Abstract
Zanthoxylum, an ancient economic crop in Asia, has a satisfying aromatic taste and immense medicinal values. A lack of genomic information and genetic markers has limited the evolutionary analysis and genetic improvement of Zanthoxylum species and their close relatives. To better understand the evolution, domestication, and divergence of Zanthoxylum, we present a de novo transcriptome analysis of an elite cultivar of Z. bungeanum using Illumina sequencing; we then developed simple sequence repeat markers for identification of Zanthoxylum. In total, we predicted 45,057 unigenes and 22,212 protein coding sequences, approximately 90% of which showed significant similarities to known proteins in databases. Phylogenetic analysis indicated that Zanthoxylum is relatively recent and estimated to have diverged from Citrus ca. 36.5–37.7 million years ago. We also detected a whole-genome duplication event in Zanthoxylum that occurred 14 million years ago. We found no protein coding sequences that were significantly under positive selection by Ka/Ks. Simple sequence repeat analysis divided 31 Zanthoxylum cultivars and landraces into three major groups. This Zanthoxylum reference transcriptome provides crucial information for the evolutionary study of the Zanthoxylum genus and the Rutaceae family, and facilitates the establishment of more effective Zanthoxylum breeding programs.
Introduction
Zanthoxylum L. of the Rutaceae family is a species-rich genus, widely distributed in tropical and subtropical areas. The Zanthoxylum genus consists of >250 species of deciduous and evergreen trees, shrubs, and dunga-runga, with 39 species and 14 varieties being found in China1,2. Zanthoxylum has a long history of cultivation and domestication for both economic and chemical value in Asia. This genus is a promising source of secondary metabolites including alkylamides3, alkaloids4, and flavonoids5. Species within this genus can be used as spices and in traditional medicine.
In contrast to studies of phytochemistry and biological activity6–8, historical and evolutionary analysis of Zanthoxylum has received little attention. Zanthoxylum has a rich genetic patrimony in China and is characterized by an abundance of cultivars, some of which have a unique botanical feature of nucellar embryony9. This process of apomixis has hindered genetic study and breeding improvement of Zanthoxylum, because the seedlings are essentially clones of the maternal parent. Moreover, empirical observations indicate that the classification of the commercial classes of Zanthoxylum cultivars is often confusing because of homonyms, synonyms, and scarce or incorrect historical records.
The lack of genetic resources and genomic information has hampered the improvement of Zanthoxylum to enhance yield and resistance to biotic and abiotic stresses. Additionally, inappropriate selection of cultivars in agricultural production of Zanthoxylum can cause economic loss. Therefore, reliable and distinguishable genetic markers are a requirement for genetic and breeding studies of Zanthoxylum, enabling the development of improved genotypes or varieties with enhanced trait values.
Complete information on transcriptomes can easily be generated via high-throughput mRNA sequencing technology, which is a powerful and cost-effective tool for gene expression profiling in non-model organisms without a reference genome10. Illumina transcriptome de novo sequencing and assembly have been successfully used for Rutaceae plants, with recent research focusing on Citrus 11–14 and Pilocarpus species15. To the best of our knowledge, no studies have reported transcriptome sequencing of Zanthoxylum.
In this study, we present the first transcriptome of a Zanthoxylum species, for Z. bungeanum, which is native to China. Z. bungeanum has become the leading commercially cultivated species in this country because of its fast growth and adaptability to adverse soil and climatic conditions16. We aimed to: i) characterize the Z. bungeanum transcriptome for gene content; ii) describe the evolutionary features of Zanthoxylum; and iii) develop simple sequence repeat (SSR) markers for Zanthoxylum species. The transcriptome analysis presented in this study yields new insights into the evolutionary origin of Zanthoxylum and provides a rich resource of genetic information for breeding and genetic improvement.
Results
Sequencing and de novo assembly of Illumina paired-end reads
In this study, we generated sequences for the transcriptome of an elite Z. bungeanum cultivar, ‘Fengxiandahongpao’. Approximately 56.2 million high-quality reads were generated, with a GC content of 43.38%. The combined sequences of high-quality reads were assembled into 65,337 individual transcripts and 45,057 unigenes, reaching a total length of 41.0 Mb and 27.5 Mb, respectively. The average length of the assembled transcripts was 627 bp, with an N50 of 874 bp. The length of the assembled unigenes ranged from 201 to 6,750 bp, with an average of 610 bp and an N50 of 846 bp (Table 1).
Table 1.
Summary for the transcriptome of Z. bungeanum cultivar.
| Total number of reads | 56,242,196 |
|---|---|
| Total number of transcripts | 65,337 |
| Total number of unigenes | 45,057 |
| Average transcript length (bp) | 627 |
| Average unigenes length (bp) | 610 |
| N50 of transcript (bp) | 874 |
| N50 of unigenes (bp) | 846 |
| Longest unigenes (bp) | 6,750 |
| Shortest unigenes (bp) | 201 |
| GC content 43.38% | |
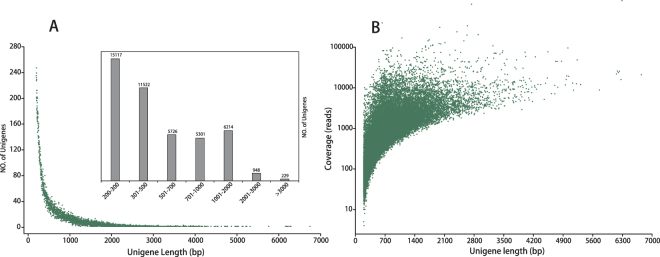
Of all the unigenes, those from 201 to 300 bp accounted for 33.55% (15,117). The unigenes ranging from 301 to 500 bp accounted for 25.57% (11,522). There were 5,726 unigenes (12.71%) with lengths ranging from 501 to 700 bp, and 5,301 unigenes (11.77%) with lengths ranging from 701 to 1,000 bp. Only 2.61% of the unigenes (1177) were longer than 2,000 bp (Fig. 1).
Figure 1.
Distribution of unigene length and coverage depth in the transcriptome assembly of Z. bungeanum. (A) Length distribution of the assembled unigenes. (B) Density scatter-plot showing the relationship between unigene length and coverage.
Sequence annotation
For validation and annotation of the sequence assembly contigs and unique singletons, all unigenes were searched against six public protein databases, including the NCBI non-redundant protein (Nr) database, protein family (Pfam), Swiss-Prot, the Kyoto Encyclopedia of Genes and Genomes pathway (KEGG) database, the EuKaryotic Orthologous Groups (KOG) database, and the Gene Ontology (GO) database. In total, 40,223 of 45,057 unigenes (89.27%) provided significant BLAST results, with 40,214 (89.25%) showing significant similarity to known proteins in the Nr database, 28,644 (63.57%) to proteins in the Swiss-Prot, and 18,128 (40.23%) to proteins in the Pfam. Only 1,715 unigenes were annotated in the six databases (Supplement Figure S1).
To further analyze the BLAST results, E-value and similarity distributions were calculated using the Nr database. The E-value distribution of the top hits revealed that 78.77% of the mapped sequences showed significant homology (E-value < 10−30), and 21.22% of the homologous sequences had E-values in the range 10−6 to 10−30 (Supplement Figure S2A). Additionally, 91.18% and 76.17% of the sequences were found to have similarities >70% and 80%, respectively (Supplement Figure S2B). These results reflect the high identities of the mapped sequences, suggesting a good assembly quality.
