Abstract
Dogs with X-linked hereditary nephropathy (XLHN) have a glomerular basement membrane defect that leads to progressive juvenile-onset renal failure. Their disease is analogous to Alport syndrome in humans, and they also serve as a good model of progressive chronic kidney disease (CKD). However, the gene expression profile that affects progression in this disease has only been partially characterized. To help fill this gap, we used RNA sequencing to identify differentially expressed genes (DEGs), over-represented pathways, and upstream regulators that contribute to kidney disease progression. Total RNA from kidney biopsies was isolated at 3 clinical time points from 3 males with rapidly-progressing CKD, 3 males with slowly-progressing CKD, and 2 age-matched controls. We identified 70 DEGs by comparing rapid and slow groups at specific time points. Based on time course analysis, 1,947 DEGs were identified over the 3 time points revealing upregulation of inflammatory pathways: integrin signaling, T cell activation, and chemokine and cytokine signaling pathways. T cell infiltration was verified by immunohistochemistry. TGF-β1 was identified as the primary upstream regulator. These results provide new insights into the underlying molecular mechanisms of disease progression in XLHN, and the identified DEGs can be potential biomarkers and therapeutic targets translatable to all CKDs.
Introduction
X-linked hereditary nephropathy (XLHN) in dogs leads to chronic kidney disease (CKD) because of a defect in type IV collagen in the glomerular basement membrane (GBM). In the XLHN dogs in this study, a naturally-occurring, 10-base-pair deletion in the COL4A5 gene located on the X chromosome results in the inability to synthesize complete α5 chains1. This alteration in the type IV collagen network compromises the structure and function of the GBM in both affected (hemizygous) males and, to a lesser extent, carrier (heterozygous) female dogs2.
XLHN in dogs is analogous to Alport syndrome (AS) in humans, as approximately 85% of people with AS have an X-linked mutation in COL4A5 3. AS is characterized by juvenile-onset CKD, ocular abnormalities, and hearing loss in affected males4. Thus far, only the renal abnormalities have been detected in XLHN dogs2. In dogs with XLHN, juvenile-onset CKD manifests as persistent proteinuria of glomerular origin as early as 3–6 months of age, followed by decreasing glomerular filtration rate and worsening azotemia, typically leading to end-stage renal failure before 1 year of age2,5.
Although XLHN has been studied as an example of canine CKD caused by glomerular disease and as an animal model of human AS2, the gene expression profile that affects progression has only been partially characterized. Furthermore, dogs with the same mutation causing XLHN display substantial variation in the rate of disease progression such that some dogs reach end-stage disease by 6 months of age and others at 12 months of age or later. Although varied times of onset and rates of progression are common among different types of mutations in people with AS6, disease progression may also vary among members of an AS family with an identical mutation7, as seen in dogs8.
While several studies have characterized gene expression in humans with CKD and in animal Alport models using microarrays9,10 or PCR8,9,11,12, studies that have incorporated high-throughput RNA sequencing (RNA-seq) with the objective of identifying differentially expressed genes (DEGs) and upstream regulators are lacking. Compared with traditional approaches in gene expression analysis, RNA-seq provides unprecedented flexibility in the discovery of DEGs13 while preserving accuracy and strong correlation with PCR14–18, even considering fold change levels19.
The objective of this study was to compare the gene expression between dogs with rapid versus slow disease progression phenotypes at 3 stages of the disease. We conducted Gene Ontology (GO) and pathway analyses to characterize DEGs among sample groups at specific time points. Since all CKDs share common pathways that lead to end-stage kidney disease20, the results help elucidate the molecular basis of CKD progression and thus may benefit canine patients and indicate potential therapeutic targets for AS patients.
Results
Histopathological evaluation of kidney biopsies
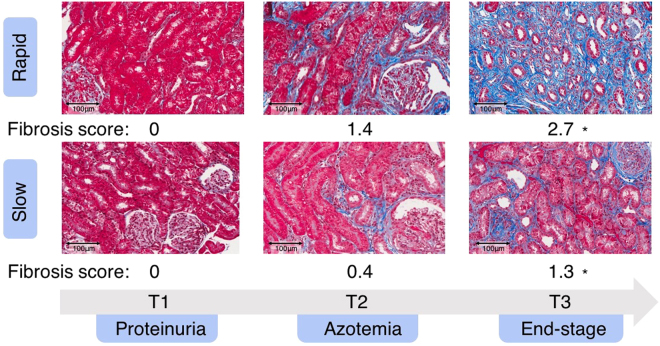
Figure 1 presents representative cortical fields of the kidney biopsies and the mean interstitial fibrosis scores comparing the rapid versus slow groups. Clinical time points in affected dogs were defined as: T1 - onset of proteinuria (the earliest time point that clinical disease can be detected); T2 - onset of azotemia (serum creatinine ≥ 1.2 mg/dL); and T3 - end-stage disease (serum creatinine ≥ 5 mg/dL). At both T2 and T3, mean fibrosis and chronic inflammation scores were significantly higher in diseased dogs than in controls (Supplementary Tables S1 and S2). The degree of fibrosis in the rapid group was more severe at T2 and T3 than that in the slow group, despite the two groups being clinically indistinguishable; however, statistical significance between the scores was reached only at T3 (Supplementary Table S1 and Fig. S1). No statistically significant difference was observed for the chronic inflammation score between the rapid and slow groups (Supplementary Table S2 and Fig. S2).
Figure 1.
Kidney biopsies from representative dogs and mean interstitial fibrosis scores (score range: 0 [normal] to 3 [severe]) for each group of affected dogs at 3 time points (T1, T2, and T3). Fibrosis scores were based on evaluation of multiple 20x fields. *Statistical significant. Scale bar: 100 μm. (Trichrome-stained).
RNA-sequencing (RNA-seq) of dog transcriptomes
The average RNA yield from the 24 kidney biopsies was 86.2 ng/μL, and the average RNA integrity number (RIN) was 3.4 (Supplementary Table S3). Because of the variable quality of RNA, the proper library preparation kit was used to compensate for the low-input samples according to the best practice for RNA with variable qualities21 (see “RNA isolation and sequencing” section in Methods). After performing quality control, we obtained an average of over 30 million paired-end reads from each sample (n = 24). Overall, 91–96% of reads were mapped to the canine genome (CanFam 3.1) by HISAT222. Among them, 70–78% of reads were uniquely mapped (Supplementary Table S3 and Fig. S3). Based on the union setting of HTSeq23, ambiguous reads that mapped to multiple genes were not included in our analysis.
