Abstract
Background
Long-lasting transcriptional changes underlie a number of adaptations that contribute to alcohol use disorders (AUD). Chromatin remodeling, including histone methylation, can confer distinct, long-lasting transcriptional changes, and histone methylases are known to play a role in the development of addiction. Conversely, little is known about the relevance of Jumonji (JmjC) domain-containing demethylases in AUDs. We systematically surveyed the alcohol-induced phenotypes of null mutations in all 13 Drosophila JmjC genes.
Methods
We used a collection of JmjC mutants, the majority of which we generated by homologous recombination, and assayed them in the Booze-o-mat to determine their naïve sensitivity to sedation and their tolerance (change in sensitivity upon repeat exposure). Mutants with reproducible phenotypes had their phenotypes rescued with tagged genomic transgenes, and/or phenocopied by nervous system-specific knock down using RNA interference (RNAi).
Results
Four of the 13 JmjC genes (KDM3, lid, NO66 and HSPBAP1) showed reproducible ethanol-sensitivity phenotypes. Some of the phenotypes were observed across doses, e.g. the enhanced ethanol-sensitivity of KDM3KO and NO66KO, but others were dose-dependent, such as the reduced ethanol sensitivity of HSPBAP1KO, or the enhanced ethanol tolerance of NO66KO. These phenotypes were rescued by their respective genomic transgenes in KDM3KO and NO66KO mutants. While we were unable to rescue lidk mutants, knock down of lid in the nervous system recapitulated the lidk phenotype, as was observed for KDM3KO and NO66KO RNAi-mediated knock down.
Conclusion
Our study reveals that the Drosophila JmjC-domain histone demethylases Lid, KDM3, NO66, and HSPBAP1 are required for normal ethanol-induced sedation and tolerance. Three of three tested of those four JmjC genes are required in the nervous system for normal alcohol-induced behavioral responses, suggesting that this gene family is an intriguing avenue for future research.
Keywords: Alcohol use disorders, histone demethylases, behavior, Drosophila, genetics
INTRODUCTION
Alcohol use disorders (AUD) are a major cause for serious health (Edenberg and Foroud, 2013) and societal problems (Topper et al., 2014). Repeat alcohol consumption leads to changes in behavior, including tolerance and addiction. These changes coincide with alterations in gene expression, associated gene networks, and cellular functions (Iancu et al., 2017; Tulisiak et al., 2016). Transcriptional changes include alterations to the organization of the chromatin structure via several epigenetic mechanisms, and alcohol exposure can modify gene expression by remodeling chromatin (Ponomarev, 2013). Drugs have been shown to alter the epigenetic landscape and affect expression of addiction-related genes (Farris et al., 2015; Maze and Nestler, 2011; Robison and Nestler, 2011), and some chromatin remodeling enzymes have been shown to be required for drug-induced behavioral changes (Maze et al., 2014).
Histone modifications modulate access to the genome, modifying patterns of gene expression and associated behaviors (Ponomarev, 2013; Ponomarev et al., 2012). One such modification mechanism involves the methylation and demethylation of histones. Histone demethylases remove methyl groups from lysine or arginine residues in the N-terminal histone tails (Shi et al., 2004). Lysines can be methylated in three different states: mono-, di- or tri-methylation, where each state can have distinct consequences on transcriptional activation or repression (Shi, 2007). This flexibility makes methylation and demethylation an effective regulatory system (Krishnan et al., 2011; Shi and Whetstine, 2007) with the potential to respond to environmental cues. For example, H3K9 di-methylation (histone H3, lysine 9) is a transcriptional marker for repression, and is induced during alcohol binge-induced neurodegeneration (Subbanna et al., 2013). Decreases in H3K9 methylation, mediated by downregulation of methyltransferases, have been associated with ethanol treatments and withdrawal (Qiang et al., 2011). While methylation is a dynamic and reversible process, little is known about the role of histone demethylases in the context of alcohol abuse and addiction.
Drosophila melanogaster in one of the model organisms used to study genes and mechanisms underlying AUDs. The vinegar fly exhibits alcohol-induced behaviors (e.g. disinhibition, locomotor hyperactivity, and sedation) similar to mammals during naïve exposure to alcohol (Lee et al., 2008; Narayanan and Rothenfluh, 2016; Prescott and Kendler, 1999; Wolf et al., 2002). Upon repeated exposures, flies develop tolerance (Scholz et al., 2000) and preference for alcohol consumption (Devineni and Heberlein, 2009; Peru y Colón de Portugal et al., 2014). Many of the genes regulating ethanol responses in Drosophila are conserved in mammals (Grotewiel and Bettinger, 2015), and a number of genes have been shown to affect alcohol responses in both flies and humans (Gonzalez et al., 2017; Ojelade et al., 2015; Schumann et al., 2016). Histone demethylase genes are also conserved between mammals and Drosophila, with the fly’s genome encoding 13 jumonji C domain-containing demethylases (JmjC-KDMs). Even though there are fewer JmjC-KDMs in flies than in humans (34), Drosophila JmjC-KDMs’ structural diversity matches the human enzymes’ diversity, with at least one protein belonging to six out of the seven JmjC known phylogenetic groups (Klose et al., 2006). Reproducible behavioral assays, high genetic conservation with human JmjC-KDM diversity, and economy of scale offer an opportunity to study the functionality of JmjC-KDMs in Drosophila and their role in alcohol-induced responses and behaviors.
We previously developed global knock outs for the 13 known JmjC-domain demethylases in two parallel studies on development and the modulation of sleep and circadian rhythms (Shalaby et al., 2017a; 2017b). Here, we investigated the role JmjC demethylases play in regulating alcohol-induced behaviors and whether or not such functions are specific to the nervous system. We performed a behavioral screen using ethanol-induced sensitivity to loss-of-righting and tolerance assays with different ethanol doses. Our results show that loss of four of the Drosophila JmjC genes (KDM3, lid, NO66, and HSPBAP1) caused reproducible sedation and tolerance phenotypes, and three of three genes tested were required in the nervous system for normal alcohol-related behaviors. Most of the remaining 9 JmjC mutants showed no significant difference vs. controls, and many effect sizes were below 0.5, suggesting that select JmjC-domain histone demethylases regulate alcohol-induced behavioral responses in the Drosophila nervous system.
MATERIALS AND METHODS
Flies husbandry and maintenance
HSPBAP1 (CG12879), JARID2 (CG3654), JMJD4 (CG7200), JMJD5 (CG13902), JMJD7 (CG10133), KDM2 (CG11033), KDM3 (CG8165), KDM4A (CG15835), KDM4B (CG33182), and NO66 (CG2982) mutants were generated using a combination of in vivo bacterial recombineering and homologous recombination (Shalaby et al., 2017b). The lidk06801 stock (BL10403 - lidk) was obtained from the Bloomington Stock Center, PSRFM1 was provided by Kristin White (Massachusetts General Hospital, Charlestown, MA), and UTX1 by Andreas Bergmann (UMass Medical School, Worcester, MA). Genomic rescues for KDM3, lid, and NO66 were constructed by recombineering combined with Gateway technology, introducing a C-terminal 6xHis-, HA-tag (Shalaby et al., 2017b). RNAi flies for KDM3 (BL32975), lid (V103830), and NO66 (V107819) were obtained from the Bloomington Drosophila Stock Center at Indiana University (BL) and Vienna Drosophila Resource Center (V). All flies were outcrossed for at least five generations to the w− Berlin genetic background (wB) prior to behavioral analyses with the exception of UTX1 and UAS-KDM3-RNAi. For those flies, we used parents of the genotype JmjC/visible marker, and then assayed the sibling progeny JmjC/+ vs. +/visible marker, which were both in an identical Berlin/unknown genetic background. For the genomic rescue experiments we crossed the {g JmjC-HA} rescue construct (Shalaby et al., 2017b) into the JmjC− background and assayed the resulting flies in parallel with the JmjC− mutant and control flies. RNAi knock down experiments were performed by crossing elav-Gal4 females to UAS-JmjC-RNAi males and to + control males in parallel (for NO66 and lid). For KDM3, we crossed nSyb-Gal4 females to UAS-KDM3-RNAiHMS/visible marker males and assayed the two sibling progeny genotypes. All flies were maintained on standard cornmeal/molasses food at 25°C and 75% humidity on a 12-hour day and 12-hour night cycle prior to any assay.
