Abstract
Purpose
Pancreatic cysts are estimated to be present 2–3% of the adult population. Unfortunately, current diagnostics do not accurately distinguish benign cysts from those that can progress into invasive cancer. Missregulated pericellular proteolysis is a hallmark of malignancy, and therefore, we used a global approach to discover protease activities that differentiate benign nonmucinous cysts from premalignant mucinous cysts.
Experimental Design
We employed an unbiased and global protease profiling approach to discover protease activities in 23 cyst fluid samples. The distinguishing activities of select proteases was confirmed in 110 samples using specific fluorogenic substrates and required less than 5 µL of cyst fluid.
Results
We determined that the activities of the aspartyl proteases gastricsin and cathepsin E are highly increased in fluid from mucinous cysts. Immunohistochemical analysis revealed that gastricsin expression was associated with regions of low-grade dysplasia, whereas cathepsin E expression was independent of dysplasia grade. Gastricsin activity differentiated mucinous from nonmucinous cysts with a specificity of 100% and a sensitivity of 93%, whereas cathepsin E activity was 92% specific and 70% sensitive. Gastricsin significantly outperformed the most widely used molecular biomarker, carcinoembryonic antigen (CEA), which demonstrated 94% specificity and 65% sensitivity. Combined analysis of gastricsin and CEA resulted in a near perfect classifier with 100% specificity and 98% sensitivity.
Conclusions
Quantitation of gastricsin and cathepsin E activities accurately distinguished mucinous from nonmucinous pancreatic cysts and has the potential to replace current diagnostics for analysis of these highly prevalent lesions.
Keywords: Proteolysis, pancreatic cyst, biomarker, aspartyl protease, pancreatic cancer
Introduction
The detection of pancreatic cysts has increased dramatically due to the rising use of high-resolution abdominal imaging. Pancreatic cysts are incidentally detected in 13–45% of patients evaluated by magnetic resonance imaging and 2% of patients evaluated by computed tomography (1–3). The most frequently detected pancreatic cysts include intraductal papillary mucinous neoplasms (IPMNs), mucinous cystic neoplasms (MCNs), pseudocysts, and serous cystadenomas (SCAs) (4). Both IPMNs and MCNs, which are collectively referred to as mucinous cysts, may develop foci of high-grade dysplasia or cancer (5). At the time of resection, ~30% of IPMNs and ~15% of MCNs contain invasive cancer (6,7). Pseudocysts and SCAs, which are both nonmucinous, rarely undergo malignant degeneration and are considered benign lesions that typically do not require resection or continued surveillance. Clinical decision making for pancreatic cysts relies largely on radiographic and clinical features, augmented by analysis of cyst fluid collected by endoscopic ultrasound with fine needle need aspiration (EUS-FNA) (8). Unfortunately, with current clinical guidelines, distinguishing nonmucinous from mucinous cysts remains a challenge. The preoperative diagnosis of mucinous cysts is incorrect in up to 30% of cases and benign cysts are often resected, exposing patients to an unnecessary risk for morbidity (9–12).
As abdominal imaging remains unable to accurately differentiate pancreatic cyst types, there has been considerable effort towards developing improved diagnostic biomarkers. Most of these biomarkers utilize cyst fluid collected by EUS-FNA. CEA is the most widely investigated biomarker and is 60–80% accurate for differentiating mucinous from nonmucinous cysts (13,14). KRAS mutations occur in more than 90% of pancreatic cancers and are frequently observed in mucinous cysts (15,16). Analysis of cyst fluid DNA revealed that KRAS mutations are 100% specific, but only 50% sensitive for diagnosing a mucinous cyst (17). Similarly, analysis of mutations in the oncogene GNAS are specific for diagnosing IPMNs, but suffer from low sensitivity (18). A variety of other cyst fluid biomarkers have also been explored (19–23); however, CEA remains the only widely applied molecular biomarker for differentiating mucinous from nonmucinous cysts.
Proteases mediate a variety of critical processes in cancer, including invasion of the basement membrane via cleavage of extracellular matrix proteins and promotion of oncogenic signaling pathways through activation of growth factors and receptor tyrosine kinases (24,25). In pancreatic cancer, members of the cathepsin family of endolysosomal proteases are upregulated and found extracellularly. Aberrant secretion leads to cleavage of extracellular substrates, driving increased cellular invasion (26). Either genetic deletion or pharmacological inhibition of cysteine cathepsin activity decreases tumor progression and invasion (27,28).
Gene expression profiling studies of IPMNs and MCNs indicate overexpression of a range of proteases (29–31). Furthermore, analysis of protein expression in cyst fluid showed substantial differences in the abundance of pancreatic proteases and their cognate inhibitors between cyst types. The serine protease inhibitor SPINK1 was recently investigated as a biomarker for differentiating benign from malignant cysts (32,33). Collectively, these results suggest that there may be altered levels of proteolytic activity between mucinous and nonmucinous cysts and that these differences could be exploited to distinguish the type of lesion and its associated malignant potential.
In the current study, we applied a global protease profiling technology to discover proteolytic activity markers for differentiating mucinous from nonmucinous cysts. Using this approach, we identified enhanced aspartyl protease activity in mucinous cysts, due to upregulation of gastricsin and cathepsin E. We characterized the localization of both aspartyl proteases within the dysplastic tissue surrounding the mucinous cysts and determined that gastricsin expression was dependent on the degree of dysplasia. Lastly, highly selective fluorescent substrates for gastricsin and cathepsin E both confirmed their upregulated activities and outperformed CEA for differentiating mucinous from nonmucinous pancreatic cystic lesions.
Materials and methods
Patients and sample acquisition
Pancreatic cyst fluid samples were collected from preconsented patients under institutional review board approved protocols and in accordance with U. S. Common Rule at the University of California San Francisco, the University of Pittsburgh Medical Center, Indiana University School of Medicine, and Stanford University School of Medicine. Patient information is summarized in Table S1. All patients included in our study underwent surgical resection of their cystic lesion and have a pathologically confirmed diagnosis. The highest grade of dysplasia observed during pathological evaluation of each cystic lesion is reported. Samples were collected either at the time of surgical resection or during diagnostic endoscopic ultrasound prior to resection of the cystic lesion. Cyst fluid samples were split into 100 µL aliquots and frozen to −80 °C within 60 minutes of collection. Samples underwent at most two freeze-thaw cycles prior to experimental analysis. Total cyst fluid protein concentration was determined by the bicinchoninic acid assay. CEA levels were evaluated for the majority of samples, but were unavailable in 21 cases due to limited cyst fluid volume.
Multiplex substrate profiling by mass spectrometry (MSP-MS) assay
The MSP-MS assay was performed as previously described (34). Cyst fluid was diluted to 100 µg/mL in assay buffer (either pH 7.5 phosphate buffer or pH 3.5 acetate buffer) and pre-incubated for 10 minutes. For analysis of protease inhibitor sensitivity, 1 mM AEBSF (Sigma, A8456), 2 µM E-64 (Sigma, E3132), 2 µM pepstatin (Sigma, P5318), 2 mM 1,10-phenanthroline (Sigma, 131337), or DMSO were included in pre-incubation. The 228 tetradecapeptide library was split into two pools and diluted in assay buffer to a concentration of 1 µM of each peptide. 75 µL of diluted cyst fluid and peptide pools were then combined and incubated at room temperature. 30 µL aliquots were removed after 15 and 60 minutes, protease activity quenched with 8 M guanidinium hydrochloride, and flash-frozen in liquid N2. For recombinant gastricsin (R&D Systems, 6186-AS), cathepsin D (R&D Systems, 1014-AS), and cathepsin E (R&D Systems, 1294-AS), the MSP-MS assay was performed as described above with slight modifications: 10 nM of recombinant protease in pH 3.5 acetate buffer was used and aliquots were removed after 15, 60, and 240 minutes.