The species distribution of the top BLASTx hits against the Nr database showed that Z. bungeanum genes had the greatest number of matches with genes of orange (Citrus), cacao (Theobroma), and cotton (Gossypium). Among these, C. sinensis ranked first, with 13.3% top BLASTx hits, followed by T. cacao (7.1%), C. clementine (6.2%), G. hirsutum (4.7%), and G. raimondii (4.6%) (Supplement Figure S2C).
Functional classification by GO analysis
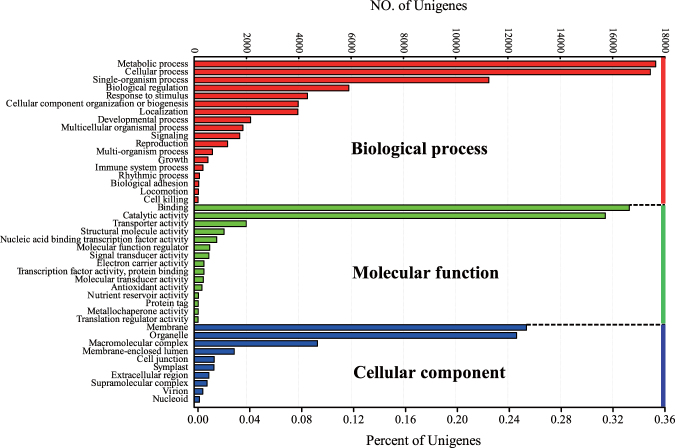
In total, 32,715 unigenes (72.61%) with BLAST matches to known proteins were assigned to GO classes with 132,647 functional terms (Fig. 2). The annotated unigenes that belonged to the biological process, cellular component, and molecular function clusters were categorized into 43 functional groups. Molecular function (47,706, 35.96%) and biological process (46,861, 35.33%) represented the largest number of unigenes, followed by cellular component (38,080, 28.71%).
Figure 2.
Gene ontology classification of assembled unigenes of Z. bungeanum. Results are summarized in three main categories: biological process, cellular component, and molecular function. The bottom x-axis indicates the percentage of a specific category of genes in that main category. The top x-axis indicates the actual number of genes in a category.
Under the classification molecular function (in total 47,706), binding (16,797, 35.21%) and catalytic activity (15,869, 33.26%) were the largest and second largest categories, respectively. Other categories, such as transporter activity, structural molecule activity, nucleic acid binding transcription factor activity, and molecular transducer activity, contained 15,040 unigenes (31.53%) in total. Under the category biological process (in total 46,861), the most frequent subcategories were metabolic process (17,831, 38.05%) and cellular process (17,619, 37.60%), followed by single-organism process (11,325, 24.17%), biological regulation (5,869, 12.52%), and response to stimulus (4,265, 9.10%). Three categories, membrane, organelle, and macromolecular complex, represented approximately 38.75% of cellular component classifications (Fig. 2).
Functional classification by KOG analysis
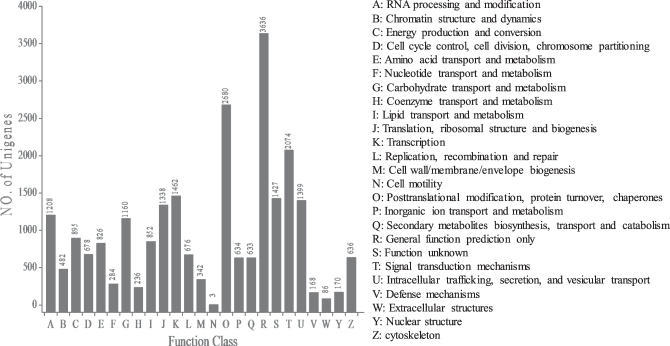
To further evaluate the completeness of our transcriptome library, all unigenes were searched against the KOG database for functional prediction and classification. In total, 31,657 of 45,057 unigenes (70.26%), showing significant homology to entries in the KOG database, were functionally classified into 25 molecular families (Fig. 3). Among these KOG categories, the largest group was “General function prediction only” containing 3,636 unigenes (15.16%), followed by “Posttranslational modification, protein turnover, chaperones” (11.17%), “Signal transduction mechanisms” (8.65%), “Transcription” (6.10%), “Function unknown” (5.95%), and “Intracellular trafficking, secretion, and vesicular transport” (5.83%). Only three unigenes were assigned to “Cell motility” (0.01%).
Figure 3.
Histogram presentation of KOG classification of Z. bungeanum unigenes. Out of 48,300 Nr database hits, 3,112 sequences have a KOG classification among the 25 categories.
Functional classification by KEGG pathways
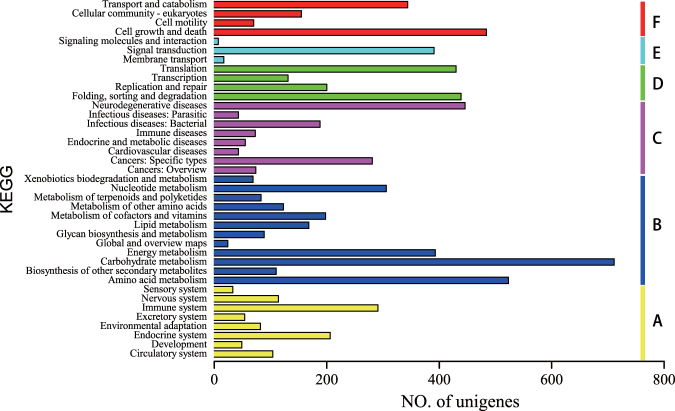
To further analyze the transcriptome of Z. bungeanum, all the unigenes were analyzed via the KEGG pathway database, where a total of 3,093 unigenes had significant matches and were assigned to six main categories in 229 pathways (Fig. 4). Among these six main categories, metabolism was the largest, containing 2,797 unigenes (90.43%), followed by genetic information processing (1,200, 38.80%), human diseases (1,076, 34.79%), cellular processes (1,047, 33.85%), organismal systems (886, 28.65%), and environmental information processing (415, 13.42%).
Figure 4.
Pathway assignment based on KEGG pathways of Z. bungeanum unigenes. (A) Classification based on organismal systems categories. (B) Classification based on metabolism categories. (C) Classification based on human diseases categories. (D) Classification based on genetic information processing categories. (E) Classification based on environmental information processing categories. (F) Classification based on cellular process categories.
