Abstract
While recent studies have revealed higher than anticipated heterogeneity of mutation rate across genomic regions, mutations in exons and introns are assumed to be generated at the same rate. Here we find fewer somatic mutations in exons than expected based on their sequence content, and demonstrate that this is not due to purifying selection. Moreover, we show that it is caused by higher mismatch repair activity in exonic than in intronic regions. Our findings have important implications for our understanding of mutational and DNA repair processes, our knowledge of the evolution of eukaryotic genes, and practical ramifications for the study of the evolution of both tumors and species.
Introduction
Genetic variation in exonic regions is lower than in intronic ones both across species and within populations. This differential exon-intron variation rate is attributed to the action of stronger purifying selection on exonic nucleotide changes, whereas the rate of generation of variants –that precedes the effect of selection– is generally assumed to be overall homogeneous between these two genic regions. This assumption lays at the heart of evolutionary biology and cancer genomics approaches that compare the rate of intronic and exonic variation to estimate the strength of selection acting on coding genes1–5.
Recent studies have shown that the rate of mutations across genomic regions is highly heterogeneous. Replication time6,7, the level of gene expression8, and the degree of chromatin compaction9,10 have been described as features that affect mutation rate at the megabase scale. Our group and others recently demonstrated that the local efficiency of DNA repair is influenced by factors that affect accessibility of the repair machinery11–14.
The assumption that introns and exons suffer a similar basal rate of mutations before the action of purifying selection is a reasonable one because both exonic and intronic regions are replicated at the same time and transcribed equally and, therefore, DNA repair mechanisms associated with the advance of the replication fork, as well as transcription-coupled repair are expected to have equivalent access to both. Nevertheless, several features of the chromatin structure –including some that have been related to the recruitment of DNA repair machineries15–17– vary widely between exons and introns18,19. This motivated us to question the long-standing assumption that introns and exons receive similar rate of mutations before selection.
Somatic mutations detected in tumors20 are an ideal ground to explore whether exonic and intronic variants appear at the same rate. Tumor cells, upon clonal expansion, accumulate somatic mutations at accelerated rates compared to the germline. We demonstrate here that even in the absence of purifying selection, exons receive fewer mutations than expected given their nucleotide composition. We show that this decrease of the exonic mutation burden is detectable across seven tumor types. We also demonstrate that the cause of this reduction is that the Mismatch Repair (MMR) system acts more efficiently in exons than introns, and we propose that this differential repair is caused by the differential positioning of histone marks in these two genic regions.
These findings imply that the differential genetic variation in exonic and intronic regions across species and within populations is caused by a combination of differential sequence context, rate of DNA repair and purifying selection. This possesses ramifications of technical nature for evolutionary methods that rely on the calculation of intronic variation to estimate the strength of selection on genes or to detect cancer driver genes1–3,5,21,22. More generally, these findings have profound implications for our knowledge of gene evolution and DNA repair mechanisms.
Results
Differential distribution of chromatin features in exons and introns
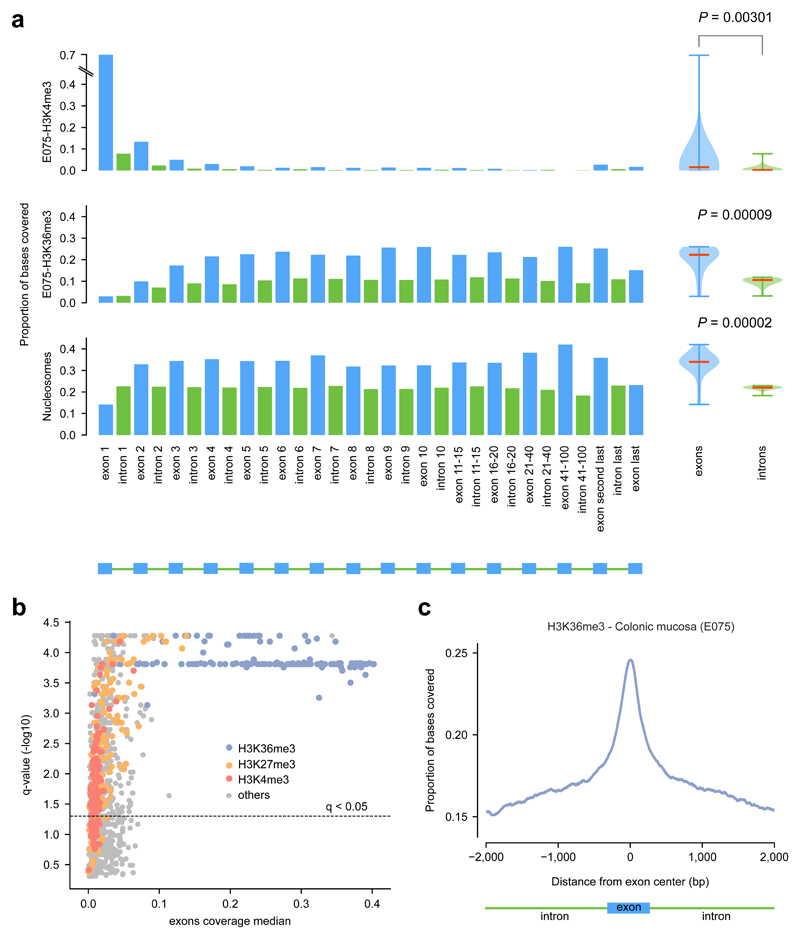
We first sought to identify chromatin features with the most differential distribution between exons and introns, using data generated by the Epigenome Roadmap23 and ENCODE24. We analyzed 32 chromatin features –comprising 30 histone modifications, the presence of a histone variant (H2A.Z) and DNase I hypersensitive site (DHS)– on 127 cell lines and primary cells from different tissue types and nucleosome density obtained in a lymphoblastoid cell line (Supp. Table 1). We computed the coverage (fraction of bases overlapping peaks) of each feature on exons and introns located at different positions along the structure of genes (Fig. 1a illustrates the results of this calculation for three chromatin features; Methods). Then, we defined the difference of the exonic and intronic coverage of each mark in each cell type, as the p-value of the two-tailed Mann-Whitney test of their comparison (boxplots in Fig. 1a). Several chromatin marks exhibited a significant overall difference of exonic and intronic coverage (Figs. 1a and b). In particular, nucleosome density and H3K36me3 are significantly higher in exons than introns across the gene, and H3K36me3 is the histone mark with higher coverage across all exons in the gene. This behavior of H3K36me3 is consistent across the majority of the 127 cell types in the Epigenome Roadmap (Fig. 1b and Supp. Tables 1 and 2). Moreover, the coverage of H3K36me3 decreases steeply at the flanking introns (Fig. 1c). Interestingly, the protein hMutSα of the MMR machinery, involved in the recognition of mismatches, has recently been described as recruited to the chromatin through the interaction of its hMSH6 subunit with the tri-methylated H3K3615,17.
Figure 1. Exonic enrichment for several histone marks.
(a) Exonic and intronic coverage of H3K4me3 and H3K36me3 peaks in primary cells of the normal colon mucosa (E075), and of nucleosome covered regions in GM12878 (lymphoblastoid cell line). Each bar represents the coverage of the mark in exons or introns at different positions of genes, depicted by the schematic structure of the genes at the bottom of the figure. The distribution of the exonic and intronic coverage of each chromatin feature across the genes structure is represented by the boxplots at the right of the panel. The pvalue of a two-tailed Mann-Whitney test comparing the two distributions is shown.
(b) Scatter plot representing the difference of exonic and intronic coverage of each histone mark (log10 two-tailed Mann-Whitney p-value corrected for multiple testing) in the y axis and the median coverage across all exons of genes in the x axis. Each dot represents one chromatin mark in one cell type, colored according to the former. All data on exons and introns coverage of all marks across cell types is available in Supplementary Tables 1 and 2.
(c) Proportion of bases covered by H3K36me3at internal exons and flanking introns in primary cells of the normal colon mucosa (E075).
We therefore hypothesized that the exonic enrichment of certain chromatin features, in particular the H3K36me3, may result in an increased recruitment of the MMR machinery to exons. This, in turn would lead to a reduction in the quantity of exonic mutations with respect to the number of mismatches expected from the exonic sequence content alone.
