oxidative phosphorylation (Ndufb8n, Dufa4), cellular response to stress and mTor1-mediated signaling (Lamtor1, Lamtor22, Hspa14, Chmp6, Dtnll2) as well as Tnfa/Nfkb signaling (Fbi, Psmd6, Smarcd1) and TCA Cycle gene (Mds2). In contrast, a total of 33 genes were significantly decreased due to alcohol feeding including genes involved in membrane biosynthesis and collagen (Cal5a1, Cal6a1) assembly, and Mapk (Flnb, Mapt, Map3k13) signaling (Figure 4Eb; TopGenesDown). The top protein hub that was enriched due to alcohol feeding was Gabarapl1:(Rps14, Prpf39, Pdcd6ip, Mdh2, Ndufa4, Rcc2, Pls3, Flnb, Ank2), which is involved in the formation of autophagosomal vacuoles. Additional RNA-seq Venn diagram and heat map analysis is described in Figure 5. The Venn diagram (Figure 5A) shows far fewer alcohol-induced changes were observed in the jejunum with only 14 genes being commonly influenced by alcohol in jejunum organoids (jO) and jejunum tissue (jT). The Venn diagram also depicts genes differentially expressed due to alcohol feeding (control vs. alcohol) that were unique or shared between the intestine sites (jejunum vs. colon) (Figure 5A). Significantly, alcohol-induced effects in the colon were much more numerous than those in the small intestine, matching the results obtained from the in vivo permeability testing and organoid staining and qPCR. Specifically, alcohol appears to have had the greatest effect by far on gene expression in colonic organoids (cO) with cO having 3634 uniquely differentially alcohol expressed genes compared to jO (102 genes), cT (433 genes) and jT (785 genes) suggesting an alcohol-specific change especially in colonic stem cells (organoids).
Figure 5. Venn diagram and additional heat maps for RNA-seq data.
(A.) A Venn diagram of RNA seq data for genes differentially expressed in each designated tissue with alcohol feeding (vs. control) was constructed as described in Methods. Groups are: jO (jejunum organoids); cO (colonic organoids); cT (colonic tissue); jT (jejunum tissue). Heat maps were also constructed from RNA seq data as described in Methods using the 206 alcohol feeding related genes found to overlap between cO and cT in the Venn diagram (5A) but without the genes already shown in heat maps Figs. 4Ea (43 genes) and 4Eb (33 genes) and thus depict 130 genes: (B.) Down in tissue (blue) Up in organoids (red); and (C.) Up in tissue (red) Down in organoids (blue).
Notch1 epigenetic suppression as a mechanism to explain alcohol-induced changes in the colon
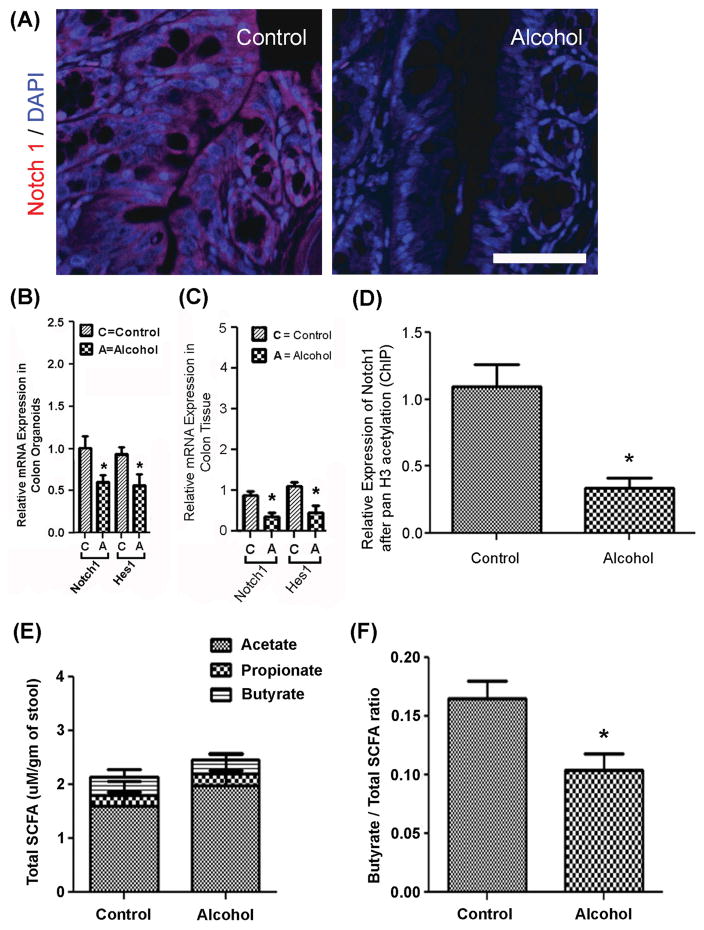
To investigate the mechanisms contributing to changes in organoid stem cell fate, colon tissue from control and alcohol-fed mice were assessed for Notch1. Notch1 receptor staining was markedly decreased in colon tissue from alcohol-fed mice compared to control-fed mice (Figure 6A). Likewise, qPCR expression of both Notch1 and its downstream effector Hes1 were significantly decreased in both colon organoids (Figure 6B) and in colon tissue obtained from alcohol-fed mice (Figure 6C) (p<0.05 for all). To investigate a possible epigenetic mechanism for suppression of Notch1 expression by chronic alcohol feeding, a ChIP-PCR analysis for Notch1 expression was performed using an antibody to pan Histone 3 acetylation (panH3ac) for ChIP followed by qPCR for Notch1. Analysis revealed that chronic alcohol feeding significantly suppressed Notch1 mRNA expression with a concurrent deacetylation of H3ac at the Notch1 locus (p<0.01, Figure 6D). These data support a role for alcohol-induced H3 deacetylation at the Notch1 locus resulting in significant suppression of Notch1 mRNA and protein expression.