Alcohol-related behaviors
We measured two alcohol-induced behaviors: naïve sensitivity to sedation and the development of tolerance to repeat exposure. The assays were done in a booze-o-mat exposure chamber (Wolf et al., 2002). In this device 20 flies are placed in a test tube (n = 1) and a predetermined flow rate of humidified air and vaporized alcohol is streamed into the tubes at a constant total flow rate 150 (arbitrary flow rate units of Flowmeter P-03219-21, Cole Parmer, Vernon Hills, IL). We used males collected one day prior to our tests. During the ethanol/air exposure we counted flies losing their righting reflex every 5 minutes, until the number equaled half of the sample. This time, the ST50, was our sedation measure. For tolerance, flies were exposed to twice the length of ST50 of the wild-type control, then removed from the Booze-o-mat and allowed to recover on standard food for 4 hours. After the recovery period, we re-exposed the flies to the same conditions and calculated a second ST50. The percent difference between the first and the second ST50 was our measurement of tolerance. Average ST50 and percent tolerance for each genotype was calculated from a total of 12 samples per genotype/experiment. For the experiments presented, we used four ethanol concentration levels: high (flow rate of ethanol/water-saturated air of 130/20 E/A), medium (110/40), low (80/70), and very low (50/100).
Ethanol absorption
We determined the internal concentration of ethanol in the flies at different times (0, 5, 10, 15 and 20 min) during an exposure to 130/20 E/A mixture. At each time point, 5 whole flies per genotype were homogenized in 50 μL of water and centrifuged at maximum speed (14,000 RPM) for 5 minutes. Then 25 μL of supernatant was mixed with 300 μL semicarbazide buffer (3.3% tetrasodium pyrophosphate, 0.84% semicarbazide hydrochloride and 0.16% Glycine), 25 μL 1.6% NAD+ and 25 μL of alcohol dehydrogenase (4000 units / mL). The solution was incubated at 40°C for 40 minutes and absorbance at 340 nm for each sample determined on a NanoDrop spectrophotometer (Ishmayana et al., 2015). We repeated the procedure at least three times for each genotype (n ≥ 3). Ethanol concentration was determined from a standard curve (0, 2.5, 3.5, 5, 10, 17 mM ethanol).
Statistical analyses
Data distributions were tested for normality using the Shapiro-Wilk test with p < 0.05 as significantly non-normal. We Bonferroni-adjusted the p-value of the Shapiro-Wilk test for multiple testing (i.e. number of genotypes), in each figure, since we were interested in maximal specificity of the test (and not sensitivity). One data set ended up being significantly not normally distributed (Fig. 4, lidk/+ sedation at 110/40 E/A; Shapiro Wilk adjusted p < 0.01; n = 12). This was caused by one apparent “outlier” point (very stringently defined as > 2× interquartile range above the third quartile). As we did not pre-define a cutoff for outliers, we left this one data point in the analysis and analyzed the comparison with w Berlin control non-parametrically. Comparisons between the wild-type and the mutant’s measures were analyzed using GraphPad Prism 7 for Mac. We used one-way ANOVA with Dunnett’s post-hoc test for multiple comparisons, and Student’s t-test for experiments with two test subjects (i.e. mutant vs. wild type). Error bars in all experiments represent the standard error of the mean (SEM). We determined effect sizes (Hedge’s g with 95% confidence intervals – a small sample size bias-adjusted version of Cohen’s d, where d = 1 signifies an effect size of a mutant being 1 standard deviation from the wild-type mean) using an Excel spreadsheet (Durham University, UK; http://www.cem.org/effect-size-calculator). An effect size of > 0.8 is generally considered a large effect, though in our experience, we would consider g > 1.3 a strong mutant.
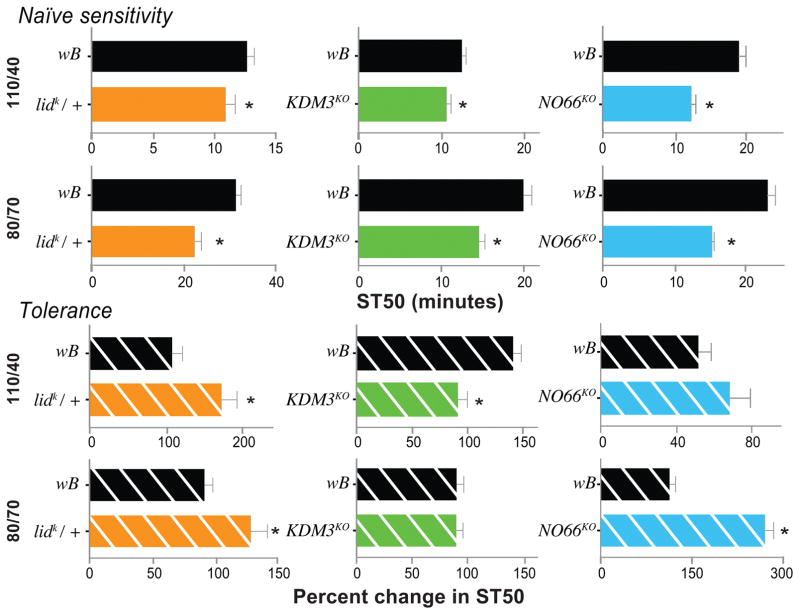
Figure 4.
Ethanol-induced sedation and tolerance phenotypes for lid, KDM3 and NO66 mutants at lower ethanol exposures. Flies were exposed to two ethanol flow rates (110/40, higher dose, and 80/70 E/A, lower dose, indicated on the left), and naïve alcohol sensitivity (top half) and tolerance upon repeat exposure (bottom half) was determined in three mutants (lidk/+, KDM3KO, and NO66KO, left to right). lidk/+ flies consistently showed enhanced sensitivity to sedation, and developed more tolerance compared to the controls (wB, run in parallel; black bars). KDM3KO and NO66KO also showed consistently increased sensitivity to sedation. KDM3KO, however, did not display a tolerance phenotype at the lowest ethanol dose (80/70) while NO66KO, on the other hand, showed only a tolerance phenotype at that low dose. (*p < 0.05, student’s t-test comparison between the control and the test flies; note that lidk/+ sedation at 110/40 was the only non-normally distributed measure, due to one “outlier”, included here. We therefore also analyzed the data using a Mann-Whitney test, which confirmed a significant difference at p = 0.023).
