Abstract
Several susceptibility loci for classical Hodgkin lymphoma have been reported. However, much of the heritable risk is unknown. Here, we perform a meta-analysis of two existing genome-wide association studies, a new genome-wide association study, and replication totalling 5,314 cases and 16,749 controls. We identify risk loci for all classical Hodgkin lymphoma at 6q22.33 (rs9482849, P = 1.52 × 10−8) and for nodular sclerosis Hodgkin lymphoma at 3q28 (rs4459895, P = 9.43 × 10−17), 6q23.3 (rs6928977, P = 4.62 × 10−11), 10p14 (rs3781093, P = 9.49 × 10−13), 13q34 (rs112998813, P = 4.58 × 10−8) and 16p13.13 (rs34972832, P = 2.12 × 10−8). Additionally, independent loci within the HLA region are observed for nodular sclerosis Hodgkin lymphoma (rs9269081, HLA-DPB1*03:01, Val86 in HLA-DRB1) and mixed cellularity Hodgkin lymphoma (rs1633096, rs13196329, Val86 in HLA-DRB1). The new and established risk loci localise to areas of active chromatin and show an over-representation of transcription factor binding for determinants of B-cell development and immune response.
Classical Hodgkin lymphoma is a cancer that originates in lymph nodes. Little is known about its genetic susceptibility. Here, the authors combined existing and new genome-wide association studies to identify risk loci for classical Hodgkin lymphoma at 6q22.33, and nodular sclerosis Hodgkin lymphoma at 3q28, 6q23.3, 10p14, 13q34, 16p13.13.
Introduction
Classical Hodgkin lymphoma (cHL) is a lymphoid malignancy of germinal centre (GC) B-cell origin1, which is characterised by Hodgkin and Reed–Sternberg (HRS) cells with a dominant background population of reactive inflammatory cells1. Of the four major subtypes of cHL, nodular sclerosis Hodgkin lymphoma (NSHL) and mixed cellularity Hodgkin lymphoma (MCHL) account for 65% and 20% of cHL, respectively2. While Epstein–Barr virus (EBV) infection is causally associated with a subset of cHL cases, proportionally higher in MCHL, no other environmental factor has thus far been robustly linked to cHL risk3.
Evidence for inherited genetic influence on susceptibility to cHL is provided by the familial risk and the high concordance between monozygotic twins4, 5. A strong HLA association for cHL risk is well established; however, our understanding of cHL heritability has been transformed by recent genome-wide association studies (GWAS), which have identified single-nucleotide polymorphisms (SNPs) at seven non-HLA loci influencing risk6–9. Although projections indicate that additional risk variants for cHL can be discovered by GWAS10, the statistical power of published studies is limited.
To gain a more comprehensive insight into cHL predisposition, we performed a meta-analysis of two previous GWAS7, 8 and a new GWAS, thereby more than doubling study power to discover risk SNPs. With replication, our study has allowed us to identify six new non-HLA risk loci. Additionally, by conducting region-specific imputation we have defined the specific HLA associations underlying NSHL and MCHL risk.
Results
Association analysis
We analysed GWAS data from three studies of European ancestry: a new GWAS from the UK National Study of Hodgkin Lymphoma Genetics (NSHLG) and two previously reported GWAS (Supplementary Table 1)7, 8. After quality control the three studies provided SNP genotypes on 3,077 cases and 13,680 controls (Supplementary Tables 2, 3, 4; Supplementary Fig. 1). To increase genomic resolution, we imputed >10 million SNPs using the 1000 Genomes Project and the UK10K data as reference11, 12. Quantile–quantile (Q–Q) plots for SNPs with minor allele frequency (MAF) > 0.05% post imputation did not show evidence of substantive over-dispersion (λ = 1.03–1.09; Supplementary Fig. 2). An overview of the analysis strategy is outlined in Supplementary Fig. 3. Meta-analysing the association test results from the three GWAS into a joint discovery set, we calculated joint odds ratios and 95% confidence intervals for each SNP and associated per-allele P-value for all cHL, NSHL and MCHL cases vs. controls (Supplementary Fig. 4). In this analysis, associations for the established non-HLA risk loci at 2p16.1, 3p24.1, 5q31.1, 6q23.3, 8q24.21, 10p14 and 19p13.3 were consistent in direction and magnitude of effect with previously reported studies (Supplementary Fig. 4; Supplementary Table 5)6–8.
We sought validation of association SNPs with a P-value from the meta-analysis under a fixed-effects model at P < 1.0 × 10−7 and P < 1.0 × 10−6 for loci not previously associated with cHL and NSHL risk, respectively, by genotyping two additional independent series (Supplementary Table 1), totalling 2,237 cases and 3,069 controls (Table 1; Supplementary Table 6). Where the strongest signal was provided by an imputed SNP, we confirmed the fidelity of imputation by genotyping (Supplementary Table 7). In the combined meta-analysis, we identified genome-wide significant associations for cHL (Table 1; Supplementary Tables 8 and 9), at 3q28 (rs4459895, P = 4.45 × 10−18), 6q22.33 (rs9482849, P = 1.52 × 10−8), 6q23.3 (rs6928977, P = 1.24 × 10−10) and 10p14 (rs3781093, P = 4.91 × 10−12), which were predominantly driven by an association with NSHL. The rs6928977 association is independent of the previously identified association at 6q23.3 marked by rs9402684 (Supplementary Table 5); respective conditional P-values, P = 1.28 × 10−8 and P = 9.80 × 10−6 (pairwise LD metrics r2 = 0.002, D’ = 0.007)8. Furthermore, the rs3781093 association is independent of the previously identified 10p14 association marked by rs2388486 (Supplementary Table 5); respective conditional P-values are P = 3.38 × 10−8 and P = 1.32 × 10−12 (pairwise LD metrics r2 = 0.002, D’ = 0.27)7. For NSHL we identified two new associations at 13q34 (rs112998813, P = 4.58 × 10−8) and 16p13.13 (rs34972832, P = 2.12 × 10−8, Table 1).
Table 1.
Summary results for newly identified risk loci
| Nearest genea | Risk allele (frequency) | Discovery GWAS meta-analysis | UK Replication 1 | UK Replication 2 | Meta-analysis | ||||||||
|---|---|---|---|---|---|---|---|---|---|---|---|---|---|
| Position (hg19, bp) | P-value | OR (95% CI) | P-value | OR (95% CI) | P-value | OR (95% CI) | P-value | OR (95% CI) | I2 (%) | P het | |||
| 3q28, rs4459895 | LPP | A (0.20) | 187954414 | ||||||||||
| cHL | 4.16 × 10−10 | 1.27 (1.18–1.36) | 6.85 × 10−9 | 1.44 (1.27–1.63) | 0.02 | 1.26 (1.04–1.52) | 4.45 × 10−18 | 1.30 (1.23–1.38) | 13 | 0.33 | |||
| NSHL | 9.16 × 10−9 | 1.37 (1.23–1.53) | 1.37 × 10−8 | 1.43 (1.26–1.62) | 0.04 | 1.30 (1.02–1.66) | 9.43 × 10−17 | 1.39 (1.28–1.50) | 0 | 0.93 | |||
| MCHL | 0.92 | 1.04 (0.91–1.19) | 0.98 | 1.00 (0.68–1.47) | 0.55 | 1.04 (0.92–1.19) | 0 | 0.82 | |||||
| 6q22.33, rs9482849 | PTPRK | C (0.17) | 128288536 | ||||||||||
| cHL | 5.02 × 10−8 | 1.24 (1.15–1.35) | 0.13 | 1.11 (0.97–1.27) | 0.13 | 1.17 (0.95–1.43) | 1.52 × 10−8 | 1.20 (1.13–1.28) | 3 | 0.39 | |||
| NSHL | 2.91 × 10−6 | 1.32 (1.17–1.48) | 0.17 | 1.10 (0.96–1.25) | 0.20 | 1.19 (0.91–1.54) | 4.13 × 10−6 | 1.21 (1.12–1.33) | 10 | 0.35 | |||
| MCHL | 0.17 | 1.11 (0.96–1.28) | 0.78 | 1.06 (0.70–1.61) | 0.16 | 1.10 (0.96–1.26) | 0 | 0.97 | |||||
| 6q23.3, rs6928977 | AHI1 | G (0.57) | 135626348 | ||||||||||
