Abstract
New methodologies for site-specifically radiolabeling proteins with 18F are required to generate high quality radiotracers for preclinical and clinical applications with positron emission tomography. Herein, we report an approach by which we use lipoic acid ligase (LplA) to conjugate [18F]-fluorooctanoic acid to an antibody fragment bearing the peptide substrate of LplA. The mild conditions of the reaction preserve antibody immunoreactivity, and the efficiency of LplA allows for >90% yield even with very small amounts of peptidic precursor (1–10 nmol). These features are advantageous compared to the current gold standard in the field. Moreover, the methodology introduces a new application for an important tool in chemical biology.
Graphical Abstract
Preclinical feasibility studies and the clinical translation of experimental radiotracers with short-lived isotopes like 18F require rapid, highly efficient reaction and purification schemes. Although protocols have been developed to translate small molecules like 2-deoxy-2-[18F]-fluoroglucose, [18F]-choline, and 3′-deoxy-3′-[18F]-fluorothymidine, the chemistry used for these C-[18F] bond formations is too severe to be applied to biomolecules with higher order structure (e.g., affibodies, diabodies, antibody fragments). Developing better radiolabeling strategies to couple 18F to small biomolecules is an important unmet need for the imaging field, as biomolecules can be evolved to surpass small molecules in potency and selectivity; they can bind proteins that small molecules cannot address, and like small molecules, their rapid equilibration into peripheral tissues in vivo allows for data collection within just hours after injection (imaging larger radiolabeled molecules like IgGs in peripheral human tissues requires waiting days after injection).
Responsive to these considerations, several groups have developed small molecule [18F]-prosthetics that can be coupled to endogenous biomolecule amino acids using mild bioconju-gation chemistry.1 The most widely used compound is N-succinimidyl-[18F]-fluorobenzoate ([18F]-SFB), an activated ester that reacts with the epsilon amino moiety on solvent-exposed lysine residues.2,3 However, radiofluorination with [18F]-SFB has several well-recognized limitations, including a time-consuming multistep synthesis to prepare and then react with the respective biomolecule (usually hours), low bio-conjugation yields (usually ~40%), an impractically large requirement of biomolecules to achieve useful quantities of radiotracer (>100 nmol), and a lack of control over which and how many lysines are labeled.
Many alternatives to [18F]-SFB that target other endogenous amino acids or engineered unnatural moieties have been developed. Engineered cysteines exploit the rarity of endogenous solvent-exposed cysteines to insert a reactive moiety for site-specific labeling. This thiol can be radiolabeled directly, for example with an [18F]-maleimide or [18F]-fluoro-2-cyanobenzothiazole,4–7 or further functionalized as part of a 2-step procedure. Common protocols ligate either an oxime8,9 or a click moiety10 to the engineered cysteine, which is then subsequently labeled with a complementary reactive [18F]-prosthetic (e.g., [18F]-fluorobenzaldehyde, [18F]-trans-cyclo-octene). Expressed protein ligation has also been used to introduce a terminal oxime moiety for subsequent radio-fluorination.11 Unnatural amino acids with orthogonally reactive moieties can also be engineered directly into a biomolecule for subsequent labeling. Examples include L-homopropargylglycine,12,13 which can subsequently be labeled with an [18F]-azide and para-iodophenylalanine,14 which can be labeled with [18F]-4-fluorophenylboronic acid. An exciting recent extension of this concept uses biorthogonal click chemistry to label biomolecules in vivo, making pretargeting with long-circulating antibodies realistic.15–17 Unfortunately, none of these approaches have overcome all of the limitations of [18F]-SFB and many require prior chemical manipulation of the protein. Direct radiofluorination of proteins premodified with [18F]-acceptor moieties (e.g., NOTA for labeling with Al[18F]) is an emerging field; however, labeling conditions are frequently harsh, and proteins must always be chemically manipulated beforehand.18,19
We hypothesized that many of the aforementioned chemical challenges could be overcome by using an enzyme to conjugate an [18F]-labeled prosthetic group to a biomolecule. The basis for our optimism was founded on several considerations. Foremost, there are many well-characterized enzymes that add small hydrocarbons to proteins, including farnesyl or myristoyl transferases, as well as several classes of enzymes that modify histones with small carbon-based moieties. These post-translational modifications resemble some of the structures of the small prosthetic groups already used to chemically conjugate 18F to biomolecules, suggesting that an enzyme from one of the aforementioned classes might couple a modestly altered [18F]-substrate to its respective target peptide. Beyond this important biological precedent, an enzymatic conjugation strategy would also provide the aqueous, mild conditions required to preserve the integrity of the [18F]-biomolecule. In addition, bioconjugation with an enzyme opens the opportunity for site specific radiolabeling, a major priority in contemporary radiotracer development.
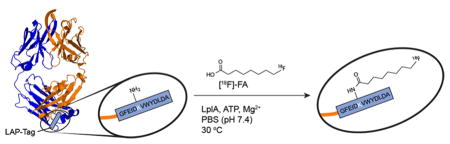
Surveying the literature, we found the bacterial enzyme lipoic acid ligase (LplA) to be a promising candidate for radiofluorination of biomolecules for several reasons. First, LplA catalyzes the formation of a stable amide bond between the ε-amine of a lysine residue and a range of structurally distinct alkyl carboxylates,20–24 which provides a substrate plasticity that suggested it might tolerate an [18F]-alkyl carboxylate only slightly different from known substrates. Second, LplA only performs biochemistry on lysines within a signature peptide motif (termed “LAP”),25 making site specific radiolabeling realistic.26 Third, the KM of LplA is relatively low (13.3 μM),25 which we anticipated would lead to high bioconjugation yields at low protein concentrations. This is an important consideration for radiofluorination because the specific activity (Ci/mmol) of an [18F]-biomolecule is related to the amount of material required for labeling, as it is generally not possible to separate radiolabeled and unlabeled material. Other investigators have recognized the virtues of LplA biochemistry for unrelated applications in chemical biology, exploiting this enzyme to conjugate fluorescent molecules to LAP-tagged proteins in live cells to study protein biology.20–22,27
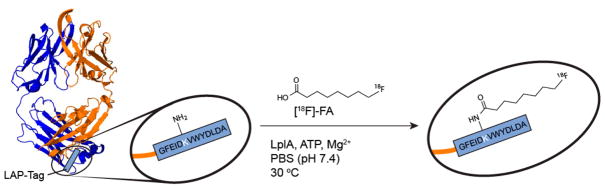
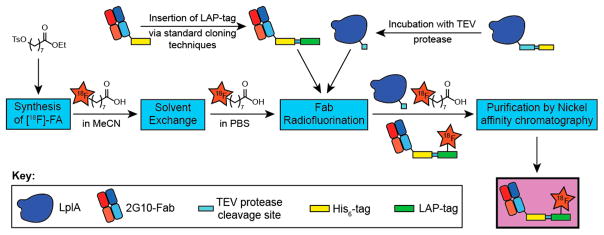
Our strategy for realizing enzymatic radiofluorination of proteins is summarized in Figure 1. [18F]-8-Fluorooctanoic acid ([18F]-FA) was chosen as a prosthetic for 2 reasons: octanoic acid is a known substrate of LplA,28 and a radiosynthesis of [18F]-FA29 was already reported. We opted to radiofluorinate 2G10, a recombinant human Fab antibody fragment we previously engineered to have high affinity (KD < 50 mM) for the urokinase plasminogen activator receptor (uPAR).30–32 We reasoned that our previous data with this biomolecule would also serve as a benchmark to contextualize the results from our new bioconjugation strategy.
