Abstract
The cathepsin family of lysosomal proteases is increasingly being recognized for their altered expression in cancer and role in facilitating tumor progression. The aspartyl protease cathepsin E is overexpressed in several cancers and has been investigated as a biomarker for pancreatic ductal adenocarcinoma (PDAC). Here we show that cathepsin E expression in mouse PDAC tumors is increased by more than 400-fold when compared to healthy pancreatic tissue. Cathepsin E accumulates over the course of disease progression and accounts for more than 3% of the tumor protein in mice with end-stage disease. Through immunoblot analysis we determined that only procathepsin E exists in mouse PDAC tumors and cell lines derived from these tumors. By decreasing the pH, this procathepsion E is converted to the mature form, resulting in an increase in proteolytic activity. Although active site inhibitors can bind procathepsin E, treatment of PDAC mice with the aspartyl protease inhibitor ritonavir did not decrease tumor burden. Lastly, we used multiplex substrate profiling by mass spectrometry to identify two synthetic peptides that are hydrolyzed by procathepsin E near neutral pH. This work represents a comprehensive analysis of procathepsin E in PDAC and could facilitate the development of improved biomarkers for disease detection.
Keywords: aspartyl protease, cathepsins, pericellular proteolysis, substrate specificity, tumor microenvironment, zymogen
Introduction
With a mean survival rate of 6 months and a 5-year survival rate of less than 5%, PDAC remains one of the most lethal cancers (Costello et al., 2012). PDAC can be distinguished from other pancreatic and non-pancreatic malignancies by a characteristic set of mutations, including activation of the oncogene Kras, which occurs in 95% of cases (Hezel et al., 2006). Kras mutation is thought to initiate the formation of pre-invasive ductal lesions, known as pancreatic intraepithelial neoplasias (PanINs) (Morris et al., 2010). Successive mutations in the tumor suppressor genes Ink4a (90%), Trp53 (75%), and Smad4 (50%) cause PanINs to undergo graded histological progression and eventual transformation into PDAC (Hezel et al., 2006). Generation of mice harboring these signature genetic mutations has yielded models that closely recapitulate the histopathogenesis of the human disease.
In cancer, dysregulation of protease activity can lead to degradation of the extracellular matrix and facilitate neoplastic progression (Mason and Joyce, 2011). Many studies have focused on the roles of matrix metalloproteases (MMPs) and serine proteases due to their localization on the exterior of the cell (Kessenbrock et al., 2010; Sevenich and Joyce, 2014). In PDAC, silencing of the metalloprotease ADAM17 markedly reduced invasiveness and migration of cancer cells (Ringel et al., 2006). Cysteine proteases of the papain subfamily, known as cysteine cathepsins, are being increasingly investigated for their role in cancer. These proteases are predominantly found within endolysosomal vesicles, but are upregulated and secreted by cancer cells and thus may play an intracellular and extracellular role in tumor progression (Mohamed and Sloane, 2006). Using a cysteine cathepsin inhibitor, Joyce and colleagues observed defects in tumor growth, invasion, and angiogenesis in a mouse model of pancreatic islet cell carcinoma (Joyce et al., 2004). This phenotype was not observed following treatment with a broad spectrum MMP inhibitor (Bergers et al., 1999). Further studies by the same group determined that deletion of cathepsins B, L, or S in this mouse model correlated with a reduction in tumor burden and invasion (Gocheva et al., 2006, 2010).
Two catalytically distinct members of the cathepsin family are the aspartyl proteases, cathepsins D and E. Cathepsin D is a ubiquitously expressed lysosomal protease (Reid et al., 1986). The proform of the enzyme is overexpressed and secreted by a number of cancer types (Laurent-Matha et al., 2001; Beaujouin et al., 2010). Secreted procathepsin D binds the cell surface and stimulates growth of breast, prostate and lung cancer cells in vitro and in vivo.
Cathepsin E is an intracellular aspartyl protease found mainly in cells of the immune system, such as lymphocytes (Sakai et al., 1989), microglia (Nishioku et al., 2002), dendritic (Chain et al., 2005), and activated B cells (Burster et al., 2008). Unlike the highly homologous cathepsin D, its intracellular localization varies with cell type. Cathepsin E has been reported to reside in endosomes, the plasma membrane, the endoplasmic reticulum, and the Golgi apparatus (Sakai et al., 1989; Zaidi and Kalbacher, 2008). While the exact physiological role of cathepsin E has yet to be elucidated, some studies indicate that it plays a role in antigen processing via the MHC class II pathway. Cathepsin E knockout mice develop atopic dermatitis-like skin lesions with increased susceptibility to bacterial infection and accumulate lysosomal membrane sialoglycoproteins that result in a novel form of lysosomal storage disorder (Tsukuba et al., 2003, 2006). Overexpression of cathepsin E has been found in gastric, cervical, lung, intestinal and pancreatic cancer (Mota et al., 1997; Ullmann et al., 2004; Cruz-Monserrate et al., 2012). Cathepsin E detection in the urine of mice with intestinal adenomas was reported as a potential marker for disease progression, while strong expression of cathepsin E in tumors of patients with lung carcinomas correlates with increased survival (Ullmann et al., 2004). In the pancreas, cathepsin E is detectable in early PanIN lesions and accumulates as cells progress to PDAC (Buchholz et al., 2005). While detection of the protein in pancreatic juice of patients has been shown to be a promising diagnostic marker to distinguish PDAC from chronic pancreatitis, no functional studies have been performed to characterize the role of cathepsin E in cancer progression (Uno et al., 2000).
Our group previously identified multiple secreted proteases, including cathepsin E, from a mouse PDAC cell line (O’Donoghue et al., 2012). In addition, we determined that cathepsin E was the dominant proteolytic activity in conditioned media from this cell line when assayed at pH 5.2. As cathepsin E is generally found intracellularly and is optimally active between pH 3.5 and pH 4.5, we decided to investigate if this enzyme could function in the pericellular space near neutral pH, namely pH 6.5 (Yasuda et al., 1999). We performed a comprehensive biochemical analysis of cathepsin E activation in both a mouse PDAC cell line and in tumors from a PDAC genetic model. Cell line and tumor-associated cathepsin E exclusively exists as a proenzyme and tumor growth was not slowed by treatment with an active site inhibitor. However, procathepsin E retains residual proteolytic activity as detected by our MSP-MS assay.
Results
Only procathepsin E exists in primary PDAC cells
In a previous study, we detected complement factor B, carboxypeptidase E, and cathepsins E, B, and L in conditioned media from a primary PDAC cell line derived from a mouse tumor (O’Donoghue et al., 2012). When this media was assayed at pH 5.2, cathepsin E activity was responsible for the majority of detected proteolysis. These studies were performed with a global and unbiased substrate-profiling assay that uses mass spectrometry to detect the proteolytic degradation of a synthetic peptide library (O’Donoghue et al., 2012). To uncover the functional role of this enzyme in PDAC, an in-depth biochemical analysis of PDAC derived cathepsin E was required.
