Abstract
The specificity of membrane traffic involves tethers at destination organelles that selectively capture incoming transport vesicles to allow SNAREs on opposing membranes to then assemble and drive fusion1,2. Tethers include both protein complexes and long coiled-coil proteins, although how they contribute to specificity remains unclear3,4. The golgin coiled-coil proteins at the Golgi apparatus capture vesicles from different origins, but the vesicle-specific molecular cues that they recognise are unknown5–8. Vesicle tethering is typically a transient process and so challenging to interrogate in vivo. We have thus used a system where an ectopic golgin causes vesicles to accumulate in a tethered state. By applying proximity biotinylation to the golgin-captured vesicles we identify TBC1D23, an apparently catalytically inactive member of a family of Rab GTPase activating proteins (GAPs), as a vesicle-golgin adaptor that is required for endosome-to-Golgi traffic. The Rab-GAP domain of TBC1D23 binds to a conserved motif at the tip of golgin-245 and golgin-97 at the trans-Golgi, while the C-terminus binds to the WASH complex on endosome-derived vesicles. Thus TBC1D23 is a specificity determinant that links vesicle to target membrane during endosome-to-Golgi trafficking.
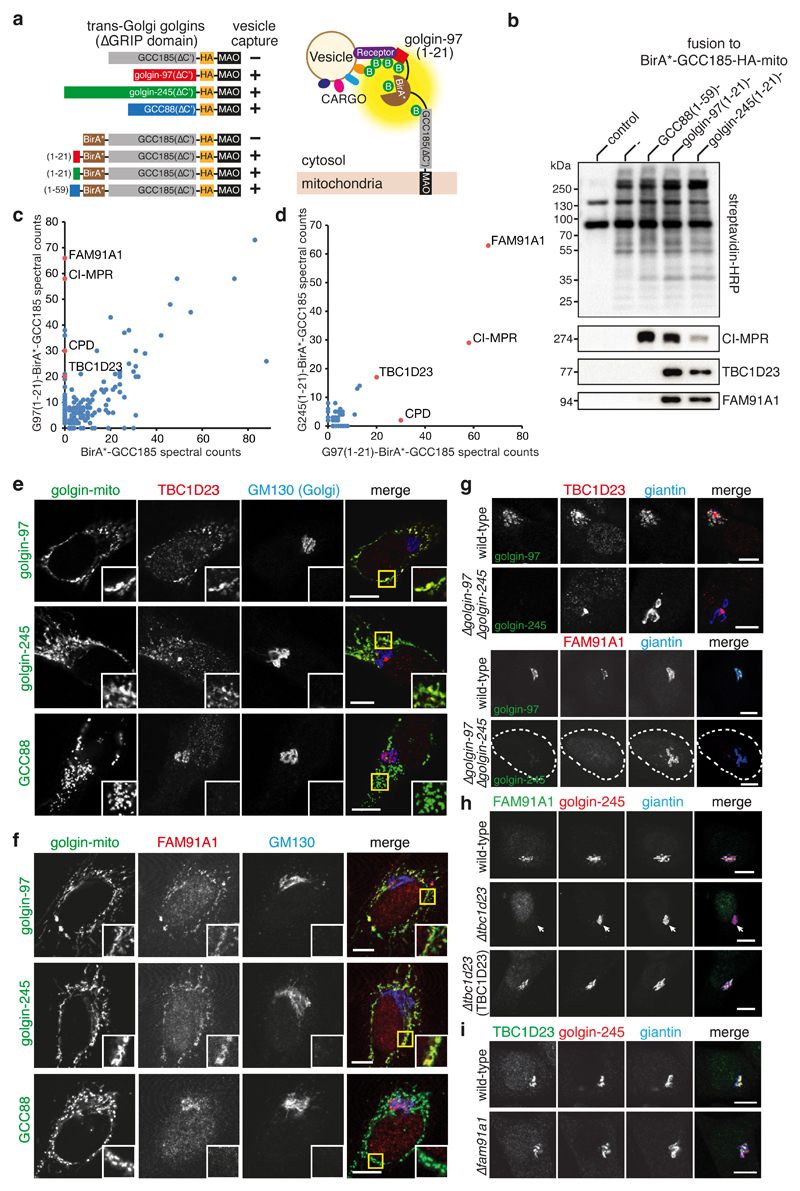
The Golgi apparatus receives transport vesicles originating from diverse compartments including the endoplasmic reticulum (ER), endosomes and from within the Golgi itself7,9,10. Studies on vesicle tethering at the Golgi have had a particular focus on the golgins, a set of long coiled-coil proteins that are anchored to the Golgi via C-terminal domains that either bind small GTPases or span the lipid bilayer5–8. Seven of the golgins are able to capture specific classes of Golgi-directed carriers when ectopically installed at mitochondria11. Of these seven, the trans-Golgi golgins golgin-97, golgin-245, and GCC88 have all been implicated in endosome-to-Golgi traffic, a pathway critical for recycling to the Golgi of the machinery that delivers lysosomal hydrolases and also part of the route used by bacterial toxins to move from endosomes to the ER to gain access to the cytoplasm12–14. For all three of golgin-97, golgin-245 and GCC88 a conserved N-terminal region is necessary and sufficient for vesicle capture, with the motifs found in golgin-97 and golgin-245 being closely related but distinct from that in GCC8815. To identify candidate vesicle-associated specificity factors for these golgins, we combined ectopic golgin-mediated vesicle capture with BioID proximity biotinylation16.
The vesicle-binding motifs from the N-termini of golgin-97, golgin-245 and GCC88 were fused to the promiscuous biotin ligase BirA*, which was appended to the N-terminus of a mitochondrial form of GCC185, a golgin that has no detectable vesicle capture activity when relocalised to mitochondria in the HeLa or 293T cell lines used in this study11,16 (Fig. 1a). These chimeras retain both biotinylation activity and the ability to capture endosome-to-Golgi vesicles, permitting the identification of golgin-proximal proteins by mass spectrometry of affinity-purified biotinylated proteins (Fig. 1b and Supplementary Fig. 1a-e). Inspecting the proteins efficiently biotinylated by the golgin-97 construct but not by the negative control revealed, as expected, cargo known to travel along the endosome-to-Golgi route, such as the cation-independent mannose 6-phosphate receptor (CI-MPR) and carboxypeptidase D (CPD)17,18 (Fig. 1c). Comparison of the strongest specific hits that were common to both golgin-97 and golgin-245 revealed in addition two poorly characterised cytosolic proteins (Fig. 1d). These are TBC1D23, a member of the TBC (Tre-2/Bub2/Cdc16) family of Rab GAPs but otherwise of unknown function19,20, and FAM91A1. FAM91A1 has no known function or clear similarity to any other protein, but has been suggested to form a trimeric complex with two other proteins of unknown function, WDR11 and C17orf75, based on cofractionation and coprecipitation21–23. Immunoblotting of streptavidin-purified biotinylated proteins from cells expressing the different BirA*-golgin chimeras validated the mass spectrometry results: CI-MPR was biotinylated by all three golgin constructs as expected for their endosomal vesicle capture activity, while TBC1D23 and FAM91A1 were biotinylated by golgin-97 and golgin-245 but not by GCC88, consistent with the latter having a N-terminal vesicle-capturing sequence which is distinct from that of the other two golgins (Fig. 1b).
Figure 1. BioID screens to identify putative vesicle receptors recognised by golgin-97 and golgin-245 reveal TBC1D23 and FAM91A1.
(a) Schematics of the BirA*-golgin-mito chimeras and of BioID where BirA* biotinylates the golgin-receptor and other vesicle proteins. The chimeras contain the transmembrane domain of monoamine oxidase (MAO) to anchor them to the outer membrane of mitochondria, and an HA-epitope tag.
(b) Immunoblot of biotinylated proteins purified by streptavidin-beads from 293T cells stably expressing either no construct (control) or the indicated doxycycline-inducible BirA*-golgin-mito chimeras. Expression was induced with 1 μg/ml doxycycline for 24 hours, and the blot labeled with streptavidin-HRP or the indicated antibodies.