Figure 1.
Schematic overview of 2G10-Fab-LAP radiofluorination with [18F]-FA catalyzed by LplA.
RESULTS AND DISCUSSION
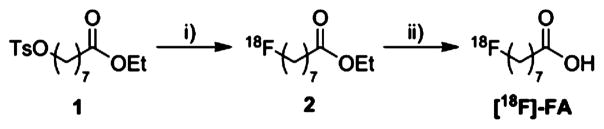
Initially, we sought to confirm that nonradioactive [19F]-FA is a viable substrate for LplA. This compound was synthesized in 4 steps from 8-hydroxyoctanoic acid as previously reported (Supporting Information (SI) Scheme 1).33 A known 13-amino acid target peptide sequence (GFEIDKVWYDLDA, “LAP” peptide) with excellent LplA coupling kinetics was used throughout these studies.25 This LAP peptide (60 μM) was incubated with LplA (500 nM) and [19F]-FA (750 μM) at 30 °C in PBS along with the required enzymatic cofactors ATP (3 mM) and Mg2+ (5 mM). At various time points, aliquots were withdrawn; LplA activity was quenched with EDTA, and reaction progress was measured via RP-HPLC (Figure 2). Conversion of LAP to a more hydrophobic species, consistent with conjugation to [19F]-FA, was complete at 30 min. The product peak was isolated via semipreparative RP-HPLC and confirmed as [19F]-FA-LAP by ESI-MS (m/z found = 1756.85; expected = 1756.92; Figure S1). As an initial assessment of the site-specificity of LplA, a “scrambled” LAP peptide (EFDDW-KYADVGLI) was also incubated with identical reaction components. No productive reaction was observed after 60 min, suggesting that only the precise amino acid sequence of the LAP-tag is recognized by LplA.
Figure 2.
(A) Schematic overview of conjugation of [19F]-FA to the LAP peptide catalyzed by LplA. (B) RP-HPLC trace demonstrating conversion of the LAP peptide to [19F]-LAP-FA. The identity of [19F]-LAP-FA was confirmed by ESI-MS (m/z = 1756.848).
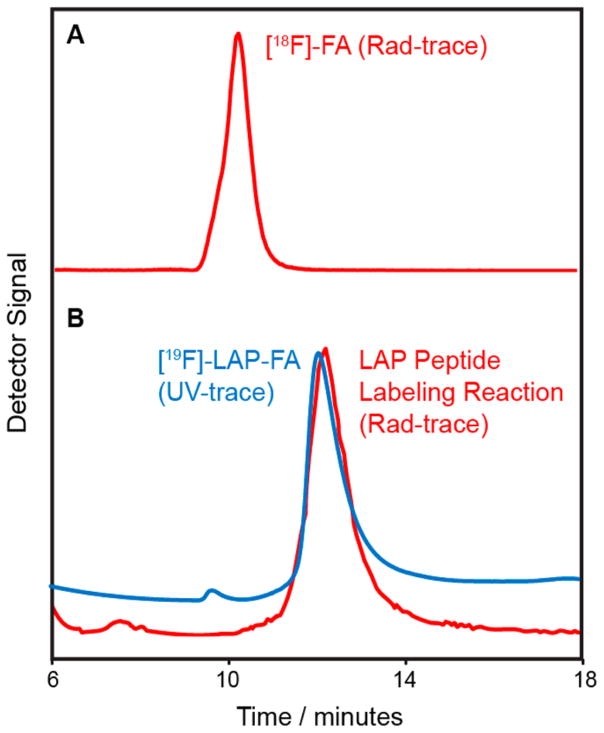
Next, we synthesized [18F]-FA in 2 steps from tosylate 1, making minor changes to a previously published protocol (Scheme 1).29 Briefly, 1 was radiofluorinated under standard conditions, and the resulting alkyl [18F]-fluoride 2 was purified by semipreparative RP-HPLC. The ethyl ester was then hydrolyzed in 5 N KOH, and [18F]-FA was immobilized on a C18 sep-pak, washed with H2O to remove all traces of KOH, and eluted with MeCN. To remove of all MeCN prior to dissolving [18F]-FA in PBS for radiolabeling, this solution was heated under reduced pressure at 50 °C for 1 h. Higher temperatures led to a significant loss of activity, presumably due to the volatility of [18F]-FA. The total time from production of [18F]-fluoride to dissolving [18F]-FA in PBS ready for peptide/ protein radiofluorination was ~180 min. Starting with ~500 mCi of [18F]-fluoride, ~40 mCi of [18F]-FA was generated (nondecay corrected yield of 8 ± 1.5%, n = 4) with sufficient activity to investigate the radiofluorination of the LAP peptide and, subsequently, 2G10-Fab-LAP. [18F]-FA was ~98% pure by RP-HPLC (Figure 3A) with no evidence of any impurities in the UV trace.
Scheme 1. Radiosynthesis of [18F]-FAa.
aReagents, conditions, and yield: (i) K[18F], K2.2.2, K2CO3, MeCN; 90 °C; 10 min (ii) 5N KOH in MeOH; 90 °C; 10 min. Total synthesis time from production of [18F] to dissolution of [18F]-FA in PBS was ~180 min. Nondecay corrected yield = 8 ± 1.5% (n = 4). Decay-corrected yield = 26 ± 4% (n = 4).
Figure 3.
RP-HPLC traces demonstrating conjugation of [18F]-FA to the LAP peptide for (A) [18F]-FA and (B) (red trace) LAP peptide (60 μM), LplA (5 μM), [18F]-FA (~200 μCi), ATP (3 mM), and Mg(OAc)2 (5 mM) with the reaction incubated at 37 °C for 10 min and quenched via addition of EDTA to a final volume of 180 mM and analyzed by RP-HPLC using Rad-detector and (blue trace) the [19F]-LAP-FA nonradioactive standard. The HPLC eluent was 45:55:0.1 v:v:v MeCN:H2O:TFA.