| cHL | 1.66 × 10−8 | 1.18 (1.12–1.26) | 0.01 | 1.14 (1.03–1.26) | 0.05 | 1.16 (1.00–1.34) | 1.24 × 10−10 | 1.17 (1.12–1.23) | 0 | 0.85 | |||
| NSHL | 9.34 × 10−10 | 1.31 (1.20–1.42) | 0.03 | 1.12 (1.01–1.24) | 0.01 | 1.30 (1.06–1.58) | 4.62 × 10−11 | 1.23 (1.16–1.31) | 26 | 0.25 | |||
| MCHL | 0.24 | 1.06 (0.96–1.18) | 0.69 | 1.06 (0.79–1.42) | 0.22 | 1.06 (0.96–1.17) | 0 | 0.22 | |||||
| 10p14, rs3781093 | GATA3 | T (0.88) | 8101927 | ||||||||||
| cHL | 4.89 × 10−12 | 1.35 (1.23–1.47) | 4.00 × 10−4 | 1.32 (1.25–1.44) | 0.11 | 1.21 (0.96–1.52) | 4.91 × 10−12 | 1.28 (1.19–1.37) | 64 | 0.01 | |||
| NSHL | 9.16 × 10−12 | 1.53 (1.36–1.75) | 2.00 × 10−4 | 1.44 (1.31–1.61) | 0.64 | 0.92 (0.68–1.26) | 9.49 × 10−13 | 1.39 (1.28–1.53) | 61 | 0.06 | |||
| MCHL | 0.03 | 1.18 (1.02–1.36) | 0.05 | 1.56 (1.02–2.40) | 0.16 | 0.91 (0.79–1.04) | 73 | 0.03 | |||||
| 13q34, rs112998813 | UPF3A | C (0.08) | 115059729 | ||||||||||
| cHL | 3.63 × 10−3 | 1.19 (1.06–1.33) | 0.03 | 1.23 (1.03–1.47) | 0.43 | 1.12 (0.84–1.50) | 2.70 × 10−4 | 1.19 (1.08–1.30) | 13 | 0.32 | |||
| NSHL | 8.43 × 10−8 | 1.58 (1.34–1.88) | 0.03 | 1.22 (1.02–1.47) | 0.28 | 1.23 (0.85–1.78) | 4.58 × 10−8 | 1.39 (1.23–1.56) | 27 | 0.24 | |||
| MCHL | 0.92 | 0.99 (0.80–1.22) | 0.27 | 1.35 (0.80–2.23) | 0.75 | 1.03 (0.85–1.25) | 0 | 0.56 | |||||
| 16p13.13, rs34972832 | CLEC16A | A (0.18) | 11198938 | ||||||||||
| cHL | 1.45 × 10−4 | 1.15 (1.07–1.23) | 6.34 × 10−3 | 1.18 (1.05–1.34) | 0.10 | 1.17 (0.97–1.42) | 8.03 × 10−7 | 1.16 (1.09–1.23) | 6 | 0.37 | |||
| NSHL | 7.47 × 10−7 | 1.24 (1.15–1.34) | 6.53 × 10−3 | 1.30 (1.17–1.45) | 0.28 | 1.15 (0.89–1.50) | 2.12 × 10−8 | 1.24 (1.15–1.34) | 37 | 0.18 | |||
| MCHL | 0.65 | 0.97 (0.85–1.11) | 0.91 | 1.02 (0.69–1.52) | 0.70 | 0.98 (0.86–1.10) | 0 | 0.94 | |||||
The risk allele is the allele corresponding to the estimated odds ratio. Frequency of the risk allele is from the CEU population from 1000 Genomes Project
cHL classical Hodgkin lymphoma, NSHL nodular sclerosis Hodgkin lymphoma, MCHL mixed cellularity Hodgkin lymphoma, bp base pair, OR odds ratio, CI confidence interval, I2 proportion of the total variation due to heterogeneity
I2 value ≥ 75% is considered to be characteristic of large heterogeneity
aNearest gene may not be the functional gene
Relationship between the new risk SNPs and phenotype
A hallmark of cHL epidemiology is the bimodal age-specific incidence and it has been argued that the disease in young adults and older adults is aetiologically different; in particular there is a low prevalence of EBV-positive disease in NSHL patients aged 16–353. Case-only analysis did not provide evidence of sex differences at newly identified risk SNPs (Supplementary Table 10) or a relationship between age in the NSHL subgroup. Albeit not significant after correction for multiple testing, we observed an association between EBV-positive disease and cHL at 6q23.3 in 796 cases analysed (rs6928977, P = 0.03, Supplementary Table 10).
Biological inference
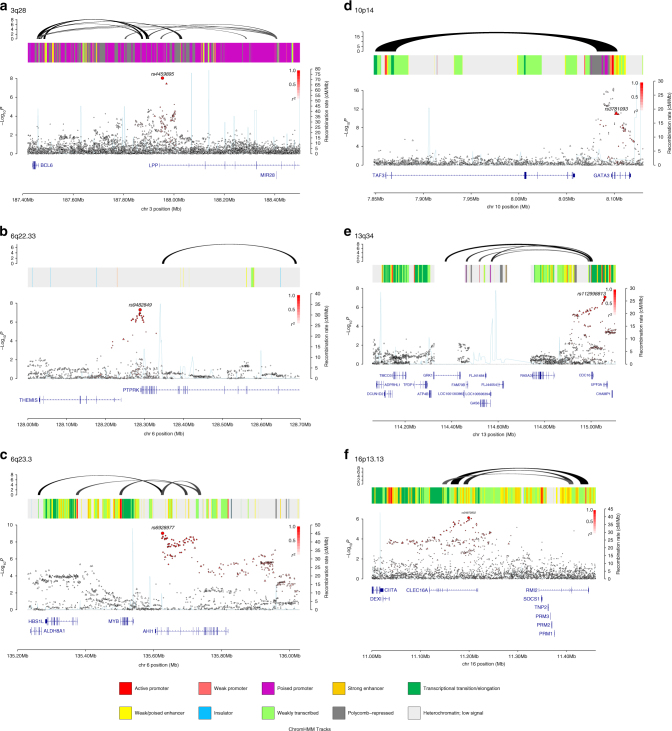
Five of the six new risk SNPs localise in or near genes which have either been previously implicated in the development of cHL or have established roles in B-cell development and are therefore strong candidates for cHL susceptibility. Specifically, the 6q22.33 association marked by rs9482849 maps intergenically to THEMIS (thymocyte-expressed molecule involved in selection) and PTPRK (receptor-type tyrosine protein phosphatase kappa) (Fig. 1). Downregulation of PTPRK by the EBV-encoded EBNA1 contributes to the growth and survival of HRS cells13. The 6q23.3 association defined by rs6928977 localises to intron 3 of AHI1 (abelson helper integration site-1) (Fig. 1) which has been implicated in the development of both B- and T-cell lymphoma14, 15. The 13q34 association marked by rs112998813 is located in intron 5 of UPF3A (Fig. 1), a regulator of nonsense transcripts16. The LD region of association also harbours CDC16 (cell division cycle protein 16). CDC16, a subunit of the anaphase-promoting complex17, targets cell cycle regulatory proteins for proteasome degradation, thereby allowing cell cycle progression, and is downregulated in HRS cells18. At 16p13.13, the rs34972832 association for NSHL maps to intron 18 of CLEC16A (C-type lectin domain family 1, Fig. 1) whose loss of function affects both B-cell number and function19. The 10p14 association marked by rs3781093 maps intronic of GATA3 (Fig. 1). Transcriptional repression of GATA3 is essential for early B-cell commitment, and aberrant GATA3 expression has been observed in HRS cells20, 21. Intriguingly, the rs3781093 risk allele for NSHL has previously been demonstrated to be protective for paediatric B-cell acute lymphoblastic leukaemia (ALL)22.
Fig. 1.
Regional plots of association results and recombination rates for the newly identified classical Hodgkin lymphoma (NSHL) risk loci. Results for a 3q28 (rs4459895) and nodular sclerosis Hodgkin lymphoma (NSHL) risk, b 6q22.33 (rs9482849) and classical Hodgkin lymphoma (cHL) risk, c 6q23.3 (rs6928977) and NSHL risk, d 10p14 (rs3781093) and NSHL risk, e 13q34 (rs112998813) and NSHL risk, and f 16p13.13 (rs34972832) and NSHL risk. Plots show association results of both genotyped (triangles) and imputed (circles) single-nucleotide polymorphisms (SNPs) in the genome-wide association study samples and recombination rates. −log10 P-values (y-axes) of the SNPs are shown according to their chromosomal positions (x-axes). The sentinel SNP in each combined analysis is shown as a large circle or triangle and is labelled by its rsID. The colour intensity of each symbol reflects the extent of LD with the top SNP, white (r2 = 0) through to dark red (r2 = 1.0). Genetic recombination rates, estimated using 1000 Genomes Project samples, are shown with a light blue line. Physical positions are based on NCBI build 37 of the human genome. Also shown are the relative positions of genes and transcripts mapping to the region of association. Genes have been redrawn to show their relative positions; therefore maps are not to physical scale. The middle track represents the chromatin state segmentation track (ChromHMM) for lymphoblastoid cells using data from the HapMap ENCODE Project. The top track represents capture Hi-C promoter contacts in GM12878 cells. The colour intensity of each contact reflects the interaction score
To the extent that they have been deciphered, many GWAS risk loci map to non-coding regions of the genome and influence gene regulation. Hence, to gain insight into the biological mechanisms for the associations of the newly identified risk SNPs, we interrogated publicly accessible expression data on lymphoblastoid cell lines (LCLs)23, 24. We used the summary data-based Mendelian randomisation (SMR) analysis to test for pleiotropy between GWAS signal and cis-expression quantitative trait (eQTL) for genes within 1 Mb of the sentinel SNP at each locus to identify a causal relationship25. At 6q23.3 and 10p14, significant eQTLs were observed with AHI1 (PSMR = 8.63 × 10−6; Supplementary Table 11 and Supplementary Fig. 5) and GATA3 (PSMR = 4.70 × 10−8; Supplementary Table 11 and Supplementary Fig. 5).
