Abstract
BACKGROUND: Bone metastasis is common in renal cell carcinoma (RCC), and the lesions are mainly osteolytic. The mechanism of bone destruction in RCC bone metastasis is unknown. METHODS: We used a direct intrafemur injection of mice with bone-derived 786-O RCC cells (Bo-786) as an in vivo model to study if inhibition of osteoblast differentiation is involved in osteolytic bone lesions in RCC bone metastasis. RESULTS: We showed that bone-derived Bo-786 cells induced osteolytic bone lesions in the femur of mice. We examined the effect of conditioned medium of Bo-786 cells (Bo-786 CM) on both primary mouse osteoblasts and MC3T3-E1 preosteoblasts and found that Bo-786 CM inhibited osteoblast differentiation. Secretome analysis of Bo-786 CM revealed that BIGH3 (Beta ig h3 protein), also known as TGFBI (transforming growth factor beta-induced protein), is highly expressed. We generated recombinant BIGH3 and found that BIGH3 inhibited osteoblast differentiation in vitro. In addition, CM from Bo-786 BIGH3 knockdown cells (786-BIGH3 KD) reduced the inhibition of osteoblast differentiation compared to CM from vector control. Intrafemural injection of mice with 786-BIGH3 KD cells showed a reduction in osteolytic bone lesions compared to vector control. Immunohistochemical staining of 18 bone metastasis specimens from human RCC showed strong BIGH3 expression in 11/18 (61%) and moderate BIGH3 expression in 7/18 (39%) of the specimens. CONCLUSIONS: These results suggest that suppression of osteoblast differentiation by BIGH3 is one of the mechanisms that enhance osteolytic lesions in RCC bone metastasis, and raise the possibilty that treatments that increase bone formation may improve therapy outcomes.
Abbreviations: RCC, renal cell carcinoma; BIGH3, Beta ig h3; TGFBI, transforming growth factor beta-induced; PMO, primary mouse osteoblasts
Introduction
The development of bone metastasis is a common and devastating feature of renal cell cancer (RCC) progression [1]. Lesions from RCC bone metastases are characteristically osteolytic, causing pathologic fractures and consequent morbidity and mortality in patients [1]. Current pharmacologic treatments are focused on blocking osteolysis with RANKL inhibitors (denosumab) and/or bisphosphonates [1]. However, these treatments are of limited efficacy, suggesting that other mechanisms are operative for developing osteolysis.
The mechanism of osteolysis in RCC bone metastasis is not fully understood. In normal bone remodeling, bone formation from osteoblasts is balanced by bone resorption from osteoclasts. The osteolytic phenotype presumably results from dysregulation of normal bone remodeling that leads to a “vicious cycle” of bone destruction and tumor growth [2]. According to this theory, tumor cells stimulate osteoclastogenesis, and the increased bone destruction releases factors that promote metastatic cell growth and invasion, which in turn causes more tumor cell proliferation and osteolysis. Since osteoclasts are present at bone metastasis sites in RCC patient samples [3], it is generally considered that osteolytic bone metastasis of RCC is due to osteoclasts. RANKL is one of the major factors that lead to osteoclast activation [4]. However, RCC bone metastasis is resistant to treatments based solely on inhibiting RANKL-mediated osteolysis.
In normal bone biology, an osteolytic response is usually compensated by enhanced bone formation [5]. A pathological osteolytic phenotype can also be enhanced by the concomitant reduction in bone formation. In multiple myeloma-induced bone disease, an inhibition of bone formation by DKK1 was found to be one of the mechanisms that contribute to multiple myeloma-induced osteolytic bone lesions [6]. Whether inhibition of bone formation is involved in RCC bone metastasis has not been examined.
In this study, we used a bone-derived RCC cell line, Bo-786, which generates osteolytic bone lesions when injected into bone, to examine the mechanisms that are involved in osteolytic bone lesions from RCC. We found that a high level of BIGH3/TGFBI was secreted from Bo-786 cells. BIGH3 (Beta ig h3 protein), also known as TGFBI (transforming growth factor beta-induced protein, TGFBI for protein and TGFBI for gene), is a transforming growth factor beta (TGFβ)–induced secreted extracellular matrix protein. BIGH3/TGFBI was initially cloned from human lung adenocarcinoma A549 cells treated with TGF-beta [7]. BIGH3/TGFBI is comprised of 683 amino acids, and its secreted form has a predicted molecular mass of 68 kDa. BIGH3 contains an N-terminal secretory signal (1-24 amino acids), a cysteine-rich domain, four internal repetitive fasciclin-1 domains (FAS1 1-4), integrin binding motifs in the C-terminus known as Arg-Gly-Asp (RGD), YH18, EPDIM, and an internal NKDIL motif [8], [9]. A number of studies have demonstrated that BIGH3 is a versatile molecule and plays a role in a wide range of physiological and pathological conditions, including diabetes [10] and corneal dystrophy [11]. BIGH3 has been reported to have dual functions as a tumor suppressor or tumor promoter depending on the tumor microenvironment [12]. Additionally, several in vitro studies found a decrease in BIGH3 levels during the differentiation of human bone marrow stromal cells towards osteogenic lineage [13], [14] and during differentiation of osteoblast, suggesting that BIGH3 acts as a negative regulator of osteogenesis. In this study, we further showed that BIGH3 plays a role in RCC bone metastasis by enhancing RCC-induced osteolytic bone lesions.
Methods
Cell Lines, Antibodies, and Reagents
The human 786-O RCC cell line, derived from a primary clear cell renal adenocarcinoma, was purchased from the American Type Culture Collection (Manassas, CA). Luciferase- and green fluorescent protein–labeled 786-O and bone-derived 786-O (Bo-786) RCC cells were generated as described previously [15]. The murine preosteoblast MC3T3-E1(clone 4) cells, (MC3T3-E1 clone 4) Caki-1 and Caki-2 cell lines were purchased from American Type Culture Collection. SN12PM6, SLR23, and SLR25 RCC cell lines were purchased from Characterized Cell Line Core Facility in MD Anderson Cancer Center. GIPZ lentiviral human BIGH3 shRNA and GIPZ nonsilencing lentiviral shRNA control plasmids were purchased from MD Anderson core facility. Lentiviral particles containing shRNA for BIGH3 or nonsilencing control were generated in HEK293 cells 48 hours after transfection using Lipofectamine 2000 (Thermo Fisher Scientific) and were used for infecting Bo-786 RCC cells. The infected cells were selected by puromycin, and the efficiency of BIGH3 knockdown was determined by Western blot and real-time reverse-transcription polymerase chain reaction (RT-PCR) analysis. All the cell lines with/without BIGH3/TGFBI knockdown in Bo-786 RCC cells were authenticated by STR analysis at MD Anderson Cancer Center Characterized Cell Line Core Facility (SET354). All cells were maintained in a humidified atmosphere with 5% CO2 at 37°C with the passages between 6 and 20. Primary mouse osteoblasts (PMO) were prepared from newborn mouse calvaria as described previously [16] and cultured in α-MEM containing 10% FBS. Mouse anti-BIGH3 and rabbit anti-BIGH3 antibodies were purchased from Proteintech (Rosemont, IL, USA). Ascorbic acid and β-glycerophosphate were purchased from Sigma.
Animal Studies
All animal procedures were performed according to an approved protocol from MD Anderson's Animal Care and Use Committee, in strict accordance with the recommendations in the Guide for the Care and Use of Laboratory Animal of the National Institutes of Health. Bo-786 cells grown to subconfluence were harvested and resuspended in PBS to a final concentration of 1 × 106 cells/5 μl. Cells were injected directly into the distal end of the right femur of male, 5-week-old SCID mice (Jackson Laboratory) using a 26-gauge needle. Tumor growth was monitored biweekly by bioluminescent imaging (BLI) using an IVIS 200 Imaging System (Xenogen). After specific time periods, mice were euthanized, and both the injected and contralateral control femurs were collected and fixed in 10% paraformaldehyde for 48 hours, followed by washing with PBS and soaking in 70% ethanol. The femurs were then subjected to micro-CT analysis.
X-Ray and Micro-CT Analysis
X-ray analysis of tumor-bearing bones was performed using MX-20 cabinet X-ray system. Micro-CT analysis was performed with an Enhanced Vision Systems hybrid specimen scanner (GE Medical Systems, London, ON, Canada) at a resolution of 20 μm. The images were reconstructed, and bone mineral density (BMD) was analysized using Microview (2.1.2) software provided by GE Healthcare.