Prior to peptide cleavage site identification by mass spectrometry, samples were desalted using C18 tips (Rainin). Mass spectrometry analysis was carried out with an LTQ Orbitrap XL mass spectrometer coupled to a 10,000 psi nanoACQUITY Ultra Performance Liquid Chromatography (UPLC) System (Waters) for peptide separation by reverse phase liquid chromatography (RPLC). Peptides were separated over a C18 column (Thermo) and eluted by applying a flow rate of 300 nL/min with a 65-minute linear gradient from 2–30% acetonitrile. Survey scans were recorded over a 325–1500 m/z range and the six most intense precursor ions were fragmented by collision-induced dissociation (CID) for MS/MS.
Raw mass spectrometry data was processed to generate peak lists using MSConvert. Peak lists were then searched in Protein Prospector v. 5.10.0 (35) against a custom database containing the sequences from the 228 tetradecapeptide library. Searches used a mass accuracy tolerance of 20 ppm for precursor ions and 0.8 Da for fragment ions. Variable modifications included N-terminal pyroglutamate conversion from glutamine or glutamate and oxidation of tryptophan, proline, and tyrosine. Searches were subsequently processed using the MSP-xtractor software (http://www.craiklab.ucsf.edu/extractor.html), which extracts the peptide cleavage site and spectral counts of the corresponding cleavage products. Spectral counts were used for the relative quantification of peptide cleavage products.
Proteomic analysis of cyst fluid samples
Cyst fluid samples were processed for proteomic analysis using a standard protocol. Briefly, 8 µg of cyst fluid protein was denatured in 40 µL of 6 M urea. Disulfide bonds were reduced with 10 mM dithiothreitol and free thiols were subsequently alkylated with 12.5 mM iodoacetamide. Samples were then diluted to with 25 mM ammonium bicarbonate to 2 M urea and digested with 100 ng trypsin for 16 hours at 37 °C. Following trypsin digestion, samples were desalted with C18 tips (Rainin), dried, and resuspended in 0.1% formic acid. Triplicate LC-MS/MS analysis of all samples was performed as described above and details for this and protein identification are provided in the Supplemental Methods and Materials.
Label-free quantitation was used to compare relative abundance of the three aspartyl proteases identified in cyst fluid samples. The Skyline software package was used to obtain extracted ion chromatograms and peak areas for precursor ions from the aspartyl proteases (36). To correct for potential differences in protein loading between runs, peak areas were normalized by the median peak area of all fragmented ions from that run. The average peak area of the precursor ions from a given aspartyl protease was then used to estimate the abundance in each cyst fluid sample.
Western blots of gastricsin and cathepsin E
Cyst fluid protein (2 µg) or recombinant protease (20 ng) was pre-incubated for 30 minutes in either pH 7.5 phosphate buffer or pH 3.5 acetate buffer. Samples were then subjected to electrophoresis on a 10% NuPAGE Bis-Tris gel. Proteins were transferred to polyvinylidene fluoride membranes and blocked in Tris-buffered saline with 0.1% Tween (TBS-T) and 5% (w/v) non-fat dry milk for 2 hours at room temperature. Membranes were then incubated with either rabbit anti-gastricsin antibody (1:500; Abcam, ab104595) or rabbit anti-cathepsin E antibody (1:1,000; Abcam, ab49800) for 1 hour at room temperature. Following a wash in TBS-T, horseradish peroxidase (HRP)-conjugated secondary antibody (1:15,000; Abcam, ab97051) was applied for 2 hours at room temperatures. Proteins were detected with the enhanced chemiluminescence (ECL) detection system (Thermo).
Animal strains
The following mice strains were used: Ptf1a-Cre (gift of Christopher Wright, Vanderbilt University, Nashville, Tennessee, USA), LSL-KrasG12D (gift of Dave Tuveson, Cold Spring Harbor Laboratory, USA), Brg1f/f (gift of David Reisman, University of Florida, USA with permission of Pierre Chambon). Mice were crossed on a mixed background. The UCSF Institutional Care and Use of Animals Committee (IACUC) approved all mouse experiments.
Immunohistochemical analysis of pancreatic tissue
Tissue samples were obtained from patients who underwent resection of pancreatic cystic lesions at UCSF. Gastricsin and cathepsin E immunohistochemistry assays were developed and performed on a Ventana Discovery Ultra automated slide stainer (Ventana Medical Systems). In brief, formalin-fixed, paraffin-embedded (FFPE) samples (4 µm sections) were deparaffinized using EZPrep (Ventana Medical Systems) followed by treatment with antigen retrieval buffer (Ventana Medical Systems, 950-124). Specimens were incubated with either goat anti-gastricsin antibody (1:300; Santa Cruz, sc-51185) or goat anti-cathepsin E antibody (1:200; Santa Cruz, sc-6508) for 32 minutes at 36 °C. OmniMap anti-goat secondary antibody (Ventana Medical Systems, 760-4647) was then applied for 16 minutes before employing a DAB detection kit (Ventana Medical Systems, 760-500). All samples were counterstained with haematoxylin and Bluing Reagent (Ventana Medical Systems, 760-2037). H&E staining of tissue sections was performed using standard protocols.
Mouse pancreatic tissue samples were collected from 8 Ptf1a-Cre; LSL-KrasG12D; Brg1f/f animals between 3 and 40 weeks of age. FFPE samples (5 µm sections) were deparaffinized with xylene and subsequently rehydrated. Sections were either subjected to H&E staining or heat-induced epitope retrieval with Citra buffer (BioGenex; HK086). Primary antibodies (goat anti-mouse) for cathepsin E (1:1,000; R&D Systems; AF1130) and gastricsin (1:1,000; Santa Cruz; sc-51188) were incubated with sections overnight at 4 °C. Anti-goat secondary antibody (1:200; Vector Labs; BA-9500) was then added to sections for 1 hour at room temperature. ABC (Vector Labs; PK-6100) and DAB kits (Vector Labs; SK-4100) were employed for detection. Sections were counterstained with haematoxylin and incubated in 0.25% ammonium hydroxide for bluing.
Peptide synthesis
Synthesis of internally quenched fluorescent peptides was conducted using standard Fmoc solid-phase peptide synthesis on a Syro II automated synthesizer (Biotage). Details are included in the Supplemental Methods and Materials.
Fluorescent protease activity assays
All fluorescent protease activity assays were performed in triplicate in black, round-bottom 384 well plates. Assays were run for 1 hour in 15 µL of acetate buffer with 0.01% Tween. The pH of the acetate buffer was adjusted to promote activity of either aspartyl protease (pH 3.5 for the cathepsin E substrate and pH 2.0 for the gastricsin substrate). 10 µM of substrate was used for all assays (unless otherwise indicated) and was incubated with either 10 nM of recombinant protease or 50 µg/mL of cyst fluid protein. For kinetic analysis of gastricsin activity, the substrate concentration ranged from 0.1–25 µM. Fluorescent substrate cleavage was monitored with a Biotek Synergy HT plate reader using excitation and emission wavelengths of 328 nm and 393 nm, respectively. Selectivity of the recombinant proteases was assessed by comparing the initial velocity of substrate hydrolysis in relative fluorescent units per second (RFU/sec). For cyst fluid samples, we compared the change in endpoint RFU relative to wells that contained substrate, but no cyst fluid.
