Abstract
The G protein-coupled receptor opsin is a phospholipid scramblase that facilitates rapid transbilayer phospholipid exchange in liposomes. The mechanism by which opsin scrambles lipids is unknown. It has been proposed that lipid translocation may occur at protein-protein interfaces of opsin dimers. To test this possibility, we rationally engineered QUAD opsin by tryptophan substitution of four lipid-facing residues in transmembrane helix 4 (TM4) that is known to be important for dimerization. Atomistic molecular dynamics simulations of wild type and QUAD opsins combined with continuum modeling revealed that the tryptophan substitutions lower the energetically unfavorable residual hydrophobic mismatch between TM4 and the membrane, reducing the drive of QUAD opsin to dimerize. We purified thermostable wild type and QUAD opsins, with or without a SNAP tag for fluorescence labeling. Single molecule fluorescence measurements of purified SNAP-tagged constructs revealed that both proteins are monomers. Fluorescence-based activity assays indicated that QUAD opsin is a fully functional scramblase. However, unlike wild type opsin which dimerizes en route to insertion into phospholipid vesicles, QUAD opsin reconstitutes as a monomer. We conclude that an engineered opsin monomer can scramble phospholipids, and that the lipid-exposed face of TM4 is unlikely to contribute to transbilayer phospholipid exchange.
Introduction
Phospholipids flip-flop rapidly across disc membranes of retinal rod photoreceptor cells in an ATP-independent manner1,2. Biochemical reconstitution studies revealed that this phenomenon is due to the phospholipid scramblase activity of the G protein-coupled receptor (GPCR) opsin3–5. Purified opsin promotes transbilayer lipid exchange at a rate >10,000 s−1 when reconstituted into phosphatidylcholine liposomes. Scrambling is a constitutive activity of opsin4 that has been suggested to be necessary for homeostasis of photoreceptor disc membranes5. While the molecular basis of opsin-mediated scrambling is not known, two distinct mechanistic models have emerged4,5 in which the structural features needed for scrambling are proposed to be located either at the protein-lipid interface, or within the protein-protein interface of opsin dimers6. Differentiation between these models has been challenging because opsin dimerizes en route to reconstitution into lipid vesicles in which its scramblase activity is measured4, necessitating strategies to disrupt dimerization. A recent report indicated that opsin dimerization can be prevented by peptides that mimic transmembrane helices7; a similar peptide-based strategy was also deployed to disrupt dimerization of β2-adrenegeric receptors8. However, such peptides are not useful in clarifying the role of dimers in lipid scrambling as opsin-peptide complexes mimic dimeric interfaces that could potentially provide a lipid translocation pathway.
To address the role of opsin dimerization in lipid scrambling, we initially tested the scramblase activity of previously reported opsin mutants bearing amino acid substitutions in transmembrane (TM) helices 1 and 5 that have been proposed to be important for dimerization of opsin as well other GPCRs9–12. To this end, we used a reconstitution-based approach that was designed to reveal not only the effect of a particular mutation on opsin’s scramblase activity, but also to indicate with considerable precision the oligomeric state of the protein as it inserts into preformed vesicles during detergent-mediated reconstitution13,14. Using this approach we found that certain rhodopsin point mutants in TM1 and TM5 that are associated with autosomal dominant retinitis pigmentosa, reconstitute into vesicles as monomers but retain wild-type (WT)-like scramblase activity13. These studies indicated that opsin dimerization is not required for lipid scrambling and suggested a novel disease mechanism based on dimerization deficiency13. However, questions remained about how point mutations in specific TM helices can globally affect opsin’s ability to dimerize, and which structural elements of opsin are necessary (or dispensable) for lipid scrambling. To address these points, we considered that it would be important to target specific segments of the protein by rational mutagenesis with the goal of altering oligomerization properties in a predictive manner and studying the scrambling properties of the resulting oligomerization-deficient constructs.
In the present study we targeted TM4, a helix implicated in dimer formation across many Class A GPCRs9–12 that had not been considered in our initial studies13. We rationally designed an opsin variant (QUAD opsin) in which four lipid-facing residues in TM4 were modified to tryptophan. Molecular dynamics simulations combined with continuum modeling of the energetics of protein-lipid interactions indicated that the effect of the tryptophan substitutions is to lower the energetically costly residual hydrophobic mismatch between TM4 and the membrane, and thus to reduce the drive of QUAD opsin to dimerize via the TM4 interface. Single molecule fluorescence microscopy experiments revealed that both WT and QUAD opsins are monomers when purified in dodecyl-β-d-maltoside after expression in HEK293 cells, and fluorescence-based activity assays indicated that QUAD opsin scrambles phospholipids similarly to WT opsin. However, unlike WT opsin which dimerizes prior to insertion into phospholipid vesicles13, QUAD opsin reconstitutes into vesicles as a monomer. We therefore conclude that an engineered opsin monomer can scramble phospholipids, i.e. a dimer interface is not required for scrambling, and that the lipid-exposed face of TM4 is unlikely to contribute to transbilayer phospholipid exchange.
Results
Design of an opsin variant with impaired dimerization
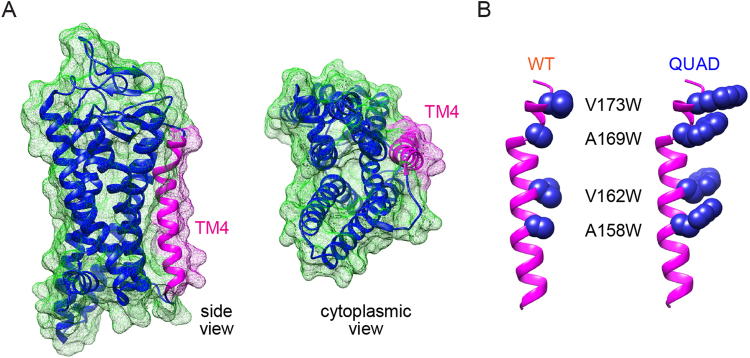
To design an opsin construct with impaired dimerization, we had the choice of modifying transmembrane (TM) helix 1 (TM1) and/or TM4, as both these helices have been strongly implicated in GPCR dimerization9. We targeted TM4 because it engages in fewer inter-TM contacts within the core helical bundle compared with other TM segments15 (Fig. 1A), suggesting that modifications to this helix would only minimally perturb the overall protein structure. Atomic force microscopy and electron cryomicroscopy studies identified TM4 residues 4.47, 4.51 and 4.58 as being important for rhodopsin dimerization16 (the residues are labeled according to the Ballesteros-Weinstein generic residue numbering scheme for GPCRs17), and crosslinking studies of the dopamine D2 receptor, another Class A (rhodopsin-like) GPCR, placed TM4 residues 4.51, 4.58 and 4.62 at the homodimer interface18. These residues correspond to amino acids on the outward-facing surface of TM4. We decided to substitute tryptophan (Trp) in place of these residues (4.47, 4.51, 4.58 and 4.62, corresponding to V173, A169, V162 and A158 in bovine opsin) (Fig. 1B). Because Trp has a bulky side-chain and is well tolerated in both hydrophobic and hydrophilic environments19–22, we reasoned that substitution of the selected residues by Trp would impact opsin dimerization without affecting the overall structure of the protein. Our expectation was that the quadruple mutant (QUAD opsin: V4.62W, A4.58W, V4.51W, A4.47W) would reconstitute into vesicles as a monomer, thereby providing us with the necessary tool to establish whether an opsin monomer can scramble lipids and potentially reveal whether TM4 plays a role in the scrambling process.