With [18F]-FA in hand, we sought to demonstrate that it remained a viable substrate for LplA even at the lower concentrations typical of radiofluorinations. Approximately 200 μCi of [18F]-FA was added to a 200 μL solution of the LAP peptide (60 μM, 12 nmol) and LplA (5 μM), and the consumption of [18F]-FA was measured by radio-TLC following quenching with EDTA (see Figure S3 for representative examples of radio-TLC analyses). After just 10 min, ~90% of the prosthetic had been consumed and converted to a more polar species that remained on the baseline of the TLC plate, consistent with the conjugation of [18F]-FA to the LAP peptide (Table 1). The formation of [18F]-LAP-FA was confirmed by RP-HPLC and comparison to [19F]-FA-LAP (Figure 3B). Interestingly, a control reaction containing LplA (10 μM) but no LAP peptide exhibited a 30% consumption of [18F]-FA by radio-TLC, suggestive of productive peptide/ protein labeling. We reasoned that [18F]-FA might bind noncovalently to LplA, generating a false-positive signal in the TLC assay. This was confirmed by diluting an aliquot from the reaction with reducing SDS-PAGE buffer and briefly heating it to 95 °C to break any noncovalent bonds. Radio- TLC analysis of this treated sample showed no labeling, confirming this hypothesis. Pleasingly, the conjugation yields measured in the presence of the LAP peptide were unchanged after this treatment due to the covalent bond formed under these conditions. Moving forward, a small sample (3 μL) of each reaction was treated in this way to ensure radio-TLC analyses accurately reflected productive bioconjugation.
Table 1.
LplA Conjugates [18F]-FA to the LAP Peptidea
| peptide | [peptide]/μM | [LplA]/μM | yield/%b |
|---|---|---|---|
| LAP | 60 | 5 | 91 ± 1.5 (n = 3) |
| LAP | 60 | 0 | 0 |
| Sc. LAP | 60 | 5 | 0 |
| 0 | 10 | 0 |
General considerations: all reactions performed in 200 μL of PBS + 3 mM ATP + 5 mM Mg(OAc)2 at 30 °C for 10 min and quenched with EDTA (180 mM final concentration) prior to analysis.
Radio-TLC yields measured after treatment of the reaction sample with SDS-PAGE buffer at 95 °C for 5 min. The eluent used was 7:3:0.1 EtOAc:hexanes:acetic acid.
We then explored the lower limits of the peptide concentration at which high conjugation yields (>80%) were retained. Keeping LplA concentration fixed at 5 μM, the LAP concentration was incrementally reduced, resulting in conjugation yields of <80% below 15 μM (Table 2). Raising LplA concentration to 10 μM restored labeling yields even at an LAP concentration of 5 μM. However, reducing the LAP concentration still further again lowered conjugation yields, which could not be improved by adding more LplA (up to 50 μM).
Table 2.
Establishing the Dependence of Conjugation Yields on LAP/LplA Concentrationa
| [LAP]/μM | [LplA]/μM | average yield/%b | range of yields/%b |
|---|---|---|---|
| 60 | 5 | 91 ± 1.5 (n = 3) | 90–93 |
| 15 | 5 | 92 | |
| 5 | 5 | 57 | |
| 5 | 10 | 83 ± 10.8 (n = 4) | 67–93 |
| 2.5 | 50 | 53 | |
| 1 | 50 | 28 |
General considerations: all reactions performed in 200 μL of PBS + 3 mM ATP + 5 mM Mg(OAc)2 at 30 °C for 10 min and quenched with EDTA (180 mM final concentration) prior to analysis.
Radio-TLC yields measured after treatment of the reaction sample with SDS-PAGE buffer at 95 °C for 5 min. The eluent used was 7:3:0.1 EtOAc:hexanes:acetic acid.
Encouraged by the rapid and high yield labeling of the isolated LAP-tag, we extended the methodology to the Fab antibody fragment 2G10-Fab. The LAP-tag was inserted at the C-terminus of the heavy chain using standard cloning methods, a site at which His6 tags had previously been placed without affecting epitope affinity. The resulting construct, 2G10-Fab-LAP, was expressed in E. coli BL21 (DE3) and purified via nickel affinity chromatography. Radio-TLC analysis was then used to measure conjugation of [18F]-FA to 2G10-Fab-LAP (Table 3). The optimal conditions identified for the LAP peptide (5 μM 2G10-Fab-LAP, 10 μM LplA) gave inconsistent conjugation yields of 49–83%. Doubling the concentration of 2G10-Fab-LAP (10 μM, 2 nmol) and slightly extending the reaction time to 15 min gave reliably high conjugation yields (92 ± 7%, n = 4). Pleasingly, 2G10-Fab-wt without an LAP-tag was barely radiofluorinated (3 ± 1%, n = 3) under identical conditions, illustrating the site-specificity of the methodology. Following a standard protocol,34 2G10-Fab-LAP (10 μM) was also radiofluorinated with [18F]-SFB. Radio-TLC measured substantially lower conjugation yields of 22 ± 1.2% (n = 3).
Table 3.
Radiofluorination of 2G10-Fab-LAPa
| protein | [protein]/ μM | [LplA]/ μM | reaction time/ min | yield/%b |
|---|---|---|---|---|
| 2G10-Fab- LAP | 5 | 10 | 10 | 49–83 |
| 2G10-Fab- LAP | 10 | 10 | 15 | 92 ± 7 (n = 4) |
| 2G10-Fab-wt | 10 | 10 | 15 | 3 ± 1 (n = 3) |
General considerations: all reactions performed in 200 μL of PBS + 3 mM ATP + 5 mM Mg(OAc)2 at 30 °C for 10 min and quenched with EDTA (180 mM final concentration) prior to analysis.
Radio-TLC yields measured after treatment of the reaction sample with SDS-PAGE buffer at 95 °C for 5 min. The eluent used was 7:3:0.1 EtOAc:hexanes:acetic acid.