Statistical analysis and data presentation
A two-tailed Mann-Whitney U test was used to compare the differences in CEA abundance, gastricsin activity, and cathepsin E activity between mucinous and nonmucinous cysts. Univariate and multivariate logistic regression models were used for cyst prediction. Receiver operating characteristic (ROC) curves and Youden’s J statistic were employed to identify the optimal cutoff. All mass spectrometry data (spectral counts and peak areas) was log2 transformed and analyzed with unpaired two-tailed t-tests. GraphPad Prism was used to fit kinetic data and generate scatter plots and bar charts. Volcano plots, heat maps, venn diagrams, ROC curves, and logistic regression models were generated in RStudio v. 0.98.1091. iceLogo software was used to visualize patterns in peptide cleavage sites at ±4 positions away from the scissile bond (37).
Results
Global protease activity profiling of patient cyst fluid
To identify differences in proteolytic activity between mucinous and nonmucinous cysts we used our MSP-MS assay, which is a global and unbiased substrate-based protease profiling approach (34). In the MSP-MS assay, a physicochemically diverse library of 228 tetradecapeptide substrates is incubated with a protease-containing sample of interest and tandem mass spectrometry is used to monitor protease-derived peptide cleavage products. We have previously validated this assay through analysis of all classes of protease and used it to develop selective substrate probes (38–40).
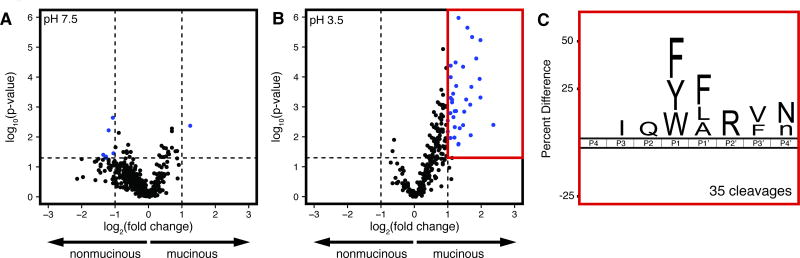
Using the MSP-MS assay, we profiled 16 mucinous and 7 nonmucinous cyst fluid samples. To capture a broad range of protease activities, we performed the assay under acidic conditions and at neutral pH. At pH 7.5, we detected a total of 1117 unique peptide cleavages among the patient sample set (Fig. 1A). Only 7 peptide cleavages met our selectivity criteria for differentiating mucinous from nonmucinous cysts (+/− 1 log2(mucinous/nonmucinous), p < 0.05). 6 of these cleavages were enriched in nonmucinous cysts, and overall, there was a slight trend toward increased proteolytic activity in fluid from nonmucinous cysts (Fig. S1A). When the same samples were assayed at pH 3.5, a total of 691 peptide cleavages were detected, and we observed increased proteolytic activity in the mucinous cysts (Fig. 1B and Fig. S1B). All 35 unique substrate cleavages that differentiated mucinous from nonmucinous cysts were enriched in the mucinous set. The degree of dysplasia within a mucinous cyst is also an important factor in determining whether surgical intervention is recommended. However, no major differences in protease activity were evident between mucinous cysts with low- or high-grade dysplasia (Fig. S2).
Fig. 1. Comparison of global proteolytic activity in mucinous and nonmucinous cysts by MSP-MS.
Volcano plots displaying the peptide cleavages generated by mucinous (n=16) and nonmucinous cysts (n=7) when assayed at pH 7.5 (A) or pH 3.5 (B). Spectral counts of peptide cleavage products were used for relative quantification of the fold change (mucinous/nonmucinous) and hypothesis testing. Cleavages that met the criteria for differentiating mucinous from nonmucinous cysts (+/− 1 log2(fold change), p < 0.05) are shown in blue. The substrate specificity of the cleavages within the red box is displayed with an iceLogo plot (C). Residues shown are statistically significant with p < 0.05.
We generated an iceLogo frequency plot to visualize the substrate specificity pattern of the 35 mucinous-specific peptide cleavages detected at pH 3.5 (Fig. 1C) (37). At the P1 and P1′ positions, which flank the cleavage site, there was a predominant enrichment of hydrophobic amino acids with the aromatic residues tyrosine and tryptophan more favored at P1. This mirrors the previously reported substrate specificity of lysosomal aspartyl proteases (41,42).
Identification of cathepsin E and gastricsin in mucinous cysts
We next sought to identify the specific proteases within the mucinous cysts that are responsible for the increased acid-optimal cleavage of the 35 mucinous-specific substrates. To aid in the characterization of protease activity, we initially focused on a single mucinous cyst fluid sample that cleaved 30 out of the 35 substrates.
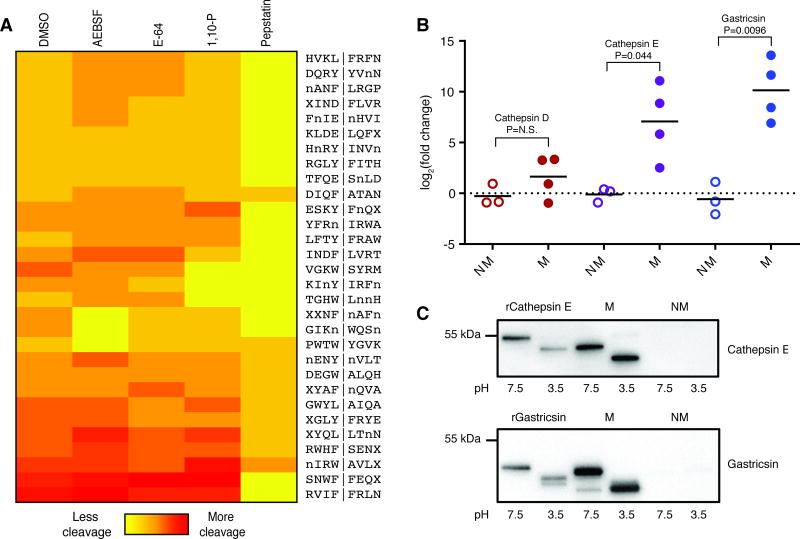
We treated the cyst fluid with broad-spectrum inhibitors against all major protease classes and analyzed changes in cleavage of the 35 mucinous-specific substrates by MSP-MS (Fig. 2A). Treatment with the aspartyl protease inhibitor pepstatin fully inhibited cleavage of 20 mucinous-specific substrates and partially inhibited cleavage of 8 additional substrates. The other broad-spectrum protease inhibitors minimally affected cleavage of the mucinous-specific substrates. The serine protease inhibitor AEBSF and the metal chelator 1,10-phenanthroline only inhibited cleavage of 3 substrates each. In a second mucinous cyst fluid sample, we confirmed that aspartyl protease inhibition with pepstatin blocks cleavage of the majority of the mucinous-specific substrates (Fig. S3).
Fig. 2. Identification of enriched aspartyl protease activity in mucinous cysts.
(A) Heatmap displaying cleavage of 30 mucinous-specific substrates following treatment of a mucinous cyst fluid sample with DMSO or various broad-spectrum protease inhibitors. Spectral counts were used for relative quantification of peptide cleavage products. Vertical bar (|) indicates the site of cleavage within substrates. (B) Label-free quantitation of aspartyl protease relative abundance in mucinous (M) and nonmucinous (NM) cysts. (C) Western blot analysis of recombinant (r) and cyst fluid-derived cathepsin E and gastricsin. Samples were pre-incubated at the indicated pH for 10 minutes.