Figure 1.
Comparison of molecular models of the TM4 helix in wild-type (WT) and QUAD opsin constructs. (A) Alternative views of WT opsin (PDBID: 4J4Q). The TM helices are shown as ribbons and green mesh represents the surface of the molecule. The TM4 helix is colored in magenta, while the rest of the protein is depicted in dark blue. (B) Views of the TM4 helix of WT and QUAD opsin highlighting (in blue space-fill) four residues, at positions 1734.62, 1694.58, 1624.51, 1584.47, that were substituted by tryptophan in the QUAD construct.
We first characterized QUAD opsin computationally by quantifying the energetics of protein-lipid interactions from analysis of atomistic molecular dynamics simulations combined with continuum mean-field modeling (CTMD)23. We analyzed how the Trp substitutions would affect residual hydrophobic mismatch (RHM), defined as the hydrophobic mismatch between a protein and lipid bilayer unalleviated by membrane deformations24. Minimization of the energetic cost related to RHM has been suggested to be a driving force underlying the association of multi-spanning proteins in lipid membranes23–25. Indeed, by identifying structural elements where the RHM energy penalty is largest, it is possible to predict specific modes of association23.
Computational analyses suggest that QUAD opsin has a reduced tendency to dimerize
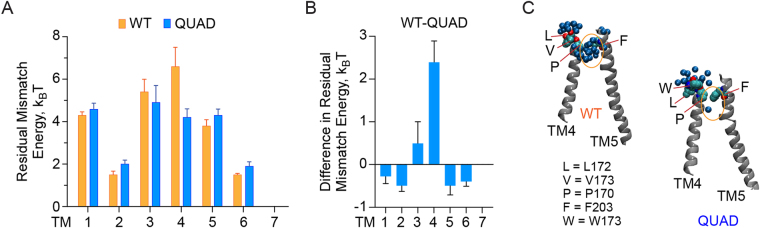
To quantify RHMs for WT and QUAD opsin, we first used the X-ray structure of WT opsin (PDBID: 4J4Q26) to build a 3D molecular model of QUAD opsin (see Methods). We then carried out extensive (totaling ~13 µs) atomistic molecular dynamics (MD) simulations of both WT and QUAD opsin in an explicit 9:1 (mol/mol) POPC/POPG lipid membrane and analyzed the resulting trajectories with a previously described CTMD protocol (see Experimental Procedures)24. The results show that TM4 has the largest RHM among all the TM helices in WT opsin (Fig. 2A), suggesting that TM4-mediated dimerization would provide a mode of association driven by RHM minimization. Furthermore, the RHM energy penalty is significantly lower at TM4 for QUAD compared to WT opsin (Fig. 2A,B). This is due to the different extent of water penetration between the extracellular ends of TM4 and TM5 (Fig. 2C) in the two constructs, resulting in larger RHM exposure at TM4 (at residue P1704.59 in particular) for WT opsin. RHM values for all other helices were similar (within kBT) for the two constructs (Fig. 2A,B). These results suggest that TM4 is an important driver of opsin dimerization in the membrane, and that the quadruple Trp substitutions in TM4 are expected to decrease the propensity for dimerization by lowering RHM. We therefore proceeded to test experimentally whether QUAD opsin (i) scrambles lipids and (ii) reconstitutes into vesicles as a monomer.
Figure 2.
Residual hydrophobic mismatch (RHM) at TM helices in WT and QUAD opsin. (A) RHM energies were calculated at TM helices for WT and QUAD opsin, using microsecond-scale atomistic MD simulations (see Methods) in an explicit 9:1 mixture of 1-palmitoyl-2-oleoyl-sn-glycero-3-phosphocholine (POPC) and 1-palmitoyl-2-oleoyl-sn-glycero-3-(1′-glycerol) (POPG); error bars represent standard deviation of RHM measurements carried out on overlapping time intervals of the MD trajectory. (B) Difference in RHM energies between WT and QUAD opsin calculated from the data in panel A. The difference in RHM at the TM4 helix was statistically highly significant (p value < 0.002 from unpaired t-test). (C) Final snapshots from the simulations illustrating the source of RHM energies at TM4. Shown are TM4 and TM5 (in cartoon), and amino acid residues as indicated (van der Waals representation). Dark blue spheres are water oxygens within 5 Å of these residues. Water accumulation at the exoplasmic ends of TM4 and TM5 in WT opsin (region within orange oval) breaks hydrophobic contacts between P170 and F203, resulting in a large RHM at TM4. This hydrophobic contact is intact in the QUAD protein, thus reducing the RHM.
Expression of QUAD opsin
Using HEK293S GnTI− cells for expression of homogeneously N-glycosylated proteins27, we obtained purified QUAD opsin in yields comparable to that of WT opsin (Fig. 3A). Analysis of GFP-tagged QUAD opsin by fluorescence size exclusion chromatography in 0.1% (w/v) dodecyl-β-d-maltoside (DDM) revealed a symmetric monodisperse profile identical to that of GFP-tagged WT opsin (Fig. 3B), indicating that the Trp substitutions in QUAD opsin do not affect the overall structure of the protein.
Figure 3.
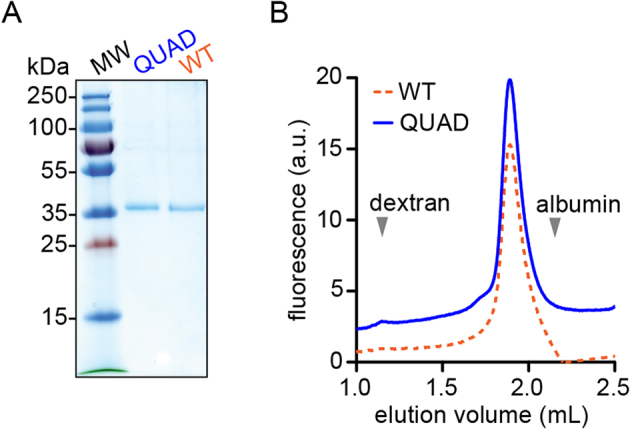
Expression of QUAD opsin. (A) Protein expression. WT and QUAD opsins were expressed as thermostable, FLAG-tagged variants in HEK293S GnTI− cells, extracted with dodecyl-β-D-maltoside (DDM) and purified by FLAG affinity chromatography. An SDS-PAGE Coomassie-stained gel is shown of samples obtained from the same number of cell equivalents. (B) Fluorescence size exclusion chromatography (FSEC) analysis. GFP-tagged WT and QUAD opsins were expressed in HEK293S GnTI− cells, and a DDM extract of the cells was analyzed (without purification of the expressed proteins) by FSEC. Both proteins display mono-disperse profiles (the trace for QUAD opsin is vertically displaced from that of WT opsin for clarity). Elution positions of dextran (void volume marker) and albumin are shown.