Principal component analysis (PCA) and hierarchical clustering analysis
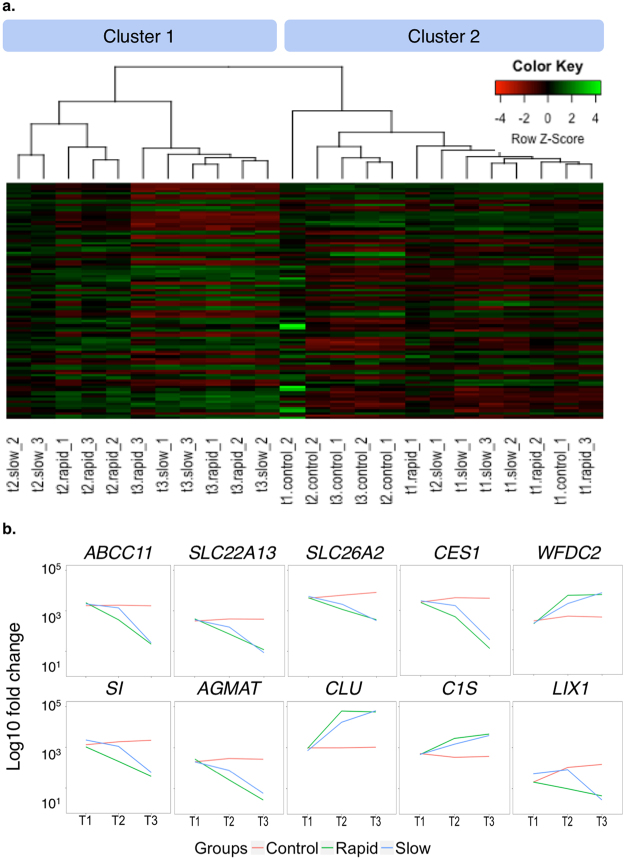
We performed PCA at each time point to determine whether samples in each group clustered with each other or other groups. First, we used HTSeq23 to count reads that uniquely aligned to one gene, and these data were then imported into DESeq224 to generate PCA plots. At T1, the PCA results demonstrated that most samples clustered together, regardless of the grouping (Fig. 2). Except for one dog, rapid and slow groups became separated from controls at T2. Although the slow group tended to be closer to the controls than the rapid group, there was no clear distinction between rapid and slow groups at T2 or T3. Furthermore, PCA scree plots confirmed that principal components 1 (PC1) and 2 (PC2) accounted for 76–90% of the total variation in gene expression at each time point (Supplementary Fig. S4). To further investigate the time-dependent nature of the DEGs, we performed hierarchical clustering of the top 100 DEGs (i.e., those with the smallest q-values identified in the time course analysis in DESeq2). In agreement with the PCA plots, this analysis demonstrated clustering of almost all sample groups at T1 (Fig. 3a). At time points T2 and T3, the rapid and slow groups clustered together (Cluster 1 in Fig. 3a) and were distinctly separated from the control group for all but one T2 sample (Cluster 2 in Fig. 3a).
Figure 2.

Principal component analysis (PCA) for all samples at 3 time points. Principal component 1 (PC1) and principal component 2 (PC2) were identified by variance stabilizing transformation in DESeq2 at the 3 time points. The percentage of variance indicates how much variance was explained by PC1 and PC2. (Red: control group; Green: rapid group; Blue: slow groups).
Figure 3.
Hierarchical clustering analysis, heatmap, and gene expression. (a) Hierarchical clustering analysis and heatmap of the 100 genes with the smallest q-values in the time course analysis in DESeq2 (Column names: t1, t2, and t3 designate 3 clinical time points; slow, rapid, and control represent grouping; _1, _2, and _3 are individual dogs in each group). (b) Trends of gene expression over time for the 10 genes with the smallest q-values (from left to right, top to bottom) (Red: control group; Green: rapid group; Blue: slow group).
Differentially expressed genes (DEGs)
In average, 20,090 genes were mapped by at least one read in each of the kidney biopsy samples (Supplementary Table S3). Overall, 1,947 DEGs with a q-value < 0.05 were detected over the 3 time points in the time course analysis of DESeq2 (Supplementary Table S4). We applied the plot counts function in DESeq2 to visualize the top 10 genes with the smallest q-values (Fig. 3b). While these genes were not differentially expressed at T1, group-specific changes were observed over time, and expression in the slow group was consistently closer to that in the control group for each gene at T2 (Fig. 3b).
To achieve the primary goal of this study, we identified 70 DEGs between the rapid and slow groups among all time points (q-value < 0.05) (Table 1 and Fig. 4b). In this comparison, T2 demonstrated the most DEGs, with 68 of the 70 DEGs unique to T2. Two DEGs were identified at T1: stearoyl-CoA desaturase 5 (SCD5) (fold change = −3.55, q-value = 1.1 × 10−05), which was also detected at T2, and thymidine kinase 1 (TK1) (fold change = 2.49, q-value = 0.02). At T3, no DEGs were identified when these 2 groups were compared.
Table 1.
Overview of 70 significant DEGs comparing rapid and slow groups (q-value < 0.05).
| Up-regulated DEGs in the rapid group | |||
|---|---|---|---|
| Gene Symbol | Full Name | Fold Change | q-value |
| COL1A1 | collagen type I alpha 1 chain | 6.52 | 2.62E-20 |
| COL3A1 | collagen type III alpha 1 chain | 5.35 | 1.05E-17 |
| COL1A2 | collagen type I alpha 2 chain | 4.01 | 4.66E-13 |
| COL5A1 | collagen type V alpha 1 chain | 3.8 | 7.21E-05 |
| COL6A3 | collagen type VI alpha 3 chain | 3.27 | 7.08E-07 |
| COL6A1 | collagen type VI alpha 1 chain | 3.26 | 1.82E-05 |
| LOX | lysyl oxidase | 3.13 | 1.42E-03 |
| COL6A2 | collagen type VI alpha 2 chain | 3.07 | 2.34E-04 |
| PAMR1 | peptidase domain containing associated with muscle regeneration 1 | 2.99 | 5.76E-03 |
| CDCA8 | cell division cycle associated 8 | 2.98 | 2.80E-02 |
| COL11A1 | collagen type I alpha 1 chain | 2.86 | 5.33E-03 |
| COL15A1 | collagen type XI alpha 1 chain | 2.82 | 6.24E-03 |
| C1QTNF6 | C1q and tumor necrosis factor related protein 6 | 2.81 | 4.98E-03 |
| FNDC1 | fibronectin type III domain containing 1 | 2.81 | 3.22E-02 |
| FN1 | fibronectin 1 | 2.8 | 4.10E-02 |
| CCDC80 | coiled-coil domain containing 80 | 2.72 | 7.98E-04 |
| MFSD7 | major facilitator superfamily domain containing 7 | 2.7 | 3.22E-02 |
| FBLN1 | fibulin 1 | 2.69 | 4.10E-02 |
| NID2 | nidogen 2 | 2.61 | 1.52E-07 |
| COL4A2 | collagen type IV alpha 2 chain | 2.6 | 1.42E-03 |
| FAM69B | family with sequence similarity 69, member B | 2.6 | 3.42E-02 |
| NDN | necdin | 2.59 | 4.39E-02 |
| COL4A1 | collagen type I alpha 1 chain | 2.59 | 5.76E-03 |
| HTR7 | 5-hydroxytryptamine (serotonin) receptor 7, adenylate cyclase-coupled | 2.58 | 2.60E-02 |
| PCOLCE | procollagen C-endopeptidase enhancer | 2.58 | 5.83E-03 |
| OLFML2B | olfactomedin like 2B | 2.56 | 6.24E-03 |
| TK1a | Thymidine kinase 1 | 2.49 | 1.98E-02 |
| C15orf39 | chromosome 15 open reading frame 39 | 2.47 | 2.76E-02 |
| RCN3 | reticulocalbin 3 | 2.46 | 6.94E-04 |
| MFAP2 | microfibrillar associated protein 2 | 2.45 | 1.42E-03 |
| HSPG2 | perlecan | 2.45 | 1.20E-02 |
| PRSS35 | protease, serine 35 | 2.45 | 4.10E-02 |
| MMP2 | matrix metallopeptidase 2 | 2.43 | 4.48E-05 |
| FBN1 | fibrillin 1 | 2.37 | 3.57E-03 |
| CD248 | CD248 molecule | 2.36 | 2.72E-02 |
| FOXRED2 | FAD dependent oxidoreductase domain containing 2 | 2.35 | 1.11E-02 |
| GXYLT2 | glucoside xylosyltransferase 2 | 2.34 | 3.62E-02 |
| FSCN1 | fascin actin-bundling protein 1 | 2.34 | 1.20E-02 |
| ENSCAFG00000008741b | novel gene | 2.29 | 1.18E-02 |
| ENSCAFG00000012963b | novel gene | 2.28 | 1.23E-03 |
| BGN | biglycan | 2.19 | 1.85E-02 |
| FAS | Fas (TNF receptor superfamily member 6) | 2.16 | 2.08E-02 |
| ADAMTS2 | ADAM metallopeptidase with thrombospondin type 1 motif 2 | 2.15 | 3.23E-02 |
| PXDN | peroxidasin | 2.14 | 4.10E-02 |
| SPARC | secreted protein acidic and cysteine rich | 2.13 | 4.48E-05 |
| THBS1 | thrombospondin 1 | 2.12 | 1.26E-04 |
| KCP | kielin/chordin-like protein | 2.12 | 4.19E-02 |
| LRP1 | LDL receptor related protein 1 | 2.11 | 7.98E-04 |
| CERCAM | cerebral endothelial cell adhesion molecule | 2.11 | 4.39E-02 |
| ITGA5 | integrin subunit alpha 5 | 2.07 | 7.03E-03 |
| BMP1 | bone Morphogenetic Protein 1 | 1.88 | 6.74E-05 |
| FSTL1 | follistatin Like 1 | 1.72 | 1.25E-05 |
| PTGFRN | prostaglandin F2 Receptor Inhibitor | 1.66 | 2.61E-05 |
| Down-regulated DEGs in the rapid group | |||
| UGT1A6 | UDP glucuronosyltransferase family 1 member A6 | −4.77 | 1.23E-03 |