(c) Mass-spectrometric analysis of biotinylated proteins purified from 293T cells expressing BirA*-golgin-mito chimeras as in (b). Plot compares golgin-97(1-21)-BirA*-GCC185-mito to the negative control BirA*-GCC185-mito that does not capture vesicles. The two golgin-97-specific proteins with more spectra than CPD are SFPQ and NONO, pre-mRNA binding proteins that are probably irrelevant as they are abundant and present in a different negative control.
(d) Comparison of the proteins biotinylated specifically by the golgin-97 chimera from (c) and by the equivalent golgin-245 chimera. Specific defined as having ten-fold more spectral counts than with GCC185 alone, or no chimera.
(e) Confocal micrographs of cells expressing the indicated golgin-mito chimeras and labeled for the HA tag in the chimera and endogenous TBC1D23 and GM130 (Golgi marker). The TBC1D23 antisera also labels centrosomes, but this is apparently nonspecific as it is unaffected by either CRISPR gene disruption or siRNA knockdown (Supplementary Fig. 2a,b).
(f) Confocal micrographs of cells expressing the indicated golgin-mito chimeras and labeled for the HA tag in the chimera and endogenous FAM91A1 and GM130 (Golgi marker).
(g) Confocal micrographs of HeLa cells showing loss of targeting of endogenous TBC1D23 and FAM91A1 to the Golgi in a ∆golgin-97/∆golgin-245 double mutant.
(h) Immunofluorescence to show loss of FAM91A1 from the Golgi when TBC1D23 is deleted.
(i) Immunofluorescence of wild-type and ∆fam91a1 HAP1 cells to show that removal of FAM91A1 does not effect the Golgi localisation of TBC1D23. Scale bars, 10 μm. Unprocessed original scans of blots are shown in Supplementary Fig. 6. Experiments were repeated 3 times except (b) which was performed twice.
TBC1D23 and FAM91A1 colocalised with golgin-97 and golgin-245 at the trans-Golgi (Supplementary Fig. 1f). Relocation to mitochondria of golgin-97 and golgin-245, but not GCC88, resulted in the relocation of TBC1D23 and FAM91A1 (Fig. 1e,f). The double deletion of golgin-97 and golgin-245 by CRISPR/Cas9, but not the single deletions, resulted in the loss of targeting of both TBC1D23 and FAM91A1 to the Golgi (Fig. 1g and Supplementary Fig. 1g,h). Thus, golgin-97 and golgin-245 are necessary and sufficient for the recruitment of TBC1D23 and FAM91A1 to membranes.
To determine if either TBC1D23 or FAM91A1 depends on the other for Golgi recruitment we used CRISPR/Cas9-generated null mutants of TBC1D23 or FAM91A1 (Supplementary Fig. 2a-d). Loss of TBC1D23 resulted in the delocalisation of FAM91A1 from the Golgi, whereas deletion of FAM91A1 did not affect the Golgi localisation of TBC1D23 (Fig. 1h,i). In neither case was the location of the golgins to the Golgi perturbed. Taken together these results suggest that golgin-97 and golgin-245 recruit TBC1D23, and that this is then required for recruitment of FAM91A1.
We next asked whether TBC1D23 binds directly to golgin-97 and golgin-245. The conserved N-terminal regions that were used for the BioID experiments were expressed in E. coli as fusions to the N-terminus of GST (Fig. 2a). Recombinant TBC1D23 expressed in E. coli bound directly in vitro to golgin-97(1-21)-GST and golgin-245(1-21)-GST but not to GCC88(1-59)-GST (Fig. 2b). These regions of golgin-97 and golgin-245 share several highly conserved residues, of which Phe2 and Leu5 were identified by alanine scanning mutagenesis to be particularly critical for binding TBC1D23 in vitro (Fig. 2a-c). The F2A and L5A mutations in full-length golgin-97 also abolished ectopic capture of TBC1D23 in vivo (Fig. 2d). Taken together these results show that TBC1D23 binds directly to the N-termini of golgin-97 and golgin-245.
Figure 2. TBC1D23 binds directly to the N-termini of golgin-97 and golgin-245 to mediate vesicle capture.
(a) Alignments of the N-terminal regions of human golgin-97 and golgin-245 with their orthologs from diverse metazoans. Red diamonds indicate the residues in golgin-97 that were mutated to alanine for in vitro binding studies.
(b) Binding of TBC1D23 expressed in E. coli (lysate) when applied to recombinant GST fusions containing the indicated golgin N-terminal sequences. Bound material analysed by Coomassie-blue staining following SDS-PAGE.
(c) Binding of TBC1D23 expressed in E.coli (lysate) when applied to recombinant GST fusions to the indicated variants of the first 21 residues of golgin-97. Bound material analysed as in (b).
(d) Confocal micrographs or transfected HeLa cells showing that mutations in golgin-97 that disrupt binding of TBC1D23 in vitro result in loss ability to relocate TBC1D23 in vivo.
(e,f) Confocal micrographs of wild-type or ∆tbc1d23 mutant cells expressing the indicated golgin-mito constructs and labeled for the HA tag in the chimera and the endogenous endosome-to-Golgi cargo TGN46 (ZFPL1, Golgi marker). Scale bars, 10 μm. Experiments were repeated 3 times except (c) which was performed twice.
We next investigated whether TBC1D23 is required for the capture of endosomal vesicles by the mitochondrial forms of golgin-97 or golgin-245. In the ∆tbc1d23 mutant, mitochondrial capture of endosomal vesicles, visualised by the endosome-to-Golgi cargo TGN4611,18, was markedly reduced, but rescued by reintroduction of TBC1D23 (Fig. 2e,f). In contrast to these findings for TBC1D23, deletion of FAM91A1 had no detectable effect on the ability of mitochondrial golgin-97 to capture vesicles or recruit TBC1D23 (Supplementary Fig. 2e). Thus TBC1D23 is required for vesicle capture by the golgins, whereas FAM91A1 is recruited by TBC1D23 but is not essential for vesicle capture by TBC1D23.
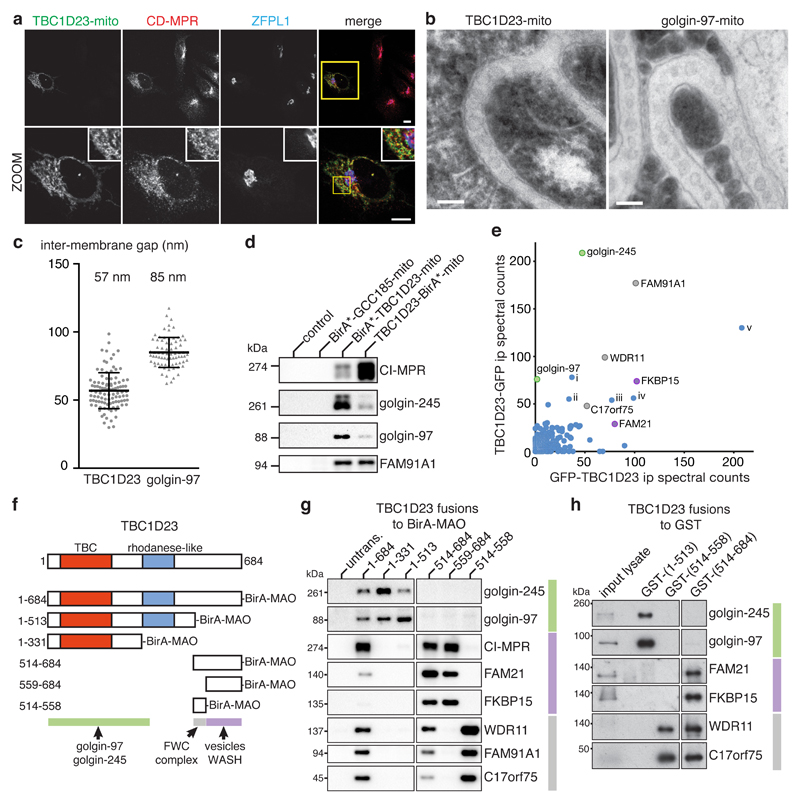
To test the hypothesis that TBC1D23 is an adaptor that links endosome-derived carriers to golgin-97 and golgin-245, we relocated TBC1D23 to mitochondria (TBC1D23-mito, Supplementary Fig. 3a). As expected, TBC1D23-mito could recruit cytosolic forms of golgin-97 and golgin-245, while GCC88 remained at the trans-Golgi (Supplementary Fig. 3a,b). Strikingly, TBC1D23-mito relocated four different endosome-to-Golgi cargo proteins to mitochondria (Fig. 3a and Supplementary Fig. 3c). Electron microscopy revealed an accumulation of circular and oval membranes tethered between the mitochondria (Fig. 3b), similar to that seen with the golgins11. Interestingly, the distance between the tethered carriers and the mitochondrial membrane was less for TBC1D23-mito, consistent with the absence of the long coiled-coil region of the golgin to act as a spacer (Fig. 3c). While TBC1D23-mito also recruited FAM91A1, deletion of FAM91A1 had no effect on carrier relocation by TBC1D23-mito (Supplementary Fig. 3d). BirA* fused to the N- or C-terminus of TBC1D23-mito specifically biotinylated golgin-97, golgin-245, FAM91A1 and the cargo of endosome-derived carriers (Fig. 3d), validating biochemically the results of the visual relocation assay.