Evolutionary pattern of protein coding sequences
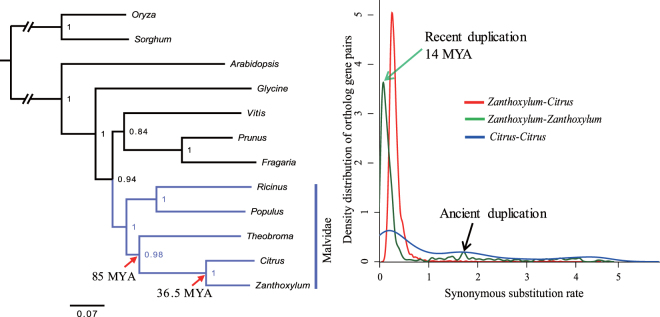
The protein coding sequences (CDS) extracted from unigenes also provided comprehensive information for phylogenetic analysis. A phylogenetic analysis was conducted based on the 34 single-copy nuclear genes (Supplement Dataset S1) shared in Zanthoxylum, sweet orange (Citrus sinensis), poplar (Populus euphratica), grape (Vitis aestivalis), strawberry (Fragaria × ananassa), soybean (Glycine max), Arabidopsis, cacao (Theobroma cacao), peach (Prunus persica), and castor bean (Ricinus communis). Zanthoxylum was found to be phylogenetically closest to sweet orange, cacao, poplar, and castor bean (Fig. 5A). For comparison, a phylogenetic tree was derived from 68 chloroplast genes (Dataset S2) in the same species, which showed that Zanthoxylum was phylogenetically closest to sweet orange, cacao, Arabidopsis, and grape (Supplement Figure S3). This indicated a significantly different evolutionary pattern between or within nuclear and plastid genomes in some plant species (e.g., grape and Arabidopsis). Assuming orange diverged from cacao 85 million years ago (MYA)17, we confidently estimated a divergence time for Zanthoxylum and Citrus of 36.5 and 37.7 MYA from the nuclear and chloroplast trees, respectively (Fig. 5A and S3).
Figure 5.
Evolutionary analysis of Z. bungeanum. (A) Zanthoxylum phylogeny on the basis of 34 single-copy nuclear genes shared by Zanthoxylum and other plant species. Rice (Oryza sativa) and sorghum (Sorghum bicolor) were used as out-groups. MYA, million years ago. (B) Distribution of synonymous substitution rates (Ks) for homologous gene groups in protein coding sequences (CDS) of Zanthoxylum and Citrus.
Meanwhile, an age distribution was calculated by comparing the synonymous substitution values (Ks) of 2,750 orthologous gene pairs between the Zanthoxylum and Citrus genomes, 794 paralogous gene pairs in the Zanthoxylum transcriptome, and 1,054 paralogous gene pairs in Citrus genomes. A Zanthoxylum whole-genome duplication (WGD) event occurred 14 MYA (Ks peaks at 0.074), more recently than the divergence of Zanthoxylum and Citrus. Interestingly, we detected three peaks in the Ks distribution in orange, suggesting that at least three ancient WGD events occurred in orange (Fig. 5B). A second large Ks peak at a synonymous substitution rate of about 1.7 was observed in both Zanthoxylum and Citrus, indicating that an ancient WGD event occurred in their common ancestor (Fig. 5B and Supplement Figure S4). We also inferred the direction and magnitude of natural selection acting on protein coding genes in Zanthoxylum by calculating the Ka/Ks through referring to Citrus orthologous genes. Only four genes with Ka/Ks > 1 but non-significant were observed, implying that most Zanthoxylum genes experienced purifying or stabilizing selection (Supplement Figure S5).
To better understand Zanthoxylum-specific biology, we identified Zanthoxylum-specific genes by comparative analyses of the Zanthoxylum protein CDS with 12 other plant genomes and rescreened them in the Nr protein database to exclude false-positive genes caused by incomplete genome annotation. In total, 22,212 CDS in the Zanthoxylum transcriptome were clustered into 8154 gene families, with 173 genes being specific to Zanthoxylum (Supplement Figure S6). One hundred of the Zanthoxylum-specific genes had unknown function. Of the 73 annotated genes, overrepresented molecular functions were oxidation-reduction, regulation of transcription, and translation processes. Exploring the function of these genes will contribute significantly to understanding of important Zanthoxylum-specific biological processes, such as the synthesis of numb-taste components.
Identification of single nucleotide polymorphisms
From the Z. bungeanum transcriptome sequencing data, we identified 170,000 single nucleotide polymorphisms (SNPs) in 37,658 unigenes. The proportions of transversions were: 10.48% A/C; 12.44% A/T; 7.04% C/G and 10.33% G/T. The proportions of transitions were: 29.61% A/G and 30.10% C/T. The percentage of transitions (59.71%) was 1.48-times higher than that of transversions (40.29%) in the Z. bungeanum transcriptome. Further analysis of the SNPs revealed that approximately 72% of them were distributed in CDS, and 20,461 CDS contained at least one SNP, leading to a rough estimate of polymorphism density of 6.16 SNPs per kb in sequences containing CDS. The large number of SNPs located in sequences containing CDS is possibly beneficial for future development of important economic and agronomic traits. These SNPs could be useful for molecular breeding programs and marker-assisted selection practices, as well as for understanding phenotypic differences between Zanthoxylum species.
Identification of simple sequence repeats from Z. bungeanum transcriptome
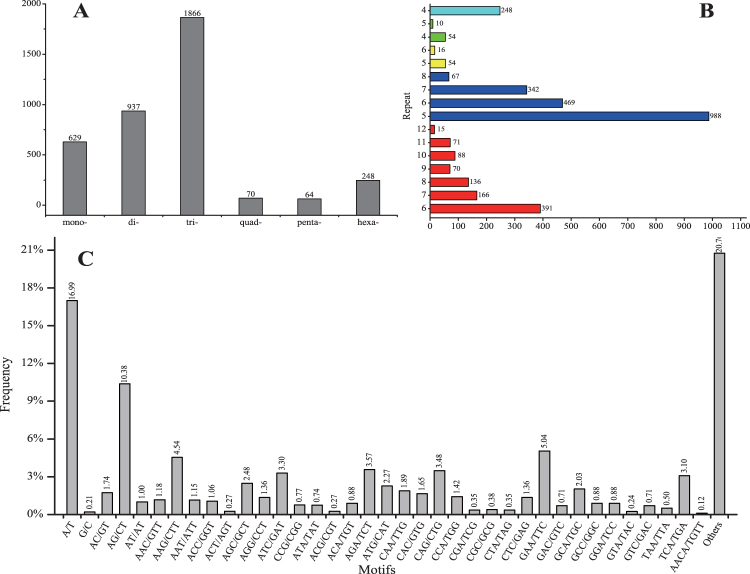
A total of 3,814 potential simple sequence repeats (SSRs) were identified across 45,057 unigenes from the transcriptome of Z. bungeanum. Of these, 488 unigenes (12.79%) contained one or more SSR sequences, 282 unigenes (7.39%) contained at least two separate SSR sequences, and 206 unigenes (5.40%) contained compound SSRs of different motifs (Fig. 6A). The number of SSRs per unigene ranged from 1 to 5. Trinucleotide repeat motifs were the most frequent type (1866, 48.93%) with 5–8 repeats, followed by 937 (24.57%) di-type motifs with 6–12 repeats, 629 (16.49%) mono-type motifs, and 248 (6.50%) hexa-type motifs with 4 repeats. Fewer quad-type motifs (70, 1.84%) and penta-type motifs (64, 1.68%) were observed, with 5–6 and 4–5 repeats, respectively (Fig. 6B).
Figure 6.
Results of microsatellite SSR analyses in Z. bungeanum. (A) Distribution of the total number of SSRs in different classes of repeat type. Di-, tri-, tetra-, penta-, and hexanucleotide repeats were analyzed. (B) Distribution of the number of SSR repeats. (C) Frequency distribution of SSRs based on motif sequence type.