Internal exons exhibit decreased exonic mutation burden in POLE-mutant tumors
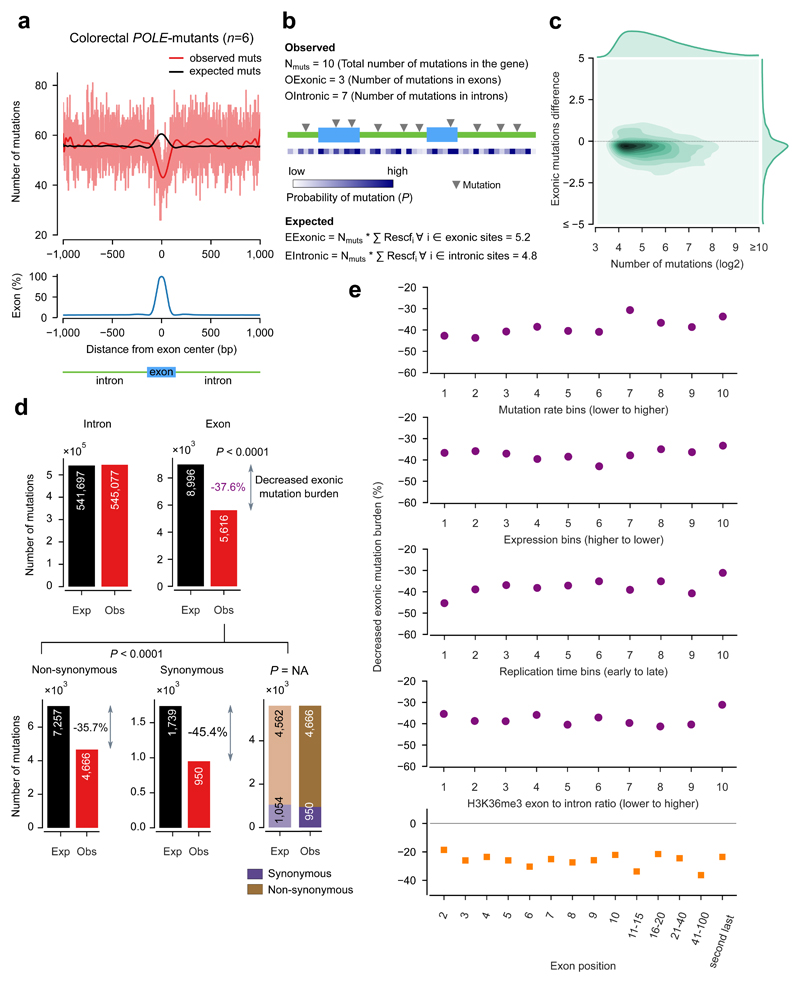
POLE-mutant tumors, due to the decreased proofreading capabilities of the DNA polymerase ε, sustain an important number of mismatches during DNA replication, which hence make a sizable part of their somatic mutations. Therefore, to determine whether the rate of somatic mutations caused by mismatches differs in exonic and intronic regions, we first explored the mutations detected across the whole genomes of 6 colorectal POLE-mutant tumors, sequenced by The Cancer Genome Atlas (TCGA). We stacked exon-centered 2001-nucleotide sequences and compute the mutation burden at each position of this window as the number of mutations overlapping the position. This analysis shows that the mutation burden in positions dominated by exonic sequences is lower than that observed along their flanking intronic regions (Fig. 2a, red line).
Figure 2. Decreased exonic mutation burden in colorectal POLE-mutant tumors.
(a) Exon-centered 2001-nucleotide wide observed and expected profiles of mutations in 6 colorectal POLE-mutant tumors. (The light red line represents the actual mutation rate at each position, while the red and black lines represents smoothed mutation rates using a polynomial fit.) The bottom panel represents the distribution of the percentage of exonic bases at each position across the 2001-nucleotide window.
(b) Schematic representation of the method used to compute observed and expected number of mutations in exons and introns at the gene level. Rescfi represent the rescaled expected frequency of mutations of each nucleotide in the gene. The conditional probability of mutation of each site (P in the figure) is proportional to this quantity. EExonic and Eintronic represent the expected number of exonic and intronic mutations, respectively. (See Methods for details.)
(c) Density plot representation of the distribution of gene-level differences of the numbers of observed and expected exonic mutations vs total mutations in POLE-mutant tumors. The distribution of exonic mutations difference (right-hand one-dimensional density plot) is biased towards negative values, indicating that a majority of genes possess lower-than-expected numbers of mutations in exons. An analogous plot,restricted to genes with at least one expected exonic mutation (Supp. Fig. 2b) shows similar results.
(d) An overall highly-significant 37.6% decreased exonic mutation burden (3368 fewer observed exonic mutations than the 8996 expected) is observed in these tumors (top panel). Both synonymous and non-synonymous mutations account for this decrease (bottom panel), with their observed values significantly below expected (left and middle plots), and no fewer non-synonymous mutations than expected when the latter are computed solely rom observed exonic mutations (right plot).
(e) The decreased exonic mutation burden is maintained around the overall computed value (37.6%) across groups of genes with different mutation rate (top panel), level of expression (second), replication time (third), the number of genic bases covered by H3K36me3 peaks (fourth) and across exons at different positions in the gene (bottom). The values in the abscissa in each graph represent the ordinal number of the bins of genes, sorted in the direction indicated in each panel.
The mutation probability at individual DNA positions is influenced by their sequence context. Therefore, differences in nucleotide composition between exons and introns could provide a plausible explanation for the observed difference of exonic and intronic mutation counts. To compute the expected mutation burden at each position of the 2001-nucleotide exon-centered window, we distributed the mutations observed in each sequence in the stack taking into account the conditional probability that each of its 2001 positions was mutated given its 5’ and 3’ bases. This sequence-wise distribution of expected mutations (details in methods) avoids potential biases resulting from aggregating genic regions with different mutation rate and exon/intron proportions (Supp. Fig. 1). The distribution of these synthetically generated ‘expected’ mutations in POLE-mutants across exons and their flanking introns shows that more mutations are expected in exons than in introns, as represented by the black line in Figure 2a.
We then set out to compare the number of observed exonic mutations to their expected quantity in POLE-mutant tumors and to assess the statistical significance of the deviation between the two (Fig. 2b; Methods). We carried out this comparison at the level of individual genes, to guarantee that its results are free of the aforementioned caveat. (Known cancer genes25,26 were excluded from this and subsequent analyses to eliminate any deviation due to positive selection.) First, we randomly distributed a number of mutations equal to that observed in each gene across its exons and introns following the probability of each nucleotide to be mutated. A second method to obtain their expected mutation burden based on permutations of observed mutations yielded similar results. (Methods, Supp. Fig. 2 and Supp. Table 3). We then computed the difference between the observed and expected mutation burden of each gene (Fig. 2c). Most genes (77%) possess fewer exonic mutations than expected from their sequence content, resulting in a negative difference. After aggregating the number of observed and expected mutations across all genes (Fig. 2d), we discovered that while internal exons bear only 5616 mutations in the six POLE-mutant tumors, 8996 exonic mutations were expected, given i) the total number of genic mutations, ii) the nucleotide composition of exons and introns, and iii) the mutational processes operating in these tumors. This represents a decrease of 37.5% of the observed exonic mutation burden with respect to the expected. Employing a likelihood-based statistical approach (details in Methods) we found this decrease to be statistically significant (p-value < 0.0001). We have named this phenomenon decreased exonic mutation burden, and globally we quantify it as the percent of decrease with respect to the expected mutation burden.