Figure 3. TBC1D23 captures vesicles, golgin-97, golgin-245, FAM91A1, and WASH complex subunits.
(a) Confocal micrographs showing TBC1D23-mito relocates endogenous CD-MPR to mitochondria in HeLa cells (ZFPL1, Golgi marker) with similar results observed for CI-MPR, TGN46 and Vti1a (Supplementary Fig. 3c). Scale bars 10 μm.
(b) Electron micrographs of HeLa cells transfected with a bicistronic plasmid expressing TBC1D23-mito or golgin97-mito along with mitochondrial-APEX to identify transfected cells. In both cases vesicles accumulate between mitochondria. Scale bars, 100 nm.
(c) Quantification of the distance between adjacent mitochondrial membranes across vesicles tethered by the indicated proteins (mean +/- s.d., golgin-97: n=76 measurements across different vesicles; TBC1D23: n=100 measurements; difference is statistically significant P <0.0001, unpaired, two-tailed Mann-Whitney test). Source data in Supplementary Table 1.
(d) Immunoblots with the indicated antibodies of biotinylated proteins isolated from 293T cells stably expressing either no construct (control) or the indicated BirA* under the control of an inducible promoter with expression induced by 1 μg/ml doxycycline and 50 μM biotin for 24 hours.
(e) Mass-spectrometric analysis of proteins co-precipitated with TBC1D23 tagged with GFP at either the N- or C-terminus. Identified are the golgins, the three subunits of the putative FAM91A1 complex, and the WASH complex subunit FAM21 and its interactor FKBP15. Other proteins found at comparable levels were abundant cytosolic proteins or nuclear proteins that are most likely nonspecific interactions (i, ZBTB33, ii fatty acid synthase, iii Grp78, iv mitoHsp70, v Hsc71, with Hsp70 being beyond the axes).
(f) Schematic of TBC1D23 and of the various truncations attached to BirA* and the MAO mitochondrial targeting signal. The rhodanese-like domain (Pfam PF00581) is related to bacterial sulphur transferases, but in eukaryotes it is in diverse proteins including protein phosphatases and ubiquitin ligases.
(g) Immunoblots of streptavidin precipitates to detect proteins biotinylated by the BirA* constructs in (f) transiently expressed in 293T cells (untrans, untransfected). Cells treated with 50 μM biotin for 24 hours.
(h) Immunoblots to detect proteins from 293T cell lysates that bind beads coated with the indicated fragments of TBC1D23 fused to GST. Unprocessed original scans of blots are shown in Supplementary Fig. 6. Experiments were repeated 3 times except (b) which was performed twice.
The above results suggest the existence of factors on the vesicles that can bind TBC1D23. To seek such factors we used mass-spectrometry to identify the proteins that could be co-precipitated with TBC1D23 tagged with GFP at either the N or C terminus. Both of the tagged forms of TBC1D23 co-precipitated golgin-97 and golgin-245 along with FAM91A1 and its putative binding partners WDR11 and C17orf75 (Fig. 3e). The only other strong hits in the precipitates that were not chaperones or abundant cytosolic enzymes were FAM21, the largest subunit of the pentameric WASH complex (Wiskott-Aldrich syndrome protein and Scar Homologue), and FKBP15, a known interaction partner of FAM2124,25. The WASH complex is recruited to carriers forming on endosomes, including those that return to the Golgi, and coordinates the recruitment of membrane traffic components and actin filaments25–27.
To understand the contribution that the golgins, FAM91A1, and the WASH complex make to the mechanism by which TBC1D23 links golgin-97 and golgin-245 to vesicles, we mapped the domains of TBC1D23 that are responsible for its different interactions. Truncated forms of TBC1D23 were fused to BirA*, ectopically localised to mitochondria, and interacting proteins identified by proximity biotinylation (Fig. 3f). The first 331 residues of TBC1D23, including the TBC domain, were sufficient to biotinylate golgin-97 and golgin-245 but not the subunits of the putative FAM91A1 complex or the vesicle resident protein CI-MPR (Fig. 3g). In contrast, residues 514-559 were specific for FAM91A1 and its associated proteins, with FAM91A1 itself binding directly to this region (Fig. 3g and Supplementary Fig. 4a). Finally, residues 559-684 were specific for the cargo of endosome-derived carriers (Fig. 3g). Thus distinct parts of TBC1D23 bind to the golgins, to FAM91A1, and to endosome-derived carriers.
Strikingly, the WASH subunit FAM21 and its known binding partner FKBP15 were biotinylated by the part of TBC1D23 that binds carriers but not by the parts that bind the golgins or FAM91A1 (Fig. 3g). Moreover, affinity chromatography of cytosol with recombinant fragments of TBC1D23 revealed that the C-terminal carrier-binding region bound to WASH subunits (Fig. 3h), and this region was sufficient to recruit to mitochondria both exogenous FAM21A-GFP and the endogenous WASH subunit WASH1 (Fig. 4a,b). In humans FAM21 is encoded by two genes that express proteins that are 99% identical (FAM21A/WASHC2A and FAM21C/WASHC2C), and for this study we have used FAM21A as it has been used in previous work and is the predominant form in HeLa cells25,28. FAM21 is responsible for linking the WASH complex to a range of different proteins by interactions it makes through a long unstructured tail that contains repeats of a conserved motif24,27,29. The entire unstructured tail of FAM21A (residues 357-1341) can be expressed in E. coli29, and we found that this recombinant form of the tail bound directly to residues 559-684 of TBC1D23, but not to the regions of TBC1D23 that bind to the golgins or to the FAM91A1 complex (Fig. 4c). A similar pattern of binding was observed for the entire WASH complex, confirming that the interaction with FAM21 can occur in the context of the WASH complex (Supplementary Fig. 4b). Finally, by using recombinant proteins expressed in E. coli we could show that the presence of TBC1D23 allowed the FAM21A tail to bind to residues 1-21 of golgin-97, indicating that TBC1D23 acts as a bridging protein by binding simultaneously to the golgins and to FAM21A (Fig. 4d).
Figure 4. TBC1D23 links golgin-97 and golgin-245 to the FAM21A subunit of the WASH complex.
(a) Confocal micrographs of HeLa cells expressing FAM21A-GFP and the indicated mitochondrial forms of TBC1D23 and labeled for endogenous WDR11 to follow recruitment of FAM21A-GFP and of the FAM91A1 complex to mitochondria.
(b) Confocal micrographs of HeLa cells expressing the indicated mitochondrial forms of TBC1D23 and labeled for endogenous WASH1 and the Golgi marker GM130 to follow recruitment of WASH1 to mitochondria. Scale bars 10 μm.
(c) Coomassie-stained gel showing the binding of His6-MBP-FAM21A(356-1341) expressed in E. coli to beads coated with the indicated fragments of TBC1D23 fused to GST.
