Abstract
Purpose
Mutation of the Kirsten ras sarcoma viral oncogene homolog (KRAS) and loss of p53 function are commonly seen in non-small cell lung cancer (NSCLC). Combining therapeutics targeting these tumor defensive pathways with cisplatin in a single nanoparticle platform are rarely developed in clinic.
Experimental Design
Cisplatin was encapsulated in liposomes which multiple polyelectrolyte layers including siKRAS and miR-34a were built on to generate multifunctional layer-by-layer nanoparticle. Structure, size, and surface charge were characterized, in addition to in vitro toxicity studies. In vivo tumor targeting and therapy was investigated in an orthotopic lung cancer model by microCT, fluorescence imaging, and immunohistochemistry.
Results
The singular nanoscale formulation, incorporating oncogene siKRAS, tumor suppressor stimulating miR-34a, and cisplatin, has shown enhanced toxicity against lung cancer cell line, KP cell. In vivo, systemic delivery of the nanoparticles indicated a preferential uptake in lung of the tumor-bearing mice. Efficacy studies indicated prolonged survival of mice from the combination treatment.
Conclusion
The combination RNA-chemotherapy in an LbL formulation provides an enhanced treatment efficacy against NSCLC, indicating promising potential in clinic.
Keywords: Combination, NSCLC, Layer-by-layer, Cisplatin, siRNA, microRNA
Introduction
Non-small cell lung cancer (NSCLC) accounts for 85% of lung cancer, a leading cause of cancer death worldwide.(1) Cisplatin and other platinum-based chemotherapeutics are front-line therapies for the treatment of NSCLC.(2,3) However, drug resistance and desensitizing, caused by complex genetic mutations of the cancer cells, limits the clinical efficacy of platinum-based chemotherapeutics against NSCLC, with a less than 20% 5-year survival rate for NSCLC patients and a 4% 5-year survival rate for patients with metastatic tumors.(4) The most common subtype of NSCLC, adenocarcinoma, is usually associated with 20–30% mutation of oncogenic Kirsten ras sarcoma viral oncogene homolog (KRAS) and ~50% loss of p53 function.(1) For these lung tumor types, and several other aggressive cancers (including colon cancer, leukemia, and pancreatic cancer), the KRAS mutation is essential for tumor formation and maintenance, thus rapid tumor regression is found with deletion of KRAS.(5–7) P53, a frequently mutated tumor suppressor gene, is involved in the cell cycle progression, proliferation, survival, and apoptosis.(8,9) Loss of p53 function can increase the function of P-glycoprotein, a membrane pump protein that causes resistance toward chemotherapeutic drugs.(10–13) More importantly, loss of P53 has been shown to accelerate KRAS driven tumorigenesis, indicating a synergistic effect between KRAS mutation and loss of p53 in promoting tumor development.(14,15) As a result, the simultaneous inhibition of the KRAS oncogene and restoration of the p53 suppressor function are appealing therapeutic strategies for lung adenocarcinoma. However, small molecule inhibitors and drugs to restore p53 function remain elusive.(16,17) Small molecule inhibitors to target the KRAS oncogene remain limited, while only a few reports have shown any potential of developing KRAS inhibitors due to the challenges of the KRAS binding pocket.(18,19) On the other hand, both of these genetic pathways can be targeted directly using RNA. It has quite recently been shown that siRNA to target the KRAS oncogene is an effective strategy to impede KRAS signaling and prevent tumor growth and progression.(20–23) Furthermore, microRNA (miRNA), small coding RNAs that regulate gene expression in the post-transcriptional stage(24), can be used to address p53 function; miR34a, one member of the miR-34 microRNA family, can activate multiple p53 downstream pathways which mediate cell proliferation, survival, and apoptosis, thus restoring antitumor effects.(24–29) MRX34, a liposomal miR-34, has been investigated in clinical trials phase I. Although it was placed on hold due to safety issue, it is still a principal proof that miR-34a could serve as a valuable anticancer drug once a promising delivery vehicle could be found to remediate the safety concern.(30) Therefore, the delivery of siKRAS and miR34a can be an effective treatment approach to mediate the genetic mutations of lung cancer cells and enhance the antitumor efficacy of cisplatin.
Despite the enormous therapeutic potential of siRNA and miRNA, systemic delivery of RNA to the target site still remains problematic when translating to clinic.(31) Some of the major issues include mononuclear phagocyte system clearance, achieving sufficient RNA loading capacity, nuclease degradation, toxicity, and establishing a prolonged blood circulation time to allow accumulation in the tumor.(32–34) Nanoscale non-viral delivery systems developed from cationic polymers, lipids and lipid-like systems, and peptides have been investigated extensively for small RNA delivery in pre-clinical studies.(32,34) Recently, co-delivery of siKRAS and miR-34a in vivo has been achieved using a lipid-based formulation, and lung tumor regression was observed in a “KRAS mutation & P53 deletion” (“KP”) adenocarcinoma model in which KRAS oncogene is mutated and p53 tumor suppressor is deleted; for this model, the chemotherapy drug was delivered separately via intravenous injection.(15) The development of a combination therapy that is truly delivered together in a synergistic fashion using nanoparticle technologies provides the potential of highly targeted therapies with lowered toxicity and a greater window of therapeutic potential; however, this mode of targeted multi-drug delivery is still missing in traditional systems such as cationic polymers or charged lipids that lack the modular design to incorporate multiple therapeutics.
Layer-by-Layer (LbL) nanoparticle is a promising drug delivery platform with great clinical translational potential.(35–43) Utilizing the process of depositing oppositely charged polyelectrolytes sequentially on a charged core, LbL nanoparticles possess hierarchical and multifunctional multilayered structure with great modularity and versatility. LbL nanoparticles have several desirable features, including precise control of size, combination therapeutics with high loading capacity, staged cargo release, enhanced stability in vivo, and tunable surfaces for modification.(35) Because of the modular nature of LbL nanoparticles, it is possible to incorporate therapeutics such as RNAs, inhibitors, or proteins in multilayers on a charged colloidal core substrate. Furthermore, the LbL platform can yield surface chemistry that enables targeting via response of the hypoxic tumor microenvironment and the presence of specific ligands that bind a number of known aggressive tumor cell types, from ovarian to lung cancer.(37) We have found that these LbL stealth coatings provide extended blood plasma half-life when applied to liposomal, quantum dot, gold and other nanoparticle systems.(35,39) In addition, the stealth layer provides an advantageous characteristic where it enables direct tumor targeting due to the outer layer design formed from hyaluronic acid (HA).(35–37) Recent work using the LbL platform has demonstrated a staged release of siRNA and a chemotherapeutic agent for treatment of triple negative breast cancer.(35) Furthermore, the LbL platform can also provide improved biocompatibility and reduced off-target toxicity of the loaded therapeutics.(36) The modularity, flexibility, and versatility of this platform make it an optimal candidate for preparing combination nano-therapeutics containing RNA-based drugs and DNA-damaging chemotherapeutics.
It is important to examine these systems in a more meaningful mouse model that better replicates advanced disease within the relevant tissue. Furthermore, we hope to show that more than one type of nucleic acid can be effectively delivered from these systems to address multiple types of gene dysregulation that are synergistic. In this study, we present a KRAS/P53 targeted LbL nanoparticle that contains a cisplatin loaded core to treat aggressive lung adenocarcinoma in vivo. Taking advantage of the modularity and versatility of LbL platform, we were able to build RNA films (siKRAS and miR-34a) with poly L-arginine (PLA) as the polycation, atop the cisplatin-containing liposomes, followed by coating of exterior layer with hyaluronic acid (HA), that possess both “stealth” and targeting properties. We demonstrated high loading capacity and controlled release of small RNA and cisplatin. In vitro studies showed efficient KRAS gene knockdown and enhanced tumor killing effect of cisplatin when combined with siKRAS, miR-34a, or siKRAS/miR-34a combination. We further demonstrated enhanced accumulation of LbL nanoparticles in the lungs of tumor-bearing mice in an orthotopic lung adenocarcinoma model utilizing tumor cells derived from genetically modified KRAS mutant, p53 deficient mice. It was found that mice treated with combination therapy demonstrated prolonged survival compared to mice treated with either cisplatin or RNA alone. Given the similarities between the lung adenocarcinoma model and human NSCLC, this study highlights the promising potential of incorporating LbL nanoparticles as a combination therapy platform to deliver RNA-based therapeutics that address common tumor mutations that enable tumor cell drug resistance and survival, in combination with chemotherapeutic agents.