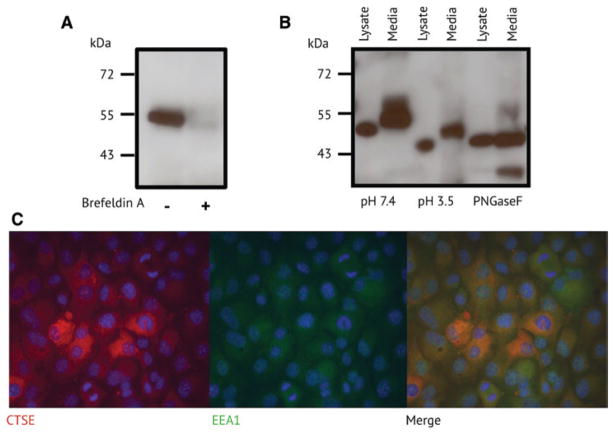
In this study, we used the PDAC cell line from our previous work. This cell line is derived from p48-Cre; KrasG12D; Trp53f/f mice, which develop PDAC that histologically mirrors the human disease (Nolan-stevaux et al., 2009). Immunoblotting analysis confirmed that cathepsin E was present in conditioned media from this cell line (Figure 1A). As cathepsin E is generally found intracellularly, we first wanted to confirm that the protein found in the media was not simply the result of cellular lysis. To test this, we treated cells with brefeldin A, an inhibitor of the secretory pathway, and confirmed that cathepsin E was no longer present in the conditioned media. Interestingly, with an apparent molecular mass of 53 kDa, the secreted cathepsin E was larger than the 46 kDa protein that was previously observed in a mouse study of atopic dermatitis (Tsukuba et al., 2003). In addition, the molecular mass of the intracellular protein was lower than that of the extracellular protein, indicating that these enzymes were differentially post-translationally modified (Figure 1B). Mouse cathepsin E is synthesized as a 397 amino acid protein, consisting of a 20 amino acid signal peptide, a 39 amino acid propeptide, and a 338 amino acid catalytic domain (Zaidi and Kalbacher, 2008). Cathepsin E also has two N-linked glycosylation sites at asparagines 91 and 323. Like other aspartyl proteases, procathepsin E can auto-activate under acidic conditions resulting in the irreversible hydrolysis of the propeptide (Richter et al., 1998). To determine if the higher molecular weight extracellular cathepsin E corresponded to the proform, conditioned media was exposed to acidic conditions. This resulted in conversion to a lower molecular weight protein of approximately 49 kDa (Figure 1B). Surprisingly, intracellular cathepsin E at 50 kDa was also converted to a lower molecular weight form (46 kDa) following acid exposure, indicating that both intracellular and extracellular proteins exist in the proform. The intracellular 46 kDa protein is likely the same cathepsin E that was detected by Tsukuba and coworkers (Tsukuba et al., 2003). Treatment of procathepsin E with the deglycosylase enzymes, PNGaseF, resulted in a protein of the same molecular weight, indicating that the mass difference was due to alternative glycosylation.
Figure 1.
Expression and localization of cathepsin E in a mouse PDAC cell line.
(A) Western blot analysis of cathepsin E in conditioned media with and without brefeldin A treatment. Size markers correspond to kilodaltons (kDa). (B) Western blot analysis of cathepsin E in the cell lysate and conditioned media treated with acid and the deglycosylase PNGaseF. (C) Immunofluorescence of cathepsin E and the endosomal marker EEA1. DAPI is shown in blue.
Sastradipura and coworkers have previously shown that procathepsin E is expressed in rat microglia cells and is rapidly converted to the mature enzyme (Sastradipura et al., 2002). Immunoelectron microscopy of these microglia cells showed that the enzyme was associated with dense endosome-like organelles. As only procathepsin E was detected in PDAC cellular lysates, we wanted to investigate if intracellular localization might be restricting acid-mediated protease activation. Using immunofluorescence, procathepsin E was detected throughout the cytoplasm and colocalized with early endosomal antigen 1 (EEA1) (Figure 1C). Late endosomes have a pH <6.0 and auto-activation of procathepsin E to the mature form occurs between pH 3.0 and pH 6.0 (Rao-Naik et al., 1995; Cappiello et al., 2004). Therefore it is unlikely that procathepsin E proceeds into late endosomes as the mature enzyme is not detected in the whole cell lysate. Instead, procathepsin E remains in early endosomes or is trafficked outside of the cell.
Acidification of tumor lysates activates cathepsin E
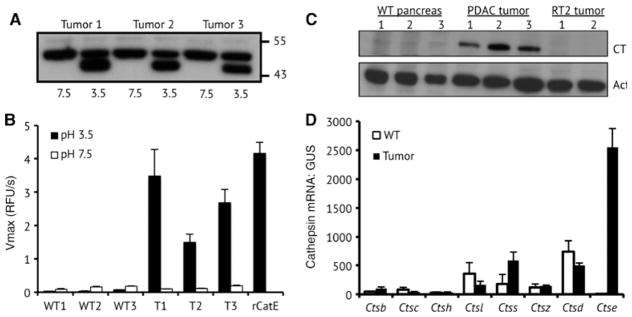
We proceeded to further analyze cathepsin E in pancreatic tumors from p48-Cre; KrasG12D; Trp53f/f mice. Tumors isolated from 13 to 14 week old mice were lysed under non-denaturing conditions and incubated at neutral or acidic pH for 15 min. Immunoblot analysis could only detect 50 kDa procathepsin E at neutral pH. Exposure to acid caused a shift in molecular weight to 46 kDa, which corresponds to removal of the cathepsin E prodomain and generation of the mature enzyme (Figure 2A). No mature cathepsin E was detected in non-acid treated tumor lysates.
Figure 2.
Cathepsin E expression and activity in mouse PDAC tumors.
(A) Western blot analysis of cathepsin E from three tumors before and after acid treatment. (B) Cathepsin E activity in tumors and pancreatic tissue from wild type (WT) littermates. Cleavage velocity is given in relative fluorescence units per second (RFU/sec). (C) Immunoblot analysis of cathepsin E expression in pancreatic tissue from WT, p48-Cre; KrasG12D; Trp53f/f, and RIP1-Tag2 mice. (D) Quantitative PCR analysis of cathepsin mRNA levels in pancreatic tissue from WT and PDAC animals. GUS mRNA levels were used for normalization.
As procathepsin E can be activated to the mature enzyme, we used an internally quenched fluorescent substrate to quantify the activity in tumor lysates. This substrate is selective for cathepsin E over cathepsin D and other acid-acting proteases (Yasuda et al., 2005). At pH 3.5, recombinant mouse cathepsin E rapidly hydrolyzed the internally quenched substrate, while no activity was detected at pH 7.4 (Figure 2B). At neutral pH, low levels of proteolytic activity were detected in tumor extracts from 10 week old PDAC mice and from exocrine pancreatic tissue isolated from non-tumor bearing littermates. This low-level activity is likely due to non-specific cleavage by proteases that are optimally active at neutral pH. When these same protein extracts were assayed at pH 3.5, a 13- to 37-fold increase in activity was detected in the tumor lysates, but not in the pancreatic extracts from healthy littermate controls. It was unclear from these results if cathepsin E activity in tumor extracts was due to the lack of an endogenous protease inhibitor or an increase in cathepsin E expression. To address this concern, procathepsin E protein levels were assessed in pancreatic tissue lysates from PDAC mice and non-tumor bearing littermates.
