Abstract
The genus Paracoccidioides comprises known fungal pathogens of humans and can be isolated from different infection sites. Metabolic peculiarities in different members of the Paracoccidioides led us to perform proteomic studies in the presence of the two-carbon molecule acetate, which predominates in the nutrient-poor environment of the phagosome. To investigate the expression rates of proteins of different members of Paracoccidioides, including one isolate of P. lutzii (Pb01) and three isolates of P. brasiliensis (Pb03, Pb339, and PbEPM83), using sodium acetate as a carbon source, proteins were quantified using label-free and data-independent liquid chromatography-mass spectrometry. Protein profiles of the isolates were statistically analyzed, revealing proteins that were differentially expressed when the fungus was cultivated in a non-preferential carbon source rather than glucose. A total of 1,160, 1,211, 1,280, and 1,462 proteins were reproducibly identified and relatively quantified in P. lutzii and the P. brasiliensis isolates Pb03, Pb339, and PbEPM83, respectively. Notably, 526, 435, 744, and 747 proteins were differentially expressed among P. lutzii and the P. brasiliensis isolates Pb03, Pb339, and PbEPM83, respectively, with a fold-change equal to or higher than 1.5. This analysis revealed that reorganization of metabolism occurred through the induction of proteins related to gluconeogenesis, glyoxylic/glyoxylate cycle, response to stress, and degradation of amino acids in the four isolates. The following differences were observed among the isolates: higher increases in the expression levels of proteins belonging to the TCA and respiratory chain in PbEPM83 and Pb01; increase in ethanol production in Pb01; utilization of cell wall components for gluconeogenesis in Pb03 and PbEPM83; and increased β-oxidation and methylcitrate cycle proteins in Pb01and PbEPM83. Proteomic profiles indicated that the four isolates reorganized their metabolism in different manners to use acetate as a carbon source.
Keywords: Paracoccidioides spp., proteomic, two-carbon source, sodium acetate, metabolism
Introduction
The genus Paracoccidioides includes fungal pathogens causing paracoccidioidomycosis, a disease geographically restricted to subtropical areas of Latin America (San-Blas et al., 2002). Approximately 80% of reported cases of paracoccidioidomycosis occurred in Brazil, followed by Colombia, Venezuela, and Argentina (Martinez, 2015). This genus comprises thermally dimorphic fungi that grow in a hyphal form in the environment, but exist as budding yeast in mammalian hosts. Infection is initiated by inhalation of fungal propagules, which differentiate into the yeast form after reaching the alveolar epithelium (Restrepo et al., 2001).
Multilocus sequencing led to the proposal of four cryptic studies in the genus Paracoccidioides: S1, PS2, and PS3 in the P. brasiliensis complex and P. lutzii, a single monophyletic population (Matute et al., 2006a; Teixeira et al., 2009). Additionally, a new lineage PS4 was proposed, belonging to a region in Venezuela (Teixeira et al., 2014). Recently, the S1 group was divided into lineages S1a and S1b. Dating time analysis to determine the separation of these lineages indicated that S1b was the earliest diverging lineage of P. brasiliensis (Muñoz et al., 2016).
The genomes of Paracoccidioides have been widely examined. A reduced number of genes was found to be involved in the metabolism of carbohydrates and proteins, as well as in the synthesis of secondary metabolites (Desjardins et al., 2011; Muñoz et al., 2014). Additionally, the genomes of 31 isolates representing lineages S1, PS2, PS3, and PS4 were sequenced, providing new reference genomes for two lineages from PS3 and PS4 (Muñoz et al., 2016). Further functional analysis can provide more data regarding fungus biology.
To grow, a pathogen must assimilate carbon and have metabolic flexibility to assimilate available nutrients in different host niches. Data from transcriptional and proteomic profiles are consistent with this assumption. It has been shown that Candida albicans requires isocitrate lyase (ICL), a key enzyme for virulence, in the glyoxylate cycle (Lorenz and Fink, 2001). Because ICL is repressed by glucose, its expression in the host is restricted to the nutrient-limited environment of the phagosome of immune cells (Barelle et al., 2006). Paracoccidioides regulate their genes by using different carbon sources. Upon carbon starvation, P. lutzii yeast cells shift the metabolism to gluconeogenesis and ethanol production, which is supported by the degradation of amino acids and fatty acids and by modulation of the glyoxylate and tricarboxylic cycles (Lima et al., 2014). During macrophage infection, P. brasiliensis positively regulates proteins involved in alternative carbon metabolism, such as those related to gluconeogenesis, beta oxidation, and amino acids catabolism (Parente-Rocha et al., 2015). During the initial stages of lung infection, P. brasiliensis alters the expression of genes related to several functional categories including energy metabolism and cell wall metabolism. Paracoccidioides brasiliensis remodels cellular lipid metabolism to catabolize its own lipid stores via beta oxidation and the glyoxylate cycle, which are strongly induced during the first 6 h of lung infection (Pigosso et al., 2017).
Proteomic characterization of Paracoccidioides was performed by 2D electrophoresis and mass spectrometry. Comparison of the protein expression profiles from yeast cells using glucose as a carbon source revealed different metabolic aspects among members of four phylogenetic species (Pigosso et al., 2013). Paracoccidioides lutzii preferentially used anaerobic pathways for energy production; P. lutzii and P. brasiliensis (Pb2 and Pb339) showed a better response to reactive oxygen species (ROS) compared to PbEPM83, and Pb2 showed a higher abundance of enzymes from the pentose phosphate pathway compared to the others. Additionally, antigenic proteins were differentially accumulated (Pigosso et al., 2013). Results of comparative proteomic analyses were confirmed by biochemical assays (Pigosso et al., 2013). The hierarchical clustering of proteins reflects the phylogenetic relationships among the studied Paracoccidioides species, where S1 is an independent species (Pb339), PS3 (PbEPM83) is phylogenetically closer to S1 than PS2 (Pb2), and P. lutzii (Pb01) is highly divergent from the other phylogenetic species (Matute et al., 2006a,b; Teixeira et al., 2009).
Macrophages are important components of the innate immune system and include neutrophils, which are front-line phagocytes that eliminate pathogens. During infection by Paracoccidioides spp., alveolar macrophages constitute one of the primary defense mechanisms (Brummer et al., 1988). It is generally thought that the phagosome is a nutrient-poor environment (Haas, 2007) whose carbon sources are derived from the breakdown of fatty acids via beta-oxidation, producing acetyl-CoA (Lorenz and Fink, 2002).
Metabolic peculiarities in different members of the Paracoccidioides genus, as described above, led us to investigate their metabolism in the presence of acetate, a two-carbon molecule. Acetate utilization requires tricarboxylic acid (TCA) shunting through the glyoxylate cycle, which allows the utilization of two-carbon sources. In this study, we examined Pb339 (ATCC 200273), Pb03, PbEPM83, and P. lutzii (ATCC-MYA-826) (Carrero et al., 2008; Teixeira et al., 2009) to investigate the metabolic utilization of two-carbon sources by these organisms.
Materials and methods
Strains and growth conditions
Paracoccidioides brasiliensis Pb339 (ATCC 200273), Pb03, PbEPM83 (Matute et al., 2006a), and P. lutzii (ATCC-MYA-826) (Carrero et al., 2008; Teixeira et al., 2009) were used in this study. Fungi were maintained in brain heart infusion (BHI) medium supplemented with 4% (w/v) glucose at 36°C to cultivate the yeast form. After 3 days of incubation in BHI liquid medium with agitation at 150 rpm, the yeast cells were washed twice with 1X phosphate-buffered saline (1X PBS; 1.4 mM KH2PO4, 8 mM Na2HPO4, 140 mM NaCl, 2.7 mM KCl; pH 7.2) and inoculated in chemically defined McVeigh/Morton medium (Restrepo and Jiménez, 1980). Glucose (100 mM) (control) or sodium acetate (100 mM) (test) was added to the defined medium as carbon sources and the cells were grown for 48 h at 36°C with agitation at 150 rpm.
ICL activity
Following Paracoccidioides Pb01, Pb03, Pb339, and PbEPM83 growth in the presence of acetate and glucose (both 100 mM), the cells were centrifuged at 1,500 × g, resuspended in a solution containing 20 mM Tris-HCl, pH 8.8, and 2 mM CaCl2 (Fonseca et al., 2001) and disrupted using glass beads and bead beater apparatus (BioSpec, Bartlesville, OK, USA) for 5 cycles of 30 s each while on ice. The cell lysate was centrifuged at 10,000 × g for 15 min at 4°C and the supernatant was quantified using Bradford reagent (Sigma-Aldrich, St. Louis, MO, USA) (Bradford, 1976). The amount of protein used was 50 μg (Lima et al., 2014).
ICL activity was determined by measuring the formation of glyoxylate as its phenylhydrazone derivative (Ebel et al., 2006). Glyoxylate-phenylhydrazone formation was determined by measuring the absorbance at 324 nm with an extinction coefficient of 16.8 mM−1 cm−1 in a reaction mixture containing 2 mM threo-D,L-isocitrate (Sigma Aldrich), 2 mM MgCl2, 10 mM phenylhydrazine HCl (Sigma Aldrich), 2 mM dithiothreitol, and 50 mM potassium phosphate at pH 7.0. Specific activity was determined as the amount of enzyme required to form 1 μmol of glyoxylate- phenylhydrazone per min per mg of total protein. Statistical comparisons were performed using the Student's t-test and p-values ≤ 0.05 were considered statistically significant.
Paracoccidioides spp. cell dry weight assay
Yeast cells of Paracoccidioides spp. were grown as described above. A total of 106 cells/50 mL was inoculated into chemically defined McVeigh/Morton medium (Restrepo and Jiménez, 1980) containing the carbon sources glucose or sodium acetate followed by incubation at 36°C. At each time-point, 10 mL aliquots of the culture were centrifuged at 1,500 × g and the supernatants were carefully removed. The cells were resuspended in 1X PBS up to 500 μL and incubated at 95°C for 1 h. The cells were centrifuged, frozen in liquid nitrogen, and lyophilized for 24 h (Lima et al., 2014). Data are expressed as the mean ± standard deviation of the triplicate independent experiments. Statistical comparisons were performed using Student's t-test, and a p-value ≤ 0.05 was considered significant.
Obtaining protein extracts
To obtain the cytoplasmic protein extract, the cells were centrifuged at 10,000 × g for 5 min, followed by washing with 1X PBS twice and the addition of Tris-Ca buffer (20 mM Tris–HCl pH 8.8; 2 mM CaCl2). This suspension was distributed in tubes containing an equal volume of glass beads (425–600 μm) to the volume of the cell pellet. The cells were disrupted by vigorous mixing in a bead beater apparatus (BioSpec) for 5 cycles for 30 s on ice. The cell lysate was centrifuged at 10,000 × g for 15 min at 4°C until no pellet formed. Protein content in the supernatant was quantified using Bradford reagent (Bradford, 1976) using bovine serum albumin as a standard.