Single molecule fluorescence experiments indicate that purified WT opsin and QUAD opsin are monomers
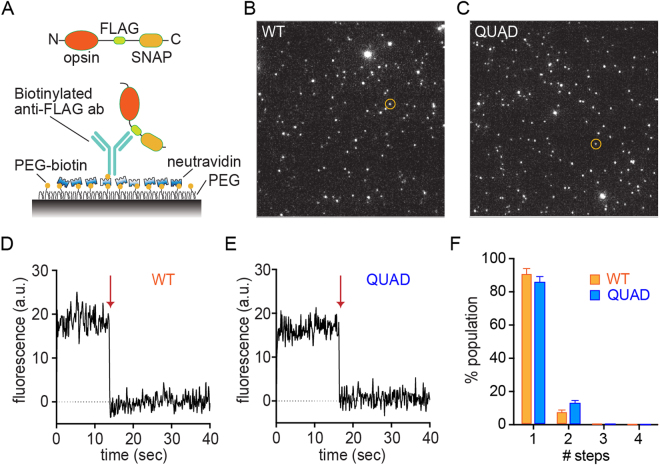
It has been previously reported that opsin is monomeric when purified in 0.1% (w/v) DDM4,13,28 and we sought to confirm that this was also the case for our opsin variants. Single molecule subunit counting using photo-bleaching step analysis has been established as a useful approach to determine the distribution of oligomeric states of proteins29,30. Recently, single molecule pulldown (SiMPull) was introduced as a means of isolating individual protein complexes on a polyethylene glycol (PEG)-passivated coverslip at low densities for single molecule imaging analysis31. SiMPull has been successfully used in conjunction with subunit counting to analyze the monomer/dimer equilibrium of membrane proteins, including GPCRs31,32. In preliminary experiments, we analyzed GFP-tagged WT opsin expressed in HEK293T cells using SiMPull of fresh lysates in 0.1% (w/v) DDM and observed that 91% of the fluorescent spots were bleached in a single step indicating that the protein is a monomer (Supplementary Fig. S1). We next used this approach with affinity purified FLAG-SNAP-tagged WT and QUAD opsin (Fig. 4A) that we labeled with a benzylguanine-tagged red fluorophore (“SNAP-Surface-549”) via SNAP-tag chemistry33. The efficiency of SNAP-tag labeling under our reaction conditions is expected to be >80%34, comparable to the fraction of mature GFP in GFP-fusion proteins29. Individual proteins were immobilized using an anti-FLAG antibody (the antibody was biotinylated to enable its capture onto neutravidin-coated cover slips (Fig. 4A)) which interacts with the FLAG epitope situated between opsin and the SNAP tag in the fusion protein (Fig. 4A). Individual spots were visualized using total internal reflection (TIRF) microscopy (see Experimental Procedures; Fig. 4B,C). For most spots fluorophore bleaching occurred within 30 s (Fig. 4D,E) and bleaching analysis revealed that ~90% of spots bleached in 1-step (Fig. 4F), similar to our preliminary observation with WT-opsin-GFP (Supplementary Fig. S1). A small population of two-step bleaching spots was observed which is likely due to occasional coincidental localization of two proteins within a diffraction limited spot and/or because of the bivalency of the antibody. This background level of 2-step bleaching observed in our experiments (Fig. 4F) is consistent with previous studies with monomeric yellow fluorescent protein31. Our results strongly indicate that both purified WT and QUAD opsins are monomers when affinity purified in 0.1% (w/v) DDM.
Figure 4.
Single molecule fluorescence measurements reveal that purified WT and Quad opsins are monomers. (A) Schematic illustration of the single molecule pulldown (SiMPull) set up. (B) Representative TIRF image of SNAP-surface 549-labeled WT opsin-FLAG-SNAP; the circled spot corresponds to the photobleaching trace in panel D. (C) Representative TIRF image of SNAP-surface 549-labeled QUAD opsin-FLAG-SNAP; the circled spot corresponds to the photobleaching trace in panel E. (D) Trace depicting 1-step photobleaching of the circled spot from panel B when exposed to a 561-nm laser beam at time 0 s. The arrow depicts the point at which photobleaching occurred. (E) Trace depicting 1-step photobleaching of the circled spot from panel C when exposed to a 561-nm laser beam at time 0 s. The arrow depicts the point at which photobleaching occurred. (F) Fraction of the total population of spots that show 1-, 2-, 3-, or 4-step photobleaching (1102 and 1083 spots were analyzed for WT and QUAD opsin constructs, respectively). Error bars indicate standard errors calculated from 5 movies for each condition (169–257 spots were analyzed per movie).
Scramblase activity of QUAD opsin
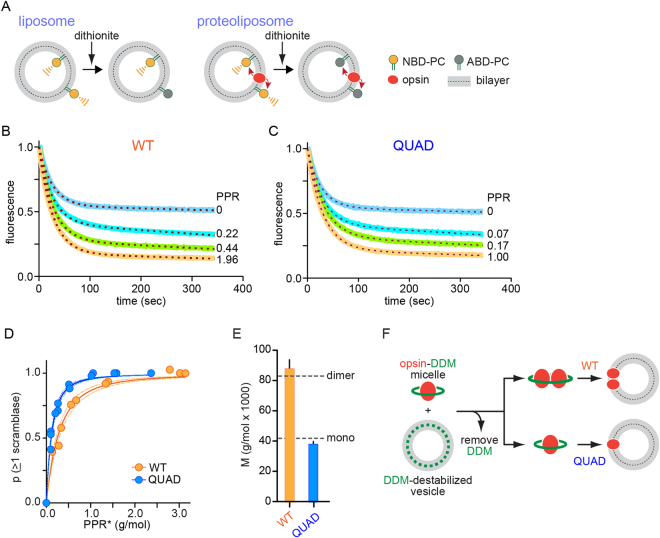
We next reconstituted QUAD opsin in large unilamellar vesicles to test its scramblase activity. For the scramblase assay (Fig. 5A), phospholipid vesicles are symmetrically reconstituted with a trace amount of fluorescent NBD-labeled phosphatidylcholine (NBD-PC) as well as the protein to be tested. NBD-PC in the outer leaflet of the vesicle is detected by dithionite, a membrane-impermeant reducing agent that eliminates NBD fluorescence. Treatment of protein-free vesicles with dithionite should lower fluorescence by 50% because NBD-PC molecules in the outer leaflet are reduced whereas those in the inner leaflet are protected. For vesicles reconstituted with a scramblase, e.g. WT opsin, dithionite treatment should result in 100% loss of fluorescence as NBD-PC can be exchanged between the inner and outer leaflet. In reality, the maximum extent of fluorescence loss does not exceed ~85% (Fig. 5B) because a fraction of vesicles is refractory to reconstitution4,13,35.
Figure 5.
QUAD opsin scrambles lipids as a monomer. (A) Schematic representation of the fluorescence-based scramblase activity assay. (B) Representative fluorescence traces of scrambling by WT opsin, reconstituted at different protein/phospholipid ratios (PPR) into POPC:POPG (9:1) vesicles containing a trace amount of NBD-PC. (C) As in panel B, for QUAD opsin. (D) The extent of fluorescence reduction in the scramblase assay was determined for vesicles reconstituted with QUAD and WT opsin over a range of PPR values, 0–3 g/mol and the data were transformed into plots of p(≥1) scramblase (the probability of a vesicle having at least one scramblase) vs PPR* (related to measured PPR, see Experimental Procedures). The solid lines are data fits (Poisson analysis, see Experimental Procedures and Table 2), and the dashed lines are the 95% confidence interval for the fits. (E) Molar mass of the functionally reconstituted scramblase deduced from the data shown in panel D (see Table 2). (F) Schematic illustration showing that whereas both WT and QUAD opsin are monomers when added to DDM-destabilized phospholipid vesicles (based on Fig. 4), WT opsin dimerizes (multimerization is not shown here for simplicity) en route to reconstitution whereas QUAD opsin reconstitutes as a monomer (based on this figure, panel E). Direct evidence for dimerization of WT opsin during detergent withdrawal was previously obtained through co-immunoprecipitation studies13.