Human Specimens
Eighteen formalin-fixed, paraffin-embedded tissues from patients with RCC bone metastases were used to determine the protein expression of BIGH3/TGFBI by immunohistochemistry (IHC). Usage of clinical specimens was approved by the Institutional Research Board (IRB protocol PA15-0225).
Mass Spectrometry Analysis of Bo-786 Cells
Bo-786 cells grown to 80% confluence in RPMI/10% FBS were washed with PBS and were further cultured in serum-free RPMI medium for 48 hours. The conditioned medium (CM) was collected, centrifuged at 8000 g for 20 minutes to remove dead cells and debris, and concentrated 10-fold by centrifugation at 4000 g for 30 minutes using Amicon Ultra Centrifuge Filter device (Ultracel 5K, REF: UFC900524). The proteins in the CM were analyzed by tandem mass spectrometry as described previously [17].
Expression and Purification of Recombinant BIGH3 Proteins
Recombinant BIGH3 protein was expressed in HEK293 cells and purified from the CM of HEK293 cells. HEK293 cells were transfected with pcDNA3.1 vector containing complementary DNA (cDNA) encoding BIGH3 with a 7-histidine tag using polyethylenimine (Sigma Aldrich). The cell culture medium was collected 48 hours after transfection and concentrated using Amicon Ultra Centrifuge Filter device, and the BIGH3 protein was purified using Ni-NTA agarose (Qiagen) as described previously [18]. Protein concentrations were determined by Coomassie Plus Protein Assay (Thermo Scientific, Waltham, MA) using BSA as the protein standard.
Cell Proliferation Assay
Cells were seeded in 6-well or 96-well plates. Treatments were started after overnight incubation. Cells in 6-well plates were trypsinized and cell numbers were counted using a hemocytometer. The viability of cells in 96-well plate was detected using Presto Blue assay kit (Thermo Fisher Scientific).
Osteoblast Differentiation
PMO or MC3T3-E1 preosteoblast cells were seeded in 12-well plates. After the cells reached confluence, the cells were cultured in differentiation medium containing ascorbic acid (100 μg/ml) and β-glycerophosphate (10 mmol/L) and continued culturing for 24 days with medium changes every 3 days.
Alkaline Phosphatase (ALP) Activity
Assays for the osteoblast differentiation marker ALP were carried out after osteoblasts were cultured in differentiation medium for 12 days. Cells were rinsed with calcium-free PBS, and the enzymatic activity was measured using substrate p-nitrophenyl phosphate (1 mg/ml in diethanolamine substrate buffer, Sigma, St Louis, MO). The absorbance at 405 nm was measured using a multiwell plate reader.
Alizarin Red S Staining
The mineralization of osteoblasts was determined using Alizarin Red S (sodium alizarin sulfonate, Sigma) staining. Differentiated osteoblasts were fixed with 10% formalin for 30 minutes followed with Alizarin Red S (2% in water, pH ranged from 4.1 to 4.3) for 30 to 45 minutes. The mineralized nodules were quantified using Image J software.
Enzyme-Linked Immunosorbent Assay (ELISA)
Osteocalcin measurement was performed using the Mouse Osteocalcin ELISA kit (BT-470; Biomedical Technologies). BIGH3 measurement was performed using human BIGH3 ELISA Kit (ab155426, Abcam, Cambridge, MA, USA).
Quantitative Real-Time PCR Analysis
Total RNA was extracted using RNeasy mini purification kit (Qiagen, Valencia, CA), and cDNA was synthesized using TaqMan Reverse Transcription Reagents (Life Technologies). Quantitative real-time PCR was conducted using 20 ng cDNA, and the relative mRNA levels for each gene were determined using glyceraldehyde-3-phosphate dehydrogenase as a control. The nucleotide sequences of primers for various genes are listed in Supplemental Table S1.
Western Blot
Total protein was extracted from cells using mammalian tissue lysis/extraction reagent (Sigma, St. Louis, MO) supplemented with protease inhibitor cocktails according to the manufacturer's protocol. Equal amounts of protein lysates were subjected to 4% to 12% SDS-PAGE, transferred to nitrocellulose membrane, and incubated with the indicated antibody. Bands were visualized with an enhanced chemiluminescence detection kit (Pierce Biotechnology).
Immunohistochemistry
Immunohistochemical analysis of formalin-fixed, paraffin-embedded tissues was performed as previously described [19], [20] using primary antibody against BIGH3 (Proteintech). Images were analyzed with a Leica confocal microscope at magnification 200×.
Tartrate-Resistant Acid Phosphatase (TRAP) Staining
TRAP staining was performed using a TRAP staining kit (Kamiya Biomedical Company, Seattle, WA).
Statistical Analysis
All data were collected from three or more independent experiments, and values were expressed as mean ± S.E. unless otherwise stated. Statistical significance was carried out using Student's t test (two-tailed, paired). The level of significance was set at P < .05.
Results
Induction of Osteolytic Bone Lesions by Bo-786 Cells In Vivo
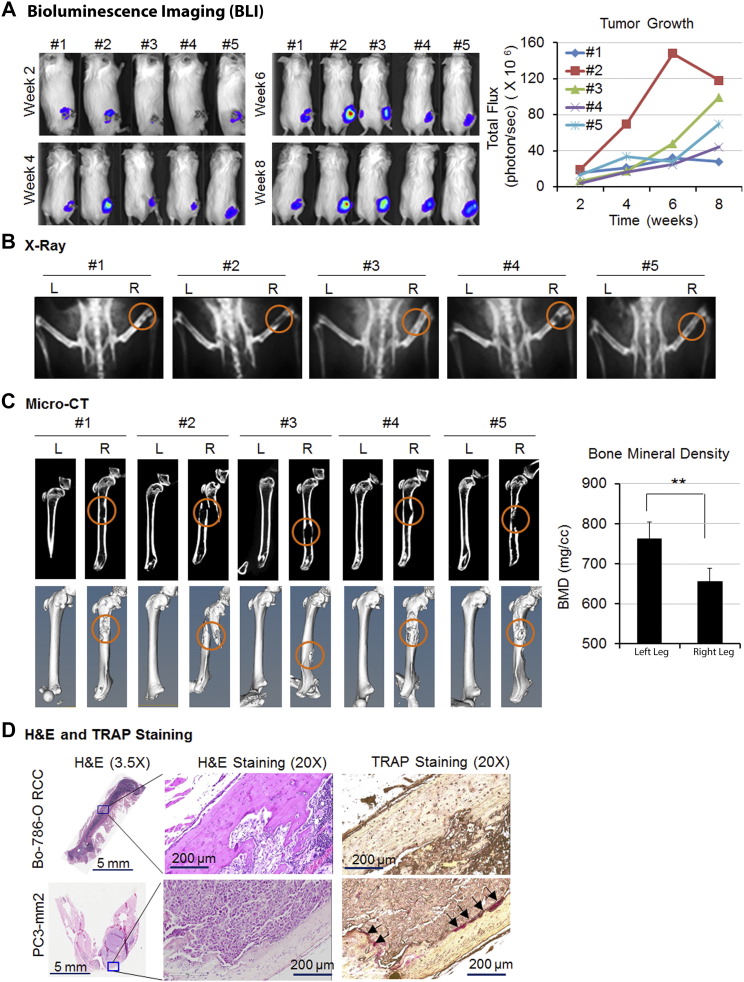
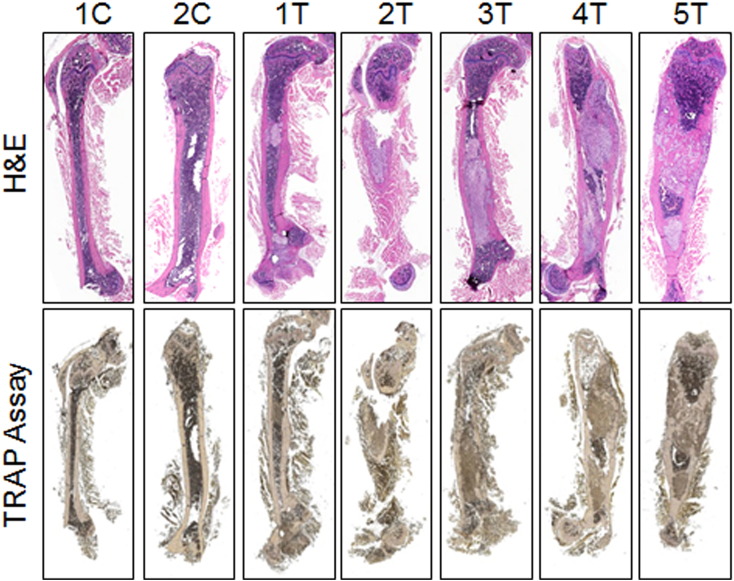
RCC bone metastases are characteristically osteolytic. In order to study this process, a mouse model of osteolytic bone lesions was created by direct intrafemur injection using Bo-786 RCC cells. Bo-786 cells were derived from 786-O, an RCC cell line from clear cell carcinoma [21], via in vivo selection for their ability to grow in bone [22]. After direct injection of Bo-786 cells into mouse femurs, we monitored tumor growth over time using BLI (Figure 1A). All five animals developed osteolytic lesions in the injected femurs as detected by both X-ray (Figure 1B) and micro-CT (Figure 1C). Quantification of the BMD from micro-CT of the entire femur showed a significant decrease of bone density in the Bo-786 cell-injected femurs compared to contralateral femur controls (Figure 1C). Histological analysis showed tumor cells invading the cortical and trabecular bone (Figure 1D). These results suggest that Bo-786 cells can grow in femurs and induce osteolytic bone lesions. To detect the presence of osteoclasts, we performed TRAP staining. Although osteolytic lesions were obvious, we did not find much TRAP positivity in the Bo-786 injected bone (Figure 1D, Supplemental Figure S1). In contrast, numerous TRAP-positive cells were detected at lytic sites from injecting PC3 cells, a prostate cancer cell line (Figure 1D). Together, these results demonstrate that Bo-786 cells induce osteolytic bone lesions without a marked increase in the TRAP positivity, raising the possibility that inhibition of osteoblast function may contribute to the osteolytic phenotype.