Protein digestion for NanoUPLC-MSE analysis
Proteins were enzymatically digested as described previously (Murad et al., 2011) with some modifications. Briefly, approximately 150 μg of protein (previous item) was added to 10 μL of 50 mM ammonium bicarbonate, pH 8.5, in a microcentrifuge tube. Next, 75 μL of RapiGESTTM SF Surfactante (0.2% v/v) (Waters Corporation, Billerica, MA, USA) was added and the sample was vortexed and incubated in a dry bath at 80°C for 15 min. Disulfide bonds were reduced by adding 2.5 μL of 100 mM dithiothreitol (GE Healthcare, Little Chalfont, UK) at 60°C for 30 min, while cysteines were alkylated by 2.5 μL of 300 mM iodoacetamide (GE Healthcare) for 30 min at room temperature in the dark. The proteins were subsequently digested with 30 μL of trypsin 0.05 μg/μL (Promega, Madison, WI, USA) at 37°C in dry bath for 16 h. To precipitate the RapiGEST, samples were acidified with 30 μL of a 5% (v/v) trifluoracetic acid solution (Sigma-Aldrich) and the mixture was incubated for 90 min at 37°C in a dry bath, followed by centrifugation at 18,000 × g at 4°C for 30 min. The supernatants were dried in a speed vacuum (Eppendorf, Hamburg, Germany). All obtained peptides were suspended in 80 μL of solution containing 20 mM of ammonium formate and 80 fmol/μL of PHB (Rabbit Phosphorylase B) (Waters Corporation) (MassPREPTM protein) as an internal standard. Samples were transferred to a Waters Total Recovery vial (Waters Corporation). The samples were then placed in an auto-sampler and stored at 4°C for nano-ultra-high-performance liquid chromatography-mass spectrometry analysis.
Mass spectrometry
Mass spectrometric experiments were performed on a Synapt G2-Si High Definition Mass Spectrometer (Waters Corporation) equipped with mass analyzers such as a hybrid quadrupole/ion mobility mass spectrometry/orthogonal acceleration time-of-flight MS geometry, and coupled to nanoAcquityTM UPLC system (Waters Corporation). The peptide mixture was loaded for 5 min at a flow rate of 8 μL/min for phase A (0.1% formic acid) using a Symmetry C18 trapping column (5 μm particles, 180 μm × 20 mm length; Waters Corporation). The mixture of trapped peptides was subsequently separated by elution over a gradient of 7–35% of phase B (0.1% formic acid in acetonitrile) through a BEH 130 C18 column (1.7 μm particles, 75 × 150 mm; Waters Corporation) in 93 min at 350 nL/min. For each measurement, the mass spectrometer was operated in resolution mode with an m/z resolving power of approximately 40,000 full width at half maximum using ion mobility with a cross-section resolving power of at least 40 Ω/ΔΩ. LC–MS multiplex data were collected using ion mobility enhanced MSE. Data were acquired in data-independent mode (UDMSE) (Distler et al., 2014) over an m/z range of 50–2,000 in resolution mode. The exact mass retention time signals from multiplexed ion-mobility DIA scanning (UDMSE) were collected in alternating low energy and elevated energy acquisition modes. In the low energy mode, data were collected at 6 eV. In the elevated collision energy, quasi m/z-specific collision energies were applied to the different drift time bins to fragment precursor ions prior to orthogonal acceleration time-of-flight analysis, applied to the transfer traveling-wave, collision-induced dissociation cell filled with argon gas with a total cycle time scan of 0.5 s (Souza et al., 2017). Nano electrospray ionization in the positive mode (nanoESI+) source was operated with a capillary voltage of 2.75 kV, block temperature of 70°C, and cone voltage of 30 V. For lock mass correction, [Glu1]-Fibrinopeptide B solution (100 fmol/ml in 50% V/V water and acetonitrile, 0.1% V/V formic acid was infused through the reference sprayer at 300 nL/min and sampled every 30 s (Garrido et al., 2016; Abreu et al., 2017). The peptides fractions were analyzed in triplicates.
Data processing and protein identification
Mass spectrometry raw data of peptide fractions were loaded in Progenesis QI for Proteomics (Nonlinear Dynamics). A reference run for the triplicates was automatically selected as the default. Precursor ion retention times were processed for alignment, peak picking, and normalized to a reference run with default parameters. The MS data were processed by the Apex3D and Peptide3D module using a low-energy threshold of 150 counts, elevated energy threshold of 50 counts, and intensity threshold of 750 counts. Databank searches were performed using Progenesis QIP and the databank of Paracoccidioides was used with decoy proteins (http://www.broadinstitute.org/annotation/genome/paracoccidioides_brasiliensis/Multiome.tml). The database was chosen according to the isolated sample set, together with reverse sequences: (i) Pb339 (S1) and PbEPM83 (PS3): P. brasiliensis Pb18 protein sequences; (ii) Pb03 (PS2): P. brasiliensis (strain Pb03) protein sequences; (iii) P. lutzii (Pb01) protein sequences.
The mass error tolerance for peptide identification was less than 10 ppm. Protein identification criteria included: (i) the detection of at least 2 fragment ions per peptide, (ii) 5 fragments per protein, (iii) the determination of at least 1 unique (proteotypic) peptide per protein, (iv) carbamidomethylation of cysteine as a fixed modification, (v) phosphorylation of serine, threonine, and tyrosine, and oxidation of methionine were considered as variable modifications, (vi) maximum protein mass (600 kDa), (vii) one missed cleavage site was allowed for trypsin, and (viii) maximum false positive ratio of 4% was allowed. Peptide ion data were exported from Progenesis QIP as *.csv files and edited in Excel (Microsoft®). Only features detected above an intensity threshold of 150 and in at least 2 of 3 replicates were considered for further analysis. To compare the ratios between the control (glucose) and test (sodium acetate), proteins displaying at least 50% differences in expression values compared to the control were considered as regulated. The Uniprot (http://www.uniprot.org) and Pedant on MIPS () database were used for functional classification. The NCBI database was employed to annotate uncharacterized proteins (https://www.ncbi.nlm.nih.gov/).
Data treatment
Quantitative data on the expression of each protein was converted to a logarithmic scale (log2) for analysis purposes. For each protein, the significance of the effects associated with isolates, carbon sources, and interaction between these two factors was evaluated by analysis of variance (ANOVA) using the factorial model, followed by the Tukey test. All comparisons were performed using the critical significance level of 0.05. All analyses were performed in R (R Core Team, 2017) software.
The Multi Experiment Viewer software V.4.8 (http://www.tm4.org) was used to group comparative proteomic data. To analyze the isolates, the up- and down-regulated proteins were combined and the expression levels estimated for each protein were used to build a heat map. Differences in proteomic profiles were examined by hierarchical clustering using Pearson correlation as a measure of similarity.
Ethanol quantification assay
Ethanol concentration was determined using an enzymatic detection kit according to the manufacturer's instructions (UV-test for ethanol, RBiopharm, Darmstadt, Germany). Briefly, ethanol was oxidized to acetaldehyde by the enzyme alcohol dehydrogenase in the presence of nicotinamide-adenine dinucleotide (NAD). Acetaldehyde was quantitatively oxidized to acetic acid in the presence of aldehyde dehydrogenase, releasing NADH, which was determined by measuring the absorbance at 340 nm. Paracoccidioides Pb01, Pb03, Pb339, and PbEPM83 yeast cells were cultivated in minimal media containing acetate (100 mM) or glucose (100 mM), and 106 cells were assayed. Briefly, the cells were centrifuged and lysed using glass beads and a bead beater apparatus (BioSpec) in 5 cycles of 30 s while keeping the samples on ice. The cell lysate was centrifuged at 10,000 × g for15 min at 4°C and the supernatant was used for enzymatic assays according to the manufacturer's instructions. The concentrations of ethanol were determined in triplicate. Statistical analysis was performed using Student's t-test, and p ≤ 0.05 was considered significant.
Analysis of glucans by fluorescence microscopy
To evaluate the beta glucan content in the cell wall, staining was performed with aniline blue solution 100% (v/v) (Sigma, catalog no. B8563). Yeast cells of different isolates, cultured in the presence of acetate or glucose, were incubated with aniline blue for 5 min while stirring and subsequently washed twice with 1X PBS (Renshaw et al., 2016; De Curcio et al., 2017). Samples stained with aniline blue were visualized under a fluorescence microscope (Zeiss Axiocam MRc-Scope A1, Oberkochen, Germany). A minimum of 100 cells on each microscope slide were used to evaluate fluorescence intensity in triplicate. The software determined the fluorescence intensity (in pixels) and standard error of each analysis. Statistical comparisons were performed using the Student's t-test and p ≤ 0.05 was considered statistically significant.
Macrophage infection assays
The survival of the Paracoccidioides isolates was determined by quantifying the number of colony-forming units (CFUs) recovered from macrophage infection. J774 1.6 macrophages (Rio de Janeiro Cell Bank–BCRJ/UFRJ, accession number 0273 were employed). Macrophages were maintained in RPMI medium (RPMI 1640, Vitrocell, Brazil) with 10% FBS ([v/v]) and MEM non-essential amino acid solution (Sigma Aldrich), at 36°C and 5% CO2 until the cells were completely confluent. The phagocytosis assay was performed in 12-well polypropylene plates (Greiner Bio-One, Kremsmünster, Austria). A total of 106 J774 macrophages were plated per well in RPMI medium containing IFN-γ (1 U/mL) (Sigma Aldrich) and incubated for 24 h at 36°C and 5% CO2 for adherence and activation. Prior to co-cultivation, Paracoccidioides yeast cells were grown in BHI liquid medium (4% [w/v] glucose, 3.7% [w/v] brain heart infusion, pH 7.2) for 48 h. Next, 5 × 106 yeast cells per well were added to the macrophages, giving a yeast:macrophage cell ratio of 5:1. The cells were incubated for 24 h at 36°C and 5% CO2 in RPMI medium containing IFN-γ (1 U/mL). Non-phagocytosed/non-adhered yeast were washed 3 times with PBS (Parente-Rocha et al., 2015). Macrophages were lysed with water and fungal cells were recovered. The number of viable cells was determined based on the number of CFUs. The lysates were plated in BHI medium supplemented with 5% FBS [v/v]. CFUs were determined after growth at 36°C in 5% CO2 for 10 days. Data were expressed as the mean value deviation from triplicate measurements and the statistical analyses were performed using ANOVA.