Procathepsin E is overexpressed in PDAC tumors
Procathepsin E was detected in all tumor extracts by immunoblot, but not in tissue from healthy littermates (Figure 2C). This suggests that malignant transformation in this mouse model is driving enhanced expression of this protein. Interestingly, immunoblot analysis failed to detect procathepsin E or mature cathepsin E in tumor lysate from the RIP1-Tag2 model of pancreatic islet carcinoma. The RIP1-Tag2 mouse model expresses the SV40 T antigen oncogenes in insulin-producing β cells and a number of prior studies have demonstrated that protease overexpression promotes tumorigenesis in this model (Folkman et al., 1989). In fact, the cysteine cathepsins H, L, C, Z, B and S genes were found to be upregulated during RIP1-Tag2 tumorigenesis and these enzymes contributed to angiogenic switching, tumor vascularity, and proliferation (Joyce and Hanahan, 2004; Joyce et al., 2004). Using the same quantitative RT-PCR approach as outlined in the RIP1-Tag2 mouse study, we compared cathepsin expression levels in PDAC tumors to healthy pancreatic tissue. Cathepsin E mRNA expression increased 441-fold in tumors relative to healthy pancreatic tissue, while cathepsins B, H, S, and Z mRNA levels increased by 3-fold or less (Figure 2D). Cathepsin L, C and D expression levels were lower in tumors than in healthy pancreatic tissue. The high expression of cathepsin E relative to the other cathepsins prompted us to further investigate the role of this protease in promoting tumorigenesis in the p48-Cre; KrasG12D; Trp53f/f PDAC mouse model. More generally, this points to specific cathepsins playing unique roles in the development of different types of pancreatic tumors.
Cathepsin E expression increases with progressive dysplasia
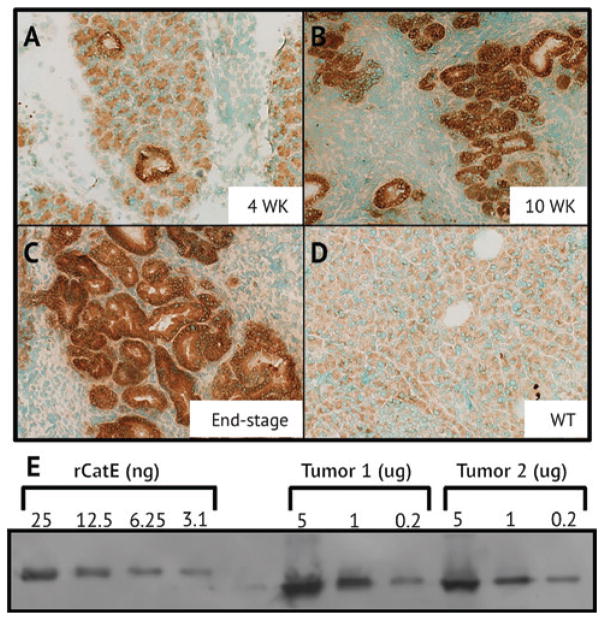
As procathepsin E was present in PDAC tumors and not in healthy tissue, we investigated the time-dependent expression and localization of this protein in the pancreas of p48-Cre; KrasG12D; Trp53f/f mice as they progress from harboring low-grade PanINs to invasive PDAC. Immunohisto-chemical staining of the exocrine pancreas of non-tumor bearing mice showed only basal levels of procathepsin E. However, by 4 weeks procathepsin E expression increased and the protein localized to neoplastic ductal structures. Staining intensity increased with progressive stages of dysplasia from 4 weeks to end-stage (Figure 3A–D). Mice succumb to the disease between 13 and 15 weeks and dense staining is evident in tumors from deceased mice. This unusually strong staining prompted us to quantity the amount of procathepsin E protein in PDAC tumor lysates using a semi-quantitative immunoblot. Surprisingly, approximately 6.25 nanograms of procathepsin E were present in 0.2 μg of total protein extracted from PDAC tumor lysates (Figure 3E). Therefore, in end-stage mouse tumors, procathepsin E accounted for approximately 3% of the total soluble protein extracted from the tumor.
Figure 3.
Immunohistochemical staining of cathepsin E in pancreatic tissue from p48-Cre; KrasG12D; Trp53f/f mice after
(A) 4 weeks, (B) 10 weeks, (C) and in end-stage (D) and WT mice. (E) Semi-quantitative western blot analysis of cathepsin E from tumor lysate of end-stage mice.
Peptide inhibitors bind to procathepsin E
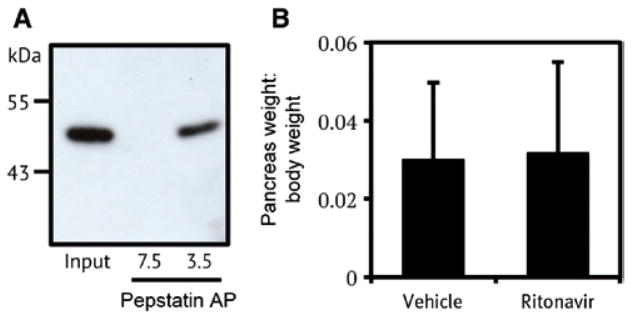
Structural studies of aspartyl protease zymogens revealed that the propeptide occupies the enzyme active site. Upon acid exposure, structural rearrangements lead to cleavage and dissociation of the propeptide (Ostermann et al., 2004). At pH 7.5 we did not observe procathepsin E activity against a fluorescent substrate (Figure 2B), suggesting that the propeptide is restricting access to the protease active site. We were curious to determine if an inhibitor of cathepsin E could compete with the propeptide for binding to the active site. Pepstatin is a potent inhibitor of cathepsin E and other pepsin-type aspartyl proteases (Dunn, 2002). We have successfully used pepstatin-agarose beads to enrich for aspartyl proteases from complex protein mixtures (Bibo-Verdugo et al., 2016). Using a similar approach, we added pepstatin-agarose beads to tumor lysate and then diluted the mixture in either pH 7.5 lysis buffer or pH 3.5 activation buffer. After 30 min incubation at pH 7.5 we were unable to recover procathepsin E. However, incubation at pH 3.5 in the presence of excess pepstatin-agarose led to recovery of procathepsin E and not the lower molecular weight mature protease (Figure 4A). This shows that the cathepsin E prodomain can be displaced prior to cleavage occurring, enabling access to the active site of the proenzyme.
Figure 4.
Cathepsin E inhibitor binding in tumor lysate and inhibition in PDAC mice.
(A) Affinity purification (AP) of cathepsin E from PDAC tumors using pepstatin-agarose beads followed by western blot analysis. Acid treatment was required for AP of cathepsin E. (B) Effect of ritonavir treatment on tumor burden in PDAC mice.
To target procathepsin E in PDAC, we used the FDA approved protease inhibitor, ritonavir. Ritonavir was approved to treat HIV and potently inhibits the viral aspartyl protease, however, it has nanomolar affinity towards cathepsin E (Kempf et al., 1998). Although it is likely that ritonavir has weaker affinity for procathepsin E we decided to treat Pdx-1-Cre; LSLKrasG12D; Trp53f/f mice with 125 mg/kg of for 28 days. After this period, tumors were removed and weighed. No significant difference in tumor burden was observed between ritonavir and vehicle treated animals (Figure 4B). There are a number of possibilities for why ritonavir failed to reduce tumor volume. PDAC tumors may be refractory to therapeutic intervention using this compound. Alternatively, procathepsin E may not play a critical enzymatic role in tumor progression and the use of an active site inhibitor will, therefore, not impact tumor progression.