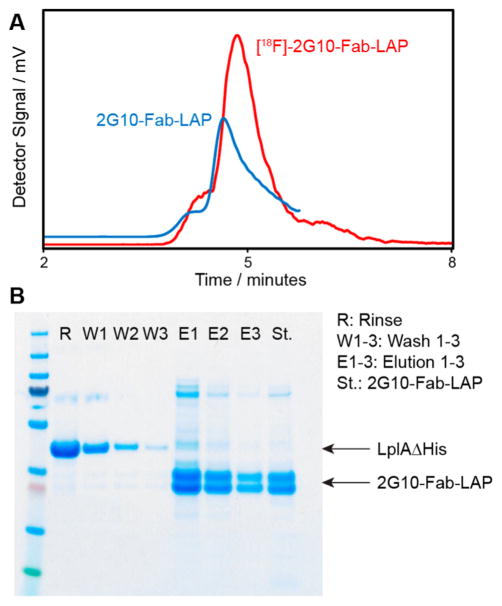
Having demonstrated efficient radiolabeling, we then sought to develop a rapid purification scheme to deliver high purity [18F]-2G10-Fab-LAP for animal studies. It was not possible to separate [18F]-2G10-Fab-LAP from LplA by size-exclusion chromatography, presumably due to their similar hydrodynamic radii, and [18F]-2G10-Fab-LAP did not bind to protein L spin columns after a 10 min incubation at RT. A Myc-epitope tag was then inserted at the C-terminus of the heavy chain immediately following the LAP-tag; however, [18F]-2G10-Fab-LAP-Myc did not bind anti-Myc beads efficiently within our stringent 10 min time window. Our previous experience with 2G10-Fab-LAP informed us that we could use its His6-tag for rapid purification; however, to do so we needed to remove the His6-tag from LplA. For this to be achieved, a TEV protease cleavage site was inserted between LplA and its His6-tag. Once LplA had been isolated from E. coli, but prior to any radiochemistry, it was incubated with TEV overnight at 4 °C to remove the His6-tag. The radiofluorination performance of the resulting enzyme, LplAΔHis, was indistinguishable from the wild type enzyme (data not shown). Following radiofluorina-tion, [18F]-2G10-Fab-LAP bound to nickel beads within the desired 10 min incubation period. Residual LplAΔHis and [18F]-FA were washed off the column, and the purified probe was subsequently eluted in PBS + 250 mM imidazole. Radiotracer purity was confirmed by SEC and SDS-PAGE (Figure 4). Serum stability of [18F]-2G10-Fab-LAP was assessed by incubation in mouse serum for 1 h, and the resulting radioactivity was analyzed by SEC (Figure S4). No release of low molecular weight material consistent with cleavage of [18F]-FA from the protein in serum was observed, suggesting that [18F]-2G10-Fab-LAP is sufficiently stable for use in vivo.
Figure 4.
Analysis of purified [18F]-2G10-Fab-LAP (A) SEC traces of purified [18F]-2G10-Fab-LAP (red, rad-trace) and 2G10-Fab-LAP (blue, UV-trace). (B) SDS-PAGE analysis of fractions from nickel affinity purification of [18F]-2G10-Fab-LAP. After a 10 min incubation, the column was eluted (R) and washed three times with PBS + 25 mM imidazole (W1–3), and then [18F]-2G10-Fab-LAP was eluted with PBS + 250 mM imidazole (E1–3).
We then characterized labeled 2G10-Fab-LAP to measure retention of epitope affinity and confirm site-specific labeling. For these experiments, μM concentrations of nonradioactive [19F]-FA were used to ensure labeling of a high percentage of the 2G10-Fab-LAP present in the sample. Comparison of the binding affinities of [19F]-2G10-Fab-LAP, measured using an Octet instrument, with unlabeled 2G10-Fab-LAP and wild type 2G10-Fab demonstrated complete retention of epitope affinity (Table 4). Pleasingly, only a single conjugation site for [19F]-FA at K245, the lysine within the LAP-tag, was identified by LC/ MS/MS analysis of the labeled peptide following tryptic digestion (Figure S6).
Table 4.
Dissociation Constants, Measured Using an Octet Instrument, for 2G10-Fabs Binding to uPAR
| protein | KD (nM) |
|---|---|
| 2G10-Fab | 38 ± 2.7 |
| 2G10-Fab-LAP | 31 ± 2.2 |
| [19F]-2G10-Fab-LAPa | 31 ± 2.5 |
Labeling conditions: 2G10-Fab-LAP (10 μM), LplAΔHis (50 μM), FA (750 μM), ATP (3 mM), Mg(OAc)2 (5 mM). The resulting solution was incubated at 30 °C for 15 h and then quenched with 100 μL of 360 mM EDTA.
To this point, we had used small quantities for [18F]-FA (~200 μCi) for the radiochemical optimization studies. To establish that the methodology can prepare enough radiotracer for an animal imaging study, we executed the radiolabeling of 2G10-Fab-LAP with 2–3 mCi of [18F]-FA. Initially, radio-TLC reported disappointing yields of 27–38% (Table 5). Increasing both the amount of 2G10-Fab-LAP (10 nmol) and LplAΔHis restored high conjugation yields (95 ± 7%, n = 4). In summary, starting with 2.78–3.02 mCi of [18F]-FA, 1.19–1.62 mCi of [18F]-2G10-Fab-LAP was isolated following purification (69 ± 12% decay-corrected yield). Analysis of the purified sample using the BCA assay demonstrated ~100% recovery of 2G10-Fab-LAP (~10 nmol protein). The total conjugation process, including purification, lasted 55–60 min, and the specific activity of the generated radiotracer was 119–162 Ci/mmol.
Table 5.
Optimization of 2G10-Fab-LAP Radiofluorination Using 2–3 mCi of [18F]-FAa
| [2G10-Fab- LAP]/μM | [LplAΔHis]/ μM | reaction volume/μL | amount of [18F]-FA/mCi | yieldb |
|---|---|---|---|---|
| 10 | 10 | 200 | 0.35–0.55 | 92 ± 7 |
| 10 | 10 | 200 | 2.0–2.3 | 27–38 |
| 10 | 50 | 200 | 2.7–2.8 | 19 ± 3 |
| 25 | 50 | 400 | 2.7 | 95 ± 7 |
General considerations: all reactions performed in PBS + 3 mM ATP + 5 mM Mg(OAc)2 at 30 °C for 15 min and quenched with EDTA (180 mM final concentration) prior to analysis.
Radio-TLC yields measured after treatment of the reaction sample with SDS-PAGE buffer at 95 °C for 5 min. The eluent used was 7:3:0.1 EtOAc:hexanes:acetic acid.
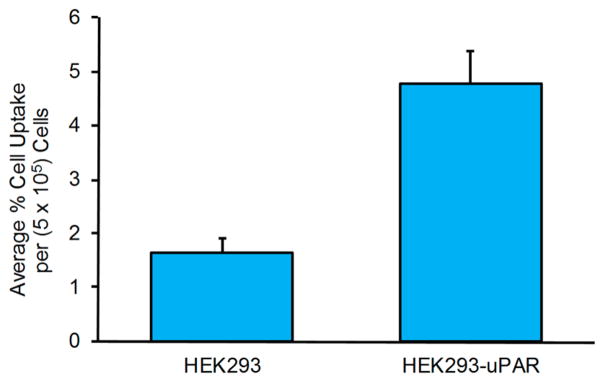
Finally, we tested the ability of [18F]-2G10-Fab-LAP to measure uPAR expression in a biologically relevant setting. We used an isogenic pair of HEK293 sublines that differed only in uPAR expression.31 The wild type cells do not express uPAR; in contrast, HEK293-uPAR cells have been transfected to stably express this epitope at their cell surface. We compared the uptake of [18F]-2G10-Fab-LAP by HEK293 and HEK293-uPAR cells in vitro. For this, ~1 μCi of the radiotracer was incubated on the cells at 37 °C for 45 min in PBS, and the percentage of radioactivity that remained bound to the cells following a washing step was measured using a gamma counter. As shown in Figure 5, the HEK293-uPAR cells had approximately 3-times more associated radioactivity than that of the wild type control (4.77 ± 0.61% vs 1.63 ± 0.23%, p = 0.0028).