Results and discussion
ICL activity
To determine the adequate time of exposure of the isolates to sodium acetate for the proteomic assays, the enzymatic activity of ICL was determined in the four isolates at different time intervals (18, 24, 48, and 72 h). Increased ICL activity was observed in all four isolates at 48 h, indicating activation of the glyoxylic/glyoxylate cycle; thus, this time was used for proteomic analysis (Figure 1). ICL is an enzyme exclusively involved in the glyoxylic/glyoxylate cycle, an anaplerotic carbon pathway that deviates in the decarboxylation steps of the TCA cycle, to produce malate and succinate using acetyl-CoA when the production of pyruvate from glycolysis is reduced (Lorenz and Fink, 2002; Fleck et al., 2011). Studies showed that ICL is an indicator of the glyoxylic/glyoxylate cycle because it allows the formation of glyoxylate from the cleavage of isocitrate, which becomes the substrate for malate synthase. ICL and other enzymes of the glyoxylate cycle are induced under conditions such as low glucose levels and low oxygen tension (Wayne and Lin, 1982; Fernandez et al., 1993) in the presence of carbon sources such as acetate and ethanol and at high temperatures (Bowyer et al., 1994). In P. brasiliensis, Pb01 carbon starvation promoted up-regulation of ICL expression, indicating up-regulation of the glyoxylic/glyoxylate cycle in yeast cells after 48 h of treatment (Lima et al., 2014). The enzymatic activity of ICL was reduced when glucose was used as a carbon source in relation to acetate in yeast cells of P. brasiliensis Pb01 (Cruz et al., 2011) and was increased under carbon starvation conditions for 48 h (Lima et al., 2014). ICL activity can also be used as an indicator of the glyoxylic/glyoxylate cycle.
Figure 1.
Isocitrate lyase (ICL) activity assay. Activity was determined by measuring the formation of glyoxylate as its phenylhydrazone derivative under each condition. A total of 50 mg of each total protein extract of Paracoccidioides isolates Pb01, Pb03, Pb339, and PbEPM83 yeast cells were cultivated in minimal media with sodium acetate (100 mM) and glucose (100 mM) for different times (18, 24, 48, and 72 h). The specific activity of ICL was determined as the amount of enzyme required to form 1 μmol of glyoxylate-phenylhydrazone per minute, per mg of total protein, and represented as U·mg−1. Errors bars represent standard deviation from three biological replicates while * represents p ≤ 0.05.
Growth of P. lutzii and isolates of P. brasiliensis in the presence of glucose and sodium acetate as carbon sources
Yeast cells of P. lutzii (Pb01) and of P. brasiliensis Pb03, Pb339, and PbEPM83 were grown in the presence of glucose and sodium acetate. Growth was evaluated by dry weight analysis at several time intervals. For all isolates, growth was significantly reduced in the presence of sodium acetate compared to glucose, as depicted in Supplementary Figure 1. The lower growth in acetate likely reflects the lower content of proteins extracted under this condition, as follows: Pb01 glucose (14.6 μg/μL), Pb01 acetate (10.76 μg/μL); Pb03 glucose (10.22 μg/μL), Pb03 acetate (8.82 μg/μL); Pb339 glucose (8.6 μg/μL), Pb339 acetate (7.04 μg/μL); PbEPM83 glucose (11.14 μg/μL), and PbEPM83 acetate (10.78 μg/μL). Similar results were observed in Escherichia coli grown on acetate or glucose (Treitz et al., 2016).
Proteome of P. lutzii and of P. brasiliensis isolates in the presence of sodium acetate
We identified differentially expressed proteins among P. lutzii (Pb01) and three members of the species P. brasiliensis, as following: Pb03, Pb339, and PbEPM83. The proteins identified in the four analyzed groups of the genus Paracoccidioides are depicted in Supplementary Table 1. A total of 1,160, 1,211, 1,280, and 1,462 proteins were identified in P. lutzii (Pb01), Pb03, Pb339, and PbEPM83, respectively, after filtering using the criterion of minimum repeat rate of 2 and a total of 5112 proteins (Supplementary Table 1).
The proteins in each isolate that grew in the presence of acetate were compared to the counterparts in yeast cells growing in the presence of glucose. Statistical analysis revealed differential protein expression in each isolate in relation to the carbon source used. Based on the differential protein expression results, the different isolates were compared using the heat map. There were 526, 442, 744, and 569 differentially expressed proteins showing fold-changes of at least 1.5, for P. lutzii (Pb01) and Pb03, Pb339, and PbEPM83, respectively, as depicted in Supplementary Table 1. The number of differentially expressed proteins when comparing the four isolates to the others were 45.3, 36.5, 58.1, and 38.9%, respectively of the total of proteins identified in each isolate, for P. lutzii (Pb01) and Pb03, Pb339, and PbEPM83. These data indicate that high metabolic flexibility exists among members of the Paracoccidioides genus. Similar results were observed by Pigosso et al. who found significant differences in the protein expression profiles among the representatives of different phylogenetic clades of Paracoccidioides Pb01, Pb02, Pb339, and PbEPM83 (Pigosso et al., 2013).
Differentially regulated proteins in analyzed Paracoccidioides isolates
Based on statistical analysis, the differentially expressed proteins were classified. Supplementary Tables 2–9 present the proteins of the analyzed isolates significantly up- or down-regulated upon growth with the two-carbon molecule acetate. Those proteins were classified according to the functional categories present in FunCat2. The functional categories for up- and down-regulated proteins were similar among the four analyzed isolates, as depicted in Supplementary Figures 2A,B, respectively. Some proteins were assigned as unclassified because of their unknown function, accounting for 20–30% of the analyzed proteins in the isolates (data not shown).
The metabolic pathways, glycolysis/gluconeogenesis and ethanol production, cell wall metabolism, glyoxylic/glyoxylate shunt and the TCA cycle, beta-oxidation and the methylcitrate cycle, electron transport chain and oxidative phosphorylation, amino acids degradation, and response to oxidative stress were analyzed among isolates, as depicted in Supplementary Table 10.
Regulation of glycolysis/gluconeogenesis and ethanol production
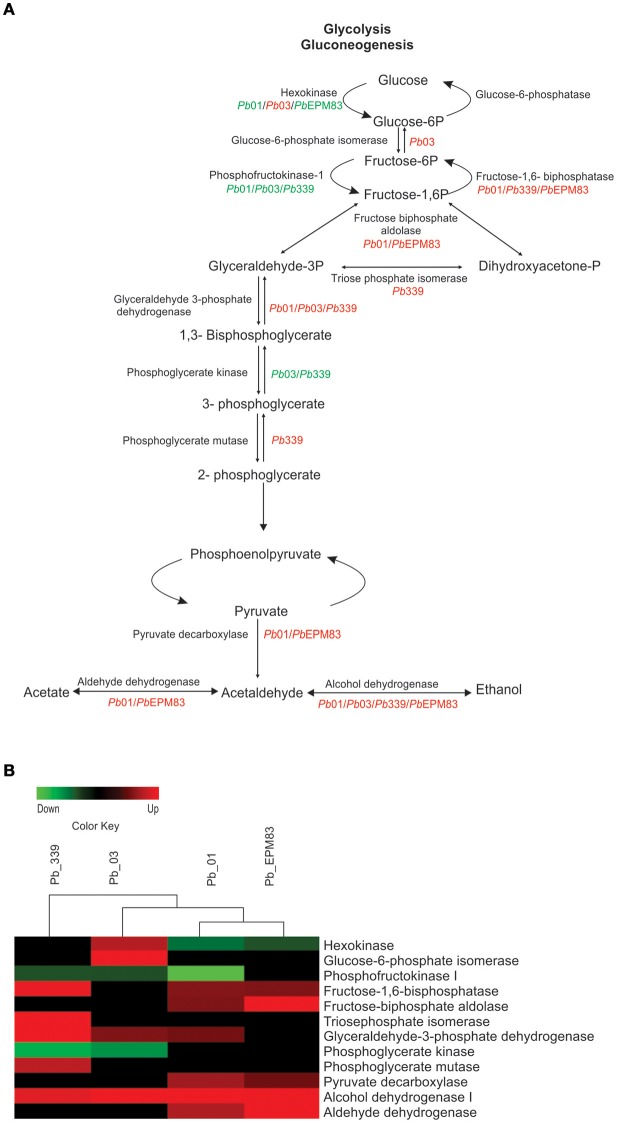
To assess changes in the expression of proteins related to glycolysis/gluconeogenesis in the isolates, we compared the levels of enzymes listed in Supplementary Tables 2–9. As depicted in Figures 2A,B, fructose 1,6-biphosphatase (PAAG_02682; PADG_01706) was significantly induced in Pb01, Pb339, and PbEPM83, suggesting increased gluconeogenesis. Regulatory enzymes of glycolysis, phosphofructokinase-I (PAAG_01583; PABG_03640; PADG_00192) and hexokinase (PADG_03813), were repressed in Pb01, Pb03, Pb339, and PbEPM83. The reciprocal regulation of phosphofructokinase-I and fructose 1,6-biphosphatase clearly indicate increased gluconeogenesis and decreased glycolysis in the isolates of P. lutzii and P. brasiliensis, as depicted in Figures 2A,B. A shift to gluconeogenesis has been described as a metabolic hallmark of fungal cells exposed to two-carbon sources. Studies of the response of C. albicans to internalization by macrophages revealed reprogramming of transcription, including gluconeogenic growth (Lorenz et al., 2004).
Figure 2.
Comparison of protein profiles related to glycolysis and gluconeogenesis in P. lutzii and isolates of P. brasiliensis. ANOVA was applied to compare expression values among isolates, applying a cut-off of 1.5-fold. Expression data were obtained using the Multi Experiment Viewer software V.4.8, which was used to group and compare expression data. (A) Representative diagram of the glycolysis/gluconeogenesis and fermentation pathways depicting down-regulated (green) and up-regulated (red) proteins in the isolates, as cited above. (B) Changes in expression levels in yeast cells incubated with acetate in the four analyzed isolates are represented in a heat map format. Mean values of experimental triplicates are shown for down-regulation (green) and up-regulation (red) of genes of isolates Pb01, Pb03, Pb339, and PbEPM83 in the presence of sodium acetate. Black indicates that no significant difference was observed.
