Abstract
The data presented here had been originally collected for a research project entitled ‘Sleep EEG spectral analysis in psychophysiological insomnia and normal sleep subjects’. This article describes the data of 11 subjects, referred to Sleep Disorders Research Center (SDRC) in Kermanshah, Iran. The data includes 14 EEG, 6 EOG, and 3 EMG channels, with a sampling ratio of 256 Hz. It includes power spectral features in segments of 30 s for each channel, and nonlinear analysis parameter. Also, the complete demographic and polysomnography specifications are attached.
Keywords: Sleep dataset, Psychophysiological, Insomnia, EEG
Specifications Table
| Subject area | Neuroscience, Neurobiology |
| More specific subject area | Psychiatry, sleep, psychophysiological insomnia |
| Type of data | Table, text file, m-file, mat file, |
| How data was acquired | Polysomnography, Matlab software |
| Data format | Filtered, analyzed, |
| Experimental factors | Sleep questionnaire were used for subjective features. Age, gender, height, weight, education, marriage, and body mass index were used as covariates. |
| Experimental features | Power spectrum includes delta, theta, alpha and beta bands. Parameters from nonlinear analysis (Poincare's map and standard descriptors). |
| Data source location | Samples were collected in the Sleep Disorders Research Center in Kermanshah University of Medical Science. |
| Data accessibility | The dataset is freely available at[1]for any academic, educational, and research purposes. |
| http://dx.doi.org/10.17632/3hx58k232n.4 |
Value of the data
-
•
The raw data could be processed using algorithms and other procedures during future researches.
-
•
The data represents 8 h of sleep signals (EEG, EOG, and EMG) from 22 subjects; including 11 psychophysiological insomniacs and 11 normal subjects.
-
•
Psychophysiological insomnia is a more prevalent sleep disorder, which leads to clinically significant impairment in social, occupational, and cognitive functions.
-
•
The data can also be used to assess the EEG Sleep Pattern in psychophysiological insomnia patients as well as good sleepers.
-
•
The diagnosis was performed by a sleep clinician, based on subjective and objective sleep features.
1. Experimental design, materials and methods
1.1. Participant
A total of 22 subjects that includes 8 males (36.36%) and 14 females (63.64%) aged between 18 and 63 years (43.2±14.2) were recruited for participation. Out of the participators, 11 patients were suffering from psychophysiological insomnia (18.18% male; age: 44±13.2 years; Body Mass Index (BMI): 26.6±3.7 kg m−2) and 11 subjects had normal sleep pattern (54.5% male; age: 42.4±15.4 years; BMI: 27.53±4.24 kg m−2).
The patients were selected from people referred to SDRC, due to insomnia complaints. Normal sleep subjects were recruited from the general population. A detailed written consent was obtained from all participants. Both patients and normal subjects completed their demographic and medical history checklists, including substance and alcohol check as well as psychiatric disorders. For selection of normal subjects, candidates had to first complete the Pittsburgh questionnaire. Preliminary selection was done based on these results. Then, they were further tested using Polysomnography. Finally, subjects who cleared the PSG test round, were selected.
1.2. Procedure
All subjects underwent a one-night polysomnography (PSG) test with the help of SOMNOscreen device called SOMNOscreen™ plus PSG produced by SOMNOmedics GmbH, Germany. The duration of the test was 8 h (23:00–07:00 h), as per standard protocol at SDRC of KUMS, Iran. A day before appointment, the subjects were invited to sleep in the laboratory of SDRC. They were advised against consuming any tea, coffee, heavy diet or cigarette. Sleeping during the day was also prohibited. Upon arriving at the laboratory, the height and weight of the subject was measured by an experienced personnel. Next, the subjects and participants completed the Pittsburgh questionnaire followed by a detailed briefing on PSG procedures. The measurement of PSG was based on the American Academy of Sleep Medicine guidelines. The polysomnography room was cleaned from artefacts like auditory and visual noises, based on standards [1].