We next tested whether the decreased exonic mutation burden was due to increased selective pressure on exons resulting in purifying selection of mutations in these regions during tumor evolution. To determine the impact of purifying selection on the exonic mutation burden, we separated exonic mutations on the basis of their consequence type. We found that the 5616 exonic mutations in the 6 POLE-mutants yielded 950 synonymous, and 4666 non-synonymous mutations. If the decreased exonic mutation burden were caused by purifying selection, we would expect it to consist mostly of a decrease of non-synonymous mutations. Nevertheless, when redistributing genic mutations across intronic, synonymous and non-synonymous sites according to their mutational probabilities, we found a 35.7% decrease of non-synonymous mutations, along with a 45.4% decrease of synonymous mutations (Fig. 2d; p-value<0.0001). On the other hand, when redistributing solely exonic mutations based on their mutational probability we found that the expected number of non-synonymous mutations is very close to their actual observed number: 4562 (with the remaining 1054 expected to yield synonymous variants). In other words, there are not significantly fewer non-synonymous mutations than expected from the potential non-synonymous sites in exons (non-significant p-value). The results of these two tests support the conclusion that the decrease of exonic mutation burden is not due to negative selection (Fig. 2d). This result is maintained across bins of genes with different mutation rate, and is observable for all individual POLE-mutant tumors (Supp. Tables 4 and 5).
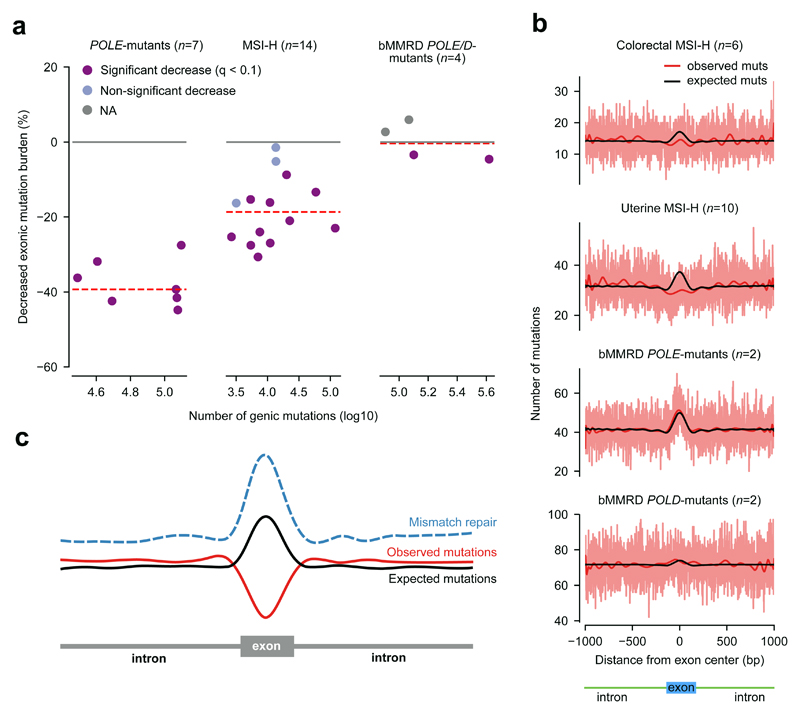
We then checked that the decreased exonic mutation rate was not driven by a subset of genes at either extreme of the mutation rate range. To do this, we binned the genes into 10 groups of increasing mutation rate (Fig. 2e, top panel). We then aggregated the mutations of the genes in each bin and confirmed that the decreased exonic mutation burden remains around 40% across all bins (top panel). Finally, we demonstrated that very similar values of decreased exonic mutation burden are observed across groups of genes with increasing replication time, expression level, H3K36me3 coverage, and also across exons at different positions along the gene (Fig. 2e, second to bottom panel). Furthermore, the decrease of exonic mutation burden is not driven by one or few POLE-mutant tumors, as it is observable and significant for each of them (Fig. 3a; this analysis includes also a POLE-mutant of uterine adenocarcinoma origin).
Figure 3. Decreased exonic mutation burden across scenarios of MMR activity.
(a) Tumor-level decreased exonic mutation burden in colorectal and uterine POLE-mutant (left), MSI-H (center), and bMMRD (right) tumors. Dots represent individual tumors. Broken red lines represent the median decreased exonic mutation burden of each group of tumors.
(b) Exon-centered 2001-nucleotide wide observed and expected profiles of mutations in 6 colorectal and 10 uterine MSI-H tumors, and 2 POLE-mutant and 2 POLD-mutant bMMRD glioblastomas.
(c) Schematic representation showing the increased efficiency of MMR at exons, and the decreased exonic mutation burden.
In summary, we found a significant decrease of the exonic mutation rate in POLE-mutant tumors. This decrease is not due to the sequence content and cannot be explained by negative selection acting on exonic mutations, and it is maintained across genes with all levels of mutation rate and across exons at different positions of the gene.
The decreased exonic mutation rate is caused by differential mismatch repair
We reasoned that the decreased exonic mutation burden observed in POLE-mutant tumors could be caused by an elevated activity of MMR in exons with respect to their neighboring introns. MMR is the main mechanism responsible for the repair of errors generated by the polymerase during DNA replication. Colorectal tumors –and other cancer types– acquire the microsatellite instability (MSI) phenotype when mismatches introduced by the DNA polymerase are not corrected, due to deficiencies in the MMR system27. MSI tumors are normally classified on the basis of the level of five biomarkers into MSI-H (high, with over 40% of the biomarkers of MSI) and MSI-L (low, with less than 40%), although the latter have recently been shown to not significantly differ from microstaellite stable (MSS) tumors in numbers of gained microsatellite alleles28. Thus, if our hypothesis were true, we would expect that tumors with an impaired MMR function (MSI-H) showed lower decreased exonic mutation burden than MMR competent tumors, such as POLE-mutants or MSS tumors.
We proceeded to compute the decreased exonic mutation burden of 6 colorectal and 8 uterine MSI-H tumors in the TCGA cohort. We found, as predicted by our hypothesis, that MSI-H tumors exhibit a decreased exonic mutation burden around 20% (Fig. 3a and b); in other words, close to half of the decrease observed in POLE-mutant MMR-proficient tumors. Several reasons may explain why the decreased exonic mutation burden does not disappear completely in MSI-H tumors. On the one hand, the impairment of the MMR system may not be complete, and it probably has not existed throughout the entire history of the tumor. On the other, alternative mutational processes may also contribute to the mutation load.
Then, we computed the decreased exonic mutation burden of 2 POLE-mutant and 2 POLD-mutant glioblastomas from children with inherited biallelic mismatch repair deficiency (bMMRD) sequenced by The International BMMRD Consortium29. These tumors have been MMR-deficient throughout their entire history and their POLE/D mutations guarantee a preponderance of mismatch-caused mutations. Their decreased exonic mutation burden is indeed close to zero (Fig. 3a and b), with independence of the mutation rate of genes (Supp. Fig. 3 and Supp. Table 4). Mismatches in these tumors are generated at comparable rate as in previously analyzed POLE-mutant tumors. However, the majority of these mismatches remain uncorrected and turn into mutations. In other words, the mutations observed in these tumors follow the pattern of generation of mismatches, corroborating our hypothesis that they appear with higher probability in exons than introns, and that it is the MMR, with its increased efficiency in the former that causes the decreased exonic mutation burden.
In summary, the decreased exonic mutation burden differs between three different scenarios of MMR activity, with higher decrease in MMR proficient tumors to none in MMR deficient tumors. These results indicate that the increased activity of the MMR in exons is the cause of the decrease exonic mutation burden in POLE-mutant tumors (Fig. 3c).