Figure 1.
Bo-786 cells generated osteolytic lesions in mouse femur. Bo-786 cells were injected into the femur of mouse right leg. (A) The growth of cancer cells in the right legs was monitored by BLI over 8 weeks. (B) Osteolytic bone lesions were detected by X-ray. (C) Micro-CT of the femurs with or without tumors. Quantification of BMD was done on whole femurs using Microview software. (D) A representative image of H&E-stained bone section of the leg shows the presence of tumor cells. A representative image of TRAP staining shows strong staining in PC3-mm2–injected femur but only weakly in Bo-786–injected femur. “L”: normal left femur; “R”: tumor cell injected right femur. **P < .01.
Figure S1.
H&E and TRAP staining on femurs injected with Bo-786-O cells. “C”: control non–tumor-bearing femur; “T”: tumor-bearing femur.
Inhibition of Osteoblast Differentiation and Mineralization by Bo-786 CM
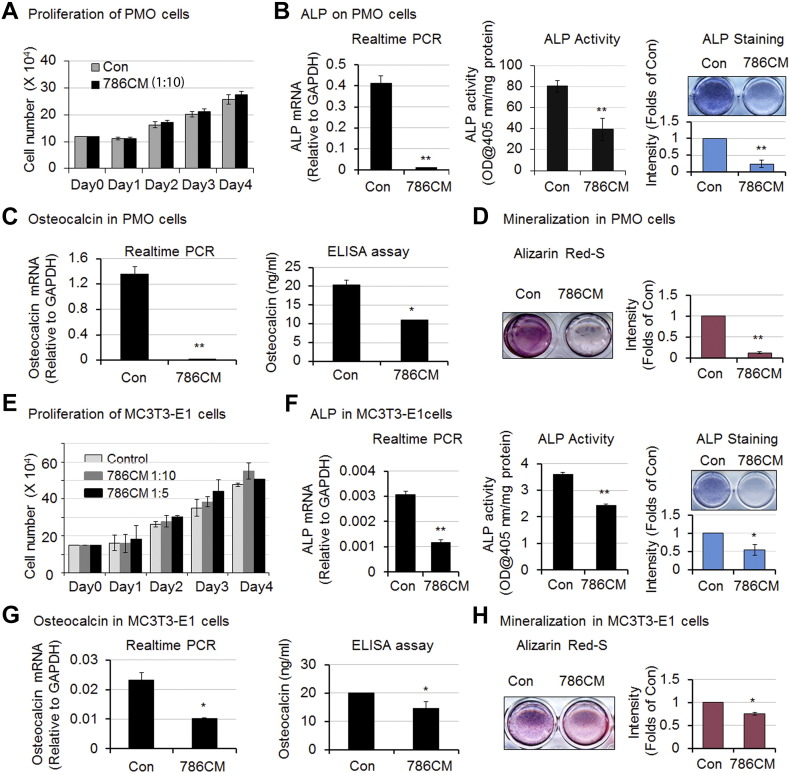
To determine whether Bo-786 cells inhibit osteoblast function, we examined the effects of Bo-786 cells on osteoblast proliferation, differentiation, and mineralization. CM from Bo-786 cells (Bo-786 CM) was concentrated 10-fold and used to treat PMO at 1:10 dilution. Treatment with Bo-786 CM did not affect the proliferation of PMO (Figure 2A). In contrast, when PMO were cultured in differentiation medium, Bo-786 CM treatment lead to a decrease in ALP mRNA, ALP activity, and ALP staining (Figure 2B). ALP is an early osteoblast differentiation marker. Treatment of PMO with Bo-786 CM also led to a decrease in both mRNA and protein levels of osteocalcin, a late osteoblast differentiation marker (Figure 2C). In addition, Bo-786 CM treatment decreased mineralization/bone nodule formation as determined by Alizarin Red S staining (Figure 2D). Similarly, treatment with 10-fold concentrated Bo-786 CM at 1:10 and 1:5 dilution did not affect the proliferation of MC3T3-E1 cells, an SV40-immortalized mouse osteoblast cell line [23] (Figure 2E). Using 10-fold concentrated Bo-786 CM at 1:10 dilution to treat MC3T3-E1 cells, we also found inhibitory effects on osteoblast differentiation (Figure 2, F and G) and mineralization (Figure 2H). However, the inhibitory effect of Bo-786 CM on osteoblast differentiation and mineralization was greater in PMO than that in MC3T3-E1 cells. This was most likely due to the fact that MC3T3-E1 is a T-antigen immortalized cell line. Taken together, these results suggest that Bo-786 CM contains factors that inhibit osteoblast differentiation and mineralization.
Figure 2.
Bo-786 culture medium inhibited osteoblast differentiation. (A) PMO cell proliferation with or without treatment with Bo-786 CM (786CM). (B) ALP gene expression (left), ALP activity (middle), and ALP staining (right) in Bo-786 CM-treated PMO cells. (C) Osteoclacin mRNA (left) and osteocalcin protein in 786-O CM-treated PMO cells (right). (D) Effect of Bo-786 CM on mineralization of PMO cells as determined by Alizarin Red S staining. (E) Proliferation of MC3T3-E1 cells with or without Bo-786 CM treatment. (F) ALP gene expression (left), ALP activity (middle), and ALP staining (right) in Bo-786 CM-treated MC3T3-E1cells. (G) Osteoclacin mRNA (left) and osteocalcin protein in 786-O CM-treated MC3T3-E1 cells (right). (H) MC3T3-E1 mineralization was determined by Alzarin Red S staining. “Con”: Control. *P < .05; **P < .01.