Materials and Methods
Materials
All lipid components were purchased from Avanti Polar Lipids, except for cholesterol which was purchased from Sigma Aldrich. Cisplatin and other polyelectrolyte were purchased from Sigma Aldrich. CCK-8 cell proliferation assay kit was purchased from Sigma-Aldrich. All siRNAs and microRNA including siKRAS 5’-CUUAGAAAAAAGAAGGUUUCC-dTdT-3’, scrambled control 5’-GCCUAAUAAUAAGGAAUACGU-dTdT-3’, and miR-34a 5’-UGGCAUGUCUAGCUGGUUGU-3’ were customized from Dharmacon. DNA primers, including siKRAS (sense 5’-GACTGAATATAAACTTGGTAGTTGGACCT-3’ and antisense 5’-TCCTCTTGACCTGCTGTGTCG-3’), β-actin (sense 5’-TGAGCGCGGCTACAGCTT-3’, antisense 5’-TCCTTAATGTCACGCACGATTT-3’), are purchased from DNA Technologies. Monoclonal antibodies, including anti-CD44 and the IgG isotype control antibody were purchased from Santa Cruz Biotechnologies. Dulbecco’s modified eagle’s medium (DMEM), fetal bovine serum, penicillin/streptomycin, and RNase-free deionized water were purchased from Invitrogen. All polymer and buffer solutions were filtered with a 0.2 µm pore size polycarbonate syringe filter before use.
Preparation of LbL nanoparticles
The protocol of LbL nanoparticles preparation was developed based on previous established method.(35) Briefly, liposomes were first formulated at a mass ratio of 7:2:1 (DSPC:POPG:Cholesterol). These three compounds were dissolved in chloroform and a thin lipid film was generated by rotary evaporation. These films were then allowed to dry in desiccator overnight to completely remove chloroform. Cisplatin was suspended in deionized water and sonicated for 1 hr to allow complete cisplatin dissolution with concentration of 8 mg/mL. The lipid film was then hydrated with cisplatin solution at 65°C for 1 hr. Following the cisplatin loading in liposomes, these drug-loaded liposomes were purified using tangential flow filtration (TFF) to remove free cisplatin and were then re-suspended in PBS for storage. For LbL assembly, liposomes at 2 mg/mL were mixed with PLA (2mg/mL) in RNase-free water which was facilitated by a brief period of bath sonication (3s). The excessive PLA was purified by TFF. To incorporate RNAs, purified nanoparticles were mixed with RNAs (siKRAS/miR-34a, 1/1 molar ratio, 10 µM) in RNase-free water, followed by purification using TFF. Another PLA layer was deposited onto the RNA terminated nanoparticles via similar mixture method and TFF purification steps. Finally, the purified PLA-terminated LbL nanoparticles (2 mg/mL) were mixed with HA (1mg/mL, in dibasic sodium phosphate buffer, pH = 7.4, 10 mM) and washed using TFF. The obtained LbL nanoparticles were stored in PBS solution at 4°C. To prepare the Cy5.5-labelled nanoparticles, Cy5.5-siRNA (10% molar ratio) was incorporated into the total RNAs to serve as the RNA layer during the LbL process. The generated Cy5.5-labelled nanoparticles share similar physicochemical properties as the unlabeled nanoparticles.
Physicochemical characterization
All size, zeta potential, and polydispersity index were measured using a Malvern Zetasizer Nano ZS90 particle analyzer (λ = 633 nm, material/dispersant RI 1.590/1.330). The loading efficiency of RNA in the LbL nanoparticles were examined by measuring the free RNA in the washed waste using Picogreen Assay (Invitrogen) against a dsRNA standard curve, by absorbance of the washed waste at 260 nm using Nanodrop, or using a fluorescent dye labelled RNA and measuring the nanoparticle-associated fluorescence intensities against a fluorescent siRNA standard curve. The stability of the LbL nanoparticles were examined in PBS or in phenol-free DMEM at room temperature. For the cisplatin loading measurement, the cisplatin-containing LbL nanoparticles were diluted to 10000-fold with DI water and the concentration of platinum was measured using automatic flameless atomic absorption spectrophotometer (Model AA-6700, Shimadzu, Kyoto, Japan). Potassium dichloroplatinate was used as a standard. A standard curve with platinum concentrations in the range of 50–250 ng/mL was performed before analysis of each sample. The RNAs release from the LbL nanoparticles were measured at 37°C by quantifying the amount of RNA released in supernatant over different time points using picogreen assays. The release of cisplatin was quantified by measuring the remaining cisplatin in the float-a-lyzers (MWCO = 3500 Spectrum) at room temperature.
In Vitro Experiments
KP cells in this study were obtained from the lung tumors of a human autochthonous mouse model developed by Jacks and coworkers.(15) The cells were grown in DMEM media supplemented with 10% fetal bovine serum, 50 units/mL of penicillin and 50 units/mL of streptomycin. KP cells were stable in expressing tdTomato.
Gene silencing of LbL nanoparticles was examined in KRAS expressing KP cells. Briefly, the cells were seeded on a 96-well plate overnight with 30% confluence, and treated with increasing concentration of LbL nanoparticles, of which the amounts were normalized to the siRNA loading. The cells were then treated with siKRAS LbL nanoparticles, and scrambled control siRNA nanoparticles for comparison. Three or five days after the treatments, RNA was purified using TRIzol (Invitrogen) and reverse-transcribed using a High-Capacity cDNA Reverse Transcription Kit (Applied Biosystems). Real-time PCR (Q-PCR) reactions were carried out using Taqman probes (Invitrogen). The KRAS mRNA levels were normalized to Actin mRNA using scrambled siRNA as the control. To quantify the miRNA expression, 10 ng of total RNA was reverse-transcribed using miRNA-specific RT primer and measured by real-time PCR using miRNA-specific probes. The miRNA expression was normalized to U6 RNA.
Cytotoxicity assays were carried out using the CCK-8 cytotoxicity assay. Briefly, the cells were first plated in a 96-well plate with 30% confluence for 24 hrs and treated with the nanoparticles at various concentrations of cisplatin. After 3 days of incubation, a fresh serum-free OptiMEM media containing 10% v/v of the CCK-8 proliferation kit was used to replace the media. After 2 hrs incubation, the absorbance at 450 nm was measured by a plate reader. Cell viabilities were measured and normalized to an untreated control group. IC 50 values of cisplatin at various combinations were calculated from the viability curve using Prism 5.
In Vivo Experiments
All animal studies were approved by the Massachusetts Institute of Technology Animal Care and Use Committee. AIN-93 purified diet was purchased from PharmaSev/Testdiets. Cohorts of KP and KP;R26LSL-Luciferase/LSL-tdTomato mice were infected with 2.5 × 10−7 pfu of Adeno-Cre by intranasal inhalation as described previously.(15) The mice were monitored weekly using a GE Healthcare microCT imaging device (45-µm resolution, 80 kV, with 450-µA current). Before the targeting studies, mice were placed on AIN-93 special diet for a week to reduce body autofluorescence.
For the tumor initiation, KP cells (1 × 10−5) were injected to nude mice via tail vein injection. The targeting and treatment were typically initiated after two weeks post-injection.
For the tumor targeting studies, both healthy and tumor-bearing mice were treated with Cy5.5-labelled LbL nanoparticles (10% Cy5.5-labelled siRNA in total RNAs) via intravenous injections with a dose of 2 mg/kg of RNAs. The whole-body imaging of mice following nanoparticle injections were carried out using IVIS imager (Xenogen) with excitation at 650 nm and emission at 750 nm. The mice were sacrificed 48 hrs post injection. The vital organs were harvested and fluorescence was quantified using IVIS imaging.