Our inhibition data demonstrated that aspartyl proteases have increased activity in mucinous cysts. Therefore, we performed shotgun proteomic analysis of a set of mucinous (n=4) and nonmucinous cysts (n=3) to determine if there were differences in the abundance of individual aspartyl proteases. This analysis identified three aspartyl proteases – cathepsin D, cathepsin E, and gastricsin (Table S2). Label-free quantitation using precursor ion abundance, revealed that cathepsin D was present at similar levels in the mucinous and nonmucinous cysts, whereas cathepsin E and gastricsin were significantly more abundant in the mucinous cysts (Fig. 2B and Table S3). Aspartyl proteases are synthesized as inactive zymogens that undergo enzymatic maturation at an acidic pH (43). As the tumor microenvironment is known to be acidic, we investigated whether cathepsin E and gastricsin were present in the pro- or mature forms. Exposure of fluid from a mucinous cyst to acidic pH induced a mass shift in cathepsin E and gastricsin that was comparable to that observed using recombinantly produced proteins (Fig. 2C), indicating that both proteases are released into cyst fluid in their proforms. As expected, no cathepsin E or gastricsin was detected in fluid from a representative nonmucinous cyst. Collectively, these results demonstrate that the proforms of cathepsin E and gastricsin are differentially expressed in mucinous cysts and that this induction is responsible for the increased proteolytic activity under acidic conditions.
Immunohistochemical analysis of gastricsin and cathepsin E in mucinous cysts
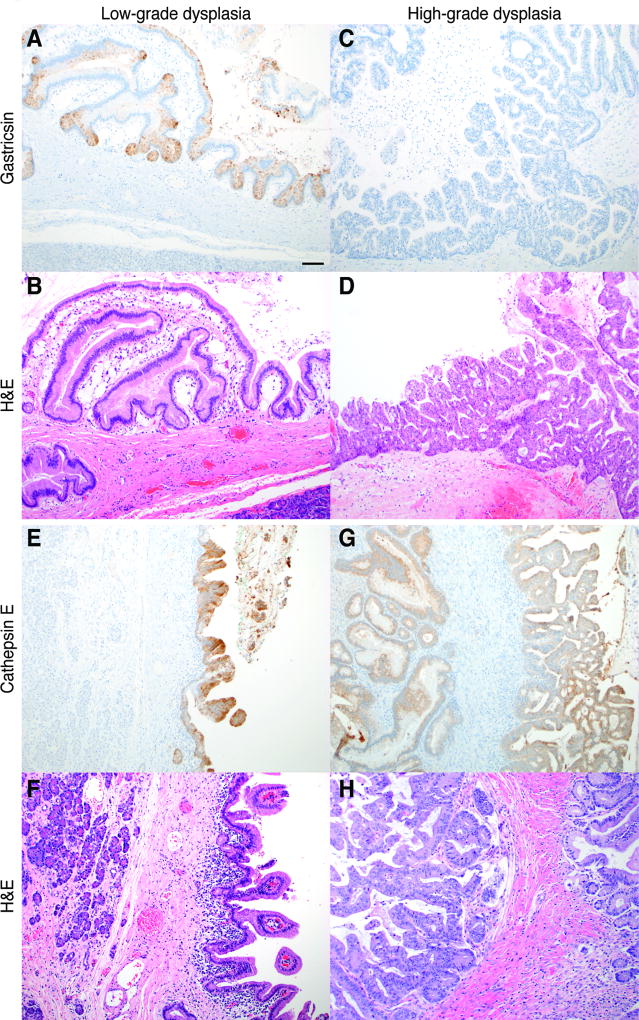
We further examined overexpression of cathepsin E and gastricsin in 14 mucinous cysts using immunohistochemical (IHC) analysis. Cytoplasmic gastricsin staining was observed in the epithelial cells lining 11 of the 14 mucinous cysts examined (Table S4). Interestingly, gastricsin expression was primarily associated with regions of low-grade dysplasia, and no staining was observed in regions of high-grade dysplasia (Figs. 3A–D). Gastricsin staining was also apparent in areas of low-grade dysplasia within mucinous cysts that contained both low- and high-grade dysplasia. Cytoplasmic cathepsin E was detected in all 14 mucinous cysts examined; however, staining did not show a dependence on the degree of dysplasia (Figs. 3E–H). No gastricsin or cathepsin E staining was evident in the neighboring normal ductal epithelium or stromal tissue. In addition, neither protease was detected in either of the two nonmucinous SCAs examined.
Fig. 3. Immunohistochemical analysis of gastricsin and cathepsin E in mucinous cysts.
Histological analysis of mucinous cysts with low-grade dysplasia (A, B, E, F) and high-grade dysplasia (C, D, G, H). Gastricsin (A, C), cathepsin E (E, G), and haematoxylin and eosin (H&E) staining (B, D, F, H) in IPMNs (A–F) and MCNs (G, H). Scale bar is 10 µm.
We also examined expression of gastricsin and cathepsin E in an IPMN genetic mouse model. Ptf1a-Cre; LSL-KrasG12D; Brg1f/f mice develop cystic lesions of the pancreas that closely resemble human IPMNs (44). Consistent with the above results, we observed cytoplasmic gastricsin and cathepsin E staining in the epithelial cells surrounding the cystic lesion (Fig. S4). Once again, there was no staining in normal pancreatic tissue.
Development of a gastricsin selective fluorescent substrate
The MSP-MS assay is ideal for discovering global differences in protease activity, but is not readily amenable for use as a diagnostic tool. Therefore, we sought to identify sensitive and selective fluorescent substrates that could be used in a standard microplate format to distinguish mucinous from nonmucinous cysts.
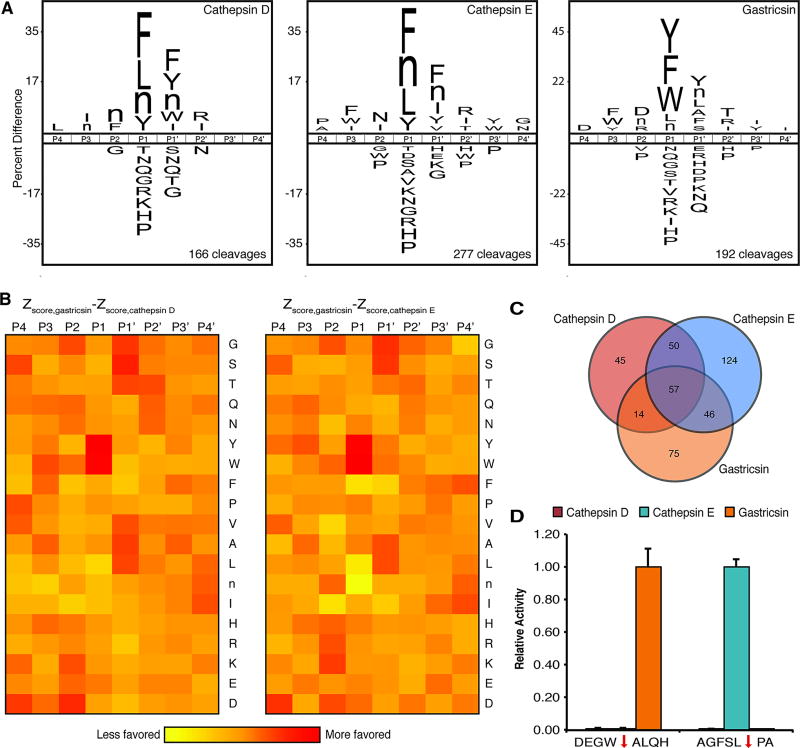
We focused on designing a gastricsin selective substrate as a cathepsin E selective substrate has been previously reported (45). We first analyzed the substrate specificity of recombinant cathepsin E and gastricsin using the MSP-MS assay (Fig. 4A). We also profiled recombinant cathepsin D, as it was detected in cyst fluid samples by our shotgun proteomic analysis (Fig. 2B), and therefore, we wanted to ensure that the synthesized substrates are not cleaved by this protease. Cathepsins E and D showed highly similar substrate specificity with a Pearson correlation coefficient of 0.81 and both proteases displayed a clear preference for hydrophobic residues in the P1 and P1′ positions. Gastricsin also preferred hydrophobic residues in the P1 and P1′ positions, however, direct comparison of the amino acid enrichment profiles revealed that gastricsin also has distinct cleavage preferences (Fig. 4B). Most notably, gastricsin shows a significantly stronger preference for tyrosine and tryptophan in the P1 position. Gastricsin also slightly favored small amino acids, such as glycine, serine, and alanine, in the P1’ position.