A role for H3K36me3 in the differential activity of MMR in exons and introns
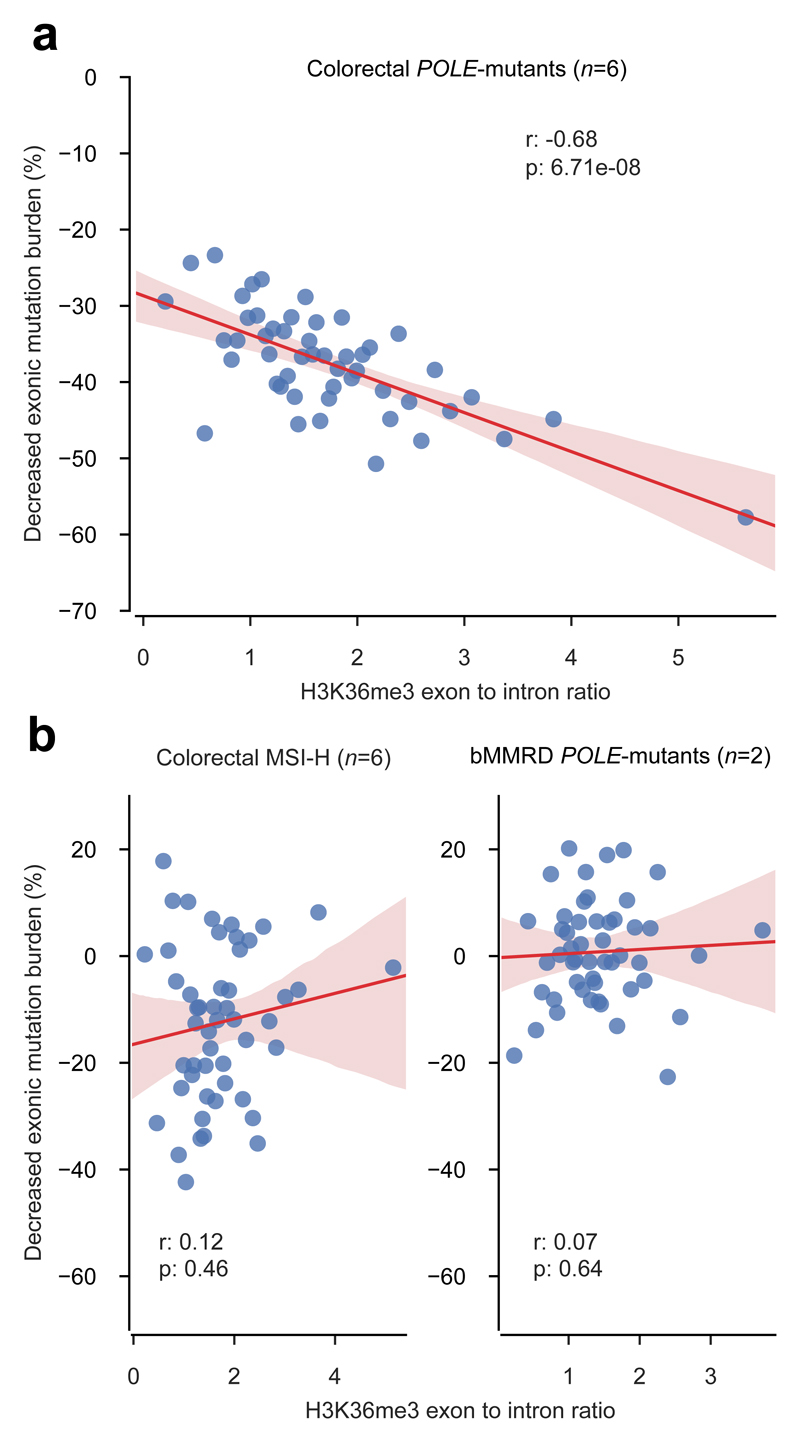
The results of the two previous sections demonstrate that the enhanced efficiency of the MMR system in exons is the cause of the observed decreased exonic mutation burden of colorectal POLE-mutant tumors. On the basis of formerly established mechanistic links between H3K36me3 and the recognition of mismatches, we then hypothesized that the decreased exonic mutation burden could be explained, at least in part, by the exonic enrichment of this histone mark in cells of the colon epithelium (see first section of Results). If true, we should be able to observe the biggest decreased exonic mutation burden in genes with the strongest exonic enrichment for H3K36me3 in MMR-proficient tumors. To test this, we first computed the exon to intron ratio of H3K36me3 readcount of primary cells from the colonic mucosa (Fig. 4a; E07523). Then, we grouped the genes into bins of increasing H3K36me3 exon to intron ratio, and we computed the aggregated decrease of exonic mutation burden of the genes in each bin for POLE-mutant colorectal tumors (Fig. 4a and Supp. Figs. 4 and 5). As predicted by our hypothesis, we found a significant negative correlation between the H3K36me3 exon to intron ratio and the decrease of exonic mutation burden (correlation coefficient, -0.68, p-value=6.7x10-8). A much lower, non-significant correlation (Supp. Fig. 5 bottom panel) is observed between the exon to intron ratio of nucleosomes and the decrease of exonic mutation burden. This suggests that the H3K36me3 histone mark and not just the presence of nucleosomes underpins the increased level of MMR in exons that results in the decreased exonic mutation burden. The correlation with other histone marks is also lower (Supp. Table 6). On the other hand, the negative correlation between H3K36me3 exon to intron ratio and the decreased exonic mutation burden disappears in MSI-H colorectal tumors (correlation coefficient, 0.12, p-value=0.46) and bMMRD tumors (exon to intron H3K36me3 readcount ratio computed from cells of the brain angular gyrus, E067; correlation coefficient, 0.07, p-value=0.64) (Fig 4b).
Figure 4. Decreased exonic mutation burden and H3K36me3 exon to intron ratio.
Decreased exonic mutation burden computed in (a) colorectal POLE-mutant tumors, (b) colorectal MSI-H, and (c) glioblastoma bMMRD tumors for 50 groups of genes with increasing exon to intron ratio of H3K36me3 coverage (in the corresponding cell-of-origin; see Methods). The trendline and its confidence intervals in graphs were added using the seaborn package of python;, while the correlation coefficient and its significance were computed using iteratively re-weighted least squares approach.
These results indicate that the exonic enrichment for H3K36me3, possibly in combination with other chromatin features, could act as a driver of the enhanced MMR activity in exons that ultimately results in the decreased exonic mutation rate of POLE-mutant tumors (Fig. 4d). When cells become MMR deficient, either during tumor evolution (as MSI-H colorectal samples), or before its emergence (like bMMRD glioblastomas), the link between the H3K36me3 exonic enrichment and the decreased exonic mutation burden is thus severed. This results in uncorrected mismatches accumulating, and ultimately mutations appearing more frequently in exons.
Tumors of other cancer types also exhibit decreased exonic mutation rate
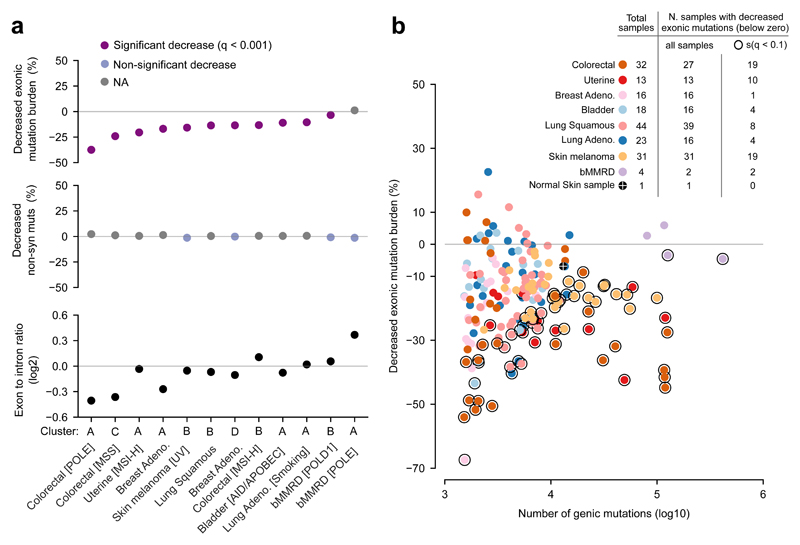
Our observations in previous sections have been limited to colorectal and uterine carcinomas, the mutational spectra of which are dominated by the interplay between the generation of mismatches in the course of DNA replication and their correction by the MMR machinery. The mutational processes of other somatic tissues are dominated by different types of damage dealt with by other DNA repair systems. Nevertheless, somatic cells in a human body, including the gametes, are the result of millions of cell divisions involved in organism development and tissue renewal. Therefore, MMR must play a role –although with different relative contribution– in shaping the mutational processes of all human tissues. We then asked whether tumors originated from other tissues exhibit a decreased exonic mutation rate. To do this, we first clustered the samples of the 8 tumor types in the studied cohort based on their mutational signatures (Supp. Fig. 6).