RESULTS
Systematic Analysis of Naïve Ethanol Sensitivity and Rapid Tolerance in JmjC Mutants
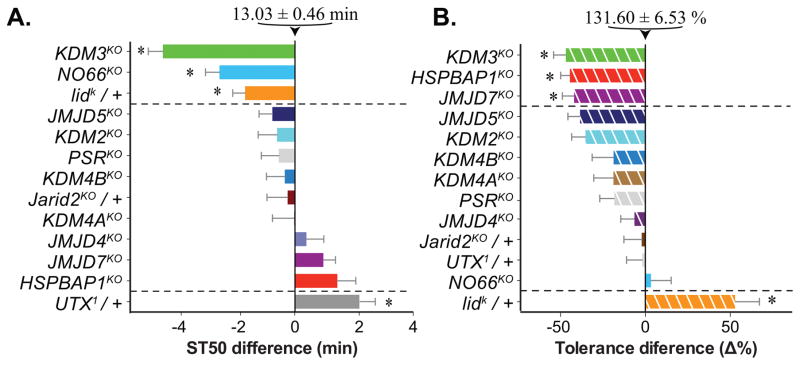
In order to determine ethanol sedation and tolerance phenotypes associated with loss of JmjC demethylases, we exposed mutants of every Drosophila JmjC gene to a high concentration of ethanol/air (130/20 E/A). Because the JARID2KO and UTX1 alleles are lethal, and lidk is semi-lethal, we tested these mutations as heterozygotes after crossing them to wild type (white− Berlin, wB). Our analysis yielded four mutants with significant sensitivity phenotypes when compared to wB. KDM3KO, NO66KO and lidk/+ displayed increased sensitivity to ethanol-induced sedation, while UTX1/+ flies showed reduced sensitivity (Fig. 1A).
Figure 1.
Alcohol-induced phenotypes in flies lacking JmjC histone demethylases. (A) Naïve sensitivity to alcohol-induced sedation, displayed as difference (in minutes) from wB controls (whose average time to 50% sedation, ST50, was 13.03±0.46 min, depicted at the top). Flies were exposed to 130/20 E/A (ethanol/water-saturated air) in test tubes, and the ST50 determined by visual inspection. In this and the following graphs, error bars represent the standard error of the mean. (B) Difference in the %tolerance induced by two ethanol exposures (4 hr apart with recovery on food) between JmjC knock out and that of the wB controls, which developed 132±7% tolerance. Tolerance was calculated as percent change in ST50 from first to second exposure to 120/30 E/A for the same cohort of flies. The first exposure was the same for all genotypes. Mutants that displayed significantly different behaviors are separated by dotted lines and highlighted with an asterisk (*p < 0.05, one-way ANOVA with Dunnett’s post hoc comparison vs. wB control).
To determine if any of the Drosophila JmjC-domain demethylases are required for the development of alcohol tolerance, we exposed all our mutants a second time to ethanol four hours after the first exposure. KDM3KO, HSPBAP1KO, and JMJD7KO showed decreased tolerance compared to wild type, while lidk/+ displayed increased tolerance (Fig. 1B). None of the other JmjC mutants showed significant tolerance phenotypes, including NO66KO and UTX1/+, which both had sensitivity phenotypes.
The functional relationship between acute sensitivity to alcohol and rapid tolerance is not understood. Tolerance is by definition a reduction of sensitivity upon ethanol re-exposure, and therefore could clearly employ mechanisms that also alter naïve sensitivity. Indeed, one could argue that mutants with reduced naïve sensitivity are “pre-tolerant” and might therefore develop less rapid tolerance upon re-exposure. Indeed, a number of mutants (dlp, mys, scb) show both increased sensitivity and tolerance phenotypes (Grotewiel and Bettinger, 2015), while others (whir, ics) display both decreased sensitivity and decreased tolerance (unpublished observation). There are, however, also contrasting examples, such as mutations in homer, Arf6 and Efa6, which all show enhanced sensitivity, but reduced tolerance (Gonzalez et al., 2017; Grotewiel and Bettinger, 2015). To test whether JmjC sensitivity and tolerance phenotypes correlated, we analyzed the relationship between these two measures and found no significant correlation between these behaviors in our set of JmjC mutants (p = 0.83; Fig. S1). These results are consistent with another set of ethanol response mutants, which also did not reveal a correlation between sensitivity and tolerance changes (Devineni et al., 2011). Therefore, even though numerous JmjC mutants affected both sensitivity and tolerance, they did so in unpredictable ways.
Changes in Ethanol Pharmacokinetics do not Correlate with Ethanol Sensitivity Phenotypes
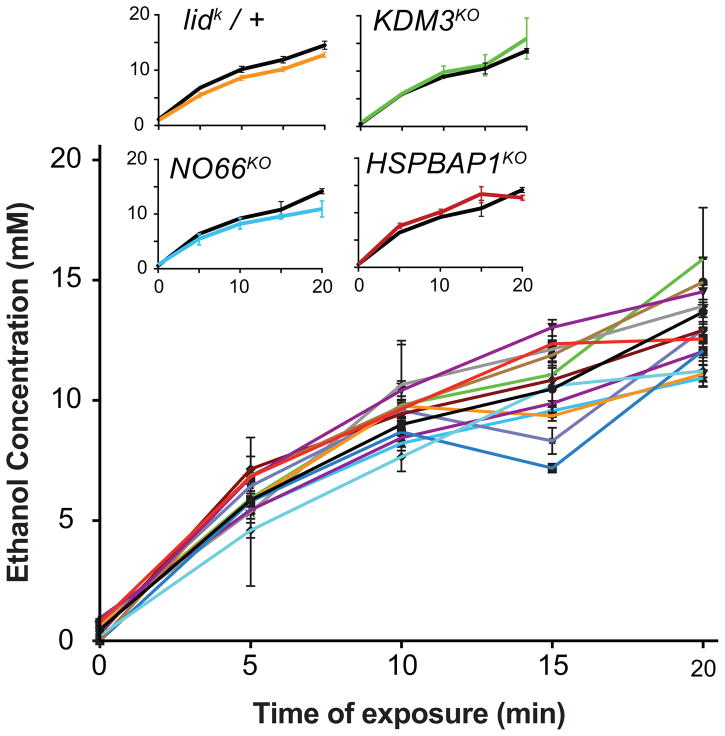
To determine whether altered ethanol pharmacokinetics underlie the observed phenotypes, we measured the concentration of ethanol on exposed flies (130/20 E/A) at different times (0, 5, 10, 15, 20 min). All JmjC mutants showed similar ethanol absorptions/metabolism curves as those seen in the wild type (Figure 2), and for each mutant, the internal ethanol concentration correlated strongly with the exposure time (R2 > 0.82 for each of the mutants). None of the slopes of the linear regressions were significantly different (pair-wise comparison with Bonferroni correction). However, two-way ANOVAs with Dunnett’s post hoc comparison suggested that NO66KO (p = 0.045) and KDM4BKO (dark blue, p = 0.047) showed a significant main effect of genotype. KDM4BKO did not display alcohol-induced behavioral phenotypes (Fig. 1A,B), while NO66KO showed increased sensitivity to ethanol-induced sedation at three ethanol doses (Figs. 1A, 4). If NO66KO was indeed an alcohol absorption and metabolism mutant with reduced internal ethanol, that would seem unlikely to be the cause of the enhanced sensitivity observed in NO66KO. Rather, NO66KO showing enhanced sensitivity to ethanol-induced sedation while having lower levels of ethanol on board makes its sedation-sensitive phenotype even more striking. Overall, these results indicate that the observed sedation and tolerance phenotypes in JmjC mutant flies are not explained by altered ethanol absorption or metabolism, but rather are caused by pharmacodynamic differences due to the lack of distinct JmjC genes.
Figure 2.