Fig. 4. Design and synthesis of gastricsin selective fluorescent substrate.
(A) Substrate specificity of cathepsin D, cathepsin E, and gastricsin as determined by MSP-MS. Residues shown in iceLogo are statistically significant with p < 0.05. (B) Heatmap comparing the amino acid enrichment Z-scores for gastricsin relative to cathepsin D and cathepsin E. (C) Venn diagram depicting the unique and overlapping cleavages detected by MSP-MS with cathepsin D, cathepsin E, and gastricsin. (D) Cleavage of the fluorescent substrates by cathepsin D, cathepsin E, and gastricsin. Activity was normalized to 1.00 based on the maximal activity against each substrate. Red arrow indicates the site of cleavage. Error bars denote standard error of the mean (SEM) from triplicate analysis.
Using the MSP- MS assay, we identified 75 peptides that were cleaved by gastricsin and not by cathepsins D or E (Fig. 4C). We used the specificity information from Fig. 4A–B to select a single peptide substrate that we expected to be highly selective for gastricsin. In particular, we chose a peptide that was cleaved by gastricsin with a tryptophan and alanine in the P1 and P1′ positions, respectively. We synthesized an internally quenched, fluorescent substrate incorporating the P4 to P4′ amino acids from this peptide. This substrate was found to be greater than 120-fold selective for gastricsin over both cathepsins D and E (Fig. 4D) and is cleaved with a kcat/Km of 4.8 × 105 M−1/s−1 (Fig. S5). We also synthesized the previously reported cathepsin E selective substrate and confirmed that it is more than 100-fold selective for cathepsin E over both cathepsin D and gastricsin (Fig. 4D) (45). Lastly, we confirmed that we could use these substrates to monitor cathepsin E and gastricsin protease activity in cyst fluid. Indeed, both substrates were cleaved in fluid from a mucinous cyst and this activity was fully inhibited by pre-incubation with pepstatin (Fig. S6).
Gastricsin and cathepsin E activity differentiate mucinous from nonmucinous cysts
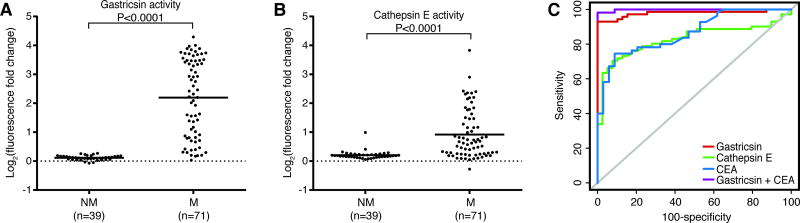
We next used the gastricsin and cathepsin E fluorescent substrates to assess their relative protease activities in cyst fluid samples to determine if these activities could be used to differentiate mucinous from nonmucinous cysts. We first analyzed the 23 cyst fluid samples that we previously profiled using the MSP-MS assay. Cleavage of both the gastricsin and cathepsin E substrate was significantly higher in mucinous relative to nonmucinous cysts (Fig. S7). This prompted us to assess cathepsin E and gastricsin activity in a validation cohort comprised of an additional 87 cyst fluid samples. Again, mucinous cysts displayed significantly increased levels of gastricsin and cathepsin E activity (Fig. S7). There were no significant differences in activity between the two patient cohorts. Analysis of all 110 patient samples revealed that gastricsin activity was on average increased more than 6-fold in mucinous cysts, while cathepsin E activity was increased only 2-fold (Fig. 5A–B). The ROC curve for gastricsin activity exhibited an area under the curve (AUC) of 0.979 for distinguishing mucinous cysts (Fig. 5C and Table S5). At the optimal cutoff of a 1.23-fold change in fluorescence, gastricsin activity demonstrated a specificity of 100% and a sensitivity of 93%. Cathepsin E activity had an AUC of 0.828 and, using this same optimal cutoff, displayed 92% specificity and 70% sensitivity for differentiating mucinous from nonmucinous cysts. Gastricsin and cathepsin E activity did not show a dependence on the type of mucinous cyst or the degree of dysplasia within a mucinous cyst (Fig. S8). Considering that gastricsin expression was only observed in regions of low-grade dysplasia (Fig. 3), we were surprised to observe that gastricsin activity was also not associated with the degree of dysplasia. This is likely because highly dysplastic and invasive mucinous lesions also often contain regions of low-grade dysplasia. We also examined whether gastricsin or cathepsin E activity were correlated with features from the revised Sendai criteria, which is a widely applied consensus guidelines for the management of mucinous cysts (8). Neither gastricsin nor cathepsin E activity showed significant differences in relation to the Sendai features we assessed (Table S6).
Fig. 5. Quantification of gastricsin and cathepsin E activity in 110 cyst fluid samples.
Analysis of gastricsin (A) and cathepsin E (B) activity in nonmucinous (NM) and mucinous (M) cysts using fluorescent substrates. (C) ROC curves comparing sensitivity and specificity of CEA, gastricsin, cathepsin E, and CEA and gastricsin in combination.
CEA levels were independently measured for 89 of the cyst fluid samples, and we compared abundance between mucinous (n=55) and nonmucinous cysts (n=34). As expected, CEA was significantly elevated in the mucinous cysts (Fig. S9). The CEA ROC curve exhibited an AUC of 0.865 for distinguishing mucinous cysts from nonmucinous cysts (Fig. 5C). CEA-based classification underperformed gastricsin activity, but was comparable to cathepsin E activity-based classification. For CEA, a cutoff level of 192 ng/mL is the commonly used clinical standard for differentiating mucinous from nonmucinous cysts (46). At this cutoff, CEA demonstrated a specificity of 94% and a sensitivity of 65%, which is consistent with what has been previously reported. All 19 of the mucinous cyst fluid samples with CEA levels below the standard cutoff of 192 ng/mL were correctly classified by gastricsin activity. Additionally, the two nonmucinous cysts with CEA levels above 192 ng/mL were also correctly classified by gastricsin activity.
We also assessed whether combined analysis of CEA with gastricsin and cathepsin E activity could better differentiate mucinous from nonmucinous cysts. Gastricsin activity with CEA evaluation resulted in a classifier with an AUC of 0.998 (Fig. 5C), exhibiting a specificity of 100% and sensitivity of 98%. Inclusion of all three markers did not lead to improved differentiation of mucinous from nonmucinous cysts (Table S5).
Discussion
Although pancreatic cysts are being detected at an increasing rate, available diagnostic tests do not accurately discriminate between cyst types. Mucinous cysts have malignant potential and may require resection, while nonmucinous cysts are considered benign and require no further evaluation if these lesions are asymptomatic. Increasing the level of certainty in this distinction would spare some patients unnecessary surgical resections and reduce the need for ongoing surveillance for many more individuals. In this study, we used an unbiased and global substrate-based profiling strategy coupled with proteomics, to identify distinguishing protease activities in cyst fluid samples. Using this approach, gastricsin and cathepsin E activities were found to be promising markers for differentiating benign nonmucinous cysts from potentially malignant mucinous cysts. Selective fluorescent substrates both confirmed induction of these proteases in mucinous cysts and enabled sensitive and specific differentiation of these lesions in 110 patient samples.