Identification of BIGH3/TGFBI in Bo-786 CM by Mass Spectrometry Analysis
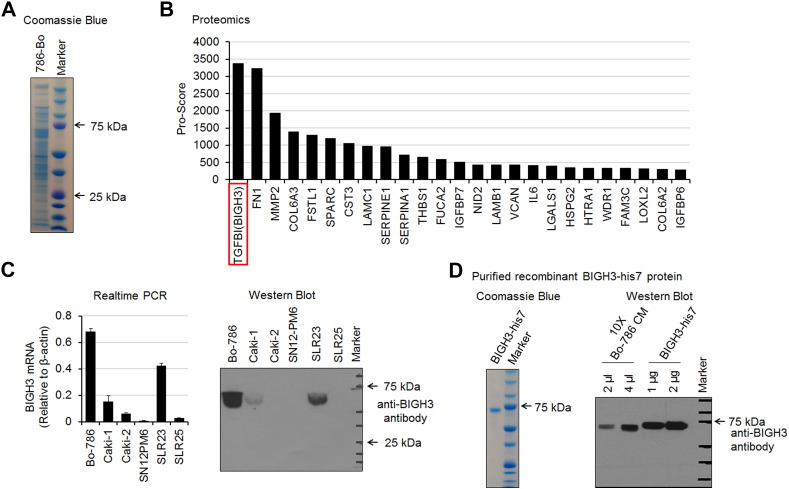
To identify the proteins in the Bo-786 CM that may affect osteoblast differentiation, Bo-786 CM was concentrated 10-fold and subjected to SDS-PAGE. The protein profile of Bo-786 CM was shown by Coomassie blue staining (Figure 3A). Mass spectrometry analysis of Bo-786 CM identified a total of 634 proteins, including 95 (15%) that were secreted (Supplemental Table S2). Among the secreted proteins, BIGH3/TGFBI [Beta Ig H3 protein, also known as transforming growth factor beta-induced protein (TGFBI)] protein had the highest protein score (>3000) (Figure 3B). BIGH3/TGFBI was previously shown to act as a negative regulator of osteogenesis [13], [14], suggesting that BIGH3/TGFBI may be one of the factors that may affect osteoblast differentiation in Bo-786 CM. We thus further determined BIGH3 mRNA and protein levels in several kidney cell lines. We found that the bone-derived Bo-786 RCC cells expressed the highest levels of BIGH3/TGFBI mRNA, as determined by real-time RT-PCR (Figure 3C, left), and highest levels of BIGH3/TGFBI protein, as determined by Western blot analysis (Figure 3C, right). Low levels of BIGH3/TGFBI expression were also detected in Caki-1 and SLR23 cell lines. To examine the effect of BIGH3/TGFBI on osteoblasts, BIGH3 with a 7-histidine tag was expressed in HEK293 cells and purified from the CM through metal affinity chromatography. Purified BIGH3-his7 has an apparent molecular mass of 68 kDa (Figure 3D, left panel). Western blot using anti-BIGH3 antibody showed that the purified BIGH3-his7 has a slightly higher molecular mass compared to that in Bo-786 CM (Figure 3D, right panel), likely due to the presence of the 7-histidine tag.
Figure 3.
Mass spectrometry analysis of Bo-786 CM. (A) SDS-PAGE analysis of Bo-786 CM. The proteins were stained with Coomassie blue. (B) Lists of 25 secretory proteins with highest protein score from tandem mass spectrometry analysis. (C) Expression of BIGH3 in kidney cell lines as determined by real-time PCR and Western blot. (D) SDS-PAGE analysis of BIGH3-his7 protein expressed and purified from HEK293 cells (left). Western blot of purified BIGH3-his7 protein and Bo-786 CM with anti-BIGH3 antibody (right).
Inhibition of Osteoblast Differentiation and Mineralization by Recombinant BIGH3/TGFBI
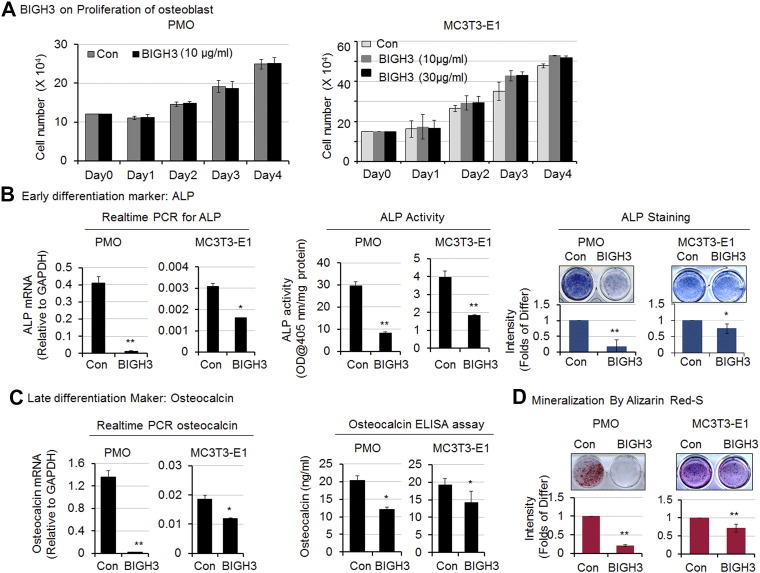
Next, we examined whether BIGH3/TGFBI can affect osteoblast differentiation. We found that treatment of PMO with 10 μg/ml recombinant BIGH3-his7 or MC3T3-E1 cells with recombinant BIGH3-his7 at either 10 μg/ml or 30 μg/ml did not lead to a significant change in cell proliferation compared to untreated controls (Figure 4A), consistent with the findings in PMO or MC3T3-E1 cells treated with Bo-786 CM (Figure 2). Moreover, treatment of PMO and MC3T3-E1 cells with recombinant BIGH3-his7 led to a decrease in ALP mRNA, ALP activity, and ALP staining compared to untreated control cells (Figure 4B). Recombinant BIGH3-his7 treatment also resulted in lower osteocalcin expression in PMO and MC3T3-E1 cells at both mRNA and protein levels (Figure 4C) and reduced mineralization (Figure 4D) compared to controls. The inhibitory effects of BIGH3/TGFBI were greater on PMO than those on MC3T3-E1 cells, similar to those observed with Bo-786 CM. Taken together, these results show that BIGH3/TGFBI alone is sufficient to inhibit differentiation and mineralization of osteoblasts.
Figure 4.
Recombinant BIGH3-his7 protein inhibited osteoblast differentiation. (A) Cell proliferation was determined on PMO (left) and MC3T3-E1 cells (right) after cells were treated with recombinant BIGH3-his7 protein. (B) Effects of BIGH3 on ALP gene expression (left), ALP activity (middle), and ALP staining (right) in PMO and MC3T3-E1 cells. (C) Effects of BIGH3 on osteoclacin mRNA (left) and osteocalcin protein (right) in PMO and MC3T3-E1 cells. (D) Effects of BIGH3 on PMO and MC3T3-E1 cell mineralization as determined by Alizarin Red S staining. “Con”: [200 mM] imidazole vehicle control; “BIGH3”: recombinant purified BIGH3-his7 protein. *P < .05; **P < .01.
Reduction of Osteoblast Inhibition by Knockdown of BIGH3/TGFBI in Bo-786 Cells
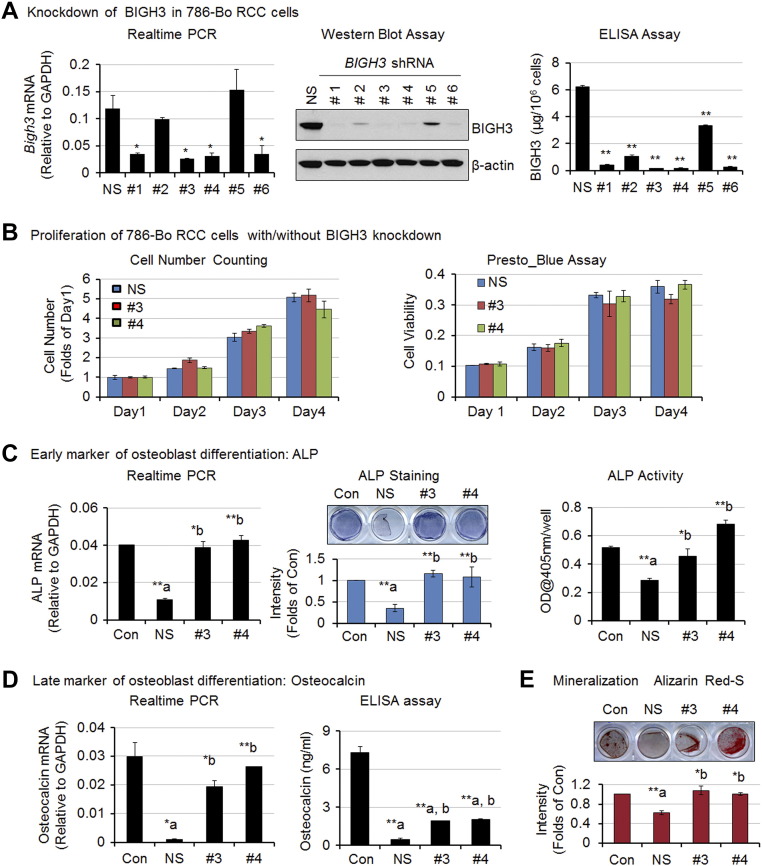
To examine whether BIGH3 plays a role in Bo-786–mediated osteoblast inhibition, we knocked down BIGH3 message in Bo-786 cells using shRNA in a lentiviral vector. Among six different shRNAs, clones #1, #3, #4, and #6 showed more reduction in both message and protein levels of BIGH3 as compared to control cells transfected with nonsilencing shRNA (786-shNS) (Figure 5A). The 786-shBG clones #3 (786-shBG #3) and #4 (786-shBG #4) were selected for further analyses.