Except for compound SSRs, A/T was the most abundant motif in the two possible types of mono-nucleotide repeat, contributing 16.31% of the total SSRs. The C/G motif was less abundant than A/T, accounting for 0.21% of the total SSRs. The dominant di-nucleotide repeat motif in SSRs was AG/CT accounting for 10.38% of the total SSRs. The AC/GT motif was slightly more abundant than the AT/AT motif, which constituted 1.74% and 1.00% of the total SSRs, respectively. Among the trinucleotide repeat motifs, the two most frequent repeats were GAA/TTC (5.04% of the total SSRs) and AAG/CTT (4.54%), followed by ATC/GAT (3.30%), AGA/TCT (3.57%), and CAG/CTG (3.48%). Other motifs among quad-, penta-, and hexa- nucleotide repeats constituted 20.88% of the total SSRs (Fig. 6C).
Development and validation of SSR markers
Out of 3,814 SSRs, a total of 1,000 primer pairs were successfully designed; 100 primer pairs were selected at random and tested to amplify the genomic DNA of Z. bungeanum (20 cultivars and two landraces) and Z. armatum (five cultivars and four landraces). Sixty primer pairs resulted in successful PCR amplification, 25 of which produced fragments of the expected size. Taking into account the specific amplification and polymorphic loci, we selected 16 primer pairs that presented clear and polymorphic loci to help evaluate polymorphism levels in the two Zanthoxylum species. After further testing, we selected 11 primer pairs for subsequent polymorphic analysis (Table 2). Compared with Z. armatum cultivars, Z. bungeanum cultivars possessed a relatively high number of alleles (N a = 4.636), effective number of alleles (N e = 3.088), observed and expected heterozygosity (H o = 0.749 and H e = 0.655), and Shannon index (I = 1.225). Additionally, the fixation index (F IS) of Z. bungeanum and Z. armatum varied from −0.485 to 0.367 and −0.385 to 0.600, with an average of −0.183 and 0.027, respectively (Supplement Table S2).
Table 2.
Characteristics of primers designed for analyzing genetic diversity of Zanthoxylum cultivars.
| Code | Primer pair | Length (bp) | Melting temperature (°C) | Product size (bp) | GC content (%) |
|---|---|---|---|---|---|
| ZB-2 | F: AACAAATGGTGCTGATGACA | 20 | 54 | 177 | 45.2 |
| R: ATTACTGCTCCTCGAAACTGG | 21 | ||||
| ZB-4 | F: TCAACAGATATGGCTCGAGT | 20 | 54 | 206 | 43.2 |
| R: TTTGAAACCCCGAACATGG | 19 | ||||
| ZB-11 | F: AGGCTTTCAATGAGTTCAAGG | 21 | 56 | 206 | 43.7 |
| R: CTCTTCTGGATTTTCCTCGT | 20 | ||||
| ZB-28 | F: ACAACAGACTGCTAAGCCAA | 20 | 54 | 139 | 39.6 |
| R: TCCTTTTGCCTTTCAACCC | 19 | ||||
| ZB-30 | F: ACACTGATATACCGGCACCT | 20 | 58 | 201 | 49.8 |
| R: ATGCCTCGTTTGGATCGCTT | 20 | ||||
| ZB-32 | F: ATTCCGAAATCCCGCCCAA | 19 | 56 | 180 | 47.2 |
| R: TTTCCGACAGCAACGGCTT | 19 | ||||
| ZB-43 | F: CAGTCTCATTAAACGCTCC | 19 | 54 | 214 | 40.2 |
| R: TCTTCTCTTCATCCGCTTC | 19 | ||||
| ZB-69 | F: CCTACGCCAACTCACC | 16 | 58 | 149 | 65.1 |
| R: ATGCGAACCTTCAACGTC | 18 | ||||
| ZB-73 | F: TTGAGGACGACGAAGATG | 18 | 56 | 201 | 51.4 |
| R: CTTCTTCCCCAGTCTCTC | 18 | ||||
| ZB-89 | F: GATGTTGCGAATGATACAAG | 20 | 54 | 100 | 46 |
| R: GACAATATTCAGGAGCAGGA | 20 | ||||
| ZB-97 | F: AGTATTGTGGAGCTAACAT | 19 | 54 | 191 | 38.2 |
| R: TGAGGCCATCTTTACACT | 18 |
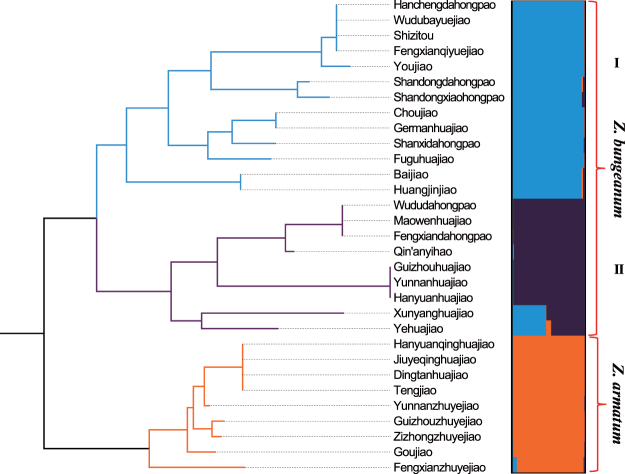
Neighbor-joining tree and STRUCTURE analyses clearly grouped the 31 cultivars and landraces into three major clusters (Fig. 7). Cluster I (cyan) included 13 cultivars of Z. bungeanum mainly distributed north of the Qinling Mountains. Cluster II (purple) included nine Z. bungeanum cultivars mainly distributed south of the Qinling Mountains. These two clusters are commonly known as “Red huajiao” for their bright red pericarp. Cluster III (orange) consisted of nine cultivars of Z. armatum, commonly known as “Green huajiao” for their bright green pericarp.
Figure 7.
Cluster analysis of 31 cultivars and landraces of Z. bungeanum and Z. armatum based on 11 polymorphic SSR markers. (A) NJ-tree genetic relationships among Zanthoxylum cultivars and landraces. (B) STRUCTURE-based clusters resolved 31 cultivars and landraces into three groups (K = 3), Z. bungeanum I and II, and Z. armatum. Color groups correspond to NJ-tree clades.
Discussion
Here we report the first transcriptome analysis of Zanthoxylum, an economic crop of great importance because of its flavor and industrial and medical applications18. The transcriptome sequence of Z. bungeanum provides a genomic resource for genetic study and breeding improvement of Zanthoxylum, and a reference for plant transcriptome-scale evolutionary analyses of the Rutaceae family.
Since Z. bungeanum has been bred for more than 4,000 years, a large number of elite cultivars have been widely dispersed, named, and released in China. Sequencing of the Z. bungeanum transcriptome provides genomic data for comprehensively assessing the heterozygosity of Zanthoxylum plants. Our transcriptome analysis indicated that Z. bungeanum might have a high degree of heterozygosity. Heterozygosity is a common feature in most plants and has important biological functions in crops. The heterozygosity of Z. bungeanum, estimated in the present study, is higher than that of sweet orange17, which suggests Z. bungeanum may have undergone an intensive inter- or intra-specific hybridization. Moreover, the high values of mean H o and H e also suggest a relatively high heterozygosity, while the negative value of F IS also indicates a significant heterozygote excess in Z. bungeanum. Therefore, it is not feasible to consider whole-genome sequencing of Zanthoxylum. Fortunately, the advent of RNA-Seq is useful for collecting genetic information from Z. bungeanum.