| NAT8 | N-acetyltransferase 8 (putative) | −4.1 | 1.09E-04 |
| R3HDML | R3H domain containing like | −4.02 | 1.42E-03 |
| LIX1 | limb and CNS expressed 1 | −3.83 | 7.21E-03 |
| PRLR | prolactin receptor | −3.73 | 4.64E-04 |
| SCD5c | stearoyl-CoA desaturase 5 | −3.55 | 1.07E-05 |
| FMO2 | flavin containing monooxygenase 2 | −3.42 | 1.78E-02 |
| OAT3/SLC22A8 | solute carrier family 22 member 8 | −3.37 | 2.45E-02 |
| SI | sucrase-isomaltase | −3.25 | 1.16E-02 |
| ENSCAFG00000003760b | novel gene | −2.92 | 3.96E-02 |
| SLC26A4 | solute carrier family 26 member 4 | −2.79 | 7.14E-03 |
| ENSCAFG00000000799b | novel gene | −2.77 | 3.25E-02 |
| HEPACAM2 | HEPACAM family member 2 | −2.66 | 8.81E-03 |
| IDO2 | indoleamine 2,3-dioxygenase 2 | −2.58 | 3.42E-02 |
| PECR | peroxisomal trans-2-enoyl-CoA reductase | −2.57 | 6.24E-03 |
| MT-ND3 | mitochondrially encoded NADH dehydrogenase 3 | −2.16 | 3.49E-03 |
| ABCA4 | retinal-specific ATP-binding cassette transporter | −1.91 | 1.08E-04 |
aDEG identified only at T1. bGenes are displyed with ensembl IDs if gene annotations are unavailable. cDEG identified at both T1 and T2.
Figure 4.
DEGs in different pairs of comparisons at the 3 time points (T1, T2, and T3). Comparing the rapid and slow groups, 70 DEGs were found. Comparing each affected group with controls, several thousand DEGs were identified, with most of the DEGs occurring at T3. For each pair, only genes with a q-value < 0.05 were considered as DEGs. The total number of DEGs found at each time point appears in parentheses.
We also compared the rapid and slow groups with the control group, both individually and combined as a single “affected” group (Fig. 4a,c, and d). In these comparisons, the number of DEGs increased with advancing disease, with the largest number of DEGs identified at T3. This phenomenon indicates that the DEGs are disease-dependent as they are more differentially expressed in the later time points (T2 and T3) than T1 (Fig. 4).
A substantial overlap of DEGs was present when comparing rapid and slow groups with the control group at T2 and T3 (Fig. 5). The overlapping DEGs between the rapid and slow groups were more numerous at T3 (2,952 DEGs) than at T2 (190 DEGs), supporting that the two groups behave similarly at the end-stage disease, as expected based on Figs 2–4. Furthermore, the number of DEGs (1,189 DEGs) identified in both T2 and T3 in the rapid group was higher than the number of DEGs (171 DEGs) identified in both T2 and T3 in the slow group. This supports the theory that rapidly-progressing dogs express end-stage DEGs at a young age. The complete lists of DEGs from the time course analysis and all pairs of comparisons appear in Supplementary Table S4.
Figure 5.

Overlapping DEGs in rapid and slow groups compared with control at T2 and T3. For each comparison, only genes with a q-value < 0.05 were considered as DEGs. The total number of DEGs found at each time point appears in parentheses.
Gene Ontology (GO) and pathway analysis of DEGs
To characterize the GO terms, including molecular functions, biological processes, cellular components, and functional pathways of DEGs, we conducted over-representation tests for all pairs of comparisons in PANTHER version 11.1 (released on October 24, 2016) (Fig. 6 and Supplementary Table S5). We used the GO-Slim PANTHER annotation data set, which represents phylogenetically inferred annotations25.
Figure 6.
Enriched pathways and GO term analyses for 10 selected comparisons. Enriched pathways, biological processes, and regulator analyses for all DEGs are presented based on T2 and T3 comparisons as well as a time course analysis incorporating all time points for all groups. The number of genes was normalized to allow comparisons between groups within the same pathway, and comparisons were color coded in pairs, with the darker color corresponding to the later time point or the rapid group.
Overlaps of GO terms among comparisons were commonly seen in the current study. We focused on “biological process” since it is the most characterized GO term. Within this category, the “immune system process” family was upregulated in all 10 comparisons presented in Fig. 6, and the “immune response” family was upregulated in all except for the rapid versus slow comparison at T2 and the time course analysis. Both immune-related GO terms appeared to be more upregulated at T2 than T3. At T2, the “biological adhesion” family, especially the cell-cell adhesion subfamily, was expressed in the rapid group more than in the slow group (Supplementary Table S5).
The most common pathway represented by the DEGs within the various comparisons was the “integrin signaling pathway” (Fig. 6). This was the only pathway identified in the comparison between rapid and slow groups. It was also the top upregulated pathway at T2 comparing rapid and control groups. Interestingly, analyzing the overlapping DEGs between T2 and T3, the integrin signaling pathway was identified as the top upregulated pathway within the rapid group, but not within the slow group, as compared with control.
The “T cell activation pathway" was another frequently detected pathway in our study (Fig. 6). It was upregulated in all comparisons with control at T2 and in overlapping DEGs between T2 and T3 within both the rapid and the slow groups. Another upregulated pathway included the “inflammation mediated by chemokine and cytokine signaling pathway” that was identified only when comparing all affected dogs with controls at T2. Because only limited numbers of DEGs were discovered, no enriched pathways were identified at T1 with any comparison.
To further explore the possible biological interaction between orthologous genes in the human, mouse, and rat, we performed Ingenuity Pathway Analysis (IPA) to discover the most prevalent pathways and upstream regulators within each comparison. The “hepatic fibrosis/hepatic stellate cell activation pathway” was identified as the top pathway in multiple comparisons (Supplementary Table S6). Transforming growth factor beta 1 (TGF-β1) was the most activated upstream regulator when the rapid and slow groups were compared at T2 and in the time course analysis (Supplementary Table S6).
Immunohistochemistry (IHC) validation of inflammatory pathways
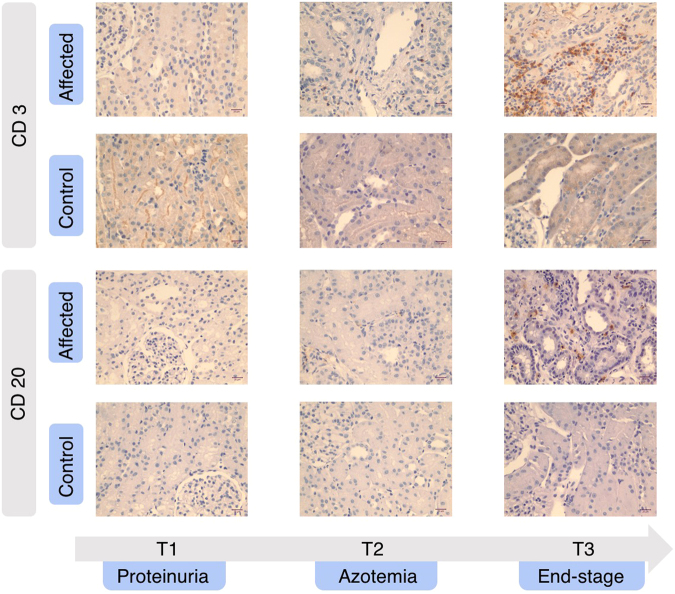
Lastly, we aimed to validate the overexpression of inflammatory pathways in kidney biopsies. We chose to identify the predominant lymphocyte subtype present (T versus B cell) since the identified inflammatory pathways are all closely related to the presence of T lymphocytes26 and antibodies for CD3 (T cell) and CD20 (B cell) are validated for use in dogs27,28. As shown in Fig. 7, we confirmed the lymphocyte infiltration in the affected dogs to be composed mostly of T cells rather than B cells. This result correlated with the previously assigned chronic inflammation scores based on the histopathological evaluation (Supplementary Table S2 and Fig. S2) and validated our RNA-seq data.