Ethanol pharmacokinetics during a 130/20 E/A exposure. Ethanol concentration was measured in whole fly extracts from 5 flies in 5 min intervals (n ≥ 3 per time point). All mutants showed highly significant correlations between internal ethanol concentration and exposure time (R2 > 0.82). None of the slopes of linear regression fits were significantly different (pair-wise comparison with Bonferroni adjustment), but two-way ANOVA suggested a genotype effect for KDM4BKO (dark blue) and NO66KO (p < 0.05, Dunnett’s post hoc comparison). Inset shows the concentration curves for the four JmjC mutants followed-up in more detail (Fig. 3–6).
Ethanol Phenotypes at Different Exposure Doses
In addition to determining statistical significance for the difference between wild-type and mutant genotypes (Fig. 1), we also determined the effect sizes of the changes and found five “large effects” (with a Hedge’s g effect size > 0.8; Fig. S2): enhanced ethanol-sensitivity in KDM3KO and NO66KO, enhanced ethanol tolerance in lidk/+, and reduced ethanol tolerance in KDM3KO and HSPBAP1KO. We re-analyzed UTX1/+, which was on the cusp of a “large effect” (Hedge’s g = 0.8; Fig. S2), and this mutant did not show a phenotype when we assayed it again (Fig. S3). Unlike most other mutants in this study, UTX1 is a pre-existing allele we did not generate ourselves by knock out and could, therefore, also not outcross. Furthermore, UTX1 is one of the few JmjC loss-of-function alleles that is homozygous lethal. For these combined reasons, we decided to focus on the other four genes with large effects for follow up.
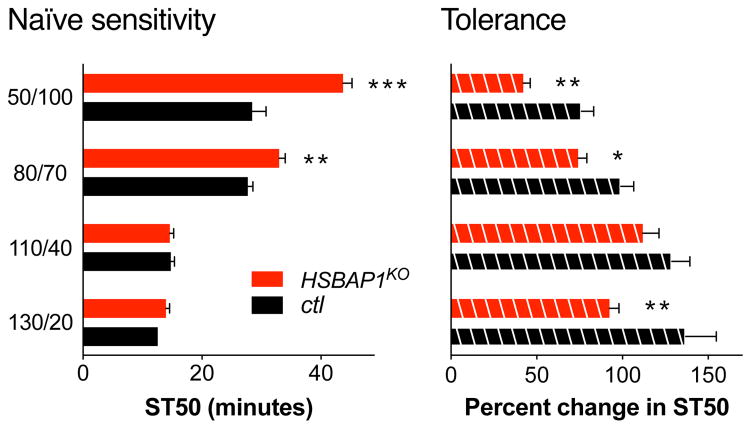
To test whether any of our observed phenotypes were dependent on the dose of ethanol used, we tested the JmjC mutants at lower doses of ethanol. HSPBAP1KO flies showed no significant sedation phenotype at 130/20 (Fig. 1A) and 110/40 (Fig. 3), but at the low dose of 80/70 it showed reduced ethanol-sensitivity (Fig. 3). Because of this low-dose specificity of the phenotype, we also determined the phenotype at an even lower dose of 50/100, and found that this reproduced the reduced ethanol-sensitivity (Fig. 3). For three out of these four doses, HSPBAP1KO flies also displayed a reduced ethanol tolerance phenotype, while there was also mild (yet non-significant) effect in the same direction at 110/40 (Fig. S4). Overall, these data indicate that HSPBAP1 is required for the normal development of ethanol tolerance, and at low doses is also required for wild-type ethanol-sensitivity (see Table S1 for a statistical analysis of the genotype × dose effects).
Figure 3.
Ethanol-induced sedation and tolerance phenotypes of HSPBAP1KO. Depicted are mutant and control values run in parallel. The data for 130/20 is replicated from Fig. 1. Two-way ANOVAs indicated significant main effects for dose and for genotype (p < 0.001), and for sedation there was also a significant genotype × dose interaction (p < 0.001). Post hoc comparisons revealing significant differences between mutant and control at the different doses are indicated (***p < 0.001, **p < 0.01, *p < 0.05; Bonferroni adjusted for multiple comparisons).
Heterozygous lidk/+ flies showed increased ethanol-sensitivity and tolerance phenotypes across all tested exposure doses (Fig. 4). KDM3KO showed increased sensitivity to sedation at both 110/40 and 80/70 compared to wild type. This mutant also developed significantly less tolerance at 110/40, but not at the lowest dose (80/70, Fig. 4). Thus, KDM3KO showed reduced tolerance phenotypes at the two highest, but not the lowest dose (see Fig. 1B). Lastly, NO66KO mutants were more sensitive at all three ethanol doses (Fig. 4), and at the lowest exposure dose these mutants also revealed an increase ethanol tolerance phenotype. Together these data revealed specific dose × genotype interactions for the alcohol responses of HSPBAP1KO and NO66KO, and KDM3KO, while lidk/+ was the only mutant that did not show a significant dose × genotype interaction (Table S1).
Rescue of KDM3KO and NO66KO Ethanol Phenotypes
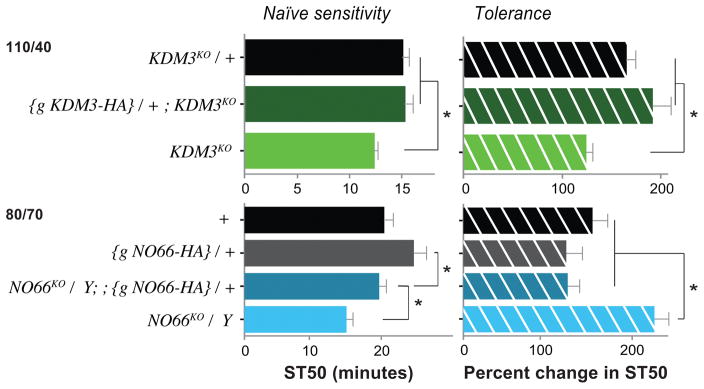
In order to ascertain that loss of a specific JmjC demethylase gene was causing the phenotype, we attempted to rescue the sedation and tolerance phenotypes for lid, KDM3, and NO66. In all cases, we used genomic rescue constructs with a C-terminal HA-tag (Shalaby et al., 2017b). Both KDM3KO and NO66KO mutant phenotypes were restored to wild-type measures upon introduction of their respective genomic rescue constructs (Fig. 5). The lid rescue construct, however, did not restore the phenotypes observed in lidk/+ flies (Fig. S5). This may be because the HA-tag interferes with Lid protein function. Alternatively, because our lidk/+ phenotype was caused by a heterozygous mutation – clearly not a complete loss-of–function genotype – it may reflect the need for exact wild-type levels of Lid protein expression to obtain normal ethanol-induced behavior. Lastly, our rescue construct may also lack an important enhancer element, guiding lid expression in ethanol response-relevant brain regions.
Figure 5.
Rescue of KDM3KO and NO66KO phenotypes. We used genomic, C-terminally HA-tagged rescue constructs (indicated as {g JmjC-HA} in the Figure). These transgenes restored both the sensitivity and tolerance phenotypes of KDM3KO and NO66KO towards wild-type levels. (*p < 0.05, one-way ANOVA with Dunnett’s post-hoc multiple comparison). Ethanol/Air flow rates for each are indicated on the left. Note that KDM3KO/+ heterozygotes are no different from +/+ flies in both ethanol-induced sedation and tolerance (Fig. S6).
