Abstract
Migratory birds use the Earth’s magnetic field as a source of navigational information. This light-dependent magnetic compass is thought to be mediated by cryptochrome proteins in the retina. Upon light activation, electron transfer between the flavin adenine dinucleotide cofactor and tryptophan residues leads to the formation of a spin-correlated radical pair, whose subsequent fate is sensitive to external magnetic fields. To learn more about the functional requirements of this complex chemical compass, we have created a family of simplified, adaptable proteins—maquettes—that contain a single tryptophan residue at different distances from a covalently bound flavin. Despite the complete absence of structural resemblance to the native cryptochrome fold or sequence, the maquettes exhibit a strong magnetic field effect that rivals those observed in the natural proteins in vitro. These novel maquette designs offer unprecedented flexibility to explore the basic requirements for magnetic sensing in a protein environment.
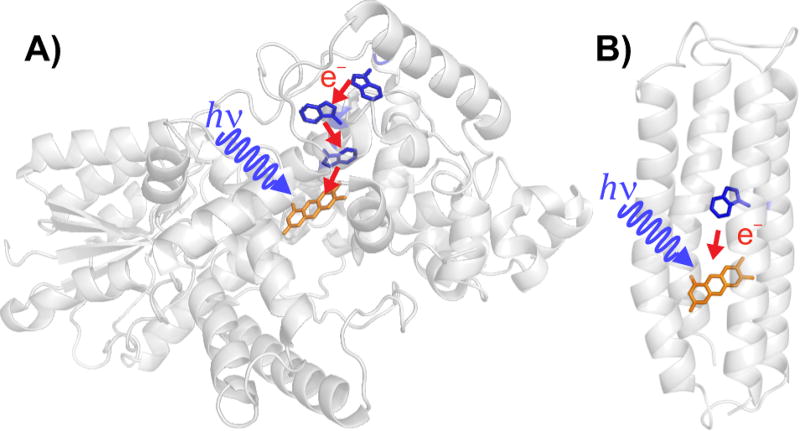
Migratory birds detect the direction of the Earth’s magnetic field (25–65 µT) for the purposes of orientation and navigation on a global scale.1 Recent evidence suggests that this light-dependent magnetic sense is mediated by cryptochromes (Cry), a class of blue-light photoreceptor proteins.2 Mostly α-helical in structure, Crys contain a noncovalently bound flavin adenine dinucleotide (FAD) cofactor and a conserved tryptophan (Trp) triad that acts as an electron transfer chain (Figure 1A).3 Excitation of the FAD with blue light (<500 nm) leads to formation of a radical pair (RP) by electron transfer from the distal Trp to the FAD along the Trp-triad.4
Figure 1.
(A) Drosophila melanogaster Cry. (B) Representation of the W16 flavomaquette demonstrating the secondary structure and the location of the Trp residue (blue) relative to the flavin (orange). Red arrows show the photoinduced electron transfers that produce magnetically sensitive RPs.
Magnetic sensitivity arises because an applied field can influence the timing and extent of interconversion of the singlet (S) and triplet (T) states of the RP.5 Charge recombination of the RP from the T state is spin-forbidden and can only occur from the S state; therefore by altering the instantaneous probability of being S or T, an external field can change the lifetime of the RP and the yields of long-lived reaction products that could act as signaling states. Such magnetic field effects (MFEs) have been observed in Cry and a related protein (DNA photolyase), albeit in fields (1–30 mT) stronger than the geomagnetic field.6
Understanding the fundamental requirements for a magnetic compass protein5,7 has been hindered by the difficulty of obtaining sufficient quantities of the structurally complex natural material for spectroscopic analysis.8 Therefore, much of what is known about the design features of RP compass sensors has come from experiments on simple model systems that, while easier to work with, are far from biologically relevant.9
To determine the biophysical properties required of a protein-based magnetoreceptor, we have designed a series of model proteins, known as maquettes. These are simple, robust, non-natural proteins that can be designed and adapted for specific functions in order to cut through the bewildering complexity of natural systems.10 Maquettes have served as excellent models for understanding the functions of other proteins, including photosystem I and hemoglobin.11 Here we demonstrate the photophysical behavior and magnetic sensitivity of a new class of flavomaquettes designed to reveal the biophysical requirements underlying the formation of a light-activated magnetically sensitive RP similar to that in Cry (Figure S1). These are the first de novo proteins capable of sensing magnetic fields.