Figure 7.
Expression of CD3 and CD20 using immunohistochemistry (IHC) in kidney biopsies from representative dogs. In affected dogs, lymphocyte infiltration present at later time points consisted mostly of CD3-positive lymphocytes with few CD20-positive lymphocytes identified. Scale bar: 20 μm.
Discussion
Dogs with XLHN have been studied as both an example of progressive canine glomerular disease and an animal model of human AS, which has CKD as a major syndrome component2. The genetic cause is well characterized; however, the gene expression and molecular pathways influencing disease progression are incompletely known. In particular, the variable rate of disease progression in dogs with the same mutation and within families affected by AS is intriguing7. We, therefore, aimed to evaluate differential gene expression, overrepresented pathways, and upstream regulators by comparing RNA-seq data in dogs that displayed a rapid clinical progression of the disease to those with relatively slow disease progression.
In this study, we examined serial biopsies from rapid and slow groups as well as healthy age-matched littermates. To understand the biological changes during the pathogenesis of CKD, we included the earliest time point at which clinical disease could be detected in these dogs (onset of proteinuria, T1), with the onset of azotemia (T2) and the advent of end-stage disease (T3). By performing renal biopsies when animals reached specific clinical markers of disease progression, we could compare the same clinical stage in the rapid and slow groups. We believe this type of approach provides more confidence in identifying DEGs that are involved in the rate of disease progression than the traditional method of using age-driven time points, as differences detected are likely to be the driving force rather than the consequence of disease progression. Our data demonstrate the dynamic changes in gene expression at different stages of the disease. It also supports that the biological processes and pathways of fibrosis/adhesion and inflammation are the likely driving forces producing different rates of disease progression in the rapid and slow groups.
While it is known that serum creatinine correlates strongly with tubulointerstitial fibrosis29, one of the most intriguing findings when comparing the rapid and slow groups was the increase in fibrosis observed in the rapid group at the same clinical stage of disease (T2 and T3; Fig. 1). This corresponds with many of the 70 DEGs identified using RNA-seq between the rapid and slow groups that are implicated in fibrosis, almost all of which were identified at T2. Several of these genes have been previously described as upregulated in XLHN dogs, Alport mice, and other kidney diseases9,11,30,31. Among these, one of the upregulated genes, CD248 (endosialin or tumor endothelial marker 1, TEM1), has been found to mediate the adhesion and migration of cells through ligand interaction with the upregulated collagen-related genes: collagen type I, collagen type IV, and fibronectin-1 (FN1)32. CD248+ stromal cells bind extracellular matrix and have been implicated in kidney33, liver34, and lung fibrosis35. In non-inflamed kidneys, CD248 is expressed by mesangial cells located in glomeruli. In fibrotic kidneys, CD248 is additionally expressed by myofibroblasts and stromal fibroblasts, and the increased expression is closely related to prognostic indicators, such as albuminuria and renal scarring33. Of note, one of the downregulated genes in the rapid group, prolactin receptor (PRLR), decreased in association with the extent of interstitial collagen I deposition in kidney transplant rejection36, suggesting that PRLR might be a protectant against renal fibrosis. In our study, the decrease in PRLR could be responsible for the more rapid development of fibrosis and consequential faster progression of disease in the rapid group.
Involvement of inflammatory components in the progression of CKD is another major finding of this study. Several inflammatory genes involved in fibrotic changes, such as biglycan (BGN), kielin/chordin-like protein (KCP), and matrix metallopeptidase-2 (MMP2) were upregulated in the rapid versus slow groups. BGN plays a role in bone growth, muscle development and regeneration, and collagen fibril assembly in multiple tissues. BGN is upregulated in renal fibrosis37, and BGN protein expression strongly correlated with chronic kidney progression in one study30, which may suggest its role in regulating inflammation and innate immunity. KCP expression is stimulated by renal stress, and it enhances the antifibrotic function of BMP7 to attenuate the profibrotic stimulus of TGF-β and to suppress proinflammatory cytokines38. In Alport mice, the administration of recombinant BMP7 reduces glomerular and interstitial fibrosis but also upregulates MMP239. This upregulation of MMP2 seems contradictory, as it is associated with renal fibrosis in several animal models, including XLHN dogs11. However, the function of MMP2 is specific to the temporal context of fibrosis. At the prefibrotic phase, increased MMP2 induces epithelial to mesenchymal transition, tubular atrophy, and fibrosis. For established fibrosis, inducing MMP2 synthesis by BMP7 promotes proteolytic removal of accumulated extracellular matrix, which is thought to be a potential therapeutic strategy40. Since both KCP and MMP2 are upregulated at T2, when fibrosis is already relatively well established in the rapid group, their downstream actions are likely skewed toward anti-fibrotic effects.
Mechanisms other than fibrosis and inflammation can also play roles in the rapid progression of CKD. NAT8, which is almost exclusively expressed by tubular cells in the renal cortex41, is a cysteine S-conjugate N-acetyltransferase that is responsible for glutathione-mediated detoxification of nephrotoxic substances. The downregulation of NAT8 in the rapid group suggests a more severe loss of normal renal function in the presence of similar serum creatinine concentrations compared with the slow group. Another downregulated gene, organic anion transporter 3 (OAT3, also known as SLC22A8), is decreased in kidney biopsies from human CKD patients and in a nephrectomized rat model of CKD42,43. Reduced protein expression of Oat3 is associated with decreased excretion of an endogenous uremic toxin; meanwhile, the accumulation of this uremic toxin further inhibits Oat3-mediated transportation, accelerating toxin accumulation in serum42,43. Therefore, downregulation of OAT3 in the rapid group may result in impaired urinary excretion that is not adequately represented by serum creatinine concentration.
Only 2 DEGs, SCD5 and TK1, were differentially expressed in the rapid group as compared with the slow group at T1. SCD5 was downregulated at both T1 and T2 in the rapid compared with the slow group. And, it was downregulated in the rapid versus control comparison throughout all 3 time points. SCD5 is an isoform of stearoyl-CoA desaturase that is responsible for the formation of monounsaturated fatty acids. Although SCD5 has not previously been described in the CKD literature, it has been proposed as a novel regulator of neural cell proliferation and differentiation, likely through β-catenin-independent (non-canonical) Wnt pathways44. In fibrotic kidneys, the canonical Wnt pathway induces myofibroblast differentiation, and the non-canonical Wnt pathway leads to cytoskeleton rearrangement, cell adhesion, and cell movement45. Thymidine kinase 1 (TK1) is the only DEG that was exclusively upregulated at T1. Thymidine kinase is responsible for producing dTMP that is later incorporated into DNA. The cytoplasmic isoform of TK1 is cell cycle-dependent, as it substantially increases in the S phase of the cell cycle. Given that unregulated proliferation is the hallmark of neoplasia, TK1 is a valuable serum marker for breast cancer, non-Hodgkin’s lymphoma, plasmacytoma, and lung cancer46. Its upregulation in our study could indicate increased cell proliferation at T1 in the rapid group. However, further investigation of SCD5 and TK1 is needed to determine their roles in CKD progression.