(d) FAM21A binds to the N-terminus of golgin-97 in the presence of TBC1D23. Lysates from E. coli expressing golgin-97(1-21)-MBP-His6 (wild-type or F2A) were applied to beads coated with GST-FAM21A(356-1341) with or without extract from E. coli expressing TBC1D23. The input lysates and the eluates from the GST-beads were analysed on a Coomassie-stained gel. Binding of the golgin-97 N-terminus to FAM21A depends on the presence of TBC1D23, and is disrupted by the F2A mutation that prevents TBC1D23 binding. Experiments were repeated 3 times except (d) which was performed twice.
The in vivo assays of TBC1D23 function described above use a direct assay of vesicle tethering, but one that occurs at mitochondria. We thus investigated whether TBC1D23 is required for endosome-to-Golgi traffic itself. TGN46 provides an ideal cargo for such analysis as it is well established to cycle between endosomes and Golgi, and when retrieval from endosomes is perturbed it is diverted to lysosomes and degraded, thus allowing efficacy of retrieval to be quantified by both immunofluorescence and blotting30. Examination of cells lacking TBC1D23 showed that the accumulation of TGN46 in the Golgi and its overall level were significantly reduced (Fig. 5a,b and Supplementary Fig. 4c,d). A similar effect on TGN46 was observed when both golgin-97 and golgin-245 were deleted, consistent with TBC1D23 and the golgins acting in the same process, and indeed rescue of the golgin double deletion by golgin-245 required the presence of the N-terminal 21 residues that bind TBC1D23 (Fig. 5c,d and Supplementary Fig. 4c-e). The endosome-to-Golgi traffic of another well established cargo, CI-MPR, can be assayed using antibody uptake, and again deletion of the golgins or of TBC1D23 perturbed this process (Fig. 5e,f). The WASH complex has been reported to be required for normal endosome-to-Golgi traffic of CI-MPR and TGN4625,26,31, and we could confirm that depletion of FAM21A also caused a reduction in the levels of TGN46 (Supplementary Fig. 4f). The defect in endosome-to-Golgi traffic in the ∆tbc1d23 cells could be restored by expression of full-length TBC1D23 but not by a truncated form lacking residues 559-684 that bind to FAM21A (Fig. 5g). In contrast, deletion of FAM91A1 did not affect TGN46 levels, suggesting that it is not essential for tethering but might instead have a regulatory or cell type-specific role (Supplementary Fig. 4c).
Figure 5. TBC1D23 is required for endosome-to-Golgi traffic.
(a) Confocal immunofluorescence of HeLa cells showing the effects of deleting TBC1D23 on endogenous TGN46 (golgin-245, trans-Golgi marker), imaged using the same settings throughout.
(b) Immunoblot of whole cell extracts showing deletion of TBC1D23 from HeLa cells reduces of TGN46 levels with reversal by stable over-expression of TBC1D23.
(c) Confocal immunofluorescence of HeLa cells showing the effects on TGN46 of deleting both golgin-97 and golgin-245 (ZFPL1, Golgi marker).
(d) Immunoblot of whole cell extracts showing effect on TGN46 levels of deleting golgin-97 and golgin-245.
(e) Confocal micrographs of the indicated HeLa cells to which anti-CI-MPR had been bound for 10 minutes at 37°C, acid washed at 4°C to remove surface material, and then chased at 37°C for 45 minutes prior to fixation and staining for IgG and the Golgi marker giantin.
(f) Quantitation of traffic of anti-CI-MPR to the Golgi in cells treated as in (e). Means +/- s.e.m.: n=100 cells. Both mutants showed a statistically significant reduction in traffic (both P values <0.0001 using unpaired, two-tailed Mann-Whitney test). Source data in Supplementary Table 1.
(g) Confocal micrographs of HeLa cells lacking TBC1D23 and transiently transfected with the indicated forms of TBC1D23-GFP. Immunofluorescent imaging of TGN46 levels in the Golgi (marked with ZFPL1) in >20 cells per construct showed that 1-558 (lacking the domain that binds FAM21A) gave no rescue, even in cells expressing ten times the level that consistently gave rescue by the full-length form. Scale bars 10 μm.
(h) A model for the role of TBC1D23 as a link between the TGN golgins and endosome-derived vesicles. To ensure specificity TBC1D23 may well bind further factors on the vesicle (X) in addition to FAM21, as the latter is also involved in generating vesicles on endosomes that are destined for the plasma membrane. The FAM91A1 complex is not required for tethering in our assays and so may augment or regulate tethering or downstream events, or act only under particular circumstances. Unprocessed original scans of blots are shown in Supplementary Fig. 6. Experiments were repeated 3 times except (e) which was performed twice.
Our findings are consistent with a model in which TBC1D23 is a modular bridging factor whose separate domains link the tips of golgins at the trans-Golgi to in-coming endosome-derived carriers (Fig. 5h). Furthermore, the carrier-binding domain of TBC1D23 interacts with the FAM21 subunit of the WASH complex and hence is directly contacting these carriers. TBC1D23 is conserved outside of metazoans indicating that it was present in the last eukaryotic common ancestor (Supplementary Fig. 5). It has been lost in non-basal fungi, and interestingly the components of the WASH complex show a closely related pattern of evolutionary conservation. However, the WASH complex is recruited to more than one of the types of carrier leaving endosomes as it interacts with retromer which is involved in transport to both the plasma membrane and to the Golgi24,25,29,32. Thus it seems likely that TBC1D23 makes additional interactions with the carrier in order to select only those carriers destined for the Golgi (Fig. 5h). Although the TBC domain in TBC1D23 lacks the arginine and glutamine residues that are conserved in most TBC proteins and are essential for Rab GAP activity20, it could conceivably bind to a Rab, although in our assays this domain was neither necessary nor sufficient for vesicle capture. In addition, the role of the rhodanese-like domain in TBC1D23 is unclear - it is found in combination with other domains in a range of proteins with diverse roles including phosphatases, heat shock proteins and ubiquitin hydrolases33. What is clear is that there is no shortage of other proteins on endosome-to-Golgi carriers that TBC1D23 could potentially bind in conjunction with the WASH complex. Indeed the question of how endosome-derived carriers are generated is currently debated as some components such as retromer and WASH apparently act in multiple routes, whereas other components have been proposed to act independently in the same route. For example both the AP-1 clathrin adaptor and the sorting nexins SNX1/2 have been reported to direct retrieval of proteins from endosomes to the Golgi34,35. Thus it seems possible that multiple partially redundant mechanisms operate in parallel, confounding experimental dissection. Indeed, the observation that GCC88 captures CI-MPR-containing vesicles independently of TBC1D23 strongly suggests that either there are at least two types of endosome-to-Golgi carrier with overlapping cargos, or there are two mechanisms for capture of the same vesicles. Interestingly, two recent studies report that truncating mutations in human TBC1D23 are not cell lethal but lead to impaired brain development, consistent with there being additional mechanisms to sustain endosome-to-Golgi traffic36,37.
Our work has also revealed that TBC1D23 interacts with FAM91A1, a protein of unknown function, but the FAM91A1 interacting domain of TBC1D23 is neither necessary nor sufficient for capture of endosome-derived carriers, at least in the context of the mitochondrial relocation assay (Fig. 3g). FAM91A1 has been proposed to form a trimer with WDR11 and C17orf75, two proteins identified in a genome-wide siRNA screen for modifiers of sensitivity to the toxin ricin as enhancing toxin transport from cell surface to ER21,22. It is possible that the FAM91A1 interaction acts to regulate or augment vesicle capture by TBC1D23, or to aid capture of a specific sub-population of vesicles that our assays do not detect. Our data also do not preclude it acting at other locations in the endocytic pathway in addition to the Golgi, such that its removal results in increased surface exposure of some recycling membrane proteins including receptors for toxins.
In conclusion TBC1D23 defines a bridge that mediates the recognition of an incoming vesicle by its destination compartment. The surprisingly extensive nature of this bridge is perhaps necessitated by the extremely crowded cytosolic environment. Indeed, large tethering complexes appear to be a common theme in most, if not all, intracellular trafficking pathways. Defining the requisite interactions that impart both specificity and efficiency to vesicle capture, as done here for two golgins, will be crucial to understand the molecular basis of membrane organisation. The strategy of ectopic tether localisation for both the functional analysis of capture and the identification of candidate specificity factors should prove widely useful in this area.