To date, CEA remains the most widely used clinical biomarker for differentiating mucinous from nonmucinous cysts. However, the performance of this marker is generally considered suboptimal. Indeed, CEA analysis was only 76% accurate in our study at the standard cutoff of 192 ng/mL. Gastricsin activity was 95% accurate, and correctly classified all 21 cysts that were misclassified by CEA, clearly demonstrating the clinical utility of this marker. Furthermore, we were able to improve classification accuracy to 99% by combining CEA with gastricsin activity analysis.
Preoperatively determining the degree of dysplasia within a mucinous cyst is another major challenge for ensuring appropriate clinical intervention. However, the protease activity markers identified in this study do not differentiate between mucinous cysts with low- or high-grade dysplasia. Although this is a limitation of our markers, correctly differentiating mucinous from nonmucinous cysts is a critical first step in deciding which cysts should undergo resection. For example, pancreatic resection of the 39 benign nonmucinous cysts included in this study could potentially have been avoided through the application of our assay. In addition, 19 mucinous cysts within our patient cohort had CEA levels below the standard cutoff of 192 ng/mL. In our high-volume pancreatic centers, radiographic and clinical features allowed experienced clinicians to correctly identify these cysts as mucinous. However, medical centers without dedicated cyst specialists may be inclined to misclassify these samples as nonmucinous and would greatly benefit from our simple and accurate diagnostic assay. A number of molecular and clinical markers have recently shown promise for distinguishing mucinous cysts based on their degree of dysplasia (19,23). A sequential diagnostic strategy may emerge in which gastricsin and cathepsin E activity are used to determine if a lesion is mucinous, followed by analysis of a secondary marker to define the degree of dysplasia. Assessing gastricsin and cathepsin E activity in combination with other promising markers will be a primary focus of future work.
Previous gene expression profiling studies of IPMNs and MCNs demonstrated overexpression of gastricsin and cathepsin E mRNA (29–31). However, the protein levels and activity of these aspartyl proteases has not been previously investigated within these lesions. Protease activity is particularly well suited to the development of a rapid and simple diagnostic test for differentiating cysts. Activity-based detection is highly sensitive because of catalytic signal amplification. Indeed, the assays described in this study use less than 5 µL of cyst fluid, whereas CEA tests often require at least 500 µL. Furthermore, unlike immunoassays, protease activity assays do not require the costly development of high-quality antibody reagents. Spectrophotometric assays can be readily adapted to the standard plate readers present in clinical laboratories, and there are already several examples of such protease activity assays in common clinical use for other indications (47,48).
We were particularly interested to observe that gastricsin expression within mucinous cysts was primarily associated with areas of low-grade dysplasia and was absent in high-grade dysplasia, although we were only able to assess four cysts containing regions of high-grade dysplasia. Previous work demonstrated that gastricsin and other foregut markers are overexpressed in other pancreatic cancer precursor lesions, reflecting a cellular dedifferentiation step prior to malignant transformation (49). Gastricsin overexpression within IPMNs and MCNs might be reflective of a similar process. In support of this hypothesis, recent work using the same IPMN genetic mouse model examined in this study showed that cellular dedifferentiation is a critical step in the development of IPMNs (44,50). Dedifferentiation within this genetic mouse model is transient and occurs prior to the development of invasive cancer, which may explain why gastricsin expression is associated with regions of low-grade dysplasia. In contrast to gastricsin, we did not observe an association between cathepsin E expression and the degree of dysplasia present within a mucinous cyst. This suggests that different processes control the expression of these two proteases and that cathepsin E levels are less reflective of cellular identity. Additional studies using the recently developed genetic mouse models of mucinous cysts are needed to characterize how the expression of these proteases is regulated and what roles – if any – gastricsin and cathepsin E are playing in neoplastic transformation (44).
In summary, our results demonstrate that gastricsin and cathepsin E activity are sensitive and specific markers for differentiating mucinous from nonmucinous pancreatic cystic lesions. In particular, gastricsin activity is a promising candidate for the development of a simple, diagnostic test with superior performance to CEA. This could provide clinical stratification to properly manage the growing problem of pancreatic cysts.
Supplementary Material
Translational relevance.
With advances in abdominal imaging technologies, the incidental detection of pancreatic cysts continues to rise. However, there remains a lack of accurate molecular diagnostics for differentiating benign cystic lesions from those that can progress into pancreatic cancer. This has led to a dramatic increase in the number of potentially unnecessary pancreatic resections, which are associated with high rates of morbidity. Using a global and unbiased protease-activity profiling approach and patient cyst fluid, we determined that the activities of the aspartyl proteases gastricsin and cathepsin E accurately differentiate premalignant mucinous cysts from benign nonmucinous cysts. In particular, analysis of gastricsin activity demonstrated 93% sensitivity and 100% specificity for differentiating mucinous lesions. Our simple and direct fluorescence-based approach for stratification of pancreatic cysts significantly outperformed the most widely used molecular biomarker – carcinoembryonic antigen (CEA) – and can be readily translated into an actionable diagnostic assay to help improve clinical management of these challenging lesions.
Acknowledgments
We would like to thank Brendan C. Visser, George A. Poultsides, and Jeffrey Norton for providing cyst fluid samples used in this study. IHC assays were performed by the UCSF Helen Diller Family Comprehensive Cancer Center (HDFCCC) IHC Core (supported by the HDFCCC support grant NIH/NCI P30 CA82103). Mass spectrometry was performed in collaboration with the UCSF Mass Spectrometry Facility (directed by Alma Burlingame and supported by NIH grant P41GM10348). We would like to thank Debbie Ngow for mouse tissue processing. We would also like to thank Arun Wiita, Hector Huang, Adam Olshen, Giselle Knudsen, Yvonne Lee, Elizabeth Gilbert, Michael B. Winter, Matthew Ravalin, and Tine Soerensen for their assistance and helpful discussions.
Grant support: This study was supported by the following grants: NIH National Heart, Lung, And Blood Institute grant U54HL119893 (to C. S. Craik), NIH/NCATS UCSF-CTSI grant UL1TR000004 (to C. S. Craik), NIH grant A119685 (to K. S. Kirkwood), NIH/NCI U01CA196403 (to K. S. Kirkwood), NIH/NCI U01CA152653-01 (to R. E. Brand), and NIH/NCI grants R01CA172045 and R01CA112537 (to M. Hebrok). W. G. Park was supported by the American College of Gastroenterology Junior Faculty Development Award. This work was also supported by two UCSF Enabling Technologies Advisory Committee awards (to C. S. Craik). S. L. Ivry was supported by NIH Pharmaceutical Sciences and Pharmacogenomics Training grant T32GM008155.