Figure 6. Analysis for colon Notch1 tissue staining, Notch1 and Hes1 qPCR, ChIP-PCR for H3ac-Notch1 and stool SCFA.
(A.) Colon tissue from Control (left) and Alcohol fed (right) mice was stained with antibody for Notch1 (red) and DAPI (blue) as described in Methods. Scale bar in Panel A represents 50μm and applies to both control and alcohol images. (B.) Colon organoids and (C.) colon tissue from control and alcohol fed mice were analyzed for expression of Notch1 and Hes1 by qPCR as described in Methods.*p<.05 (t-test) (n = 4 mice/group). (D.) ChIP-PCR for Notch1 was carried out using whole colon tissue from control and alcohol fed mice with ChIP antibody to pan acetylated Histone 3 (panH3ac) followed by qPCR for Notch1 as described in Methods.* p<.01(t-test) (n = 4 mice/group). (E.) Stool samples from control and alcohol mice described in Fig. 2 were analyzed at the end of the study for short chain fatty acids (SCFA) using gas chromatography as described in Methods. SCFA quantitated included acetate, propionate and butyrate. Units shown are uM SCFA/gm stool means ± SEM. (n = 7 mice/group). (F.) Values were determined from the results of the SCFA measures for total SCFA and total butyrate and the ratio of butyrate/total SCFA is shown as means ± SEM. *p<.01 (t-test) (n = 7 mice/group).
Potential role for SCFA and butyrate
Alcohol-induced Notch1 suppression through deacetylation of H3ac in the colon could be due to the alteration in the colonic environment secondary to alcohol intake. Our group and others showed that chronic alcohol consumption causes intestinal dysbiosis (Mutlu et al., 2012, Mutlu et al., 2009), specifically an alcohol-induced decrease in the bacteria (e.g., Lachnospiracea) that produce the short-chain fatty acids (SCFAs) acetate, propionate and butyrate.(Engen et al., 2015) SCFAs are produced due to fermentation of dietary complex carbohydrates by the intestinal microbiota and these SCFA, especially butyrate, play an important role in the maintenance of the intestinal barrier.(Hamer et al., 2008, Ploger et al., 2012) In addition, butyrate is a potent histone deacetylase (HDAC) inhibitor (Boffa et al., 1978) and contributes to epigenetic regulation of gene expression. Thus, alcohol-induced changes in the intestinal microbiota and resulting changes in butyrate/SCFA production may be one mechanism by which alcohol feeding influences colonic crypt stem cell fate. While the total acetate/propionate/butyrate amounts for control- (0.4/0.2/1.2 uM/gm) and alcohol-fed mice (0.3/0.3/1.8 uM/gm) were similar (Figure 6E), the ratio of butyrate to total SCFA was significantly decreased in alcohol-fed (0.10±0.02) vs. control-fed (0.17 ± .03) mice (p<0.01) (Figure 6F).(Weaver et al., 1988) Together these data suggest that decreased relative butyrate production, could be contributing to the observed changes in stem cell fate as a result of decreased H3ac acetylation at the Notch1 locus.
DISCUSSION
The current study shows that chronic alcohol consumption in mice increased intestinal permeability and steatohepatitis demonstrating the injurious effects of alcohol in our model. Colonic hyperpermeability, rather than small intestinal hyperpermeability, primarily accounted for the alcohol-induced gut leakiness. Specifically, the L/M ratio (a measure of small intestinal permeability) was not altered by chronic alcohol feeding whereas permeability to sucralose and the sucralose/lactulose ratio (both measures of whole gut permeability) were increased (Figure 2). Taken together these data support the colon as the primary site of alcohol-induced intestinal hyperpermeability.(Arrieta et al., 2006, Summa et al., 2013, Shaikh et al., 2015)
Alcohol-induced decreased Notch1 expression is a viable mechanism that may be contributing to long-term changes in intestinal barrier integrity. There are numerous studies demonstrating that Notch1 regulates the intestinal barrier.(Turgeon et al., 2013, Mathern et al., 2014, Min et al., 2014, Obata et al., 2012, Dahan et al., 2011) siRNA knockdown of Notch1 in vitro (i.e., Caco-2 cells) is associated with impaired intestinal cell monolayer integrity and decreased expression of the tight junction protein occludin.(Dahan et al., 2011) Downregulation of Notch1 in vivo is associated with gut leakiness and alterations in tight junction protein expression and colon inflammation.(Mathern et al., 2014, Dahan et al., 2011, Min et al., 2014) Notch1 cleavage product NICD pairs with the transcription factor Rbpj in mice to activate canonical Notch1 target genes and in vivo knockout of Rbpj in mice causes intestinal hyperpermeability and colitis.(Obata et al., 2012) A recent study in diabetic mice showed that decreased NICD/Hes1 expression resulted in intestinal hyperpermeability.(Min et al., 2014) Significantly, a rectal enema containing siRNA to Notch1 administered to mice reduces colonic stem cell Notch1 expression and results in colonic hyperpermeability.(Mathern et al., 2014) Taken together, these data demonstrate that Notch1 regulates intestinal/colonic barrier integrity and supports alcohol-induced suppression of Notch1 as a potential mechanism contributing to alcohol-induced colonic hyperpermeability.