Figure 5.
In vitro testing of [18F]-2G10-Fab-LAP shows preferential uptake by uPAR-positive cells; ~1 μCi of [18F]-2G10-Fab-LAP was incubated on either HEK293 (uPAR-negative) or HEK293-uPAR (uPAR-positive) cells at 37 °C for 45 min in PBS prior to measuring cell-associated radioactivity. Significantly higher uptake was observed in the HEK293-uPAR cell line (p = 0.0028).
Discussion
In this manuscript, we report a new approach for the efficient radiofluorination of a biomolecule using the enzyme LplA. We have shown that LplA recognizes the unnatural substrate [18F]-FA and conjugates it site-specifically to its peptide substrate LAP (Figure 6). The biochemistry was highly efficient, proceeding under mild conditions (pH 7.4, 30 °C) with conjugation yields of >80% after short 10–15 min incubations. Moreover, we defined a rapid scheme to purify the radiolabeled Fab away from LplA. We found that the LAP and the 19F-LAP prosthesis did not diminish the immunoreactivity and affinity of an Fab for its epitope on uPAR. Finally, the radiochemistry was adaptable to the scale required for animal imaging studies, and the radiolabeled construct was stable in mouse serum ex vivo. Pleasingly, [18F]-2G10-Fab-LAP was highly effective at detecting cell surface uPAR expression in cellulo, as demonstrated by the uptake in HEK293-uPAR cells being nearly 3-fold higher compared to that in the wild-type negative control.
Figure 6.
Schematic overview of enzymatically catalyzed radiofluorination of protein using LplA and [18F]-FA.
Several aspects of our methodology represent an improvement over the chemical radiolabelings that have dominated the field to date. For instance, we observed significantly lower yields (22%) of radiolabeled 2G10-Fab-LAP using [18F]-SFB, the current gold standard, as the fluorinating agent. Our yield with [18F]-SFB is also very typical of what has been previously reported in the field.34,35 Beyond this consideration, our enzymatic approach results in site-specific bioconjugation, which greatly improves the reproducibility of the radiotracer preparation and reduces the likelihood that radiolabeling will compromise the pharmacology of the respective biomolecule. Our studies also highlighted some limitations of the current methodology, in particular, the lengthy radiosynthesis of [18F]-FA. This likely limited the specific activity of [18F]-2G10-Fab-LAP we were able to achieve (119–162 Ci/mmol), although it should be noted that these values are consistent with those previously reported with [18F]-SFB.34 A degree of expertise in molecular biology is also required to generate the necessary LAP-tagged antibody fragments and LplAΔHis, which may partially limit the methodology’s accessibility.
Enzymes have been incorporated into radiofluorination schemes recently reported by other groups. Rashidian et al. used a sortase to introduce a tetrazine moiety to an antibody fragment, which subsequently enabled radiolabeling with an [18F]-trans-cyclooctene prosthetic.36 The tetrazine ligation is one of the most efficient bioconjugation reactions known, and yields of ~90% were reported using 6 nmol of protein. Encouragingly, the conjugation yields herein are comparable (95% using 10 nmol of protein). Thompson et al. used a fluorinase enzyme to radiofluorinate a nucleotide-tagged RGD short-peptide, also achieving excellent yields albeit with relatively large amounts of peptide precursor (~80 nmol).37 A more systematic comparison between these technologies is warranted to establish their relative strengths and weaknesses, and we are currently working toward this end.
Being a close structural analogue of a known LplA substrate, [18F]-FA was a logical choice for proof-of-concept. However, we found the radiosynthesis to be relatively lengthy, and [18F]-FA can be challenging to work with given its volatility. On this basis, we are now actively working to refine its structure to reduce radiosynthesis time and improve the ease of handling. Because LplA is tolerant of structural variation in its substrates, we are optimistic that we can improve upon [18F]-FA without impairing enzymatic activity and conjugation yields. Another implication of LplA’s substrate plasticity is that we may be able to incorporate molecules bearing larger radioisotopes with longer half-lives (e.g., Iodine-124, radiometals), which would allow for radiolabeling and imaging studies with full length IgGs.
CONCLUSIONS
We have developed an enzymatic radiofluorination that uses LplA to directly conjugate an [18F]-prosthetic site specifically to a protein. Our methodology has several advantages compared to traditional chemical [18F]-bioconjugations. The labeling is rapid and high yielding under mild, aqueous conditions and with minimal amounts of protein substrate (1–10 nmol). The mild conditions and site specificity preserve the epitope affinity of delicate proteins. In addition, the serum stability of the construct and the ability to scale to mCi amounts suggest animal and human imaging is feasible.
EXPERIMENTAL SECTION
Labeling of LAP Peptide with Nonradioactive [19F]-FA
The following stock solutions were generated in PBS: LAP peptide (600 μM), [19F]-FA (7.5 mM), LplA (~40–100 μM), ATP (30 mM), and Mg(OAc)2 (50 mM). Each reagent was diluted to the appropriate final concentration in PBS, and the resulting solution was incubated at 30 °C. At specified time points, 100 μL aliquots were withdrawn and diluted with 100 μL of 360 mM EDTA. Then, 99 μL of this solution was analyzed via RP-HPLC using a 20 min 30–60% gradient of MeCN in H2O (plus 0.1% TFA). For preparing a sample of [19F]-LAP-FA for ESI-MS analysis, a 1 mL labeling reaction was purified via SP-HPLC using the same gradient. ESI-MS (m/z): calcd (C84H118N15O25F.H+) 1756.92; found 1756.85.
Synthesis of [18F]-FA
[18F]-Fluoride (100–500 mCi) was eluted off an ORTG cartridge using 0.5 mL of a K2.2.2/K2CO3 solution (12.6 mg mL−1 of K2.2.2, 2 mg mL−1 of K2CO3, 9:1 v:v MeCN:H2O). The resulting solution was subjected to 3× drying cycles at 110 °C under a gentle stream of nitrogen. Tosylate 1 (~2 mg) was dissolved in anhydrous MeCN (300 μL) and added to the dried [18F]-fluoride mixture, and the resulting solution was sealed and heated at 90 °C for 10 min. It was then diluted to ~5 mL with H2O and purified via semipreparative RP-HPLC (60–90% gradient of MeCN in H2O plus 0.1% TFA; product eluted at ~17 min). Purified 2 was diluted to ~30 mL with H2O and loaded onto a C18-light sep-pak. The sep-pak was washed with H2O (10 mL), and then the activity was eluted in 5 N KOH (1 mL). The resulting solution was heated at 90 °C for 10 min and then cooled for 1–2 min over ice. The solution was neutralized with acetic acid (750 μL) and diluted to ~30 mL with H2O, and [18F]-FA was loaded onto an Oasis HLB sep-pak. The sep-pak was then washed with H2O (10 mL), and [18F]-FA was then eluted in MeCN (2 mL). This solution was then concentrated for 1 h at 50 °C under reduced pressure, and [18F]-FA was then dissolved in PBS for use in subsequent radiolabeling studies. RP-HPLC analysis of [18F]-FA used 45:65:0.1 v:v:v MeCN:H2O:TFA as the eluent (1 mL/min).