In addition to abovementioned genes, many genes that are rarely described in CKD progression were found differentially expressed in the rapid versus slow groups at T2 (e.g., LOX, PAMR1, CDCA8, C1QTNF6, FNDC1, CCDC80, MFSD7, FMO2, SI, SLC26A4). The protein product of the upregulated gene lysyl oxidase (LOX) is an extracellular enzyme that is essential for covalent cross-linking of collagen in irreversible extracellular matrix deposition47. Despite reports of its upregulation in liver fibrosis48,49 and cardiomyopathy50, the upregulation of LOX in CKD has only been described in one study using a glomerulonephritis mouse model51. Simtuzumab, a monoclonal antibody that inhibits one of the LOX family members, lysyl oxidase homologue 2 (LOXL2), has recently been a focus of research as a possible new treatment for lung, liver, and kidney fibrosis due to a similar pathogenesis52. Another gene minimally described in nephrology is the downregulated sucrase-isomaltase (SI). SI is an α-glucosidase that commonly appears on the brush border of small intestinal enterocytes and is involved in glucose digestion. SI is also present in small amounts in non-intestinal cells such as blood leukocytes and kidney cells53. Little is known about the function and significance of SI in the renal tubule. However, the decreased expression of SI could indicate renal damage.
To characterize DEGs in functional groups, we performed GO terms and pathway analyses by comparing affected dog groups, both individually and collectively, with control dogs at each time point. GO terms showed that the functions of identified DEGs were associated with “biological adhesion,” “immune system processes,” and “immune response,” representing a common mechanism of disease progression in the early stages of CKD54. Consistent with the GO terms, the “integrin signaling pathway” was the most upregulated pathway, and the “T cell activation pathway” and the “chemokine and cytokine signaling pathway” were also upregulated in multiple comparisons. The exclusive early expression of the “integrin signaling pathway” in the rapid group and universal expression at a later time point could indicate that it is an essential pathway driving rapid progression of disease in these dogs. The “integrin signaling pathway” consists mainly of collagen and integrin genes. The integrin subunit alpha 2 gene (ITGA2) was increased in the rapid versus control group at both T2 and T3. The COL4A3−/−/ITGA2−/− double knockout Alport mouse model has delayed renal fibrosis compared with COL4A3−/−/ITGA2+/+ Alport mice, which express significantly higher levels of MMP2, MMP9, MMP12, and TIMP155. Upregulation of MMP2 is consistent with our Alport dog model, suggesting that the “integrin signaling pathway” is involved in matrix accumulation.
To verify the pathway analysis results, we used IHC to identify the infiltrating lymphocyte population. The “T cell activation,” “integrin signaling,” and the “inflammation mediated by chemokine and cytokine signaling” pathways are all closely related to the presence of T lymphocytes26. Consistent with this, T lymphocytes were identified as the predominant inflammatory cell population present in the affected dogs, as well as in a study of canine end-stage renal disease56. Moreover, T cell infiltration inversely correlates with renal function at the time of renal biopsies in AS patients57. Overall, IHC validated the results of GO terms and pathway analyses, which showed that inflammatory pathways and corresponding biological processes are altered during the progression of CKD.
Ingenuity Pathway Analysis (IPA) allows for characterizing orthologous genes, and results identify possible mechanisms that have been validated in humans, mice, and rats. The top upregulated pathway identified in multiple comparisons was the “hepatic fibrosis/hepatic stellate cell activation pathway.” The IPA identified enriched pathways based on the over-represented DEGs, and the principle is the same as that used in the GO terms analysis via Panther. There was an extensive overlap between the DEGs we identified and the genes involved in the “hepatic fibrosis/hepatic stellate cell activation pathway” in the IPA, including collagen genes, cytokine-related genes, matrix metalloproteinases, tissue inhibitor of metalloproteinase, and the TNF receptor superfamily. Therefore, the upregulation of this pathway in our study supports common mechanisms involved in hepatic and renal fibrosis. It could also indicate contributing genes beyond those identified using known canine gene pathways.
The upstream regulator analysis of IPA identified the TGF-β group, especially TGF-β1, as the top upstream regulator, with both the highest activated z-score and the lowest p-value across multiple comparisons. Previous studies in canine8 and murine58 models of AS demonstrated the expression of TGF-β mRNA in kidney tissue. However, TGF-β was not differentially expressed in our comparisons, and IHC staining for TGF-β did not demonstrate appreciable differences in XLHN dogs compared to controls in a previous study8. The IPA Upstream Regulator Analysis predicts the upstream regulator of gene expression changes based on the knowledge of this regulatory cascade in the literature compiled in the Ingenuity Knowledge Base. Thus, the identified upstream regulator may not be identified as a DEG despite its importance in gene regulation.
A limiting factor of this study is that the kidney biopsies were immediately placed into RNALater to preserve RNA integrity, so microscopic evaluation could not be performed to determine whether the biopsy used for RNA isolation was representative of the cortex as a whole. Thus, one of the samples in the control group at T1 could have represented an area affected by a clinically insignificant insult. Another limiting factor is that expression data represents the mean expression by many cell types. Because the kidney has many cell types, all with different roles in CKD progression, it would be ideal to study gene expression changes in individual cells using laser-capture microdissection to further elucidate the progression of CKD. Last, extensive validation of the RNA-seq results was not performed; however, previous studies have shown RNA-seq to be a robust tool that highly correlates with qPCR results14–19. RNA-seq may even be more reliable than qPCR due to its higher sensitivity and lower probe bias16. We did perform IHC to further characterize the inflammatory population, which supported the pathways identified through the RNA-seq results.
In what appears to be the first RNA-seq study of a canine CKD model, we identified several previously described and novel genes and enriched pathways involved in the pathogenesis and development of CKD. The approach of acquiring biopsies at time points determined by the clinical stage of disease was an attempt to target causative gene expression starting at the clinical onset of proteinuria rather than secondary changes. Regardless of initial insult, CKD has common pathways that lead to end-stage kidney disease20. Therefore, many genes found in the current study may serve as predictive or diagnostic biomarkers for early detection of CKD in dogs and people. They may also be potential targets for drug development for this condition.
Methods
Animals
The dogs in this study were part of a colony with XLHN maintained at Texas A&M University5. XLHN is caused by a 10-base deletion in the gene encoding the α5 chain of type IV collagen. Affected males develop juvenile-onset CKD that progresses to end-stage renal disease as previously described5. Overall, 6 affected dogs and 2 unaffected littermates were studied. All dogs were raised according to standardized protocols, and no treatments were given to these dogs. All protocols were approved by the Texas A&M University Institutional Animal Care and Use Committee.
Clinical phenotypes
For this study, dogs were selected to represent both extremes in the speed of disease progression in this family of dogs (rapid versus slow progression). Clinical progression was determined by serial monitoring of serum and urine biomarkers of kidney disease, which allowed us to establish specific progression time points29: T1 (onset of proteinuria: defined as the presence of microalbuminuria for 2 consecutive weeks (E.R.D. HealthScreen Canine Urine Test Strips, Loveland, CO, USA)); T2 (onset of azotemia: serum creatinine ≥ 1.2 mg/dL); and T3 (end-stage disease: serum creatinine ≥ 5 mg/dL). Rapidly-progressing (rapid) dogs (n = 3) reached each time point at an earlier age than slowly-progressing (slow) dogs (n = 3). On average, the rapid group reached T3 at 26.3 weeks of age (range: 26–27 weeks), while the slow group reached the last clinical time point (T3) at 49 weeks of age (range: 46–52 weeks) (Supplementary Table S7).
Tissue collection
Kidney cortex was serially collected from each dog at the aforementioned 3 clinical time points (independent of age). Control dogs (n = 2) were biopsied to correspond with an affected littermate. All samples were collected by ultrasound-guided needle biopsy. This technique was appropriate for the current study as it is unlikely to induce changes that might be confused with those of CKD progression59. Samples for pathology evaluation and immunohistochemistry were placed in formalin and embedded in paraffin. Samples for RNA sequencing were immediately placed in RNAlater Stabilization Solution (Life Technologies, Foster City, CA, USA) and stored at −80 °C until RNA isolation.