To explore the molecular mechanism by which Notch1 impacts alcohol-induced intestinal barrier integrity we investigated the impact of Notch1 signaling on intestinal stem cell fate. While little is known about alcohol-induced effects on intestinal stem cells, the impact of alcohol on neural stem cells has been extensively studied. Alcohol affects differentiation of neural stem cells into neurons, glial cells, and oligodendrocytes and Notch1 plays an essential role in cell fate determination in both neural and intestinal stem cells. In rat fetal neural ex vivo stem cell neurospheres (analogous to intestinal organoids), alcohol promotes the formation of neurons at the expense of glial cells.(Vemuri and Chetty, 2005) Similarly, exposure of ex vivo human fetal neural stem cells to alcohol promotes neurons at the expense of other cell types.(Vangipuram and Lyman, 2010) Importantly, decreased Notch1 signaling in the brain favors the development of neurons whereas in the intestine decreased Notch1 signaling favors the development of cells of secretory lineage. Thus alcohol effects are similar for brain and intestinal stem cell fate and both are characteristic of Notch1 suppression.
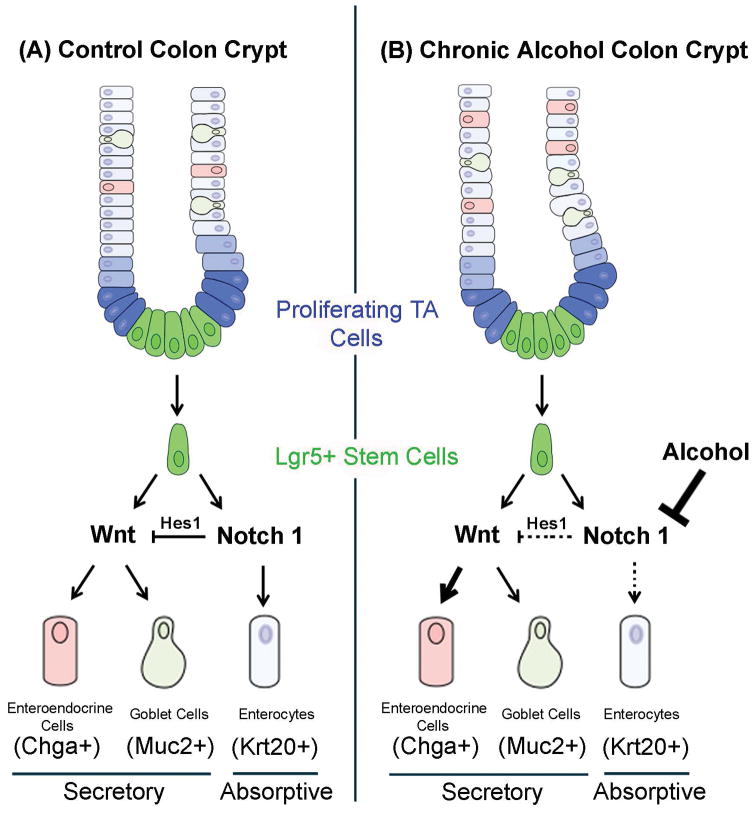
High levels of Notch1 signaling in intestinal stem cells promote the development of cells of absorptive cell lineage whereas low levels of Notch1 signaling promote secretory lineage cells.(Barker, 2014, Noah and Shroyer, 2013) Specifically, Notch1 signaling activates the transcription factor Hes1 to promote the absorptive cell development and repression of Atoh1 (Math1) the transcription factor that is required for secretory cell development.(Noah and Shroyer, 2013, van der Flier and Clevers, 2009, VanDussen et al., 2012). In the current study, Notch1 was decreased by alcohol in both colon tissue as well as in colon organoids with a concurrent decrease in Krt20 and increase in Chga supporting a decrease in absorptive cell lineage and an increase in secretory cell fate by alcohol. The alcohol-induced reduction in Notch1 and Hes1 was observed in colon organoids that were grown in vitro in the absence of alcohol for over three weeks, suggesting that alcohol-induced effects persist beyond the period of alcohol exposure and may be the consequence of epigenetic changes affecting stem cell fate. These data are summarized in Figure 7.
Figure 7. Model for alcohol effects on colonic stem cell fate determination.