The basic flavomaquette fold is a single chain, four-α-helical bundle connected by three hydrophilic loops (Figure 1B).13 Each helix contains 26 amino acids and is binary patterned, with an exposed hydrophilic face and a hydrophobic face buried in the core of the α-helical bundle. For structural simplicity, the scaffold was not designed with a binding pocket for the flavin. Instead, 8-bromo-riboflavin was covalently attached to the maquette using a core cysteine at position 9 on helix 2. A core residue was replaced with Trp on helix 2 at varying distances, either one (0.56 nm, W13), two (1.12 nm, W16), or three (1.68 nm, W20) helical turns away from the site of flavin attachment (Figure S1; distances between β-carbons of the amino acid residues of an α-helix). To avoid unwanted photochemistry, amino acids that could react with flavins were excluded (other than the unique Trp). After attachment of the flavin, the protein molecular weight was ca. 15 kDa, the exterior charge was −12 at pH 7.5, and the pI was 4.22. A flavomaquette with no Trp was developed as a control. The maquettes were expressed with high yield in E. coli; for details see the Supporting Information.
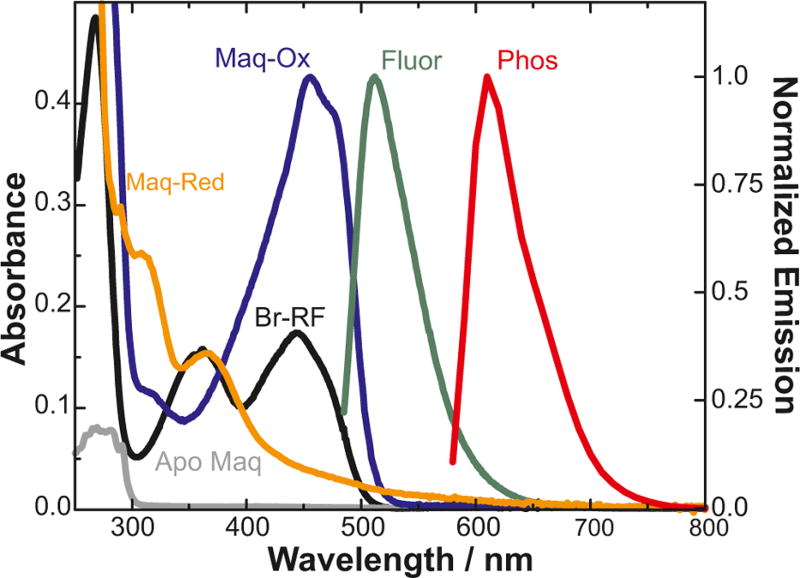
Circular dichroism showed that all four flavinated maquettes are highly helical in structure and thermally stable (Table 1, Figure S4). Covalent attachment of the flavin red-shifts its absorption band from 450 to 475 nm (ε = 24 600 cm−1 mol−1). Two-electron reduction of the flavin results in loss of the feature at 475 nm and formation of a weak absorption band centered at 367 nm (Figure 2). The fluorescence of the covalently bound flavin showed no solvato-chromic shift in different maquette designs, suggesting that the cofactor experiences similar environments (Figure S6). Additionally, the Trp fluorescence maxima are blue-shifted relative to the free amino acid, implying that the Trp residues are located well within the hydrophobic core of the maquette (Table 1). The redox potentials of the flavin and Trp indicate a favorable driving force for light-activated electron transfer in the maquette (Table 1, Figure S8).12 The fluorescence quantum yield is greatest for the Trp-free control and W20 and decreases with decreasing distance between the chromophore and the Trp. This is consistent with time-correlated single photon counting measurements (TCSPC, Figure S7): the initial fluorescence intensity was comparable in all designs, but the fluorescence lifetimes decreased with decreasing flavin-Trp separation. This indicates distance-dependent quenching of the flavin fluorescence in the presence of Trp, consistent with electron transfer involving the excited singlet state of the flavin.