References
- 1.Moris M, Bridges MD, Pooley RA, Raimondo M, Woodward TA. Association Between Advances in High-Resolution Cross-Section Imaging Technologies and Increase in Prevalence of Pancreatic Cysts From 2005 to 2014. Clin Gastroenterol Hepatol. 2016;14:585–93. doi: 10.1016/j.cgh.2015.08.038. [DOI] [PubMed] [Google Scholar]
- 2.Lee KS, Sekhar A, Rofsky NM, Pedrosa I. Prevalence of incidental pancreatic cysts in the adult population on MR imaging. Am J Gastroenterol. 2010;105:2079–84. doi: 10.1038/ajg.2010.122. [DOI] [PubMed] [Google Scholar]
- 3.Laffan TA, Horton KM, Klein AP, Berlanstein B, Siegelman SS, Kawamoto S, et al. Prevalence of unsuspected pancreatic cysts on MDCT. Am J Roentgenol. 2008;191:802–7. doi: 10.2214/AJR.07.3340. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Volkan Adsay N. Cystic lesions of the pancreas. Mod Pathol. 2007;20:71–93. doi: 10.1038/modpathol.3800706. [DOI] [PubMed] [Google Scholar]
- 5.Matthaei H, Schulick RD, Hruban RH, Maitra A. Nat Rev Gastroenterol Hepatol. Vol. 8. Nature Publishing Group; 2011. Cystic precursors to invasive pancreatic cancer; pp. 141–50. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Crippa S, Del Castillo CF, Salvia R, Finkelstein D, Bassi C, Dominguez I, et al. Mucin-Producing Neoplasms of the Pancreas: An Analysis of Distinguishing Clinical and Epidemiologic Characteristics. Clin Gastroenterol Hepatol. 2011;8:213–9. doi: 10.1016/j.cgh.2009.10.001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Jang K-T, Park SM, Basturk O, Bagci P, Bandyopadhyay S, Stelow EB, et al. Clinicopathologic characteristics of 29 invasive carcinomas arising in 178 pancreatic mucinous cystic neoplasms with ovarian-type stroma: implications for management and prognosis. Am J Surg Pathol. 2015;39:179–87. doi: 10.1097/PAS.0000000000000357. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Tanaka M, Fernández-del Castillo C, Adsay V, Chari S, Falconi M, Jang J-Y, et al. Pancreatology. Vol. 12. Elsevier; 2012. International consensus guidelines 2012 for the management of IPMN and MCN of the pancreas; pp. 183–97. [DOI] [PubMed] [Google Scholar]
- 9.Correa-Gallego C, Ferrone CR, Thayer SP, Wargo JA, Warshaw AL, Fernandez-Del Castillo C. Incidental pancreatic cysts: Do we really know what we are watching? Pancreatology. 2010;10:144–50. doi: 10.1159/000243733. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Parra-herran CE, Garcia MT, Herrera L, Bejarano PA. Cystic Lesions of the Pancreas : Clinical and Pathologic Review of Cases in a Five Year Period. J Pancreas. 2010;11:358–64. [PubMed] [Google Scholar]
- 11.Quan SY, Visser BC, Poultsides GA, Norton JA, Chen AM, Banerjee S, et al. Predictive Factors for Surgery Among Patients with Pancreatic Cysts in the Absence of High-Risk Features for Malignancy. J Gastrointest Surg. 2015;19:1101–5. doi: 10.1007/s11605-015-2786-3. [DOI] [PubMed] [Google Scholar]
- 12.Cho CS, Russ AJ, Loeffler AG, Rettammel RJ, Oudheusden G, Winslow ER, et al. Preoperative classification of pancreatic cystic neoplasms: the clinical significance of diagnostic inaccuracy. Ann Surg Oncol. 2013;20:3112–9. doi: 10.1245/s10434-013-2986-6. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Park WG, Mascarenhas R, Palaez-Luna M, Smyrk TC, Kane DO, Ph D, et al. Diagnostic Performance Of Cyst Fluid Carcinoembryonic Antigen And Amylase In Histologically Confirmed Pancreatic Cysts. Pancreas. 2011;40:42–5. doi: 10.1097/MPA.0b013e3181f69f36. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Ngamruengphong S, Bartel MJ, Raimondo M. Dig Liver Dis. Vol. 45. Editrice Gastroenterologica Italiana; 2013. Cyst carcinoembryonic antigen in differentiating pancreatic cysts : A; pp. 920–6. [DOI] [PubMed] [Google Scholar]
- 15.Almoguera C, Shibata D, Forrester K, Martin J, Arnheim N, Perucho M. Most Human Carcinomas of the Exocrine Contain Mutant c-K-ras Genes. Cell. 1988;53:549–54. doi: 10.1016/0092-8674(88)90571-5. [DOI] [PubMed] [Google Scholar]
- 16.Hezel AF, Kimmelman AC, Stanger BZ, Bardeesy N, Depinho Ra. Genetics and biology of pancreatic ductal adenocarcinoma. Genes Dev. 2006;20:1218–49. doi: 10.1101/gad.1415606. [DOI] [PubMed] [Google Scholar]
- 17.Khalid A, Zahid M, Finkelstein SD, Leblanc JK. Pancreatic cyst fluid DNA analysis in evaluating pancreatic cysts : a report of the PANDA study. Gastrointest Endosc. 2009;69:1095–102. doi: 10.1016/j.gie.2008.07.033. [DOI] [PubMed] [Google Scholar]
- 18.Wu J, Matthaei H, Maitra A, Dal Molin M, Wood LD, Eshleman JR, et al. Recurrent GNAS mutations define an unexpected pathway for pancreatic cyst development. Sci Transl Med. 2011;3:92ra66. doi: 10.1126/scitranslmed.3002543. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Hata HT, Dal Molin M, Suenaga M, Yu J, Pittman M, Weiss M, Canto MI, Wolfgang C, Lennon AM, Hruban RH, Goggins M. Cyst fluid telomerase activity predicts the histologic grade of cystic neoplasms of the pancreas. Clin Cancer Res. 2016;20:5141–41. doi: 10.1158/1078-0432.CCR-16-0311. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Zikos T, Pham K, Bowen R, Chen AM, Banerjee S, Friedland S, et al. Cyst Fluid Glucose is Rapidly Feasible and Accurate in Diagnosing Mucinous Pancreatic Cysts. Am J Gastroenterol. 2015;110:909–14. doi: 10.1038/ajg.2015.148. [DOI] [PubMed] [Google Scholar]
- 21.Yip-Schneider MT, Wu H, Dumas RP, Hancock BA. Vascular Endothelial Growth Factor, a Novel and Highly Accurate Pancreatic Fluid Biomarker for Serous Pancreatic Cysts. J Am Coll Surg. 2014;218:608–17. doi: 10.1016/j.jamcollsurg.2013.12.019. [DOI] [PubMed] [Google Scholar]
- 22.Cao Z, Maupin K, Curnutte B, Fallon B, Feasley CL, Brouhard E, et al. Specific glycoforms of MUC5AC and endorepellin accurately distinguish mucinous from nonmucinous pancreatic cysts. Mol Cell Proteomics. 2013;12:2724–34. doi: 10.1074/mcp.M113.030700. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Maker AV, Katabi N, Qin L, Klimstra DS, Schattner M, Brennan F, et al. Cyst fluid interleukin-1b (IL1b) levels predict the risk of carcinoma in intraductal papillary mucinous neoplasms of the pancreas. Clin Cancer Res. 2012;17:1502–8. doi: 10.1158/1078-0432.CCR-10-1561. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Kessenbrock K, Plaks V, Werb Z. Matrix metalloproteinases: regulators of the tumor microenvironment. Cell. 2010;141:52–67. doi: 10.1016/j.cell.2010.03.015. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Sevenich L, Joyce JA. Pericellular proteolysis in cancer. Genes Dev. 2014:2331–47. doi: 10.1101/gad.250647.114. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Sobotič B, Vizovišek M, Vidmar R, Van Damme P, Gocheva V, Joyce JA, et al. Proteomic Identification of Cysteine Cathepsin Substrates Shed from the Surface of Cancer Cells. Mol Cell Proteomics. 2015;14:2213–28. doi: 10.1074/mcp.M114.044628. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Joyce Ja, Baruch A, Chehade K, Meyer-Morse N, Giraudo E, Tsai F-Y, et al. Cathepsin cysteine proteases are effectors of invasive growth and angiogenesis during multistage tumorigenesis. Cancer Cell. 2004;5:443–53. doi: 10.1016/s1535-6108(04)00111-4. [DOI] [PubMed] [Google Scholar]