Since spatial proximity between specific genomic regions and chromatin looping interactions are central for the regulation of gene expression, we identified patterns of chromatin interactions at candidate causal SNPs by analysing promoter capture Hi-C data on GM12878 as a source of B-cell information26. Looping chromatin interactions were shown at 3q28 (rs4459895), 6q23.3 (rs6928977), 10p14 (rs3781093) and 16p13.13 (rs34972832). While no significant eQTL was shown for these chromatin looping interactions they involved a number of genes with biological relevance to cHL development (Fig. 1). At 3q28, the looping interaction implicates BCL6 and mir-28, which have well documented roles in B-cell tumour biology and GC B-cell development27, 28. At 6q23.3, we observed interactions with promoter sequences upstream in MYB and ALDH8A1. At 10p14, both risk SNPs encompass a region that interacts with TAF3, which encodes transcription initiation factor TFIID subunit 3. TAF3 forms part of the transcription initiation factor TFIID and is necessary for haematopoiesis29. Finally, we observed interactions at the 16p13.13 risk locus with RMI2 (encoding RecQ mediated genome instability 2) (Fig. 1). RMI2 is an essential component of the Bloom helicase-double Holliday junction dissolvasome and is responsible for genomic stability30.
Across the new and established risk loci for cHL we confirmed a significant enrichment of DNase hypersensitivity in GM12878 cells (false discovery rate (FDR) adjusted P-value = 0.0035), as well as enhancer elements in primary B-cells (FDR adjusted P-value = 0.00064) and GM12878 cells (FDR adjusted P-value = 0.015)31. Analysis of ChIP-seq data on 82 transcription factors (TFs) showed an over-representation of the binding of TFs that play a central role in B-cell signalling-networks such as RELA (nuclear factor NF-kappa-B p65), EBF1 (early B-cell factor 1), RUNX3 (runt-related transcription factor 3) and BATF (basic leucine zipper transcription factor, ATF-like) (Fig. 2). Collectively, these observations support the assertion that risk loci for cHL mediate their effects through B-cell developmental networks, and are strongly involved in transcriptional initiation and enhancement.
Fig. 2.
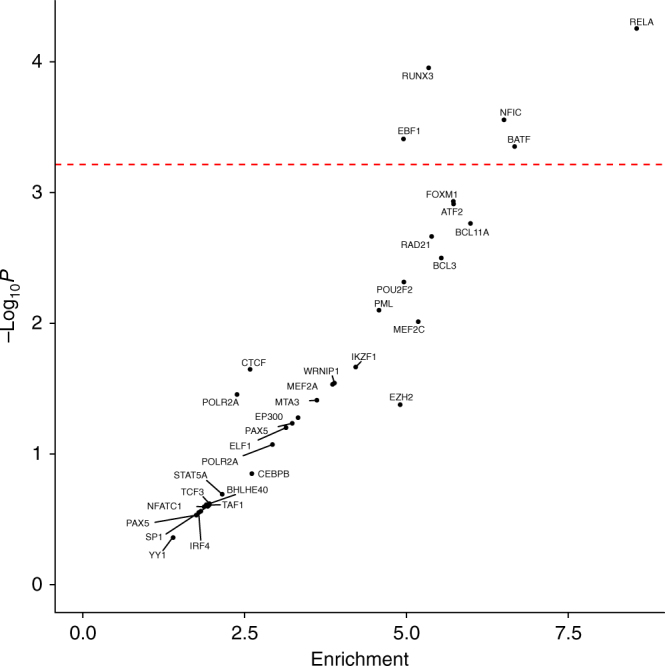
Enrichment of transcription factors binding at classical Hodgkin lymphoma risk loci. The enrichment and over-representation of transcription factors binding at all cHL risk loci. The red line represents the Bonferroni corrected P-value threshold
The HLA region
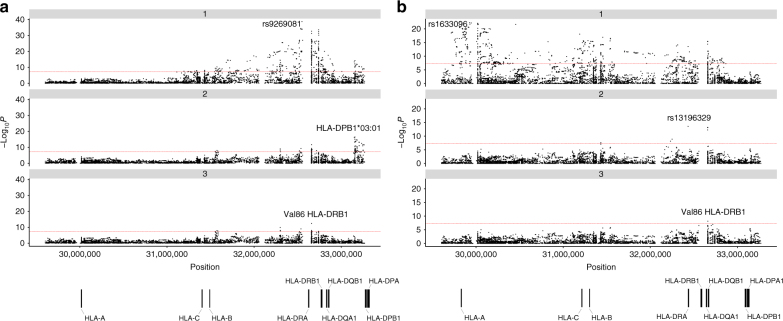
To obtain additional insight into plausible functional variants within the major histocompatibility region at 6p21, we imputed the classical HLA alleles, amino-acid residues and SNPs using SNP2HLA32. To isolate independent associations for NSHL and MCHL, we performed conditional step-wise logistic regression on the strongest associated SNP, amino acid or allele, until no further variants attained genome-wide significance (Fig. 3; Supplementary Table 12). For NSHL, we identified the strongest association at rs9269081 (P = 1.74 × 10−39), which maps within the class II HLA region, 3’of HLA-DRA. Additional class II associations were shown by HLA-DPB1*03:01 (P = 3.35 × 10−17) and Val86 in HLA-DRB1 (P = 3.52 × 10−13) (Fig. 3). In contrast, the strongest association for MCHL was provided by rs1633096, a class I HLA association 3’ of HLA-F (P = 2.72 × 10−23). Additional class II associations for MCHL were observed at rs13196329, located intronic of C6orf10 (P = 2.58 × 10−14) and Val86 in HLA-DRB1 (P = 7.10 × 10−9) (Fig. 3).
Fig. 3.
Manhattan plot representation of the step-wise conditional logistic regression of risk of a nodular sclerosis Hodgkin lymphoma and b mixed cellularity Hodgkin lymphoma within the human leukocyte antigen (HLA) region. (a1) Unconditioned test of the HLA region. (a2) Results of the HLA region after conditioning on rs9269081. (a3) Results of the HLA region after conditioning on rs9269081 and HLA-DPB1*03:01. (b1) Unconditioned test of the HLA region. (b2) Results of the HLA region after conditioning on rs1633096. (b3) Results of the HLA region after conditioning on rs1633096 and rs13196329. Physical positions are based on NCBI build 36 of the human genome. The −log10 of the combined logistic regression test P-values are plotted against their physical chromosomal position. The broken red line represents the genome-wide level of significance (P < 5 × 10−8)
Heritability of cHL
By fitting all SNPs from GWAS simultaneously using Genome-wide Complex Trait Analysis33, the estimated heritability of cHL, NSHL and MCHL attributable to all common variation is 24.0% (±2.3%), 25.2% (±3.4%) and 21.9% (±2.4%), respectively. This estimate represents the additive variance, and therefore does not include the potential impact of dominance effects or gene–environment interactions having an impact on cHL risk. The currently identified non-HLA risk SNPs thus only account for around 12% of the additive heritable risk.
Co-heritability with autoimmune disease
Although not universal, some epidemiological studies have reported associations between cHL and various autoimmune diseases, raising the possibility of common genetic susceptibility and hence common biological pathways34. Variation at a number of the cHL risk loci, including 3p24.1, 5q31.1 and 6q23.3 has previously been implicated as determinants of autoimmune disease risk supporting such an assertion (Supplementary Data 1).
To investigate co-heritability globally between cHL and autoimmune disease, we implemented cross-trait LD score regression35. Using summary-level GWAS data we estimated genetic correlations between cHL and six autoimmune diseases curated by ImmunoBase; specifically rheumatoid arthritis36, systemic lupus erythematosus37, multiple sclerosis (MS)38, primary biliary cirrhosis39, ulcerative colitis (UC)40 and coeliac disease41 GWAS data (Supplementary Table 13). We observed a positive genetic correlation between cHL and MS (rg = 0.35, P = 0.04) and a negative correlation between cHL and UC (rg = −0.23, P = 0.01).
Discussion
To our knowledge, we have performed the largest GWAS of cHL to date, identifying six new non-HLA risk loci. The availability of comprehensive reference panels for the HLA region has allowed us to delineate class I and class II associations for NSHL and MCHL, substantiating recent documented differences between these cHL histologies9.
Although functional analyses are required to determine the biological basis of cHL association signals, we have demonstrated that these risk loci are enriched for regulatory elements in B-cells. Moreover, they feature an over-representation of key B-cell TF binding, notably RELA, RUNX3, EBF1 and BATF. RELA is a TF involved in NF-κB heterodimer formation. HRS cells show high constitutive activity of NF-κB (both canonical and non-canonical pathways)42, which promotes cell survival and growth through inducing anti-apoptotic and pro-proliferative gene programs43, 44. Inhibition of NF-κB in HRS cells leads to caspase-independent apoptosis43. EBF1 cooperates with E2A and PAX5 to regulate B-cell maturation45. Its expression in HRS cells is low46, which is thought to contribute to the loss of normal B-cell phenotype47. RUNX3 has important roles in B-cell maturation48 and downregulation of RUNX1 by RUNX3 is required for EBV-driven LCL growth49. BATF also appears to co-ordinate B-cell maturation50, and is highly expressed in HRS cells51.