Tumor-bearing mice were treated with four groups including vehicle control without any therapeutics, cisplatin only, RNA only, and cisplatin/RNA combination. Each group contains 8 mice. The dose of cisplatin was 12 mg/kg, while for the RNA only (with dosing at 2 mg/kg RNA only) and vehicle control groups, doses were equivalent to 12 mg/kg, given to the weight ratio of cisplatin to lipid. The mice were dosed repeatedly once a week, for a total of 4 weeks. At indicated time points, the up-chest area of the mice were scanned for 5 min with a microCT while mice were under isoflurane anesthesia and acquired images were then processed using GE MicroView software. The total tumor volumes in the lungs before and after treatment was calculated using GE MicroView software.(15,44) The weights of mice were monitored daily, with 20% body weight loss, mice were euthanized and lung tissues were recovered for analysis. The survival curve was calculated and illustrated as Kaplan-Meier curve using GraphPad software.
Q-PCR analyses of KRAS and miR-34a were performed by isolating RNA from the lung tumors using PARIS kit (Invitrogen) according to the manufacturer’s protocol. The iScript cDNA synthesis kit was used to synthesize the cDNA. Q-PCR was carried out using iQ SYBR Green Supermix (Bio-Rad) along with the selected DNA primers. The amplification was performed by incubation at 95°C for 10 min, followed by 40 cycles of 95°C for 15 s and 60°C for 1 min. The relative gene expression was normalized to either U6 RNA or β actin.
Western blots were performed using standard methods. After the treatments, tumored lung tissues were prepared by radioimmuno-precipitation assay buffer (150 mM sodium chloride, 50 mM Tris pH 8.0, 1% Triton X-100, 0.5% sodium deoxycholate, 0.1% SDS) supplemented with 10 mM NaF, 1 mM Na3VO4, 5 mM EDTA, 1 mM EGTA, 5 µg ml−1 leupeptin, 1 µg ml−1 pepstatin A, 1 mM phenylmethylsulfonyl fluoride, and protease and phosphatase inhibitor cocktail (Calbiochem). Lysates were centrifuged at 12,000 r.p.m. for 20 min at 4 °C. Protein concentration of the supernatants was determined using a BCA protein Assay Kit (Thermo Scientific). Equal amounts (30–50 µg) of the proteins were resolved by 8–12% SDS–polyacrylamide gel electrophoresis gels and transferred to nitrocellulose membranes (Millipore). Membranes were blocked in 5% bovine serum albumin for 1 h at room temperature, and then incubated with specific antibodies for different western blot analyses at 4 °C overnight. The bound primary antibodies were detected by secondary conjugates compatible with infrared detection at 700 and 800 nm, and membranes were scanned using the Odyssey Infrared Imaging System (Odyssey, LI-COR). The following antibodies were used: anti-KRAS antibody (ab84573, 1:2,000 dilution) was obtained from Abcam. Anti-β-actin antibody was purchased from Sigma (A5441, 1:10,000 dilution). Secondary antibodies were from LI-COR Biosciences, including IRDye 800CW Goat anti-Mouse IgG (926–32210, 1:10,000 dilution) and IRDye 800CW Goat anti-Rabbit IgG (926–32211, 1:10,000 dilution).
For immunohistochemistry (IHC) assays, mice were euthanized with CO2 asphyxiation and lungs were inflated with 4% formalin. The tissue samples are collected and processed after harvesting the fresh organs. Overnight fixation was performed. The lungs were then embedded in paraffin based on standard procedures. The lungs were then sectioned at 4 µm and stained with specific antibodies for detecting the biomarkers of interests. The following antibodies were used: anti-CC3 (1:200, Cell signaling), anti-pErk Thr202/Tyr204 (1:300, Cell Signaling), anti-SIRT (1:500, Cell Signaling), anti-CDK6 (1:300, Cell Signaling), and anti-Ki67 (1:100, Cell Signaling). The number of positive cells per tumor area was quantified.
Results and Discussion
Combination Nano-therapeutics Construction and Characterization
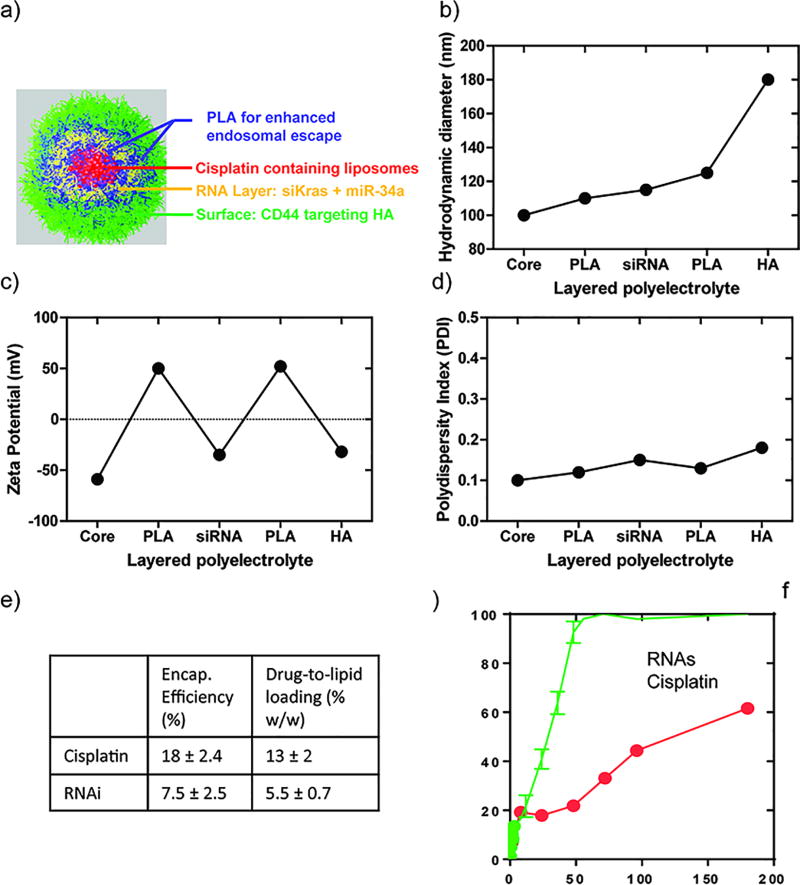
Multilayered LbL nanoparticles containing RNA therapeutics and cisplatin are illustrated in Figure 1a. To construct the LbL-based nano-therapeutics, cisplatin was first encapsulated in the hydrophilic core of negatively charged phospholipid liposomes. Positively charged poly-L-arginine, (PLA), the two negatively charged therapeutic RNAs combined together (siKRAS and miR-34a), and PLA were sequentially assembled on top of the liposome by sonication and tangential flow titration (TFF) to remove free polyelectrolyte.(45) Hyaluronic acid (HA, 40 kDa), a negatively charged natural polysaccharide, was deposited as the outermost layer due to its capability to extend blood circulation time and its ability to target CD44, an overexpressed receptor on lung adenocarcinoma cells.(46) Sequential build-up of an LbL film around the nanoparticle was confirmed at each step by dynamic light scattering (DLS) measurements that indicated a 10 nm growth in diameter following the deposition of PLA and the RNAs, and a 50 nm growth following deposition of the terminal HA layer (Figure 1b). This final charged HA layer is thought to be highly hydrated and loop-like in nature, thus yielding a thicker, but possibly higher water content outer layer when measured hydrodynamically. Further validation of the coating of each layer was provided by the electrophoretic measurements that indicated a complete charge reversal following each layer deposition (Figure 1c). The completed LbL nanoparticle, with a multilayered structure of liposomes/PLA/RNA/PLA/HA, possessed a zeta potential of approximately −30 mV and hydrodynamic diameter of ~180 nm (Figure 1b). The uniformity of LbL deposition was evidenced by the low polydispersity index (PDI) value (< 0.20) (Figure 1d). The RNA combination system was adsorbed onto the nanoparticle from a solution with a 1:1 molar ratio of siRNA to miRNA. To examine the total RNA loading in the LbL nanoparticle, waste solution generated from the TFF after each wash was quantified for free RNA and the resulting value was subtracted from the total RNA prior to LbL assembly (Figure S1). Approximately 3000 siRNA molecules per nanoparticle was layered by the PLA film, implying a conformal coating of RNAs on the nanoparticle with ~90% surface coverage (assuming 6 nm × 2 nm cross-sectional surface area per siRNA molecule).(47) A similar approach was applied to measure the quantity of loaded cisplatin in liposomes. Total RNA loading was determined at 5.5% of drug-to-lipid weight loading, comparable to our previously reported siRNA loading efficiency.(35) The weight loading of cisplatin in the LbL nanoparticle (13%) is comparable to cisplatin loading achieved in LipoplatinTM, a lipid-based cisplatin formulation in clinical trials.(48–51) Furthermore, release of both RNA molecules and cisplatin were measured over an extended period of time. It was found that the PLA/RNA film was stable at pH 7.4 in PBS, with a net release of less than 30% at 24 hrs (Figure 1f) and 37°C; furthermore, we observed a sustained release of cisplatin that was more delayed than the RNAi, with less than 50% release at 96 hrs. This controlled release kinetics of RNA allows an initial downregulation of the KRAS oncogene expression and restoration of p53 tumor suppression, both of which should lower the tumor’s drug resistance, while the release of cisplatin introduces tumor cell killing via DNA damage once the RNAs affected the tumor resistance pathways. The staged release profile shown in Figure 1f is therefore desirable for this dual therapeutic approach.