JmjC Demethylases are Required in the Brain for Normal Responses to Alcohol
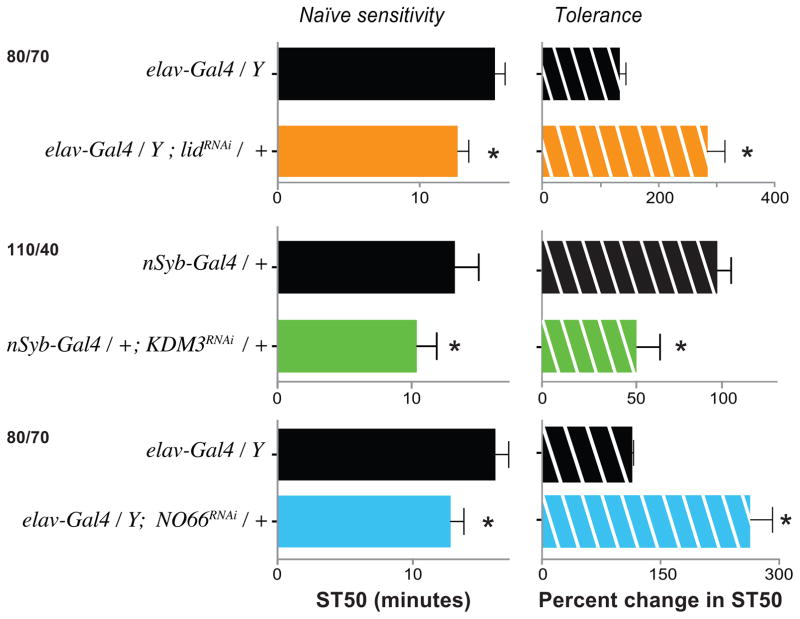
Because we were unable to rescue the lidk/+ phenotype, we sought a different independent confirmation that the observed lidk/+ phenotype was indeed caused by reduction of lid activity. We therefore used an RNA interference (RNAi) line targeted against the lid gene to test whether we could recapitulate the phenotypes observed in Figures 1 and 4. As our primary interest is in ethanol’s effects in the nervous system, we also wanted to test whether nervous system-specific knock down of lid – and of KDM3 and NO66 – would be sufficient to phenocopy the knockout phenotypes.
Similar to the global loss of function phenotype, panneuronal knock down of KDM3 (nSyb-Gal4/+; UAS-KDM3-RNAi/+) resulted in a reduction in sedation and tolerance to ethanol exposure compared to control (Fig. 6), suggesting a requirement for KDM3 in neurons for normal ethanol-induced behaviors. Similarly, driving RNAi against NO66 in neurons (elavC155-Gal4; UAS-NO66-RNAi/+) recapitulated the NO66KO mutant’s enhanced sensitivity and tolerance phenotypes (Fig. 6), indicating a requirement of NO66 in the nervous system. Lastly, when driven panneuronaly (elavC155-Gal4; UAS-lid-RNAi/+) lid knock down flies showed significantly enhanced sensitivity to ethanol-induced sedation and more tolerance than the control (Fig. 6). This phenocopies the lidk/+ results (Fig. 1 and 4), and suggests that i) loss of lid gene function is indeed resulting in ethanol behavior phenotypes, and ii) as for KDM3 and NO66, lid function is required in the nervous system for normal behavioral responses to ethanol.
Figure 6.
JmjC demethylases are required in the nervous system for normal alcohol-induced behaviors. Driving panneuronal Gal4s (nSyb- and elavc155-) to express of RNAi against lid, KDM3, and NO66 recapitulated the sedation-sensitivity (left column) and tolerance phenotypes (right column) of these genes (cf. Fig 1 and 3). (*p < 0.05 Student’s t-test comparison between the control and the test flies). These experiments were carried out under the same conditions as the knockout assays, with the ethanol/air flow rates indicated on the left. Ethanol/Air flow rates are indicated on the left. Note that none of the RNAi transgenes caused a phenotype in the absence of a Gal4 driver (Fig. S7).
DISCUSSION
Alcohol consumption alters the epigenetic landscape and subsequent gene transcription (Krishnan et al., 2014; Ponomarev, 2013; Ponomarev et al., 2012). Histone methylation can play a pivotal role in gene expression, and the methylation state of different lysines on histones can lead to silencing and/or activation of gene transcription (Krishnan et al., 2014). Little is known about the role and relevance of histone demethylases in alcohol abuse disorders and addition. Here we systematically examined loss-of-function mutations in all 13 Drosophila JmjC-domain histone demethylases, and found four genes that reproducibly affected ethanol-induced sedation and tolerance. Loss of NO66 led to increased sensitivity to sedation, and also to enhanced tolerance at low ethanol doses. Interestingly, a stress response study in worms reported a decreased recovery rate to acute ethanol exposure in NO66 mutants (Kirienko and Fay, 2010). This result is similar to our findings with NO66KO flies, which showed increased sensitivity to naïve alcohol exposure. Additionally, the mouse ortholog of NO66 was upregulated in whole brain tissue of a mouse strain predisposed to prefer alcohol (Mulligan et al., 2006), and the human ortholog was downregulated in the amygdala of alcoholics (Ponomarev et al., 2012), suggesting a role for NO66 in ethanol-related behaviors across phyla. Furthermore, we found that KDM3KO flies also showed enhanced sensitivity to sedation, but unlike NO66KO, the KDM3KO flies showed decreased ethanol tolerance. Again, mouse KDM3A was upregulated in whole brain tissue of a mouse strain predisposed to prefer alcohol (Mulligan et al., 2006), and together with our data, this supports an in vivo role for KDM3 genes in ethanol-relevant behaviors. We also found that lid−/+ heterozygous flies showed enhanced sedation sensitivity and increased tolerance. Once more, KDM5B, the mouse lid ortholog, was upregulated in whole brain tissue of a mouse strain predisposed to prefer alcohol (Mulligan et al., 2006). The mouse brain expression differences of these KDM genes, in combination with our findings that all three genes – NO66, KDM3, and lid – are required in the nervous system, suggest that activity levels of numerous JmjC-domain histone demethylase in the brain can predispose animals to show distinct ethanol-induced behavioral differences. Lastly, HSPBAP1KO flies showed reduced ethanol tolerance and, at low doses, reduced ethanol sensitivity. Little is known about this gene, or its mammalian ortholog. Interestingly, HSPBAP1 mutants did not alter phenotypes caused by chromatin rearrangement, and the protein was localized to the cytoplasm (Shalaby et al., 2017b). This is in contrast to Lid, NO66, and KDM3, which all localized to the nucleus, and altered chromatin organization (Shalaby et al., 2017b).