The strong HLA associations we identified for NSHL and MCHL support recent observations for distinct class I and class II relationships for these cHL subtypes9. Specifically, the class II NSHL association marked by rs9269081 is in strong LD with the previously identified risk SNP rs6903608 (r2 = 0.92, D’ = 1.0) for EBV-negative NSHL9. For MCHL the class I association rs1633096 shows correlation with the previously identified marker SNP rs2734986 (pairwise r2 = 0.41, D’ = 0.97) for EBV-positive cHL9. A class I association for MCHL is consistent with a high EBV positivity and supports the notion of defective cytotoxic T-cell lymphocyte responses in EBV-infected HRS cells52. Variation within the class II HLA region alters the risk of autoimmune diseases53, but the underlying biological mechanism of these associations has yet to be fully defined. The class II HLA association for NSHL and MCHL risk, comprising both coding variants and non-coding SNPs, may explain the importance of CD4+ T follicular helper (TFH) cells in cHL pathogenesis. In the GC, there is a requirement for CD4+ TFH cells to interact with GC B-cells through the T-cell antigen receptor (TCR) and HLA class II proteins for normal plasma and memory cells formation54. It is therefore plausible that variation in peptide binding and expression of the HLA class II proteins contributes to cHL pathogenesis through interaction with CD4+ TFH cells. Such a model is supported by the observation of variation at position 86 of HLA-DRB1 influencing TCR Vα gene expression55, the predominance of CD4+ T-cells in cHL tumours56, the reliance of HRS cells on the micro-environment for survival1, and the loss of MHC class II expression on HRS cells57, the last of which is associated with adverse prognosis. An alternative explanation for the class II HLA association in cHL is the involvement of an unidentified pathogen playing a causative role in cHL. Amino-acid variants and SNPs within HLA-DRB1 modulate humoral immune responses to common viruses, such as influenza A and JC polyomavirus58. Consistent with such a model is dimorphic variation at position 86 of HLA-DRB159, which we identify as influencing risk of NSHL and MCHL, modulating the anchoring pocket of the antigen binding site, and influencing the conformation of peptide–DR protein complexes while maintaining a T-cell response60.
In our analysis we noted a reciprocal relationship between NSHL risk and ALL risk at 10p14 (GATA3)22. Since GATA3 plays a key role in B-cell development and both ALL and NSHL are malignancies derived from B-cells at different stages of maturation, our observation leads to speculation of a significant temporal effect of genetic variation at this locus in response to an environmental or mutational insult.
Although supported by a contemporaneous study and requiring further validation61, we found evidence for common genetic susceptibility between cHL and MS, thus raising the possibility of shared environmental risk factors. A potential biological basis for such a relationship may encompass aberrant immune activation and cell proliferation.
In conclusion, our study provides further evidence for inherited susceptibility to cHL and supports a model whereby risk loci influence disease through effects on B-cell regulatory networks, providing a mechanistic link between susceptibility and biology. Our findings also emphasise the differences between the major subtypes, which are reflective of differences in tumour aetiology.
Methods
Ethics
Collection of patient samples and associated clinico-pathological information was undertaken with written informed consent. Relevant ethical review boards approved the individual studies in accordance with the tenets of the Declaration of Helsinki (UK-GWAS MREC 03/1/096, German-GWAS University of Heidelberg 104/2004 and UK-GWAS-NSHLG MREC 09/MRE00/72). The diagnosis of cHL (i.e., excluding cases with nodular lymphocyte predominant HL), NSHL and MCHL (ICD-10-CM C81.1-3) in all cases was established in accordance with World Health Organisation guidelines.
Genome-wide association studies
Primary study: We analysed constitutional DNA from 1,717 cases ascertained through the NSHLG (http://www.public.ukcrn.org.uk) from 2010 to 2013. These are detailed in Supplementary Table 1. Cases were genotyped using the Illumina Oncoarray (Illumina Inc.). Controls which were also genotyped using the oncoarray comprised: (1) 2,976 cancer-free men recruited by the PRACTICAL Consortium—the UK Genetic Prostate Cancer Study (UKGPCS) (age < 65 years), a study conducted through the Royal Marsden NHS Foundation Trust and SEARCH (Study of Epidemiology & Risk Factors in Cancer), recruited via GP practices in East Anglia (2003–2009), (2) 4,446 cancer-free women from across the UK via the Breast Cancer Association Consortium (BCAC).
Published studies: We used GWAS data generated on two non-overlapping case–control series of Northern European ancestry, which have been the subject of previous analyses that are summarised in Supplementary Table 1. Briefly: (1) The UK-GWAS was based on 622 cases ascertained through the Royal Marsden Hospital National Health Service Trust Family History study during 2004–20087, and 5,677 controls from the UK Wellcome Trust Case Control Consortium 2 (WTCCC2)62. (2) The German-GWAS comprised 1,001 cases ascertained by the German Hodgkin Study Group during 1998–20078, and 1,226 controls from the Heinz Nixdorf Recall (HNR) study.
GWAS and meta-analysis
Standard quality control measures were applied to each of the three GWAS (Supplementary Tables 2, 3 and 4)7, 8, 63. Specifically, individuals with a low call rate (< 95%) as well as all individuals evaluated to be of non-European ancestry (using the HapMap version III CEU, JPT/CHB and YRI populations as a reference, Supplementary Fig. 1) were excluded. For apparent first-degree relative pairs, we excluded the control from a case–control pair or the individual with the lower call rate. SNPs with a call rate < 95% were excluded as were those with a MAF < 0.01 or displaying deviation from Hardy–Weinberg equilibrium (HWE) (i.e., P < 10−6, Supplementary Table 4). GWAS data were imputed to >10 million SNP with IMPUTE2 v2.364 software, using a merged reference panel consisting of data from 1000 Genomes Project (phase 1 integrated release 3, March 2012)11 and UK10K (ALSPAC, EGAS00001000090/EGAD00001000195 and TwinsUK EGAS00001000108/EGAS00001000194 studies)12. HLA imputation was conducted using SNP2HLA and the Type I Diabetes Genetics Consortium reference panel of 5,225 individuals of European descent32. The number of variants in the HLA imputation recovered with an information measure of > 0.80 were 8,436 (94% of total variants), 8506 (95% of total variants) and 8599 (96% of total variants) in the UK-GWAS, German-GWAS and UK-NSHLG-GWAS data sets, respectively. Imputation was conducted separately for each study, and in each, the data were pruned to a common set of SNPs between cases and controls prior to imputation. Poorly imputed SNPs defined by an information measure <0.80 were excluded. Tests of association between SNPs and cHL were performed using logistic regression under an additive genetic model in SNPTESTv2.565. The adequacy of the case–control matching was evaluated using Q–Q plots of test statistics (Supplementary Fig. 2). The inflation factor λ was based on the 90% least-significant SNP66. Where appropriate, principal components, generated using common SNPs, were included in the analysis to limit the effects of cryptic population stratification that otherwise might cause inflation of test statistics. Eigenvectors for the GWAS data sets were inferred using smartpca (part of EIGENSOFT) by merging cases and controls with Phase III HapMap samples. LD metrics were calculated in vcftools v0.1.12b67, using UK10K merged 1000 Genomes Project data and plotted using visPIG68.
Replication studies and technical validation
The eight SNPs in the most promising loci (Table 1; Supplementary Table 6), were taken forward for de novo replication in an additional 1,284 cases from the NSHLG and 2,504 controls from the UK replication 1 series (Supplementary Table 1). After this six SNPs were genotyped in an additional replication series, (UK replication 2 series) comprising 953 cases and 565 controls from the Scotland and Newcastle Epidemiological Study of Hodgkin Disease (SNEHD), the Young Adult Hodgkin Case–Control Study (YHCCS) and the Epidemiology and Cancer Statistics Group Lymphoma Case–Control Study (ELCCS; http://www.elccs.info) (Supplementary Table 1). Full details of the SNEHD, YHCCS and ELCCS studies have been previously reported69–71. Briefly, SNEHD involved ascertainment of incident cases from Scotland and Northern England during 1993–1997. YHCCS was based on newly diagnosed cases aged 16–24 years from Northern England during 1991–1995. ELCCS comprised cases residing in the north or parts of southwest of England aged 16–69 years with newly diagnosed, non-human immunodeficiency virus-related cHL during 1998–2003. UK population controls matched to cases on age, sex and area of residence were obtained from SNEHD, YHCCS and ELCCS. The EBV status of cHL tumours in the UK replication 2 series was determined by immunohistochemical staining for EBV latent membrane antigen-1 and/or EBV EBV-encoded RNA in situ hybridisation using sections of paraffin-embedded material72, 73.
The fidelity of GWAS imputation was assessed by the concordance between imputed and directly genotyped SNP in a subset of samples (Supplementary Table 7). Replication genotyping of UK samples was performed using competitive allele-specific PCR KASP chemistry (LGC, Hertfordshire, UK). Primers, probes and conditions are detailed in Supplementary Table 14. Call rates for SNP genotypes were > 95% in each of the replication series. To ensure the quality of genotyping in assays, at least two negative controls and a set of duplicates were genotyped (concordance > 99%).
Meta-analysis
Meta-analyses were performed under a fixed-effects model using META v1.674. Cochran’s Q-statistic to test for heterogeneity and the I2 statistic to quantify the proportion of the total variation due to heterogeneity were calculated; an I2 value ≥ 75% is considered to be characteristic of large heterogeneity75. We used the test-based method of Higgins et al.76 to derive 95% CIs for I2 values (Supplementary Table 9). To estimate study power of the discovery GWAS phase, we made use of the CaTS online calculator77, assuming a risk allele frequency of 0.2 and genotype relative risk of 1.20.