Table 1.
Physical, Spectroscopic, and Magnetic Field-Sensing Properties of the Single-Trp Series of Flavomaquettes
| design |
Tm (°C) |
ΦFa |
τF (ns)b |
Trp Fluor. max. (nm) |
Flavin Emc (mV vs SHE) |
Trp Emc (V vs SHE) |
Predicted ET rate (s−1)d |
MFE (%, 520–610 nm) |
|---|---|---|---|---|---|---|---|---|
| control | 49.0 | 0.11 | 1.150 | – | −110 | – | – | 0 |
| W13 | 50.5 | 0.04 | 0.606 | 335 | −137 | 1.10 | 2.55 × 1011 | +16.0 |
| W16 | 53.2 | 0.07 | 0.846 | 338 | −128 | 1.08 | 1.11 × 108 | +17.5 |
| W20 | 55.3 | 0.10 | 0.974 | 337 | −130 | 1.06 | 4.86 × 104 | 0 |
Quantum yield of flavin fluorescence.
Amplitude-averaged lifetime of flavin fluorescence obtained from a triexponential fit to TCSPC data.
Midpoint potentials.
Rate constants for forward electron transfer to the triplet state flavin predicted using the Moser–Dutton ruler.12
Figure 2.
Absorption spectra of the apo-maquette (gray), oxidized 8-bromo-riboflavin precursor (black), and oxidized (blue) and fully reduced (orange) control flavomaquette. Normalized emission spectra of the same maquette (green, fluorescence; red, phosphorescence) are also shown.
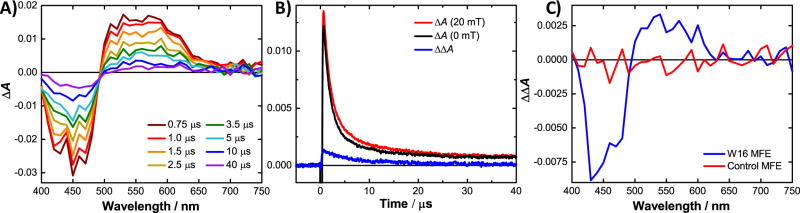
The photochemistry of the maquettes was investigated further by nanosecond transient absorption (TA) spectroscopy. Figure 3A shows the TA spectrum of W16; that of W13 is similar (Figure S14). There is a broad ground state bleach in the range 400–500 nm and a strong excited state absorption at 500–650 nm consistent with the rapid formation (≪1 µs), by photoinduced electron transfer, of a [F•− TrpH•+] RP,6a in which the F•− radical is covalently bound to a protein at the 8-position of the flavin ring system.14 In the control maquette and in W20, only a weak absorption band was observed at 500–800 nm with a lifetime of 17 µs (Figure S14), assigned to the triplet excited state of the flavin.15 Formation of the RP should be possible in W20; however, intraprotein electron tunneling rates usually drop by a factor of ca. 2300 for every 0.56 nm added by the turn of the α-helix.12 Electron transfer in W20 (estimated in Table 1 for a donor–acceptor separation of 3 α-helical turns) is unlikely to compete with other excited state deactivation pathways, and was not observed.
Figure 3.
(A) Transient absorption spectra of ca. 30 µM flavomaquette W16 at 298 K in a pH 7.4 phosphate buffer. (B) Kinetic traces obtained from TA experiments at 520 nm demonstrating the difference (blue) between measurements with the field on (red) and field off (black). Data for other wavelengths may be found in Figure S13. (C) Comparison of the action spectra [ΔΔA = ΔA(20 mT) − ΔA(0)] showing the MFE for W16 and the control at a pump–probe delay of 1 µs.