Figure 5.
Inhibition of osteoblast differentiation by Bo-786 CM was partially mediated through BIGH3. (A) Generation of Bo-786 RCC cells with knockdown of BIGH3. Knockdown of BIGH3 was confirmed by real-time PCR, Western blot assay, and ELISA. (B) Cell proliferation and cell viability of BIGH3 knockdown cell lines. (C) Effects of CM from BIGH3 knockdown cell lines on PMO differentiation. ALP mRNA (left), ALP staining (middle), and ALP activity (right). (D) Osteocalcin mRNA and protein levels. (E) Mineralization. “NS”: 786-shNS nonsilencing control cells; “#3”: 786-shBG#3 cells; “#4”: 786-shBG#4 cells. *P < .05; **P < .01; “a”: compared to “Con”; “b”: compared to “NS”.
There was no significant difference in cell proliferation between 786-shNS and 786-shBG #3 or 786-shBG #4 cells as determined by cell numbers and Presto Blue assay when cells were grown in RPMI medium with 10% FBS (Figure 5B), suggesting that BIGH3 does not play a role in the growth of 786-O cells in vitro. Next, we determined the effects of CM from BIGH3 knockdown cell lines on osteoblast differentiation. Treatment of PMO with CM from 786-shNS cells reduced ALP mRNA and inhibited ALP activity when compared to control untreated cells (Figure 5C). In contrast, CM from 786-shBG #3 and 786-shBG #4 cells was unable to reduce ALP expression and activity as observed in 786-shNS cells (Figure 5C). CM from 786-shBG #3 and 786-shBG #4 showed only a partial effect in the inhibition of osteocalcin mRNA and osteocalcin relative to 786-shNS cells (Figure 5D) and a lack of ability to prevent mineralization compared to 786-shNS cells (Figure 5E). These results suggest that BIGH3 is one of the factors that inhibit osteoblast differentiation and mineralization in Bo-786 CM.
Decrease of Bo-786 Cell-Mediated Osteolytic Bone Lesions by Knockdown of BIGH3/TGFBI In Vivo
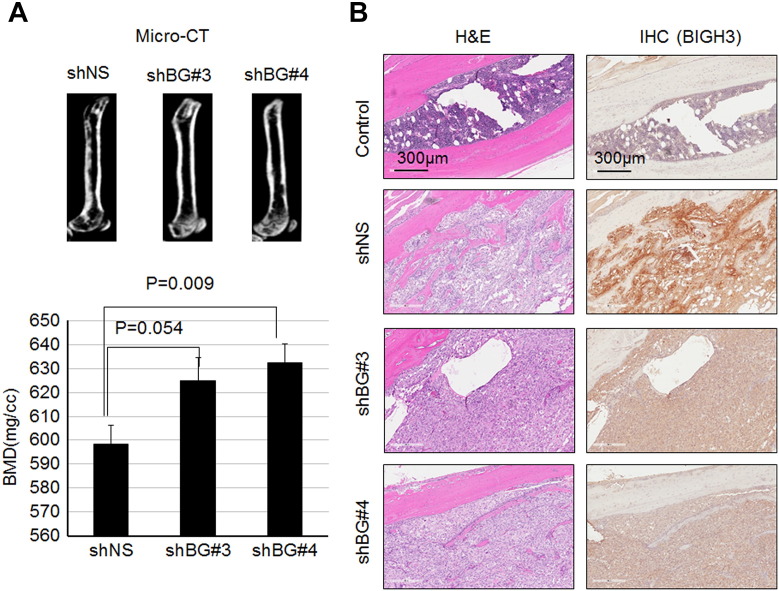
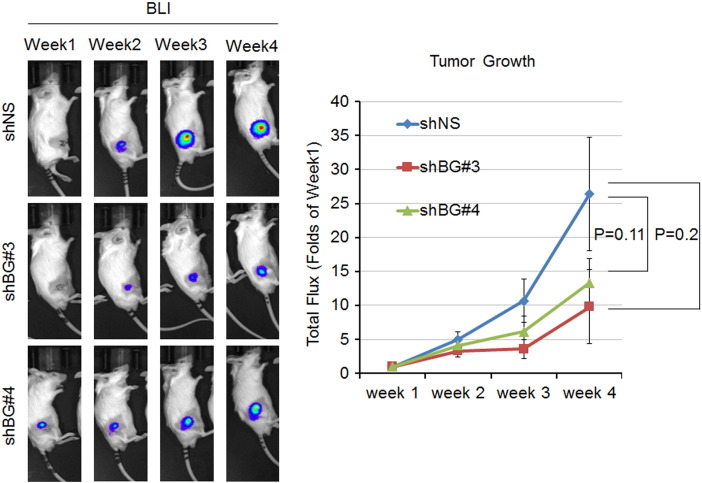
We next examined the effects of BIGH3 knockdown on Bo-786-mediated osteolysis in vivo. 786-shBG #3, 786-shBG #4, or 786-shNS cells were injected into mouse femurs. After 4 weeks, the BMDs of the tumor-bearing femurs were analyzed by micro-CT imaging. Micro-CT images showed less osteolysis from knockdown cell lines (786-shBG#3 and 786-shBG#4) than that from the control cell line (786-shNS) (Figure 6A, upper). Consistently, BMDs in femurs injected with the knockdown cell lines were higher compared to control 786-shNS cell-injected femurs (Figure 6A, lower), indicating less osteolysis. BLI of the tumor volume showed that there is a trend, although it did not reach statistical significance, towards slower growth in the knockdown cell lines than the control cell line (Supplemental Figure S2). This is likely due to a decrease in osteolysis that leads to a reduction in the “vicious cycle” of bone destruction and tumor growth [2]. IHC for BIGH3 in injected femurs showed less staining in knockdown cells compared with control 786-shNS cells (Figure 6B), suggesting that the knockdown of BIGH3 was maintained in 786-shBG #3 and 786-shBG #4 in vivo. Taken together, these data suggest that BIGH3 knockdown reduces Bo-786–induced osteolysis.
Figure 6.
Knockdown of BIGH3 gene in Bo-786 cells reduced Bo-786-induced osteolytic lesions. (A) Representative micro-CT images of mouse femur 4 weeks after being injected with 786-shNS, 786-shBG#3, or 786-shBG#4 cells (upper). BMD of whole femur was quantified using Microview software (lower). (B) H&E-stained bone section of an injected femur showed the presence of tumor cells and IHC of BIGH3 on the adjacent sections of H&E. “shNS”: 786-shNS cells (nonsilencing control cells); “shBG#3”: 786-shBG#3 cells; “shBG#4”: 786-shBG#4 cells.
Figure S2.
Growth of 786-shNS and BIGH3 knockdown cells in mouse femurs as monitored by BLI. Mice were injected with 786-shNS or BIGH3 knockdown 786-shBG#3 and 786-shBG#4 cells at right femurs. 786-shNS cells were used as control. The tumor growth was monitored by BLI over a 4-week time period, and the total flux was expressed as folds of week 1.