Three enzymes involved in ethanol production were differentially expressed in the presence of acetate. Pyruvate decarboxylase (PAAG_02050; PADG_00714) and aldehyde dehydrogenase (PAAG_03910) were induced in Pb01 and PbEPM83 and alcohol dehydrogenase (PAAG_00403; PABG_04316; PADG_11405) was induced in all studied isolates. Acetate may provide ethanol because of the increase in aldehyde dehydrogenase and alcohol dehydrogenase. A high level of enzymes involved in ethanol production was observed, particularly for Pb01 and PbEPM83, which presumably produces ethanol from pyruvate, acetate, and acetaldehyde, as depicted in Figures 2A,B. Ethanol measurement revealed that up to 48 h in presence of sodium acetate, a significantly higher level of ethanol was produced compared to in cells cultured in glucose in all isolates, confirming the proteomic analysis data (Figure 3). In P. lutzii during carbon starvation, enzymes related to the production of ethanol were induced, such as alcohol dehydrogenase and pyruvate decarboxylase and ethanol production was increased in cells cultured with glucose (Lima et al., 2014). Comparative proteomic studies among isolates Pb01, Pb2 Pb339, and PbEPM83 in nutrient-rich media revealed a high level of ethanol production in Pb01 compared to in the other isolates, suggesting differential production of ethanol among isolates (Pigosso et al., 2013). Ethanol production has been related to fungi virulence. The influence of alcohol dehydrogenase in fungal pathogenesis was observed in invasive pulmonary aspergillosis with an increase in the inflammatory response in the lung of mice infected with an alcohol dehydrogenase null mutant strain, and reduction in fungal burden (Grahl et al., 2011).
Figure 3.
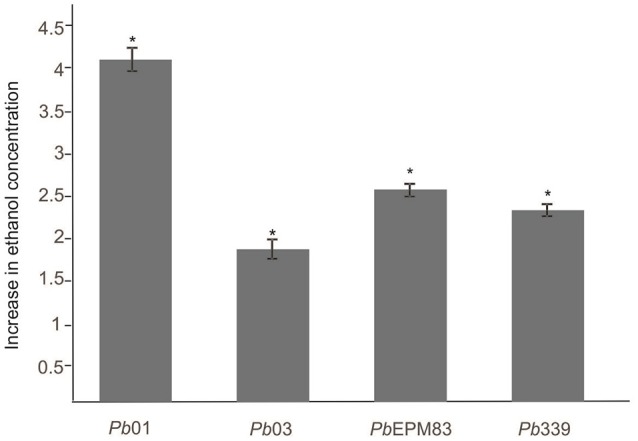
Ethanol measurement in P. lutzii and P. brasiliensis. The concentration of ethanol (g/L) was determined in yeast cells of the isolates Pb01, Pb03, Pb339, and PbEPM83 upon growth in minimal media with sodium acetate (100 mM) and glucose (100 mM) for 48 h. A total of 106 cells was used for each sample, and the ethanol compound was quantified using an enzymatic detection kit (UV-test for ethanol, RBiopharm, Darmstadt, Germany). Data are expressed as the mean ± standard deviation of biological triplicates in independent experiments. The data were analyzed by two-way ANOVA and Tukey's multiple comparisons test. *Indicate significant differences among isolates at p-value of ≤0.05.
Regulation of cell wall metabolism
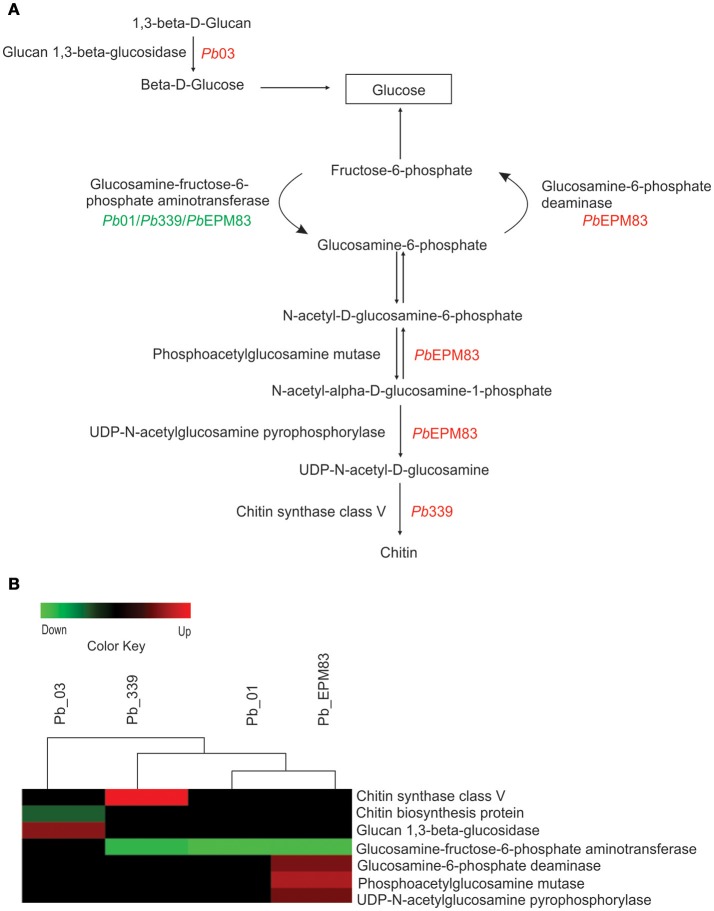
Figure 4 depicts differences in the expression of cell wall metabolic enzymes in the presence of acetate in the analyzed isolates. Only Pb03 showed up-regulated glucan 1,3-beta-glucosidase, which catalyzes successive hydrolysis of the beta-D-glucose units from the non-reducing ends of (1-3)-beta-D-glucans, releasing beta glucose. Aniline blue, which selectively stains 1,3-beta-D-glucan, was employed to estimate the amount of this polymer in the cell wall, as depicted in Supplementary Figure 3. As depicted in Supplementary Figure 3A, fluorescence was visibly reduced in Pb03 grown with acetate. Quantitative analyses of the fluorescence intensity (in pixels) in yeast cells of isolate Pb03 in the presence of sodium acetate showed a significant decrease in fluorescence in cells (p ≤ 0.05), Supplementary Figure 3B, strongly suggesting a decrease in the glucan content, as indicated by proteomic analysis. Glucosamine-fructose-6-phosphate aminotransferase (PAAG_00850; PADG_03984) was down-regulated in PbEPM83, Pb01, and Pb339. This enzyme catalyzes the formation of glucosamine 6-phosphate and is the first and rate-limiting enzyme in the hexosamine biosynthetic pathway controlling the flux of glucose into the hexosamine pathway. The final product of the hexosamine pathway, UDP-N-acetyl-glucosamine, is an active precursor of numerous macromolecules containing amino sugars, including chitin in fungi and arthropods (Badet et al., 1987). In our analysis, only PbEPM83 showed up-regulation of the enzymes glucosamine-6-phosphate-deaminase (PADG_00401), phosphoacetylglucosamine mutase (PADG_00604), and UDP-N-acetylglucosamine pyrophosphorylase (PADG_04312). These enzymes participate in amino sugar metabolism and cell wall biosynthesis. Phosphoacetylglucosamine mutase reversibly converts N-acetyl-alpha-D-glucosamine-1-phosphate to N-acetyl-D-glucosamine-6-phosphate. The products from this reaction are substrates to the enzymes glucosamine-6-phosphate-deaminase and UDP-N-acetylglucosamine pyrophosphorylase, respectively. The first converts glucosamine-6-phosphate and H2O to NH3 and fructose-6-phosphate, which is used in glycolysis and gluconeogenesis; the second converts N-acetyl-alpha-D-glucosamine-1-phosphate to UDP-N-acetyl-D-glucosamine, which is used to produce chitin for the cell wall. Down-regulation of glucosamine: fructose-6-phosphate-aminotransferase (PADG_03984) and up-regulation of glucosamine-6-phosphate-deaminase (PADG_03984) suggests that PbEPM83 mainly uses glucosamine-6-phosphate for glucose production. These results suggest that PbEPM83 can use polymeric carbohydrates precursors to obtain glucose by gluconeogenesis and synthesize chitin polymer into the cell wall.
Figure 4.
Comparison of protein profiles related to cell wall metabolism in P. lutzii and isolates of P. brasiliensis. Dataset comparisons were carried out as cited in the Figure 2 legend. (A) Representative diagram of the cell wall metabolism pathways depicting down-regulated (green) and up-regulated (red) proteins in the isolates, as cited above. (B) Changes in expression levels upon yeast cells incubated with acetate in Pb01, Pb03, Pb339, and PbEPM83 are represented in a heat map format. Black indicates that no significant difference was observed.
Regulation of the glyoxylic/glyoxylate shunt and TCA cycle
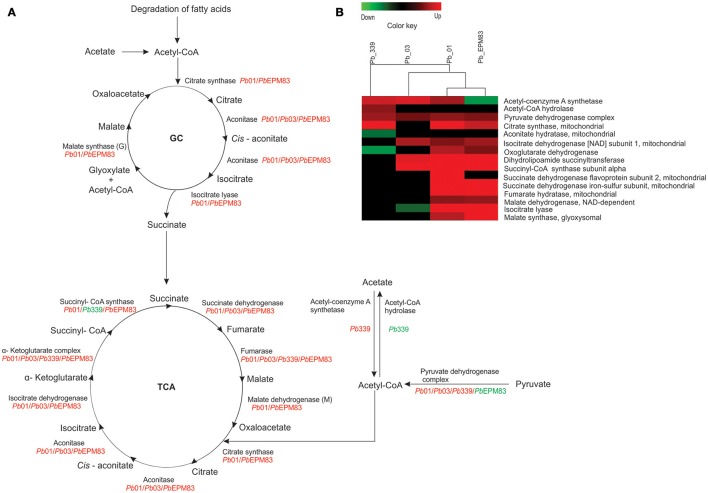
Acetyl-CoA synthase (PADG_01677) and pyruvate dehydrogenase (PAAG_11035; PABG_03494; PADG_07213) can promote the synthesis of acetyl-CoA from acetate and pyruvate, respectively, as depicted in Figures 5A,B. The major fate of acetyl-CoA is the TCA cycle, in which most of enzymes were induced in the analyzed isolates, mainly in Pb01 and PbEPM83 (Figures 5A,B). The glyoxylate shunt, which is a shortcut for the decarboxylation steps of the TCA cycle, can allow the use of acetyl-CoA in the synthesis of cellular components (Chung et al., 1988). ICL (PAAG_06951; PADG_01483) and malate synthase (PAAG_04542; PADG_04702) were increased in isolates Pb01 and PbEPM83. This aspect is of relevance, as ICL and malate synthase allow the use of two-carbon compounds from the glyoxylic/glyoxylate cycle. Induction of transcripts encoding ICL and malate synthase was observed in phagocytized C. albicans upon macrophage infection, enhancing the importance of acquiring non-preferential carbon sources and glyoxylate cycle function in glucose synthesis (Lorenz and Fink, 2001). Thus, activation of this metabolic pathway enables the pathogen to survive and persist in the host in tissues with limited nutrients such as inside macrophages. As described above, the glyoxylic/glyoxylate cycle is induced in fungi and bacteria during phagocytosis as a response to nutrient deprivation in the phagolysosome (Brock, 2009). Genes encoding the enzymes of the glyoxylate cycle, ICL and malate synthase, were induced in C. albicans upon phagocytosis by murine macrophages (Lorenz et al., 2004) and human neutrophils (Fradin et al., 2015). This metabolic pathway enables the fungus to utilize two-carbon molecules as a carbon source, and the induction of this pathway likely reflects the nutrient-deprived environment inside phagocytes (Miramón et al., 2012). The role of the glyoxylate cycle in virulence has been described in organisms, such as Aspergillus fumigatus (Olivas et al., 2007), C. albicans (Lorenz and Fink, 2001, 2002), and Mycobacterium tuberculosis (McKinney et al., 2000; Munõz-Elias and McKinney, 2005).