The flavin radical dominates the transient spectra of W13 and W16 because the extinction coefficients of Cys-bound flavins are substantially higher than those for free flavins. From a comparison with electronically similar flavins (Figure S11), the spectrum in Figure 3A appears to be the result of partial protonation of the flavin radical to form the neutral FH• species around pH 7 (Figure S12). The absorption of the Trp radicals occurs at very similar wavelengths and cannot be resolved independently. However, based on the pKa for TrpH•+ (4.3),16 deprotonation is expected to form the neutral Trp• radical. Both W13 and W16 have similar lifetimes for the RP (Table S3), on the order of several microseconds. This is shorter than observed in Cry proteins,6b but sufficiently long that MFEs can be generated and observed.
To measure the MFE, TA spectra were taken in the presence and absence of a 20 mT magnetic field at 298 K. The action spectra [ΔΔA = ΔA(20 mT) − ΔA(0)] of W16 (Figure 3C) and W13 match the spectral shape of the excited state absorption between 500 and 650 nm. The negative field effect observed in the ground state bleach and positive effect in the excited state absorption are expected if the RP forms via the triplet excited state of the flavin. No MFE was observed for W20, consistent with the absence of RP signals in the TA spectrum. Likewise, the control maquette showed no measurable MFE, further supporting the view that there is no magnetically sensitive intra- or intermolecular chemistry in the absence of a Trp residue.
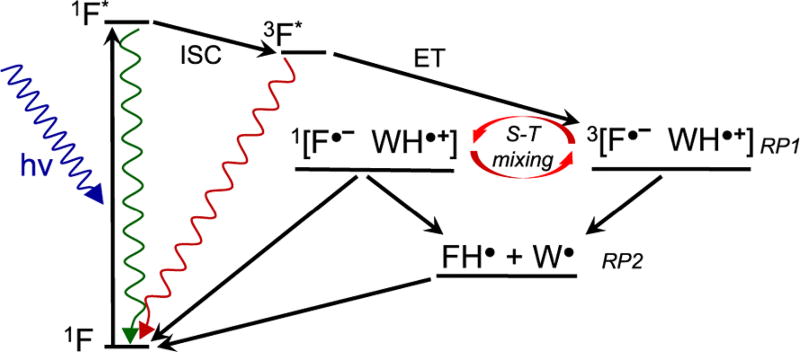
The spectra and MFEs observed for these flavomaquettes are consistent with Scheme 1. After excitation, a fraction of the excited flavin (1F*) undergoes intersystem crossing (ISC) to form the excited triplet state (3F*). The primary spin-correlated RP, denoted RP1, is formed as a triplet by electron transfer from the Trp to the excited triplet flavin. RP1 can undergo S–T interconversion induced by electron–nuclear hyperfine interactions. In the presence of a strong field, the Zeeman interaction removes the degeneracy of the triplet RP energy levels, reduces the efficiency of spin-mixing, and leads to a MFE. Protonation of the flavin radical or deprotonation of the Trp radical creates the secondary radical pair, RP2, observed in our TA measurements, which eventually returns to the ground state.
Scheme 1.
Proposed Photochemistry of the Flavomaquettes Leading to the Observed Triplet-Born MFE
The fractional MFEs [(ΔA(20 mT) − ΔA(0))/ΔA(0)] for W13 and W16 are comparable, but the absolute magnitude [ΔA(20 mT) − ΔA(0)] is smaller for W13 (Figure S14). This probably results from competition between singlet- and triplet-born RP formation in W13, where the smaller flavin-Trp distance leads to a faster forward electron transfer that competes with ISC. Rapid formation of a singlet-born RP is consistent with the increased quenching and shorter fluorescence lifetimes observed by TCSPC. However, the positive MFE indicates that formation of the RP must also occur via the triplet route. Singlet photochemistry is not observed on the microsecond time scale because charge recombination rapidly returns the singlet RPs to the ground state. Charge recombination is initially spin-forbidden for triplet-born RPs which, therefore, have a longer lifetime and contribute more strongly to the MFE.