Expression of BIGH3/TGFBI in Human RCC Bone Metastasis Specimens
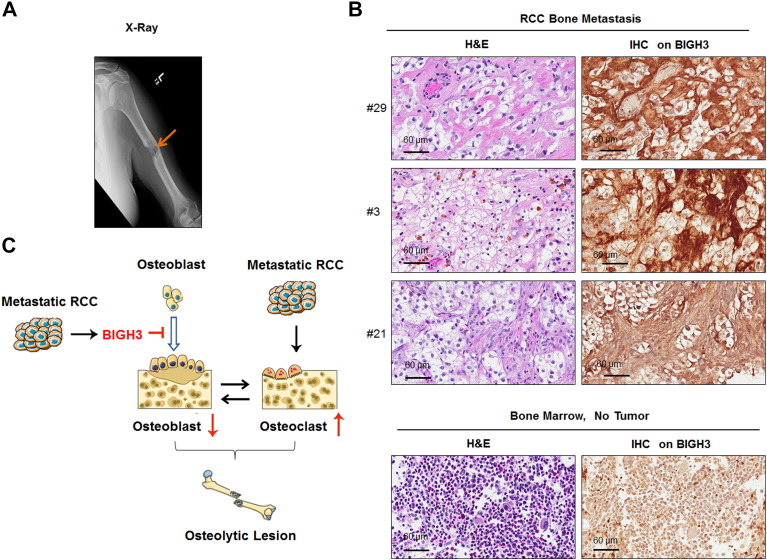
We further examined the expression of BIGH3/TGFBI in 18 specimens collected from patients undergoing surgery for bone metastasis from RCC (Figure 7). These patients required surgery due to an impending pathologic fracture or subsequent to a pathologic fracture caused by RCC bone metastasis. Figure 7A shows an x-ray from a patient with osteolytic appearance of RCC bone metastasis. Bone metastasis samples were also obtained from several other patients for analysis. We detected BIGH3/TGFBI expression (strong to moderately positive) in all 18 bone metastasis samples (Figure 7B and Supplemental Figure S3). BIGH3/TGFBI was found to be secreted into the extracellular space, resulting in BIGH3 staining in the matrix adjacent to tumor cells (Figure 7B, upper). In contrast, BIGH3/TGFBI expression was low/absent in the bone marrow in adjacent areas where there was no tumor present (Figure 7B, lower). These results indicate that high levels of BIGH3/TGFBI expression were present in bone metastatic RCC. Together, we propose that BIGH3/TGFBI may enhance osteolytic bone lesions from RCC bone metastasis through its inhibition of osteoblast differentiation (Figure 7C).
Figure 7.
BIGH3 expression in RCC bone metastasis. (A) A representative X-ray from a patient showing a pathologic fracture due to bone metastasis from RCC. (B) BIGH3 expression in three specimens of human RCC bone metastasis samples (upper) and normal bone marrow (lower) was determined by IHC staining. Both H&E staining and IHC staining for BIGH3 are shown at the magnification of 40×. (C) The proposed role of BIGH3 in enhancing osteolytic bone lesions from RCC bone metastasis. RCC cells in bone secrete BIGH3 that inhibits osteoblast differentiation that would normally counterbalance osteoclast activity, thus enhancing RCC-mediated osteolytic bone destruction.
Figure S3.
H&E and IHC of BIGH3 in human RCC bone metastasis tissue samples.
Discussion
We have identified a novel mechanism that contributes to the osteolytic bone metastasis in RCC. While RCC is known to activate osteoclasts to induce osteolytic bone lesions, other mechanisms are likely involved as treatments that focused on blocking osteolysis with RANKL inhibitors are of limited efficacy. We showed that BIGH3/TGFBI secreted by an RCC cell line, Bo-786, inhibits osteoblast differentiation, further tipping bone homeostasis towards osteolysis. Our in vivo data linked BIGH3/TGFBI expression to tumor associated bone lysis, showing that its knockdown reduced the osteolytic bone lesions induced from Bo-786 cells. We also showed that human RCC bone metastasis specimens expressed high levels of BIGH3/TGFBI. Our study suggests that suppression of osteoblast differentiation is one of the mechanisms that contribute to the osteolytic progression of renal cell carcinoma in bone. Furthermore, our studies imply that therapies that enhance osteoblast differentiation might be a promising strategy for improving treatments for osteolytic bone metastasis.
BIGH3/TGFBI is a protein induced by TGF-β1 in human lung adenocarcinoma cells but is not structurally related to the TGF-β family [7], [24]. BIGH3/TGFBI is known to bind to integrins and other extracellular matrix proteins and is thought to mediate cell adhesion and migration [24]. The clinical significance of BIGH3/TGFBI has been noted in other studies. BIGH3/TGFBI was reported to be upregulated in 40% of kidney, 90% of colorectal, and 60% of intestinal cancers [25]. BIGH3/TGFBI is also highly expressed in brain tumors but not in normal brain tissue [25]. Strong BIGH3/TGFBI expression was shown to be associated with a poor prognosis in clear cell RCC [26], [27].
In addition to playing a role in tumor progression, BIGH3/TGFBI has been shown to be involved in bone homeostasis. BIGH3/TGFBI was previously shown to inhibit differentiation of mature osteoblasts in vitro [28]. Thapa et al. [28] showed that BIGH3/TGFBI binding to osteoblasts is mediated via αvβ3 and αvβ5 integrins. Disruption of interactions by function-blocking antibody specific for these integrins abolished the inhibitory effect of BIGH3/TGFBI on osteoblast differentiation. Similarly, BIGH3/TGFBI is downregulated in melorheostosis, a disease characterized by hyperostosis, implying that loss of BIGH3/TGFBI promotes sclerosis [29]. Thus, BIGH3/TGFBI appears to be a negative regulator of bone formation. However, in the study by Lee et al. [30], the skeletal size in Bigh3/Tgfbi knockout mice was smaller than in wild-type and heterozygous mice. Because BIGH3/TGFBI is expressed in a wide variety of tissues and plays important roles in cell adhesion and migration, the changes in skeletal size may be the outcomes of the multiple effects of BIGH3/TGFBI on various developmental processes. Our data agree with previous studies demonstrating an inhibitory effect of BIGH3/TGFBI on the differentiation of mature osteoblasts. Our study is one of the first to specifically demonstrate an effect of BIGH3 on the tumor bone microenvironment.
Our studies raised interesting issues on the molecular mechanisms that upregulate BIGH3/TGFBI during RCC. Using Northern blot analysis, Ivanov et al. [25] showed that BIGH3/TGFBI messages are upregulated in paired normal versus tumor samples from many cancer types, especially renal, colon, lung, rectal, small intestine, and pancreatic cancers. They also showed that BIGH3/TGFBI messages are induced by stress-related stimuli, including chemotherapeutic agents, UV irradiation, heat shock, hydrogen peroxide, and gamma irradiation, in 26 cancer cell lines. Analysis of the expression database cBioportal (http://www.cbioportal.org/) showed that BIGH3/TGFBI mRNA is amplified in 13.4% of 448 clear RCC cells examined. In RCC, the von Hippel–Lindau tumor suppressor (VHL) gene is frequently mutated in clear cell RCC [31], [32]. Ivanov et al. [25] also showed that BIGH3/TGFBI expression was downregulated by wild-type but not by mutant VHL in 786-O cells, suggesting that upregulation of BIGH3/TGFBI in 786-O cells may be due to a VHL mutation. In our studies, we found that BIGH/TGFBI is expressed in all of the tissue samples collected from patients undergoing surgery for bone metastasis from RCC (Supplemental Figure S3, Figure 7). Whether the increase in BIGH3/TGFBI expression in RCC bone metastasis is due to VHL mutation or due to tumor response to stress in bone microenvironment requires further investigation.
Factors that promote bone destruction via osteoclast activation are likely involved in RCC-induced osteolytic bone lesions. Factors that increase osteoclastic activity, e.g., IL-6, were also found in the CM of Bo-786 cells (Figure 3B, Supplemental Table S2). IL-6 has been shown to stimulate osteoblasts to secrete IGFBP5, which enhances osteoclast formation and activity [33]. Thus, the bone destruction in RCC may have certain similarities with multiple myeloma, which is characterized by the presence of osteolytic components accompanied by the suppression of osteoblast differentiation and function [34].
Osteolytic bone lesions associated with renal carcinoma bone metastasis are difficult to treat and have significant impact on patient mobility and survival. Current therapy modality mainly inhibits osteoclast activity. In breast and prostate cancer, RANKL and PTHrP have been shown to mediate bone destruction by increasing osteoclastic activity [35], [36]. For these patients, current therapies target osteoclast activity by using bisphosphonates or denosumab, a monoclonal antibody that inhibits RANKL. RCC bone metastases are resistant to these treatments. In our studies, we did not detect significant secretion of RANKL or PTHrP in Bo-786 CM by mass spectrometry or by real-time RT-PCR [22]. Moreover, in our study, TRAP staining for osteoclasts in Bo-786 cell-injected bone was significantly lower than that produced by PC3 prostate cancer cell line. Together, these observations support an additional mechanism for bone destruction that is more influenced by the inhibition of bone formation, and suggests that therapies that enhance osteoblast differentiation may synergize with current therapy modalities to improve clinical outcomes. BMP family proteins have been shown to stimulate osteoblast differentiation [37]. Interestingly, several tyrosine kinase inhibitors, including dasatinib [38] and cabozantinib [39], [40], [41], [42], have also been shown to stimulate osteoblast differentiation. Whether these tyrosine kinase inhibitors are suitable for treating RCC bone metastasis remains to be studied.