Radiolabeling of LAP Peptide and 2G10-Fab-LAP
The stock solutions of LAP peptide or 2G10-Fab-LAP, LplA or LplAΔHis, and ATP/Mg(OAc)2 were diluted to the appropriate concentration in PBS (200 μL final volume). [18F]-FA (0.2–3 mCi) in PBS was added, and the resulting solution was incubated at 30 °C for 10–15 min. EDTA (360 mM, 200 μL) was added to quench LplA activity. An aliquot (3 μL) was withdrawn from the quenched reaction and added to SDS-PAGE reducing buffer (1 μL) and then heated at 95 °C for 5 min. This solution was then analyzed via radio-TLC (eluent of 7:3:0.1 EtOAc:hexanes:acetic acid). RP-HPLC analysis of reaction mixtures used 45:65:0.1 v:v:v MeCN:H2O:TFA as the eluent (1 mL/min). SEC analysis used aqueous solutions of 100 mM sodium phosphate (pH 6.8) and 300 mM NaCl (2 mL/min).
Radiolabeling of 2G10-Fab-LAP with [18F]-SFB
A solution of 2G10-Fab-LAP (10 μM) and [18F]-SFB (~200 μCi) in 50 mM sodium borate buffer (pH 8.5) was heated at 40 °C for 10 min. The reaction mixture was then analyzed via radio-TLC (eluent of 7:3:0.1 EtOAc:hexanes:acetic acid) to determine the conjugation yield.
Purification of [18F]-2G10-Fab-LAP
The reaction solution was diluted with 100 mM imidazole in PBS to a final concentration of 10 mM imidazole. The solution was then loaded onto a nickel-affinity spin column (Thermo Fisher Scientific, Waltham MA). The column was washed 3 times with 25 mM imidazole in PBS (400 μL each) before elution with 3 × 250 mM imidazole in PBS (400 μL each). [18F]-2G10-Fab-LAP was present in each of the three elutions but was most concentrated in the first one. The concentration of 2G10-Fab in each sample was determined using the BCA protein assay kit (Thermo Fisher Scientific Life Technologies, Waltham, MA) following 5-fold dilution with PBS.
Serum Stability Studies
[18F]-2G10-Fab-LAP (~400 μCi) was added to mouse serum (1 mL) and incubated at 37 °C for 1 h. MeCN (1 mL) was added, and the resulting suspension was centrifuged at 2000 rpm for 5 min. Then, ~1 mL of the supernatant was filtered through a 0.45 μM filter, and the resulting solution was analyzed via SEC and radio-TLC using conditions previously described (vide supra).
Measurement of [19F]-2G10-Fab-LAP Affinity for uPAR
Stocks solutions were diluted to the following concentrations in 100 μL of PBS: 2G10-Fab-LAP (10 μM), LplAΔHis (50 μM), FA (750 μM), ATP (3 mM), and Mg(OAc)2 (5 mM). The resulting solution was incubated at 30 °C for 15 h and then quenched with 100 μL of 360 mM EDTA. Kinetic constants for this sample, along with 2G10 and 2G10-Fab-LAP, were determined using an Octet RED384 instrument (ForteBio). Four concentrations of each Fab (500, 250, 100, and 50 nM) were tested for binding to the biotinylated antigen (human uPAR) immobilized on ForteBio streptavidin SA biosensors. All measurements were performed at RT in 384-well microplates, and the running buffer was PBS with 0.1% (w/v) bovine serum albumin (BSA) and 0.02% (v/v) Tween 20. Biotinylated human uPAR was loaded for 180 s from a solution of 150 nM; the baseline was equilibrated for 60 s, and then the Fabs were associated for 120 s followed by 300 s dissociation. Between each Fab sample, the biosensor surfaces were regenerated three times by exposing them to 10 mM glycine pH 1.5 for 5 s followed by PBS for 5 s. Data were analyzed using a 1:1 interaction model on the ForteBio data analysis software 8.2.
Mass Spectrometry Analysis of [19F]-2G10-Fab-LAP
Stocks solutions were diluted to the following concentrations in 100 μL of PBS: 2G10-Fab-LAP (10 μM), LplAΔHis (50 μM), FA (10 μM), ATP (3 mM), and Mg(OAc)2 (5 mM). The resulting solution was incubated at 30 °C for 30 min and then quenched with 100 μL of 360 mM EDTA. [19F]-2G10-Fab-LAP was purified by SDS-PAGE and analyzed by the Bio-Organic Biomedical Mass Spectrometry Resource at UCSF (see Supporting Information for experimental details of analysis).
Cell Culture
HEK293 cells were purchased from the UCSF cell culture facility. HEK293-GPI-uPAR cells were provided by the Craik laboratory. Both cell lines were maintained at 37 °C and 5% CO2 in DMEM media supplemented with 10% FBS, 100 U/ml of penicillin, and 100 μg/mL of streptomycin.
Cell Uptake Studies
Cells were seeded at a density of 4 × 105 cells per well in 12-well plates (Corning, USA) and grown at 37 °C for 24 h. To dissociate endogenously produced uPA from uPAR, the cells were subjected to a mild acid wash protocol as follows: Earle’s balanced salt solution (EBSS), 10 mM HEPES pH 7.4 for 2 min; 50 mM glycine-HCl, 100 mM NaCl pH 3.0 for 15 min; and 0.5 M HEPES, 0.1 M NaCl, pH 7.5 for 2 min. The cells were then washed three times with EBSS, 10 mM HEPES, pH 7.4. All of the steps in the acid wash procedure were executed at 4 °C. Then, ~1 μCi of [18F]-2G10-Fab-LAP in PBS (1 mL) was added to each well, and the cells were incubated at 37 °C for 45 min. Following incubation, the cells were washed twice with ice cold PBS, which was retained for analysis (externalized fraction). The cells were lysed with 1 M NaOH (1 mL), which was also collected and retained for analysis (cell associated fraction). The radioactivity associated with all fractions was measured using a γ-counter (Wizard2, PerkinElmer), and the percentage of cell uptake was calculated from these values (cell associated fraction/ externalized fraction). Experiments were performed in triplicate.
Statistical Analysis
All statistical analysis of significance between 2 cohorts of data was performed using 1-tailed Student’s t-test assuming 2 separate populations with unequal variances.