Triplet-dominated photochemistry in the maquettes is in direct contrast to Cry, which forms an RP from the singlet excited state. This is not surprising: ISC is certainly faster in flavomaquettes than in Cry because of increased spin–orbit coupling due to the Cys sulfur atom attached directly to the flavin ring system. Additionally, for W16 and W20, electron transfer rates are probably slower than those observed in Cry. However, the difference in the spin state of the excited flavin leading to the RP does not affect the ability of the maquettes to serve as a model for elucidating the principles of efficient protein-based sensing of magnetic fields. In fact, triplet-born photochemistry is arguably advantageous for such a simple model system. Given the short distances and large driving force in W13 and W16, the singlet RP is expected to undergo rapid charge recombination. The triplet spin state stabilizes the RP in the maquettes, while natural proteins must rely on physical separation of the charges using an electron transfer chain or conformational changes to achieve the same end. The extension of the RP lifetime in the maquettes due to its triplet spin state allows the observation of a room temperature (298 K) MFE of ~16% at 20 mT on a microsecond time scale, comparable in magnitude to that of Arabidopsis thaliana Cry-1 or E. coli photolyase (12–17% at 28 mT).6b Such large MFEs in natural systems are only observed at reduced temperatures (278 K and below) and in the presence of large amounts of glycerol.
This work represents the first report of an artificial protein-based magnetic sensor. The ease of production and modification of maquettes make them an ideal model system for investigating the basic requirements of protein-based magnetoreception. Developing a thorough understanding of these principles would be an important step toward the elucidation of the biophysical mechanism of the avian magnetic compass. Future work will focus on applying cavity-enhanced and transient absorption spectroscopies17 to these novel proteins to fully resolve the kinetic and spectra properties of the RP. These early findings are being used to design maquettes that contain two and three Trp residues, with the intention of creating a light-activated electron transport chain akin to that found in photolyases and cryptochromes.
Supplementary Material
Acknowledgments
We are grateful to the following for financial support: the European Research Council (under the European Union’s 7th Framework Programme, FP7/2007-20013/ERC Grant Agreement No. 340451 to P.J.H.), and the Air Force Office of Scientific Research (Air Force Materiel Command, USAF Award No. FA9550-14-1-0095 to P.J.H., C.R.T., and S.R.M.). C.B. is grateful to the NIH for a graduate fellowship (T32 GM008275 - Structural Biology & Molecular Biophysics Training Program). We thank G. A. Logsdon for help with molecular biology, S. A. Vinogradov for the use of his phosphorimeter and useful discussions, and T. Troxler of the Ultrafast Optical Processes Laboratory at the University of Pennsylvania (NIH Resource Grant P41-104605) for use of his TCSPC equipment.
Footnotes
ASSOCIATED CONTENT
- Detailed materials and methods, HPLC, mass spectroscopy, peptide sequence, circular dichroism, spectroelectrochemistry, fluorescence, TCSPC, and transient absorption data including pH dependence, kinetic traces, and spectra and MFEs for the control, W13, and W20 (PDF)
The authors declare no competing financial interest.
References
- 1.Mouritsen H. Magnetoreception in Birds and Its Use for Long-Distance Migration. In: Scanes CG, editor. Sturkie’s Avian Physiology. 6. Elsevier; New York: 2015. pp. 113–133. [Google Scholar]