Figure 1.
Physicochemical characterization of liposome/PLA/RNA/PLA/HA LbL nanoparticles: (a) Schematic illustration, red denotes the core liposomes, encapsulating cisplatin, blue denotes the two PLA layers with yellow RNA layer in-between, green layer denotes the tumor targeting agent, HA, which is the outer-layer. (b) Hydrodynamic diameters of nanoparticles during LbL fabrication. Charge reversal in zeta potential (c) indicates the successful layer deposition. (d) Polydispersity index of LbL nanoparticles indicates the narrow dispersity in solution. (e) Encapsulation efficiency and weight loading of both cisplatin and RNAs. (f) Staged release of RNA and cisplatin in PBS (pH 7.4) at 37°C. The results represent mean ± Standard Deviation (S.D.) n = 3.
In Vitro Studies
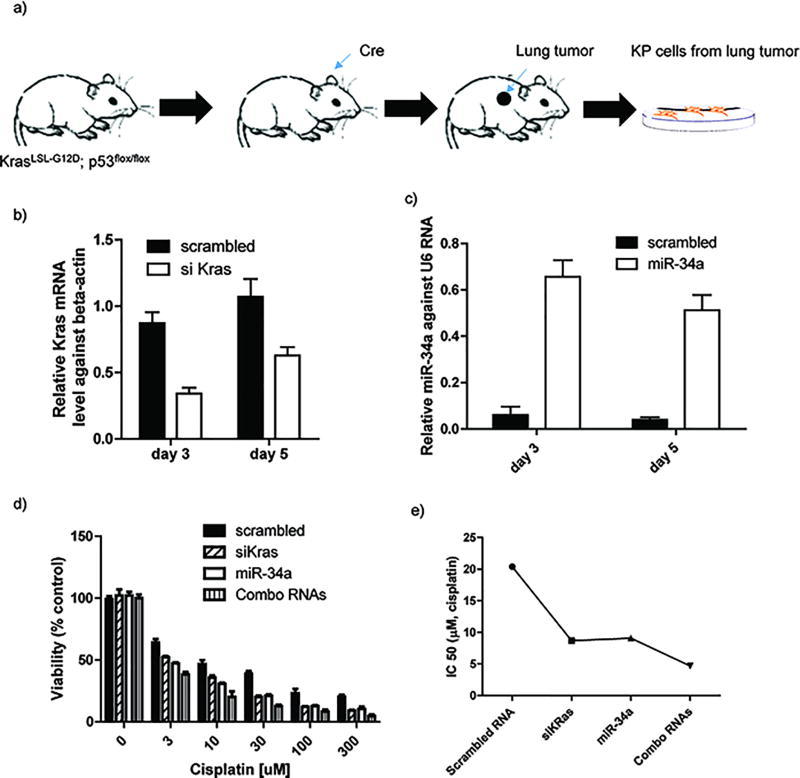
To investigate the efficacy of combination therapy in vitro using tissue culture experiments, lung adenocarcinoma cells were first obtained from an autochthonous murine lung cancer (KP) mouse model with an activatable KRAS mutation and p53 loss developed by the Jacks laboratory.(15) The KP mice were crossed with two strains carrying Lox-STOP-Lox reporter alleles, R26LSL-tdTomato and R26LSL-Lusiferace, to generate KRASLSL-G12D/wt; p53flox/flox;R26LSL-Lusiferase/LSL-tdTomato mice. In this model, mice are treated via intranasal inhalation with Adeno-Cre which causes deletion of p53 and activation of KRASG12D (Figure 2a). Ten weeks after the tumor initiation, aggressive tumors were isolated and KP cells cultured for in vitro assays. To confirm the loss of p53 and KRAS oncogene activation, we assessed the miR-34 and KRAS expression in both healthy and tumored lungs. It was found that miR-34 expression was significantly decreased in the tumored lungs, whereas KRAS expression is elevated in tumored lungs, compared with healthy lungs (Figure S2a, S2b, and S2d). Furthermore, it was confirmed that CD44 is overexpressed in the tumored lungs using immunostaining (Figure S2c), indicating that the HA outer layer should serve as a targeting moiety for this tumor cell type. We first examined whether the LbL nanoparticles can deliver siKRAS to lung adenocarcinoma cells and effectively knockdown KRAS. The KP cells were treated with LbL nanoparticles containing siKRAS, and gene knockdown was monitored at days 3 and 5. It was found that siKRAS was successfully delivered to cells; ~70% and 40% reduction of KRAS expression was observed at day 3 and 5, respectively (Figure 2b). We also demonstrated the successful delivery of miR-34a to KP cells, with miR-34a expression levels at ~60% and ~50% at day 3 and 5, respectively (Figure 2c). The extended period of transfection observed over multiple days is achieved due to the controlled siRNA release from the LbL nanoparticle inner layers, thus providing sustained oncogene suppression. To further investigate the effectiveness of the combination therapy, cell viability was monitored by varying the concentration of cisplatin, co-delivered with scrambled siRNA, siKRAS, miR-34a, or siKRAS/miR-34a combination. We observed enhanced cytotoxicity against KP cells using combinations with either siRKAS, miR-34a, or siKRAS/miR-34a combo, compared with combinations with scrambled RNA as a control after 3 days (Figure 2d). This finding is further supported by calculating the half maximum inhibitory concentrations (IC50) of cisplatin at various conditions. Combining cisplatin with either siKRAS or miR-34a in the LbL nanoparticle formulations significantly decreased the IC50 of cisplatin, and the combination of the two RNA molecules together with cisplatin yielded a 5-fold decrease compared to cisplatin with scrambled RNA (Figure 2e), confirming the enhanced efficacy of cisplatin in killing lung adenocarcinoma cells by suppressing the tumor cell survival pathways; the KRAS oncogene was knocked down by siKRAS and the p53 functional pathway was stimulated by miR-34a.
Figure 2.
In vitro Characterization of combination therapeutics against lung adenocarcinoma cells. (a) KP cells were derived from lung tumor, generated through inhalation of Cre. (b) KRAS mRNA expression in KP cells after 3 or 5 days treated with combination LbL nanoparticles. (c) miR-34a mRNA expression in KP cells after 3 or 5 days treated with combination LbL nanoparticles. (d) Examination of RNA enhanced cytotoxicities against KP cells at 72 hrs. (e) Calculated IC 50 value of cisplatin while combined with RNA therapeutics using GraphPad software. The results represent mean ± Standard Deviation (S.D.) n = 3.