CBC Lgr5+ stem cells are in green while transit amplifying (TA) cells are in blue. The left hand side of the panel shows a simplified version of the Control (normal) colonic crypt and how Notch1-Hes1 signaling regulates cell fate determination. High Notch1 levels stimulate Hes1 expression which promotes Krt20+ enterocytes and inhibits Wnt/Atoh1 stimulation of secretory lineage cells. In the right hand Chronic Alcohol panel, the presence of alcohol results in stable decreased Notch1 expression (heavy inhibitor bar) and thus lower Hes1 expression resulting in less absorptive lineage enterocyte signaling (dotted line) and fewer Krt20+ enterocytes. This lower Hes1 expression also results in less suppression of Wnt/Atoh1 (shown as dotted line inhibitor bar) and thus increased secretory lineage Wnt/Atoh1 signaling (heavy arrow) resulting in increased Chga+ enteroendocrine cells.
The next question was how alcohol suppresses Notch1 signaling in the colon intestinal stem cells. In the current study, stable changes in gene expression/cell fate were observed in tissue from alcohol-fed mice and in organoid tissue that had not been exposed to alcohol in at least 21 days (time required to grow colon organoids), suggesting an epigenetic mechanism may contribute to changes in intestinal stem cells. Alcohol has a number of epigenetic effects on a variety of tissues (Basavarajappa and Subbanna, 2016, Shukla and Lim, 2013) including increased histone deacetylase (HDAC) activity resulting in silencing of gene expression.(Palmisano and Pandey, 2017, Kirpich et al., 2013) For example, alcohol craving is associated with brain H3ac deacetylation with a concurrent increase in brain HDAC expression (Palmisano and Pandey, 2017) that can be reversed with administration of oral sodium butyrate.(Simon-O’Brien et al., 2014, Legastelois et al., 2013) Indeed, in the current study H3ac at the Notch1 locus was deacetylated resulting in Notch1 suppression in alcohol-fed mice as shown by ChiP-PCR (Figure 6D). In fact, the Clever’s lab recommends the addition of the HDAC inhibitor valproic acid as a way to activate Notch1 signaling in colonic organoids (Yin et al., 2014) with other studies also demonstrating that HDACi activate Notch1 signaling.(Stockhausen et al., 2005, Greenblatt et al., 2008) A recent study demonstrated that knocking out HDAC1 and HDAC2 (thereby promoting acetylation) in colonic stem cells upregulates Notch1 activation and increases absorptive lineage markers (Turgeon et al., 2013) exactly the opposite pattern of cell fate of what was observed in alcohol-fed mice. As would be expected, the pattern of increased enterocytes and decreased secretory cells is also observed in mice overexpressing colonic activated Notch1 NICD.(Stanger et al., 2005) Thus, our data are not the first to show regulation of colonic Notch1 by histone acetylation but our data are the first to show this effect due to alcohol feeding. Future studies will focus on identifying specific HDAC in colonic stem cells that are increased with chronic alcohol. Thus, alcohol feeding may promote suppression of Notch1 through deacetylation of H3ac.
Short-chain fatty acids (SCFAs) are produced as a byproduct of bacterial fermentation by specific types of intestinal microbiota and have multiple beneficial effects on the host. The SCFA butyrate is a HDAC inhibitor (HDACi) and impacts H3 acetylation.(Vidali et al., 1978) Chronic alcohol consumption causes intestinal dysbiosis (Mutlu et al., 2012, Mutlu et al., 2009) and SCFA/butyrate-producing bacteria are decreased in alcoholics.(Engen et al., 2015) Oral supplementation with tributyrin, a butyrate precursor, was recently shown to prevent alcohol-induced intestinal permeability and damage.(Cresci et al., 2014) In the current study, alcohol feeding reduced the stool butyrate ratio to total SCFA content and this may contribute to H3ac deacetylation at the Notch1 locus along with alcohol stimulation of HDAC as the potential mechanism for colonic Notch1 suppression. Such low butyrate/total SCFA ratios are also associated with increased polyps in colon cancer subjects compared to healthy subjects.(Weaver et al., 1988)
qPCR analysis revealed that colon organoids generated from alcohol-fed mice had significantly higher Chga and lower Krt20 expression compared to control-fed mice, matching the results obtained from staining. However, qPCR results for Lgr5 and Ctnnb did not match the staining data. Our data show that in the colon no significant change was seen in Lgr5 expression by qPCR but Lgr5 staining was elevated in colon tissue and organoids. The reason for the discrepancy between staining and qPCR data are not clear. While some studies show that intestinal Notch1 repression results in loss of Lgr5, these studies looked at the short term effects of chemical Notch1 inhibition.(VanDussen et al., 2012) A recent study using chronic Notch1 blocking antibodies observed a short term loss of small intestinal Lgr5 staining but over several days Lgr5 staining rebounded to become greater than initial values.(Tian et al., 2015) In another study repression of intestinal Notch1 was associated with increased Lgr5 protein without change in Lgr5 mRNA. The authors attributed this effect to post-translational regulation of Lgr5.(Srinivasan et al., 2016) This may also be true in our model. The mechanism for loss of β-catenin staining in colon organoids without loss of mRNA expression is also not clear but β-catenin has also been linked to Notch1 expression and directly interacts with Notch1 NICD.(Andersen et al., 2012, Gopalakrishnan et al., 2014) Thus, further studies are required in colonic tissue and organoids to investigate the effects of alcohol feeding on Lgr5 and β-catenin.