Methods
Plasmids
BioID plasmids were as follows: BirA* followed by GSGSGS linker attached to the N terminus of GCC185∆Cterm-HA-MAO11, to generate BirA*-GCC185∆Cterm-HA-MAO in pcDNA3.1+ (pJJS127) or pcDNA5/FRT/TO (pJJS138). Golgin-97(1-21), golgin-245(1-21), and GCC88(1-59) followed by linker GGGGSLVPRGSGGGGG attached to the N terminus of BirA*-GCC185∆Cterm-HA-MAO in both pcDNA3.1+ and pcDNA5/FRT/TO versions to generate golgin-97(1-21)-BirA*-GCC185∆Cterm-HA-MAO (pJJS129 and pJJS140), golgin-245(1-21)-BirA*-GCC185∆Cterm-HA-MAO (pJJS130 and pJJS141), and GCC88(1-59)-BirA*-GCC185∆Cterm-HA-MAO (pJJS131 and pJJS142) respectively. All TBC1D23 plasmids for mammalian cell expression were based on a mouse cDNA encoding the 684 residue form that lacks alternatively spliced exon 15. TBC1D23-BirA*-HA-MAO plasmid in pcDNA3.1+ (pJJS249) and pcDNA5/FRT/TO (pJJS239) consist of TBC1D23(1-684) followed by a linker GSGSGS, BirA*, linker GGSGGGGG and HA-MAO. The BirA*-TBC1D23-HA-MAO plasmid in the pcDNA5/FRT/TO (pJJS237) vector consists of BirA*, a GSGSGSG linker, TBC1D23(1-684), a GGSGGGGG linker and HA-MAO. For the truncated TBC1D23-BirA*-MAO plasmids, TBC1D23(1-684) in pJJS249 was replaced with TBC1D23(1-331), (1-513), (272-684), (332-684), (514-684), (559-684), and (514-558) to generate pJJS255, pJJS256, pJJS257, pJJS258, pJJS259, pJJS273, and pJJS272 respectively.
CRISPR/Cas9 used plasmids eSpCas9 (Addgene #71814) and pIRESpuro3 (Clontech). eSpCas9 was digested with BbsI and the following gRNA sequences inserted by annealed oligo cloning: CTGCCAACGTCGAGCGGCGA targeting human TBC1D23 (pJJS291); TGAACAGATACGGAAGTTAG targeting human GOLGA1 (pJJS288); GTCCTGCTCCTCATTCTTGT targeting human GOLGA4 (pJJS290); and CACCACACAAATGAGTGATG targeting human WASHC2C (FAM21A and FAM21C) (pJJS335).
AF120, GAG-POL, and VSVG plasmids for retroviral transduction were provided by Felix Randow (MRC-LMB, Cambridge, UK). Mouse TBC1D23 was cloned into AF120 between NcoI and NotI (pJJS271). For the bicistronic construct expressing mitochondrial APEX and TBC1D23-HA-MAO (pJJS243), was based on golgin in Mito-APEX golgin-97∆Cterm-HA-MAO (pMX0167) 11.
Bacterial expression used E. coli codon-optimised cDNAs for human TBC1D23 (684 residue form, Uniprot Q9NUY8-2) or FAM21A. For N-terminal GST fusions regions of TBC1D23 were amplified and cloned into pGEX6p2 using EcoR1 and XhoI (GE Healthcare Life Sciences). For C-terminal GST fusions pOPT (Olga Perisic, MRC-LMB, Cambridge, UK) was modified to insert residues 1-21 of golgin-97, 1-21 of golgin-245 and 1-59 of GCC88 upstream of a GSGSGS linker and GST using NdeI and SacII. Site-directed mutagenesis and DpnI digestion was used to mutagenise the golgin N-termini. For TBC1D23-His6 the E. coli codon-optimised form was amplified with a GAGA linker and His6 tag in the reverse primer and cloned into pOPT using NdeI and BamH1. E. coli optimised FAM21A 356-1341 was cloned into pETM-41 between NcoI and BamHI. Golgin-97ΔCterm (1-Val681) and golgin-245ΔC-term (1-Gly2163) were expressed in pcDNA3.1+ (Clontech) upstream of a GAGAGA linker, an HA tag and a mitochondrial targeting sequence (MAO, residues 481-527); Myc-golgin-245 pSVT7 and FAM21A-GFP were as published 38,39.
Cell culture, transfection and immunofluorescence
Cell lines HeLa (ATCC), 293T (ATCC), and 293T Flp-In T-REx (ThermoFisher Scientific, R78007) were cultured in Dulbecco’s modified Eagle’s medium (DMEM; Invitrogen) supplemented with 10% fetal calf serum (FCS) and penicillin/streptomycin at 37 °C and 5% CO2 unless otherwise stated. The ∆fam91a1 mutant and its WT parent HAP1 cell line were purchased from Horizon Discovery, with FAM91A1-001 having been targeted using the guide GTTGATAAGATCATCGATTC. Cells were tested regularly to ensure that they were mycoplasma free (MycoAlert, Lonza). For immunofluorescence, cells were transfected with plasmid DNA using FuGENE 6 according to the manufacturer’s instructions (Promega). Cells were fixed with 4% formaldehyde in PBS and permeabilised in 0.5% (v/v) Triton-X-100 in PBS. Cells were blocked for one hour in PBS containing 20% FCS and 0.25% Tween-20 and probed in the same buffer with primary antibodies (Supplementary Table 2) and then Alexa-labeled secondary antibodies (Molecular Probes). The cells were mounted in Vectashield (Vector Laboratories) and imaged using LSM 780 (Zeiss) or TCS SP8 (Leica) confocal microscopes. For quantification of TGN46, cells and the Golgi were defined with DAPI and ZFPL1 respectively (NIS-Elements, Nikon). The Golgi mask had a minimum threshold of 18% of the mean ZFPL1 Golgi labeling to exclude out of focus Golgi.
Generation of CRISPR knock out and rescued cell lines
HeLa cells grown in 6 well plates to ~70% confluence were transfected with 0.5 μg pIRESpuro3 and 1.5 μg of pJJS288, pJJS290, or pJJS291 using 6 μl FuGENE 6 in 200 μl Opti-MEM. The cells were trypsinised one day later and diluted into 96 well plates in medium containing 1 μg/ml puromycin, which was replaced with non-selective medium 24 h later. Single colonies were expanded and analysed by immunofluorescence and immunoblotting. To rescue the ∆tbc1d23 mutant HeLa cell line, plasmid pJJS271 was used for retroviral-based packaging from 293T cells, and the supernatant used to transduce ∆tbc1d23 cells grown to ~50% confluence. The medium was changed one day later, and the cells trypsinised two days after transduction and diluted into 96 well plates in medium containing 5 μg/ml blasticidin. Single colonies were expanded and analysed by immunofluorescence and immunoblotting.
siRNA knockdown of TBC1D23
ON-TARGETplus Human TBC1D23 (55773) siRNA SMARTpool was purchased from Dharmacon GE, which targeted AGAGAUCCUUCAAGCGAAU, GGGAGAUUGUUUCACGGAA, GCGCUGAAUUCUGUAGUUA, and CCGUUAAUGUCAGGGAAAA of TBC1D23. HeLa cells at ~30% confluence were resuspended in DMEM medium lacking FCS and antibiotics and transfected with 2 μM of siRNA using Oligofectamine (ThermoFisher Scientific). This step was repeated one day, and after two further days cells tested by immunofluorescence and immunoblotting.
Generation of doxycycline inducible cell lines
293T Flp-In T-REx cell lines containing doxycycline inducible BirA*-GCC185∆Cterm-HA-MAO, golgin-97(1-21)-BirA*-GCC185∆Cterm-HA-MAO, golgin-245(1-21)-BirA*-GCC185∆Cterm-HA-MAO, GCC88(1-59)-BirA*-GCC185∆Cterm-HA-MAO, BirA*-TBC1D23-MAO, and TBC1D23-BirA*-MAO were generated according to the manufacturer’s instructions. Briefly, 293T Flp-In T-REx cells were grown in 6 well plates to ~50% confluence and transfected with 1 μg pOG44 and 1 μg pcDNA5/FRT/TO based-plasmid using 6 μL FuGENE 6 in 200 μl of Opti-MEM. The medium was replaced after one day, and after two days cells were trypsinised, expanded in medium containing 100 μg/ml hygromycin, and verified by induction of expression with 1 μg/ml doxycycline and immunoblotting for the HA epitope.