The extent of fluorescence loss in vesicles reconstituted with QUAD opsin was greater than that seen for protein-free vesicles (Fig. 5C) indicating that QUAD opsin - like WT opsin - is a scramblase. A control experiment using encapsulated NBD-Glucose4,35 confirmed that dithionite cannot cross the membrane of reconstituted vesicles (Supplementary Fig. S2), and that the only explanation for the greater extent of reduction of NBD-PC in WT-opsin and QUAD-opsin-containing vesicles is scrambling of the lipid reporter from the inner leaflet of the vesicles to the dithionite-accessible outer leaflet. The kinetics of NBD-PC fluorescence loss (t1/2 ~20 s) was the same for both opsin-containing vesicles and protein-free vesicles (Fig. 5B,C, Table 1) indicating that the rate at which dithionite reduces the NBD fluorophore is slow compared with the rate of scrambling. Within this limit of the time resolution of the assay we conclude that QUAD opsin and WT opsin have similar scramblase activity.
Table 1.
Fit parameters for fluorescence reduction traces.
| n | Half-life (sec) | Magnitude of line slope S (sec−1 × 104) | |
|---|---|---|---|
| No protein | 6 | 20.23 ± 1.79 | 1.20 ± 0.22 |
| WT | 20 | 22.63 ± 1.26 | 1.40 ± 0.16 |
| QUAD | 15 | 23.37 ± 0.87 | 1.40 ± 0.13 |
Fluorescence traces from scramblase assays performed over a range of PPR values, including those shown in Fig. 5, were fit to the equation F(t) = (1 − Plateau)*exp(−K*t) + Plateau − S*t, where F(t) = fluorescence at time t, t = 0 sec is the time of dithionite addition, and S = absolute value of the slope of the linear component. The results are provided as mean ± SEM (n = number of independent vesicle reconstitution samples). The standard error of individual fits was at least an order of magnitude lower than the SEM. Values of half-life (=0.69/K) and S are provided. The similar half-life values obtained irrespective of the PPR of the vesicles confirms that the dithionite reaction is rate-limiting in all cases. Only the plateau values (not shown here) vary with PPR – these are used to calculate the data fits described in Table 2.
QUAD opsin reconstitutes into phospholipid vesicles as a monomer
We observed that we needed to reconstitute less QUAD opsin compared with WT opsin in order to obtain the maximum level of fluorescence reduction in the scramblase assay (Fig. 5B,C). As both proteins were reconstituted with similar efficiency (~70%, see Experimental Procedures) this observation suggested that on a per protein molecule basis, QUAD opsin could populate more vesicles and render them scramblase-competent than WT opsin. Because a single reconstitution event confers scramblase activity to a liposome4, the relationship between the fraction of scramblase-active liposomes and the protein to phospholipid ratio (PPR) of the reconstituted samples can be used to determine the molar mass of the functionally reconstituted protein. Using this approach, we showed previously that WT opsin reconstitutes into vesicles as a dimer, and to a small extent as a higher order oligomer4,13. We reinvestigated the reconstitution behavior of WT opsin in order to make a side-by-side comparison with QUAD opsin. Analysis of the data shown in Fig. 5D,E indicates that WT opsin functionally reconstitutes with molar mass 87.9 ± 5.79 kDa (±indicates standard error associated with the data fit), slightly higher than the mass of an opsin dimer (83.4 kDa), consistent with previous results (Table 2). In contrast (Fig. 5D,E), QUAD opsin functionally reconstitutes with molar mass 38.0 ± 1.73 kDa, equivalent to that of an opsin monomer (41.7 kDa) (Table 2).
Table 2.
Analysis of p(≥1 scramblase) vs PPR* plots.
| α (x 104) | M (g/mol) | ~n (Opsin n-mer) | |
|---|---|---|---|
| WT | 8.17 ± 0.54 | 87,905 ± 5, 810 | ≥2 |
| QUAD | 18.90 ± 0.86 | 38,000 ± 1, 730 | 1 |
The data were fit to equation 5 (Experimental Procedures) where α = 16/Mε2 is the fit constant, with units of mol.g−1.nm−2 and x is PPR*. See Experimental Procedures for details. M is the molar mass of the functional scramblase deduced from the fit, using ε = 0.472 nm as the cross-sectional radius of a phospholipid. The standard error associated with the fit is indicated. The molar mass of an opsin monomer is 41,700 g/mol.
Discussion
Our data indicate that QUAD opsin scrambles phospholipids as a monomer (Fig. 5F) and that, within the limit of the time resolution of the scramblase assay, both WT and QUAD opsins are equivalently active in scrambling lipids (Table 1). Because the protein dependence plot in Fig. 5D has no inflection point we can rule out a scenario in which two or more QUAD opsins reconstitute independently into the same vesicle and subsequently dimerize in order to generate a functional scramblase; in this scenario, the protein-dependence plot would be sigmoidal, indicative of cooperativity, which is not the case here. Our observations clearly rule out a model of scrambling where lipid translocation necessarily occurs at the interface between protomers in an opsin dimer. This conclusion supports our recent report in which we showed that certain rhodopsin mutants associated with autosomal dominant retinitis pigmentosa also reconstitute into vesicles as monomers while retaining WT-like scramblase activity13. The combined results of the present and previous13 studies suggest that mutagenesis of any of the three TM helices implicated in dimerization (TM1 and TM5 (ref.13) and TM4 (present study)) globally affects the ability of opsin to dimerize via any of its potential dimerization interfaces as it transitions from a DDM-soluble monomer to a membrane-integrated protein during reconstitution (Fig. 5F). The mechanistic basis for this effect remains to be elucidated.
Importantly, while the bulky tryptophan substitutions of TM4 in QUAD opsin exert a profound effect on the magnitude of the residual hydrophobic mismatch between TM4 and the membrane (Fig. 2), thereby reducing the drive of QUAD opsin to dimerize via the TM4 interface and, indeed, affecting opsin dimerization via any interface (see above), the same substitutions have no detectable effect on the ability of the protein to scramble lipids. Thus, we suggest that an opsin monomer scrambles phospholipids without direct participation of TM4. Despite this advance, the molecular mechanism by which opsin translocates phospholipids remains elusive. Apart from several class-A GPCR proteins and the retinylidene protein bacteriorhodopsin that display opsin-like phospholipid scramblase activity when reconstituted into phospholipid vesicles4,36, the only other known phospholipid scramblases belong to the TMEM16 family of Ca2+-dependent ion channels and/or scramblases35,37,38. These proteins are homodimers, but each monomer possesses a membrane facing hydrophilic groove that likely provides the path for lipid headgroup and ion translocation37–40. Likewise, bacteriorhodopsin presents a series of membrane-exposed polar residues that could provide a transbilayer path for lipid translocation36. As opsin lacks these features, it may generate a translocation path dynamically, or scramble lipids by a mechanism that is distinct from that suggested for the TMEM16 scramblases and bacteriorhodopsin. For example, features of the opsin-lipid interface might create disturbances in the membrane that could promote scrambling. These disturbances could cause local thinning of the bilayer or create lipid packing defects. Interestingly, previous computational studies41 showed that rhodopsin is permissive to a surprisingly high degree of water permeation between its TM helices. These deeply penetrant water molecules may provide the route for lipid headgroups to negotiate the otherwise hydrophobic milieu of the interior of the membrane. Detailed evaluation of this concept by both computational and experimental approaches will be the subject of future work.