Conclusions
We showed that suppression of osteoblast differentiation by BIGH3/TGFBI is one of the mechanisms for the osteolytic bone metastasis of RCC. Our studies provide a plausible explanation for the limited efficacy of therapies that target osteoclasts only. The identification of this new mechanism expands the possibility of combination therapy strategies for RCC bone metastasis.
The following are the supplementary data related to this article.
Sequence of Primers
Extracellular Proteins in Bo-786-O RCC CM
Acknowledgments
Acknowledgements
The authors would like to thank Sarah Rizvi for preparing bone metastasis tissue bank samples.
Footnotes
Funding Sources: This work was supported by grants from the National Institutes of Health[CA174798, 5P50 CA140388, and P30CA16672], by grants from the Cancer Prevention and Research Institute of Texas [CPRIT RP110327, CPRIT RP150179, CPRIT RP150282], by funds from the University Cancer Foundation via the Sister Institute Network Fund at the MD Anderson Cancer Center, and by the Institutional Research Grant (IRG-Knowledge Gap Grant) Program at the MD Anderson Cancer Center.
Authors' Contributions: R. L. S. and S. H. L. developed study concepts and design and supervise the study. T. P., S. C. L., K. J. Y., G. Y., and J. H. S. developed methodology and performed experiments, R. L. S., V. O. L., J. E. B., B. M., P. P. L., N. M. T., E. J., C. G. W., G. E. G., and S. H. L. acquired and managed patient specimens, provided animals, provided facilities, etc. T. P., R. L. S., S. H. L., G. E. G., and L. Y. Y. L. performed data analysis and interpretation. T. P., R. L. S., S. H. L., G. E. G., and L. Y. Y. L. wrote and revised the manuscript.
Contributor Information
Sue-Hwa Lin, Email: slin@mdanderson.org.
Robert L. Satcher, Email: rlsatcher@mdanderson.org.
References
- 1.Chen SC, Kuo PL. Bone metastasis from renal cell carcinoma. Int J Mol Sci. 2016;17 doi: 10.3390/ijms17060987. pii: E987. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Guise TA. The vicious cycle of bone metastases. J Musculoskelet Neuronal Interact. 2002;2:570–572. [PubMed] [Google Scholar]
- 3.Mikami S, Katsube K, Oya M, Ishida M, Kosaka T, Mizuno R, Mochizuki S, Ikeda T, Mukai M, Okada Y. Increased RANKL expression is related to tumour migration and metastasis of renal cell carcinomas. J Pathol. 2009;218:530–539. doi: 10.1002/path.2567. [DOI] [PubMed] [Google Scholar]
- 4.Takahashi N, Udagawa N, Suda T. A new member of tumor necrosis factor ligand family, ODF/OPGL/TRANCE/RANKL, regulates osteoclast differentiation and function. Biochem Biophys Res Commun. 1999;256:449–455. doi: 10.1006/bbrc.1999.0252. [DOI] [PubMed] [Google Scholar]
- 5.Raggatt LJ, Partridge NC. Cellular and molecular mechanisms of bone remodeling. J Biol Chem. 2010;285:25103–25108. doi: 10.1074/jbc.R109.041087. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Tian E, Zhan F, Walker R, Rasmussen E, Ma Y, Barlogie B, Shaughnessy JD., Jr. The role of the Wnt-signaling antagonist DKK1 in the development of osteolytic lesions in multiple myeloma. N Engl J Med. 2003;349:2483–2494. doi: 10.1056/NEJMoa030847. [DOI] [PubMed] [Google Scholar]
- 7.Skonier J, Neubauer M, Madisen L, Bennett K, Plowman GD, Purchio AF. cDNA cloning and sequence analysis of beta ig-h3, a novel gene induced in a human adenocarcinoma cell line after treatment with transforming growth factor-beta. DNA Cell Biol. 1992;11:511–522. doi: 10.1089/dna.1992.11.511. [DOI] [PubMed] [Google Scholar]
- 8.LeBaron RG, Bezverkov KI, Zimber MP, Pavelec R, Skonier J, Purchio AF. Beta IG-H3, a novel secretory protein inducible by transforming growth factor-beta, is present in normal skin and promotes the adhesion and spreading of dermal fibroblasts in vitro. J Invest Dermatol. 1995;104:844–849. doi: 10.1111/1523-1747.ep12607024. [DOI] [PubMed] [Google Scholar]
- 9.Kim JE, Kim SJ, Lee BH, Park RW, Kim KS, Kim IS. Identification of motifs for cell adhesion within the repeated domains of transforming growth factor-beta-induced gene, betaig-h3. J Biol Chem. 2000;275:30907–30915. doi: 10.1074/jbc.M002752200. [DOI] [PubMed] [Google Scholar]
- 10.Han B, Luo H, Raelson J, Huang J, Li Y, Tremblay J, Hu B, Qi S, Wu J. TGFBI (betaIG-H3) is a diabetes-risk gene based on mouse and human genetic studies. Hum Mol Genet. 2014;23:4597–4611. doi: 10.1093/hmg/ddu173. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Kannabiran C, Klintworth GK. TGFBI gene mutations in corneal dystrophies. Hum Mutat. 2006;27:615–625. doi: 10.1002/humu.20334. [DOI] [PubMed] [Google Scholar]
- 12.Thapa N, Lee BH, Kim IS. TGFBIp/betaig-h3 protein: a versatile matrix molecule induced by TGF-beta. Int J Biochem Cell Biol. 2007;39:2183–2194. doi: 10.1016/j.biocel.2007.06.004. [DOI] [PubMed] [Google Scholar]
- 13.Dieudonne SC, Kerr JM, Xu T, Sommer B, DeRubeis AR, Kuznetsov SA, Kim IS, Gehron Robey P, Young MF. Differential display of human marrow stromal cells reveals unique mRNA expression patterns in response to dexamethasone. J Cell Biochem. 1999;76:231–243. doi: 10.1002/(sici)1097-4644(20000201)76:2<231::aid-jcb7>3.0.co;2-x. [DOI] [PubMed] [Google Scholar]
- 14.Monticone M, Liu Y, Tonachini L, Mastrogiacomo M, Parodi S, Quarto R, Cancedda R, Castagnola P. Gene expression profile of human bone marrow stromal cells determined by restriction fragment differential display analysis. J Cell Biochem. 2004;92:733–744. doi: 10.1002/jcb.20120. [DOI] [PubMed] [Google Scholar]
- 15.Satcher RL, Pan T, Bilen MA, Li X, Lee YC, Ortiz A, Kowalczyk AP, Yu-Lee LY, Lin SH. Cadherin-11 endocytosis through binding to clathrin promotes cadherin-11-mediated migration in prostate cancer cells. J Cell Sci. 2015;128:4629–4641. doi: 10.1242/jcs.176081. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Lin SH, Cheng CJ, Lee YC, Ye X, Tsai WW, Kim J, Pasqualini R, Arap W, Navone NM, Tu SM. A 45-kDa ErbB3 secreted by prostate cancer cells promotes bone formation. Oncogene. 2008;27:5195–5203. doi: 10.1038/onc.2008.156. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Heinemann ML, Ilmer M, Silva LP, Hawke DH, Recio A, Vorontsova MA, Alt E, Vykoukal J. Benchtop isolation and characterization of functional exosomes by sequential filtration. J Chromatogr A. 2014;1371:125–135. doi: 10.1016/j.chroma.2014.10.026. [DOI] [PubMed] [Google Scholar]
- 18.Liang AK, Liu J, Mao SA, Siu VS, Lee YC, Lin SH. Expression of recombinant MDA-BF-1 with a kinase recognition site and a 7-histidine tag for receptor binding and purification. Protein Expr Purif. 2005;44:58–64. doi: 10.1016/j.pep.2005.03.025. [DOI] [PubMed] [Google Scholar]