Supplementary Material
Acknowledgments
The plasmid pYFJ16 for the expression of LplA-His6 was a kind gift from John Cronan (University of Illinois). [18F]-SFB was synthesized by Dr. Kiel Neumann (UCSF). Mass spectrometry analysis of [19F]-2G10-Fab-LAP was provided by the Bio-Organic Biomedical Mass Spectrometry Resource at UCSF (A.L. Burlingame, Director) supported by the Biomedical Technology Research Centers program of the NIH National Institute of General Medical Sciences, NIH NIGMS 8P41GM103481 and Howard Hughes Medical Institute. C.R.D., C.T., and M.J.E. were supported by the National Cancer Institute (R00CA172695, R01CA176671) and the Resource Allocation Program at UCSF. M.J.E. was supported by a Young Investigator Award from the Prostate Cancer Foundation. C.R.D. was supported by a seed grant from the Department of Radiology and Biomedical Imaging at UCSF. C.S.C. and N.S. were supported by the National Institute of Health (1P41CA1962276-01). C.S.C. and H.V.B. were supported by the Department of Defense (W81XWH-12-1-0440). H.V.B. was supported by the Department of Energy (DOE DE-SC002061).
Footnotes
Notes
The authors declare no competing financial interest.
The Supporting Information is available free of charge on the ACS Publications website at DOI: 10.1021/acschem-bio.6b00172.
Generic experimental considerations, synthetic chemistry, 1H NMR data, protein expression and purification information, RP-HPLC, ESI-MS, radio-TLC, SEM, and LC/MS/MS analyses, and kinetic binding data (PDF)
References
- 1.Jacobson O, Kiesewetter DO, Chen X. Fluorine-18 Radiochemistry, Labeling Strategies and Synthetic Routes. Bioconjugate Chem. 2015;26:1–18. doi: 10.1021/bc500475e. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Vaidyanathan G, Zalutsky MR. Labeling Proteins with Fluorine- 18 Using 4-[18F] Fluorobenzoate. Nucl Med Biol. 1992;19:275–281. doi: 10.1016/0883-2897(92)90111-b. [DOI] [PubMed] [Google Scholar]
- 3.Vaidyanathan G, Zalutsky MR. Synthesis of N-succinimidyl 4-[18F]fluorobenzoate, an agent for labeling proteins and peptides with 18F. Nat Protoc. 2006;1:1655–1661. doi: 10.1038/nprot.2006.264. [DOI] [PubMed] [Google Scholar]
- 4.Jeon J, Shen B, Xiong L, Miao Z, Lee KH, Rao J, Chin FT. Efficient method for site-specific 18F-labeling of biomolecules using the rapid condensation reaction between 2-cyanobenzothiazole and cysteine. Bioconjugate Chem. 2012;23:1902–1908. doi: 10.1021/bc300273m. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Boutureira O, Bernardes GJL, D’Hooge F, Davis BG. Direct radiolabeling of proteins at cysteine using [18F]-fluorosugars. Chem Commun. 2011;47:10010–10012. doi: 10.1039/c1cc13524d. [DOI] [PubMed] [Google Scholar]
- 6.Berndt M, Pietzsch J, Wuest F. Labeling of low-density lipoproteins using the 18F-labeled thiol-reactive reagent N-[6-(4-[18F]fluorobenzylidene)aminooxyhexyl]maleimide. Nucl Med Biol. 2007;34:5–15. doi: 10.1016/j.nucmedbio.2006.09.009. [DOI] [PubMed] [Google Scholar]
- 7.Gill HS, Tinianow JN, Ogasawara A, Flores JE, Vanderbilt AN, Raab H, Scheer JM, Vandlen R, Williams SP, Marik J. A modular platform for the rapid site-specific radiolabeling of proteins with 18F exemplified by quantitative positron emission tomography of human epidermal growth factor receptor 2. J Med Chem. 2009;52:5816–5825. doi: 10.1021/jm900420c. [DOI] [PubMed] [Google Scholar]
- 8.Namavari M, Padilla De Jesus O, Cheng Z, De A, Kovacs E, Levi J, Zhang R, Hoerner JK, Grade H, Syud FA, Gambhir SS. Direct site-specific radiolabeling of an affibody protein with 4-[18F]fluorobenzaldehyde via oxime chemistry. Mol Imaging Biol. 2008;10:177–181. doi: 10.1007/s11307-008-0142-7. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Cheng Z, De Jesus OP, Namavari M, De A, Levi J, Webster JM, Zhang R, Lee B, Syud Fa, Gambhir SS. Small-animal PET imaging of human epidermal growth factor receptor type 2 expression with site-specific 18F-labeled protein scaffold molecules. J Nucl Med. 2008;49:804–813. doi: 10.2967/jnumed.107.047381. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Liu S, Hassink M, Selvaraj R, Yap L, Park R, Wang H, Chen X, Fox JM, Li Z, Conti PS. Efficient 18F Labeling of Cysteine-Containing Peptides and Proteins Using Tetrazine–Trans-Cyclooctene Ligation. Mol Imaging. 2013;12:121–128. [PMC free article] [PubMed] [Google Scholar]
- 11.Flavell RR, Kothari P, Bar-Dagan M, Synan M, Vallabhajosula S, Friedman JM, Muir TW, Ceccarini G. Site-specific (18)F-labeling of the protein hormone leptin using a general two-step ligation procedure. J Am Chem Soc. 2008;130:9106–9112. doi: 10.1021/ja801666z. [DOI] [PubMed] [Google Scholar]
- 12.Way JD, Bergman C, Wuest F. Sonogashira cross-coupling reaction with 4-[18F]fluoroiodobenzene for rapid 18F-labelling of peptides. Chem Commun. 2015;51:3838–3841. doi: 10.1039/c5cc00182j. [DOI] [PubMed] [Google Scholar]
- 13.Boutureira O, D’Hooge F, Fernández-Gonzaález M, Bernardes GJL, Sánchez-Navarro M, Koeppe JR, Davis BG. Fluoroglycoproteins: ready chemical site-selective incorporation of fluorosugars into proteins. Chem Commun (Cam-bridge, U K) 2009;46:8142–8144. doi: 10.1039/c0cc01576h. [DOI] [PubMed] [Google Scholar]