Expression quantitative trait locus analysis
To examine the relationship between SNP genotype and gene expression, we carried out SMR analysis as per Zhu et al., 201625. Briefly, if bxy is the effect size of x (gene expression) on y (slope of y regressed on the genetic value of x), bzx is the effect of z on x, and bzy is the effect of z on y. Therefore bxy (bzy/bzx) is the effect of x on y. To distinguish pleiotropy from linkage where the top associated cis-eQTL is in LD with two causal variants, one affecting gene expression the other affecting trait, we tested for heterogeneity in dependent instruments, using multiple SNPs in each cis-eQTL region. Under the hypothesis of pleiotropy bxy values for SNPs in LD with the causal variant will be identical. Thus testing against the null hypothesis that there is a single causal variant is equivalent to testing heterogeneity in the bxy values estimated for the SNPs in the cis-eQTL region. For each probe that passed significance threshold for the SMR test, we tested the heterogeneity in the bxy values estimated for multiple SNPs in the cis-eQTL region using HEIDI.
We used publicly available LCL expression data from the MuTHER (n = 825)23 and GTEx consortium (n = 114)24. Briefly, GWAS summary statistics files were generated from the meta-analysis. Reference files were generated from merging 1000 Genomes Project phase 3 and UK10K (ALSPAC and TwinsUK) vcfs11, 12. As previously advocated, only probes with at least one eQTL P-value of < 5.0 × 10−8 were considered for SMR analysis25. We set a threshold for the SMR test of PSMR < 5.49 × 10−4 corresponding to a Bonferroni correction for 91 tests (91 probes with a top eQTL P < 5.0 × 10−8 across the 12 loci and two LCL eQTL data sets). For all genes passing this threshold, we generated plots of the eQTL and GWAS associations at the locus, as well as plots of GWAS and eQTL effect sizes (i.e., corresponding to input for the HEIDI heterogeneity test). HEIDI test P-values < 0.05 were considered as being reflective of heterogeneity. This threshold is conservative for gene discovery because it retains fewer genes than when correcting for multiple testing. SMR plots for significant eQTLs are shown in Supplementary Fig. 5.
Chromatin state dynamics
Enrichment of cHL risk SNPs with DNAse and enhancers is conducted using Haploreg v431. The overlap of cHL risk SNPs with enhancers in GM12878 cell is compared to a background model of all 1000 Genomes Project variants with a frequency above 5% in any population. The enrichment relative to these background frequencies was performed using a binomial test and a FDR P-value was subsequently calculated; we considered an FDR < 0.05 as being significant.
To examine enrichment in specific TF binding across risk loci, we adapted the variant set enrichment method of Cowper-Sal lari et al.78. For each risk locus, a region of strong LD (defined as r2 > 0.8 and D′ > 0.8) was determined, and these SNPs were termed the associated variant set (AVS). TF ChIP-seq uniform peak data were obtained from ENCODE for the GM12878 cell line, and included data for 82 TFs. For each of these marks, the overlap of the SNP in the AVS and the binding sites was determined to produce a mapping tally. A null distribution was produced by randomly selecting SNP with the same LD structure (generated from 1000 Genomes Project and UK10K data) as the risk associated SNP, and the null mapping tally calculated. This process was repeated 10,000 times, and approximate P-values were calculated as the proportion of permutations where the null mapping tally was greater or equal to the AVS mapping tally. An enrichment score was calculated by the tallies to the median of the null distribution. Thus the enrichment score is the number of standard deviations of the AVS mapping tally from the mean of the null distribution tallies.
Promoter capture Hi-C data
To map risk SNPs to interactions involving promoter contacts and identify genes involved in cHL susceptibility, we analysed promoter capture Hi-C data on the LCL cell line GM12878 as a model B-cell26. Reads from technical replicates (E-MTAB-2323) were combined before processing with HiCUP79. Significant interactions (i.e., score ≥ 5) on two biological replicates were determined using CHiCAGO80.
Co-heritability of Hodgkin lymphoma with autoimmune disease
We utilised LD regression to estimate genetic correlation between individual autoimmune diseases and cHL, NSHL and MCHL35. Summary statistics for published studies of coeliac disease41, systemic lupus erythematosus37, primary biliary cirrhosis39, rheumatoid arthritis36, MS38 and UC40 were downloaded from the ImmunoBase website (http://www.immunobase.org/).
Heritability analysis
We used genome-wide complex trait analysis to estimate the polygenic variance (i.e., heritability) ascribable to all genotyped and imputed GWAS SNPs33. SNPs were excluded based on low MAF < 0.01, poor imputation (info score < 0.9) and evidence of departure from HWE (P < 0.05). Individuals were excluded for poor imputation and where two individuals were closely related. A genetic relationship matrix of pairs of samples was used as input for the restricted maximum likelihood analysis to estimate the heritability explained by the selected set of SNPs. To transform the estimated heritability to the liability scale, we used the lifetime risk, for cHL, which is estimated to be 0.002 by SEER (https://seer.cancer.gov/statfacts/html/hodg.html).
Data availability
Genotype data that support the findings of this study have been deposited in the European Genome-phenome Archive (EGA) under accession codes EGAD00000000022 and EGAD00000000024.
Sequencing data, which forms the reference panel for imputation, have been deposited in the European Genome-phenome Archive (EGA) under accession codes EGAS00001000090, EGAD00001000195, EGAS00001000108.
Transcriptional profiling data from the MuTHER consortium that support the findings of this work have been deposited in the European Bioinformatics Institute (Part of the European Molecular Biology Laboratory, EMBL-EBI) under accession code E-TABM-1140.
Transcriptional profiling data from the Genotype-Tissue Expression (GTEx) project, that support the findings of this work are available here: https://www.gtexportal.org/
Transcription factor binding data that support the findings of this work are available here: http://genome.ucsc.edu/ENCODE/downloads.html.
Promoter capture Hi-C data in GM12878 cells that support the findings of this work have been deposited in the European Bioinformatics Institute (Part of the European Molecular Biology Laboratory, EMBL-EBI) under accession code E-MTAB-2323.
The remaining data contained within the paper and supplementary files are available from the author upon request.
Electronic supplementary material
Acknowledgements
In the United Kingdom, Bloodwise (LLR; 10021) provided principal funding for the study. Support from Cancer Research UK (C1298/A8362 supported by the Bobby Moore Fund) and the Lymphoma Research Trust is also acknowledged. A.S. is supported by a clinical fellowship from Cancer Research UK. For the UK-GWAS, sample and data acquisition were supported by Breast Cancer Now, the European Union and the Lymphoma Research Trust. The UK-GWAS made use of control genotyping data generated by the WTCCC. We acknowledge use of genotype data from the British 1958 Birth Cohort DNA collection, which was funded by the Medical Research Council Grant G0000934 and the Wellcome Trust Grant 068545/Z/02. A full list of the investigators who contributed to the generation of the data is available from http://www.wtccc.org.uk. Funding for this project was provided by the Wellcome Trust under awards 076113 and 085475. Patients for the new GWAS were ascertained through the National Study of Hodgkin Lymphoma Genetics (http://www.public.ukcrn.org.uk) and we thank the High-Throughput Genomics Group at the Wellcome Trust Centre for Human Genetics (funded by Wellcome Trust grant reference 090532/Z/09/Z) for the generation of Genotyping data. The BCAC study would not have been possible without the contributions of the following: Manjeet K. Bolla, Qin Wang, Kyriaki Michailidou and Joe Dennis. BCAC is funded by Cancer Research UK (C1287/A10118, C1287/A16563). For the BBCS study, we thank Eileen Williams, Elaine Ryder-Mills, Kara Sargus. The BBCS is funded by Cancer Research UK and Breast Cancer Now and acknowledges NHS funding to the National Institute of Health Research (NIHR) Biomedical Research Centre (BRC) and the National Cancer Research Network (NCRN). We thank the participants and the investigators of EPIC (European Prospective Investigation into Cancer and Nutrition). The coordination of EPIC is financially