Figure 5.
Comparison of protein profiles related to TCA and glyoxylate cycle in P. lutzii and isolates of P. brasiliensis. Dataset comparisons were carried out as cited in Figure 2 legend. (A) Representative diagram of the TCA cycle and glyoxylate cycle depicting down-regulated (green) and up-regulated (red) proteins in the isolates. (B) Changes in expression levels upon yeast cells incubated with acetate in Pb01, Pb03, Pb339, and PbEPM83 are represented in a heat map format. Black indicates that no significant difference was observed.
Regarding the genus Paracoccidioides, the induction of ICL was observed in Pb01 during the transition from mycelium to yeast cells (Bastos et al., 2007) and in yeast cells recovered from the kidneys of infected mice (Costa et al., 2007). The glyoxylate cycle was positively regulated in yeast cells of P. lutzii (Felipe et al., 2005) and the level of malate synthase transcript was higher in yeast cells in the presence of a two-carbon source in relation to glucose (Zambuzzi-Carvalho et al., 2009). Increased expression of ICL was also observed in Pb01 from 6 to 12 h under carbon starvation (Lima et al., 2014).
Regulation of β-oxidation and methyl citrate cycle
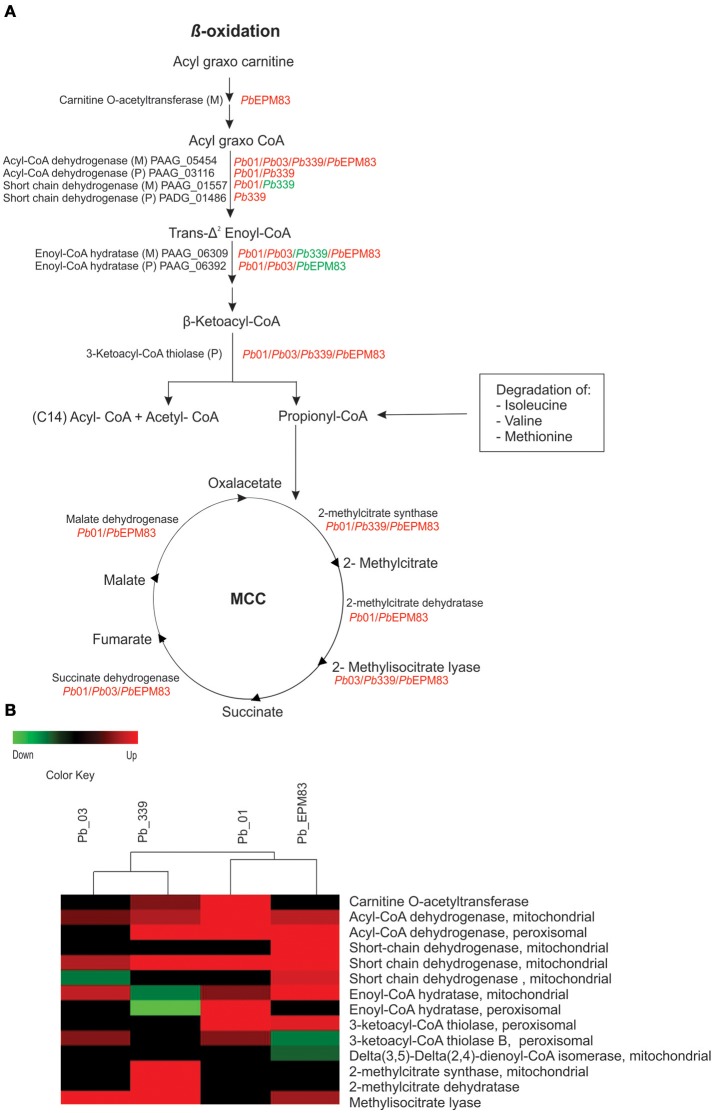
In general, the beta-oxidation of fatty acids mainly functions in Pb01 and PbEPM83 (PAAG_06329; PAAG_02664; PAAG_05454; PAAG_03116; PAAG_06309; PAAG_06392; PAAG_01557; PADG_01228; PADG_01687; PADG_06805; PADG_07023; PADG_06721; PADG_01209; PADG_02527), producing acetyl-CoA and propionyl-CoA. Acetyl-CoA can be consumed in the glyoxylate cycle for biosynthetic purposes or in the TCA cycle to generate cellular energy as reduced cofactors. Beta-oxidation was induced in the mitochondria and peroxisomes, as shown in Figure 6, with the up-regulation of enzymes in both compartments. Carnitine O-acetyl transferase (PADG_07023), which is related to the entry of acyl fatty acid into the mitochondria, and acylCoA dehydrogenase mitochondrial (PAAG_05454; PABG_01791; PADG_06805), which catalyzes the initial step in each cycle of fatty acid β-oxidation, were up-regulated in Pb01, Pb03, Pb339, and PbEPM83, suggesting higher fatty acid degradation in those isolates in the presence of acetate.
Figure 6.
Comparison of protein profiles related to β-oxidation and methyl citrate cycle in P. lutzii and isolates of P. brasiliensis. Dataset comparisons were carried out as cited in Figure 2, legend. (A) Representative diagram of the β-oxidation and methyl citrate cycle pathways depicting down-regulated (green) and up-regulated (red) proteins in the isolates (B) Changes in expression levels in yeast cells incubated with acetate in Pb01, Pb03, Pb339, and PbEPM83 are represented in a heat map format. Black indicates that no significant difference was observed.
Enzymes levels in the methyl citrate cycle were induced or maintained, as shown in Figure 6, indicating that this cycle was active in the presence of acetate. Notably, all identified enzymes in the cycle were increased in Pb339 (PADG_04709; PADG_04710; PADG_04718; PADG_05281). Methyl citrate synthase is a key enzyme in the methyl citrate cycle and is essential for the degradation of propionyl-CoA in fungi and some bacteria, avoiding accumulation of propionyl, which is toxic (Ibrahim-Granet et al., 2008). The accumulation of propionyl in Aspergillus nidulans and A. fumigatus inhibits growth and secondary metabolism, and occurs when there are defects in the enzyme methyl citrate synthase (Brock and Buckel, 2004; Zhang et al., 2004; Maerker et al., 2005). Methyl citrate synthase is essential in A. fumigatus for the manifestation of the invasive properties of aspergillosis, suggesting that amino acids serve as nutritional support for the growth and invasion of A. fumigatus (Ibrahim-Granet et al., 2008).
Electron transport chain and oxidative phosphorylation
Additionally, in PbEPM83, the electron transport chain and ATP synthase complex were predominantly induced but no classes of proteins were repressed, as depicted in Supplementary Figure 4. Proteins of the different respiratory chain complexes were differentially regulated among isolates (Supplementary Figure 4). PbEMP83 showed the highest number of induced respiratory chain proteins/enzymes, whereas in Pb01, most components of the electron transport system and membrane-associated energy conservation were not induced, corroborating the results of Pigosso and colleagues who demonstrated that P. lutzii (Pb01) is metabolically more anaerobic than P. brasiliensis (Pigosso et al., 2013).
Regulation of amino acids degradation
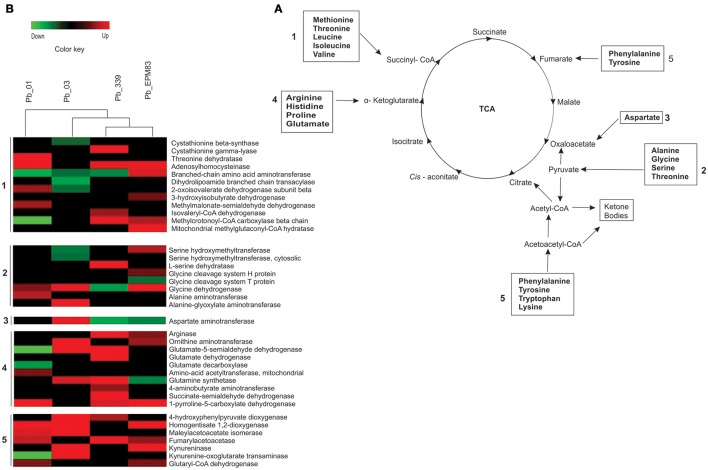
The degradation of amino acids providing precursors for the TCA cycle, pyruvate, or acetyl-CoA was predominantly induced in the analyzed isolates (Figure 7). Higher production of succinyl-CoA appeared to occur in Pb01 from the degradation of methionine and threonine because of the induction of adenosyl homocysteinase (PAAG_02859) and threonine dehydratase (PAAG_03168) (Figure 7 and Supplementary Figure 5). Glyoxylate production from the degradation of glycine was observed in Pb01, Pb03, and PbEPM83 with the induction of the enzyme glycine dehydrogenase (PAAG_1568; PABG_04990) (Figure 7 and Supplementary Figure 5). Pb01 also induced alanine aminotransferase (PAAG_08207), producing pyruvate from alanine. Notably, Pb03 appeared to induce the interconversion of alanine and glyoxylate to pyruvate because of the up-regulation of alanine glyoxylate aminotransferase (PABG_00589), allowing pyruvate synthesis from the transamination of alanine and glyoxylate synthesized by the glyoxylate cycle (Figure 7 and Supplementary Figure 5). Pb03 may be the most efficient producer of oxaloacetate from asparagine because aspartate aminotransferase (PABG_02806) was up-regulated in this strain (Figure 7 and Supplementary Figure 5).
Figure 7.