For the tumors in each cluster, we next computed the decreased exonic mutation burden (Fig. 5a, top panel). All clusters, except the one grouping POLE bMMRD glioblastomas exhibited significant decreased exonic mutation burden. This global trend was corroborated for individual samples (Fig. 5b). Interestingly, we found that the decreased exonic mutation rate is apparent also in the somatic mutations detected in a normal skin sample (Figure 5b, orange dot)30, indicating that this phenomenon is not a pathologic effect caused by tumorigenesis. In none of the clusters could the decreased exonic mutation burden be attributed to negative selection acting on exonic mutations (Fig. 5a, middle panel). We also computed the exon to intron mutation rate ratio as explained in the first section for the chromatin features (see Methods). In coherence with the decreased exonic mutation burden, in most clusters exons showed fewer mutations than their intronic counterparts (bottom panel).
Figure 5. Decreased exonic mutation burden across cancer types.
(a) Top panel: decreased exonic mutation burden of groups of tumors clustered according to their underlying mutational processes. (Dots represent clusters of tumors denoted at the bottom panel.) Middle panel: decreased non-synonymous mutation burden of the same groups of tumors, computed as in Figure 2d. Bottom panel: ratio of exonic and intronic mutation rates of the same groups of tumors. Clusters of tumors in the three panels are sorted following their decreased exonic mutation burden.
(b) Decreased exonic mutation burden of individual tumors vs their mutational burden. Dots representing individual tumors are colored according to their cancer type; dots of tumors with significant decreased exonic mutation burden are encircled by a black ring. The table at the top left corner of the panel presents the total number of samples, the subset of them with decreased exonic mutation burden, and the subset of these with significant decrease.
Strikingly, even melanomas and lung carcinomas, whose mutations arise mostly as a consequence of DNA damage caused by UV light or tobacco, respectively, repaired via the Nucleotide Excision Repair (NER)31,32, exhibit a clear decreased exonic mutation burden. Two explanations are plausible for the pervasive identified decreased exonic mutation rate. The first –as pointed out above– is that, although modest, in relative terms, the MMR still plays a role in DNA repair in these tumors. Nevertheless, a second intriguing possibility is that other DNA repair machineries, also acting with higher efficiency in exons, contribute to the reduced exonic mutation rate. Exploring this prospect in the case of NER in melanomas, we indeed found higher activity in exonic regions (Supp. Fig. 7), although we cannot rule out that this is due to a higher exonic rate of UV-induced damage.
To sum up, the decrease of somatic mutation burden in exonic regions with respect to the expectations and to neighboring introns is apparent across cancer types. While we have demonstrated that the MMR plays a role in shaping this decrease, other DNA repair mechanisms may also contribute to it.
Discussion
In this work we provide, to the best of our knowledge, the first demonstration that the generation of somatic mutations –in the absence of negative selection– is lower in exons than expected given their nucleotide composition. In other words, somatic cells exhibit a decreased exonic mutation burden. We have also shown that the reason is that mismatches in exonic DNA are repaired more efficiently than their intronic counterparts. These results represent a significant contribution to the body of research that in recent years has revealed a higher than anticipated heterogeneity in the mutation rate across different regions of the genome. Several recent seminal studies exploiting whole-genome germline and somatic mutations and the availability of nucleotide resolution maps of DNA repair33,34 have provided glimpses at a complex relationship between chromatin conformation, basic cellular processes like gene expression, DNA replication, the binding of transcription factors, and DNA repair5,7,10–12,14,32,35–39. It is the complicated interplay between these processes which determines that mutations accumulate heterogeneously across the genome. The results we present here reveal that the interaction of the most basic structural feature of eukaryotic genes, namely their segmentation in exons and introns –and its correlative chromatin structural differences– results in these two regions being repaired at very different rates.
As a possible explanation of the mechanisms through which the segmented structure of genes influences the activity of the DNA repair machinery, we have shown a pervasive enrichment of the tri-methylation of H3K36 in exons of normal tissues, which correlates with the decrease of exonic mutation burden in the corresponding tumors. As we show here H3K36me3, possibly in combination with other chromatin features, may participate in shaping the observed depletion of exonic mutations. The enrichment of H3K36me3 for exonic regions –which appears in both germline and somatic tissues– has been proposed to be ultimately responsible for the correct recognition of exon-intron boundaries by the splicing machinery18,19,40. Nevertheless, the H3K36me3 is bound by the MutSα protein via the PWWP domain of its MSH6 subunit15. A concomitant factor may thus have acted in the evolution of H3K36me3-enriched exons. Indeed, our results suggest that the increased recruitment of the MMR machinery to exonic regions as a result of higher levels of this histone mark would result in a reduction of the exonic mutation burden after DNA replication and ultimately, in an increase of fitness.
Our results demonstrate that the decreased exonic mutation burden is not due to negative selection in the generation of cancer somatic mutations across all tumor types analyzed. This finding suggests that the mutational landscape of cancer genes is not strongly influenced by negative selection, in agreement with a recent report41. Nevertheless, we expect that, in the germline, purifying selection plays a predominant role, filtering out all variants that prevent the development of a viable individual42. Given that MMR components are highly conserved across evolution –and that the exonic enrichment for H3K36me3 and other chromatin marks has been observed across species18,43–, it is reasonable to assume that the enhanced exonic MMR observed in human somatic cells is also present in germline cells and in other organisms. Therefore intronic regions could accumulate more nucleotide changes across evolution as a result not only of intense purifying selection on exonic variants but also of this differential repair. This, in turn would bring into question the use of the rates of intronic substitutions (Ki) as a proxy for neutral evolution44–46, with important implications for our understanding of the evolution of genes. Further implications may be extracted for methods aimed at detecting cancer driver genes that model the background mutation rate of exonic elements from their surrounding areas to identify signals of positive selection in the coding region of genes. Some of these methods5,21,22, which use intronic mutations as estimators of the exonic background mutation rate, may be strongly affected by the differential generation of mutations in these regions.
In summary, we demonstrate that the differential MMR in exons and introns in somatic cells causes the former to harbor fewer mutations than expected from their nucleotide composition. This finding advances our knowledge of the interplay between mutational processes and the DNA repair machinery. Moreover, our results have important implications regarding the way we study the forces that shape the development of tumors and our understanding of the evolution of the genome.
Online Methods
Whole-genome expression and mutation data
Whole-genome somatic mutations and expression data of 38 skin cutaneous melanomas (SKCM), 46 lung adenocarcinomas (LUAD), 45 lung squamous cell carcinomas (LUSC), 42 colorectal adenocarcinomas (CRC), 96 breast carcinomas (BRCA), 21 bladder carcinomas (BLCA), 47 uterine corpus squamous cell carcinomas (UCEC), 27 glioblastomas (GBM), 18 low grade gliomas (LGG), 20 prostate adenocarcinomas (PRAD), 34 thyroid carcinomas (THCA) and 27 head and neck squamous cell carcinomas (HNSC) probed by TCGA were obtained from Fredriksson et al. (2014)20. Cohorts of tumors with fewer than 5000 genic mutations or fewer than 1000 exonic mutations (HNSC, GBM, KIRC, THCA, LGG and PRAD) were discarded from the analysis. The somatic mutations detected in four bi-allelic mismatch repair deficient (bMMRD) pediatric glioblastomas sequenced by The International BMMRD Consortium29 were obtained through personal communication from the authors. Finally, we obtained the somatic mutations detected across the whole genome of a normal skin sample from Martincorena et al. (2015)30.
Genomic coordinates of exons and introns
GENCODE47 v19 coordinates for 20,345 protein-coding genes were retrieved. Genes without introns, overlapping genes, and cancer driver genes, according to the Cancer Gene Census and other sources25,26, were discarded, thus obtaining a filtered set of 12,104 genes. All transcripts per gene were merged, generating meta exon and meta intron coordinates. Finally, the 5’ and the 3’ exons were removed, as well as all UTR regions (except for the analysis shown in Fig. 1), thus leaving only internal exons and their flanking introns. We then identified all genic regions where mutation calling would be technically challenging due to low sequence complexity, ambiguous mappability of sequencing reads, or low sequencing coverage. Regions of low complexity or low mappability were obtained from the UCSC genome browser (http://genome.ucsc.edu/cgi-bin/hgFileUi?db=hg19&g=wgEncodeMapability). The former included repetitive regions defined by RepeatMasker, while the latter comprised low unique mappability regions for 36-mer sequences (CRG Alignability 36' Track, score <1). Finally regions covered by fewer than 8 reads in any of five randomly selected tumor samples of each tumor type (requirement to make somatic calling in Fredriksson et al. 201420) were considered of low coverage. Regions of any of these three types were removed from introns and exons.