Tumor targeting of LbL nanoparticles
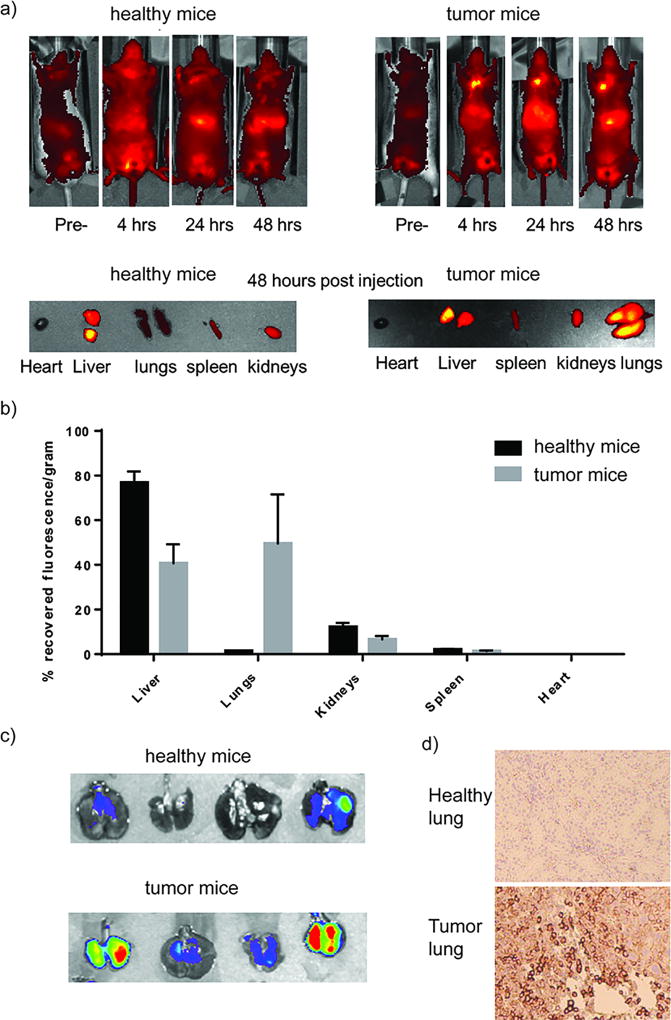
Prior to in vivo efficacy investigations, we first assessed the capability of LbL nanoparticles to actively target the lung adenocarcinoma using the orthotopic KP adenocarcinoma model. Compared to the autochthonous model, this orthotopic KP model maintains the same genetic mutations and requires only two weeks for tumor initiation, whereas ten weeks are required for the autochthonous model. Furthermore, by controlling the number of KP cells implanted, an optimal therapeutic window of one month survival time after tumor initiation for non-treated mice can be obtained to observe the therapeutic effect of combination therapies. After tumor initiation by intravenous injection of KP cells, multiple tumors, with volumes ranging from 1 mm3 to 20 mm3, were formed in the lung area (Figure S3). Both healthy and tumor-bearing mice were intravenously injected with Cy5.5-labelled LbL nanoparticles which are generated by embedding Cy5.5-labelled siRNA (10% molar ratio) within the RNA layer (Figure S4), and whole-body imaging was completed at certain time intervals (4 hrs, 24 hrs, and 48 hrs). As shown in the whole-body images of Figure 3a, taken at the 48 hour time point, nanoparticles mainly accumulated in the livers of healthy mice, as is typically observed for nanoparticle systems due to filtration through the liver, but there was no significant accumulation in the lung; whereas the nanoparticles in tumor-bearing mice were detected by fluorescence in both the lung and liver with comparable intensities. It was also observed that the nanoparticles accumulated in the kidneys of both healthy and tumor-bearing mice, as can be seen from the excised organ images (Figure 3a). The quantitative analysis of the recovered fluorescent intensities for each organ is also provided (Figure 3b). The HA terminated LbL nanoparticles provided clear evidence of selective targeting in this orthotopic model, with the accumulation in the lungs of tumor-bearing mice approximately 22-fold greater than in the lungs of healthy mice (Figure 3b). Approximately 45% of the initial dose (based on net recovered fluorescence) was co-localized in the lungs of tumor-bearing mice, whereas only 2% of the initial dose was detected in the lungs of healthy mice (Figure 3b). The enhanced accumulation in the lungs of tumor-bearing mice was accompanied by a reduction of nanoparticles in the liver by 2-fold. Compared to healthy mice, in which 80% of recovered fluorescence is detected in the liver, approximately 40% of recovered fluorescence was observed in the liver of tumor-bearing mice (Figure 3b). Given that these LbL nanoparticles are terminated with an HA layer, the endogenous ligand for CD44 receptor, the enhanced lung accumulation in tumor-bearing mice is attributed to active targeting of HA to the CD44 receptor on KP cells. To validate this, we first demonstrated the existence of KP cells in the lungs of tumor-bearing mice by tracking the fluorescence of tdTomato, a red fluorescent protein inserted in the KP cells (Figure 3c). Additionally, CD44 overexpression in the lungs of tumor-bearing mice was validated by immunohistochemistry, whereas negligible CD44 expression is observed in the lungs of healthy mice (Figure 3d). It is also noted that these HA-terminated LbL nanoparticles can undergo an enhanced cellular uptake via response to hypoxic tumor microenvironment.(37) Furthermore, this enhanced accumulation of LbL nanoparticles in tumor-bearing mice can also be attributed to passive targeting due to defective tumor vasculature, and an extended blood circulation time.(35) In summary, we achieved significantly enhanced lung tumor targeting using the modular LbL platform while existing RNA combination therapy for NSCLC using lipids do not show this enhanced lung tumor targeting properties as typical lipids lack the modularity, responsive behavior and native ligand binding achieved with the outer LbL bilayer for multi-modal targeting.(15)
Figure 3.
Targeting of HA-terminated LbL nanoparticles to orthotopic lung cancer model using tumor cells derived from KP autochthonous mouse model. (a) Both healthy and tumored mice were treated with Cy-5.5 labelled LbL nanoparticles and whole-body fluorescence imaging were taken at 4 hrs, 24 hrs, & 48 hrs and tissues were harvested and imaged at 48 hrs. (b) Quantified recovered fluorescence intensities of different organs after harvest at 48 hours. The results represent mean ± Standard Deviation (S.D.) n = 4. (c) Tomato fluorescence of the KP tumor cells in harvested lungs and (d) CD44 immunohistochemistry staining confirmed tumor formation and tumor cell overexpression of CD44 in the lungs of the orthotopic model.