Comparison of protein profiles related to amino acid catabolism in P. lutzii and isolates of P. brasiliensis. Dataset comparisons were carried out as cited in Figure 2 legend. (A) Changes in expression levels in yeast cells incubated with acetate in Pb01, Pb03, Pb339, and PbEPM83 are represented in a heat map format. (B) Representative diagram of the amino acid catabolism pathways depicting down-regulated (green) and up-regulated (red) proteins in the isolates, as cited above. Black indicates that no significant difference was observed.
The degradation of phenylalanine and tyrosine to acetoacetyl-CoA was expected to be very similar in all isolates because they upregulate at least two enzymes in the pathway (Figure 7; Supplementary Figure 6). The degradation of tryptophan rendering acetyl-CoA may be higher in PbEPM83, and Pb03 because of up-regulation of kyneurinase (PADG_00349; PABG_01965) (Figure 7 and Supplementary Figure 6).
The conversion of arginine and ornithine to alpha-ketoglutarate appeared to predominate in Pb339, Pb03, and PbEPM83, as the enzymes in this pathway such as arginase (PADG_00637) and ornithine aminotransferase (PABG_02827; PADG_01328) were up-regulated in the three isolates (Figure 7 and Supplementary Figure 7).
The isolates PbEPM83 and Pb339 may efficiently produce acetyl-CoA from leucine because of the induction of the enzyme methylcrotonyl-CoA carboxylase (PADG_07370) in both isolates, as depicted in Figure 7 and Supplementary Figure 8. Related to this pathway, the enzymes isovaleryl-CoA dehydrogenase (PADG_07369) and methylglutaconyl-CoA hydratase (PADG_00643) were induced in Pb339 and PbEPM83, respectively. However, Pb01 and Pb03 may be less efficient in producing acetoacetate from leucine, while the enzymes in this pathway were not up-regulated and the methylcrotonyl-CoA carboxylase (PAAG_04103) was repressed (Figure 7 and Supplementary Figure 8). The conversion of valine and isoleucine to succinyl-CoA may be more predominant in PbEPM83 and Pb01 because of up-regulation of the enzymes acylCoA dehydrogenase (PAAG_05454; PADG_06805) and enoyl-CoA hydratase (PAAG_06309; PADG_01209) in both (Figure 7, Supplementary Figure 8), as well as the up-regulation of 2-oxovalerate dehydrogenase (PAAG_01194) and methylmalonate-semialdehyde dehydrogenase (PAAG_07036) in Pb01 and 3-hydroxyisobutyrate dehydrogenase (PADG_03466) and branched-chain amino acid aminotransferase (PADG_04570) in PbEPM83 (Figure 7, Supplementary Figure 8).
The catabolism of amino acids is highly induced in Paracoccidioides. In the presence of a non-preferential carbon source, there was a metabolic shift to the catabolism of amino acids to produce TCA intermediates. Similar results were obtained in E. coli, in which acetate as the carbon source activates amino acid degradation (Treitz et al., 2016). Upon nutrient deprivation, C. albicans utilizes amino acids as the main carbon source, leading to ammonia excretion which increases the environmental pH and contributes to morphogenesis (Vylkova et al., 2011). Paracoccidioides lutzii, upon carbon deprivation, activates the degradation of amino acids to provide precursors for gluconeogenesis (Lima et al., 2014). Additionally, upon phagocytosis, P. brasiliensis induces the enzymes glutamate dehydrogenase, alanine glyoxylate aminotransferase, and aspartate aminotransferase, indicating the importance of the catabolism of amino acids for obtaining glucose precursors and energy in the harsh environment of the phagolysosome (Parente-Rocha et al., 2015).
Regulation of stress response
We observed the induction of several proteins and enzymes related to ROS detoxification in pathogenic microorganisms in the different isolates of P. brasiliensis and P. lutzii when yeast cells were cultured in the presence of sodium acetate, as depicted in Figure 8. Detoxifying molecules are involved in the pathogenesis and virulence of pathogens in phagocytes (Enjalbert et al., 2007; Youseff et al., 2012). In all analyzed isolates, we detected increases in superoxide dismutases (SODs), Cu/Zn-containing SOD1 (PAAG_04164; PABG_03954; PADG_07418), Fe/Mn-containing SOD2 (PAAG_02725; PABG_03204; PADG_01755), SOD5 (PAAG_02926; PABG_03387; PADG_01954), and peroxisomal catalase (PAAG_01454; PADG_00324; PABG_01943). SOD is a primary antioxidant defense against ROS which dismutates the superoxide radical () into molecular oxygen and H2O2 (Turrens, 2003). It has been previously suggested that one of the mechanisms used by P. brasiliensis for eliminating ROS is the expression of proteins from the antioxidant system, such as SOD enzymes, to neutralize superoxide and convert them into less harmful molecules such as hydrogen peroxide and oxygen molecules (Campos et al., 2005; de Arruda Grossklaus et al., 2013). The Paracoccidioides genome database lists six SOD isoforms. Analysis of gene expression between phylogenetic lineages of Paracoccidioides revealed the induction of SOD1 and SOD3 during the transition from mycelium-to-yeast, exposure to oxidative agents, and interaction with phagocytic cells (Tamayo et al., 2016). Catalase and peroxidase catabolize H2O2. Paracoccidioides isolates encode three catalases, PbCatA, PbCatP, and PbCatB, which catalyze the decomposition of H2O2 into oxygen and water (Chagas et al., 2008; Tamayo et al., 2017). CATP was up-regulated in all isolates of Paracoccidioides (Figure 8). Cytochrome C peroxidase (PABG_00720) and glutathione peroxidase (PABG_04219) were increased only in Pb03 compared to the other isolates upon incubation when acetate was the only carbon source. It was previously demonstrated that Pb01, Pb339, and PbEPM83 are less resistant to incubation with menadione compared to Pb02, which belongs to the same group of Pb03 evaluated in this study (Pigosso et al., 2013; Muñoz et al., 2016), corroborating our results. In proteomic analysis, during infection of macrophages, P. brasiliensis induced the expression of gamma glutamyltranspeptidase, cytochrome c peroxidase, Cu/Zn SOD, and thioredoxins. Cytochrome C peroxidase knockdown mutant strains show reduced survival upon macrophage interactions and during infection in mouse (Parente-Rocha et al., 2015). All isolates analyzed in this study showed similar survival rates upon macrophage phagocytosis (Supplementary Figure 9), suggesting that they possess equivalent mechanisms for avoiding oxidative stress caused by phagocytes.
Figure 8.
Comparison of protein profiles related to detoxification mechanisms against oxidative stress in P. lutzii and isolates of P. brasiliensis. Dataset comparisons were carried out as cited in Figure 2 legend. (A) Changes in expression levels in yeast cells incubated with acetate in Pb01, Pb03, Pb339, and PbEPM83 are represented in a heat map format. (B) Representative diagram of the detoxification mechanisms against oxidative stress pathways depicting down-regulated (green) and up-regulated (red) proteins in the isolates as cited above. Black indicates that no significant difference was observed. Glutathione (GSH); glutathione reductase (GR); glutathione disulfide (GSSG).
Induced processes in P. lutzii and P. brasiliensis isolates
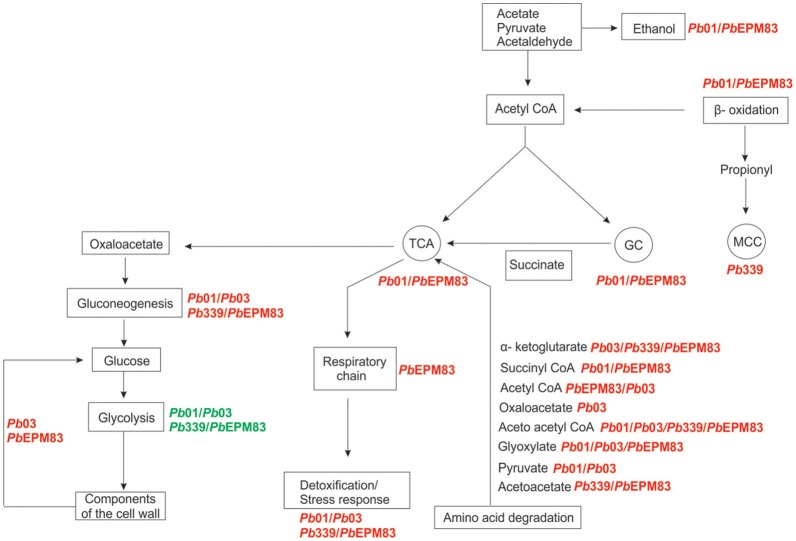
By comparing the metabolic pathways that reflect carbon flux distribution, preferential metabolic strategies can be proposed for the analyzed Paracoccidioides isolates in the presence of acetate as the only carbon source. Figure 9 depicts the variations of in vivo carbon flux distribution of enzymes involved in central metabolism. Gluconeogenesis is thought to occur at similar rates in P. brasiliensis and P. lutzii because of similar induction of the enzymes in the pathway (Figure 2). Pb03 and PbEPM83 are thought to use components or intermediates of cell wall catabolism to produce glucose, as depicted in Figures 4, 9. Pb01 and PbEPM83 may increase the glyoxylate cycle and TCA cycle to a greater extent than Pb03 and Pb339, as depicted in Figures 5, 9. β-Oxidation predominates in Pb01 and PbEPM83, as depicted in Figures 5, 9. The methyl citrate cycle predominates in Pb339, as shown in Figures 6, 9. Regarding the oxidative phosphorylation and ATP synthesis, PbEPM83 showed greater induction than other isolates of the Paracoccidioides genus. The catabolism of amino acids producing intermediates of the TCA cycle was prominent among the isolates. Pb03 and PbEPM8 were the most efficient producers of precursors to for the TCA cycle from amino acid metabolism. For the detoxification process, all analyzed isolates in the Paracoccidioides genus can detoxify H2O2 through catalase P. Pb03 showed induction of glutathione peroxidase and cytochrome C peroxidase, which may account for the higher potential for eliminating ROS (Figures 8, 9). Taken together, we suggest that PbEPM83 uses aerobic metabolism, including the TCA cycle and glyoxylate shunt, to produce ATP through oxidative phosphorylation.
Figure 9.
Overview of metabolic features of members of the Paracoccidioides genus in sodium acetate as carbon source. The metabolic pathways were identified by analyzing up-regulated proteins in P. lutzii (Pb01) and P. brasiliensis Pb03, Pb339, and PbEPM83. The main metabolic pathways were represented: gluconeogenesis, fermentation, cycles of the tricarboxylic acid (TCA), glyoxylate (GC) and methyl citrate (MCC), respiratory chain, β- oxidation, amino acid degradation, detoxification/response to stress, and cell wall components.