Affinity capture of biotinylated proteins
The protocol for isolating proteins biotinylated by BirA* was adapted from the BioID method16. Briefly, 293T Flp-In T-REx cell lines containing the BirA* constructs were grown in two 175 cm2 flasks in DMEM supplemented with 10% tetracycline-free FCS to ~70% confluence. 50 μM biotin (10 mg/ml stock in DMSO) and 1 μg/ml doxycycline (1 mg/ml stock in DMSO) were added, and the cells incubated for 24 hours before lysing. For transient transfections, 293T cells were grown in two 175 cm2 flasks to ~50% confluence and transfected with 25 μg plasmid and 75 μl FuGENE 6 in 5 ml Opti-MEM. One day after transfection, biotin was added to 50 μM, and the cells incubated for a further day. For lysis cells were trypsinised, pelleted by centrifugation (3000 rpm, 10 min), washed twice in PBS and resuspended in 1 ml lysis buffer (50 mM Tris pH 7.4, 0.1 M NaCl, 1 mM EDTA, 1% Triton X-100, 1 mM PMSF, cOmplete protease inhibitor cocktail (Roche), and incubated on ice for 20 min. After centrifugation at 13,000 rpm for 5 min at 4 °C, the supernatants were mixed with 1 ml of 50 mM Tris pH 7.4. 5% of this sample was saved for immunoblotting, and the rest added to 500 μl Dynabeads MyOne Streptavidin C1 beads (Invitrogen) that had been pre-washed twice in the same buffer. The beads were incubated at 4°C overnight, washed twice in Wash-Buffer 1 (2% SDS PAGE, cOmplete inhibitors), thrice in Wash-Buffer 2 (1% TritonX-100, 0.1% deoxycholate, 500 mM NaCl, 1 mM EDTA, 50 mM HEPES, cOmplete inhibitors, pH 7.5), and thrice in Wash-Buffer 3 (50 mM Tris pH 7.4, 50 mM NaCl, cOmplete inhibitors). Finally, the beads were incubated in 50 μl SDS sample buffer containing 10% β-mercaptoethanol and 3 mM biotin at 98°C for 5 min to dissociate the biotinylated proteins from the beads, 30 μl was run on a gel for mass spectrometry with the remainder reserved for immunoblotting.
In vitro binding assays using recombinant proteins
Golgin N-termini fused to GST, TBC1D23-His6 and His6-MBP-FAM21(356-1341) were expressed in the E. coli strain BL21-GOLD (DE3; Agilent Technologies). Bacteria were grown at 37°C to an OD600 of 0.7 and induced with 100 μM IPTG overnight at 15°C. Cells were harvested by centrifugation, dounce homogenised and sonicated in lysis buffer (50 mM Tris-HCl, pH 7.4, 150 mM NaCl, 1 mM EDTA, 1% Triton X-100, 1 mM PMSF and cOmplete inhibitors). Lysates were clarified by centrifugation at 12,000 g for 15 min and for golgin NT-GST applied at saturating concentrations to glutathione-Sepharose beads (GE Healthcare) for 1 h with rotation at 4 °C. Beads were washed once in high salt buffer (lysis buffer containing 500 mM NaCl), followed by several washes in lysis buffer. Lysate from bacteria expressing TBC1D23-His6 was also incubated with glutathione-Sepharose beads to pre-clear the sample prior to incubation with 50 μl golgin NT-GST coated beads for 1 h with rotation at 4°C. Beads were washed extensively in lysis buffer. In some experiments an additional incubation with pre-cleared lysate from bacteria expressing His6-MBP-FAM21(356-1341) was also performed for 1 h at 4°C before further washes in lysis buffer. In all cases samples were eluted in SDS sample buffer containing 1 mM β-mercaptoethanol. 5% of each eluate and 1% of the total lysates were separated on SDS-PAGE gels and stained with Coomassie InstantBlue (Expedeon). The interaction between GST-FAM21(356-1341) expressed in pGEX6p2, TBC1D23 expressed without a tag, and golgin-97 and golgin-245 (residues 1-21) fused to MBP-His6 was performed essentially as described above. Likewise for the interaction between FAM91A1(1-328)-His6 and GST fusions to TBC1D23 fragments expressed in pGEX6p2.
Affinity chromatography using GST-TBC1D23 fragments
Glutathione-Sepharose beads saturated with GST-TBC1D23 fusion proteins were incubated with lysates from two confluent 175 cm2 flasks of 293T cells. 293T cells were collected by centrifugation, washed once in ice-cold PBS and lysed in lysis buffer (25 mM Tris-HCl, pH 7.4, 150 mM NaCl, 1 mM EDTA, 1% (v/v) Triton X-100, 1 mM PMSF, cOmplete inhibitors) followed by clarification by centrifugation. Lysates were pre-cleared on glutathione-Sepharose beads prior to a two hour incubation with TBC1D23-coated beads. Beads were washed extensively in lysis buffer and proteins eluted first with high salt elution buffer (25 mM Tris-HCl, 1.5 M NaCl, 1 mM EDTA) to release interacting proteins, and then SDS sample buffer to release the GST-fusion proteins. Proteins were chloroform/methanol precipitated from the high salt eluate and resuspended in SDS sample buffer containing 1 mM β-mercaptoethanol. Lysates and eluates were separated by SDS-PAGE and analysed either by mass spectrometry or transfered to nitrocellulose for immunoblotting.
Immunoprecipitation of GFP-tagged TBC1D23
Two 175 cm2 flasks of 293T cells at ~75% confluency were transfected with plasmids encoding GFP-tagged TBC1D23 using FuGENE 6 according to the manufacturer’s instructions. Cells were harvested by centrifugation, washed once in ice-cold PBS and lysed in lysis buffer (25 mM Tris-HCl pH 7.4 150 mM NaCl, 0.5 mM EDTA, 0.5% (V/V) Triton X-100, 1mM PMSF cOmplete inhibitors) for 30 min at 4 °C. Lysates were clarified by centrifugation before GFP-tagged proteins were isolated by incubation with 25 μL of packed GFP-Trap beads (Chromotek) for two hours with rotation at 4 °C. Beads were washed in lysis buffer and bound proteins eluted by the addition of 100 μL 2 x SDS sample buffer. Eluates were analysed by mass spectrometry and immunoblotting.
Purification of the WASH complex from mWASH 293 cells
The WASH complex was purified from 293 cells stably expressing His6-Protein C tagged murine WASH1 as described previously40. Briefly cells from eight T150 flasks were pelleted by centrifugation at 500 x g for 5 min at 4°C, washed in PBS and resuspended in lysis buffer (200 mM NaCl, 1 mM CaCl2, 1% Triton X-100, 50 mM Na-HEPES, pH 7.4). Cells were lysed at 4°C for one hour, centrifuged at 13000 x g for 15 min and the supernatant rotating with 100 μl PC affinity resin for 4 h at 4°C. The beads were washed once in lysis buffer, once in WHB1 buffer (200 mM NaCl, 1 mM CaCl2, 0.02% methyl-cellulose, 50 mM Na-HEPES, pH 7.4) and once in WHB2 buffer (WHB1 without CaCl2) prior to elution by Ca2+ chelation using WHB2 supplemented with 5 mM EGTA. The eluate was incubated with 100 μl glutathione Sepharose resin saturated with GST-TBC1D23 fusions and affinity purification performed as described above.