GO functional classification may help us to understand the distribution of gene functions at the macro level, and to predict the physiological role of each unigene. KOG is a database in which orthologous gene products are classified19. In the present study, the annotated unigenes were classified into 25 KOG and 43 GO subterms or subcategories, suggesting that our transcriptome data for Z. bungeanum represented a wide diversity of transcripts20,21. Meanwhile, the results revealed that the assembled unigenes had diverse molecular functions and were involved in many metabolic pathways. Under the biological process category following GO classification, cellular process and metabolic process were prominent, indicating the occurrence of important cellular and metabolic activities in Z. bungeanum.
KEGG pathways are helpful to better understand biological function and gene interaction. Functional classification based on KEGG pathways indicated that numerous important metabolic pathways in Z. bungeanum were unknown and worth investigation. Obviously, “metabolic pathways” represented the largest category, suggesting Z. bungeanum invests in cell maintenance and defense. Moreover, unigenes associated with metabolism of cofactors and vitamins, terpenoids and polyketides, and biosynthesis of other secondary metabolites were identified in the present study, with evidence that numerous biologically active secondary metabolites have been isolated in Zanthoxylum species18. ‘Human diseases’ represented another dominant pathway and contained six different subcategories, indicating a complex composition of the Zanthoxylum genome, which makes it an important target for genomic study.
Phylogenetic analysis based on nuclear and chloroplast genes indicated that Zanthoxylum is relatively recent and estimated to have diverged from Citrus 36.5–37.7 MYA. Interestingly, the nuclear phylogeny showed that Arabidopsis was not linked to Malvidae with full bootstrap value support, whereas the chloroplast tree topology demonstrated that Arabidopsis was included in the Malvidae. This contradiction may be due to the difference in evolutionary pattern between or within nuclear and chloroplast genes, and the sensitivity of the topology to the maximum likelihood methods.
The WGD is predominant in most flowering plant species during evolution22, as gene duplication has provided a necessary source of material for the origin of evolutionary novelties. Due to functional redundancy, genes are rapidly silenced or lost from the duplicated genomes23. The detection method of plant WGDs is derived from genomic structure (e.g., syntenic analysis), phylogenetic tree construction, and evolutionary distances (e.g., synonymous mutation rate, Ks)24,25. To investigate the genome evolutionary history of Zanthoxylum, we estimated the WGD time in the Zanthoxylum genome by calculating the Ks age distribution using the formula T = Ks/2r. Here we chose r = 2.6 × 10−9 as the neutral substitution rate, because Zanthoxylum species are long-lived perennials and the generation time is negatively correlated with molecular evolutionary rates26. The synonymous mutation rates of several plants have been estimated using a small set of genes (2.6 × 10−9–1.5 × 10−8)27. It is likely that the slower rates may be more appropriate for Zanthoxylum. As for the transformation of the Ks values to absolute ages, low levels of Ks produce a more accurate age estimation than high Ks values because of possible substitution-saturation effects and molecular rate heterogeneity among lineages. In the present study, we deduced a recent WDG event in Zanthoxylum 14 MYA. We also calculated the Ks of each pair of three-copy and four-copy genes in Zanthoxylum and orange. Strikingly, they presented a similar but flatter curve than that of two-copy gene pairs, providing support for our confident estimation of Ks distribution and a more ancient duplication for multicopy genes.
In the present study, trinucleotide repeats were the most abundant type of SSR motifs in the transcriptome of Z. bungeanum, followed by the di-type motifs. This finding is similar to the results reported for pummelo28 and cotton29. Two hundred and six SSRs were present in the compound types, meaning they contained stretches of two or more different repeats. Among mononucleotide repeats, as in most plants, the A/T repeats were far more abundant than G/C repeats30,31. For the dinucleotide repeat category, the most abundant motif was AG/CT, which is similar to previous results in Hemarthria compressa “Yaan” and H. altissima “1110”32, Ipomoea batatas 33, Brachypodium distachyon, and Oryza sativa 34. Thirty-one motifs were found within the trinucleotide repeats; GAA/TTC and AAG/CTT were the most abundant, similar to results for colored calla lily35. These results support previous observations that dinucleotide AG/CT motifs and trinucleotide AAG/CTT motifs are the most abundant SSR motifs in dicots36.
Mining transcriptome data is an efficient way for SSR marker development and offers great flexibility in selecting markers for different applications at different resolutions37. Moreover, SSRs derived from a transcriptome database have advantages over other strategies, such as low cost and short time. In the present study, we verified and obtained 11 variety-specific EST-SSR markers (Table 2), which provides a simple, informative, reproducible, and suitable approach for evaluation of genetic relationships in Zanthoxylum species. The neighbor-joining tree and STRUCTURE analyses divided 31 Zanthoxylum cultivars and landraces into three clear clusters. The availability of the Z. bungeanum transcriptome and deep exploitation of Zanthoxylum-specific genes with known or unknown biochemical functions will facilitate a better understanding of agronomically and economically important traits, including yield, numb-taste components, oil, and stress resistance in subsequent research.
Materials and Methods
Plant material
Z. bungeanum cultivar “Fengxiandahongpao” with excellent pericarp was used in this study. At the sprouting stage, we collected 40 stem tips (1 cm) from five 7-year-old individual plants of Z. bungeanum from the Research Centre for Engineering and Technology of Zanthoxylum, State Forestry Administration, Northwest A&F University (Fengxian, Shaanxi Province, China). All stem tips were immediately frozen in liquid nitrogen and stored at −80 °C to preserve RNA for transcriptome sequencing. Currently, approximately 30 cultivars of Z. bungeanum and nine cultivars of Z. armatum are planted in China. A total of 31 cultivars and landraces, including Z. bungeanum (20 cultivars and two landraces) and Z. armatum (five cultivers and four landraces, Supplement Table S1), were collected from the main cultivated regions and used for estimating the SSR markers and cluster analysis.
RNA isolation, integrity examination and RNA-Seq library preparation
Total RNA was extracted from each group using TRIzol Regent (Invitrogen, Life Technologies, Carlsbad, CA, USA) according to the manufacturer’s instructions. The extracted RNA was treated with a Total RNA Purification Kit (LC science, USA) for 30 min at 37 °C to remove any residual DNA. The quality and quantity of RNA were detected using a 2100 Bioanalyzer (Agilent Technologies, Santa Clara, CA, USA).
After total RNA extraction, the mRNA in each group was purified and enriched using oligo (dT) magnetic beads. Following purification, a fragmentation buffer and elevated temperatures were used to break the mRNA into fragments of 200–300 bp using divalent cations. Subsequently, the cleaved RNA fragments were employed for first-strand cDNA synthesis using reverse transcriptase and random primers, and then the second-strand cDNA was synthesized using DNA polymerase I and RNaseH (Invitrogen). Double stranded cDNAs then underwent an end repair process with T4 DNA polymerase and Klenow DNA polymerase. These repaired cDNA fragments were then subjected to 3ʹ- single adenylation and ligated with sequencing adapters. These products were purified and amplified via PCR to construct a cDNA library for transcriptome sequencing. Finally, the cDNA library products were sequenced on a paired-end flow cell using an Illumina HiSeq™ 2500 platform (Illumina Inc., San Diego, CA, USA).