Concluding remarks
Paracoccidioides respond to the presence of acetate in the growth medium by increasing the expression of systems devoted to utilization of this compound, which requires changes in the abundance of a large number of proteins. Although this general metabolic reprogramming which includes a shift to the glyoxylate shunt induced gluconeogenesis and catabolism of amino acids, it was possible to determine the quantitative and qualitative differences in the metabolism of different Paracoccidioides species when acetate was the only carbon source. Whether these differential changes reflect the biological fitness of Paracoccidioides within host niches remains unclear.
Author contributions
CdAS conceived and finalized the manuscript. LB, FdM, and LP performed the experiments. GdS carried out proteomic data. AC carried out statistical analyzes. LB, FdM, LP, MP, and CdAS designed the study, discussed, analyzed, interpreted the data and writing the manuscript.
Conflict of interest statement
The authors declare that the research was conducted in the absence of any commercial or financial relationships that could be construed as a potential conflict of interest. The reviewer EL-R and handling Editor declared their shared affiliation.
Acknowledgments
This work at Universidade Federal de Goiás was supported by grants from Conselho Nacional de Desenvolvimento Científico e Tecnológico (CNPq), Fundação de Amparo à Pesquisa do Estado de Goiás (FAPEG), and Programa Nacional de Pós Doutorado - Coordenação de Aperfeiçoamento de Pessoal de Nível Superior (PNPD-CAPES). This work is part of the INCT program of Strategies of Host Pathogen Interaction (HPI).
Supplementary material
The Supplementary Material for this article can be found online at: https://www.frontiersin.org/articles/10.3389/fmicb.2017.02308/full#supplementary-material
References
- Abreu T. F., Sumitomo B. N., Nishiyama M. Y., Oliveira U. C., Souza G. H., Kitano E. S., et al. (2017). Peptidomics of Acanthoscurria gomesiana spider venom reveals new toxins with potential antimicrobial activity. J. Proteomics 151, 232–242. 10.1016/j.jprot.2016.07.012 [DOI] [PubMed] [Google Scholar]
- Badet B., Vermoote P., Haumont P. Y., Lederer F., Legoffic F. (1987). Glucosamine synthetase from Escherichia coli: purification, properties, and glutamine-utilizing site location. Biochemistry 26, 1940–1948. [DOI] [PubMed] [Google Scholar]
- Barelle C. J., Priest C. L., Maccallum D. M., Gow N. A., Odds F. C., Brown A. J. (2006). Niche-specific regulation of central metabolic pathways in a fungal pathogen. Cell Microbiol. 8, 961–971. 10.1111/j.1462-5822.2005.00676.x [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bastos K. P., Bailão A. M., Borges C. L., Faria F. P., Felipe M. S., Silva M. G., et al. (2007). The transcriptome analysis of early morphogenesis in Paracoccidioides brasiliensis mycelium reveals novel and induced genes potentially associated to the dimorphic process. BMC Microbiol. 7:29. 10.1186/1471-2180-7-29 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bowyer P., De Lucas J. R., Turner G. (1994). Regulation of the expression of the isocitrate lyase gene (acuD) of Aspergillus nidulans. Mol. Gen. Genet. 242, 484–489. [DOI] [PubMed] [Google Scholar]
- Bradford M. M. (1976). A rapid and sensitive method for the quantitation of microgram quantities of protein utilizing the principle of protein-dye binding. Anal. Biochem. 72, 248–254. [DOI] [PubMed] [Google Scholar]
- Brock M. (2009). Fungal metabolism in host niches. Curr. Opin. Microbiol. 12, 371–376. 10.1016/j.mib.2009.05.004 [DOI] [PubMed] [Google Scholar]
- Brock M., Buckel W. (2004). On the mechanism of action of the antifungal agent propionate. Eur. J. Biochem. 271, 3227–3241. 10.1111/j.1432-1033.2004.04255.x [DOI] [PubMed] [Google Scholar]
- Brummer E., Hanson L. H., Restrepo A., Stevens D. A. (1988). In vivo and in vitro activation of pulmonary macrophages by IFN-gamma for enhanced killing of Paracoccidioides brasiliensis or Blastomyces dermatitidis. J. Immunol. 140, 2786–2789. [PubMed] [Google Scholar]
- Campos E. G., Jesuíno R. S., Dantas A. S., Brígido M. M., Felipe M. S. (2005). Oxidative stress response in Paracoccidioides brasiliensis. Genet. Mol. Res. 4, 409–429. [PubMed] [Google Scholar]
- Carrero L. L., Niño-Vega G., Teixeira M. M., Carvalho M. J., Soares C. M., Pereira M., et al. (2008). New Paracoccidioides brasiliensis isolate reveals unexpected genomic variability in this human pathogen. Fungal Genet. Biol. 45, 605–612. 10.1016/j.fgb.2008.02.002 [DOI] [PubMed] [Google Scholar]
- Chagas R. F., Bailão A. M., Pereira M., Winters M. S., Smullian A. G., Deepe G. S., Jr., et al. (2008). The catalases of Paracoccidioides brasiliensis are differentially regulated: protein activity and transcript analysis. Fungal Genet. Biol. 45, 1470–1478. 10.1016/j.fgb.2008.08.007 [DOI] [PubMed] [Google Scholar]
- Chung T., Klumpp D. J., Laporte D. C. (1988). Glyoxylate bypass operon of Escherichia coli: cloning and determination of the functional map. J. Bacteriol. 170, 386–392. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Costa M., Borges C. L., Bailão A. M., Meirelles G. V., Mendonça Y. A., Dantas S. F., et al. (2007). Transcriptome profiling of Paracoccidioides brasiliensis yeast-phase cells recovered from infected mice brings new insights into fungal response upon host interaction. Microbiol. 153 (Pt 12), 4194–4207. 10.1099/mic.0.2007/009332-0 [DOI] [PubMed] [Google Scholar]
- Cruz A. H., Brock M., Zambuzzi-Carvalho P. F., Santos-Silva L. K., Troian R. F., Góes A. M., et al. (2011). Phosphorylation is the major mechanism regulating isocitrate lyase activity in Paracoccidioides brasiliensis yeast cells. FEBS J. 278, 2318–2332. 10.1111/j.1742-4658.2011.08150.x [DOI] [PubMed] [Google Scholar]
- de Arruda Grossklaus D., Bailão A. M., Vieira Rezende T. C., Borges C. L., Oliveira M. A. P., Parente J. A., et al. (2013). Response to oxidative stress in Paracoccidioides yeast cells as determined by proteomic analysis. Microbes Infect. 15, 347–364. 10.1016/j.micinf.2012.12.002 [DOI] [PubMed] [Google Scholar]
- De Curcio J. S., Silva M. G., Siva-Bailão M. G., Báo S. N., Casaletti L., Bailão A. M., et al. (2017). Identification of membrane proteome of Paracoccidioides lutzii and its regulation by zinc. Fut. Sci. 3:FSO232. 10.4155/fsoa-2017-0044 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Desjardins C. A., Champion M. D., Holder J. W., Muszewska A., Goldberg J., Bailão A. M., et al. (2011). Comparative genomic analysis of human fungal pathogens causing paracoccidioidomycosis. PLoS Genet. 7:e1002345. 10.1371/journal.pgen.1002345 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Distler U., Kuharev J., Navarro P., Levin Y., Schild H., Tenzer S. (2014). Drift time-specific collision energies enable deep-coverage data-independent acquisition proteomics. Nat. Methods 11, 167–170. 10.1038/nmeth.2767 [DOI] [PubMed] [Google Scholar]
- Ebel F., Schwienbacher M., Beyer J., Heesemann J., Brakhage A. A., Brock M. (2006). Analysis of the regulation. expression. and localisation of the isocitrate lyase from Aspergillus fumigatus. A potential target for antifungal drug development. Fungal Genet. Biol. 43, 476–489. 10.1016/j.fgb.2006.01.015 [DOI] [PubMed] [Google Scholar]
- Enjalbert B., MacCallum D. M., Odds F. C., Brown A. J. (2007). Niche-specific activation of the oxidative stress response by the pathogenic fungus Candida albicans. Infect. Immun. 75, 2143–2151. 10.1128/IAI.01680-06 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Felipe M. S., Andrade R. V., Arraes F. B., Nicola A. M., Maranhão A. Q., Torres F. A., et al. (2005). Transcriptional profiles of the human pathogenic fungus Paracoccidioides brasiliensis in mycelium and yeast cells. J. Biol. Chem. 280, 24706–24714. 10.1074/jbc.M500625200 [DOI] [PubMed] [Google Scholar]
- Fernandez E., Fernandez M., Moreno F., Rodicio R. (1993). Transcriptional regulation of the isocitrate lyase encoding gene in Saccharomyces cerevisiae. FEBS Lett. 333, 238–242. [DOI] [PubMed] [Google Scholar]
- Fleck C. B., Schöbel F., Brock M. (2011). Nutrient acquisition by pathogenic fungi: nutrient availability. pathway regulation. and differences in substrate utilization. Int. J. Med. Microbiol. 301, 400–407. 10.1016/j.ijmm.2011.04.007 [DOI] [PubMed] [Google Scholar]
- Fonseca C. A., Jesuíno R. S., Felipe M. S., Cunha D. A., Brito W. A., Soares C. M. (2001). Two-dimensional electrophoresis and characterization of antigens from Paracoccidioides brasiliensis. Microb. Infect. 3, 535–342. 10.1016/S1286-4579(01)01409-5 [DOI] [PubMed] [Google Scholar]
- Fradin C., Bernardes E. S., Jouault T. (2015). Candida albicans phospholipomannan: a sweet spot for controlling host response/inflammation. Semin. Immunopathol. 37, 123–130. 10.1007/s00281-014-0461-5 [DOI] [PubMed] [Google Scholar]
- Garrido B. C., Souza G. H., Lourenço D. C., Fasciotti M. (2016). Proteomics in quality control: whey protein-based supplements. J. Proteomics 147, 48–55. 10.1016/j.jprot.2016.03.044 [DOI] [PubMed] [Google Scholar]
- Grahl N., Puttikamonkul S., Macdonald J. M., Gamcsik M. P., Ngo L. Y., Hohl T. M., et al. (2011). In vivo hypoxia and a fungal alcohol dehydrogenase influence the pathogenesis of invasive pulmonary aspergillosis. PLoS Pathog. 7:e1002145. 10.1371/journal.ppat.1002145 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Haas A. (2007). The phagosome: compartment with a license to kill. Traffic 8, 311–330. 10.1111/j.1600-0854.2006.00531.x [DOI] [PubMed] [Google Scholar]