Experimental Procedures
Computational methods (protein constructs)
All computations were based on the X-ray structure of retinal-free opsin (PDBID 4J4Q). This structure also contains opsin-bound synthetic GαCT peptide26 which was not considered here. A quadruple mutant opsin (QUAD) with V1734.62W, A1694.58W, V1624.51W and A1584.47W mutations in TM4 helix (the numbers in superscript correspond to residue identity based on the Ballesteros-Weinstein generic residue numbering scheme for GPCRs17) was modeled using the homology modeling module of modeller software and the final model was chosen based on the best DOPE score42,43. The model for the QUAD mutant was energy minimized using TINKER molecular modeling software (http://dasher.wustl.edu/tinker/) and the OPLSAA force field44, before being used in molecular dynamics simulations (see below). RMSD of the QUAD model with respect to the WT structure, as calculated using Chimera software, was 0.07 Å for the backbone atoms.
Computational methods (membrane-protein complexes)
Using CHARMM-GUI web facility45–47 the wild type (WT) and QUAD opsin molecular models were embedded into a lipid membrane consisting of a 9:1 mixture of POPC (1-palmitoyl-2-oleoyl-sn-glycero-3-phosphocholine) and POPG (1-palmitoyl-2-oleoyl-sn-glycero-3-phospho-(1′-rac-glycerol)) lipids. The protein to lipid ratio was 1:330. After adding a solvation box containing 100 mM NaCl the total system size was ~131,000 atoms.
Molecular dynamics (MD) simulations
All-atom MD simulations of the WT and the QUAD constructs in the corresponding membrane environments were initiated with a previously established multi-step equilibration protocol48. During this stage, the backbone of the protein was first harmonically constrained and the constraints on the protein backbone were released gradually in three steps of 5 ns each, changing the restrain force constants from 1, to 0.5, and 0.1 kcal/ (mol Å2), respectively. This step was followed by unbiased MD simulations of the two proteins, ~250 ns for the WT protein and ~315 ns for QUAD opsin. The simulations were carried out using the NAMD 2.10 package and the latest CHARMM36 force field parameters for proteins, lipids, and ions47,49. The simulations implemented rigidbonds all option, 2fs integration time-step, PME for electrostatics interactions, and were carried out in NPT ensemble under semi-isotropic pressure coupling conditions, at a temperature of 298 K. The Nose-Hoover Langevin piston algorithm was used to control the target P = 1 atm pressure with the LangevinPistonPeriod set to 100 fs and LangevinPistonDecay set to 50 fs. The van der Waals interactions were calculated applying a cutoff distance of 12 Å and switching the potential from 10 Å.
After this equilibration phase, the velocities of all the atoms in the two systems (i.e. WT and QUAD opsin) were reset (at T = 298 K using random number seed) and 4 independent unbiased MD simulations per construct were carried out using ACEMD software50 resulting in a cumulative MD simulation time of ~13 µs (1.8 µs, 1.4 µs, 1.4 µs, and 1.5 µs for WT; 1.9 µs, 1.5 µs, 2.0 µs, and 1.4 µs for QUAD opsin). The simulations with ACEMD implemented CHARMM36 force fields, the PME method for electrostatic calculations, and were carried out according to the protocol developed at Acellera and implemented by us50,51 with 4 fs integration time-step and the standard mass repartitioning procedure for hydrogen atoms implemented in ACEMD. The computations were conducted under the NVT ensemble (at T = 298 K), using the Langevin Thermostat with Langevin Damping Factor set to 0.1.
Energetics of membrane-protein interactions: calculation of Residual Hydrophobic Mismatch (RHM)
To quantify the energetics of hydrophobic mismatch between opsin and the lipid bilayer we used the Continuum-Molecular Dynamics (CTMD) approach described previously24. Briefly, CTMD calculates RHM, the hydrophobic mismatch unalleviated by membrane deformation. The RHM energies were computed as described24,52–55 from the surface area (SA res,i) of the i th residue participating in unfavorable RHM interactions. For hydrophobic residues, SA res,i is the area of the residue that is exposed outside the hydrophobic core of the lipid bilayer. For polar residues, SA res,i is the part of the residue that is exposed on the surface of the protein, but is situated within the hydrophobic core of the lipid bilayer. Practically, SA res,i is quantified from residue-specific solvent accessible surface areas (SASA) obtained with NACCESS considering the solute as follows: i) the protein and the hydrophobic core of lipid bilayer (defined as the bilayer region within C2 lipid atoms), SA mem,i; ii) the protein only, SA prot,i. For hydrophobic residues
| 1 |
and for polar residues
| 2 |
The corresponding RHM energy penalty is directly proportional to SA res,i, and at a particular TM is given by
| 3 |
where N res is the number of residues in the TM and σ res proportionality coefficient is taken to be 0.0028 kcal/(mol.Å2). As described previously56, interfacial Trp is not penalized as it can be favorably accommodated at the interface. RHM for Arg and Lys located close to the membrane headgroups is not considered as well since these amino acids alleviate hydrophobic mismatch by snorkeling57. Lastly, Ser and Thr are not penalized as their polar parts form H-bonds with the helix backbone of the protein58.
Statistical analysis of the RHM results
RHM data for each protein construct were generated from the CTMD analyses performed on 4 independent MD simulation replicates (see above). Error bars reported in Fig. 2A,B are standard deviations from the mean calculated from a bootstrapping method involving calculating the mean from three randomly chosen replicates and repeating the procedure four times. Statistical significance of differences in RHM values at each TM helix between the WT and QUAD opsins was assessed by unpaired t-test.
Vectors for opsin expression
We previously described a construct encoding C-terminal 3X FLAG-tagged thermostable opsin4. Modifications to this construct were accomplished by two-step overlap extension PCR and cloned into the NotI/EcoRI sites of the pMT3 vector. The sequence for each construct was verified at the Cornell University Life Sciences Core Laboratories Center. For GFP-tagged WT and QUAD opsins we used the pEGFP-N3 plasmid containing mouse opsin cDNA (kindly provided by Adam Smith (University of Akron))59. Mutations were introduced by a single two-step overlap extension PCR. PCR fragments were restricted with EcoRI/BamHI and inserted into pEGFP-N3. We previously described the SNAP-tagged wild type opsin construct13; the corresponding QUAD opsin construct was generated using the Gibson assembly cloning kit from New England Biolabs60,61.