Mass spectrometry
Samples were loaded on 4-20% Tris-glycine SDS-PAGE gels and run for 1-2 centimeters. Proteins were stained with Coomassie InstantBlue, the entire gel lane cut into eight slices that were placed in a 96-well plate and destained with 50% v/v acetonitrile and 50 mM ammonium bicarbonate, reduced with 10 mM DTT, and alkylated with 55 mM iodoacetamide. Digestion was with 6 ng/μl trypsin (Promega, UK) overnight at 37°C, and peptides extracted in 2% v/v formic acid 2% v/v acetonitrile, and analysed by nano-scale capillary LC-MS/MS (Ultimate U3000 HPLC, Thermo Scientific Dionex) at a flow of ~ 300 nL/min. A C18 Acclaim PepMap100 5 μm, 100 μm x 20 mm nanoViper (Thermo Scientific Dionex), trapped the peptides prior to separation on a C18 Acclaim PepMap100 3 μm, 75 μm x 250 mm nanoViper. Peptides were eluted with an acetonitrile gradient. The analytical column outlet was interfaced via a nano-flow electrospray ionisation source with a linear ion trap mass spectrometer (Orbitrap Velos, Thermo Scientific). Data dependent analysis was performed using a resolution of 30,000 for the full MS spectrum, followed by ten MS/MS spectra in the linear ion trap. MS spectra were collected over a m/z range of 300–2000. MS/MS scans were collected using a threshold energy of 35 for collision-induced dissociation. LC-MS/MS data were searched against the UniProt KB database using Mascot (Matrix Science), with a precursor tolerance of 5 ppm and a fragment ion mass tolerance of 0.8 Da. Two missed enzyme cleavages and variable modifications for oxidised methionine, carbamidomethyl cysteine, pyroglutamic acid, phosphorylated serine, threonine and tyrosine were included. MS/MS data were validated using the Scaffold programme (Proteome Software Inc).
Immunoblotting
One 75 cm2 flask of cells at ~90% confluence was harvested by scraping and centrifugation, washed twice with ice-cold PBS, resuspended in 300 to 500 μl lysis buffer (50 mM Tris pH 7.4, 0.1 M NaCl, 1 mM EDTA, 1% Triton X-100, 1 mM PMSF, cOmplete inhibitors) for 30 min on ice, clarified by centrifugation, and protein concentration determined (Bio-Rad). Lysate concentrations were normalised, boiled in NuPAGE SDS Sample Buffer containing 10% β-mercaptoethanol at 90°C for 5 min, run on a gel and transferred to nitrocellulose. All blots, except for those with Streptavidin-HRP, were blocked in 5% (w/v) milk in PBS-T (PBS with 0.1% (v/v) Tween-20) for 1 hour, incubated overnight at 4°C with primary antibody in the same blocking solution (Supplementary Table 2), washed thrice with PBS-T for 5 minutes, incubated with HRP-conjugated secondary antibody in 0.1% (w/v) milk in PBS-T for one hour, washed five times with PBS, and detected with either Immobilon Western HRP substrate or Amersham ECL detection reagent. For Streptavidin-HRP blocking (one hour), incubation (one hour) and washing (five times) were all in PBS 2.5% FCS 0.4% Triton X-100.
Electron microscopy
HeLa cells were grown on glass bottom petri dishes (MatTek) to 70% confluency and transfected with pJJS243 or pMX0167. After 36 hours fixed at room temperature in 2% glutaraldehyde (EM grade, Agar Scientific) in 0.1 M cacodylate buffer pH 7.4 (CB) for 30-60 min on ice. Cells rinsed five times with CB for two minutes, blocked in 50 mM glycine in CB for 5 min on ice, and rinsed in CB again. For APEX-catalysed oxidation of 3,3-diaminobenzidine (DAB), freshly prepared 0.5 mg/ml of DAB freebase (Sigma) in HCL and 0.03 % H2O2 (10 mM; Sigma) was added and upon a brown precipitate (~15 minutes) the reaction halted by rinsing with CB fives times. Samples were post-fixed with 1% OsO4 in CB for one hour in the dark and on ice, rinsed 5x ten minutes in chilled deionised water, dehydrated in a chilled ascending ethanol series (30%, 50%, 70%, 90%, 100%) with 100% ethanol at room temperature, then infiltrated with CY212 epoxy resin and polymerised. Areas containing dark granules were excised, mounted onto dummy resin blocks and sectioned using a Leica ultramicrotome parallel to the substratum. Pale gold 70 nm sections were contrasted with saturated aqueous uranyl acetate and Reynold’s lead citrate. Electron micrographs were recorded at 80kV on an FEI Technai Spirit TEM. The distance between mitochondrial membranes was measured using the segmented lines tool in ImageJ.
CI-MPR transport assay
To monitor retrograde CI-MPR traffic, cells were incubated at 37°C with 10 μg/ml anti-CI-MPR antibody in serum-free DMEM for 10 minutes, rinsed with ice cold PBS, then low-pH solution (300 mM NaCl, 5mM KCl, 1 mM CaCl2, 1 mM MgCl2, 0.2 M acetic acid, pH 2.5), then PBS, then chased with antibody-free medium for 45 min at 37 °C, and then fixed and immunostained for the internalised antibodies. DAPI, giantin were used to define individual cells and the Golgi respectively using NIS-Elements. The Golgi mask had a minimum threshold of 30% of the mean giantin Golgi labeling to exclude out of focus Golgi. The ratio of the mean intensity of CI-MPR within the Golgi over total CI-MPR inside the cell was quantified to obtain the final CI-MPR internalisation ratio.
Statistics and reproducibility
Statistical differences between data sets were analysed using unpaired, two-tailed Mann-Whitney tests using Prism 7, with n and P values stated in the figure legends. All microscopy experiments were repeated independently three times with similar results with the exception of Figs. 3b and 5e, and Supplementary Figs 1b and 3b,d which were repeated twice with similar results. All biochemical experiments were repeated independently three times with similar results with the exception of Figs 1b, 2c, 4d and Supplementary Figs 4a,b,f which were repeated twice with similar results.
Data availability
Source data for Figs 3c, 5f and Supplementary Fig. 4d have been provided in Supplementary Table 1. All data supporting the findings of this study are available from the corresponding author on reasonable request.
Supplementary Material
Acknowledgements
We are indebted to Ramanujan Hegde and Ben Nichols for comments on the manuscript, to Tim Stevens for phylogenetic profiling, to Emmanuel Derivery, Alexis Gautreau, Olga Perisic, Felix Randow and Matthew Seaman for reagents, and to Margaret Robinson for communicating results prior to publication. Funding was from the Medical Research Council (MRC file reference number MC_U105178783), and by a European Molecular Biology Organisation long-term fellowship to John Shin.
Footnotes
Author contributions
S.M., A.G. and J.S. devised and planned the study. A.G. and J.S. performed all the experiments except for electron microscopy (J.C.) and mass spectrometric protein identification (F.B.). S.M. wrote the manuscript.
Competing financial interests
The authors declare no competing financial interests.