- Ibrahim-Granet O., Dubourdeau M., Latgé J. P., Ave P., Huerre M., Brakhage A. A., et al. (2008). Methylcitrate synthase from Aspergillus fumigatus is essential for manifestation of invasive aspergillosis. Cell Microbiol. 10, 134–148. 10.1111/j.1462-5822.2007.01025.x [DOI] [PubMed] [Google Scholar]
- Lima P. d. S., Casaletti L., Bailão A. M., Vasconcelos A. T., Fernandes Gda R., Soares C. M. (2014). Transcriptional and proteomic responses to carbon starvation in Paracoccidioides. PLoS Negl. Trop. Dis. 8:e2855 10.1371/journal.pntd.0002855 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lorenz M. C., Fink G. R. (2001). The glyoxylate cycle isrequired for fungal virulence. Nature 412, 83–86. 10.1038/35083594 [DOI] [PubMed] [Google Scholar]
- Lorenz M. C., Fink G. R. (2002). Life and death in a macrophage: role of the glyoxylate cycle in virulence. Eukaryot. Cell 1, 657–662. 10.1128/EC.1.5.657-662.2002 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lorenz M. C., Bender J. A., Fink G. R. (2004). Transcriptional response of Candida albicans upon internalization by macrophages. Eukaryot Cell 3, 1076–1087. 10.1128/EC.3.5.1076-1087.2004 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Maerker C., Rohde M., Brakhage A. A., Brock M. (2005). Methylcitrate synthase from Aspergillus fumigatus. Propionyl-CoA affects polyketide synthesis, growth and morphology of conidia. FEBS J. 272, 3615–3630. 10.1111/j.1742-4658.2005.04784.x [DOI] [PubMed] [Google Scholar]
- Martinez R. (2015). Epidemiology of Paracoccidioidomycoses. Rev. Soc. Bras. Med. Trop. 57, 11–20. 10.1590/S0036-46652015000700004 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Matute D. R., McEwen J. G., Puccia R., Montes B. A., San-Blas G., Bagagli E., et al. (2006a). Cryptic speciation and recombination in the fungus Paracoccidioides brasiliensis as revealed by gene genealogies. Mol. Biol. Evol. 23, 65–73. 10.1093/molbev/msj008 [DOI] [PubMed] [Google Scholar]
- Matute D. R., Sepulveda V. E., Quesada L. M., Goldman G. H., Taylor J. W., Restrepo A., et al. (2006b). Microsatellite analysis of three phylogenetic species of Paracoccidioides brasiliensis. J. Clin. Microbiol. 44, 2153–2157. 10.1128/JCM.02540-05 [DOI] [PMC free article] [PubMed] [Google Scholar]
- McKinney J. D., Höner zu Bentrup K., Muñoz-Elías E. J., Miczak A., Chen B., Chan W. T., et al. (2000). Persistence of Mycobacterium tuberculosis in macrophages and mice requires the glyoxylate shunt enzyme isocitrate lyase. Nature 406, 735–738. 10.1038/35021074 [DOI] [PubMed] [Google Scholar]
- Miramón P., Dunker C., Windecker H., Bohovych I. M., Brown A. J., Kurzai O., et al. (2012). Cellular responses of Candida albicans to phagocytosis and the extracellular activities of neutrophils are critical to counteract carbohydrate starvation, oxidative and nitrosative stress. PLoS ONE 7:e52850. 10.1371/journal.pone.0052850 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Muñoz J. F., Farrer R. A., Desjardins C. A., Gallo J. E., Sykes S., Sakthikumar S., et al. (2016). Genome diversity, recombination, and virulence across the major lineages of Paracoccidioides. mSphere 1:e00213-16. 10.1128/mSphere.00213-16 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Muñoz J. F., Gallo J. E., Misas E., Priest M., Imamovic A., Young S., et al. (2014). Genome update of the dimorphic human pathogenic fungi causing paracoccidioidomycosis. PLoS Negl. Trop. Dis. 8:e3348. 10.1371/journal.pntd.0003348 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Munõz-Elías E. J., McKinney J. D. (2005). Mycobacterium tuberculosis isocitrate lyases 1 and 2 are jointly required for in vivo growth and virulence. Nat. Med. 11, 638–644. 10.1038/nm1252 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Murad A. M., Souza G. H., Garcia J. S., Rech E. L. (2011). Detection and expression analysis of recombinant proteins in plant-derived complex mixtures using nanoUPLC-MS(E). J. Sep. Sci. 34, 2618–2630. 10.1002/jssc.201100238 [DOI] [PubMed] [Google Scholar]
- Olivas I., Royuela M., Romero B., Monteiro M. C., Mínguez J. M., Laborda F., et al. (2007). Ability to grow on lipids accounts for the fully virulent phenotype in neutropenic mice of Aspergillus fumigatus null mutants in the key glyoxylate cycle enzymes. Fungal Genet. Biol. 45, 45–60. 10.1016/j.fgb.2007.05.002 [DOI] [PubMed] [Google Scholar]
- Parente-Rocha J. A., Parente A. F., Baeza L. C., Bonfim S. M., Hernández O., McEwen J. G., et al. (2015). Macrophage interaction with Paracoccidioides brasiliensis yeast cells modulates fungal metabolism and generates a response to oxidative stress. PLoS ONE 10:e0137619. 10.1371/journal.pone.0137619 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Pigosso L. L., Baeza L. C., Tomazett M. V., Batista M. R. F., Moura V. M. B. D., Bailão A. M., et al. (2017). Paracoccidioides brasiliensis presents metabolic reprogramming and secretes a serine proteinase during murine infection. Virulence. [Epub ahead of print]. 10.1080/21505594.2017.1355660 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Pigosso L. L., Parente A. F., Coelho A. S., Silva L. P., Borges C. L., Bailão A. M., et al. (2013). Comparative proteomics in the genus Paracoccidioides. Fungal Genet. Biol. 60, 87–100. 10.1016/j.fgb.2013.07.008 [DOI] [PubMed] [Google Scholar]
- Renshaw H., Vargas-Muñiz J. M., Richards A. D., Asfaw Y. G., Juvvadi P. R., Steinbach W. J. (2016). Distinct roles of myosins in Aspergillus fumigatus hyphal growth and pathogenesis. Infect Immun. 84, 1556–1564. 10.1128/IAI.01190-15. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Restrepo A., Jiménez B. E. (1980). Growth of Paracoccidioides brasiliensis yeast phase in a chemically defined culture medium. J. Clin. Microbiol. 12, 279–281. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Restrepo A., McEwen J. G., Castañeda E. (2001). The habitat of Paracoccidioides brasiliensis: how far from solving the riddle? Med. Mycol. 39, 233–241. 10.1080/714031028 [DOI] [PubMed] [Google Scholar]
- San-Blas G., Niño-Vega G., Iturriaga T. (2002). Paracoccidioides brasiliensis and paracoccidioidomycosis: molecular approaches to morphogenesis, diagnosis, epidemiology, taxonomy and genetics. Med. Mycol. 40, 225–242. 10.1080/mmy.40.3.225.242 [DOI] [PubMed] [Google Scholar]
- Souza G. H., Guest P. C., Martins-de-Souza D. (2017). LC-MSE, multiplex MS/MS, ion mobility, and label-free quantitation in clinical proteomics. Methods Mol. Biol. 1546, 57–73. 10.1007/978-1-4939-6730-8_4 [DOI] [PubMed] [Google Scholar]
- Tamayo D., Muñoz J. F., Almeida A. J., Puerta J. D., Restrepo Á., Cuomo C. A., et al. (2017). Paracoccidioides spp. catalases and their role in antioxidant defense against host defense responses. Fungal Genet. Biol. 100, 22–32. 10.1016/j.fgb.2017.01.005 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tamayo D., Muñoz J. F., Lopez A., Urán M., Herrera J., Borges C. L., et al. (2016). Identification and analysis of the role of superoxide dismutases isoforms in the pathogenesis of Paracoccidioides spp. PLoS Negl. Trop. Dis. 10:e0004481. 10.1371/journal.pntd.0004481 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Teixeira M. M., Theodoro R. C., Carvalho M. J., Fernandes L., Paes H. C., Hahn R. C., et al. (2009). Phylogenetic analysis reveals a high level of speciation in the Paracoccidioides genus. Mol. Phylogenet. Evol. 52, 273–283. 10.1016/j.ympev.2009.04.005 [DOI] [PubMed] [Google Scholar]
- Teixeira M. M., Theodoro R. C., Niño-Vega G., Bagagli E., Felipe M. S. (2014). Paracoccidioides species complex: ecology, phylogeny, sexual reproduction, and virulence. PLoS Pathog. 10:e1004397. 10.1371/journal.ppat.1004397 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Treitz C., Enjalbert B., Portais J. C., Letisse F., Tholey A. (2016). Differential quantitative proteome analysis of Escherichia coli grown on acetate versus glucose. Proteomics 16, 2742–2746. 10.1002/pmic.201600303 [DOI] [PubMed] [Google Scholar]
- Turrens J. F. (2003). Mitochondrial formation of reactive oxygen species. J. Physiol. 552 (Pt 2), 335–344. 10.1113/jphysiol.2003.049478 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Vylkova S., Carman A. J., Danhof H. A., Collette J. R., Zhou H., Lorenz M. C. (2011). The fungal pathogen Candida albicans autoinduces hyphal morphogenesis by raising extracellular pH. MBio 2:e00055-11. 10.1128/mBio.00055-11 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wayne L. G., Lin K. Y. (1982). Glyoxylate metabolism and adaptation of Mycobacterium tuberculosis to survival under anaerobic conditions. Infect. Immun. 37, 1042–1049. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Youseff B. H., Holbrook E. D., Smolnycki K. A., Rappleye C. A. (2012). Extracellular superoxide dismutase protects Histoplasma yeast cells from host-derived oxidative stress. PLoS Pathog. 8:e1002713. 10.1371/journal.ppat.1002713 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zambuzzi-Carvalho P. F., Cruz A. H., Santos-Silva L. K., Goes A. M., Soares C. M. A., Pereira M. (2009). The malate synthase of Paracoccidioides brasiliensis Pb01 is required in the glyoxylate cycle and in the allantoin degradation pathway. Med. Mycol. 47, 734–744. 10.3109/13693780802609620 [DOI] [PubMed] [Google Scholar]
- Zhang Y. Q., Brock M., Keller N. P. (2004). Connection of propionyl-CoA metabolism to polyketide biosynthesis in Aspergillus nidulans. Genetics 168, 785–794. 10.1534/genetics.104.027540 [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.