Procathepsin E retains minimal activity at pH 6.5
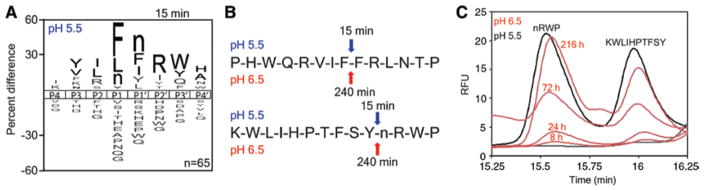
Cruz-Monserrate and colleagues have shown that the PDAC tumor microenvironment is acidic and therefore it is possible that conditions exist within the tumor that facilitate procathepsin E activity, but are insufficient to promote auto-activation to the mature enzyme (Cruz-Monserrate et al., 2014). Our studies with pepstatin-agarose showed that the cathepsin E active site is accessible prior to prodomain cleavage. We were unable to detect cleavage using a standard cathepsin E fluorescent substrate, but other substrates may be able to compete with the cathepsin E propeptide and allow us to detect activity. Therefore, we investigated procathepsin E activity and specificity using the highly sensitive MSP-MS assay. This assay uses tandem mass spectrometry to monitor proteolytic cleavage of 228 synthetic tetradecapeptides. At pH 5.5, procathepsin E was converted to the mature enzyme and 65 unique cleavages were detected following 15 min of incubation (Figure 5A, Supplementary Figure 1). The substrate specificity profile under these conditions was similar to what has been previously reported for cathepsin E (Impens et al., 2010; O’Donoghue et al., 2012). When we performed the same assay at pH 6.5, auto-activation of procathepsin E did not occur (Supplementary Figure 1). However, cleavage of two peptides – PHWQRVIFFRLNTP and KWLIHPTFSYnRWP – in the library was evident after 4 h incubation (Figure 5B). The same peptides were also hydrolyzed by mature cathepsin E at pH 5.5 within 15 min.
Figure 5.
Analysis of procathepsin E activity.
(A) iceLogo generated from the cathepsin E cleavage events detected after 15 min using the MSP-MS assay. To profile mature cathepsin E the MSP-MS assay was performed at pH 5.5. In the iceLogo n corresponds to norleucine. (B) The cleavage sites within two peptides by mature cathepsin E (pH 5.5 – blue) and procathepsin E (pH 6.5 – red). The time at which cleavage was first observed is shown above the arrows indicating the site of cathepsin E cleavage. (C) HPLC chromatograms of peptide cleavage products, which have amino acid sequences shown above each peak, following incubation with procathepsin E (pH 6.5 – red) and mature cathepsin E (pH 5.5 – black). Abundance of cleavage products was monitored using tryptophan fluorescence with excitation at 280 nm and emission at 330 nm.
The mass spectrometry based assay is largely qualitative and therefore we decided to use reversed-phase high performance liquid chromatography (RP-HPLC) to quantify the rate of hydrolysis. One of the peptide substrates is cleaved into two products that each contains a tryptophan residue, allowing for quantitation via tryptophan fluorescence during RP-HPLC analysis. At pH 5.5, 2 nM of mature cathepsin E completely cleaved 100 μM of substrate within 1 h (Supplementary Figure 2). The two cleavage products eluted from the C18 column between 15.25 and 16.25 min. Analysis of the mass of the cleavage products by MALDI-MS demonstrated that cathepsin E cut at the predicted site (Supplementary Figure 3). At pH 6.5, time-dependent product formation was evident using 200 nM of procathepsin E (Figure 5C). After 216 h incubation, only 2.6% of the parent peptide was hydrolyzed.
Discussion
Previous work by our group determined that a PDAC cell line isolated from a Pdx-1-Cre; LSLKrasG12D; Trp53f/f mouse secretes several endolysosomal proteases, such as cathepsin B, L, and E (O’Donoghue et al., 2012). Several studies have looked at the role of the cysteine cathepsins B and L in pancreatitis and pancreatic cancer (Gocheva et al., 2006; Lyo et al., 2012). In fact, these enzymes have been implicated in playing multiple roles in various cancers (Joyce and Hanahan, 2004; Sevenich et al., 2014). However, few studies have focused on the role of the aspartyl cathepsins in cancer, with the notable exception of cathepsin D in breast cancer (Masson et al., 2010). We were particularly intrigued to detect high levels of cathepsin E in conditioned media from PDAC cells and sought to investigate further the expression, localization, and enzymatic function of this enzyme. However, in this study we show that only procathepsin E is secreted from the primary cells in culture and activity is only detectable after the media becomes sufficiently acidic to induce protease auto-activation.
A previous immunohistochemical study by Buchholz and colleagues demonstrated that human cathepsin E is absent from the normal pancreatic duct, but abundant in PDAC and precursor lesions (Buchholz et al., 2005). In agreement with this, we show that cathepsin E in the Pdx-1-Cre; LSLKrasG12D; Trp53f/f mouse model is hyperexpressed in PDAC tumors and accumulates in high abundance in the cytoplasm. In fact, it is likely to be one of the most abundant proteins in the mouse tumor extract and accounts for approximately 3% of the total soluble protein. This intracellular protein exists only as procathepsin E but can be activated upon brief exposure to acid. The absence of mature cathepsin E in the tumor lysate indicates that the protein does not traffic through the normal endolysosomal pathway with exposure to a sufficiently acidic environment to cause activation. Therefore, the accumulation of excess intracellular procathepsin E may indicate endolysosomal dysfunction in these cells.
The excess procathepsin E identified in PDAC is analogous to procathepsin D secreted by breast cancer cells (Laurent-Matha et al., 2001). Procathepsin D is secreted at low levels by normal mammary epithelial cells, but secretion increases by up to 40-fold in breast cancer cell lines. Several studies have shown that procathepsin D binds to prosaposin in the ER and these proteins are co-secreted (Laurent-Matha et al., 2002; Gopalakrishnan et al., 2004). In addition, procathepsin D can bind to cystatin-C and will degrade this protein upon activation to the mature enzyme at pH 3.5 (Laurent-Matha et al., 2012; Khalkhali-Ellis and Hendrix, 2014). Degradation of this macromolecular inhibitor ultimately results in an increase in cysteine protease activity, which promotes tumor progression and metastasis. A recent study showed that secretion of procathepsin D from the MCF-7 breast cancer cell line is increased under hypoxia (Achour et al., 2016). Hypoxic tumors cause acidification of the tumor microenvironment and Cruz-Monserrate and colleagues demonstrated that an acidic microenvironment clearly exists within PDAC tumors (Cruz-Mon-serrate et al., 2014). Interestingly, the microenvironment conditions appear to favor the functioning of acid-acting proteases such as those found in the endosomes and lysosomes. However, it appears to be insufficiently acidic to activate procathepsin E in pancreatic tumors and procathepsin D in breast tumors.
While procathepsin E may mediate its functions through binding partners, we decided to investigate if this protein was enzymatically active under pH conditions that are relevant to the tumor. We show that the aspartyl pro-tease inhibitor, pepstatin, can compete with the propeptide sequence for binding to the enzyme active site. This prompted us to treat mice with the HIV protease inhibitor ritonavir; however, no reduction in tumor burden was evident. While ritonavir is a potent inhibitor of the mature enzyme (Kempf et al., 1998), it is likely to be much less potent against the zymogen. Therefore, it was unclear if the ineffectiveness of the inhibitor was the result of cathepsin E activity not being important for tumor progression or if the inhibitor was unable to sufficiently engage with the enzyme.
Although it is well established that protease zymogens can be catalytically active, this has not been demonstrated for procathepsin E. Previous studies have shown that rat cathepsin E alters its substrate specificity at neutral pH and becomes more ‘trypsin-like’, cleaving on the C-terminal side of arginine residues. However, Zaida and coworkers predicted that rat cathepsin E was likely to be contaminated with trace amounts of a ‘trypsin-like’ proteases that are optimally active at neutral pH (Zaidi et al., 2007). Here we used mass spectrometry and a library of 228 tetradecapeptides to detect proteolytic activity of procathepsin E (O’Donoghue et al., 2012). This highly sensitive assay identified two peptide substrates for procathepsin E that are also hydrolyzed by the mature enzyme. The cleavage site within these peptides mirrors the known specificity of cathepsin E. Taken together, these results show that procathepsin E has weak pro-teolytic activity at pH 6.5 and suggests that the highly abundant zymogen may play an enzymatic role in tumor progression.