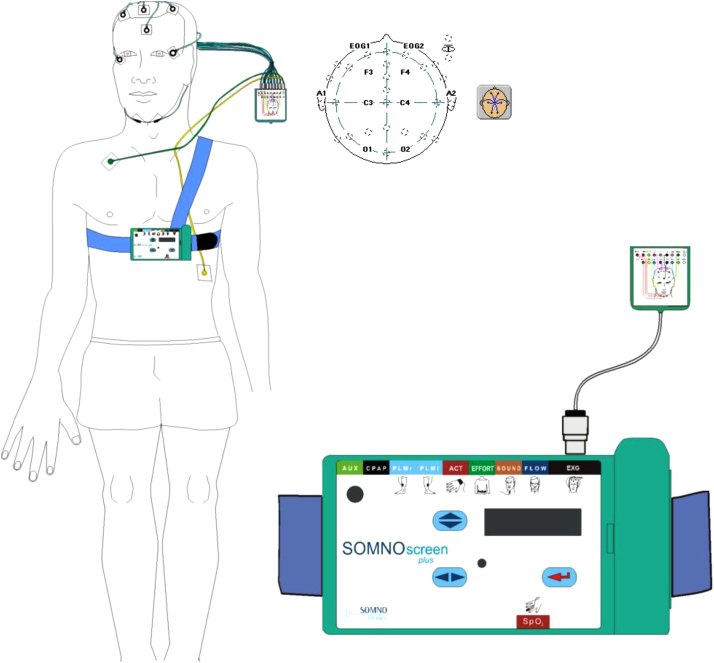
24 recording electrodes were prepared, including 14 electroencephalogram channels (C4A1, C3A2, F3, F4, C3, C4, A1, A2, O1, O2, F3A2, F4A1, O1A2, O2A1), 6 electrooculogram channels (EOG1, EOG2, EOG1A1, EOG2A1, EOG1A2, EOG2A2), 3 electromyogram channels (EMG, EMG1, EMG2), and ECG channel. All the recordings were sampled at the rate of 256 Hz. EEG was recorded using Ag/AgCl electrodes, as per the International 10–20 System of Electrode Placement, as shown in Fig. 1. Recorded EEG signals is also shown in Fig. 2.
Fig. 1.
Electrode montage corresponding to the international 10–20: Sites included frontal (F3, F4), central (C3, C4), and occipital (01, 02) placements of the International 10–20 System.
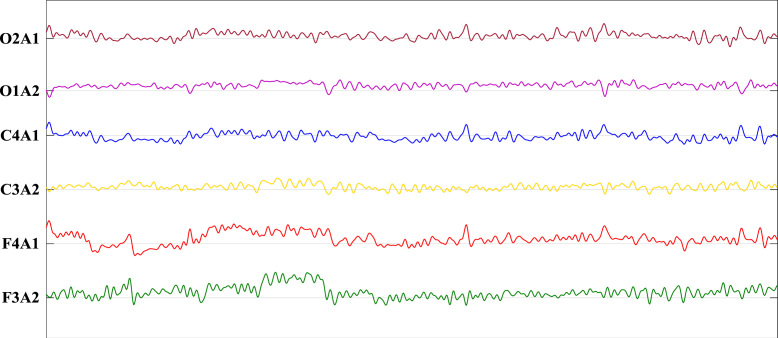
Fig. 2.
6 EEG channels sampled at 256 Hz using SOMNOscreen PSG (SOMNOscreen TM plus PSG+, SOMNOmedics GmbH, Germany). This shows the voltage in microvolts on each channel referenced to the left or right earlobe (A1, A2) over a 4 s window.
Both psychophysiological insomnia and normal sleep were determined after clinical interview and careful study of the data obtained from PSG test by an experienced psychiatrist, trained in sleep medicine and PSG. Subjective information was obtained from clinical interview using Pittsburgh Sleep Quality Index (PQSL) [2], [3].
All raw datasets were stored in the European Data Format (EDF) format, one file per subject. For example, the file ‘Normal_Subject_xx’, contains the raw data from normal subject number ‘xx’. Similarly, for a patient suffering from psychophysiological insomnia, the file would be ‘Raw_Signal_Psychophysiological_ Insomnia_xx’. Description of all participants were recorded in ‘PSG_Psycho_Normal.xlsx’. The outputs from polysomnography were collected at ‘PSG_Outputs.rar’. Data file names and their descriptions are listed in Table 1.
Table 1.