supported by the European Commission (DG-SANCO) and the International Agency for Research on Cancer. The national cohorts are supported by: Ligue Contre le Cancer, Institut Gustave Roussy, Mutuelle Générale de l’Education Nationale, Institut National de la Santé et de la Recherche Médicale (INSERM) (France); German Cancer Aid, German Cancer Research Center (DKFZ), Federal Ministry of Education and Research (BMBF) (Germany); the Hellenic Health Foundation, the Stavros Niarchos Foundation (Greece); Associazione Italiana per la Ricerca sul Cancro-AIRC-Italy and National Research Council (Italy); Dutch Ministry of Public Health, Welfare and Sports (VWS), Netherlands Cancer Registry (NKR), LK Research Funds, Dutch Prevention Funds, Dutch ZON (Zorg Onderzoek Nederland), World Cancer Research Fund (WCRF), Statistics Netherlands (The Netherlands); Health Research Fund (FIS), PI13/00061 to Granada, PI13/01162 to EPIC-Murcia, Regional Governments of Andalucía, Asturias, Basque Country, Murcia and Navarra, ISCIII RETIC (RD06/0020) (Spain); Cancer Research UK (14136 to EPIC-Norfolk; C570/A16491 and C8221/A19170 to EPIC-Oxford), Medical Research Council (1000143 to EPIC-Norfolk, MR/M012190/1 to EPIC-Oxford) (United Kingdom). We thank the SEARCH and EPIC teams, which were funded by a programme grant from Cancer Research UK (C490/A10124) and supported by the UK NIHR BRC at the University of Cambridge. We thank Breast Cancer Now and the Institute of Cancer Research (ICR) for support and funding of the UKBGS, and the study participants, study staff, and the doctors, nurses and other health-care providers and health information sources who have contributed to the study. We acknowledge NHS funding to the Royal Marsden/ICR NIHR BRC. UKGPCS would like to thank The Institute of Cancer Research and The Everyman Campaign for funding support. The UKGPCS acknowledges The Prostate Cancer Research Foundation, Prostate Action, The Orchid Cancer Appeal, The National Cancer Research Network UK, The National Cancer Research Institute (NCRI), the NIHR funding to the NIHR Biomedical Research data managers and consultants for their work in the UKGPCS study and urologists and other persons involved in the planning, and data collection of the CAPS study. Genotyping of the OncoArray was funded by the US National Institutes of Health (NIH) (U19 CA 148537 for ELucidating Loci Involved in Prostate cancer SuscEptibility (ELLIPSE) project and X01HG007492 to the Center for Inherited Disease Research (CIDR) under contract number HHSN268201200008I). Additional analytic support was provided by NIH NCI U01 CA188392 (PI: Schumacher). The PRACTICAL consortium was supported by Cancer Research UK Grants C5047/A7357, C1287/A10118, C1287/A16563, C5047/A3354, C5047/A10692, C16913/A6135, European Commission's Seventh Framework Programme grant agreement no. 223175 (HEALTH-F2-2009-223175), and The National Institute of Health (NIH) Cancer Post-Cancer GWAS initiative grant: No. 1 U19 CA 148537-01 (the GAME-ON initiative). We would also like to thank the following for funding support: The Institute of Cancer Research and The Everyman Campaign, The Prostate Cancer Research Foundation, Prostate Research Campaign UK (now Prostate Action), The Orchid Cancer Appeal, The National Cancer Research Network UK, The National Cancer Research Institute (NCRI) UK. We are grateful for support of NIHR funding to the NIHR Biomedical Research Centre at The Institute of Cancer Research and The Royal Marsden NHS Foundation Trust. The APBC BioResource, which form part of the PRACTICAL consortium, consists of the following members: Wayne Tilley, Gail Risbridger, Renea Taylor, Judith A Clements, Lisa Horvath, Vanessa Hayes, Lisa Butler, Trina Yeadon, Allison Eckert, Pamela Saunders, Anne-Maree Haynes, Melissa Papargiris. At the MRC University of Glasgow Centre for Virus Research, funding was provided by Leukaemia Lymphoma Research (12022). The Scotland and Newcastle Epidemiological Study of Hodgkin Disease (SNEHD) was funded by the Kay Kendall Leukaemia Fund and the Young Adult Hodgkin Case–Control Study (YHCCS) and the Epidemiology and Cancer Statistics Group Lymphoma Case–Control Study (ELCCS) were funded by Bloodwise. German funding was provided by the German Cancer Aid, the Harald Huppert Foundations, The German Federal Ministry of Education and Research (eMed, Cliommics 01ZX1309B), the Multiple Myeloma Research Foundation, the Heinz Nixdorf Foundation (Germany), the Ministerium für Innovation, Wissenschaft und Forschung des Landes Nordrhein-Westfalen and the Faculty of Medicine University Duisburg–Essen. For their help with UK sample collection we thank Hayley Evans, James Griffin, Joanne Micic, Susan Blackmore, Beverley Smith, Deborah Hogben, Alison Butlin, Jill Wood, Margot Pelerin, Alison Hart, Katarzyna Tomczyk and Sarah Chilcott-Burns. Finally, we are grateful to all the patients and individuals for their participation and the clinicians, investigators, other staff who contributed to sample and data collection.
Author contributions
A.S., R.S.H. and K.H. designed and provided overall project management. A.S. and R.S.H. drafted the manuscript. In the UK, R.S.H. and A.S. performed database development and oversaw laboratory analyses; R.H.A.J.S. and N.O. provided samples for UK-GWAS and UK-NSHLG-GWAS, L.W. and R.C. provided data on samples for UK-GWAS and UK-NSHLG-GWAS. D.E., P.P., A.D., J.P., F.C., R.E., Z.K.-J, K.M. and N.P. provided control samples for the UK-NSHLG-GWAS. A.S. performed bioinformatic and statistical analysis. In the UK P.L., G.O. and O.L. performed additional bioinformatic analyses. P.B. and A.S. performed sample and laboratory coordination. A.H. performed validation genotyping. For UK Replication 2, A.L. and D.M. prepared samples, T.L., E.K., E.R., D.M. and R.F.J. provided samples and data. In Germany, A.F., H.T. and M.I.d.S.F. performed bioinformatic and statistical analyses; P.H. and M.M.N. were responsible for German-GWAS analysis; K.-H.J. provided the German control samples; E.P.v.S. and A.E. were responsible for German cHL patients. All authors contributed to the final paper.
Competing interests
The authors declare no competing financial interests.
Footnotes
A full list of consortium members appears at the end of the paper.
Australian Prostate Cancer Research Centre-Qld, Institute of Health and Biomedical Innovation and School of Biomedical Science, Queensland University of Technology, Brisbane, QLD 4059, Australia
Publisher's note: Springer Nature remains neutral with regard to jurisdictional claims in published maps and institutional affiliations.
Amit Sud and Hauke Thomsen contributed equally to this work.
Kari Hemminki and Richard S. Houlston jointly supervised this work.
Change history
1/8/2019
The original version of this Article contained an error in the spelling of a member of the PRACTICAL Consortium, Manuela Gago-Dominguez, which was incorrectly given as Manuela Gago Dominguez. This has now been corrected in both the PDF and HTML versions of the Article.
Contributor Information
Richard S. Houlston, Email: richard.houlston@icr.ac.uk
The PRACTICAL consortium:
Brian E. Henderson, Christopher A. Haiman, Sara Benlloch, Fredrick R. Schumacher, Ali Amin Al Olama, Sonja I. Berndt, David V. Conti, Fredrik Wiklund, Stephen Chanock, Victoria L. Stevens, Catherine M. Tangen, Jyotsna Batra, Judith Clements, Henrik Gronberg, Johanna Schleutker, Demetrius Albanes, Stephanie Weinstein, Alicja Wolk, Catharine West, Lorelei Mucci, Géraldine Cancel-Tassin, Stella Koutros, Karina Dalsgaard Sorensen, Lovise Maehle, David E. Neal, Ruth C. Travis, Robert J. Hamilton, Sue Ann Ingles, Barry Rosenstein, Yong-Jie Lu, Graham G. Giles, Adam S. Kibel, Ana Vega, Manolis Kogevinas, Kathryn L. Penney, Jong Y. Park, Janet L. Stanford, Cezary Cybulski, Børge G. Nordestgaard, Hermann Brenner, Christiane Maier, Jeri Kim, Esther M. John, Manuel R. Teixeira, Susan L. Neuhausen, Kim De Ruyck, Azad Razack, Lisa F. Newcomb, Davor Lessel, Radka Kaneva, Nawaid Usmani, Frank Claessens, Paul A. Townsend, Manuela Gago-Dominguez, Monique J. Roobol, and Florence Menegaux
Electronic supplementary material
Supplementary Information accompanies this paper at doi:10.1038/s41467-017-00320-1.
References
- 1.Kuppers R. The biology of Hodgkin’s lymphoma. Nat. Rev. Cancer. 2009;9:15–27. doi: 10.1038/nrc2542. [DOI] [PubMed] [Google Scholar]
- 2.Smith A, et al. Lymphoma incidence, survival and prevalence 2004-2014: sub-type analyses from the UK’s Haematological Malignancy Research Network. Br. J. Cancer. 2015;112:1575–1584. doi: 10.1038/bjc.2015.94. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Hjalgrim, H. On the Aetiology of Hodgkin Lymphoma (Statens Serum Institut, 2011).