Taken together, data in our study show that chronic alcohol consumption causes colonic intestinal hyperpermeability and this is associated with changes in colon (but not small intestinal) organoid markers of stem cell fate associated with H3 deacetylation and suppression of stem cell Notch1 and downstream Hes1 expression and a decrease in apical junctional proteins. Notch1 expression in the colon may act as a sensitive rheostat responding to intestinal dysbiosis associated with alcohol consumption (or other stimuli) and may explain why the subset of alcoholics with the worse intestinal dysbiosis exhibit the greatest gut leakiness and ALD.(Mutlu et al., 2012) Future studies will investigate the mechanism for H3 deacetylation at the Notch1 locus including identification of the specific HDAC involved and how dysbiosis due to other stimuli may affect intestinal stem cell Notch1 expression and thereby regulate gut leakiness via this Notch1 epigenetic mechanism.
Supplementary Material
Acknowledgments
Grant support: NIH/NIAAA grants: AA020216 (A.K., C.B.F., R.M.V.) and AA023417 (A.K.); grants from Mr.& Mrs. Larry Field (A.K.) and Mr.& Mrs. Glass (A.K.).
Footnotes
Disclosures. The authors declare no conflicts of interest.
Author contributions: C.B.F., M.S., F.B., G.S., R.M.V. and A.K. designed research; M.S., H.D., B.T. and S.S. performed research; C.B.F., M.S., R.M.V., H.D., P.W., B.T. and S.S. analyzed data; C.B.F., M.S., F.B., G.S., R.M.V., P.W. and A.K. wrote the paper.
References
- ANDERSEN P, UOSAKI H, SHENJE LT, KWON C. Non-canonical Notch signaling: emerging role and mechanism. Trends Cell Biol. 2012;22:257–65. doi: 10.1016/j.tcb.2012.02.003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- ARRIETA MC, BISTRITZ L, MEDDINGS JB. Alterations in intestinal permeability. Gut. 2006;55:1512–20. doi: 10.1136/gut.2005.085373. [DOI] [PMC free article] [PubMed] [Google Scholar]
- BARKER N. Adult intestinal stem cells: critical drivers of epithelial homeostasis and regeneration. Nat Rev Mol Cell Biol. 2014;15:19–33. doi: 10.1038/nrm3721. [DOI] [PubMed] [Google Scholar]
- BASAVARAJAPPA BS, SUBBANNA S. Epigenetic Mechanisms in Developmental Alcohol-Induced Neurobehavioral Deficits. Brain Sci. 2016:6. doi: 10.3390/brainsci6020012. [DOI] [PMC free article] [PubMed] [Google Scholar]
- BJARNASON I, PETERS TJ, WISE RJ. The leaky gut of alcoholism: possible route of entry for toxic compounds. Lancet. 1984;1:179–82. doi: 10.1016/s0140-6736(84)92109-3. [DOI] [PubMed] [Google Scholar]
- BOFFA LC, VIDALI G, MANN RS, ALLFREY VG. Suppression of histone deacetylation in vivo and in vitro by sodium butyrate. J Biol Chem. 1978;253:3364–6. [PubMed] [Google Scholar]
- CLEVERS H. The intestinal crypt, a prototype stem cell compartment. Cell. 2013;154:274–84. doi: 10.1016/j.cell.2013.07.004. [DOI] [PubMed] [Google Scholar]
- CRESCI GA, BUSH K, NAGY LE. Tributyrin supplementation protects mice from acute ethanol-induced gut injury. Alcohol Clin Exp Res. 2014;38:1489–501. doi: 10.1111/acer.12428. [DOI] [PMC free article] [PubMed] [Google Scholar]
- DAHAN S, RABINOWITZ KM, MARTIN AP, BERIN MC, UNKELESS JC, MAYER L. Notch-1 signaling regulates intestinal epithelial barrier function, through interaction with CD4+ T cells, in mice and humans. Gastroenterology. 2011;140:550–9. doi: 10.1053/j.gastro.2010.10.057. [DOI] [PMC free article] [PubMed] [Google Scholar]
- ELAMIN EE, MASCLEE AA, DEKKER J, JONKERS DM. Ethanol metabolism and its effects on the intestinal epithelial barrier. Nutr Rev. 2013;71:483–99. doi: 10.1111/nure.12027. [DOI] [PubMed] [Google Scholar]
- ENGEN PA, GREEN SJ, VOIGT RM, FORSYTH CB, KESHAVARZIAN A. The Gastrointestinal Microbiome: Alcohol Effects on the Composition of Intestinal Microbiota. Alcohol Res. 2015;37:223–36. [PMC free article] [PubMed] [Google Scholar]