Clusters of tumors with different somatic mutational processes
To group the tumors of each cancer type in the cohort according to their underlying mutational processes, we first built a matrix of the frequencies of the 96 tri-nucleotide changes across tumors, as in a previous work12. Then, we carried out a hierarchical clustering (using Euclidean distance and average method for computing the similarity between clusters) of this matrix. We then manually separated the clusters of tumors and identified their underlying mutational processes through visual comparison with previously obtained32 mutational signatures across cancer types. Clusters of tumors with fewer than 5000 genic mutations or fewer than 1000 exonic mutations were discarded for downstream analyses.
Chromatin features
We downloaded peak (narrow) coordinates and genome-wide read-coverage of 32 chromatin features presented in Supplementary Table 1 across 127 cell lines and primary cell types from the Epigenome Roadmap23 and the nucleosome density obtained from ENCODE24. Peaks and reads (see below) obtained from http://egg2.wustl.edu/roadmap/data/byFileType/peaks/consolidated/narrowPeak and http://egg2.wustl.edu/roadmap/data/byFileType/alignments/consolidated, respectively of each feature were mapped to intronic and exonic regions of genes. The primary cell closest to colorectal tumors and glioblastomas from the Roadmap were selected to represent the exon-intron distribution of chromatin features. Genome-wide nucleosome positioning signals (density graph) of ENCODE cell line GM12878 (lymphoblastoid cell line) were obtained via the UCSC genome browser (http://hgdownload.cse.ucsc.). Further, by using the bwtool find program (with parameters local-extrema -maxima -min-sep=150) the nucleosome peak regions were identified across the genome and 146bp flanking the peak (73bp per side) were considered as regions covered by a nucleosome.
We numbered the exons and introns in each gene according to their position with respect to the TSS. Exons and introns that occupy different positions in different transcripts, and those in the lower quartile of length were discarded. We then stacked all exons –and introns separately– and computed the aggregated coverage (fraction of bases covered by peaks of each mark) at the center of the stack corresponding to the number of bases of the shortest exon or intron remaining after the filtering. Finally, the difference between the exonic and intronic coverage was computed via the two-tailed Mann-Whitney p-value of the comparison of both distributions.
Classification of colorectal tumors according to the MMR level
Colorectal samples were separated into four subtypes on the basis of their level of MMR. MSIH (N=6), MSIL (N=4) and MSS (N=26) groups were defined based on clinical information from TCGA (https://portal.gdc.cancer. The rescaled frequency (Rescf) of each nucleotide in the genegov/query). The POLE group (N=6) was defined by identifying samples with missense mutations of the DNA polymerase epsilon (POLE) gene.
Exon-centered mutational analyses
We stacked 2001-wide sequences centered at the middle position of internal exons. In this analysis we did not exclude regions that overlap any of the three types of technically challenging regions. Thus, we obtained a stack of 95164 sequences centered at exons. We then counted either observed or expected (distributed across each sequence of the 2001-wide window following the mutational probability of each nucleotide, as explained below for individual genes) mutations associated to each nucleotide of this 2001-wide sequences. With these counts across the selected windows, we produced exon-centered plots as those shown in Figures 2a, and 3b.
Computing the decrease of exonic mutation burden
We first computed the relative frequencies of the 192 tri-nucleotide changes, f(AiXjCk→AiXlCk) across each cluster of tumors, as:
where N(AiXjCk→AiXlCk) is the number of such changes within all mutations observed in the tumors, and T is the total number of substitutions observed across tumors. Then, we made f relative to the abundance of each tri-nucleotide in the genome, G(AiXjCk).
Next, for each genic site, we summed the relative frequency of its three possible changes given its 5’ and 3’ flanking bases:
Then, we rescaled the relative frequency of change of each site to one, by multiplying each by the factor:
The rescaled frequency (Rescf) of each nucleotide in the gene is proportional to the conditional probability that the reference nucleotide changes to the alternative given its 5’ and 3’ nucleotides. Finally, for each independent gene we redistributed all observed mutations (Nmuts) across exonic and intronic sites following these summed rescaled frequencies of each site to be mutated as:
Note that this redistribution process could be done equally for the mutations observed in one tumor (for single-tumor analysis, Fig. 5b) or across a group of tumors (for groups or cluster analyses, Figs. 2,3,4 and 5a). This yielded the number of expected exonic (EExonic) and intronic (EIntronic) mutations in the gene. (We employed a second method to compute the expected number of exonic mutations, based on the average of 1000 random permutations of the observed mutations in each gene following the probability of each site to suffer a mutation: Supp. Table 3). Summing the observed and expected exonic mutations over all genes, we computed the difference between the observed and expected number of exonic mutations, which we refer to as the decrease of exonic mutation burden (since in most tumors it yielded a negative difference). Throughout the paper, we express this decrease as percent of the total number of observed exonic mutations.
To compute the significance of this decrease, we employed two tests: i) a G-test of independence comparing the number of observed and expected mutations in exons and introns, under the null hypothesis that the observed and theoretical distributions of the variables are equal; ii) for the expected number of exonic mutations computed using the permutations approach (see above), we computed an empirical p-value as the fraction of the iterations with fewer expected than observed exonic mutations.
Test for negative selection on exonic mutations
The consequence type of all observed exonic mutations was obtained using the Ensembl Variant Effect Predictor48 (VEP, v. 70). We subsequently separated exonic mutations into two groups: those with synonymous consequence, and those with a consequence ranking higher than synonymous in the Ensembl Variation hierarchy (www.ensembl.org/info/genome/variation/predicted_data.html), which were collectively deemed non-synonymous. All possible nucleotide changes in a gene were then divided into three categories: i) synonymous; ii) non-synonymous (with the consequences defined above); iii) intronic. We redistributed the mutations observed in each gene across these three types of sites following the probability of occurrence of each change computed as explained above. Through the difference of observed and expected synonymous and non-synonymous mutations we were able to compute the decrease of the burden of both types of mutations (expressed as percentage of the expected number, as explained above for all exonic mutations). Finally a G-test of independence (see above) was used on the null hypothesis that fewer non-synonymous mutations should be observed than expected.
We also redistributed only the exonic mutations across synonymous and non-synonymous sites according to the probability of change of each type of sites. In this case, we used the G-test of independence on the null hypothesis that the number of expected non-synonymous mutations was not smaller than their observed number.
Stratification of genes by mutation rate and several co-variates
The mutation rate of each gene was computed as the quotient between the number of observed mutations and the number of bases in the gene. Genes were subsequently grouped into 10 bins according to their mutation rate.
We computed the 75th percentile of the expression of each gene across the tumors in each cohort. Genes with a 75th percentile of expression equal to 0 were considered to be non-expressed and were grouped together. All other genes were sorted on the basis of their previously computed expression percentile and divided into 9 bins of equal size. Non-expressed genes were subsequently added as a tenth bin.
Replication time data across the human genome measured in lymphoblastoid cell lines was obtained from Koren et al. 20126. Using this data, a mean replication time per gene was computed. Next, genes were sorted on the basis of this value and divided into 10 groups of equal size.
Finally, we also grouped the genes into 10 bins according to their H3K36me3 peak coverage.