- 19.Chu K, Cheng CJ, Ye X, Lee YC, Zurita AJ, Chen DT, Yu-Lee LY, Zhang S, Yeh ET, Hu MC. Cadherin-11 promotes the metastasis of prostate cancer cells to bone. Mol Cancer Res. 2008;6:1259–1267. doi: 10.1158/1541-7786.MCR-08-0077. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Jin JK, Tien PC, Cheng CJ, Song JH, Huang C, Lin SH, Gallick GE. Talin1 phosphorylation activates beta1 integrins: a novel mechanism to promote prostate cancer bone metastasis. Oncogene. 2015;34:1811–1821. doi: 10.1038/onc.2014.116. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Williams RD, Elliott AY, Stein N, Fraley EE. In vitro cultivation of human renal cell cancer. II. Characterization of cell lines. In Vitro. 1978;14:779–786. doi: 10.1007/BF02617972. [DOI] [PubMed] [Google Scholar]
- 22.Satcher RL, Pan T, Cheng CJ, Lee YC, Lin SC, Yu G, Li X, Hoang AG, Tamboli P, Jonasch E. Cadherin-11 in renal cell carcinoma bone metastasis. PLoS One. 2014;9:e89880. doi: 10.1371/journal.pone.0089880. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Wang D, Christensen K, Chawla K, Xiao G, Krebsbach PH, Franceschi RT. Isolation and characterization of MC3T3-E1 preosteoblast subclones with distinct in vitro and in vivo differentiation/mineralization potential. J Bone Miner Res. 1999;14:893–903. doi: 10.1359/jbmr.1999.14.6.893. [DOI] [PubMed] [Google Scholar]
- 24.Klamer SE, Kuijk CG, Hordijk PL, van der Schoot CE, von Lindern M, van Hennik PB, Voermans C. BIGH3 modulates adhesion and migration of hematopoietic stem and progenitor cells. Cell Adh Migr. 2013;7:434–449. doi: 10.4161/cam.26596. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Ivanov SV, Ivanova AV, Salnikow K, Timofeeva O, Subramaniam M, Lerman MI. Two novel VHL targets, TGFBI (BIGH3) and its transactivator KLF10, are up-regulated in renal clear cell carcinoma and other tumors. Biochem Biophys Res Commun. 2008;370:536–540. doi: 10.1016/j.bbrc.2008.03.066. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Yamanaka M, Kimura F, Kagata Y, Kondoh N, Asano T, Yamamoto M, Hayakawa M. BIGH3 is overexpressed in clear cell renal cell carcinoma. Oncol Rep. 2008;19:865–874. [PubMed] [Google Scholar]
- 27.Lebdai S, Verhoest G, Parikh H, Jacquet SF, Bensalah K, Chautard D, Rioux Leclercq N, Azzouzi AR, Bigot P. Identification and validation of TGFBI as a promising prognosis marker of clear cell renal cell carcinoma. Urol Oncol. 2015;33(69):e11–e68. doi: 10.1016/j.urolonc.2014.06.005. [DOI] [PubMed] [Google Scholar]
- 28.Thapa N, Kang KB, Kim IS. Beta ig-h3 mediates osteoblast adhesion and inhibits differentiation. Bone. 2005;36:232–242. doi: 10.1016/j.bone.2004.08.007. [DOI] [PubMed] [Google Scholar]
- 29.Kim JE, Kim EH, Han EH, Park RW, Park IH, Jun SH, Kim JC, Young MF, Kim IS. A TGF-beta-inducible cell adhesion molecule, betaig-h3, is downregulated in melorheostosis and involved in osteogenesis. J Cell Biochem. 2000;77:169–178. doi: 10.1002/(sici)1097-4644(20000501)77:2<169::aid-jcb1>3.0.co;2-l. [DOI] [PubMed] [Google Scholar]
- 30.Lee JM, Lee EH, Kim IS, Kim JE. Tgfbi deficiency leads to a reduction in skeletal size and degradation of the bone matrix. Calcif Tissue Int. 2015;96:56–64. doi: 10.1007/s00223-014-9938-4. [DOI] [PubMed] [Google Scholar]
- 31.Banks RE, Tirukonda P, Taylor C, Hornigold N, Astuti D, Cohen D, Maher ER, Stanley AJ, Harnden P, Joyce A. Genetic and epigenetic analysis of von Hippel–Lindau (VHL) gene alterations and relationship with clinical variables in sporadic renal cancer. Cancer Res. 2006;66:2000–2011. doi: 10.1158/0008-5472.CAN-05-3074. [DOI] [PubMed] [Google Scholar]
- 32.Young AC, Craven RA, Cohen D, Taylor C, Booth C, Harnden P, Cairns DA, Astuti D, Gregory W, Maher ER. Analysis of VHL gene alterations and their relationship to clinical parameters in sporadic conventional renal cell carcinoma. Clin Cancer Res. 2009;15:7582–7592. doi: 10.1158/1078-0432.CCR-09-2131. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Peruzzi B, Cappariello A, Del Fattore A, Rucci N, De Benedetti F, Teti A. c-Src and IL-6 inhibit osteoblast differentiation and integrate IGFBP5 signalling. Nat Commun. 2012;3:630–639. doi: 10.1038/ncomms1651. [DOI] [PubMed] [Google Scholar]
- 34.Christoulas D, Terpos E, Dimopoulos MA. Pathogenesis and management of myeloma bone disease. Expert Rev Hematol. 2009;2:385–398. doi: 10.1586/ehm.09.36. [DOI] [PubMed] [Google Scholar]
- 35.Azim HA, Kamal NS, Azim HA., Jr. Bone metastasis in breast cancer: the story of RANK-ligand. J Egypt Natl Canc Inst. 2012;24:107–114. doi: 10.1016/j.jnci.2012.06.002. [DOI] [PubMed] [Google Scholar]
- 36.Brown JM, Corey E, Lee ZD, True LD, Yun TJ, Tondravi M, Vessella RL. Osteoprotegerin and rank ligand expression in prostate cancer. Urology. 2001;57:611–616. doi: 10.1016/s0090-4295(00)01122-5. [DOI] [PubMed] [Google Scholar]
- 37.Lee YC, Cheng CJ, Bilen MA, Lu JF, Satcher RL, Yu-Lee LY, Gallick GE, Maity SN, Lin SH. BMP4 promotes prostate tumor growth in bone through osteogenesis. Cancer Res. 2011;71:5194–5203. doi: 10.1158/0008-5472.CAN-10-4374. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Lee YC, Huang CF, Murshed M, Chu K, Araujo JC, Ye X, deCrombrugghe B, Yu-Lee LY, Gallick GE, Lin SH. Src family kinase/abl inhibitor dasatinib suppresses proliferation and enhances differentiation of osteoblasts. Oncogene. 2010;29:3196–3207. doi: 10.1038/onc.2010.73. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Nguyen HM, Ruppender N, Zhang X, Brown LG, Gross TS, Morrissey C, Gulati R, Vessella RL, Schimmoller F, Aftab DT. Cabozantinib inhibits growth of androgen-sensitive and castration-resistant prostate cancer and affects bone remodeling. PLoS One. 2013;8:e78881. doi: 10.1371/journal.pone.0078881. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Dai J, Zhang H, Karatsinides A, Keller JM, Kozloff KM, Aftab DT, Schimmoller F, Keller ET. Cabozantinib inhibits prostate cancer growth and prevents tumor-induced bone lesions. Clin Cancer Res. 2014;20:617–630. doi: 10.1158/1078-0432.CCR-13-0839. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Varkaris A, Corn PG, Parikh NU, Efstathiou E, Song JH, Lee YC, Aparicio A, Hoang AG, Gaur S, Thorpe L. Integrating murine and clinical trials with cabozantinib to understand roles of MET and VEGFR2 as targets for growth inhibition of prostate cancer. Clin Cancer Res. 2016;22:107–121. doi: 10.1158/1078-0432.CCR-15-0235. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Graham TJ, Box G, Tunariu N, Crespo M, Spinks TJ, Miranda S, Attard G, de Bono J, Eccles SA, Davies FE. Preclinical evaluation of imaging biomarkers for prostate cancer bone metastasis and response to cabozantinib. J Natl Cancer Inst. 2014;106:dju033. doi: 10.1093/jnci/dju033. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
Sequence of Primers
Extracellular Proteins in Bo-786-O RCC CM