References
- 1.Baker RW, Hughson FM. Chaperoning SNARE assembly and disassembly. Nat Rev Mol Cell Biol. 2016;17:465–479. doi: 10.1038/nrm.2016.65. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Hong W, Lev S. Tethering the assembly of SNARE complexes. Trends Cell Biol. 2014;24:35–43. doi: 10.1016/j.tcb.2013.09.006. [DOI] [PubMed] [Google Scholar]
- 3.Waters MG, Pfeffer SR. Membrane tethering in intracellular transport. Curr Opin Cell Biol. 1999;11:453–459. doi: 10.1016/s0955-0674(99)80065-9. [DOI] [PubMed] [Google Scholar]
- 4.Yu I-M, Hughson FM. Tethering factors as organizers of intracellular vesicular traffic. Annu Rev Cell Dev Biol. 2010;26:137–156. doi: 10.1146/annurev.cellbio.042308.113327. [DOI] [PubMed] [Google Scholar]
- 5.Gillingham AK, Munro S. Finding the Golgi: golgin coiled-coil proteins show the way. Trends Cell Biol. 2016;26:399–408. doi: 10.1016/j.tcb.2016.02.005. [DOI] [PubMed] [Google Scholar]
- 6.Munro S. The golgin coiled-coil proteins of the Golgi apparatus. Cold Spring Harb Perspect Biol. 2011;3 doi: 10.1101/cshperspect.a005256. a005256. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Goud B, Gleeson PA. TGN golgins, Rabs and cytoskeleton: regulating the Golgi trafficking highways. Trends Cell Biol. 2010;20:329–336. doi: 10.1016/j.tcb.2010.02.006. [DOI] [PubMed] [Google Scholar]
- 8.Witkos TM, Lowe M. Recognition and tethering of transport vesicles at the Golgi apparatus. Curr Opin Cell Biol. 2017;47:16–23. doi: 10.1016/j.ceb.2017.02.003. [DOI] [PubMed] [Google Scholar]
- 9.Papanikou E, Glick BS. Golgi compartmentation and identity. Curr Opin Cell Biol. 2014;29:74–81. doi: 10.1016/j.ceb.2014.04.010. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Gomez-Navarro N, Miller E. Protein sorting at the ER-Golgi interface. J Cell Biol. 2016;215:769–778. doi: 10.1083/jcb.201610031. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Wong M, Munro S. Membrane trafficking. The specificity of vesicle traffic to the Golgi is encoded in the golgin coiled-coil proteins. Science. 2014;346:1256898. doi: 10.1126/science.1256898. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Lu L, Tai G, Hong W. Autoantigen Golgin-97, an effector of Arl1 GTPase, participates in traffic from the endosome to the trans-Golgi network. Mol Biol Cell. 2004;15:4426–4443. doi: 10.1091/mbc.E03-12-0872. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Cheung P-YP, Pfeffer SR. Transport vesicle tethering at the trans Golgi network: coiled coil proteins in action. Front Cell Dev Biol. 2016;4:18. doi: 10.3389/fcell.2016.00018. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Bonifacino JS, Rojas R. Retrograde transport from endosomes to the trans-Golgi network. Nat Rev Mol Cell Biol. 2006;7:568–579. doi: 10.1038/nrm1985. [DOI] [PubMed] [Google Scholar]
- 15.Wong M, Gillingham AK, Munro S. The golgin coiled-coil proteins capture different types of transport carriers via distinct N-terminal motifs. BMC Biol. 2017;15:3. doi: 10.1186/s12915-016-0345-3. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Roux KJ, Kim DI, Raida M, Burke B. A promiscuous biotin ligase fusion protein identifies proximal and interacting proteins in mammalian cells. J Cell Biol. 2012;196:801–810. doi: 10.1083/jcb.201112098. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Hirst J, et al. Contributions of epsinR and gadkin to clathrin-mediated intracellular trafficking. Mol Biol Cell. 2015;26:3085–3103. doi: 10.1091/mbc.E15-04-0245. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Pfeffer SR. Entry at the trans-face of the Golgi. Cold Spring Harb Perspect Biol. 2011;3 doi: 10.1101/cshperspect.a005272. a005272. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Fukuda M. TBC proteins: GAPs for mammalian small GTPase Rab? Biosci Rep. 2011;31:159–168. doi: 10.1042/BSR20100112. [DOI] [PubMed] [Google Scholar]
- 20.Frasa MAM, Koessmeier KT, Ahmadian MR, Braga VMM. Illuminating the functional and structural repertoire of human TBC/RABGAPs. Nat Rev Mol Cell Biol. 2012;13:67–73. doi: 10.1038/nrm3267. [DOI] [PubMed] [Google Scholar]
- 21.Borner GHH, et al. Fractionation profiling: a fast and versatile approach for mapping vesicle proteomes and protein-protein interactions. Mol Biol Cell. 2014;25:3178–3194. doi: 10.1091/mbc.E14-07-1198. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Bassik MC, et al. A systematic mammalian genetic interaction map reveals pathways underlying ricin susceptibility. Cell. 2013;152:909–922. doi: 10.1016/j.cell.2013.01.030. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Huttlin EL, et al. The BioPlex Network: a systematic exploration of the human interactome. Cell. 2015;162:425–440. doi: 10.1016/j.cell.2015.06.043. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Harbour ME, Breusegem SY, Seaman MNJ. Recruitment of the endosomal WASH complex is mediated by the extended ‘tail’ of Fam21 binding to the retromer protein Vps35. Biochem J. 2012;442:209–220. doi: 10.1042/BJ20111761. [DOI] [PubMed] [Google Scholar]
- 25.Gomez TS, Billadeau DD. A FAM21-containing WASH complex regulates retromer-dependent sorting. Dev Cell. 2009;17:699–711. doi: 10.1016/j.devcel.2009.09.009. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Derivery E, et al. The Arp2/3 activator WASH controls the fission of endosomes through a large multiprotein complex. Dev Cell. 2009;17:712–723. doi: 10.1016/j.devcel.2009.09.010. [DOI] [PubMed] [Google Scholar]
- 27.Seaman MNJ, Gautreau A, Billadeau DD. Retromer-mediated endosomal protein sorting: all WASHed up! Trends Cell Biol. 2013;23:522–528. doi: 10.1016/j.tcb.2013.04.010. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Kulak NA, Pichler G, Paron I, Nagaraj N, Mann M. Minimal, encapsulated proteomic-sample processing applied to copy-number estimation in eukaryotic cells. Nat Meth. 2014;11:319–324. doi: 10.1038/nmeth.2834. [DOI] [PubMed] [Google Scholar]
- 29.Jia D, Gomez TS, Billadeau DD, Rosen MK. Multiple repeat elements within the FAM21 tail link the WASH actin regulatory complex to the retromer. Mol Biol Cell. 2012;23:2352–2361. doi: 10.1091/mbc.E11-12-1059. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Roquemore EP, Banting G. Efficient trafficking of TGN38 from the endosome to the trans-Golgi network requires a free hydroxyl group at position 331 in the cytosolic domain. Mol Biol Cell. 1998;9:2125–2144. doi: 10.1091/mbc.9.8.2125. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Hao Y-H, et al. Regulation of WASH-dependent actin polymerization and protein trafficking by ubiquitination. Cell. 2013;152:1051–1064. doi: 10.1016/j.cell.2013.01.051. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Varandas KC, Irannejad R, von Zastrow M. Retromer endosome exit domains serve multiple trafficking destinations and regulate local G protein activation by GPCRs. Curr Biol. 2016;26:3129–3142. doi: 10.1016/j.cub.2016.09.052. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Bordo D, Bork P. The rhodanese/Cdc25 phosphatase superfamily. Sequence-structure-function relations. EMBO Rep. 2002;3:741–746. doi: 10.1093/embo-reports/kvf150. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Rojas R, Kametaka S, Haft CR, Bonifacino JS. Interchangeable but essential functions of SNX1 and SNX2 in the association of retromer with endosomes and the trafficking of mannose 6-phosphate receptors. Mol Cell Biol. 2007;27:1112–1124. doi: 10.1128/MCB.00156-06. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Meyer C, et al. mu1A-adaptin-deficient mice: lethality, loss of AP-1 binding and rerouting of mannose 6-phosphate receptors. EMBO J. 2000;19:2193–2203. doi: 10.1093/emboj/19.10.2193. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Marin-Valencia I, et al. Homozygous mutations in TBC1D23 lead to a non-degenerative form of pontocerebellar hypoplasia. Am J Hum Genet. 2017 doi: 10.1016/j.ajhg.2017.07.015. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Ivanova EL, et al. Homozygous truncating variants in TBC1D23 cause pontocerebellar hypoplasia and alter cortical development. Am J Hum Genet. 2017 doi: 10.1016/j.ajhg.2017.07.010. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Kjer-Nielsen L, van Vliet C, Erlich R, Toh BH, Gleeson PA. The Golgi-targeting sequence of the peripheral membrane protein p230. J Cell Sci. 1999;112:1645–1654. doi: 10.1242/jcs.112.11.1645. [DOI] [PubMed] [Google Scholar]
- 39.Harbour ME, et al. The cargo-selective retromer complex is a recruiting hub for protein complexes that regulate endosomal tubule dynamics. J Cell Sci. 2010;123:3703–3717. doi: 10.1242/jcs.071472. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Derivery E, Gautreau A. Assaying WAVE and WASH complex constitutive activities toward the Arp2/3 complex. Meth Enzymol. 2010;484:677–695. doi: 10.1016/B978-0-12-381298-8.00033-2. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
Data Availability Statement
Source data for Figs 3c, 5f and Supplementary Fig. 4d have been provided in Supplementary Table 1. All data supporting the findings of this study are available from the corresponding author on reasonable request.