In vivo treatment efficacy
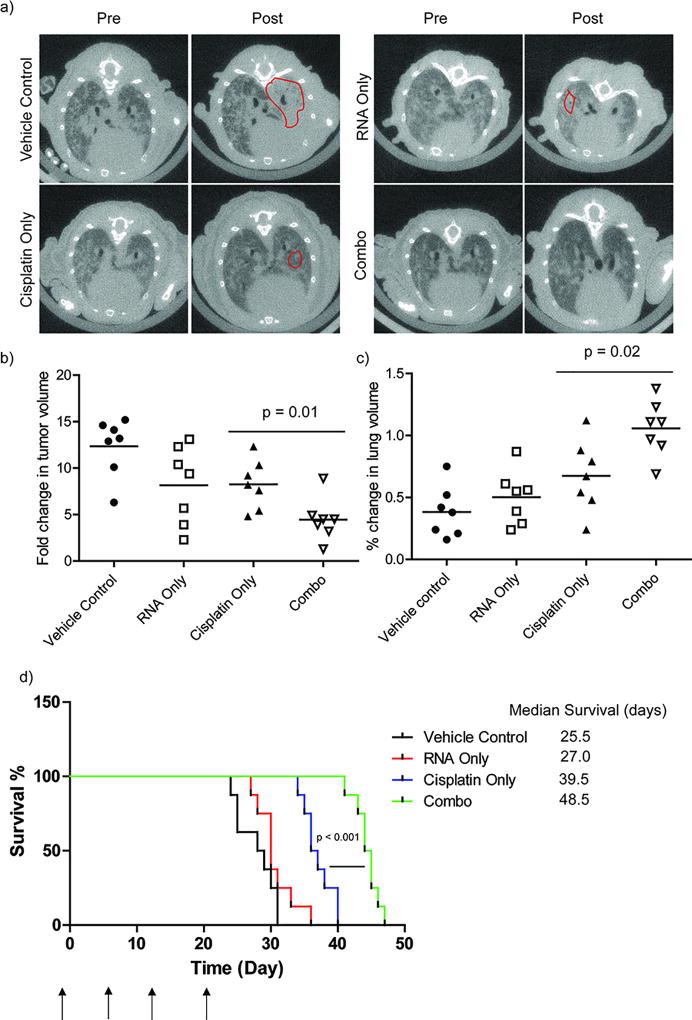
Upon successful targeting to lung adenocarcinoma using LbL nanoparticles, we further investigated treatment efficacy in the KP lung adenocarcinoma orthotopic mouse model. We anticipated that the treatment efficacy from the combination therapy would be enhanced due to the knockdown of KRAS and restoration of p53 regulated downstream pathways. For this proof-of-concept study, we included LbL nanoparticles that were loaded with either RNA therapeutics alone, cisplatin alone, or empty LbL nanoparticles as the control to demonstrate the effect of RNA and cisplatin combination therapies. The established lung adenocarcinoma mice were treated every week for 4 weeks with a total of 4 tail vein injections at 12 mg/kg of cisplatin. This specific dose is chosen based on the balance between toxicity and efficacy: dosing of 16 mg/kg shows severe toxicities, with major fatalities observed during the treatment phase (Figure S5a); dosing of 8 mg/kg shows no treatment efficacy in all groups (Figure S5b). This finding is supported by additional histology analysis, in which swollen tubules, an indication of severe kidney damage, were observed in mice treated with cisplatin dosing of 16 mg/kg (Figure S6). Additionally, serum chemistry analysis showed elevated creatine and blood urea nitrogen (BUN) levels, the indicators of kidney malfunction, in both free cisplatin (8 mg/kg) and nanoparticles with cisplatin dosing of 16 mg/kg treated groups (Figure S6). It is also noteworthy that free cisplatin (8 mg/kg) showed elevated toxicities compared to cisplatin in LbL nanoparticles (8 mg/kg), indicating that the LbL platform significantly lowers the nephrotoxicity of this chemotherapeutic (Figure S7). The lung areas were monitored through computed tomography (µCT) over the course of treatment and illustrated in the 2D axial images taken pre- and post- treatment (Figure 4a). As expected, the empty vehicle control group showed unregulated tumor growth, demonstrated by the increased lighter shaded areas which are populated with tumor cells (red circles, Figure 4a). Compared to the empty vehicle control group, both of the cisplatin and RNA only treated group showed a smaller increase in the light shaded tumor area (red circles, Figure 4a). The cisplatin/RNA combination therapy exhibited a better controlled tumor growth with no apparent increase of the lighter shaded tumor area (Figure 4a). This finding demonstrated an enhanced treatment response of combination LbL nanoparticles. It was supported by quantitative analysis of the tumor volume, indicating that combination nanoparticle therapy significantly controlled tumor growth compared to the single therapeutic treated groups or empty nanoparticles (p = 0.01, Figure 4b). We further calculated the volumes of healthy lung tissue and showed that tumors in the vehicle control group continued to grow, and displaced the healthy lung tissue, while combination therapy showed the smallest decrease in healthy lung volume, compared with either RNA or cisplatin only treated groups (Figure 4c). More importantly, the combination LbL nanoparticle therapy prolonged mouse survival significantly (p < 0.001) compared to the singular therapeutic groups, with median survival of 23.5 days versus 15.5 days and 9.0 days for cisplatin and RNA, respectively. Taken together, we not only achieved prolonged survival with the combination LbL nanoparticles but also remediated nephrotoxicity that typically arises from cisplatin treatment.
Figure 4.
Combination therapy in LbL nanoparticle promote the treatment efficacy in vivo. (a) Representative axial images of mouse lungs harboring KP tumors. The darker areas represents healthy lung whereas the lighter shades indicate areas populated by tumor cells. (b) Quantification of tumor volumes. (c) Quantification of healthy lung volumes using GE eXplore software. One or two independent lung tumors from each mouse were quantified. (d) A Kaplan-Meier curve comparing survival of mice treated with different groups. The median survival days were calculated from the Kaplan-Meier curve using GraphPad Prism software. N (number of mice in each group) = 7
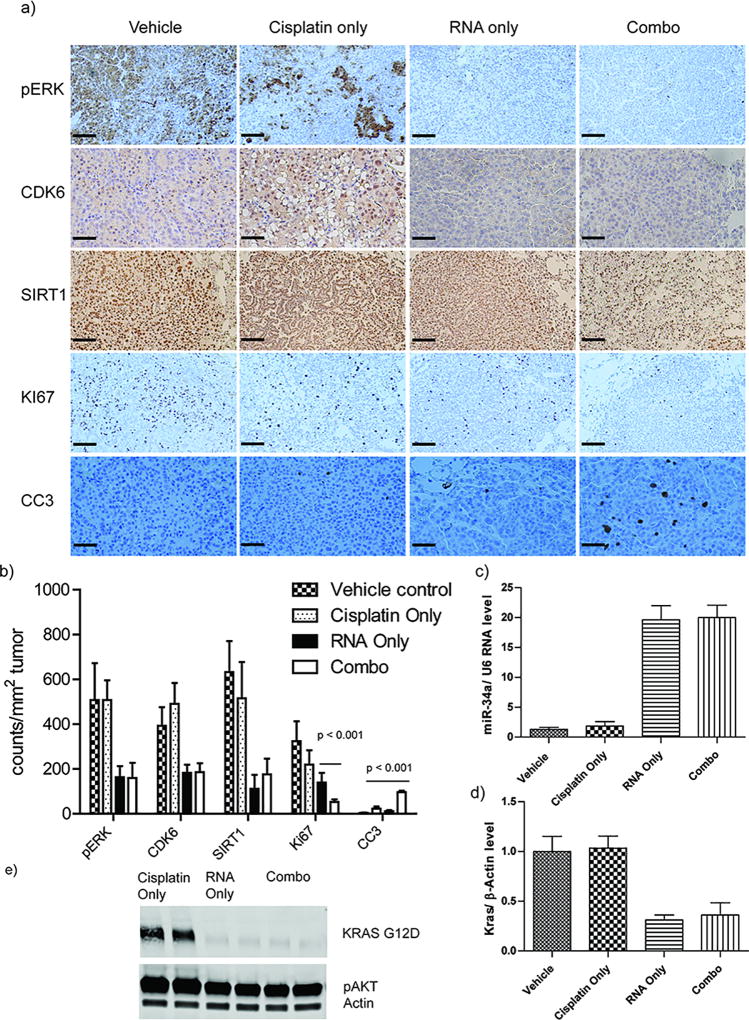
In order to examine the mechanism of combination therapy, we further excised all the lung tissues after treatment, and performed immunohistochemistry analysis to screen the biomarkers that are responsible for cell apoptosis (CC3), cell proliferation (Ki67), the molecular signaling downstream of the KRAS pathway (i.e. phospho-ERK) and miR34a pathways (i.e. CDK6 and SIRT1). The expression of pERK, CDK6, and SIRT1 were effectively suppressed in the treated mice of either the RNA-only or combination therapy, indicating the effective delivery and transfection of RNA therapeutics to the tumors. The targeted pathways were sufficiently impacted by the RNA therapeutics in these tumors. However, in the mice treated with vehicle or cisplatin only, we did not observe similar decreases in the expression of these biomarkers. We further evaluated the therapeutic effects on tumor cell proliferation using Ki67 staining. Compared to other treatment groups, the combination therapy effectively inhibited tumor cell proliferation (Figure 5b). Furthermore, we stained the cleaved products of caspase 3 (CC3), a cell apoptosis biomarker, to evaluate the therapeutic effects on tumor cells. We observed more cleaved caspase 3 products in the combination therapy treated mice, indicating greater cell death, while other treatment groups gave negligible levels of the cell apoptosis biomarker (Figure 5b). To confirm these effects were due to the successful delivery of RNA therapeutics rather than off-target effects, we tested the gene silencing against KRAS and the miR-34a content in isolated tumors. We observed higher content of miR-34a in RNA-only and combination therapy treated groups, while cisplatin and vehicle treated groups produced significantly less miR-34a (Figure 5c). For KRAS expression, it was found that both RNA and combination therapy treated groups had significantly lower gene expression relative to treated groups without RNA therapeutics (Figure 5d). Western-blot analysis was also performed in excised tumors, and confirmed the downregulation of the KRAS primary protein in RNA and combination treated groups (Figure 5e). Therefore, both immunohistochemistry analysis, q-PCR, and Western-blot results indicated successful delivery of payloads that correlate with the outcomes from the microCT evaluations. Collectively, cisplatin/RNAi combination therapy, co-packaged and delivered from a singular LbL nanoparticle formulation, appeared to provide the most effective therapeutic effect, most likely due to the enhanced effects of the targeted RNA therapeutics. More importantly, based on the similarities between this animal model and human NSCLC, these results suggest this LbL platform holds great potential for clinical translation. It is noteworthy that we are the first to report the packaging of multiple genes and chemotherapeutics in a singular platform to treat NSCLC in a highly physiologically relevant animal model.