- 14.Gao Z, Gouverneur V, Davis BG. Enhanced aqueous suzuki-miyaura coupling allows site-specific polypeptide (18)F-labeling. J Am Chem Soc. 2013;135:13612–13615. doi: 10.1021/ja4049114. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Rahim MK, Kota R, Haun JB. Enhancing Reactivity for Bioorthogonal Pretargeting by Unmasking Antibody Conjugated trans-Cyclooctenes. Bioconjugate Chem. 2015;26:352–360. doi: 10.1021/bc500605g. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Zeglis BM, Brand C, Abdel-Atti D, Carnazza KE, Cook BE, Carlin S, Reiner T, Lewis JS. Optimization of a Pretargeted Strategy for the PET Imaging of Colorectal Carcinoma via the Modulation of Radioligand Pharmacokinetics. Mol Pharmaceutics. 2015;12:3575–3587. doi: 10.1021/acs.molpharmaceut.5b00294. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Zeglis BM, Sevak KK, Reiner T, Mohindra P, Carlin SD, Zanzonico P, Weissleder R, Lewis JS. A pretargeted PET imaging strategy based on bioorthogonal Diels-Alder click chemistry. J Nucl Med. 2013;54:1389–1396. doi: 10.2967/jnumed.112.115840. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Glaser M, Iveson P, Hoppmann S, Indrevoll B, Wilson A, Arukwe J, Danikas A, Bhalla R, Hiscock D. Three methods for 18F labeling of the HER2-binding affibody molecule Z(HER2:2891) including preclinical assessment. J Nucl Med. 2013;54:1981–1988. doi: 10.2967/jnumed.113.122465. [DOI] [PubMed] [Google Scholar]
- 19.Su X, Cheng K, Jeon J, Shen B, Venturin GT, Hu X, Rao J, Chin FT, Wu H, Cheng Z. Comparison of Two Site-Specifically 18F-Labeled Affibodies for PET Imaging of EGFR Positive Tumors. Mol Pharmaceutics. 2014;11:3947–3956. doi: 10.1021/mp5003043. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Cohen JD, Zou P, Ting AY. Site-specific protein modification using lipoic acid ligase and bis-aryl hydrazone formation. ChemBioChem. 2012;13:888–894. doi: 10.1002/cbic.201100764. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Cohen JD, Thompson S, Ting AY. Structure-guided engineering of a Pacific Blue fluorophore ligase for specific protein imaging in living cells. Biochemistry. 2011;50:8221–8225. doi: 10.1021/bi201037r. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Fernández-Suaárez M, Baruah H, Martínez-Hernández L, Xie KT, Baskin JM, Bertozzi CR, Ting AY. Redirecting lipoic acid ligase for cell surface protein labeling with small-molecule probes. Nat Biotechnol. 2007;25:1483–1487. doi: 10.1038/nbt1355. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Liu DS, Tangpeerachaikul A, Selvaraj R, Taylor MT, Fox JM, Ting AY. Diels-Alder cycloaddition for fluorophore targeting to specific proteins inside living cells. J Am Chem Soc. 2012;134:792–795. doi: 10.1021/ja209325n. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Uttamapinant C, White KA, Baruah H, Thompson S, Fernández-suárez M, Puthenveetil S, Ting AY. A fluorophore ligase for site-specific protein labeling inside living cells. Proc Natl Acad Sci U S A. 2010;107:10914–10919. doi: 10.1073/pnas.0914067107. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Puthenveetil S, Liu DS, White KA, Thompson S, Ting AY. Yeast display evolution of a kinetically efficient 13-amino acid substrate for lipoic acid ligase. J Am Chem Soc. 2009;131:16430–16438. doi: 10.1021/ja904596f. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Perham RN. Biochemistry. 1991;30:8501–8512. doi: 10.1021/bi00099a001. [DOI] [PubMed] [Google Scholar]
- 27.Baruah H, Puthenveetil S, Choi Y, Shah S, Ting AY. An Engineered Aryl Azide Ligase for Site-Specific Mapping of Protein – Protein Interactions through Photo-Cross-Linking. Angew Chem, Int Ed. 2008;47:7018–7021. doi: 10.1002/anie.200802088. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Green DE, Morris TW, Green J, Cronan JE, Guest JR. Purification and properties of the lipoate protein ligase of Escherichia coli. Biochem J. 1995;309(Pt 3):853–862. doi: 10.1042/bj3090853. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Nagatsugi F, Sasaki S, Maeda M. 8-[18F]-fluorooctanoic acid and its beta-substituted derivatives as potential agents for cerebral fatty acid studies: synthesis and biodistribution. Nucl Med Biol. 1994;21:809–817. doi: 10.1016/0969-8051(94)90160-0. [DOI] [PubMed] [Google Scholar]
- 30.Lebeau AM, Duriseti S, Murphy ST, Pepin F, Hann B, Gray JW, Vanbrocklin HF, Craik CS. Targeting uPAR with Antagonistic Recombinant Human Antibodies in Aggressive Breast Cancer. Cancer Res. 2013;73:2070–2081. doi: 10.1158/0008-5472.CAN-12-3526. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Duriseti S, Goetz DH, Hostetter DR, LeBeau AM, Wei Y, Craik CS. Antagonistic anti-urokinase plasminogen activator receptor (uPAR) antibodies significantly inhibit uPAR-mediated cellular signaling and migration. J Biol Chem. 2010;285:26878–26888. doi: 10.1074/jbc.M109.077677. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.LeBeau AM, Sevillano N, King ML, Duriseti S, Murphy ST, Craik CS, Murphy LL, VanBrocklin HF. Imaging the urokinase plasminongen activator receptor in preclinical breast cancer models of acquired drug resistance. Theranostics. 2014;4:267–279. doi: 10.7150/thno.7323. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Nagatsugi F, Sasaki S, Maeda M. Synthesis of ω-fluorinated octanoic acid and its β-substituted derivatives. J Fluorine Chem. 1992;56:373–383. [Google Scholar]
- 34.Cai W, Olafsen T, Zhang X, Cao Q, Gambhir SS, Williams LE, Wu AM, Chen X. PET Imaging of Colorectal Cancer in Xenograft-Bearing Mice by Use of an. J Nucl Med. 2014;48:304–310. [PubMed] [Google Scholar]
- 35.Olafsen T, Sirk SJ, Olma S, Shen CKF, Wu AM. ImmunoPET using engineered antibody fragments: fluorine-18 labeled diabodies for same-day imaging. Tumor Biol. 2012;33:669–677. doi: 10.1007/s13277-012-0365-8. [DOI] [PubMed] [Google Scholar]
- 36.Rashidian M, Keliher EJ, Bilate AM, Duarte JN, Wojtkiewicz GR, Jacobsen JT, Cragnolini J, Swee LK, Victora GD, Weissleder R, Ploegh HL. Noninvasive imaging of immune responses. Proc Natl Acad Sci U S A. 2015;112:6146–6151. doi: 10.1073/pnas.1502609112. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Thompson S, Zhang Q, Onega M, McMahon S, Fleming I, Ashworth S, Naismith JH, Passchier J, O’Hagan D. A localized tolerance in the substrate specificity of the fluorinase enzyme enables “last-step” 18F fluorination of a RGD peptide under ambient aqueous conditions. Angew Chem, Int Ed. 2014;53:8913–8918. doi: 10.1002/anie.201403345. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.