- 4.Mack TM, et al. Concordance for Hodgkin’s disease in identical twins suggesting genetic susceptibility to the young-adult form of the disease. N. Engl. J. Med. 1995;332:413–419. doi: 10.1056/NEJM199502163320701. [DOI] [PubMed] [Google Scholar]
- 5.Kharazmi E, et al. Risk of familial classical Hodgkin lymphoma by relationship, histology, age, and sex: a joint study from five Nordic countries. Blood. 2015;126:1990–1995. doi: 10.1182/blood-2015-04-639781. [DOI] [PubMed] [Google Scholar]
- 6.Cozen W, et al. A meta-analysis of Hodgkin lymphoma reveals 19p13.3 TCF3 as a novel susceptibility locus. Nat. Commun. 2014;5:3856. doi: 10.1038/ncomms4856. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Enciso-Mora V, et al. A genome-wide association study of Hodgkin’s lymphoma identifies new susceptibility loci at 2p16.1 (REL), 8q24.21 and 10p14 (GATA3) Nat. Genet. 2010;42:1126–1130. doi: 10.1038/ng.696. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Frampton M, et al. Variation at 3p24.1 and 6q23.3 influences the risk of Hodgkin’s lymphoma. Nat. Commun. 2013;4:2549. doi: 10.1038/ncomms3549. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Urayama KY, et al. Genome-wide association study of classical Hodgkin lymphoma and Epstein–Barr virus status–defined subgroups. J. Natl. Cancer Inst. 2012;104:240–253. doi: 10.1093/jnci/djr516. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Thomsen H, et al. Heritability estimates on Hodgkin/‘s lymphoma: a genomic- versus population-based approach. Eur. J. Hum. Genet. 2015;23:824–830. doi: 10.1038/ejhg.2014.184. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.The 1000 Genomes Project Consortium. A map of human genome variation from population-scale sequencing Nature. 2010;467:1061–1073. doi: 10.1038/nature09534. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Huang J, et al. Improved imputation of low-frequency and rare variants using the UK10K haplotype reference panel. Nat. Commun. 2015;6:8111. doi: 10.1038/ncomms9111. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Flavell JR, et al. Down-regulation of the TGF-beta target gene, PTPRK, by the Epstein-Barr virus encoded EBNA1 contributes to the growth and survival of Hodgkin lymphoma cells. Blood. 2008;111:292–301. doi: 10.1182/blood-2006-11-059881. [DOI] [PubMed] [Google Scholar]
- 14.Jiang X, et al. Deregulated expression in Ph + human leukemias of AHI-1, a gene activated by insertional mutagenesis in mouse models of leukemia. Blood. 2004;103:3897–3904. doi: 10.1182/blood-2003-11-4026. [DOI] [PubMed] [Google Scholar]
- 15.Kennah E, et al. Identification of tyrosine kinase, HCK, and tumor suppressor, BIN1, as potential mediators of AHI-1 oncogene in primary and transformed CTCL cells. Blood. 2009;113:4646–4655. doi: 10.1182/blood-2008-08-174037. [DOI] [PubMed] [Google Scholar]
- 16.Chan W-K, et al. A UPF3-mediated regulatory switch that maintains RNA surveillance. Nat. Struct. Mol. Biol. 2009;16:747–753. doi: 10.1038/nsmb.1612. [DOI] [PubMed] [Google Scholar]
- 17.Schreiber A, et al. Structural basis for the subunit assembly of the anaphase-promoting complex. Nature. 2011;470:227–232. doi: 10.1038/nature09756. [DOI] [PubMed] [Google Scholar]
- 18.Steidl C, et al. Gene expression profiling of microdissected Hodgkin Reed-Sternberg cells correlates with treatment outcome in classical Hodgkin lymphoma. Blood. 2012;120:3530–3540. doi: 10.1182/blood-2012-06-439570. [DOI] [PubMed] [Google Scholar]
- 19.Li, J. et al. Association of CLEC16A with human common variable immunodeficiency disorder and role in murine B cells. Nat. Commun. 6, 6804 (2015). [DOI] [PMC free article] [PubMed]
- 20.Banerjee A, Northrup D, Boukarabila H, Jacobsen SE, Allman D. Transcriptional repression of Gata3 is essential for early B cell commitment. Immunity. 2013;38:930–942. doi: 10.1016/j.immuni.2013.01.014. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Stanelle J, Döring C, Hansmann M-L, Küppers R. Mechanisms of aberrant GATA3 expression in classical Hodgkin lymphoma and its consequences for the cytokine profile of Hodgkin and Reed/Sternberg cells. Blood. 2010;116:4202–4211. doi: 10.1182/blood-2010-01-265827. [DOI] [PubMed] [Google Scholar]
- 22.Vijayakrishnan J, et al. A genome-wide association study identifies risk loci for childhood acute lymphoblastic leukemia at 10q26.13 and 12q23.1. Leukemia. 2017;31:573–579. doi: 10.1038/leu.2016.271. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Grundberg E, et al. Mapping cis- and trans-regulatory effects across multiple tissues in twins. Nat. Genet. 2012;44:1084–1089. doi: 10.1038/ng.2394. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Lonsdale J, et al. The Genotype-Tissue Expression (GTEx) project. Nat. Genet. 2013;45:580–585. doi: 10.1038/ng.2653. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Zhu Z, et al. Integration of summary data from GWAS and eQTL studies predicts complex trait gene targets. Nat. Genet. 2016;48:481–487. doi: 10.1038/ng.3538. [DOI] [PubMed] [Google Scholar]
- 26.Mifsud B, et al. Mapping long-range promoter contacts in human cells with high-resolution capture Hi-C. Nat. Genet. 2015;47:598–606. doi: 10.1038/ng.3286. [DOI] [PubMed] [Google Scholar]
- 27.Ramachandrareddy H, et al. BCL6 promoter interacts with far upstream sequences with greatly enhanced activating histone modifications in germinal center B cells. Proc. Natl Acad. Sci. USA. 2010;107:11930–11935. doi: 10.1073/pnas.1004962107. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Schneider C, et al. microRNA 28 controls cell proliferation and is down-regulated in B-cell lymphomas. Proc. Natl Acad. Sci. USA. 2014;111:8185–8190. doi: 10.1073/pnas.1322466111. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Hart DO, Santra MK, Raha T, Green MR. Selective interaction between Trf3 and Taf3 required for early development and hematopoiesis. Dev. Dyn. 2009;238:2540–2549. doi: 10.1002/dvdy.22083. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Singh TR, et al. BLAP18/RMI2, a novel OB-fold-containing protein, is an essential component of the Bloom helicase–double Holliday junction dissolvasome. Genes Dev. 2008;22:2856–2868. doi: 10.1101/gad.1725108. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Ward LD, Kellis M. HaploReg v4: systematic mining of putative causal variants, cell types, regulators and target genes for human complex traits and disease. Nucleic Acids Res. 2016;44:D877–D881. doi: 10.1093/nar/gkv1340. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Jia X, et al. Imputing amino acid polymorphisms in human leukocyte antigens. PLoS ONE. 2013;8:e64683. doi: 10.1371/journal.pone.0064683. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Yang J, Lee SH, Goddard ME, Visscher PM. GCTA: a tool for genome-wide complex trait analysis. Am. J. Hum. Genet. 2011;88:76–82. doi: 10.1016/j.ajhg.2010.11.011. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Kristinsson SY, et al. Autoimmunity and risk for Hodgkin’s lymphoma by subtype. Haematologica. 2009;94:1468–1469. doi: 10.3324/haematol.2009.010512. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Bulik-Sullivan BK, et al. LD score regression distinguishes confounding from polygenicity in genome-wide association studies. Nat. Genet. 2015;47:291–295. doi: 10.1038/ng.3211. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Okada Y, et al. Genetics of rheumatoid arthritis contributes to biology and drug discovery. Nature. 2014;506:376–381. doi: 10.1038/nature12873. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Bentham J, et al. Genetic association analyses implicate aberrant regulation of innate and adaptive immunity genes in the pathogenesis of systemic lupus erythematosus. Nat. Genet. 2015;47:1457–1464. doi: 10.1038/ng.3434. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Sawcer S, et al. Genetic risk and a primary role for cell-mediated immune mechanisms in multiple sclerosis. Nature. 2011;476:214–219. doi: 10.1038/nature10251. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Cordell HJ, et al. International genome-wide meta-analysis identifies new primary biliary cirrhosis risk loci and targetable pathogenic pathways. Nat. Commun. 2015;6:8019. doi: 10.1038/ncomms9019. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Anderson CA, et al. Meta-analysis identifies 29 additional ulcerative colitis risk loci, increasing the number of confirmed associations to 47. Nat. Genet. 2011;43:246–252. doi: 10.1038/ng.764. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Dubois PCA, et al. Multiple common variants for celiac disease influencing immune gene expression. Nat. Genet. 2010;42:295–302. doi: 10.1038/ng.543. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Chen FE, Huang DB, Chen YQ, Ghosh G. Crystal structure of p50/p65 heterodimer of transcription factor NF-kappaB bound to DNA. Nature. 1998;391:410–413. doi: 10.1038/34956. [DOI] [PubMed] [Google Scholar]
- 43.Izban KF, et al. Characterization of NF-kappaB expression in Hodgkin’s disease: inhibition of constitutively expressed NF-kappaB results in spontaneous caspase-independent apoptosis in Hodgkin and Reed-Sternberg cells. Mod. Pathol. 2001;14:297–310. doi: 10.1038/modpathol.3880306. [DOI] [PubMed] [Google Scholar]