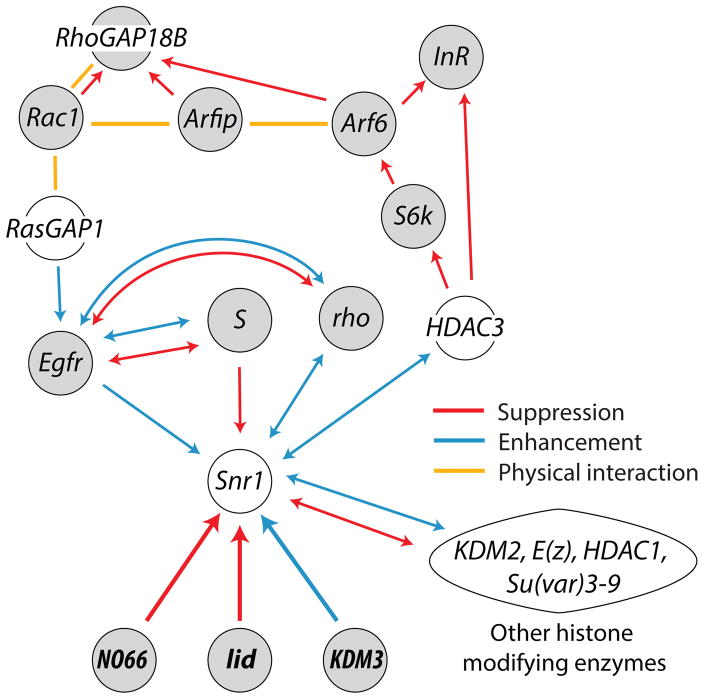
Consistent with a role in Drosophila chromatin organization for latter three genes, lid and NO66 suppress, while KDM3 enhances ectopic wing vein phenotypes caused by mutations in Snf5-related 1 (Snr1) (Curtis et al., 2011). In flies, worms and humans, SNR1 is part of the SWI/SNF chromatin-remodeling complex (Curtis et al., 2011), which regulates RNA polymerase II and gene transcription (Armstrong et al., 2002). The KDM-Snr1 interactions (Fig. 6) suggest these JmjC-KDMs modulate SWI/SNF, thereby affecting gene expression at the transcription level. In addition to KDM3, lid and NO66, Snr1 interacts with other histone modifying enzymes (e.g. HDAC3, KDM2 and E(z) – Fig. 6), suggesting a central role of the SWI/SNF complex in gene regulation. Interestingly, at least two genes (swsn-7 and swsn-9) coding for SWI/SNF complex members in worms are required for the proper development of functional alcohol tolerance (Mathies et al., 2015). Furthermore, variants in one human SWI/SNF complex gene (BRD7) associated significantly with alcohol dependence in a human genomewide association study (Mathies et al., 2015). Our results suggest that JmjC-KDMs might participate in the regulation of alcohol behaviors by modulating transcription via the SWI/SNF complex.
Genes, whose transcription is regulated by SWI/SNF and that regulate alcohol-induced behaviors are not known. However, Snr1 also genetically interacts with genes in the epidermal growth factor receptor (Egfr) pathway (Marenda et al., 2004), which has been shown to affect responses to ethanol (Corl et al., 2009). The EGFR signaling component RasGAP1 directly binds to RhoGAP18B (Friedman et al., 2011), a protein that is involved in alcohol-induced behavioral responses (Rothenfluh et al., 2006). RhoGAP18B, in turn, is connected (genetically and physically) to the insulin receptor (InR) signaling pathway, including Ar6 GTPase and S6 kinase (S6k; Acevedo et al., 2015; Peru y Colón de Portugal et al., 2012). This pathway, which is involved in the regulation of neuronal activity (Acevedo et al., 2015), is in turn linked to HDAC3 (Lv et al., 2012), a histone-modifying enzyme also linked to Snr1 (Zraly et al., 2004). Thus, while the direct transcriptional targets of JmjC and SWI/SNF that regulate alcohol-induced behaviors remain to be determined, there are already a number of suggestive links to previously identified alcohol-response pathways (Figure 6).
In summary, our results suggest that loss of many JmjC genes does not affect the ethanol responses we tested, whereas four of these genes are required for normal alcohol-induced behavioral responses. Furthermore, three of three are required in the nervous system to regulate ethanol-induced behavioral responses. This confirms that histone demethylation plays a relevant role in the regulation of alcohol-induced behavioral responses. Many KDM genes show differential expression between mouse strains predisposed to drink alcohol, or not (Mulligan et al., 2006), yet we found that only a subset of these genes is required to regulate ethanol-induced behaviors. Functional redundancy between related genes is rarely observed in Drosophila, and many unique fly genes have 2–4 vertebrate orthologs, as is the case for most JmjC genes. Our systemic analysis of developmental phenotypes suggests that of the 13 Drosophila JmjC genes, only KDM4A and KDM4B showed some redundant function (Shalaby et al., 2017b). A genome-wide study in humans found that three SNPs within the KDM4C gene, a demethylase related to Drosophila KDM4B, were associated with the presence of alcohol withdrawal symptoms (Wang et al., 2012). We did not observe altered alcohol-induced behavioral responses in KDM4AKO and KDM4BKO mutants, and this may be a reflection of aforementioned redundancy. Alternatively, Drosophila KDM4 phenotypes might be found when testing the more specific phenotype of alcohol withdrawal in flies (Ghezzi et al., 2012). Such a hypothesis – that specific JmjC genes affect specific phenotypes – is consistent with our findings here. It is also what we observed in our circadian rhythm and sleep studies (Shalaby et al., 2017a), where numerous – but not all – JmjC loss-of-function mutants displayed specific behavioral phenotypes. These phenotypes showed specificity for certain measures, and the direction of these changes (Shalaby et al., 2017a), just as we found here. Overall, our data underline the in vivo relevance of JmjC demethylases in the regulation of ethanol-induced behaviors, and make this gene family, and their targets, an intriguing avenue for future research.
Supplementary Material
Figure 7.
Model network showing the interactions of lid, KDM3, and NO66 with Snf5-related 1 (Snr1), encoding a component of the SWI/SNF chromatin-remodeling complex. Shown are genes that are involved in ethanol responses (grey), and some of their interaction partners. Genetic interactions are in blue (enhancement) and red (suppression), while physical associations (determined e.g. by co-immunoprecipitation) are in yellow (see text for details). Note that the arrows on the genetic interactions do not imply signal flow, in a biochemical sense, they merely indicate which gene enhanced/suppressed an initial mutation (arrowhead pointed towards that initial gene). Abbreviations: S: Star, transmembrane protein facilitating Egfr trafficking; Egfr: epidermal growth-factor receptor; rho: rhomboid, intra-membrane serine protease; RasGAP1: GTPase-activating protein of the Ras family; RhoGAP18B: GTPase-activating protein of the Rho-family; intg: integrin cell adhesion molecule; Rsu1: Ras suppressor 1; Rac1: small GTPase of the Rho family, regulating actin dynamics; Arfip: Arfaptin scaffolding protein; Arf6: small GTPase regulating plasma membrane trafficking; InR: insulin receptor; S6k: S6 kinase, growth and protein translation regulator; HDAC3: histone deacetylase 3; Snr1: Snf5-related 1, part of the SWI/SNF complex; and NO66, lid, KDM3: JmjC domain-containing histone demethylases described herein (see text/discussion for details).
Acknowledgments
Funding was provided by the NIH: T32 DA007290 (JHP), R21AA022404 (MB and AR), R01DK110358 (ARRo) and the American Heart Association DK7745-17 (NAS), 16CSA28530002 (ARRo).
Footnotes
The authors declare no conflict of interest.