Data file names and their descriptions.
| Name | Description |
|---|---|
| Arousal.txt | Time of arousal events |
| REM.txt | Time of REM events |
| Sleep Profile Reliability.txt | Reliability of the sleep profile |
| Sleep Profile.txt | Time of events including Stage 4, Stage 3, Stage 2, Stage 1,Rem,Wake,Movement. |
| Spindle K.txt | Time of events sleep spindle and K_Complex. |
Two files, namely PSD_Normal_Subjects and PSD_Psycophysiological_Insomnia, contain the power spectral density analyses for all subjects. In these files, the data is arranged in the form of cell arrays, where each row represents a channel and each column represents a 30 s epoch. Also, each cell contains a two-column matrix; the first column represents frequency and the second, power. The power spectrum analysis is commonly used in the study of biological signals, to calculate the frequency power [4]. Power spectrum analysis was conducted using Fast Fourier transform (FFT) in the range of 0.1–35 Hz, continually [5]. This transform is defined as follows:
| (1) |
In this data collection project, frequency resolution in power spectrum analysis was 0.5 Hz, which is defined as Fs/N in FFT [6]. As polysomnography analysis is separated by 30 s time intervals, the band's power was also extracted from these 30 s epochs.
The EEG data was filtered using the band pass method between 0.1 Hz and 35 Hz. It should be noted that signal filtering was done by SOMNOscreen™ plus instrument and the filtered data was exported only, and not completely raw data. In the next step, large artefacts due to electromyography activity, horizontal eye movement or ECG artefact were removed using independent component analysis (ICA). This method was considered in the past decade. Independent signal extraction from mixed signals is one of its applications. The ICA literature is divided into two major categories: practical algorithms and theoretical analyses. ICA could separate activities stemming from the most favourable parts of the brain by using their independent components [7].
With further spectral features, the present data has some important benefits to sleep clinicians and researchers, who are unaware of spectral analysis. They would have considerable findings by download and statistical analysis of spectral features.
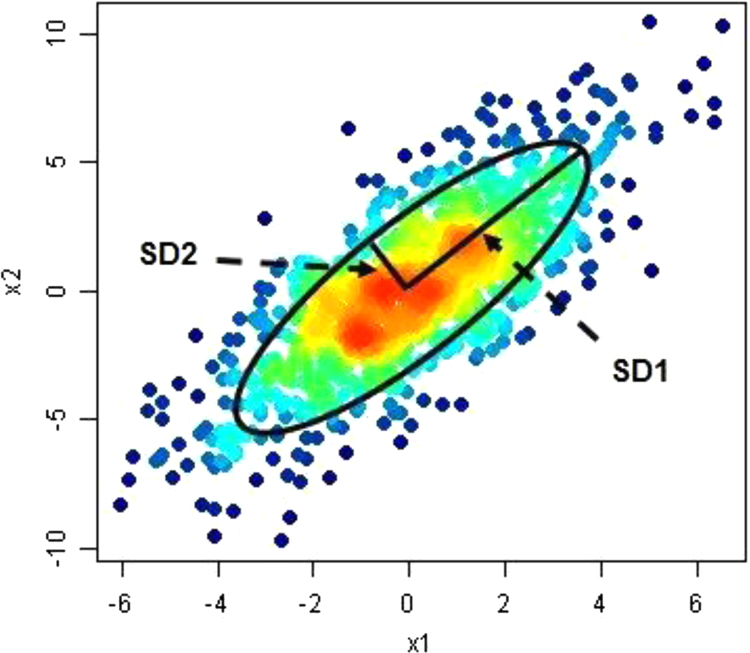
1.3. Poincare map
The Poincare's map, as a nonlinear analysis, is valuable, as it could reveal the nonlinear aspects of the data collection [8]. Therefore, the challenge lies in recording temporal information of the plot quantitatively. The standard descriptors used for quantifying the Poincare's map, measure the impure variability of the time series data. Standard deviations across the line of identification (SD2) and perpendicular to the line of identification (SD1), represent the magnitude of the major and minor axes of the ellipse, respectively. SD1 represents the SD of the instantaneous short term variability. SD2 represents the SD of long-term variability. SD1 and SD2 support the potent data and also, the proportion of SD1 to SD2 has been suggested as strongly indicator [9]. Descriptors SD1 and SD2 can be defined as:
| (2) |
| (3) |
where SD is a standard deviation of the time series.