- 44.Bargou RC, et al. Constitutive nuclear factor-kappaB-RelA activation is required for proliferation and survival of Hodgkin’s disease tumor cells. J. Clin. Invest. 1997;100:2961–2969. doi: 10.1172/JCI119849. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Lin YC, et al. A global network of transcription factors, involving E2A, EBF1 and Foxo1, that orchestrates B cell fate. Nat. Immunol. 2010;11:635–643. doi: 10.1038/ni.1891. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Tiacci E, et al. Analyzing primary Hodgkin and Reed-Sternberg cells to capture the molecular and cellular pathogenesis of classical Hodgkin lymphoma. Blood. 2012;120:4609–4620. doi: 10.1182/blood-2012-05-428896. [DOI] [PubMed] [Google Scholar]
- 47.Hertel CB, Zhou XG, Hamilton-Dutoit SJ, Junker S. Loss of B cell identity correlates with loss of B cell-specific transcription factors in Hodgkin/Reed-Sternberg cells of classical Hodgkin lymphoma. Oncogene. 2002;21:4908–4920. doi: 10.1038/sj.onc.1205629. [DOI] [PubMed] [Google Scholar]
- 48.Whiteman HJ, Farrell PJ. RUNX expression and function in human B cells. Crit. Rev. Eukaryot. Gene Expr. 2006;16:31–44. doi: 10.1615/CritRevEukarGeneExpr.v16.i1.30. [DOI] [PubMed] [Google Scholar]
- 49.Brady G, Whiteman HJ, Spender LC, Farrell PJ. Downregulation of RUNX1 by RUNX3 Requires the RUNX3 VWRPY sequence and is essential for Epstein-Barr virus-driven B-cell proliferation. J. Virol. 2009;83:6909–6916. doi: 10.1128/JVI.00216-09. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50.Betz BC, et al. Batf coordinates multiple aspects of B and T cell function required for normal antibody responses. J. Exp. Med. 2010;207:933–942. doi: 10.1084/jem.20091548. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 51.Lorenzo Y, et al. Differential genetic and functional markers of second neoplasias in Hodgkin’s disease patients. Clin. Cancer Res. 2009;15:4823–4828. doi: 10.1158/1078-0432.CCR-08-3224. [DOI] [PubMed] [Google Scholar]
- 52.Murray PG, Constandinou CM, Crocker J, Young LS, Ambinder RF. Analysis of major histocompatibility complex class I, TAP expression, and LMP2 epitope sequence in Epstein-Barr virus–positive Hodgkin’s disease. Blood. 1998;92:2477–2483. [PubMed] [Google Scholar]
- 53.Lenz TL, et al. Widespread non-additive and interaction effects within HLA loci modulate the risk of autoimmune diseases. Nat. Genet. 2015;47:1085–1090. doi: 10.1038/ng.3379. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 54.Pratama A, Vinuesa CG. Control of TFH cell numbers: why and how? Immunol. Cell. Biol. 2014;92:40–48. doi: 10.1038/icb.2013.69. [DOI] [PubMed] [Google Scholar]
- 55.Sharon E, et al. Genetic variation in MHC proteins is associated with T cell receptor expression biases. Nat. Genet. 2016;48:995–1002. doi: 10.1038/ng.3625. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 56.Greaves P, et al. Defining characteristics of classical Hodgkin lymphoma microenvironment T-helper cells. Blood. 2013;122:2856–2863. doi: 10.1182/blood-2013-06-508044. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 57.Diepstra A, et al. HLA class II expression by Hodgkin Reed-Sternberg Cells is an independent prognostic factor in classical Hodgkin’s lymphoma. J. Clin. Oncol. 2007;25:3101–3108. doi: 10.1200/JCO.2006.10.0917. [DOI] [PubMed] [Google Scholar]
- 58.Hammer C, et al. Amino acid variation in HLA class II proteins is a major determinant of humoral response to common viruses. Am. J. Hum. Genet. 2015;97:738–743. doi: 10.1016/j.ajhg.2015.09.008. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 59.Apple RJ, Erlich HA. Two new HLA DRB1 alleles found in African Americans: implications for balancing selection at positions 57 and 86. Tissue Antigens. 1992;40:69–74. doi: 10.1111/j.1399-0039.1992.tb01962.x. [DOI] [PubMed] [Google Scholar]
- 60.Busch R, Hill CM, Hayball JD, Lamb JR, Rothbard JB. Effect of natural polymorphism at residue 86 of the HLA-DR beta chain on peptide binding. J. Immunol. 1991;147:1292–1298. [PubMed] [Google Scholar]
- 61.Khankhanian P, et al. Meta-analysis of genome-wide association studies reveals genetic overlap between Hodgkin lymphoma and multiple sclerosis. Int. J. Epidemiol. 2016;45:728–740. doi: 10.1093/ije/dyv364. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 62.The Wellcome Trust Case Control Consortium. Genome-wide association study of 14,000 cases of seven common diseases and 3,000 shared controls Nature. 2007;447:661–678. doi: 10.1038/nature05911. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 63.Anderson CA, et al. Data quality control in genetic case-control association studies. Nat. Protoc. 2010;5:1564–1573. doi: 10.1038/nprot.2010.116. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 64.Howie BN, Donnelly P, Marchini J. A flexible and accurate genotype imputation method for the next generation of genome-wide association studies. PLoS Genet. 2009;5:e1000529. doi: 10.1371/journal.pgen.1000529. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 65.Marchini J, Howie B, Myers S, McVean G, Donnelly P. A new multipoint method for genome-wide association studies by imputation of genotypes. Nat. Genet. 2007;39:906–913. doi: 10.1038/ng2088. [DOI] [PubMed] [Google Scholar]
- 66.Clayton DG, et al. Population structure, differential bias and genomic control in a large-scale, case-control association study. Nat. Genet. 2005;37:1243–1246. doi: 10.1038/ng1653. [DOI] [PubMed] [Google Scholar]
- 67.Danecek P, et al. The variant call format and VCFtools. Bioinformatics. 2011;27:2156–2158. doi: 10.1093/bioinformatics/btr330. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 68.Scales M, Jäger R, Migliorini G, Houlston RS, Henrion MYR. visPIG - a web tool for producing multi-region, multi-track, multi-scale plots of genetic data. PLoS ONE. 2014;9:e107497. doi: 10.1371/journal.pone.0107497. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 69.Alexander FE, et al. Risk factors for Hodgkin’s disease by Epstein-Barr virus (EBV) status: prior infection by EBV and other agents. Br. J. Cancer. 2000;82:1117–1121. doi: 10.1054/bjoc.1999.1049. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 70.Jarrett RF, et al. The Scotland and Newcastle epidemiological study of Hodgkin’s disease: impact of histopathological review and EBV status on incidence estimates. J. Clin. Pathol. 2003;56:811–816. doi: 10.1136/jcp.56.11.811. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 71.Willett EV, Roman E. Obesity and the risk of Hodgkin lymphoma (United Kingdom) Cancer Causes Control. 2006;17:1103–1106. doi: 10.1007/s10552-006-0042-6. [DOI] [PubMed] [Google Scholar]
- 72.Lake A, et al. Mutations of NFKBIA, encoding IkappaB alpha, are a recurrent finding in classical Hodgkin lymphoma but are not a unifying feature of non-EBV-associated cases. Int. J. Cancer. 2009;125:1334–1342. doi: 10.1002/ijc.24502. [DOI] [PubMed] [Google Scholar]
- 73.Hjalgrim H, Engels EA. Infectious aetiology of Hodgkin and non-Hodgkin lymphomas: a review of the epidemiological evidence. J. Intern. Med. 2008;264:537–548. doi: 10.1111/j.1365-2796.2008.02031.x. [DOI] [PubMed] [Google Scholar]
- 74.Liu JZ, et al. Meta-analysis and imputation refines the association of 15q25 with smoking quantity. Nat. Genet. 2010;42:436–440. doi: 10.1038/ng.572. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 75.Higgins JP, Thompson SG. Quantifying heterogeneity in a meta-analysis. Stat. Med. 2002;21:1539–1558. doi: 10.1002/sim.1186. [DOI] [PubMed] [Google Scholar]
- 76.Thorlund K, et al. Evolution of heterogeneity (I2) estimates and their 95% confidence intervals in large meta-analyses. PLoS ONE. 2012;7:e39471. doi: 10.1371/journal.pone.0039471. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 77.Skol AD, Scott LJ, Abecasis GR, Boehnke M. Joint analysis is more efficient than replication-based analysis for two-stage genome-wide association studies. Nat. Genet. 2006;38:209–213. doi: 10.1038/ng1706. [DOI] [PubMed] [Google Scholar]
- 78.Cowper-Sallari R, et al. Breast cancer risk-associated SNPs modulate the affinity of chromatin for FOXA1 and alter gene expression. Nat. Genet. 2012;44:1191–1198. doi: 10.1038/ng.2416. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 79.Wingett S, et al. HiCUP: pipeline for mapping and processing Hi-C data. F1000Res. 2015;4:1310. doi: 10.12688/f1000research.7334.1. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 80.Cairns J, et al. CHiCAGO: robust detection of DNA looping interactions in capture Hi-C data. Genome Biol. 2016;17:127. doi: 10.1186/s13059-016-0992-2. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
Data Availability Statement
Genotype data that support the findings of this study have been deposited in the European Genome-phenome Archive (EGA) under accession codes EGAD00000000022 and EGAD00000000024.
Sequencing data, which forms the reference panel for imputation, have been deposited in the European Genome-phenome Archive (EGA) under accession codes EGAS00001000090, EGAD00001000195, EGAS00001000108.
Transcriptional profiling data from the MuTHER consortium that support the findings of this work have been deposited in the European Bioinformatics Institute (Part of the European Molecular Biology Laboratory, EMBL-EBI) under accession code E-TABM-1140.
Transcriptional profiling data from the Genotype-Tissue Expression (GTEx) project, that support the findings of this work are available here: https://www.gtexportal.org/
Transcription factor binding data that support the findings of this work are available here: http://genome.ucsc.edu/ENCODE/downloads.html.
Promoter capture Hi-C data in GM12878 cells that support the findings of this work have been deposited in the European Bioinformatics Institute (Part of the European Molecular Biology Laboratory, EMBL-EBI) under accession code E-MTAB-2323.
The remaining data contained within the paper and supplementary files are available from the author upon request.