Assembly and functional annotation
The raw reads of cDNA libraries were cleaned by a stringent filtering process. All adapter fragments as well as ambiguous reads containing >5% unknown nucleotides and low-quality reads with more than 20% Q < 10 bases were removed to ensure the accuracy of de novo assembly and subsequent analyses. The clean reads were assembled using Trinity software10 with the parameters set at K-mer length of 25 and a similarity of 80%, with the other parameters set to their default values. The clean reads with a certain length of overlap were first joined to form longer fragments without N, known as contigs. Subsequently, the clean reads were mapped back to the contigs, and paired-end reads were used to calculate the distance and relation among these contigs. Trinity connected these contigs to obtain consensus sequences that could not be extended on either end, called transcripts. Finally, the transcripts were clustered into unigenes using TGI Clustering Tools38. Assembled sequences of less than 200 bp were discarded due to a low annotation rate.
All unigenes were subjected to BLASTx searches against the NCBI Nr database, the Swiss-Prot database, KOG, and KEGG with an E-value threshold of 10−5. According to the Nr annotation, the functional annotations of unigenes were obtained based on GO terms (GO annotation: http://www.geneontology.org) by employing Blast2GO39.
Evolutionary analysis of CDS
Genescan40 was employed for gene prediction from unigenes. In total, 22,212 open reading frames were detected. The searching of single-copy homologous genes was performed by comparing the protein sequences of Zanthoxylum, sweet orange (Citrus sinensis), poplar (Populus euphratica), grape (Vitis aestivalis), strawberry (Fragaria × ananassa), soybean (Glycine max), Arabidopsis, cacao (Theobroma cacao), peach (Prunus persica), and castor bean (Ricinus communis), using all-to-all BLASTP analysis (E-value < 1E−5). Based on the species distribution of the top BLASTx hits against the Nr database, we selected sweet orange, poplar, grape, strawberry, soybean, Arabidopsis, cacao, peach, and castor bean for comparative phylogenetic analysis. Of these, Zanthoxylum, sweet orange, Arabidopsis, cacao, poplar, and castor bean belong to the Malvidae, a related phytogroup of Angiosperm Phylogeny Group classification. Rice (Oryza sativa) and sorghum (Sorghum bicolor) were used as out-groups. The OrthoMCL41 method was adopted to construct gene families for the 12 plant species. The translated putative amino acid sequences of the 34 shared single-copy genes were aligned in MAFFT42 and adjusted manually. Based on the concatenated sequences, phylogenetic analysis using the maximum-likelihood method was conducted in FastTree43 with the JTT model plus gamma-distributed rates and 1,000 bootstrap replications. For comparison, we also used peptide sequences of 68 genes from chloroplast genomes from the same plant species and performed maximum-likelihood analysis with the same parameters.
To detect the signature of genome duplications, an OrthoMCL clustering program was run on the protein CDS from the Zanthoxylum and Citrus genomes. One-to-one orthologous genes were then extracted from the clustering results. Subsequently, we set up a stringent criterion of at least 300 nucleotide alignments and 70% identity to reliably define two sequences as orthologous. Gene pairs with a BLAST identity of ≥99.0% were defined as identical sequences because near identical sequences occasionally are present in clustered transcript data due to sequencing errors and the fragmentary nature of transcripts. All identical gene pairs that contained the shorter sequence were discarded. The codon based alignment of the nucleotide sequence pairs was used to calculate the Ks by KaKs_Calculator44. The duplication time of the genome was estimated using the formula T = Ks/2r, where ‘r’ is the neutral substitution rate. A neutral substitution rate of 2.6 × 10−9 was used in this study27. The Ks of gene pairs of three-copy and four-copy genes families were also calculated.
Variant analysis
Only reliable clean reads were considered for SNP calling. Indels were not considered because alternative splicing impedes reliable indel discovery17. SNPs were called using the SAMtools software package45. The assembled transcript sequences were used as templates to BLAST the clean reads.
SSR locus detection and primer design
SSR locus screening was performed using the Perl script MIcroSAtellite (MISA, http://pgrc.ipk-gatersleben.de/misa/) based on the flanking sequences. SSR loci consist of repeated core sequences of two to six nucleotide motifs with minimum repeats of 6, 5, 4, 3 and 3, respectively. Mononucleotide repeats and complex SSRs were excluded from further analyses, because mononucleotide repeats may not be accurate due to sequencing errors and assembly mistakes, whereas complex SSRs show the least polymorphism. Primers could not be designed for those SSR loci whose flanking sequences were either too short or where the sequence did not meet appropriate criteria for primer design.
The SSR primer pairs were designed using the Oligo 7.0 program46 based on the following parameters: 1) primer length of 17–25 bp with 20 bp as the optimum; 2) PCR product size ranging from 100 to 500 bp; 3) GC content of 40–70% with an optimum of 50%; 4) melting temperature (Tm) between 52 °C and 60 °C with 55 °C as the optimum annealing temperature, with a difference of no greater than 4 °C between the Tm values of the forward and reverse primers; 5) primer pairs devoid of secondary structure or consecutive tracts of a single nucleotide; 6) the designed primer sequence was limited to the middle region, with 30 bp being removed from the ends of the contig sequence47. In total, 100 primers for SSR markers were synthesized.
DNA isolation, PCR amplification and SSR validation
The total genomic DNA was extracted from dried-leaf tissue of 31 cultivars and landraces of Z. bungeanum and Z. armatum (Supplement Table S1) using a modified CTAB protocol48. All SSR primer pairs were tested for amplification and polymorphism using the mixed DNA49 from five cultivars: ‘Fengxiandahongpao’, ‘Hanchengdahongpao’, ‘Huangjinjiao’, ‘Dingtanhuajiao’, and ‘Qinghuajiao’ (Supplement Table S1). In total, 100 pairs of primers were designed and validated by PCR and electrophoresis. Amplification by PCR was performed using an A200 Gradient Thermal cycler (LongGene, China), with reactions consisting of 10 µL of 2× Taq MasterMix (CWBIO Biotechnology, Beijing, China); 0.5 µL of each primer (100 µmol/L), 8 µL of double distilled water, and 1.0 µL of genomic DNA in a final reaction volume of 20 µL. The PCR conditions were: initial denaturation at 94 °C for 4 min, followed by 30 cycles of 1 min at 94 °C, 50 s at 52 to 60 °C (depending on the Tm of the primer pair used), and 1 min at 72 °C; then a final extension for 7 min at 72 °C. The resulting PCR products were subjected to electrophoresis on 12% non-denaturing polyacrylamide gels at 250 V for 2–2.5 h and stained using a silver-staining protocol50. Cluster analysis was conducted and displayed using the neighbor-joining algorithm and STRUCTURE analysis as implemented in MEGA ver. 6.051 and STRUCTURE ver. 2.3.452. The genetic diversity parameters (N a, N e, H o, H e, I, and F IS) of selected SSR primer pairs were calculated using the POPGENE software53.
Data Availability
Transcriptome datasets supporting the conclusions of this article are available in the NCBI SRA repository under the accession number SRR5114371. The full dataset is also available from Shijing Feng upon request (shijing0554010112@163.com).
Electronic supplementary material
Acknowledgements
This research was supported by grants from the Special Fund for Forestry Research in the Public Interest (No.: 201304706).
Author Contributions
Conceived and designed the experiments: A.Z.W., S.J.F. Aided in study design: T.X.Y., Y.L.L. Performed the experiments: S.J.F., L.L.Z. Analyzed the data: Z.S.L., S.J.F. Contributed materials: S.J.F. Wrote the paper: S.J.F., Z.S.L. All authors read and approved the final manuscript.