- FORSYTH CB, TANG Y, SHAIKH M, ZHANG L, KESHAVARZIAN A. Alcohol Stimulates Activation of Snail, Epidermal Growth Factor Receptor Signaling, and Biomarkers of Epithelial-Mesenchymal Transition in Colon and Breast Cancer Cells. Alcohol Clin Exp Res. 2010;34:19–31. doi: 10.1111/j.1530-0277.2009.01061.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- FRE S, HANNEZO E, SALE S, HUYGHE M, LAFKAS D, KISSEL H, LOUVI A, GREVE J, LOUVARD D, ARTAVANIS-TSAKONAS S. Notch lineages and activity in intestinal stem cells determined by a new set of knock-in mice. PLoS One. 2011;6:e25785. doi: 10.1371/journal.pone.0025785. [DOI] [PMC free article] [PubMed] [Google Scholar]
- GOPALAKRISHNAN N, SARAVANAKUMAR M, MADANKUMAR P, THIYAGU M, DEVARAJ H. Colocalization of beta-catenin with Notch intracellular domain in colon cancer: a possible role of Notch1 signaling in activation of CyclinD1-mediated cell proliferation. Mol Cell Biochem. 2014;396:281–93. doi: 10.1007/s11010-014-2163-7. [DOI] [PubMed] [Google Scholar]
- GREENBLATT DY, CAYO MA, ADLER JT, NING L, HAYMART MR, KUNNIMALAIYAAN M, CHEN H. Valproic acid activates Notch1 signaling and induces apoptosis in medullary thyroid cancer cells. Ann Surg. 2008;247:1036–40. doi: 10.1097/SLA.0b013e3181758d0e. [DOI] [PMC free article] [PubMed] [Google Scholar]
- HAMER HM, JONKERS D, VENEMA K, VANHOUTVIN S, TROOST FJ, BRUMMER RJ. Review article: the role of butyrate on colonic function. Aliment Pharmacol Ther. 2008;27:104–19. doi: 10.1111/j.1365-2036.2007.03562.x. [DOI] [PubMed] [Google Scholar]
- JUNG P, SATO T, MERLOS-SUAREZ A, BARRIGA FM, IGLESIAS M, ROSSELL D, AUER H, GALLARDO M, BLASCO MA, SANCHO E, CLEVERS H, BATLLE E. Isolation and in vitro expansion of human colonic stem cells. Nat Med. 2011;17:1225–7. doi: 10.1038/nm.2470. [DOI] [PubMed] [Google Scholar]
- KESHAVARZIAN A, FARHADI A, FORSYTH CB, RANGAN J, JAKATE S, SHAIKH M, BANAN A, FIELDS JZ. Evidence that chronic alcohol exposure promotes intestinal oxidative stress, intestinal hyperpermeability and endotoxemia prior to development of alcoholic steatohepatitis in rats. J Hepatol. 2009;50:538–47. doi: 10.1016/j.jhep.2008.10.028. [DOI] [PMC free article] [PubMed] [Google Scholar]
- KESHAVARZIAN A, HOLMES EW, PATEL M, IBER F, FIELDS JZ, PETHKAR S. Leaky gut in alcoholic cirrhosis: a possible mechanism for alcohol-induced liver damage. Am J Gastroenterol. 1999;94:200–7. doi: 10.1111/j.1572-0241.1999.00797.x. [DOI] [PubMed] [Google Scholar]
- KIRPICH I, ZHANG J, GOBEJISHVILI L, KHAREBAVA G, BARKER D, GHARE S, JOSHI-BARVE S, MCCLAIN CJ, BARVE S. Binge ethanol-induced HDAC3 down-regulates Cpt1alpha expression leading to hepatic steatosis and injury. Alcohol Clin Exp Res. 2013;37:1920–9. doi: 10.1111/acer.12172. [DOI] [PMC free article] [PubMed] [Google Scholar]
- KOLDE R. R package version 1.0.8. 2015. pheatmap: Pretty Heatmaps. [Google Scholar]
- LAUKOETTER MG, BRUEWER M, NUSRAT A. Regulation of the intestinal epithelial barrier by the apical junctional complex. Curr Opin Gastroenterol. 2006;22:85–9. doi: 10.1097/01.mog.0000203864.48255.4f. [DOI] [PubMed] [Google Scholar]
- LEGASTELOIS R, BOTIA B, NAASSILA M. Blockade of ethanol-induced behavioral sensitization by sodium butyrate: descriptive analysis of gene regulations in the striatum. Alcohol Clin Exp Res. 2013;37:1143–53. doi: 10.1111/acer.12088. [DOI] [PubMed] [Google Scholar]
- LEUSHACKE M, BARKER N. Ex vivo culture of the intestinal epithelium: strategies and applications. Gut. 2014;63:1345–54. doi: 10.1136/gutjnl-2014-307204. [DOI] [PubMed] [Google Scholar]
- MATHERN DR, LAITMAN LE, HOVHANNISYAN Z, DUNKIN D, FARSIO S, MALIK TJ, RODA G, CHITRE A, IUGA AC, YERETSSIAN G, BERIN MC, DAHAN S. Mouse and human Notch-1 regulate mucosal immune responses. Mucosal Immunol. 2014;7:995–1005. doi: 10.1038/mi.2013.118. [DOI] [PubMed] [Google Scholar]
- MIN XH, YU T, QING Q, YUAN YH, ZHONG W, CHEN GC, ZHAO LN, DENG N, ZHANG LF, CHEN QK. Abnormal differentiation of intestinal epithelium and intestinal barrier dysfunction in diabetic mice associated with depressed Notch/NICD transduction in Notch/Hes1 signal pathway. Cell Biol Int. 2014;38:1194–204. doi: 10.1002/cbin.10323. [DOI] [PubMed] [Google Scholar]