To plot the Poincare's map — summation, maximum, and standard deviation of the band's power in each epoch was calculated and saved, as time series data. Then, the standard descriptors were calculated and the Poincare plots were extracted from the statics result of the band's power. Each Poincare plot was constructed with the X-axis illustrating the statics result, at a specific epoch (x(k)), and the Y-axis illustrating the statics result after a specified epoch delay (x(k+1)).
We calculated the SD of statics result dispersion along with the diametric line and the SD of the power dissipation perpendicular to the diametric line, to present more quantitative data on the repartition of statics result in the Poincare plots (Fig. 3). The SD perpendicular to the diametric line was defined as SD1 and the SD along the diametric line was defined as SD2. SD1 is the level of instantaneous variability and demonstrates the variability from one epoch to the next. In contrast, SD2 demonstrates the power variability across all the epochs. We then defined the SD1/SD2 ratio that is a potent candidate, reflecting the psychophysiological disorder depth, which has been used to specify the clarity and linearity of the scatter pattern.
Fig. 3.
A typical Poincare plot. The horizontal axes demonstrate the illustrating the statics result at a specific epoch (x(k)) and the vertical axis illustrating the statics result after a specified epoch delay (x(k+1)). An ellipse is fitted to the data points and the Poincare plot descriptors are measured by estimating the SD perpendicular to the diametric line was defined as SD1, and the SD along the diametric line was defined as SD2 and ratio of the SD1/SD2 of the fitted ellipse.
Acknowledgements
The authors gratefully acknowledge the Research Council of Kermanshah University of Medical Sciences (Grant Number: 96373) for the financial support.
Footnotes
Transparency document associated with this article can be found in the online version at doi:10.1016/j.dib.2017.09.033.
Transparency document. Supplementary material
Transparency document
References
- 1.M. Rezaei, H. Mohammadi, H. Khazaie, EEG/EOG/EMG data from a cross sectional study on psychophysiological insomnia and normal sleep subjects, Mendeley Data, v1, 2017. 〈http://dx.doi.org/10.17632/3hx58k232n.1〉 (DOI is reserved but not active). [DOI] [PMC free article] [PubMed]
- 2.Mollayeva T., Thurairajah P., Burton K., Mollayeva S., Shapiro C.M., Colantonio A. The Pittsburgh Sleep Quality Index as a screening tool for sleep dysfunction in clinical and non-clinical samples: a systematic review and meta-analysis. Sleep Med. Rev. 2016;25:52–73. doi: 10.1016/j.smrv.2015.01.009. [DOI] [PubMed] [Google Scholar]
- 3.Buysse D.J., Reynolds C.F., Monk T.H., Berman S.R., Kupfer D.J. The Pittsburgh Sleep Quality Index: a new instrument for psychiatric practice and research. Psychiatry Res. 1989;28:193–213. doi: 10.1016/0165-1781(89)90047-4. [DOI] [PubMed] [Google Scholar]
- 4.Rezaei M., Amiri M., Mohajeri P., Rezaei M. A new algorithm for lane detection and tracking on pulsed field gel electrophoresis images. Chemom. Intell. Lab. Syst. 2016;157:1–6. [Google Scholar]
- 5.M. Frigo, S.G. Johnson, FFTW: an adaptive software architecture for the FFT, in: Proceedings of the International Conference on Acoustics, Speech, and Signal Processing, vol. 3, 1998, pp. 1381–1384.
- 6.Fenet S., Badeau R., Richard G. Reassigned time–frequency representations of discrete time signals and application to the Constant-Q Transform. Signal Process. 2017;132:170–176. [Google Scholar]
- 7.Seifzadeh S., Rezaei M., Faez K., Amiri M. Fast and efficient four‑class motor imagery electroencephalography signal analysis using common spatial pattern–ridge regression algorithm for the purpose of brain–computer interface. J. Med. Signals Sens. 2017;7(2):80–85. [PMC free article] [PubMed] [Google Scholar]
- 8.Sadeghi Bajestani G., Hashemi Golpayegani M.R., Sheikhani A., Ashrafzadeh F. Poincaré section analysis of the electroencephalogram in autism spectrum disorder using complement plots. Kybernetes. 2017;46 [Google Scholar]
- 9.Hoshi R.A., Pastre C.M., Vanderlei L.C.M., Godoy M.F. Poincaré plot indexes of heart rate variability: relationships with other nonlinear variables. Auton. Neurosci. 2013;177:271–274. doi: 10.1016/j.autneu.2013.05.004. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
Transparency document