Opsin expression, purification and fluorescence labeling
Details of cell culture, protein expression, purification and quantification are provided elsewhere4,13. Briefly, WT and QUAD opsin were expressed as thermostable (N2C/D282C), C-terminally FLAG-tagged variants in HEK293S GnTI− cells, purified by FLAG affinity chromatography4 and quantified by Coomassie-staining after SDS-PAGE, using an in-gel bovine serum albumin standard. The average yield for WT and QUAD opsin was ~6 µg per 107 transfected cells. Thermostable, FLAG-tagged WT and QUAD opsins were also expressed with a C-terminal SNAP tag. These proteins were fluorescently labeled using SNAP-tag chemistry33; labeling was performed after capture of the protein on FLAG affinity resin, prior to elution of the protein with FLAG peptide. Briefly, after capture of the protein, the resin was washed with wash buffer (0.1% (w/v) DDM, 50 mM HEPES pH 7.4, 100 mM NaCl). SNAP-surface-549 dye diluted in wash buffer (~0.2 nmol per µg of protein) was then added and the sample was incubated overnight at 4 °C with end-over-end mixing. The resin was washed to remove unbound dye before eluting the labeled protein with FLAG peptide. Purified labeled protein was characterized by SDS-PAGE, using a Typhoon imager to visualize fluorescence and Coomassie staining to quantify protein as described above. SNAP labeling efficiency is expected to be >80% under our conditions34, comparable to the fraction of mature (fluorescent) GFP in preparations of WT-opsin-GFP29.
Fluorescence size exclusion chromatography of GFP-tagged opsins
C-terminally GFP-tagged WT and QUAD opsins were expressed in HEK293S GnTI− cells and extracted in buffer (50 mM HEPES, pH 7.4, 100 mM NaCl) containing 0.1% (w/v) DDM. After centrifugation to remove insoluble material, the supernatant was filtered and analyzed by fluorescence size exclusion chromatography on a Superdex 200 Increase 5/150 GL column (GE Healthcare Life Sciences) using a Shimadzu LC-20AD Prominence liquid chromatograph equipped with an RF-20A Prominence fluorescence detector (excitation λ = 395 nm, emission λ = 507 nm).
Single molecule subunit counting
For subunit counting experiments, a passivated glass flow chamber was prepared as previously described31. Briefly, chambers were prepared using mPEG/biotin PEG-passivated quartz slides and coverslips. 0.2 mg/mL Neutravidin was added for 2 minutes at RT followed by washing with T50 buffer (10 mM Tris-HCL pH 8.0, 50 mM NaCl). To the Neutravidin coated chambers, 10–20 nM of biotinylated monoclonal anti-FLAG antibody (Sigma, cat. no. F9291) was added and incubated for 30 mins at RT followed by washing with T50 buffer. The flow chamber was placed on an inverted TIRF microscope (Olympus IX73 with cellTIRF system) and purified SNAP-surface549 (New England BioLabs) labeled QUAD-FLAG-SNAP or WT-FLAG-SNAP was immobilized at a density that allowed clear resolution of individual spots. The fluorophore was excited using a DPSS 561 nm laser, imaged through a 100x objective (NA = 1.49) and the data was collected at room temperature. Images were acquired with a scMOS detector camera (Hamamatsu ORCA-flash4.0 V3) at 20 Hz using Olympus cellSens software. Spots were analyzed using a previously described program29. Briefly, individual, immobile spots were identified and, after background subtraction and application of Gaussian weighting, manually characterized as bleaching in 1, 2, 3, 4, or 5 steps (or deemed uncountable). Data in bar graphs represent averages of the step distributions for different movies.
Liposome and proteoliposome preparation
Unilamellar liposomes were prepared from a 9:1 (mol/mol) mixture of POPC and POPG (from Avanti Polar Lipids) as described13,14. The vesicles were reconstituted with opsin and C6NBD-PC (Avanti Polar Lipids) after detergent-mediated destabilization as described4,13,14. For experiments with NBD-Glucose (Invitrogen), NBD-PC was omitted. Recovery of protein and phospholipid after reconstitution was ~70%13.
Scramblase assays and analysis of scramblase reconstitution
Scramblase activity was measured as previously reported, by adding dithionite to NBD-PC-containing vesicles and measuring the extent of fluorescence loss at the end point of the experiment (after >300 sec)4,13. Data were obtained for protein-free liposomes as well as proteoliposomes with a range of protein to phospholipid ratio (PPR, mg/mmol) values. The kinetics of fluorescence loss were evaluated as described in Table 1. End-point fluorescence reduction data were transformed as follows:
| 4 |
where F is the end point percentage fluorescence reduction, Fo is the percentage reduction obtained with liposomes (typically ~45%), Fmax is the maximum percentage reduction observed for samples with high PPR (82.5%)4,13 and p(≥1 scramblase) is the probability that a particular vesicle in the ensemble contains at least one functional scramblase. The dependence of p(≥1 scramblase) on PPR follows Poisson statistics. Taking into account that a fraction of the vesicles is refractory to reconstitution (~35%), and that the vesicle population has a range of sizes described by a Gaussian distribution ( and standard deviation σ = 28 nm), p(≥1 scramblase) can be written as13:
| 5 |
where α is a fit constant that is inversely proportional to M, the molecular weight of the functional scramblase and x is PPR*, derived from the measured PPR after taking into account the fraction of vesicles that is refractory to reconstitution (PPR* = PPR/0.65). Fitting data sets of p(≥1 scramblase) versus PPR* yields α and therefore the molecular weight of the functionally reconstituted scramblase (Table 2)13.
Data availability
The datasets generated during and/or analysed during the current study are available from the corresponding author on reasonable request.
Electronic supplementary material
Acknowledgements
We thank Harel Weinstein for insightful discussions and Deo Singh for help with the single molecule fluorescence microscopy experiments. A.K.M. thanks Sam Canis and Sidse Babett Knudsen for stimulation. This work was supported by the National Institutes of Health (EY024207 (A.K.M.) and EY027969 (A.K.M. and G.K.)), Velux Stiftung (project 881 (A.K.M.)), and the Austrian Science Foundation (FWF project J3686-B21 (B.P.)). The following computational resources are gratefully acknowledged: i) an allocation at the National Energy Research Scientific Computing Center (NERSC, repository m1710, used for carrying out MD simulations) supported by the Office of Science of the U.S. Department of Energy under Contract No. DE-AC02-05CH11231; and ii) the computational resources of the David A. Cofrin Center for Biomedical Information in the HRH Prince Alwaleed Bin Talal Bin Abdulaziz Alsaud Institute for Computational Biomedicine at Weill Cornell Medical College.
Author Contributions
K.P. performed computational analyses, FSEC and scramblase assays, generated the SNAP-tagged QUAD opsin construct and designed, performed and analyzed single molecule fluorescence microscopy experiments. B.P. generated the GFP-tagged WT and QUAD-opsin constructs and performed scramblase assays. M.A.G. conceived the project, generated the QUAD opsin and SNAP-tagged WT opsin constructs, and performed scramblase assays. J.L. designed, performed and analyzed single molecule fluorescence microscopy experiments. G.K. designed and performed computational analyses. A.K.M. conceived the project and analyzed scramblase activity data. All authors contributed to writing the paper.
Competing Interests
The authors declare that they have no competing interests.
Footnotes
Kalpana Pandey, Birgit Ploier and Michael A. Goren contributed equally to this work.
Electronic supplementary material
Supplementary information accompanies this paper at 10.1038/s41598-017-16842-z.
Publisher's note: Springer Nature remains neutral with regard to jurisdictional claims in published maps and institutional affiliations.