- MUTLU E, KESHAVARZIAN A, ENGEN P, FORSYTH CB, SIKAROODI M, GILLEVET P. Intestinal dysbiosis: a possible mechanism of alcohol-induced endotoxemia and alcoholic steatohepatitis in rats. Alcohol Clin Exp Res. 2009;33:1836–46. doi: 10.1111/j.1530-0277.2009.01022.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- MUTLU EA, GILLEVET PM, RANGWALA H, SIKAROODI M, NAQVI A, ENGEN PA, KWASNY M, LAU CK, KESHAVARZIAN A. Colonic microbiome is altered in alcoholism. Am J Physiol Gastrointest Liver Physiol. 2012;302:G966–78. doi: 10.1152/ajpgi.00380.2011. [DOI] [PMC free article] [PubMed] [Google Scholar]
- NOAH TK, SHROYER NF. Notch in the intestine: regulation of homeostasis and pathogenesis. Annu Rev Physiol. 2013;75:263–88. doi: 10.1146/annurev-physiol-030212-183741. [DOI] [PubMed] [Google Scholar]
- OBATA Y, TAKAHASHI D, EBISAWA M, KAKIGUCHI K, YONEMURA S, JINNOHARA T, KANAYA T, FUJIMURA Y, OHMAE M, HASE K, OHNO H. Epithelial cell-intrinsic Notch signaling plays an essential role in the maintenance of gut immune homeostasis. J Immunol. 2012;188:2427–36. doi: 10.4049/jimmunol.1101128. [DOI] [PubMed] [Google Scholar]
- PALMISANO M, PANDEY SC. Epigenetic mechanisms of alcoholism and stress-related disorders. Alcohol. 2017;60:7–18. doi: 10.1016/j.alcohol.2017.01.001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- PLOGER S, STUMPFF F, PENNER GB, SCHULZKE JD, GABEL G, MARTENS H, SHEN Z, GUNZEL D, ASCHENBACH JR. Microbial butyrate and its role for barrier function in the gastrointestinal tract. Ann N Y Acad Sci. 2012;1258:52–9. doi: 10.1111/j.1749-6632.2012.06553.x. [DOI] [PubMed] [Google Scholar]
- ROSE DJ, VENEMA K, KESHAVARZIAN A, HAMAKER BR. Starch-entrapped microspheres show a beneficial fermentation profile and decrease in potentially harmful bacteria during in vitro fermentation in faecal microbiota obtained from patients with inflammatory bowel disease. Br J Nutr. 2010;103:1514–24. doi: 10.1017/S0007114509993515. [DOI] [PubMed] [Google Scholar]
- SATO T, CLEVERS H. Growing self-organizing mini-guts from a single intestinal stem cell: mechanism and applications. Science. 2013;340:1190–4. doi: 10.1126/science.1234852. [DOI] [PubMed] [Google Scholar]
- SATO T, STANGE DE, FERRANTE M, VRIES RG, VAN ES JH, VAN DEN BRINK S, VAN HOUDT WJ, PRONK A, VAN GORP J, SIERSEMA PD, CLEVERS H. Long-term expansion of epithelial organoids from human colon, adenoma, adenocarcinoma, and Barrett’s epithelium. Gastroenterology. 2011;141:1762–72. doi: 10.1053/j.gastro.2011.07.050. [DOI] [PubMed] [Google Scholar]
- SATO T, VRIES RG, SNIPPERT HJ, VAN DE WETERING M, BARKER N, STANGE DE, VAN ES JH, ABO A, KUJALA P, PETERS PJ, CLEVERS H. Single Lgr5 stem cells build crypt-villus structures in vitro without a mesenchymal niche. Nature. 2009;459:262–5. doi: 10.1038/nature07935. [DOI] [PubMed] [Google Scholar]
- SHAIKH M, RAJAN K, FORSYTH CB, VOIGT RM, KESHAVARZIAN A. Simultaneous gas-chromatographic urinary measurement of sugar probes to assess intestinal permeability: Use of time course analysis to optimize its use to assess regional gut permeability. Clin Chim Acta. 2015;442C:24–32. doi: 10.1016/j.cca.2014.12.040. [DOI] [PMC free article] [PubMed] [Google Scholar]
- SHUKLA SD, LIM RW. Epigenetic effects of ethanol on the liver and gastrointestinal system. Alcohol Res. 2013;35:47–55. [PMC free article] [PubMed] [Google Scholar]
- SIMON-O’BRIEN E, ALAUX-CANTIN S, WARNAULT V, BUTTOLO R, NAASSILA M, VILPOUX C. The histone deacetylase inhibitor sodium butyrate decreases excessive ethanol intake in dependent animals. Addict Biol. 2014 doi: 10.1111/adb.12161. [DOI] [PubMed] [Google Scholar]
- SRINIVASAN T, THAN EB, BU P, TUNG KL, CHEN KY, AUGENLICHT L, LIPKIN SM, SHEN X. Notch signalling regulates asymmetric division and inter-conversion between lgr5 and bmi1 expressing intestinal stem cells. Sci Rep. 2016;6:26069. doi: 10.1038/srep26069. [DOI] [PMC free article] [PubMed] [Google Scholar]