While our studies could not detect mature cathepsin E in whole tumor extracts, it is possible that a fraction of the 53 kDa procathepsin E gets converted to the 49 kDa mature form, but is undetectable by immunoblot. A number of imaging agents targeting cathepsin E activity have recently been developed to monitor PDAC status in vivo (Cruz-Monserrate et al., 2012; Keliher et al., 2013; Li et al., 2014). These imaging agents were designed to target the mature enzyme active site or be specifically cleaved by cathepsin E. In addition, Abd-Elgaliel and colleagues have developed a cathepsin E activated prodrug that is toxic to cells expressing cathepsin E (Abd-Elgaliel et al., 2013). From our work it is clear that in PDAC, cathepsin E primarily exists as a zymogen with minimal enzymatic activity. It is likely that developing probes to target procathepsin E may therefore be more effective. For example, radio-labeled antibodies have been successfully used to image proteases that are overexpressed in tumors and could be developed against procathepsin E for non-invasive detection of PDAC (LeBeau et al., 2013).
In conclusion, our analysis of cathepsin E activation in PDAC revealed that the protease is highly overexpressed and exists exclusively in its proform. Cathepsin E may play a role in PDAC progression that is independent of proteolytic activity. Cathepsin E is now recognized as an exciting biomarker for PDAC and this information may facilitate the development of novel imaging agents for monitoring disease status.
Materials and methods
Mouse strains, tissue culture, and ritonavir administration
The p48-Cre; KrasG12D; Trp53f/f mouse strain was used and cells were isolated from PDAC tumors as previously described (Nolan-stevaux et al., 2009). Cells were maintained in complete DMEM with 10% FBS. Mice were treated for 28 days with 125 mg/kg ritonavir or ethanol by oral gavage. The mouse pancreas was removed and weighed and tumor burden was assessed using the ratio of tumor weight to body weight. All animal studies were conducted in compliance with University of California Institutional Animal Care and Use Committee guidelines.
Histological analysis, immunochemistry, and immunofluorescence
Pancreatic tissue from p48-Cre; KrasG12D; Trp53f/f mice was harvested after 4 and 10 weeks and from animals with end-stage disease. Tissue was fixed overnight in zinc-buffered formalin and embedded in paraffin. 5-mm thick sections were subjected to either H&E staining or an antigen retrieval procedure (BioGenex, San Ramon, CA, USA). Following inhibition of endogenous peroxidases and blocking the slides, goat anti-mouse cathepsin E antibody (1:100; R&D Systems AF1130, Minneapolis, MN, USA) was applied overnight at 4°C. Biotinylated or fluorochrome-conjugated antibodies were used as secondary antibodies (1:200; Jackson Immunoresearch, West Grove, PA, USA). 3-39-DAB tetrahydrochloride (Sigma D4293, St. Louis, MO, USA) was used as a chromogen.
Cathepsin E isolation and western blotting
Mouse tumors and cell lines were lysed in 20 mM Tris-HCl pH 7.5, 150 mM NaCl, 1% Triton X-100 buffer. Protein lysates were diluted to 1.5 mg/ml in lysis buffer or 1.25 mM sodium acetate pH 3.5 and incubated on ice for 30 min. Where appropriate, 20% (v/v) pepstatin-agarose (Sigma) was added to protein lysates and incubated on ice for 30 min. Agarose beads were washed in lysis buffer or sodium acetate buffer and protein was eluted in 1X LDS sample buffer containing 20 mM TCEP solution (ThermoFisher Scientific). Secreted cathepsin E was isolated by immunoprecipitation with 2.5 μg of goat anti-mouse cathepsin E antibody (R&D Systems AF1130) attached to Protein G Dynabeads. All samples were subjected to electrophoresis on denaturing 10% NuPAGE Bis-Tris gels (Invitrogen). Proteins were transferred to polyvinylidene fluoride membranes and blocked in Tris-buffered saline Triton X-100 (TBST) containing 5% (w/v) milk. Membranes were incubated with a polyclonal rabbit anti-mouse cathepsin E antibody (1:1000; Thermo Scientific PA3-16821), washed and incubated with goat-derived HRP-conjugated secondary antibody (Bio-Rad). Immunoblots were developed on film with the ECL Plus detection system (GE Healthcare). To verify that equal amounts of protein were being compared across samples, actin levels were quantified in parallel with an anti-β-actin antibody (1:10 000; Sigma). Where indicated, samples were treated with the deglycosylase PNGaseF (New England Biolabs) according to the manufacturers protocol prior to gel electrophoreses.
Fluorescent protease assays
All assays were performed at room temperature in either PBS containing 0.1% Triton X-100 or 50 mM sodium acetate, pH 3.5, 100 mM NaCl, 0.1% Triton X-100 using 40 μM substrate (Bachem M-2625, Bubendorf, Switzerland). Assays were performed in triplicate in black round-bottom 96-well plates on a SpectraMax Gemini fluorescence spectrometer (Molecular Devices, Sunnyvale, CA, USA) using excitation and emission wavelengths of 328 nm and 393 nm, respectively.
Quantitative PCR
Total RNA was prepared from tumors and cell lines using the RNeasy Mini (Qiagen, Hilden, Germany) according to the manufacturer’s recommendations. Total RNA from microdissected samples was prepared using the RNeasy Micro (Qiagen). DNase treatment and RNA cleanup were performed with the DNA-Free RNA Kit (Zymo Research, Irvine, CA, USA). cDNA synthesis was performed using iScript (Bio-Rad, Hercules, CA, USA). PCRs were performed using the following TaqMan assays (Applied Biosystems, Foster City, CA, USA): Ctsb, Ctsc, Ctsh, Ctsl, Ctss, Ctsx, Ctsd, and Ctse. The mGus assay was obtained from Integrated DNA Technologies (F) CTC ATC TGG AAT TTC GCC GA; (R) GGC GAG TGA AGA TCC CCT TC; (Probe) fam-CGA ACC AGT CAC CGC TGA GAG TAA TCG-bhq1 was used to normalize expression. Quantitative PCR reactions were performed on an ABI7900HT Sequence Detection System (Applied Biosystems). Ct values were determined and subtracted to obtain the ΔCt [ΔCt=Ct(test locus)–Ct(control locus)]. Relative fold difference was calculated as 100*2−ΔCt.
Multiplex peptide cleavage assay
Recombinant mouse cathepsin E proteolytic activity was analyzed using the MSP-MS assay as described previously (O’Donoghue et al., 2012). For all assays, an expanded 228 peptide library was used and split into two pools containing 114 peptides at 500 nM each. Assays were performed at pH 6.5 for procathepsin E and pH 5.5 for mature cathepsin E. To promote proteolytic activity of procathepsin E, 250 nM of enzyme was incubated with peptide pools at 37°C. Mature cathepsin E was assayed at 50 nM. Aliquots were removed and acid-quenched to pH 2 or less with formic acid after 15, 60, 240, and 1200 min. To avoid acid-mediated activation of cathepsin E, 10 μM of the aspartyl protease inhibitor pepstatin A (Sigma) was added to assays directly before the acid-quench. Control samples without recombinant cathepsin E were prepared under identical conditions to account for non-enzymatic degradation of the substrates. Prior to LC-MS/MS peptide sequencing, samples were desalted using C18 Zip-Tips (Millipore, Billerica, MA, USA).