References
- 1.Wu G, Hubbell WL. Phospholipid asymmetry and transmembrane diffusion in photoreceptor disc membranes. Biochemistry. 1993;32:879–888. doi: 10.1021/bi00054a020. [DOI] [PubMed] [Google Scholar]
- 2.Hessel E, Herrmann A, Müller P, Schnetkamp PP, Hofmann KP. The transbilayer distribution of phospholipids in disc membranes is a dynamic equilibrium evidence for rapid flip and flop movement. Eur. J. Biochem. 2000;267:1473–1483. doi: 10.1046/j.1432-1327.2000.01147.x. [DOI] [PubMed] [Google Scholar]
- 3.Menon I, et al. Opsin is a phospholipid flippase. Curr. Biol. 2011;21:149–153. doi: 10.1016/j.cub.2010.12.031. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Goren MA, et al. Constitutive phospholipid scramblase activity of a G protein-coupled receptor. Nat Commun. 2014;5:5115. doi: 10.1038/ncomms6115. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Ernst OP, Menon AK. Phospholipid scrambling by rhodopsin. Photochem. Photobiol. Sci. 2015;14:1922–1931. doi: 10.1039/C5PP00195A. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Mansoor SE, Palczewski K, Farrens DL. Rhodopsin self-associates in asolectin liposomes. Proc. Natl. Acad. Sci. USA. 2006;103:3060–3065. doi: 10.1073/pnas.0511010103. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Jastrzebska B, et al. Disruption of Rhodopsin Dimerization with Synthetic Peptides Targeting an Interaction Interface. J Biol Chem. 2015;290:25728–25744. doi: 10.1074/jbc.M115.662684. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Hebert TE, et al. A peptide derived from a beta2-adrenergic receptor transmembrane domain inhibits both receptor dimerization and activation. J Biol Chem. 1996;271:16384–16392. doi: 10.1074/jbc.271.27.16384. [DOI] [PubMed] [Google Scholar]
- 9.Filizola M. Increasingly accurate dynamic molecular models of G-protein coupled receptor oligomers: Panacea or Pandora’s box for novel drug discovery? Life Sci. 2010;86:590–597. doi: 10.1016/j.lfs.2009.05.004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Periole X, Knepp AM, Sakmar TP, Marrink SJ, Huber T. Structural determinants of the supramolecular organization of G protein-coupled receptors in bilayers. J Am Chem Soc. 2012;134:10959–10965. doi: 10.1021/ja303286e. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Johnston JM, Wang H, Provasi D, Filizola M. Assessing the relative stability of dimer interfaces in g protein-coupled receptors. PLoS Comput Biol. 2012;8:e1002649. doi: 10.1371/journal.pcbi.1002649. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Jastrzebska B, et al. A G Protein-Coupled Receptor Dimerization Interface in Human Cone Opsins. Biochemistry. 2017;56:61–72. doi: 10.1021/acs.biochem.6b00877. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Ploier B, et al. Dimerization deficiency of enigmatic retinitis pigmentosa-linked rhodopsin mutants. Nat Commun. 2016;7:12832. doi: 10.1038/ncomms12832. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Ploier, B. & Menon, A. K. A Fluorescence-based Assay of Phospholipid Scramblase Activity. J Vis Exp, 10.3791/54635 (2016). [DOI] [PMC free article] [PubMed]
- 15.Venkatakrishnan AJ, et al. Molecular signatures of G-protein-coupled receptors. Nature. 2013;494:185–194. doi: 10.1038/nature11896. [DOI] [PubMed] [Google Scholar]
- 16.Liang Y, et al. Organization of the G protein-coupled receptors rhodopsin and opsin in native membranes. J. Biol. Chem. 2003;278:21655–21662. doi: 10.1074/jbc.M302536200. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Ballesteros JA, Weinstein H. Integrated methods for the construction of three-dimensional models and computational probing of structure-function relations in G protein-coupled receptors. Methods in Neurosciences. 1995;25:366–428. doi: 10.1016/S1043-9471(05)80049-7. [DOI] [Google Scholar]
- 18.Guo W, Shi L, Filizola M, Weinstein H, Javitch JA. Crosstalk in G protein-coupled receptors: changes at the transmembrane homodimer interface determine activation. Proc. Natl. Acad. Sci. USA. 2005;102:17495–17500. doi: 10.1073/pnas.0508950102. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Wolfenden R. Experimental measures of amino acid hydrophobicity and the topology of transmembrane and globular proteins. J. Gen. Physiol. 2007;129:357–362. doi: 10.1085/jgp.200709743. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Hessa T, et al. Recognition of transmembrane helices by the endoplasmic reticulum translocon. Nature. 2005;433:377–381. doi: 10.1038/nature03216. [DOI] [PubMed] [Google Scholar]
- 21.Choe S, Stevens CF, Sullivan JM. Three distinct structural environments of a transmembrane domain in the inwardly rectifying potassium channel ROMK1 defined by perturbation. Proc. Natl. Acad. Sci. USA. 1995;92:12046–12049. doi: 10.1073/pnas.92.26.12046. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Monks SA, Needleman DJ, Miller C. Helical structure and packing orientation of the S2 segment in the Shaker K+channel. J. Gen. Physiol. 1999;113:415–423. doi: 10.1085/jgp.113.3.415. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Mondal S, et al. Membrane driven spatial organization of GPCRs. Sci Rep. 2013;3:2909. doi: 10.1038/srep02909. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Mondal S, Khelashvili G, Shan J, Andersen OS, Weinstein H. Quantitative modeling of membrane deformations by multihelical membrane proteins: application to G-protein coupled receptors. Biophys J. 2011;101:2092–2101. doi: 10.1016/j.bpj.2011.09.037. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Mondal S, Khelashvili G, Weinstein H. Not Just an Oil Slick: How the Energetics of Protein-Membrane Interactions Impacts the Function and Organization of Transmembrane Proteins. Biophys J. 2014;106:2305–2316. doi: 10.1016/j.bpj.2014.04.032. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Park JH, et al. Opsin, a structural model for olfactory receptors? Angew. Chem. Int. Ed. Engl. 2013;52:11021–11024. doi: 10.1002/anie.201302374. [DOI] [PubMed] [Google Scholar]
- 27.Reeves PJ, Callewaert N, Contreras R, Khorana HG. Structure and function in rhodopsin: high-level expression of rhodopsin with restricted and homogeneous N-glycosylation by a tetracycline-inducible N-acetylglucosaminyltransferase I-negative HEK293S stable mammalian cell line. Proc. Natl. Acad. Sci. USA. 2002;99:13419–13424. doi: 10.1073/pnas.212519299. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Ernst OP, Gramse V, Kolbe M, Hofmann KP, Heck M. Monomeric G protein-coupled receptor rhodopsin in solution activates its G protein transducin at the diffusion limit. Proc. Natl. Acad. Sci. USA. 2007;104:10859–10864. doi: 10.1073/pnas.0701967104. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Ulbrich MH, Isacoff EY. Subunit counting in membrane-bound proteins. Nat Methods. 2007;4:319–321. doi: 10.1038/NMETH1024. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Aggarwal V, Ha T. Single-molecule fluorescence microscopy of native macromolecular complexes. Curr Opin Struct Biol. 2016;41:225–232. doi: 10.1016/j.sbi.2016.09.006. [DOI] [PubMed] [Google Scholar]