- 2.(a) Liedvogel M, Mouritsen H. J. R. Soc., Interface. 2010;7:S147–S162. doi: 10.1098/rsif.2009.0411.focus. [DOI] [PMC free article] [PubMed] [Google Scholar]; (b) Hore PJ, Mouritsen H. Annu. Rev. Biophys. 2016;45:299–344. doi: 10.1146/annurev-biophys-032116-094545. [DOI] [PubMed] [Google Scholar]
- 3.Chaves I, Pokorny R, Byrdin M, Hoang N, Ritz T, Brettel K, Essen L-O, van der Horst GTJ, Batschauer A, Ahmad M. Annu. Rev. Plant Biol. 2011;62:335–364. doi: 10.1146/annurev-arplant-042110-103759. [DOI] [PubMed] [Google Scholar]
- 4.Immeln D, Weigel A, Kottke T, Lustres JLP. J. Am. Chem. Soc. 2012;134:12536–12546. doi: 10.1021/ja302121z. [DOI] [PubMed] [Google Scholar]
- 5.Rodgers CT, Hore PJ. Proc. Natl. Acad. Sci. U. S. A. 2009;106:353–360. doi: 10.1073/pnas.0711968106. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.(a) Henbest KB, Maeda K, Hore PJ, Joshi M, Bacher A, Bittl R, Weber S, Timmel CR, Schleicher E. Proc. Natl. Acad. Sci. U. S. A. 2008;105:14395–14399. doi: 10.1073/pnas.0803620105. [DOI] [PMC free article] [PubMed] [Google Scholar]; (b) Maeda K, Robinson AJ, Henbest KB, Hogben HJ, Biskup T, Ahmad M, Schleicher E, Weber S, Timmel CR, Hore PJ. Proc. Natl. Acad. Sci. U. S. A. 2012;109:4774–4779. doi: 10.1073/pnas.1118959109. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Brocklehurst B. Chem. Soc. Rev. 2002;31:301–311. doi: 10.1039/b107250c. [DOI] [PubMed] [Google Scholar]
- 8.Evans EW, Dodson CA, Maeda K, Biskup T, Wedge CJ, Timmel CR. Interface Focus. 2013;3:20130037. doi: 10.1098/rsfs.2013.0037. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Maeda K, Henbest KB, Cintolesi F, Kuprov I, Rodgers CT, Liddell PA, Gust D, Timmel CR, Hore PJ. Nature. 2008;453:387–390. doi: 10.1038/nature06834. [DOI] [PubMed] [Google Scholar]
- 10.Moser CC, Sheehan MM, Ennist NM, Kodali G, Bialas C, Englander MT, Discher BM, Dutton PL. Methods Enzymol. 2016;580:365–388. doi: 10.1016/bs.mie.2016.05.048. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.(a) Lichtenstein BR, Bialas C, Cerda JF, Fry BA, Dutton PL, Moser CC. Angew. Chem. Int. Ed. 2015;54:13626–13629. doi: 10.1002/anie.201507094. [DOI] [PMC free article] [PubMed] [Google Scholar]; (b) Goparaju G, Fry BA, Chobot SE, Wiedman G, Moser CC, Dutton PL, Discher BM. Biochim. Biophys. Acta Bioenerg. 2016;1857:503–512. doi: 10.1016/j.bbabio.2015.12.002. [DOI] [PMC free article] [PubMed] [Google Scholar]; (c) Koder RL, Anderson JLR, Solomon LA, Reddy KS, Moser CC, Dutton PL. Nature. 2009;458:305–309. doi: 10.1038/nature07841. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Page CC, Moser CC, Chen X, Dutton PL. Nature. 1999;402:47–52. doi: 10.1038/46972. [DOI] [PubMed] [Google Scholar]
- 13.Farid TA, Kodali G, Solomon LA, Lichtenstein BR, Sheehan MM, Fry BA, Bialas C, Ennist NM, Siedlecki JA, Zhao Z, Stetz MA, Valentine KG, Anderson JLR, Wand AJ, Discher BM, Moser CC, Dutton PL. Nat. Chem. Biol. 2013;9:826–833. doi: 10.1038/nchembio.1362. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Raibekas AA, Fukui K, Massey V. Proc. Natl. Acad. Sci. U. S. A. 2000;97:3089–3093. doi: 10.1073/pnas.040559597. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Mansurova M, Simon J, Salzmann S, Marian CM, Gärtner W. ChemBioChem. 2013;14:645–654. doi: 10.1002/cbic.201200670. [DOI] [PubMed] [Google Scholar]
- 16.Posener ML, Adams GE, Wardman P, Cundall RB. J. Chem. Soc., Faraday Trans. 1. 1976;72:2231–2239. [Google Scholar]
- 17.(a) Neil SRT, Li J, Sheppard DMW, Storey J, Maeda K, Henbest KB, Hore PJ, Timmel CR, Mackenzie SR. J. Phys. Chem. B. 2014;118:4177–4184. doi: 10.1021/jp500732u. [DOI] [PubMed] [Google Scholar]; (b) Maeda K, Neil SRT, Henbest KB, Weber S, Schleicher E, Hore PJ, Mackenzie SR, Timmel CR. J. Am. Chem. Soc. 2011;133:17807–17815. doi: 10.1021/ja206783t. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.