For LC-MS/MS, an LTQ Orbitrap XL mass spectrometer (Thermo, Waltham, MA, USA) equipped with a 10 000 psi system nanoAC-QUITY UPLC instrument (Waters, Milford, MA, USA) was used for reversed phase chromatography with a C18 column (1.7 μm bead size, 100 μm×100 mm). The LC was operated at 600 nl/min flow rate and peptides were separated using a linear gradient over 65 min from 2% B to 30% B, with solvent A being 0.1% formic acid in water and solvent B being 0.1% formic acid in 70% acetonitrile. Survey scans were recorded over 350–1800 m/z range and MS/MS was performed with CID fragmentation on the six most intense precursor ions. Mass spectrometry peak lists were generated using in-house software called PAVA. Peak lists were searched in Protein Prospector v. 5.10.0 against a database containing the 228 peptide sequences from the MSP-MS library. For database searching, peptide sequences were allowed to contain the following variable modifications: oxidation of tryptophan, proline, and phenylalanine and N-terminal pyroglutamic acid from glutamine or glutamic acid. Cleavage site data was extracted from Protein Prospector using the MSP-extractor software. iceLogo software was used to visualize conserved patterns in amino acid sequence at ±4 positions adjacent to the identified site of cleavage (Colaert et al., 2009).
Analysis of cathepsin E activity by HPLC
Activity assays with recombinant cathepsin E and individual substrates were performed in pH 6.5 PBS and pH 5.5 sodium acetate buffer. For pH 6.5 assays, 250 nM of cathepsin E was incubated with 100 μM substrate at 37°C. After 4, 8, 24, 72, and 216 h, protease activity was heat inactivated. For pH 5.5 assays, 2 nM of cathepsin E was incubated with 100 μM substrate at 37°C. Cathepsin E was heat inactivated after 2 and 60 min incubation. All samples were compared to a control containing 100 μM substrate and no enzyme. Following heat inactivation, digestion products were analyzed using a 1100 HPLC system (Agilent, Santa Clara, CA, USA) with a C18 column (10 μm bead size, 4.6 mm×250 mm, Vydac). The HPLC was operated at a flow rate of 1 ml/min with solvent A: 0.1% trifluoroacetic acid in water and solvent B: 0.1% trifluoroacetic acid in 95% acetonitrile. A linear gradient from 5% B to 95% B over 17 min was used for peptide separation. Tryptophan fluorescence with excitation at 280 nM and emission at 330 nM was used for detection of the full-length substrate and cleavage products. For quantification of substrate conversion, peak heights were compared between samples and controls.
Peptide cleavage site confirmation by mass spectrometry
Cleavage sites within individual peptides were identified by matrix-assisted laser desorption ionization mass spectrometry (MALDI-MS). 0.5 μl of reaction mixtures were mixed with 0.5 μl of α-Cyano-4-hydroxycinnamic acid (CHCA) matrix and spotted on a MALDI plate. Spectra were then acquired over a mass range of 500–2500 Daltons using an Axima Performance MALDI-time-of-flight/time-of-flight (TOF/TOF) mass spectrometer (Shimadzu, Kyoto, Japan). An average of 200 shots was used for each spectra. All spectra were analyzed using the Shimadzu Biotech Launchpad.
Supplementary Material
Acknowledgments
A.J.O. and C.S.C. were supported by the Program for Breakthrough Biomedical Research (PBBR) and the Sandler Foundation. We gratefully acknowledge additional funds that were provided from NIH CA 185689, NIH CA 186077, and NIH CA 196276 to C.S.C. S.L.I was supported by NIH Pharmaceutical Sciences and Pharmacogenomics Training Grant T32GM008155.
Footnotes
Supplemental Material: The online version of this article (DOI: 10.1515/hsz-2016-0138) offers supplementary material, available to authorized users.
Contributor Information
Anthony J. O’Donoghue, Department of Pharmaceutical Chemistry, University of California San Francisco, 600 16th Street, San Francisco, CA 94158, USA.
Sam L. Ivry, Department of Pharmaceutical Chemistry, University of California San Francisco, 600 16th Street, San Francisco, CA 94158, USA
Chaity Chaudhury, Diabetes Center and Helen Diller Family Comprehensive Cancer Center, University of California, San Francisco, CA 94143, USA.
Daniel R. Hostetter, Department of Pharmaceutical Chemistry, University of California San Francisco, 600 16th Street, San Francisco, CA 94158, USA
Douglas Hanahan, Diabetes Center and Helen Diller Family Comprehensive Cancer Center, University of California, San Francisco, CA 94143, USA.
Charles S. Craik, Department of Pharmaceutical Chemistry, University of California San Francisco, 600 16th Street, San Francisco, CA 94158, USA
References
- Abd-Elgaliel WR, Cruz-Monserrate Z, Wang H, Logsdon CD, Tung CH. Pancreatic cancer-associated Cathepsin E as a drug activator. J Control Release. 2013;167:221–227. doi: 10.1016/j.jconrel.2013.02.007. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Achour O, Ashraf Y, Bridiau N, Kacem M, Poupard N, Bordenave-Juchereau S, Sannier F, Lamerant-Fayel N, Kieda C, Liaudet-Coopman E, et al. Alteration of cathepsin D trafficking induced by hypoxia and extracellular acidification in MCF-7 breast cancer cells. Biochimie. 2016;121:123–130. doi: 10.1016/j.biochi.2015.11.007. [DOI] [PubMed] [Google Scholar]
- Beaujouin M, Prébois C, Derocq D, Laurent-Matha V, Masson O, Pattingre S, Coopman P, Bettache N, Grossfield J, Hollingsworth RE, et al. Procathepsin D interacts with the extracellular domain of the β chain of LRP1 and promotes LRP1-dependent fibroblast outgrowth. J Cell Sci. 2010;123:3336–3346. doi: 10.1242/jcs.070938. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bergers G, Javaherian K, Lo K, Folkman J, Hanahan D. Effects of angiogenesis inhibitors on multistage carcinogenesis in mice. Science. 1999;284:808–811. doi: 10.1126/science.284.5415.808. [DOI] [PubMed] [Google Scholar]
- Bibo-Verdugo B, O’Donoghue AJ, Rojo-Arreola L, Craik CS, García-Carreño F. Complementary proteomic and biochemical analysis of peptidases in lobster gastric juice uncovers the functional role of individual enzymes in food digestion. Mar Biotechnol. 2016;18:201–214. doi: 10.1007/s10126-015-9681-5. [DOI] [PubMed] [Google Scholar]
- Buchholz M, Braun M, Heidenblut A, Kestler HA, Klöppel G, Schmiegel W, Hahn SA, Lüttges J, Gress TM. Transcriptome analysis of microdissected pancreatic intraepithelial neoplastic lesions. Oncogene. 2005;24:6626–6636. doi: 10.1038/sj.onc.1208804. [DOI] [PubMed] [Google Scholar]
- Burster T, Reich M, Zaidi N, Voelter W, Boehm BO, Kalbacher H. Cathepsin E regulates the presentation of tetanus toxin C-fragment in PMA activated primary human B cells. Biochem Biophys Res Commun. 2008;377:1299–1303. doi: 10.1016/j.bbrc.2008.10.162. [DOI] [PubMed] [Google Scholar]