- 28.Gocheva V, Zeng W, Ke D, Klimstra D, Reinheckel T, Peters C, et al. Distinct roles for cysteine cathepsin genes in multistage tumorigenesis. Genes Dev. 2006:543–56. doi: 10.1101/gad.1407406. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Terris B, Blaveri E, Crnogorac-Jurcevic T, Jones M, Missiaglia E, Ruszniewski P, et al. Characterization of gene expression profiles in intraductal papillary-mucinous tumors of the pancreas. Am J Pathol. 2002;160:1745–54. doi: 10.1016/S0002-9440(10)61121-2. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Sato N, Fukushima N, Maitra A, Iacobuzio-Donahue Ca, van Heek NT, Cameron JL, et al. Gene expression profiling identifies genes associated with invasive intraductal papillary mucinous neoplasms of the pancreas. Am J Pathol. 2004;164:903–14. doi: 10.1016/S0002-9440(10)63178-1. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Fukushima N, Sato N, Prasad N, Leach SD, Hruban RH, Goggins M. Characterization of gene expression in mucinous cystic neoplasms of the pancreas using oligonucleotide microarrays. Oncogene. 2004;23:9042–51. doi: 10.1038/sj.onc.1208117. [DOI] [PubMed] [Google Scholar]
- 32.Ke E, Patel BB, Liu T, Li X, Haluszka O, Hoffman JP, et al. Proteomic Analyses of Pancreatic Cyst Fluids. Pancreas. 2009;38:1–21. doi: 10.1097/MPA.0b013e318193a08f. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Räty S, Sand J, Laukkarinen J, Vasama K, Bassi C, Salvia R, et al. Cyst fluid SPINK1 may help to differentiate benign and potentially malignant cystic pancreatic lesions. Pancreatology. 2013;13:530–3. doi: 10.1016/j.pan.2013.06.008. [DOI] [PubMed] [Google Scholar]
- 34.O’Donoghue AJ, Eroy-reveles AA, Knudsen GM, Ingram J, Zhou M, Statnekov JB, et al. Global identification of peptidase specificity by multiplex substrate profiling. Nat Methods. 2012;9:1095–100. doi: 10.1038/nmeth.2182. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Chalkley RJ, Baker PR, Medzihradszky KF, Lynn AJ, Burlingame AL. In-depth Analysis of Tandem Mass Spectrometry Data from Disparate Instrument Types. Mol Cell Proteomics. 2008:2386–98. doi: 10.1074/mcp.M800021-MCP200. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Schilling B, Rardin MJ, MacLean BX, Zawadzka aM, Frewen BE, Cusack MP, et al. Platform-Independent And Label-Free Quantitation Of Proteomic Data Using MS1 Extracted Ion Chromatograms In Skyline: Application To Protein Acetylation And Phosphorylation. Mol Cell Proteomics. 2012;11:202–14. doi: 10.1074/mcp.M112.017707. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Colaert N, Helsens K, Martens L, Vandekerckhove J, Gevaert K. Improved visualization of protein consensus sequences by iceLogo. Nat Methods. 2009;6:786–7. doi: 10.1038/nmeth1109-786. [DOI] [PubMed] [Google Scholar]
- 38.Winter MB, Salcedo EC, Lohse MB, Hartooni N, Gulati M, Sanchez H, et al. Global Identification of Biofilm-Specific Proteolysis in Candida albicans. MBio. 2016;7:1–13. doi: 10.1128/mBio.01514-16. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.O’Donoghue AJ, Knudsen GM, Beekman C, Perry Ja, Johnson AD, DeRisi JL, et al. Destructin-1 is a collagen-degrading endopeptidase secreted by Pseudogymnoascus destructans, the causative agent of white-nose syndrome. Proc Natl Acad Sci. 2015;112:7478–83. doi: 10.1073/pnas.1507082112. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Small JL, O’Donoghue AJ, Boritsch EC, Tsodikov OV, Knudsen GM, Vandal O, et al. Substrate specificity of MarP, a periplasmic protease required for resistance to acid and oxidative stress in Mycobacterium tuberculosis. J Biol Chem. 2013;288:12489–99. doi: 10.1074/jbc.M113.456541. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Impens F, Colaert N, Helsens K, Ghesquière B, Timmerman E, De Bock P-J, et al. A quantitative proteomics design for systematic identification of protease cleavage events. Mol Cell Proteomics. 2010;9:2327–33. doi: 10.1074/mcp.M110.001271. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Donoghue AJO, Ivry SL, Chaudhury C, Hostetter DR, Hanahan D, Craik CS. Procathepsin E is highly abundant but minimally active in pancreatic ductal adenocarcinoma tumors. Biol Chem. 2016;397:871–81. doi: 10.1515/hsz-2016-0138. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Dunn BM. Structure and Mechanism of the Pepsin-Like Family of Aspartic Peptidases. Chem Rev. 2002;102:4431–58. doi: 10.1021/cr010167q. [DOI] [PubMed] [Google Scholar]
- 44.von Figura G, Fukuda A, Roy N, Liku ME, Morris JP, Kim GE, et al. The chromatin regulator Brg1 suppresses formation of intraductal papillary mucinous neoplasm and pancreatic ductal adenocarcinoma. Nat Cell Biol. 2014;16:255–67. doi: 10.1038/ncb2916. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Abd-Elgaliel WR, Tung C-H. Selective detection of Cathepsin E proteolytic activity. Biochim Biophys Acta. 2010;1800:1002–8. doi: 10.1016/j.bbagen.2010.06.005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Brugge WR, Lewandrowski K, Lee-Lewandrowski E, Centeno BA, Szydlo T, Regan S, et al. Diagnosis of Pancreatic Cystic Neoplasms: A Report of the Cooperative Pancreatic Cyst Study. Gastroenterology. 2004;126:1330–6. doi: 10.1053/j.gastro.2004.02.013. [DOI] [PubMed] [Google Scholar]
- 47.Kremer Hovinga JA, Mottini M, Lämmle B. Measurement of ADAMTS-13 activity in plasma by the FRETS-VWF73 assay: Comparison with other assay methods. J Thromb Haemost. 2006;4:1146–8. doi: 10.1111/j.1538-7836.2006.01904.x. [DOI] [PubMed] [Google Scholar]
- 48.Moll S, Ortel TL. Monitoring Warfarin Therapy in Patients with Lupus Anticoagulants. Ann Intern Med. 1997;127:177–85. doi: 10.7326/0003-4819-127-3-199708010-00001. [DOI] [PubMed] [Google Scholar]
- 49.Prasad NB, Biankin AV, Fukushima N, Maitra A, Dhara S, Elkahloun AG, et al. Gene Expression Profiles in Pancreatic Intraepithelial Neoplasia Reflect the Effects of Hedgehog Signaling on Pancreatic Ductal Epithelial Cells Gene Expression Profiles in Pancreatic Intraepithelial Neoplasia Reflect the Effects of Hedgehog Signaling on. Cancer Res. 2005:1619–26. doi: 10.1158/0008-5472.CAN-04-1413. [DOI] [PubMed] [Google Scholar]
- 50.Roy N, Malik S, Villanueva KE, Urano A, Lu X, Von Figura G, et al. Brg1 promotes both tumor-suppressive and oncogenic activities at distinct stages of pancreatic cancer formation. Genes Dev. 2015;29:658–71. doi: 10.1101/gad.256628.114. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.