References
- Acevedo SF, Peru y Colón de Portugal RL, Gonzalez DA, Rodan AR, Rothenfluh A. S6 Kinase reflects and regulates ethanol-induced sedation. J Neurosci. 2015;35:15396–15402. doi: 10.1523/JNEUROSCI.1880-15.2015. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Armstrong JA, Papoulas O, Daubresse G, Sperling AS, Lis JT, Scott MP, Tamkun JW. The Drosophila BRM complex facilitates global transcription by RNA polymerase II. The EMBO Journal. 2002;21:5245–5254. doi: 10.1093/emboj/cdf517. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Corl AB, Berger KH, Ophir-Shohat G, Gesch J, Simms JA, Bartlett SE, Heberlein U. Happyhour, a Ste20 family kinase, implicates EGFR signaling in ethanol-induced behaviors. Cell. 2009;137:949–960. doi: 10.1016/j.cell.2009.03.020. [DOI] [PubMed] [Google Scholar]
- Curtis BJ, Zraly CB, Marenda DR, Dingwall AK. Histone lysine demethylases function as co-repressors of SWI/SNF remodeling activities during Drosophila wing development. Developmental Biology. 2011;350:534–547. doi: 10.1016/j.ydbio.2010.12.001. [DOI] [PubMed] [Google Scholar]
- Devineni AV, Heberlein U. Preferential ethanol consumption in Drosophila models features of addiction. Curr Biol. 2009;19:2126–2132. doi: 10.1016/j.cub.2009.10.070. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Devineni AV, McClure KD, Guarnieri DJ, Corl AB, Wolf FW, Eddison, Heberlein U. The genetic relationships between ethanol preference, acute ethanol sensitivity, and ethanol tolerance in Drosophila melanogaster. Fly. 2011;5:191–199. doi: 10.4161/fly.5.3.16987. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Edenberg HJ, Foroud T. Genetics and alcoholism. Nat Rev Gastroenterol Hepatol. 2013;10:487–494. doi: 10.1038/nrgastro.2013.86. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Farris SP, Harris RA, Ponomarev I. Epigenetic modulation of brain gene networks for cocaine and alcohol abuse. Front Neurosci. 2015;9:1–10. doi: 10.3389/fnins.2015.00176. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Friedman AA, Tucker G, Singh R, Yan D, Vinayagam A, Hu Y, Binari R, Hong P, Sun X, Porto M, Pacifico S, Murali T, Finley RL, Jr, Asara JM, Berger B, Perrimon N. Proteomic and functional genomic landscape of receptor tyrosine kinase and ras to extracellular signal–regulated kinase signaling. Science Signaling. 2011;4:rs10–rs10. doi: 10.1126/scisignal.2002029. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ghezzi A, Krishnan HR, Atkinson NS. Susceptibility to ethanol withdrawal seizures is produced by BK channel gene expression. Addic Biol. 2012;19:332–337. doi: 10.1111/j.1369-1600.2012.00465.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gonzalez DA, Jia T, Pinzón JH, Acevedo SF, Ojelade SA, Xu B, Tay N, Desrivières S, Hernandez J, Banaschewski T, Büchel C, Bokde ALW, Conrod PJ, Flor H, Frouin Gallinat J, Garavan H, Gowland PA, Heinz A, Ittermann B, Lathrop M, Martinot J-L, Paus T, Smolka MN, Rodan AR, Schumann G, Rothenfluh A IMAGEN Consortium. The Arf6 activator Efa6/PSD3 confers regional specificity and modulates ethanol consumption in Drosophila and humans. Mol Psychiatry. 2017 doi: 10.1038/mp.2017.112. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Grotewiel M, Bettinger JC. Drosophila and Caenorhabditis elegans as discovery platforms for genes involved in human alcohol use disorder. Alcoholism Clin Exp Res. 2015;39:1292–1311. doi: 10.1111/acer.12785. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Iancu OD, Colville A, Walter NAR, Darakjian P, Oberbeck DL, Daunais JB, Zheng CL, Searles RP, McWeeney SK, Grant KA, Hitzemann R. On the relationships in rhesus macaques between chronic ethanol consumption and the brain transcriptome. Addic Biol. 2017;2:72. doi: 10.1111/adb.12501. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ishmayana S, Fadhlillah M, Kristia YY, Budiman H. Validation of a modified alcohol dehydrogenase assay for ethanol determination. Curr Chem Let. 2015;4:77–84. [Google Scholar]
- Kirienko NV, Fay DS. SLR-2 and JMJC-1 regulate an evolutionarily conserved stress-response network. The EMBO Journal. 2010;29:727–739. doi: 10.1038/emboj.2009.387. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Klose RJ, Kallin EM, Zhang Y. JmjC-domain-containing proteins and histone demethylation. Nature Reviews Genetics. 2006;7:715–727. doi: 10.1038/nrg1945. [DOI] [PubMed] [Google Scholar]
- Krishnan HR, Sakharkar AJ, Teppen TL, Berkel TDM, Pandey SC. Epigenetics, International Review of Neurobiology. Elsevier; 2014. The epigenetic landscape of alcoholism; pp. 75–116. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Krishnan S, Horowitz S, Trievel RC. Structure and function of Histone H3 Lysine 9 methyltransferases and demethylases. ChemBioChem. 2011;12:254–263. doi: 10.1002/cbic.201000545. [DOI] [PubMed] [Google Scholar]
- Lee H-G, Kim Y-C, Dunning JS, Han K-A. Recurring ethanol exposure induces disinhibited courtship in Drosophila. PLoS ONE. 2008;3:e1391. doi: 10.1371/journal.pone.0001391. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lv W-W, Wei H-M, Wang D-L, Ni J-Q, Sun F-L. Depletion of histone deacetylase 3 antagonizes PI3K-mediated overgrowth of Drosophila organs through the acetylation of histone H4 at lysine 16. J Cell Sci. 2012;125:5369–5378. doi: 10.1242/jcs.106336. [DOI] [PubMed] [Google Scholar]
- Marenda DR, Zraly CB, Dingwall AK. The Drosophila Brahma (SWI/SNF) chromatin remodeling complex exhibits cell-type specific activation and repression functions. Developmental Biology. 2004;267:279–293. doi: 10.1016/j.ydbio.2003.10.040. [DOI] [PubMed] [Google Scholar]
- Mathies LD, Blackwell GG, Austin MK, Edwards AC, Riley BP, Davies AG, Bettinger JC. SWI/SNF chromatin remodeling regulates alcohol response behaviors in Caenorhabditis elegans and is associated with alcohol dependence in humans. Proc Natl Acad Sci USA. 2015;112:3032–3037. doi: 10.1073/pnas.1413451112. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Maze I, Chaudhury D, Dietz DM, Schimmelmann Von M, Kennedy PJ, Lobo MK, Sillivan SE, Miller ML, Bagot RC, Sun H, Turecki G, Neve RL, Hurd YL, Shen L, Han M-H, Schaefer A, Nestler EJ. G9a influences neuronal subtype specification in striatum. Nat Neurosci. 2014;17:533–539. doi: 10.1038/nn.3670. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Maze I, Nestler EJ. The epigenetic landscape of addiction. Annals of the New York Academy of Sciences. 2011;1216:99–113. doi: 10.1111/j.1749-6632.2010.05893.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mulligan MK, Ponomarev I, Hitzemann RJ, Belknap JK, Tabakoff B, Harris RA, Crabbe JC, Blednov YA, Grahame NJ, Phillips TJ, Finn DA, Hoffman PL, Iyer VR, Koob GF, Bergeson SE. Toward understanding the genetics of alcohol drinking through transcriptome meta-analysis. Proc Natl Acad Sci USA. 2006;103:6368–6373. doi: 10.1073/pnas.0510188103. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Narayanan AS, Rothenfluh A. I believe I can fly!: use of Drosophila as a model organism in neuropsychopharmacology research. Neuropsychopharmacology. 