Figure 5.
Immunohistochemistry analysis against a panel of biomarkers that regulate in cell proliferation, apoptosis, KRAS signaling pathway, and miR-34a pathways were tested. a) Representative images of IHCs of various biomarkers. (b) Quantification of biomarker expression was performed by counting positive stained cells at randomly picked area of 1 mm2 and average number and standard deviation were obtained by triplicate. (c) Q-PCR analysis of miR-34a expression level in isolated lung tumors. (d) Q-PCR analysis of KRAS expression in isolated lung tumors. The results represent mean ± Standard Deviation (S.D.). n (number of tumors in each group) = 10 (d) Western-blot results of KRAS primary protein in isolated lung tumors.
Conclusion
We have developed a modular LbL nanoparticle platform incorporating oncogene siRNA, tumor suppressor stimulating miRNA, and chemotherapeutic in a singular formulation to target NSCLC. These therapeutics were released in a staggered fashion to enable synergistic timing of gene silencing and chemotherapy treatment. Given the intervention of tumorigenic gene mutations achieved in this case, the efficacy of cisplatin to kill tumor cells was enhanced in vitro. In a physiologically relevant orthotopic model of lung adenocarcinoma, enhanced accumulation of nanoparticles in tumor-bearing lungs was achieved via both passive and active targeting of HA to the CD44 receptor. We further demonstrated successful delivery of the combination LbL nanoparticles to the lungs of tumored mice in this model. Enhanced antitumor efficacy and prolonged survival rate were observed in the mice treated with combination therapy, compared with any RNA or cisplatin treatment alone. Molecular evaluations confirmed the successful regulation of oncogene KRAS and restoration of p53 function by delivery of siKRAS and miR-34a, respectively. Therefore, the tumor defense pathways were blocked and the efficacy of DNA damage chemotherapeutics was facilitated, while maintaining a greatly lowered liver toxicity. Because a large number of tumors, including lung adenocarcinoma, carry mutations of KRAS and loss of p53 function, this combination approach has direct translational potential, and promise that can lead toward clinical trials. Due to the modularity of the LbL nanoparticle, a broad range of therapeutics, including inhibitors and nucleic acid, can be incorporated to target a variety of oncogene pathways, thus presenting a versatile platform for personalized medicine. Moreover, given the tunable features of the outer surface layer, this LbL RNA nanoparticle approach can be tailored to target different organs of interest, including primary or metastatic tumor sites.
Statistical Analysis
Experiments were performed in triplicates, or otherwise indicated. Data were analyzed using descriptive statistics, single-factor analysis of variance (ANOVA), and presented as mean values ± standard deviation (SD) from 3 to 10 independent measurements. Statistical comparisons between different treatments were assessed by two-tailed t tests or one-way ANOVA.
Supplementary Material
Statement of Translational Relevance.
Adenocarcinoma, the most common form of NSCLC, is associated with a mutation of the KRAS and loss of p53 function. Together these genetic mutations open pathways toward resistance of tumor cells to the therapeutic response of cisplatin, one of the main clinical chemotherapeutic drugs for NSCLC. A promising approach to overcome this limitation is the design of a combination therapy that is comprised of a chemotherapeutic drug, cisplatin, and RNA-based therapeutics that specifically target both the KRAS mutation and loss of p53 function. We designed a nano-therapeutic that uses the electrostatic layer-by-layer (LbL) approach to effectively package both RNA therapeutics and cisplatin simultaneously in a manner that enables optimal timing of each. We observed enhanced treatment efficacy using the LbL approach indicating a promising potential in the clinic.
Acknowledgments
The authors acknowledge the David H. Koch Institute for Integrative Cancer Research at MIT for providing facilities to support this work, as well as DCM (Department of Comparative Medicine, MIT) and the Koch Institute Swanson Biotechnology Center for assistance with animal experiments especially the ATWAI, microscopy, and Tang Histology facility. We also thank Abigail Powell for assistance with tail vain injections, Dr. Rod Bronson for assistance with pathological analysis, Dr. Jeffery Wycoff for assisting with microscopy.
Financial Support. This work was supported by Janssen-MIT Transcend Grant, Ortho-McNeil Janssen Pharmaceuticals, Inc (PTH, ZJD, SR) DoD Congressionally Directed Medical Research Program (CDMRP) Ovarian Cancer Research Program (PTH, W81XWH-13-1-0151), the Misrock Foundation (LG), Koch Institute Support (core) Grant P30-CA14051 from the NCI, the MIT MRSEC Grant DMR-0819762 from the NSF, the National Research Foundation (NRF-NRFF2011-01)
Footnotes
Conflict of Interest: The authors declare no competing financial interest.