Competing Interests
The authors declare that they have no competing interests.
Footnotes
Shijing Feng and Lili Zhao contributed equally to this work.
Electronic supplementary material
Supplementary information accompanies this paper at 10.1038/s41598-017-15911-7.
Publisher's note: Springer Nature remains neutral with regard to jurisdictional claims in published maps and institutional affiliations.
References
- 1.Huang C. Reipublicae popularis sinicae. In: Delectis Florae Reipublicae Popularis Sinicae agendae academiae sinicae edita. Beijing: Science Press. 1997;43:8–10. [Google Scholar]
- 2.Kubitzki, K., Kallunki, J. A., Duretto, M. & Wilson, P. G. Rutaceae. In: Kubitzki, K. (Ed.), The Families and Genera of Vascular Plants, Flowering Plants Eudicots:Sapindales, Cucurbitales, Myrtaceae. Berlin: Springer Verlag, pp. 276–356 (2011).
- 3.Yang X. Aroma constituents and alkylamides of red and green huajiao (Zanthoxylum bungeanum and Zanthoxylum schinifolium) Journal of Agricultural & Food Chemistry. 2008;56:1689–1696. doi: 10.1021/jf0728101. [DOI] [PubMed] [Google Scholar]
- 4.Ahsan M, et al. Cytotoxic dimeric quinolone–terpene alkaloids from the root bark of Zanthoxylum rhetsa. Phytochemistry. 2014;103:8–12. doi: 10.1016/j.phytochem.2014.03.008. [DOI] [PubMed] [Google Scholar]
- 5.Zhang Y, Wang D, Yang L, Zhou D, Zhang J. Purification and characterization of flavonoids from the leaves of Zanthoxylum bungeanum and correlation between their structure and antioxidant activity. Plos One. 2014;9:e105725. doi: 10.1371/journal.pone.0105725. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Xia L, You J, Li G, Sun Z, Suo Y. Compositional and antioxidant activity analysis of Zanthoxylum bungeanum seed oil obtained by supercritical CO2 fluid extraction. Journal of the American Oil Chemists’ Society. 2011;88:23–32. doi: 10.1007/s11746-010-1644-4. [DOI] [Google Scholar]
- 7.Artaria C, Maramaldi G, Bonfigli A, Rigano L, Appendino G. Lifting properties of the alkamide fraction from the fruit husks of Zanthoxylum bungeanum. International Journal of Cosmetic Science. 2011;33:328–333. doi: 10.1111/j.1468-2494.2010.00629.x. [DOI] [PubMed] [Google Scholar]
- 8.Tezuka Y, et al. Screening of Chinese herbal drug extracts for inhibitory activity on nitric oxide production and identification of an active compound of Zanthoxylum bungeanum. Journal of Ethnopharmacology. 2001;77:209–217. doi: 10.1016/S0378-8741(01)00300-2. [DOI] [PubMed] [Google Scholar]
- 9.Liu Y, Wang F, Qin N. Apomixis in Zanthoxylum bungeanum and Z. simulans. Journal of Genetics & Genomics. 1987;14:107–113. [Google Scholar]
- 10.Grabherr MG, et al. Full-length transcriptome assembly from RNA-Seq data without a reference genome. Nature Biotechnology. 2011;29:644–652. doi: 10.1038/nbt.1883. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Wang M, Zhang X, Liu JH. Deep sequencing-based characterization of transcriptome of trifoliate orange (Poncirus trifoliata (L.) Raf.) in response to cold stress. BMC Genomics. 2015;16:555. doi: 10.1186/s12864-015-1629-7. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Ma Y, Li Q, Hu G, Qin Y. Comparative transcriptional survey between self-incompatibility and self-compatibility in Citrus reticulata Blanco. Gene. 2017;609:52–61. doi: 10.1016/j.gene.2017.01.033. [DOI] [PubMed] [Google Scholar]
- 13.Liu XY, Li J, Liu MM, Yao Q, Chen JZ. Transcriptome profiling to understand the effect of Citrus rootstocks on the growth of ‘Shatangju’ Mandarin. Plos One. 2017;12:e0169897. doi: 10.1371/journal.pone.0169897. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Lu X, et al. Comparative transcriptome analysis reveals a global insight into molecular processes regulating citrate accumulation in sweet orange (Citrus sinensis) Physiologia Plantarum. 2016;158:463–482. doi: 10.1111/ppl.12484. [DOI] [PubMed] [Google Scholar]
- 15.Franco MC, Schimpl FC, Mazzafera P. Reference genes for quantitative RT-PCR of the pilocarpine producer Pilocarpus microphyllus and two other Pilocarpus species. Theoretical & Experimental Plant Physiology. 2017;29:51–59. doi: 10.1007/s40626-016-0080-8. [DOI] [Google Scholar]
- 16.Cheng J, et al. Biomass accumulation and carbon sequestration in an age-sequence of Zanthoxylum bungeanum plantations under the Grain for Green Program in karst regions, Guizhou province. Agricultural & Forest Meteorology. 2015;203:88–95. doi: 10.1016/j.agrformet.2015.01.004. [DOI] [Google Scholar]
- 17.Xu Q, et al. The draft genome of sweet orange (Citrus sinensis) Nature Genetics. 2013;45:59–66. doi: 10.1038/ng.2472. [DOI] [PubMed] [Google Scholar]
- 18.PatiñO, L. O. J., Prieto, R. J. A. L. & Cuca, S. L. E. Bioactive compounds in phytomedicine: Zanthoxylum genus as potential source of bioactive compounds, pp. 185–218. In: Rasooli I. (ed.), InTech, Rijeka Croatia (2008).
- 19.Kumar S, Shah N, Garg V, Bhatia S. Large scale in-silico identification and characterization of simple sequence repeats (SSRs) from de novo assembled transcriptome of Catharanthus roseus (L.) G. Don. Plant Cell Reports. 2014;33:905–918. doi: 10.1007/s00299-014-1569-8. [DOI] [PubMed] [Google Scholar]
- 20.Zhou T, et al. Transcriptome sequencing and development of genic SSR markers of an endangered Chinese endemic genus Dipteronia Oliver (Aceraceae) Molecules. 2016;21:166. doi: 10.3390/molecules21030166. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Yang Y, et al. Phenotype and transcriptome analysis reveals chloroplast development and pigment biosynthesis together influenced the leaf color formation in mutants of Anthurium andraeanum ‘Sonate’. Frontiers in Plant Science. 2015;6:139. doi: 10.3389/fpls.2015.00139. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Jiao Y, et al. Ancestral polyploidy in seed plants and angiosperms. Nature. 2011;473:97–100. doi: 10.1038/nature09916. [DOI] [PubMed] [Google Scholar]
- 23.Shi T, Huang HW, Barker MS, Heslopharrison JSP. Ancient genome duplications during the evolution of kiwifruit (Actinidia) and related Ericales. Annals of Botany. 2010;106:497–504. doi: 10.1093/aob/mcq129. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Peer YVD, Fawcett JA, Proost S, Sterck L, Vandepoele K. The flowering world: a tale of duplications. Trends in Plant Science. 2009;14:680–688. doi: 10.1016/j.tplants.2009.09.001. [DOI] [PubMed] [Google Scholar]
- 25.Tang H, et al. Synteny and collinearity in plant genomes. Science. 2008;320:486–488. doi: 10.1126/science.1153917. [DOI] [PubMed] [Google Scholar]
- 26.Gaut, B. S. Molecular clocks and nucleotide substitution rates in higher plants. pp. 93–120 in Hecht, M. K. ed. Evolutionary biology. Plenum Press, New York (1998).