Relationship between the decrease of exonic mutation rate and exonic enrichment of nucleosomes and histone marks
For each gene, we computed the readcount-based exonic enrichment of any chromatin feature as the ratio between the exonic and intronic readcounts (total number of bases covered by reads of the chromatin feature). (This readcount-based exonic enrichment was used to compute the correlations shown in Fig. 4 and Supp. Fig. 4.) We computed the peak-based exonic enrichment of any chromatin feature as the ratio of exonic and intronic bases covered by peaks of the feature (to compute the correlation shown in Supp. Fig. 5). The exonic and intronic number of bases covered by reads or peaks of the chromatin feature for colorectal and bMMRD glioblastoma tumors were computed from colonic mucosa (E075) and brain angular gyrus (E067) cells, respectively, both obtained from the Epigenome Roadmap. (In the case of nucleosomes, their peaks were obtained from their occupancy values as explained above.) Genes were grouped into 10, 25 and 50 bins according to their exonic H3K36me3 enrichment, and the aggregated decrease of the exonic mutation rate of the genes in each bin was computed as explained above for colorectal POLE-mutants, MSI-H and bMMRD tumors. We then computed the correlation between the median exonic chromatin feature enrichment and the decreased exonic mutation rate across the bins. The trend line and its confidence intervals were added using the bootstrapping functions of the python seaborn package, which confers equivalent weights in the regression to all points. In order to guarantee that the trend is not the results of few outliers, the correlation coefficient and its significance were computed using iteratively re-weighted least squares approach, letting the variance of exonic H3K36me3 enrichment of the bins influence the weight of each point.
Exon to intron mutation rate ratio
As described above, we stacked all exon-centered and intron-centered sequences, and averaged the total number of mutations observed at each of the 41 central positions of each stack. The selection of 41 central positions guaranteed both a vast majority of exonic sequences contributing mutations and enough mutations for the calculation across all clusters at exon-centered stacks. The exon-centered and the intron-centered mutation burden averages were then divided by the number of sequences included in each stack (see above) to make them comparable. Finally, we computed the exon to intron mutation rate ratio, as the quotient between the corrected exon-centered and intron-centered mutation burden averages.
Computing the activity of NER from XR-seq data
The genome-wide maps of nucleotide excision repair (NER) of two UV-induced photo-products, namely cyclobutane pyrimidine dimers (CPDs) and pyrimidine–pyrimidone (6–4) photoproducts (PP64s), in irradiated skin fibroblast cell lines were obtained from Hu et al., 201534. This data set comprises the NER maps for the following three cell lines: (i) wild-type NHF1 skin fibroblasts, which contain active global and transcription-coupled repair mechanisms; (ii) XP-C mutants, which are deficient in the global repair mechanism; and (iii) CS-B mutants, which are deficient in transcription-coupled repair. For each of these cell lines, we extracted the sequencing reads, processed and mapped to the human genome, by following the steps mentioned in Hu et al., 2015. Further, we selected the mapped reads that are of size 26nts, which is typically the size of NER excised oligomers, and classified the reads based on the presence of dipyrimdines (TT, CT, TC, CC) at positions 19-20 or 20-21nts of the reads. In addition, we recorded the mapped genomic location of the nucleotides in positions 19-20 or 20-21 of the reads. This way, we can predict the damage site based on the excised fragments. We mapped this information to the XR-seq exon-centered plot together with the frequency of dipyrimidines observed in each columns (see Supplementary Figure 76).
Statistics
We used the G-test described above to compute p-values to test the significance of the decreased exonic mutation burden for groups of genes, or all genes in a tumor or across groups or clusters of tumors. (All p-values computed for all comparisons are provided in Supp. Table 3, which includes as well p-values computed using a permutations-based test also described above.) When appropriate, p-values computed with this test were corrected using the Benjamini-Hochberg approach. In Figure 1 we used the two tailed Mann-Whitney test to compare the exonic and intronic distributions of chromatin features (and corrected them when appropriate). Above, we describe the approach employed to compute the correlation coefficient (and its associated p-value) of regression lines shown in Fig. 4 and Supplementary Figures 4 and 5.
Code availability
All code needed to reproduce the analyses described in the paper are available at the Bitbucket repository (https://bitbucket.org/bbglab/intron_exon_mutrate).
Data availability
Mutational data employed in the analyses described in the paper was obtained from three papers cited at the first section of Methods20,29,30. The mutations identified in four bMMRD glioblastomas were provided by the authors upon request (see acknowledgements section). All other data was obtained from public repositories included at each relevant Methods section. Pre-processed data needed to reproduce all analyses described here is provided together with the code (see above) at https://bitbucket.org/bbglab/intron_exon_mutrate.
Supplementary Material
Acknowledgments
We acknowledge funding from Spanish Ministry of Economy and Competitiveness (SAF2015-66084-R, MINECO/FEDER, UE), La Fundació la Marató de TV3, EU H2020 Programme 2014-2020 under grant agreements no. 634143 (MedBioinformatics) and by the European Research Council (Consolidator Grant 682398). IRB Barcelona is a recipient of a Severo Ochoa Centre of Excellence Award from the Spanish Ministry of Economy and Competitiveness (MINECO; Government of Spain) and is supported by CERCA (Generalitat de Catalunya). R. Sabarinathan is supported by an EMBO Long-Term Fellowship (ALTF 568-2014) co-funded by the European Commission (EMBOCOFUND2012, GA-2012-600394) support from Marie Curie Actions. A.Gonzalez-Perez is supported by a Ramón y Cajal contract. We acknowledge the contribution of Iker Reyes-Salazar to refactoring and cleaning all code produced in the study for publication. We thank the suggestions by three anonymous referees, which improved the manuscript. We are grateful to B. Campbell and U. Tabori for help in obtaining the mutation calls for bMMRD samples sequenced by The International BMMRD Consortium. The results published here are in part based upon data generated by the TCGA Research Network: http://cancergenome.nih.gov/.
Footnotes
Author Contributions
J.F. and R.S. participated in the design and execution of analyses, produced the figures, participated in the interpretation of results, and edited the manuscript. L.M. developed computational code employed in the analyses. F.M. developed the statistical framework to compute the significance of the decreased exonic mutation burden and its correlation with chromatin features. A.G-P. participated in the design of analyses, the interpretation of results, the oversight of analyses and drafted and edited the manuscript. N.L-B. conceived the study, participated in the design of analyses, oversaw the study and the interpretation of results, and drafted and edited the manuscript.
Competing Financial Interests Statement
The authors declare no competing financial interests.