- Cappiello MG, Wu Z, Scott BB, McGeehan GM, Harrison RK. Purification and characterization of recombinant human cathepsin E expressed in human kidney cell line 293. Protein Expres Purif. 2004;37:53–60. doi: 10.1016/j.pep.2004.05.013. [DOI] [PubMed] [Google Scholar]
- Chain BM, Free P, Medd P, Swetman C, Tabor AB, Terrazzini N. The expression and function of cathepsin E in dendritic cells. J Immunol. 2005;174:1791–1800. doi: 10.4049/jimmunol.174.4.1791. [DOI] [PubMed] [Google Scholar]
- Colaert N, Helsens K, Martens L, Vandekerckhove J, Gevaert K. Improved visualization of protein consensus sequences by iceLogo. Nat Methods. 2009;6:786–787. doi: 10.1038/nmeth1109-786. [DOI] [PubMed] [Google Scholar]
- Costello E, Greenhalf W, Neoptolemos JP. New biomarkers and targets in pancreatic cancer and their application to treatment. Nat Rev Gastroenterol. 2012;9:435–444. doi: 10.1038/nrgastro.2012.119. [DOI] [PubMed] [Google Scholar]
- Cruz-Monserrate Z, Abd-Elgaliel W, Logsdon C. Detection of pancreatic cancer tumours and precursor lesions by cathepsin E activity in mouse models. Gut. 2012;61:1315–1322. doi: 10.1136/gutjnl-2011-300544. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Cruz-Monserrate Z, Roland CL, Deng D, Arumugam T, Moshnikova A, Andreev OA, Reshetnyak YK, Logsdon CD. Targeting pancreatic ductal adenocarcinoma acidic microenvironment. Sci Rep. 2014;4:4410. doi: 10.1038/srep04410. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Dunn BM. Structure and mechanism of the pepsin-like family of aspartic peptidases. Chem Rev. 2002;102:4431–4458. doi: 10.1021/cr010167q. [DOI] [PubMed] [Google Scholar]
- Folkman J, Watson K, Ingber D, Hanahan D. Induction of angiogenesis during the transition from hyperplasia to neoplasia. Nature. 1989;339:58–61. doi: 10.1038/339058a0. [DOI] [PubMed] [Google Scholar]
- Gocheva V, Zeng W, Ke D, Klimstra D, Reinheckel T, Peters C, Hanahan D, Joyce JA. Distinct roles for cysteine cathepsin genes in multistage tumorigenesis. Genes Dev. 2006;24:543–556. doi: 10.1101/gad.1407406. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gocheva V, Wang H, Gadea BB, Shree T, Hunter KE, Gar-fall AL, Berman T, Joyce JA. IL-4 induces cathepsin protease activity in tumor-associated macrophages to promote cancer growth and invasion. Genes Dev. 2010;24:241–255. doi: 10.1101/gad.1874010. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gopalakrishnan MM, Grosch HW, Locatelli-Hoops S, Werth N, Smolenová E, Nettersheim M, Hasilik A. Purified recombinant human prosaposin forms oligomers that bind procathepsin D and affect its autoactivation. Biochem J. 2004;383:507–515. doi: 10.1042/BJ20040175. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hezel AF, Kimmelman AC, Stanger BZ, Bardeesy N, Depinho RA. Genetics and biology of pancreatic ductal adenocarcinoma. Genes Dev. 2006;20:1218–1249. doi: 10.1101/gad.1415606. [DOI] [PubMed] [Google Scholar]
- Impens F, Colaert N, Helsens K, Ghesquière B, Timmerman E, De Bock PJ, Chain BM, Vandekerckhove J, Gevaert K. A quantitative proteomics design for systematic identification of protease cleavage events. Mol Cell Proteomics. 2010;9:2327–2333. doi: 10.1074/mcp.M110.001271. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Joyce JA, Hanahan D. Multiple roles for cysteine cathepsins in cancer. Cell Cycle. 2004;3:1516–1519. doi: 10.4161/cc.3.12.1289. [DOI] [PubMed] [Google Scholar]
- Joyce JA, Baruch A, Chehade K, Meyer-Morse N, Giraudo E, Tsai FY, Greenbaum DC, Hager JH, Bogyo M, Hanahan D. Cathepsin cysteine proteases are effectors of invasive growth and angiogenesis during multistage tumorigenesis. Cancer Cell. 2004;5:443–453. doi: 10.1016/s1535-6108(04)00111-4. [DOI] [PubMed] [Google Scholar]
- Keliher EJ, Reiner T, Earley S, Klubnick J, Tassa C, Lee AJ, Ramaswamy S, Bardeesy N, Hanahan D, Depinho RA, et al. Targeting cathepsin E in pancreatic cancer by a small molecule allows in vivo detection. Neoplasia. 2013;15:684–693. doi: 10.1593/neo.13276. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kempf DJ, Sham HL, Marsh KC, Flentge CA, Betebenner D, Green BE, McDonal E, Vasavanonda S, Saldivar A, Wideburg NE, et al. Discovery of ritonavir, a potent inhibitor of HIV protease with high oral bioavailability and clinical efficacy. J Med Chem. 1998;2623:602–617. doi: 10.1021/jm970636+. [DOI] [PubMed] [Google Scholar]
- Kessenbrock K, Plaks V, Werb Z. Matrix metalloproteinases: regulators of the tumor microenvironment. Cell. 2010;141:52–67. doi: 10.1016/j.cell.2010.03.015. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Khalkhali-Ellis Z, Hendrix MJC. Two faces of cathepsin D: physiological guardian angel and pathological demon. Biol Med. 2014;6:1000206. doi: 10.4172/0974-8369.1000206. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Laurent-Matha V, Garcia M, Rochefort H, Glondu M, Coopman P. A mutated cathepsin-D devoid of its catalytic activity stimulates the growth of cancer cells. Oncogene. 2001;20:6920–6929. doi: 10.1038/sj.onc.1204843. [DOI] [PubMed] [Google Scholar]
- Laurent-Matha V, Lucas A, Huttler S, Sandhoff K, Garcia M, Rochefort H. Procathepsin D interacts with prosaposin in cancer cells but its internalization is not mediated by LDL receptor-related protein. Exp Cell Res. 2002;277:210–219. doi: 10.1006/excr.2002.5556. [DOI] [PubMed] [Google Scholar]
- Laurent-Matha V, Huesgen PF, Masson O, Derocq D, Prebois C, Gary-Bobo M, Lecaille F, Rebière B, Meurice G, Oréar C, et al. Proteolysis of cystatin C by cathepsin D in the breast cancer microenvironment. FASEB J. 2012;26:5172–5181. doi: 10.1096/fj.12-205229. [DOI] [PubMed] [Google Scholar]
- LeBeau AM, Lee M, Murphy ST, Hann BC, Warren RS, Delos Santos R, Kurhanewicz J, Hanash SM, VanBrocklin HF, Craik CS. Imaging a functional tumorigenic biomarker in the transformed epithelium. Proc Natl Acad Sci USA. 2013;110:93–98. doi: 10.1073/pnas.1218694110. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Li H, Li Y, Cui L, Wang B, Cui W, Li M, Cheng Y. Monitoring pancreatic carcinogenesis by the molecular imaging of cathepsin E in vivo using confocal laser endomicroscopy. PLoS One. 2014;9:106566. doi: 10.1371/journal.pone.0106566. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lyo V, Cattaruzza F, Kim TN, Walker AW, Paulick M, Cox D, Cloyd J, Kirkwood KS. Active cathepsins B, L, and S in murine and human pancreatitis. Am J Physiol Gastr L. 2012;303:894–903. doi: 10.1152/ajpgi.00073.2012. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mason SD, Joyce JA. Proteolytic networks in cancer. Trends Cell Biol. 2011;21:228–237. doi: 10.1016/j.tcb.2010.12.002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Masson O, Bach AS, Derocq D, Prébois C, Laurent-Matha V, Pattingre S, Liaudet-Coopman E. Pathophysiological functions of cathepsin D: targeting its catalytic activity versus its protein binding activity? Biochimie. 2010;92:1635–1643. doi: 10.1016/j.biochi.2010.05.009. [DOI] [PubMed] [Google Scholar]