- 27.Senchina DS, et al. Rate variation among nuclear genes and the age of polyploidy in Gossypium. Molecular Biology & Evolution. 2003;20:633–643. doi: 10.1093/molbev/msg065. [DOI] [PubMed] [Google Scholar]
- 28.Mei L, et al. De Novo transcriptome assembly of pummelo and molecular marker development. Plos One. 2015;10:e0120615. doi: 10.1371/journal.pone.0120615. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Zhang X, et al. Characterization of the global transcriptome for cotton (Gossypium hirsutum L.) anther and development of SSR marker. Gene. 2014;551:206–213. doi: 10.1016/j.gene.2014.08.058. [DOI] [PubMed] [Google Scholar]
- 30.Yue H, et al. De novo assembly and characterization of the transcriptome of broomcorn millet (Panicum miliaceum L.) for gene discovery and marker development. Frontiers in Plant Science. 2016;7:1083. doi: 10.3389/fpls.2016.01083. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Gao Z, et al. Rapid microsatellite development for tree peony and its implications. BMC Genomics. 2013;14:886. doi: 10.1186/1471-2164-14-886. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Huang X, et al. De novo transcriptome analysis and molecular marker development of two Hemarthria species. Frontiers in Plant Science. 2016;7:496. doi: 10.3389/fpls.2016.00496. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Wang Z, et al. De novo assembly and characterization of root transcriptome using Illumina paired-end sequencing and development of cSSR markers in sweet potato (Ipomoea batatas) BMC Genomics. 2010;11:726. doi: 10.1186/1471-2164-11-726. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Asp T, Frei UK, Didion T, Nielsen KK, Lübberstedt T. Frequency, type, and distribution of EST-SSRs from three genotypes of Lolium perenne, and their conservation across orthologous sequences of Festuca arundinacea, Brachypodium distachyon, and Oryza sativa. BMC Plant Biology. 2007;127:36. doi: 10.1186/1471-2229-7-36. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Wei Z, et al. Transcriptome analysis of colored calla lily (Zantedeschia rehmannii Engl.) by Illumina sequencing: de novo assembly, annotation and EST-SSR marker development. Peer j. 2016;4:e2378. doi: 10.7717/peerj.2378. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Chen LY, et al. Characterization of transcriptome and development of novel EST-SSR makers based on next-generation sequencing technology in Neolitsea sericea (Lauraceae) endemic to East Asian land-bridge islands. Molecular Breeding. 2015;35:187. doi: 10.1007/s11032-015-0379-1. [DOI] [Google Scholar]
- 37.Jin YQ, et al. Genetic evaluation of the breeding population of a valuable reforestation conifer Platycladus orientalis (Cupressaceae) Scientific Reports. 2016;6:34821. doi: 10.1038/srep34821. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Pertea G, et al. TIGR Gene Indices clustering tools (TGICL): a software system for fast clustering of large EST datasets. Bioinformatics. 2003;19:651–652. doi: 10.1093/bioinformatics/btg034. [DOI] [PubMed] [Google Scholar]
- 39.Conesa A, et al. Blast2GO: a universal tool for annotation, visualization and analysis in functional genomics research. Bioinformatics. 2005;21:3674–3676. doi: 10.1093/bioinformatics/bti610. [DOI] [PubMed] [Google Scholar]
- 40.Tiwari S, Ramachandran S, Bhattacharya A, Ramaswamy R. Prediction of probable genes by Fourier analysis of genomic sequences. Bioinformatics. 1997;13:263–270. doi: 10.1093/bioinformatics/13.3.263. [DOI] [PubMed] [Google Scholar]
- 41.Li L, Stoeckert CJ, Roos DS. OrthoMCL: Identification of ortholog groups for eukaryotic genomes. Genome Research. 2003;13:2178–2189. doi: 10.1101/gr.1224503. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Katoh K, Standley DM. MAFFT multiple sequence alignment software version 7: improvements in performance and usability. Molecular Biology & Evolution. 2013;30:772–780. doi: 10.1093/molbev/mst010. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Price MN, Dehal PS, Arkin AP. FastTree 2-approximately maximum-likelihood trees for large alignments. Plos One. 2010;5:e9490. doi: 10.1371/journal.pone.0009490. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Wang D, Zhang Y, Zhang Z, Jiang Z, Yu J. KaKs_Calculator 2.0: A toolkit incorporating Gamma-Series methods and sliding window strategies. Genomics, Proteomics & Bioinformatics. 2010;8:77–80. doi: 10.1016/S1672-0229(10)60008-3. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Li H, et al. The Sequence alignment/map format and SAMtools. Bioinformatics. 2009;25:2078–2079. doi: 10.1093/bioinformatics/btp352. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Rychlik W. OLIGO 7 primer analysis software. Methods in Molecular Biology. 2007;402:35–60. doi: 10.1007/978-1-59745-528-2_2. [DOI] [PubMed] [Google Scholar]
- 47.Chen W, Liu YX, Jiang GF. De novo assembly and characterization of the testis transcriptome and development of EST-SSR markers in the cockroach Periplaneta americana. Scientific Reports. 2014;5:11144. doi: 10.1038/srep11144. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48.Porebski S, Bailey LG, Baum BR. Modification of a CTAB DNA extraction protocol for plants containing high polysaccharide and polyphenol components. Plant Molecular Biology Reporter. 1997;15:8–15. doi: 10.1007/BF02772108. [DOI] [Google Scholar]
- 49.Liu G. Studies on quick screening of EST-SSR primers based on genomic DNA mixing pool. Journal of Central South University of Forestry & Technology. 2012;32:124–129. [Google Scholar]
- 50.Li Y, Wang SH, Li ZQ, Jin CF, Liu MH. Genetic diversity and relationships among Chinese Eucommia ulmoides cultivars revealed by sequence-related amplified polymorphism, amplified fragment length polymorphism, and inter-simple sequence repeat markers. Genetics & Molecular Research. 2014;13:8704–8713. doi: 10.4238/2014.October.27.11. [DOI] [PubMed] [Google Scholar]
- 51.Tamura K, Stecher G, Peterson D, Filipski A, Kumar S. MEGA6: Molecular Evolutionary Genetics Analysis version 6.0. Systematic Biology. 2013;30:2725–2729. doi: 10.1093/molbev/mst197. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 52.Falush D, Stephens M, Pritchard JK. Inference of population structure using multilocus genotype data: dominant markers and null alleles. Molecular Ecology Notes. 2007;7:574–578. doi: 10.1111/j.1471-8286.2007.01758.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 53.Yeh, F. C., Yang, R. C. & Boyle, T. Population genetic analysis (POPGENE 1.31). A Joint Project of University of Alberta and Center for International Forestry Research, Alberta, Canada (1999).
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
Data Availability Statement
Transcriptome datasets supporting the conclusions of this article are available in the NCBI SRA repository under the accession number SRR5114371. The full dataset is also available from Shijing Feng upon request (shijing0554010112@163.com).