2016;41:1439–1446. doi: 10.1038/npp.2015.322. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ojelade SA, Acevedo SF, Kalahasti G, Rodan AR, Rothenfluh A. RhoGAP18B isoforms act on distinct Rho-Family GTPases and regulate behavioral responses to alcohol via cofilin. PLoS ONE. 2015;10:e0137465. doi: 10.1371/journal.pone.0137465. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Peru y Colón de Portugal RL, Acevedo SF, Rodan AR, Chang LY, Eaton BA, Rothenfluh A. Adult neuronal Arf6 controls ethanol-induced behavior with Arfaptin downstream of Rac1 and RhoGAP18B. J Neurosci. 2012;32:17706–17713. doi: 10.1523/JNEUROSCI.1944-12.2012. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Peru y Colón de Portugal RL, Ojelade SA, Penninti PS, Dove RJ, Nye MJ, Acevedo SF, Lopez A, Rodan AR, Rothenfluh A. Long-lasting, experience-dependent alcohol preference in Drosophila. Addic Biol. 2014;19:392–401. doi: 10.1111/adb.12105. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ponomarev I. Epigenetic control of gene expression in the alcoholic brain. Alcohol Res. 2013;35:69–76. [PMC free article] [PubMed] [Google Scholar]
- Ponomarev I, Wang S, Zhang L, Harris RA, Mayfield RD. Gene coexpression networks in human brain identify epigenetic modifications in alcohol dependence. J Neurosci. 2012;32:1884–1897. doi: 10.1523/JNEUROSCI.3136-11.2012. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Prescott CA, Kendler KS. Genetic and environmental contributions to alcohol abuse and dependence in a population-based sample of male twins. Am J Psychiatry. 1999;156:34–40. doi: 10.1176/ajp.156.1.34. [DOI] [PubMed] [Google Scholar]
- Qiang M, Denny A, Lieu M, Carreon S, Li J. Histone H3K9 modifications are a local chromatin event involved in ethanol-induced neuroadaptation of the NR2B gene. Epigenetics. 2011;6:1095–1104. doi: 10.4161/epi.6.9.16924. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Robison AJ, Nestler EJ. Transcriptional and epigenetic mechanisms of addiction. Nature Reviews Neuroscience. 2011;12:623–637. doi: 10.1038/nrn3111. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Rothenfluh A, Threlkeld RJ, Bainton RJ, Tsai LTY, Lasek AW, Heberlein U. Distinct behavioral responses to ethanol are regulated by alternate RhoGAP18B isoforms. Cell. 2006;127:199–211. doi: 10.1016/j.cell.2006.09.010. [DOI] [PubMed] [Google Scholar]
- Scholz H, Ramond J, Singh CM, Heberlein U. Functional ethanol tolerance in Drosophila. Neuron. 2000;28:261–271. doi: 10.1016/s0896-6273(00)00101-x. [DOI] [PubMed] [Google Scholar]
- Schumann G, Liu C, O’Reilly P, Gao H, Song P, Xu B, Ruggeri B, Amin N, Jia T, Preis S, Segura Lepe M, Akira S, Barbieri C, Baumeister S, Cauchi S, Clarke T-K, Enroth S, Fischer K, Hällfors J, Harris SE, Hieber S, Hofer E, Hottenga J-J, Johansson Å, Joshi PK, Kaartinen N, Laitinen J, Lemaitre R, Loukola A, Luan J, Lyytikäinen L-P, Mangino M, Manichaikul A, Mbarek H, Milaneschi Y, Moayyeri A, Mukamal K, Nelson C, Nettleton J, Partinen E, Rawal R, Robino A, Rose L, Sala C, Satoh T, Schmidt R, Schraut K, Scott R, Smith AV, Starr JM, Teumer A, Trompet S, Uitterlinden AG, Venturini C, Vergnaud A-C, Verweij N, Vitart V, Vuckovic D, Wedenoja J, Yengo L, Yu B, Zhang W, Zhao JH, Boomsma DI, Chambers J, Chasman DI, Daniela T, de Geus E, Deary I, Eriksson JG, Esko T, Eulenburg V, Franco OH, Froguel P, Gieger C, Grabe HJ, Gudnason V, Gyllensten U, Harris TB, Hartikainen A-L, Heath AC, Hocking L, Hofman A, Huth C, Jarvelin M-R, Jukema JW, Kaprio J, Kooner JS, Kutalik Z, Lahti J, Langenberg C, Lehtimäki T, Liu Y, Madden PAF, Martin N, Morrison A, Penninx B, Pirastu N, Psaty B, Raitakari O, Ridker P, Rose R, Rotter JI, Samani NJ, Schmidt H, Spector TD, Stott D, Strachan D, Tzoulaki I, van der Harst P, van Duijn CM, Marques-Vidal P, Vollenweider P, Wareham NJ, Whitfield JB, Wilson J, Wolffenbuttel B, Bakalkin G, Evangelou E, Liu Y, Rice KM, Desrivières S, Kliewer SA, Mangelsdorf DJ, Müller CP, Levy D, Elliott P. KLBis associated with alcohol drinking, and its gene product β-Klotho is necessary for FGF21 regulation of alcohol preference. Proc Natl Acad Sci USA. 2016;113:14372–14377. doi: 10.1073/pnas.1611243113. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Shalaby NA, Pinzón JH, Narayanan AS, Jin EJ, Ritz MP, Dove RJ, Wolfenberger H, Buszczak M, Rothenfluh A. JmjC domain proteins modulate circadian behaviors in Drosophila. Sci Rep. 2017a doi: 10.1038/s41598-017-18989-1. in revision. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Shalaby NA, Sayed R, Zhang Q, Scoggin S, Eliazer S, Rothenfluh A, Buszczak M. Systematic discovery of genetic modulation by Jumonji histone demethylases in Drosophila. Sci Rep. 2017b;78:5240. doi: 10.1038/s41598-017-05004-w. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Shi Y. Histone lysine demethylases: emerging roles in development, physiology and disease. Nature Reviews Genetics. 2007;8:829–833. doi: 10.1038/nrg2218. [DOI] [PubMed] [Google Scholar]
- Shi Y, Lan F, Matson C, Mulligan P, Whetstine JR, Cole PA, Casero RA, Shi Y. Histone demethylation mediated by the nuclear amine oxidase homolog LSD1. Cell. 2004;119:941–953. doi: 10.1016/j.cell.2004.12.012. [DOI] [PubMed] [Google Scholar]
- Shi Y, Whetstine JR. Dynamic regulation of histone lysine methylation by demethylases. Molecular Cell. 2007;25:1–14. doi: 10.1016/j.molcel.2006.12.010. [DOI] [PubMed] [Google Scholar]
- Subbanna S, Shivakumar M, Umapathy NS, Saito M, Mohan PS, Kumar A, Nixon RA, Verin AD, Psychoyos D, Basavarajappa BS. G9a-mediated histone methylation regulates ethanol-induced neurodegeneration in the neonatal mouse brain. Neurobiol Dis. 2013;54:475–485. doi: 10.1016/j.nbd.2013.01.022. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Topper SM, Aguilar SC, Topper VY, Elbel E, Pierce-Shimomura JT. Alcohol disinhibition of behaviors in C. elegans. PLoS ONE. 2014;9:e92965. doi: 10.1371/journal.pone.0092965. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tulisiak CT, Harris RA, Ponomarev I. DNA modifications in models of alcohol use disorders. Alcohol. 2016 doi: 10.1016/j.alcohol.2016.11.004. in press. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wang K-S, Liu X, Zhang Q, Wu L-Y, Zeng M. Genome-wide association study identifies 5q21 and 9p24.1 (KDM4C) loci associated with alcohol withdrawal symptoms. J Neural Transm. 2012;119:425–433. doi: 10.1007/s00702-011-0729-z. [DOI] [PubMed] [Google Scholar]
- Wolf FW, Rodan AR, Tsai LTY, Heberlein U. High-resolution analysis of ethanol-induced locomotor stimulation in Drosophila. J Neurosci. 2002;22:11035–11044. doi: 10.1523/JNEUROSCI.22-24-11035.2002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zraly CB, Marenda DR, Dingwall AK. SNR1 (INI1/SNF5) mediates important cell growth functions of the Drosophila Brahma (SWI/SNF) chromatin remodeling complex. Genetics. 2004;168:199–214. doi: 10.1534/genetics.104.029439. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.