References
- 1.Chapman MA, et al. Initial genome sequencing and analysis of multiple myeloma. Nature. 2011;471 doi: 10.1038/nature09837. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Dulak AM, et al. Exome and whole-genome sequencing of esophageal adenocarcinoma identifies recurrent driver events and mutational complexity. Nat Genet. 2013;45:478–486. doi: 10.1038/ng.2591. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Pleasance ED, et al. A comprehensive catalogue of somatic mutations from a human cancer genome. Nature. 2010;463:191–196. doi: 10.1038/nature08658. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Li J, et al. A Dual Model for Prioritizing Cancer Mutations in the Non-coding Genome Based on Germline and Somatic Events. PLOS Comput Biol. 2015;11:e1004583. doi: 10.1371/journal.pcbi.1004583. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Lawrence MS, et al. Mutational heterogeneity in cancer and the search for new cancer-associated genes. Nature. 2013;499:214–8. doi: 10.1038/nature12213. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Koren A, et al. Differential relationship of DNA replication timing to different forms of human mutation and variation. Am J Hum Genet. 2012;91:1033–40. doi: 10.1016/j.ajhg.2012.10.018. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Stamatoyannopoulos JA, et al. Human mutation rate associated with DNA replication timing. Nat Genet. 2009;41:393–395. doi: 10.1038/ng.363. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Hodgkinson A, Eyre-Walker A. Variation in the mutation rate across mammalian genomes. Nat Rev Genet. 2011;12:756–766. doi: 10.1038/nrg3098. [DOI] [PubMed] [Google Scholar]
- 9.Polak P, et al. Cell-of-origin chromatin organization shapes the mutational landscape of cancer. Nature. 2015;518:360–364. doi: 10.1038/nature14221. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Schuster-Böckler B, Lehner B. Chromatin organization is a major influence on regional mutation rates in human cancer cells. Nature. 2012 doi: 10.1038/nature11273. [DOI] [PubMed] [Google Scholar]
- 11.Morganella S, et al. The topography of mutational processes in breast cancer genomes. Nat Commun. 2016;7:11383. doi: 10.1038/ncomms11383. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Perera D, et al. Differential DNA repair underlies mutation hotspots at active promoters in cancer genomes. Nature. 2016;532:259–263. doi: 10.1038/nature17437. [DOI] [PubMed] [Google Scholar]
- 13.Polak P, et al. Reduced local mutation density in regulatory DNA of cancer genomes is linked to DNA repair. Nat Biotechnol. 2013;32:71–75. doi: 10.1038/nbt.2778. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Sabarinathan R, Mularoni L, Deu-Pons J, Gonzalez-Perez A, López-Bigas N. Nucleotide excision repair is impaired by binding of transcription factors to DNA. Nature. 2016;532:264–267. doi: 10.1038/nature17661. [DOI] [PubMed] [Google Scholar]
- 15.Li F, et al. The histone mark H3K36me3 regulates human DNA mismatch repair through its interaction with MutSα. Cell. 2013;153:590–600. doi: 10.1016/j.cell.2013.03.025. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Tatum D, Li S. Evidence That the Histone Methyltransferase Dot1 Mediates Global Genomic Repair by Methylating Histone H3 on Lysine 79. J Biol Chem. 2011;286:17530–17535. doi: 10.1074/jbc.M111.241570. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.House NCM, Koch MR, Freudenreich CH. Chromatin modifications and DNA repair: beyond double-strand breaks. Front Genet. 2014;5:296. doi: 10.3389/fgene.2014.00296. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Schwartz S, Meshorer E, Ast G. Chromatin organization marks exon-intron structure. Nat Struct Mol Biol. 2009;16:990–995. doi: 10.1038/nsmb.1659. [DOI] [PubMed] [Google Scholar]
- 19.Huff JT, Plocik AM, Guthrie C, Yamamoto KR. Reciprocal intronic and exonic histone modification regions in humans. Nat Struct Mol Biol. 2010;17 doi: 10.1038/nsmb.1924. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Fredriksson NJ, Ny L, Nilsson JA, Larsson E. Systematic analysis of noncoding somatic mutations and gene expression alterations across 14 tumor types. Nat Genet. 2014;46:1258–1263. doi: 10.1038/ng.3141. [DOI] [PubMed] [Google Scholar]
- 21.Hodis E, et al. A landscape of driver mutations in melanoma. Cell. 2012;150:251–63. doi: 10.1016/j.cell.2012.06.024. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Lanzós A. Discovery of Cancer Driver Long Noncoding RNAs across 1112 Tumour Genomes: New Candidates and Distinguishing Features. Sci Rep. 2017 doi: 10.1038/srep41544. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Kundaje A, et al. Integrative analysis of 111 reference human epigenomes. Nature. 2015;518:317–330. doi: 10.1038/nature14248. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Bernstein BE, et al. An integrated encyclopedia of DNA elements in the human genome. Nature. 2012;489:57–74. doi: 10.1038/nature11247. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Futreal A, et al. A census of human cancer genes. Nat Rev Cancer. 2004;4:177–183. doi: 10.1038/nrc1299. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Rubio-Perez C. In Silico Prescription of Anticancer Drugs to Cohorts of 28 Tumor Types Reveals Targeting Opportunities. Cancer Cell. 2015;27:382–396. doi: 10.1016/j.ccell.2015.02.007. [DOI] [PubMed] [Google Scholar]
- 27.Li G-M. Mechanisms and functions of DNA mismatch repair. Cell Res. 2008;18:85–98. doi: 10.1038/cr.2007.115. [DOI] [PubMed] [Google Scholar]
- 28.Hause RJ, Pritchard CC, Shendure J, Salipante SJ. Classification and characterization of microsatellite instability across 18 cancer types. Nat Med. 2016;22:1342–1350. doi: 10.1038/nm.4191. [DOI] [PubMed] [Google Scholar]
- 29.Shlien A, et al. Combined hereditary and somatic mutations of replication error repair genes result in rapid onset of ultra-hypermutated cancers. Nat Genet. 2015;47:257–262. doi: 10.1038/ng.3202. [DOI] [PubMed] [Google Scholar]
- 30.Martincorena I, et al. High burden and pervasive positive selection of somatic mutations in normal human skin. Science (80-.) 2015;348:880–886. doi: 10.1126/science.aaa6806. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Marteijn JA, Lans H, Vermeulen W, Hoeijmakers JHJ. Understanding nucleotide excision repair and its roles in cancer and ageing. Nat Rev Mol Cell Biol. 2014;15:465–81. doi: 10.1038/nrm3822. [DOI] [PubMed] [Google Scholar]
- 32.Alexandrov LB, et al. Signatures of mutational processes in human cancer. Nature. 2013;500:415–21. doi: 10.1038/nature12477. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Adar S, Hu J, Lieb JD, Sancar A. Genome-wide kinetics of DNA excision repair in relation to chromatin state and mutagenesis. Proc Natl Acad Sci. 2016;113:E2124–E2133. doi: 10.1073/pnas.1603388113. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Hu J, Adar S, Selby CP, Lieb JD, Sancar A. Genome-wide analysis of human global and transcription-coupled excision repair of UV damage at single-nucleotide resolution. Genes Dev. 2015;29:948–60. doi: 10.1101/gad.261271.115. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Haradhvala NJ, et al. Mutational Strand Asymmetries in Cancer Genomes Reveal Mechanisms of DNA Damage and Repair. Cell. 2016;164:538–549. doi: 10.1016/j.cell.2015.12.050. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Tolstorukov MY, Volfovsky N, Stephens RM, Park PJ. Impact of chromatin structure on sequence variability in the human genome. Nat Struct Mol Biol. 2011;18:510–515. doi: 10.1038/nsmb.2012. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Francioli LC, et al. Genome-wide patterns and properties of de novo mutations in humans. Nat Genet. 2015;47:822–826. doi: 10.1038/ng.3292. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Supek F, Lehner B. Differential DNA mismatch repair underlies mutation rate variation across the human genome. Nature. 2015;521:81–84. doi: 10.1038/nature14173. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Supek F, et al. Clustered Mutation Signatures Reveal that Error-Prone DNA Repair Targets Mutations to Active Genes. Cell. 2017;170:534–547.e23. doi: 10.1016/j.cell.2017.07.003. [DOI] [PubMed] [Google Scholar]
- 40.Kim S, Kim H, Fong N, Erickson B, Bentley DL. Pre-mRNA splicing is a determinant of histone H3K36 methylation. Proc Natl Acad Sci U S A. 2011;108:13564–9. doi: 10.1073/pnas.1109475108. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Martincorena I, et al. Universal Patterns Of Selection In Cancer And Somatic Tissues. bioRxiv. 2017 doi: 10.1016/j.cell.2017.09.042. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Lynch M. Rate, molecular spectrum, and consequences of human mutation. Proc Natl Acad Sci. 2010;107:961–968. doi: 10.1073/pnas.0912629107. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Kolasinska-Zwierz P, et al. Differential chromatin marking of introns and expressed exons by H3K36me3. Nat Genet. 2009;41:376–381. doi: 10.1038/ng.322. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Chamary JV, Parmley JL, Hurst LD. Hearing silence: non-neutral evolution at synonymous sites in mammals. Nat Rev Genet. 2006;7:98–108. doi: 10.1038/nrg1770. [DOI] [PubMed] [Google Scholar]
- 45.The Chimpanzee Sequencing and Analysis Consortium. Initial sequence of the chimpanzee genome and comparison with the human genome. Nature. 2005;437:69–87. doi: 10.1038/nature04072. [DOI] [PubMed] [Google Scholar]
- 46.Hoffman MM, Birney E. Estimating the Neutral Rate of Nucleotide Substitution Using Introns. Mol Biol Evol. 2006;24:522–531. doi: 10.1093/molbev/msl179. [DOI] [PubMed] [Google Scholar]
- 47.Harrow J, et al. GENCODE: the reference human genome annotation for The ENCODE Project. Genome Res. 2012;22:1760–74. doi: 10.1101/gr.135350.111. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48.McLaren W, et al. The Ensembl Variant Effect Predictor. doi: 10.1186/s13059-016-0974-4. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
Data Availability Statement
Mutational data employed in the analyses described in the paper was obtained from three papers cited at the first section of Methods20,29,30. The mutations identified in four bMMRD glioblastomas were provided by the authors upon request (see acknowledgements section). All other data was obtained from public repositories included at each relevant Methods section. Pre-processed data needed to reproduce all analyses described here is provided together with the code (see above) at https://bitbucket.org/bbglab/intron_exon_mutrate.