Histopathological evaluation
Paraffin-embedded samples were processed and stained as previously described29. To determine the severity of interstitial fibrosis and chronic inflammation, a board-certified veterinary anatomic pathologist (REC) evaluated 5 or 20 randomly chosen 20x fields of renal cortex based on core size for each biopsy. For interstitial fibrosis, a score of 0 to 3 was assigned for each field based on the degree of tubulointerstitial architecture distortion caused by fibrosis: 0 - no fibrosis, 1 - fibrosis present but no distortion, 2 – moderate distortion, and 3 – severe distortion. For chronic inflammation, 0 – no inflammatory cells, 1 – scattered inflammatory cells, 2 – aggregates of inflammatory cells that separate or replace tubules, and 3 – diffusely distributed inflammatory cells. Statistical analysis comparing the average scores between groups was performed using bootstrap in R (version 3.2.4) to construct simultaneous 95% confidence intervals for all 3 pairwise comparisons of mean fibrosis and chronic inflammation scores at each of the latter 2 disease stages.
RNA isolation and sequencing
The MirVana miRNA Isolation Kit (Ambion, Austin, TX, USA) was used to isolate total RNA from homogenized kidney tissue according to the manufacturer’s instructions. The library preparation, sequencing, and initial quality check were performed by the Texas A&M AgriLife Genomics and Bioinformatics Service (http://www.txgen.tamu.edu/). RNA integrity was assessed by the Agilent 2100 Bioanalyzer (Agilent Technologies, Santa Clara, CA, USA). The average RIN was 3.4, and the average RNA yield was 86.2 ng/μL (Supplementary Table S3). To compensate for the low-input samples, we use the TruSeq Stranded Total RNA Library Prep Kit with Ribo-zero Gold (Illumina, San Diego, CA, USA), based on the best practice for RNA with variable qualities21 (to remove both cytoplasmic and mitochondrial rRNA) and its compatibility with canine samples. Samples were then sequenced using the Illumina Genome Analyzer (HiSeq. 2500v4 High Output). Raw sequencing data were submitted to the NCBI sequence read archive (SRA) (Accession: SRP101707; Samples: SAMN06560417, SAMN06560429-51; BioProject: PRJNA378728).
Data analysis
FastQC (version 0.11.2) was used for quality control to ensure that the quality value was above Q30. The canine (Canis lupus familiaris) genome FASTA file (ftp://ftp.ensembl.org/pub/current_fasta/canis_familiaris/dna/Canis_familiaris.CanFam3.1.dna.toplevel.fa.gz) and gene annotation GTF file (CanFam 3.1 assembly; ftp://ftp.ensembl.org/pub/current_gtf/canis_familiaris/) were obtained from Ensembl. Although RNA-seq is a popular research tool, there is no gold standard for analyzing RNA-seq data. Among the available tools, we chose up-to-date open source tools for mapping, retrieving read counts, and differential analysis. We used HISAT222 (version 2.0.3-beta) to generate indexes and to map reads to the canine genome. For assembly, we chose SAMtools (version 1.2) and the “union” mode of HTSeq23 (version 0.6.1), as the gene-level read counts could provide more flexibility in the differential expression analysis. Both HISAT2 and HTSeq analyses were conducted using the high performance research computing resources provided by Texas A&M University (http://hprc.tamu.edu) in the Linux operating system (version 2.6.32). Differential expression and statistical analysis were performed using DESeq2 (release 3.3) in R (version 3.2.4). DESeq224 was chosen as it is a popular parametric tool that provides a descriptive and continually updated user manual (https://bioconductor.org/packages/release/bioc/vignettes/DESeq2/inst/doc/DESeq2.html). DESeq2 internally corrects for library size, so it is important to provide un-normalized raw read counts as input. We used variance stabilizing transformation to account for differences in sequencing depth. P-values were adjusted for multiple testing using the Benjamini-Hochberg procedure60. A false discovery rate adjusted p-value (i.e., q-value) < 0.05 was set for the selection of DE genes.
Gene ontology (GO), pathway, and upstream regulator analysis
Gene ontology and PANTHER pathway analyses were performed with the PANTHER Overrepresentation Test (released on July 15, 2016) in PANTHER25 version 11.1 (http://www.pantherdb.org/, released on October 24, 2016). This program supports the canine genome. Also, QIAGEN’s Ingenuity Pathway Analysis (IPA, QIAGEN Redwood City, www.qiagen.com/ingenuity) was used to provide overrepresented orthologous genes in human, mouse, and rat databases and to identify orthologous pathways and upstream regulators in our data. PANTHER used the binomial test and Bonferroni correction for multiple testing, while IPA used the right-tailed Fisher Exact test and displayed z-scores to indicate whether a potential regulator was activated or inhibited. We used the default settings for statistical analysis in both the PANTHER pathway and IPA. In PANTHER, only pathways and GO terms with fold enrichment > 0.2 were listed. In IPA, p-value < 0.05 and fold change > 2 were set as cutoff values.
Immunohistochemistry (IHC)
Three-micrometer, formalin-fixed, paraffin-embedded renal cortex sections of both affected and control dogs were stained for CD3 (n = 24) and CD20 (n = 9). After deparaffinization, the sections were placed in citrate buffer (pH 6.0) for antigen retrieval, using a pressure cooker (Decloaking Chamber, Biocare Medical). Endogenous peroxidase activity and non-specific protein binding were blocked with 3% hydrogen peroxide and Sniper protein block (Biocare Medical), respectively. After blocking, the sections were incubated with primary antibodies CD3 (1:300 dilution; Dakocytomation, Carpinteria, CA) and CD20 (1:500 dilution; Thermo Scientific, Fremont, CA) for 1 hour at room temperature, then incubated with MACH 2 polymer for 30 minutes at room temperature. DAB (Vector Laboratories, Burlingame, CA) was used as the chromogen to demonstrate sites of antibody-antigen reaction. Mayer’s hematoxylin was used for the counterstain. Photographs were obtained using a SPOT Insight 2Mp FW Color Mosaic Camera (Diagnostic Instruments, Inc., Sterling Heights, MI) and the SPOT software (version 5.2).
Data Availability
Raw sequencing reads generated from this study are deposited at the NCBI sequence read archive (SRA) under accession SRP101707 (Samples: SAMN06560417, SAMN06560429-51; BioProject: PRJNA378728).
Electronic supplementary material
Acknowledgements
This work was funded by the National Institutes of Health (DK57676 and DK64273) and the Texas A&M Genomics Seed Grant Program. This study was presented in part at the 2016 American College of Veterinary Pathologists/American Society for Veterinary Clinical Pathology Annual Meeting in New Orleans, LA, December 2016. We would like to thank Dr. Ivan Ivanov for offering valuable suggestions for this study and providing RNA-seq analysis training through the Center for Translational Environmental Health Research (http://www.ctehr.org), Dr. Michael Dicken and all the personnel in the TAMU High Performance Research Computing (http://hprc.tamu.edu) for providing useful resources and instant help during data analysis, Dr. Michael Love for his rapid response in the Bioconductor online forum (https://support.bioconductor.org/), Dr. Andy Ambrus for the immunohistochemistry staining, Mia Aguilar and Mary Sanders for their technical assistance with RNA isolation and sample collection from dogs, respectively, and Cheng-An Richard Chang for guiding us through the first step of RNA-seq analysis. Special thanks to Dr. Barbara Gastel and Leah Poffenberger for their editing assistance on this manuscript.
Author Contributions
C.P.C. performed data analysis and prepared the manuscript; J.A.H. performed RNA isolations, revised the manuscript, and co-designed this project; R.E.C. evaluated the histopathology results and revised the manuscript; A.R.D. refined the statistical analyses; C.B.L. co-designed this project; G.E.L. collected samples and contributed to the study idea; and M.B.N. collected samples, designed and supervised this project, and revised the manuscript. All authors reviewed the manuscript.