- Mohamed MM, Sloane BF. Cysteine cathepsins: multifunctional enzymes in cancer. Nat Rev Cancer. 2006;6:764–775. doi: 10.1038/nrc1949. [DOI] [PubMed] [Google Scholar]
- Morris JP, Wang SC, Hebrok M. KRAS, Hedgehog, Wnt and the twisted developmental biology of pancreatic ductal adenocarcinoma. Nat Rev Cancer. 2010;10:683–695. doi: 10.1038/nrc2899. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mota F, Kanan J, Rayment N, Mould T, Singer A, Chain BM. Cathepsin E expression by normal and premalignant cervical epithelium. Am J Pathol. 1997;150:1223–1229. [PMC free article] [PubMed] [Google Scholar]
- Nishioku T, Hashimoto K, Yamashita K, Liou SY, Kagamiishi Y, Maegawa H, Katsube N, Peters C, von Figura K, Saftig P, et al. Involvement of cathepsin E in exogenous antigen processing in primary cultured murine microglia. J Biol Chem. 2002;277:4816–4822. doi: 10.1074/jbc.M108382200. [DOI] [PubMed] [Google Scholar]
- Nolan-stevaux O, Lau J, Truitt ML, Chu GC, Hebrok M, Hanahan D, Ferna ME. GLI1 is regulated through Smoothened- independent mechanisms in neoplastic pancreatic ducts and mediates PDAC cell survival and transformation. Genes Dev. 2009;1:24–36. doi: 10.1101/gad.1753809. [DOI] [PMC free article] [PubMed] [Google Scholar]
- O’Donoghue AJ, Eroy-reveles AA, Knudsen GM, Ingram J, Zhou M, Statnekov JB, Greninger AL, Hostetter DR, Qu G, Maltby DA, et al. Global identification of peptidase specificity by multiplex substrate profiling. Nat Methods. 2012;9:1095–1100. doi: 10.1038/nmeth.2182. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ostermann N, Gerhartz B, Worpenberg S, Trappe J, Eder J. Crystal structure of an activation intermediate of cathepsin E. J Mol Biol. 2004;342:889–899. doi: 10.1016/j.jmb.2004.07.073. [DOI] [PubMed] [Google Scholar]
- Rao-Naik C, Guruprasad K, Batley B, Rapundalo S, Hill J, Blundell T, Kay J, Dunn BM. Exploring the binding preferences/specificity in the active site of human cathepsin E. Proteins. 1995;181:168–181. doi: 10.1002/prot.340220209. [DOI] [PubMed] [Google Scholar]
- Reid A, Valler M, Kay J. Immunolocalisation of cathepsin D in normal and neoplastic human tissues. J Clin Pathol. 1986;39:1323–1330. doi: 10.1136/jcp.39.12.1323. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Richter C, Tanaka T, Yada RY. Mechanism of activation of the gastric aspartic proteinases: pepsinogen, progastricsin and prochymosin. Biochem J. 1998;490:481–490. doi: 10.1042/bj3350481. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ringel J, Jesnowski R, Moniaux N, Lüttges J, Ringel J, Choudhury A, Batra SK, Klöppel G, Löhr M. Aberrant expression of a disintegrin and metalloproteinase 17/tumor necrosis factor-α converting enzyme increases the malignant potential in human pancreatic ductal adenocarcinoma. Cancer Res. 2006;66:9045–9053. doi: 10.1158/0008-5472.CAN-05-3287. [DOI] [PubMed] [Google Scholar]
- Sakai H, Saku T, Yuzo K, Yamamolo K. Quantitation and immunohistochemical localization of cathepsins E and D in rat tissues and blood cells. Biochim Biophys Acta. 1989;991:367–375. doi: 10.1016/0304-4165(89)90130-x. [DOI] [PubMed] [Google Scholar]
- Sastradipura DF, Nakanishi H, Tsukuba T, Nishishita K, Sakai H, Kato Y, Gotow T, Yamamoto K. Identification of cellular compartments involved in processing of cathepsin E in primary cultures of rat microglia. J Neurochem. 2002;70:2045–2056. doi: 10.1046/j.1471-4159.1998.70052045.x. [DOI] [PubMed] [Google Scholar]
- Sevenich L, Joyce JA. Pericellular proteolysis in cancer. Genes Dev. 2014;28:2331–2347. doi: 10.1101/gad.250647.114. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sevenich L, Bowman RL, Mason SD, Quail DF, Rapaport F, Elie BT, Brogi E, Brastianos PK, Hahn WC, Holsinger LJ, et al. Analysis of tumour- and stroma-supplied proteolytic networks reveals a brain-metastasis-promoting role for cathepsin S. Nat Cell Biol. 2014;16:876–888. doi: 10.1038/ncb3011. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tsukuba T, Okamoto K, Okamoto Y, Yanagawa M, Kohmura K, Yasuda Y, Uchi H, Nakahara T, Furue M, Nakayama K, et al. Association of cathepsin E deficiency with development of atopic dermatitis. J Biochem. 2003;134:893–902. doi: 10.1093/jb/mvg216. [DOI] [PubMed] [Google Scholar]
- Tsukuba T, Yamamoto S, Yanagawa M, Okamoto K, Okamoto Y, Nakayama KI, Kadowaki T, Yamamoto K. Cathepsin E-deficient mice show increased susceptibility to bacterial infection associated with the decreased expression of multiple cell surface Toll-like receptors. J Biochem. 2006;140:57–66. doi: 10.1093/jb/mvj132. [DOI] [PubMed] [Google Scholar]
- Ullmann R, Morbini P, Halbwedl I, Bongiovanni M, Gogg-Kammerer M, Papotti M, Gabor S, Renner H, Popper HH. Protein expression profiles in adenocarcinomas and squamous cell carcinomas of the lung generated using tissue microarrays. J Pathol. 2004;203:798–807. doi: 10.1002/path.1584. [DOI] [PubMed] [Google Scholar]
- Uno K, Azuma T, Nakajima M, Yasuda K, Hayakumo T, Mukai H, Sakai T, Kawai K. Clinical significance of cathepsin E in pancreatic juice in the diagnosis of pancreatic ductal adenocarcinoma. J Gastroenterol Hepatol. 2000;15:1333–1338. [PubMed] [Google Scholar]
- Yasuda Y, Kageyama T, Akamine A, Shibata M, Kominami E, Uchiyama Y, Yamamoto K. Characterization of new fluorogenic substrates for the rapid and sensitive assay of cathepsin E and cathepsin D. J Biochem. 1999;125:1137–1143. doi: 10.1093/oxfordjournals.jbchem.a022396. [DOI] [PubMed] [Google Scholar]
- Yasuda Y, Kohmura K, Kadowaki T, Tsukuba T, Yamamoto K. A new selective substrate for cathepsin E based on the cleavage site sequence of α2-macroglobulin. Biol Chem. 2005;386:299–305. doi: 10.1515/BC.2005.036. [DOI] [PubMed] [Google Scholar]
- Zaidi N, Kalbacher H. Cathepsin E: a mini review. Biochem Biophys Res Commun. 2008;367:517–522. doi: 10.1016/j.bbrc.2007.12.163. [DOI] [PubMed] [Google Scholar]
- Zaidi N, Herrmann T, Voelter W, Kalbacher H. Recombinant cathepsin E has no proteolytic activity at neutral pH. Biochem Biophys Res Commun. 2007;360:51–55. doi: 10.1016/j.bbrc.2007.05.187. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.