- 31.Jain A, et al. Probing cellular protein complexes using single-molecule pull-down. Nature. 2011;473:484–488. doi: 10.1038/nature10016. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Levitz J, et al. Mechanism of Assembly and Cooperativity of Homomeric and Heteromeric Metabotropic Glutamate Receptors. Neuron. 2016;92:143–159. doi: 10.1016/j.neuron.2016.08.036. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Keppler A, Pick H, Arrivoli C, Vogel H, Johnsson K. Labeling of fusion proteins with synthetic fluorophores in live cells. Proc. Natl. Acad. Sci. USA. 2004;101:9955–9959. doi: 10.1073/pnas.0401923101. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Juanes, M. A. et al. Adenomatous polyposis coli nucleates actin assembly to drive cell migration and microtubule-induced focal adhesion turnover. J Cell Biol, 10.1083/jcb.201702007 (2017). [DOI] [PMC free article] [PubMed]
- 35.Malvezzi M, et al. Ca2+-dependent phospholipid scrambling by a reconstituted TMEM16 ion channel. Nat Commun. 2013;4:2367. doi: 10.1038/ncomms3367. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Verchere A, et al. Light-independent phospholipid scramblase activity of bacteriorhodopsin from Halobacterium salinarum. Sci Rep. 2017;7:9522. doi: 10.1038/s41598-017-09835-5. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Brunner JD, Lim NK, Schenck S, Duerst A, Dutzler R. X-ray structure of a calcium-activated TMEM16 lipid scramblase. Nature. 2014;516:207–212. doi: 10.1038/nature13984. [DOI] [PubMed] [Google Scholar]
- 38.Brunner JD, Schenck S, Dutzler R. Structural basis for phospholipid scrambling in the TMEM16 family. Curr Opin Struct Biol. 2016;39:61–70. doi: 10.1016/j.sbi.2016.05.020. [DOI] [PubMed] [Google Scholar]
- 39.Yu K, et al. Identification of a lipid scrambling domain in ANO6/TMEM16F. Elife. 2015;4:e06901. doi: 10.7554/eLife.06901. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Whitlock JM, Hartzell HC. A Pore Idea: the ion conduction pathway of TMEM16/ANO proteins is composed partly of lipid. Pflugers Arch. 2016;468:455–473. doi: 10.1007/s00424-015-1777-2. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Grossfield A, Pitman MC, Feller SE, Soubias O, Gawrisch K. Internal hydration increases during activation of the G-protein-coupled receptor rhodopsin. J Mol Biol. 2008;381:478–486. doi: 10.1016/j.jmb.2008.05.036. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Sali A, Blundell TL. Comparative protein modelling by satisfaction of spatial restraints. J Mol Biol. 1993;234:779–815. doi: 10.1006/jmbi.1993.1626. [DOI] [PubMed] [Google Scholar]
- 43.Shen MY, Sali A. Statistical potential for assessment and prediction of protein structures. Protein Sci. 2006;15:2507–2524. doi: 10.1110/ps.062416606. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Jorgensen WL, Julian T-R. The OPLS (optimized potentials for liquid simulations) potential functions for proteins, energy minimizations for crystals of cyclic peptides and crambin. J. Am. Chem. Soc. 1988;110:1657–1666. doi: 10.1021/ja00214a001. [DOI] [PubMed] [Google Scholar]
- 45.Jo S, Kim T, Iyer VG, Im W. CHARMM-GUI: a web-based graphical user interface for CHARMM. J Comput Chem. 2008;29:1859–1865. doi: 10.1002/jcc.20945. [DOI] [PubMed] [Google Scholar]
- 46.Brooks BR, et al. CHARMM: the biomolecular simulation program. J Comput Chem. 2009;30:1545–1614. doi: 10.1002/jcc.21287. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.Lee J, et al. CHARMM-GUI Input Generator for NAMD, GROMACS, AMBER, OpenMM, and CHARMM/OpenMM Simulations Using the CHARMM36 Additive Force Field. J Chem Theory Comput. 2016;12:405–413. doi: 10.1021/acs.jctc.5b00935. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48.Shi L, Quick M, Zhao Y, Weinstein H, Javitch JA. The mechanism of a neurotransmitter:sodium symporter–inward release of Na+and substrate is triggered by substrate in a second binding site. Mol Cell. 2008;30:667–677. doi: 10.1016/j.molcel.2008.05.008. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49.Phillips JC, et al. Scalable molecular dynamics with NAMD. J Comput Chem. 2005;26:1781–1802. doi: 10.1002/jcc.20289. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50.Harvey MJ, Giupponi G, Fabritiis GD. ACEMD: Accelerating Biomolecular Dynamics in the Microsecond Time Scale. J Chem Theory Comput. 2009;5:1632–1639. doi: 10.1021/ct9000685. [DOI] [PubMed] [Google Scholar]
- 51.Khelashvili G, et al. Spontaneous inward opening of the dopamine transporter is triggered by PIP2-regulated dynamics of the N-terminus. ACS Chem Neurosci. 2015;6:1825–1837. doi: 10.1021/acschemneuro.5b00179. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 52.Marsh D. Energetics of hydrophobic matching in lipid-protein interactions. Biophys J. 2008;94:3996–4013. doi: 10.1529/biophysj.107.121475. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 53.Mouritsen OG, Bloom M. Mattress model of lipid-protein interactions in membranes. Biophys J. 1984;46:141–153. doi: 10.1016/S0006-3495(84)84007-2. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 54.Choe S, Hecht KA, Grabe M. A continuum method for determining membrane protein insertion energies and the problem of charged residues. J Gen Physiol. 2008;131:563–573. doi: 10.1085/jgp.200809959. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 55.Ben-Tal N, Ben-Shaul A, Nicholls A, Honig B. Free-energy determinants of alpha-helix insertion into lipid bilayers. Biophys J. 1996;70:1803–1812. doi: 10.1016/S0006-3495(96)79744-8. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 56.Yau WM, Wimley WC, Gawrisch K, White SH. The preference of tryptophan for membrane interfaces. Biochemistry. 1998;37:14713–14718. doi: 10.1021/bi980809c. [DOI] [PubMed] [Google Scholar]
- 57.Strandberg E, Killian JA. Snorkeling of lysine side chains in transmembrane helices: how easy can it get? FEBS Lett. 2003;544:69–73. doi: 10.1016/S0014-5793(03)00475-7. [DOI] [PubMed] [Google Scholar]
- 58.Gray TM, Matthews BW. Intrahelical hydrogen bonding of serine, threonine and cysteine residues within alpha-helices and its relevance to membrane-bound proteins. J Mol Biol. 1984;175:75–81. doi: 10.1016/0022-2836(84)90446-7. [DOI] [PubMed] [Google Scholar]
- 59.Comar WD, Schubert SM, Jastrzebska B, Palczewski K, Smith AW. Time-resolved fluorescence spectroscopy measures clustering and mobility of a G protein-coupled receptor opsin in live cell membranes. Journal of the American Chemical Society. 2014;136:8342–8349. doi: 10.1021/ja501948w. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 60.Gibson DG, et al. Enzymatic assembly of DNA molecules up to several hundred kilobases. Nat Methods. 2009;6:343–345. doi: 10.1038/nmeth.1318. [DOI] [PubMed] [Google Scholar]
- 61.Gibson DG, et al. Creation of a bacterial cell controlled by a chemically synthesized genome. Science. 2010;329:52–56. doi: 10.1126/science.1190719. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
Data Availability Statement
The datasets generated during and/or analysed during the current study are available from the corresponding author on reasonable request.