Competing Interests
The authors declare that they have no competing interests.
Footnotes
Electronic supplementary material
Supplementary information accompanies this paper at 10.1038/s41598-017-16603-y.
Publisher's note: Springer Nature remains neutral with regard to jurisdictional claims in published maps and institutional affiliations.
References
- 1.Cox ML, Lees GE, Kashtan CE, Murphy KE. Genetic cause of X-linked Alport syndrome in a family of domestic dogs. Mamm. Genome. 2003;14:396–403. doi: 10.1007/s00335-002-2253-9. [DOI] [PubMed] [Google Scholar]
- 2.Lees GE. Kidney diseases caused by glomerular basement membrane type IV collagen defects in dogs. J. Vet. Emerg. Crit. Care (San Antonio) 2013;23:184–193. doi: 10.1111/vec.12031. [DOI] [PubMed] [Google Scholar]
- 3.Chiang CK, Inagi R. Glomerular diseases: genetic causes and future therapeutics. Nat. Rev. Nephrol. 2010;6:539–554. doi: 10.1038/nrneph.2010.103. [DOI] [PubMed] [Google Scholar]
- 4.Alport AC. Hereditary familial congenital haemorrhagic nephritis. Br. Med. J. 1927;1:504–506. doi: 10.1136/bmj.1.3454.504. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Lees GE, et al. New form of X-linked dominant hereditary nephritis in dogs. Am. J. Vet. Res. 1999;60:373–383. [PubMed] [Google Scholar]
- 6.Hashimura Y, et al. Milder clinical aspects of X-linked Alport syndrome in men positive for the collagen IV alpha5 chain. Kidney Int. 2014;85:1208–1213. doi: 10.1038/ki.2013.479. [DOI] [PubMed] [Google Scholar]
- 7.Pierides A, Voskarides K, Kkolou M, Hadjigavriel M, Deltas C. X-linked, COL4A5 hypomorphic Alport mutations such as G624D and P628L may only exhibit thin basement membrane nephropathy with microhematuria and late onset kidney failure. Hippokratia. 2013;17:207–213. [PMC free article] [PubMed] [Google Scholar]
- 8.Benali SL, et al. X-Linked hereditary nephropathy in Navasota dogs: clinical pathology, morphology, and gene expression during disease progression. Vet. Pathol. 2016;53:803–812. doi: 10.1177/0300985815624494. [DOI] [PubMed] [Google Scholar]
- 9.Greer KA, et al. Gene expression analysis in a canine model of X-linked Alport syndrome. Mamm. Genome. 2006;17:976–990. doi: 10.1007/s00335-005-0179-8. [DOI] [PubMed] [Google Scholar]
- 10.Woroniecka KI, et al. Transcriptome analysis of human diabetic kidney disease. Diabetes. 2011;60:2354–2369. doi: 10.2337/db10-1181. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Rao VH, et al. Increased expression of MMP-2, MMP-9 (type IV collagenases/gelatinases), and MT1-MMP in canine X-linked Alport syndrome (XLAS) Kidney Int. 2003;63:1736–1748. doi: 10.1046/j.1523-1755.2003.00939.x. [DOI] [PubMed] [Google Scholar]
- 12.Abrahamson DR, et al. Laminin compensation in collagen alpha3(IV) knockout (Alport) glomeruli contributes to permeability defects. J. Am. Soc. Nephrol. 2007;18:2465–2472. doi: 10.1681/ASN.2007030328. [DOI] [PubMed] [Google Scholar]
- 13.Byron SA, Van Keuren-Jensen KR, Engelthaler DM, Carpten JD, Craig DW. Translating RNA sequencing into clinical diagnostics: opportunities and challenges. Nat. Rev. Genet. 2016;17:257–271. doi: 10.1038/nrg.2016.10. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Marioni JC, Mason CE, Mane SM, Stephens M, Gilad Y. RNA-seq: an assessment of technical reproducibility and comparison with gene expression arrays. Genome Res. 2008;18:1509–1517. doi: 10.1101/gr.079558.108. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Mortazavi A, Williams BA, McCue K, Schaeffer L, Wold B. Mapping and quantifying mammalian transcriptomes by RNA-Seq. Nat. Methods. 2008;5:621–628. doi: 10.1038/nmeth.1226. [DOI] [PubMed] [Google Scholar]
- 16.Wang Z, Gerstein M, Snyder M. RNA-Seq: a revolutionary tool for transcriptomics. Nat. Rev. Genet. 2009;10:57–63. doi: 10.1038/nrg2484. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Griffith M, et al. Alternative expression analysis by RNA sequencing. Nat. Methods. 2010;7:843–847. doi: 10.1038/nmeth.1503. [DOI] [PubMed] [Google Scholar]
- 18.Nagalakshmi U, et al. The transcriptional landscape of the yeast genome defined by RNA sequencing. Science. 2008;320:1344–1349. doi: 10.1126/science.1158441. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Camarena L, Bruno V, Euskirchen G, Poggio S, Snyder M. Molecular mechanisms of ethanol-induced pathogenesis revealed by RNA-sequencing. PLoS Pathog. 2010;6:e1000834. doi: 10.1371/journal.ppat.1000834. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Yamaguchi, J., Tanaka, T. & Nangaku, M. Recent advances in understanding of chronic kidney disease. F1000Res. 4, doi:10.12688/f1000research.6970.1 (2015). [DOI] [PMC free article] [PubMed]
- 21.Adiconis X, et al. Comparative analysis of RNA sequencing methods for degraded or low-input samples. Nat. Methods. 2013;10:623–629. doi: 10.1038/nmeth.2483. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Kim D, Langmead B, Salzberg SL. HISAT: a fast spliced aligner with low memory requirements. Nat. Methods. 2015;12:357–360. doi: 10.1038/nmeth.3317. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Anders S, Pyl PT, Huber W. HTSeq–a Python framework to work with high-throughput sequencing data. Bioinformatics. 2015;31:166–169. doi: 10.1093/bioinformatics/btu638. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Love MI, Huber W, Anders S. Moderated estimation of fold change and dispersion for RNA-seq data with DESeq2. Genome Biol. 2014;15:550. doi: 10.1186/s13059-014-0550-8. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Mi H, Poudel S, Muruganujan A, Casagrande JT, Thomas PD. PANTHER version 10: expanded protein families and functions, and analysis tools. Nucleic Acids Res. 2016;44:D336–342. doi: 10.1093/nar/gkv1194. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Zhang Y, Wang H. Integrin signalling and function in immune cells. Immunology. 2012;135:268–275. doi: 10.1111/j.1365-2567.2011.03549.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Jubala CM, et al. CD20 expression in normal canine B cells and in canine non-Hodgkin lymphoma. Vet. Pathol. 2005;42:468–476. doi: 10.1354/vp.42-4-468. [DOI] [PubMed] [Google Scholar]
- 28.Vernau W, Moore PF. An immunophenotypic study of canine leukemias and preliminary assessment of clonality by polymerase chain reaction. Vet. Immunol. Immunopathol. 1999;69:145–164. doi: 10.1016/S0165-2427(99)00051-3. [DOI] [PubMed] [Google Scholar]
- 29.Nabity MB, et al. Urinary biomarkers of renal disease in dogs with X-linked hereditary nephropathy. J. Vet. Intern. Med. 2012;26:282–293. doi: 10.1111/j.1939-1676.2012.00891.x. [DOI] [PubMed] [Google Scholar]