- STANGER BZ, DATAR R, MURTAUGH LC, MELTON DA. Direct regulation of intestinal fate by Notch. Proc Natl Acad Sci U S A. 2005;102:12443–8. doi: 10.1073/pnas.0505690102. [DOI] [PMC free article] [PubMed] [Google Scholar]
- STOCKHAUSEN MT, SJOLUND J, MANETOPOULOS C, AXELSON H. Effects of the histone deacetylase inhibitor valproic acid on Notch signalling in human neuroblastoma cells. Br J Cancer. 2005;92:751–9. doi: 10.1038/sj.bjc.6602309. [DOI] [PMC free article] [PubMed] [Google Scholar]
- SUMMA KC, VOIGT RM, FORSYTH CB, SHAIKH M, CAVANAUGH K, TANG Y, VITATERNA MH, SONG S, TUREK FW, KESHAVARZIAN A. Disruption of the Circadian Clock in Mice Increases Intestinal Permeability and Promotes Alcohol-Induced Hepatic Pathology and Inflammation. PLoS One. 2013;8:e67102. doi: 10.1371/journal.pone.0067102. [DOI] [PMC free article] [PubMed] [Google Scholar]
- TIAN H, BIEHS B, CHIU C, SIEBEL CW, WU Y, COSTA M, DE SAUVAGE FJ, KLEIN OD. Opposing activities of notch and wnt signaling regulate intestinal stem cells and gut homeostasis. Cell Rep. 2015;11:33–42. doi: 10.1016/j.celrep.2015.03.007. [DOI] [PMC free article] [PubMed] [Google Scholar]
- TURGEON N, BLAIS M, GAGNE JM, TARDIF V, BOUDREAU F, PERREAULT N, ASSELIN C. HDAC1 and HDAC2 restrain the intestinal inflammatory response by regulating intestinal epithelial cell differentiation. PLoS One. 2013;8:e73785. doi: 10.1371/journal.pone.0073785. [DOI] [PMC free article] [PubMed] [Google Scholar]
- VAN DER FLIER LG, CLEVERS H. Stem cells, self-renewal, and differentiation in the intestinal epithelium. Annu Rev Physiol. 2009;71:241–60. doi: 10.1146/annurev.physiol.010908.163145. [DOI] [PubMed] [Google Scholar]
- VANDUSSEN KL, CARULLI AJ, KEELEY TM, PATEL SR, PUTHOFF BJ, MAGNESS ST, TRAN IT, MAILLARD I, SIEBEL C, KOLTERUD A, GROSSE AS, GUMUCIO DL, ERNST SA, TSAI YH, DEMPSEY PJ, SAMUELSON LC. Notch signaling modulates proliferation and differentiation of intestinal crypt base columnar stem cells. Development. 2012;139:488–97. doi: 10.1242/dev.070763. [DOI] [PMC free article] [PubMed] [Google Scholar]
- VANGIPURAM SD, LYMAN WD. Ethanol alters cell fate of fetal human brain-derived stem and progenitor cells. Alcohol Clin Exp Res. 2010;34:1574–83. doi: 10.1111/j.1530-0277.2010.01242.x. [DOI] [PubMed] [Google Scholar]
- VEMURI MC, CHETTY CS. Alcohol impairs astrogliogenesis by stem cells in rodent neurospheres. Neurochem Int. 2005;47:129–35. doi: 10.1016/j.neuint.2005.04.019. [DOI] [PubMed] [Google Scholar]
- VIDALI G, BOFFA LC, BRADBURY EM, ALLFREY VG. Butyrate suppression of histone deacetylation leads to accumulation of multiacetylated forms of histones H3 and H4 and increased DNase I sensitivity of the associated DNA sequences. Proc Natl Acad Sci U S A. 1978;75:2239–43. doi: 10.1073/pnas.75.5.2239. [DOI] [PMC free article] [PubMed] [Google Scholar]
- WANG HJ, ZAKHARI S, JUNG MK. Alcohol, inflammation, and gut-liver-brain interactions in tissue damage and disease development. World J Gastroenterol. 2010;16:1304–13. doi: 10.3748/wjg.v16.i11.1304. [DOI] [PMC free article] [PubMed] [Google Scholar]
- WEAVER GA, KRAUSE JA, MILLER TL, WOLIN MJ. Short chain fatty acid distributions of enema samples from a sigmoidoscopy population: an association of high acetate and low butyrate ratios with adenomatous polyps and colon cancer. Gut. 1988;29:1539–43. doi: 10.1136/gut.29.11.1539. [DOI] [PMC free article] [PubMed] [Google Scholar]
- WOOD S, PITHADIA R, REHMAN T, ZHANG L, PLICHTA J, RADEK KA, FORSYTH C, KESHAVARZIAN A, SHAFIKHANI SH. Chronic alcohol exposure renders epithelial cells vulnerable to bacterial infection. PLoS One. 2013;8:e54646. doi: 10.1371/journal.pone.0054646. [DOI] [PMC free article] [PubMed] [Google Scholar]
- YIN X, FARIN HF, VAN ES JH, CLEVERS H, LANGER R, KARP JM. Niche-independent high-purity cultures of Lgr5+ intestinal stem cells and their progeny. Nat Methods. 2014;11:106–12. doi: 10.1038/nmeth.2737. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.