- 30.Ju W, et al. Renal gene and protein expression signatures for prediction of kidney disease progression. Am. J. Pathol. 2009;174:2073–2085. doi: 10.2353/ajpath.2009.080888. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Gomez IG, et al. Anti-microRNA-21 oligonucleotides prevent Alport nephropathy progression by stimulating metabolic pathways. J. Clin. Invest. 2015;125:141–156. doi: 10.1172/JCI75852. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Tomkowicz B, et al. Interaction of endosialin/TEM1 with extracellular matrix proteins mediates cell adhesion and migration. Proc. Natl. Acad. Sci. USA. 2007;104:17965–17970. doi: 10.1073/pnas.0705647104. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Smith SW, et al. CD248+ stromal cells are associated with progressive chronic kidney disease. Kidney Int. 2011;80:199–207. doi: 10.1038/ki.2011.103. [DOI] [PubMed] [Google Scholar]
- 34.Wilhelm A, et al. CD248/endosialin critically regulates hepatic stellate cell proliferation during chronic liver injury via a PDGF-regulated mechanism. Gut. 2016;65:1175–1185. doi: 10.1136/gutjnl-2014-308325. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Bartis D, et al. Role of CD248 as a potential severity marker in idiopathic pulmonary fibrosis. BMC Pulm. Med. 2016;16:51. doi: 10.1186/s12890-016-0211-7. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Eikmans M, et al. Genetic factors in progressive renal disease: the good ones, the bad ones and the ugly ducklings. Nephrol. Dial. Transplant. 2006;21:257–260. doi: 10.1093/ndt/gfi325. [DOI] [PubMed] [Google Scholar]
- 37.LeBleu VS, et al. Identification of human epididymis protein-4 as a fibroblast-derived mediator of fibrosis. Nat. Med. 2013;19:227–231. doi: 10.1038/nm.2989. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Lin J, et al. Kielin/chordin-like protein, a novel enhancer of BMP signaling, attenuates renal fibrotic disease. Nat. Med. 2005;11:387–393. doi: 10.1038/nm1217. [DOI] [PubMed] [Google Scholar]
- 39.Zeisberg M, et al. Bone morphogenic protein-7 inhibits progression of chronic renal fibrosis associated with two genetic mouse models. Am. J. Physiol. Renal Physiol. 2003;285:F1060–1067. doi: 10.1152/ajprenal.00191.2002. [DOI] [PubMed] [Google Scholar]
- 40.Cheng S, Pollock AS, Mahimkar R, Olson JL, Lovett DH. Matrix metalloproteinase 2 and basement membrane integrity: a unifying mechanism for progressive renal injury. FASEB J. 2006;20:1898–1900. doi: 10.1096/fj.06-5898fje. [DOI] [PubMed] [Google Scholar]
- 41.Chambers JC, et al. Genetic loci influencing kidney function and chronic kidney disease. Nat. Genet. 2010;42:373–375. doi: 10.1038/ng.566. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Sakurai Y, et al. Pharmacokinetic significance of renal OAT3 (SLC22A8) for anionic drug elimination in patients with mesangial proliferative glomerulonephritis. Pharm. Res. 2005;22:2016–2022. doi: 10.1007/s11095-005-8383-5. [DOI] [PubMed] [Google Scholar]
- 43.Deguchi T, et al. Renal clearance of endogenous hippurate correlates with expression levels of renal organic anion transporters in uremic rats. J. Pharmacol. Exp. Ther. 2005;314:932–938. doi: 10.1124/jpet.105.085613. [DOI] [PubMed] [Google Scholar]
- 44.Sinner DI, Kim GJ, Henderson GC, Igal RA. StearoylCoA desaturase-5: a novel regulator of neuronal cell proliferation and differentiation. PLoS One. 2012;7:e39787. doi: 10.1371/journal.pone.0039787. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Edeling M, Ragi G, Huang S, Pavenstadt H, Susztak K. Developmental signalling pathways in renal fibrosis: the roles of Notch, Wnt and Hedgehog. Nat. Rev. Nephrol. 2016;12:426–439. doi: 10.1038/nrneph.2016.54. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Xu Y, et al. High thymidine kinase 1 (TK1) expression is a predictor of poor survival in patients with pT1 of lung adenocarcinoma. Tumour Biol. 2012;33:475–483. doi: 10.1007/s13277-011-0276-0. [DOI] [PubMed] [Google Scholar]
- 47.Kagan HM, Li W. Lysyl oxidase: properties, specificity, and biological roles inside and outside of the cell. J. Cell. Biochem. 2003;88:660–672. doi: 10.1002/jcb.10413. [DOI] [PubMed] [Google Scholar]
- 48.Liu SB, et al. Lysyl oxidase activity contributes to collagen stabilization during liver fibrosis progression and limits spontaneous fibrosis reversal in mice. FASEB J. 2016;30:1599–1609. doi: 10.1096/fj.14-268425. [DOI] [PubMed] [Google Scholar]
- 49.Murawaki Y, Kusakabe Y, Hirayama C. Serum lysyl oxidase activity in chronic liver disease in comparison with serum levels of prolyl hydroxylase and laminin. Hepatology. 1991;14:1167–1173. doi: 10.1002/hep.1840140635. [DOI] [PubMed] [Google Scholar]
- 50.Sivakumar P, Gupta S, Sarkar S, Sen S. Upregulation of lysyl oxidase and MMPs during cardiac remodeling in human dilated cardiomyopathy. Mol. Cell. Biochem. 2008;307:159–167. doi: 10.1007/s11010-007-9595-2. [DOI] [PubMed] [Google Scholar]
- 51.Goto Y, et al. Transforming growth factor-beta1 mediated up-regulation of lysyl oxidase in the kidneys of hereditary nephrotic mouse with chronic renal fibrosis. Virchows Arch. 2005;447:859–868. doi: 10.1007/s00428-005-0001-8. [DOI] [PubMed] [Google Scholar]
- 52.Breyer MD, Susztak K. The next generation of therapeutics for chronic kidney disease. Nat. Rev. Drug Discov. 2016;15:568–588. doi: 10.1038/nrd.2016.67. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 53.Quezada-Calvillo R, Simsek M, Juarez J, Nichols B. Protein synthesis controls the activity of maltase-glucoamylase and sucrase-isomaltase in non-intestinal tissues. FASEB J. 2015;29:596.518. [Google Scholar]
- 54.Zhou Q, et al. Identification of genes associated with Smad3-dependent renal injury by RNA-seq-based transcriptome analysis. Sci. Rep. 2015;5:17901. doi: 10.1038/srep17901. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 55.Rubel D, et al. Collagen receptors integrin alpha2beta1 and discoidin domain receptor 1 regulate maturation of the glomerular basement membrane and loss of integrin alpha2beta1 delays kidney fibrosis in COL4A3 knockout mice. Matrix Biol. 2014;34:13–21. doi: 10.1016/j.matbio.2014.01.006. [DOI] [PubMed] [Google Scholar]
- 56.Yhee JY, Yu CH, Kim JH, Sur JH. Effects of T lymphocytes, interleukin-1, and interleukin-6 on renal fibrosis in canine end-stage renal disease. J. Vet. Diagn. Invest. 2008;20:585–592. doi: 10.1177/104063870802000508. [DOI] [PubMed] [Google Scholar]
- 57.Jedlicka J, et al. Interstitial inflammation in Alport syndrome. Hum. Pathol. 2010;41:582–593. doi: 10.1016/j.humpath.2009.08.024. [DOI] [PubMed] [Google Scholar]
- 58.Sayers R, et al. Role for transforming growth factor-beta1 in Alport renal disease progression. Kidney Int. 1999;56:1662–1673. doi: 10.1046/j.1523-1755.1999.00744.x. [DOI] [PubMed] [Google Scholar]
- 59.Groman RP, Bahr A, Berridge BR, Lees GE. Effects of serial ultrasound-guided renal biopsies on kidneys of healthy adolescent dogs. Vet. Radiol. Ultrasound. 2004;45:62–69. doi: 10.1111/j.1740-8261.2004.04010.x. [DOI] [PubMed] [Google Scholar]
- 60.Benjamini Y, Hochberg Y. Controlling the false discovery rate - a practical and powerful approach to multiple testing. J. R. Stat. Soc. Series B Stat. Methodol. 1995;57:289–300. [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
Data Availability Statement
Raw sequencing reads generated from this study are deposited at the NCBI sequence read archive (SRA) under accession SRP101707 (Samples: SAMN06560417, SAMN06560429-51; BioProject: PRJNA378728).