Authors' Contributions
Conception, design, and experimentation: L. Gu, J. Deng, P. Hammond
Acquisition of data (provided animals, acquired and managed patients, provided facilities, etc.): L. Gu, J. Deng, S. Roy, P. Hammond
Analysis and interpretation of data (e.g., statistical analysis, biostatistics, computational analysis): L. Gu, J. Deng, S. Roy, P. Hammond
Writing, review, and/or revision of the manuscript: L. Gu, J. Deng, S. Roy, P. Hammond
References
- 1.Herbst RS, Heymach JV, Lippman SM. Lung cancer. The New England journal of medicine. 2008;359(13):1367–80. doi: 10.1056/NEJMra0802714. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Butts CA, Ding K, Seymour L, Twumasi-Ankrah P, Graham B, Gandara D, et al. Randomized phase III trial of vinorelbine plus cisplatin compared with observation in completely resected stage IB and II non-small-cell lung cancer: updated survival analysis of JBR-10. Journal of clinical oncology : official journal of the American Society of Clinical Oncology. 2010;28(1):29–34. doi: 10.1200/JCO.2009.24.0333. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Goss GD, Arnold A, Shepherd FA, Dediu M, Ciuleanu TE, Fenton D, et al. Randomized, double-blind trial of carboplatin and paclitaxel with either daily oral cediranib or placebo in advanced non-small-cell lung cancer: NCIC clinical trials group BR24 study. Journal of clinical oncology : official journal of the American Society of Clinical Oncology. 2010;28(1):49–55. doi: 10.1200/JCO.2009.22.9427. [DOI] [PubMed] [Google Scholar]
- 4.Finlay GA, Joseph B, Rodrigues CR, Griffith J, White AC. Advanced presentation of lung cancer in Asian immigrants: a case-control study. Chest. 2002;122(6):1938–43. doi: 10.1378/chest.122.6.1938. [DOI] [PubMed] [Google Scholar]
- 5.Chin L, Tam A, Pomerantz J, Wong M, Holash J, Bardeesy N, et al. Essential role for oncogenic Ras in tumour maintenance. Nature. 1999;400(6743):468–72. doi: 10.1038/22788. [DOI] [PubMed] [Google Scholar]
- 6.Fisher GH, Wellen SL, Klimstra D, Lenczowski JM, Tichelaar JW, Lizak MJ, et al. Induction and apoptotic regression of lung adenocarcinomas by regulation of a K-Ras transgene in the presence and absence of tumor suppressor genes. Genes & development. 2001;15(24):3249–62. doi: 10.1101/gad.947701. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Jechlinger M, Podsypanina K, Varmus H. Regulation of transgenes in three-dimensional cultures of primary mouse mammary cells demonstrates oncogene dependence and identifies cells that survive deinduction. Genes & development. 2009;23(14):1677–88. doi: 10.1101/gad.1801809. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Feldser DM, Kostova KK, Winslow MM, Taylor SE, Cashman C, Whittaker CA, et al. Stage-specific sensitivity to p53 restoration during lung cancer progression. Nature. 2010;468(7323):572–5. doi: 10.1038/nature09535. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Levine AJ, Oren M. The first 30 years of p53: growing ever more complex. Nature reviews Cancer. 2009;9(10):749–58. doi: 10.1038/nrc2723. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.He S, Liu F, Xie Z, Zu X, Xu W, Jiang Y. P-Glycoprotein/MDR1 regulates pokemon gene transcription through p53 expression in human breast cancer cells. International journal of molecular sciences. 2010;11(9):3309–051. doi: 10.3390/ijms11093039. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Linn SC, Honkoop AH, Hoekman K, van der Valk P, Pinedo HM, Giaccone G. p53 and P-glycoprotein are often co-expressed and are associated with poor prognosis in breast cancer. British journal of cancer. 1996;74(1):63–8. doi: 10.1038/bjc.1996.316. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Miyatake K, Gemba K, Ueoka H, Nishii K, Kiura K, Tabata M, et al. Prognostic significance of mutant p53 protein, P-glycoprotein and glutathione S-transferase-pi in patients with unresectable non-small cell lung cancer. Anticancer research. 2003;23(3C):2829–36. [PubMed] [Google Scholar]
- 13.Schneider J, Rubio MP, Barbazan MJ, Rodriguez-Escudero FJ, Seizinger BR, Castresana JS. P-glycoprotein, HER-2/neu, and mutant p53 expression in human gynecologic tumors. Journal of the National Cancer Institute. 1994;86(11):850–5. doi: 10.1093/jnci/86.11.850. [DOI] [PubMed] [Google Scholar]
- 14.Kasinski AL, Slack FJ. miRNA-34 prevents cancer initiation and progression in a therapeutically resistant K-ras and p53-induced mouse model of lung adenocarcinoma. Cancer research. 2012;72(21):5576–87. doi: 10.1158/0008-5472.CAN-12-2001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Xue W, Dahlman JE, Tammela T, Khan OF, Sood S, Dave A, et al. Small RNA combination therapy for lung cancer. Proceedings of the National Academy of Sciences of the United States of America. 2014;111(34):E3553–61. doi: 10.1073/pnas.1412686111. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Heist RS, Engelman JA. SnapShot: non-small cell lung cancer. Cancer cell. 2012;21(3):448, e2. doi: 10.1016/j.ccr.2012.03.007. [DOI] [PubMed] [Google Scholar]
- 17.Pylayeva-Gupta Y, Grabocka E, Bar-Sagi D. RAS oncogenes: weaving a tumorigenic web. Nature reviews Cancer. 2011;11(11):761–74. doi: 10.1038/nrc3106. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Lito P, Solomon M, Li LS, Hansen R, Rosen N. Allele-specific inhibitors inactivate mutant KRAS G12C by a trapping mechanism. Science. 2016;351(6273):604–8. doi: 10.1126/science.aad6204. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Ostrem JM, Peters U, Sos ML, Wells JA, Shokat KM. K-Ras(G12C) inhibitors allosterically control GTP affinity and effector interactions. Nature. 2013;503(7477):548–51. doi: 10.1038/nature12796. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Baumer S, Baumer N, Appel N, Terheyden L, Fremerey J, Schelhaas S, et al. Antibody-mediated delivery of anti-KRAS-siRNA in vivo overcomes therapy resistance in colon cancer. Clinical cancer research : an official journal of the American Association for Cancer Research. 2015;21(6):1383–94. doi: 10.1158/1078-0432.CCR-13-2017. [DOI] [PubMed] [Google Scholar]
- 21.Lakshmikuttyamma A, Sun Y, Lu B, Undieh AS, Shoyele SA. Stable and efficient transfection of siRNA for mutated KRAS silencing using novel hybrid nanoparticles. Molecular pharmaceutics. 2014;11(12):4415–24. doi: 10.1021/mp500525p. [DOI] [PubMed] [Google Scholar]
- 22.Yuan TL, Fellmann C, Lee CS, Ritchie CD, Thapar V, Lee LC, et al. Development of siRNA payloads to target KRAS-mutant cancer. Cancer discovery. 2014;4(10):1182–97. doi: 10.1158/2159-8290.CD-13-0900. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Zeng L, Li J, Wang Y, Qian C, Chen Y, Zhang Q, et al. Combination of siRNA-directed Kras oncogene silencing and arsenic-induced apoptosis using a nanomedicine strategy for the effective treatment of pancreatic cancer. Nanomedicine : nanotechnology, biology, and medicine. 2014;10(2):463–72. doi: 10.1016/j.nano.2013.08.007. [DOI] [PubMed] [Google Scholar]
- 24.He L, He X, Lowe SW, Hannon GJ. microRNAs join the p53 network--another piece in the tumour-suppression puzzle. Nature reviews Cancer. 2007;7(11):819–22. doi: 10.1038/nrc2232. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Junttila MR, Karnezis AN, Garcia D, Madriles F, Kortlever RM, Rostker F, et al. Selective activation of p53-mediated tumour suppression in high-grade tumours. Nature. 2010;468(7323):567–71. doi: 10.1038/nature09526. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Hermeking H. The miR-34 family in cancer and apoptosis. Cell death and differentiation. 2010;17(2):193–9. doi: 10.1038/cdd.2009.56. [DOI] [PubMed] [Google Scholar]
- 27.Donzelli S, Strano S, Blandino G. microRNAs: short non-coding bullets of gain of function mutant p53 proteins. Oncoscience. 2014;1(6):427–33. doi: 10.18632/oncoscience.52. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Feng Z, Zhang C, Wu R, Hu W. Tumor suppressor p53 meets microRNAs. Journal of molecular cell biology. 2011;3(1):44–50. doi: 10.1093/jmcb/mjq040. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Hermeking H. MicroRNAs in the p53 network: micromanagement of tumour suppression. Nature reviews Cancer. 2012;12(9):613–26. doi: 10.1038/nrc3318. [DOI] [PubMed] [Google Scholar]
- 30.Agostini M, Knight RA. miR-34: from bench to bedside. Oncotarget. 2014;5(4):872–81. doi: 10.18632/oncotarget.1825. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Brower V. RNA interference advances to early-stage clinical trials. Journal of the National Cancer Institute. 2010;102(19):1459–61. doi: 10.1093/jnci/djq405. [DOI] [PubMed] [Google Scholar]
- 32.Leng Q, Woodle MC, Lu PY, Mixson AJ. Advances in Systemic siRNA Delivery. Drugs of the future. 2009;34(9):721. doi: 10.1358/dof.2009.034.09.1413267. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Pan X, Thompson R, Meng X, Wu D, Xu L. Tumor-targeted RNA-interference: functional non-viral nanovectors. American journal of cancer research. 2011;1(1):25–42. [PMC free article] [PubMed] [Google Scholar]
- 34.Zhang Y, Wang Z, Gemeinhart RA. Progress in microRNA delivery. Journal of controlled release : official journal of the Controlled Release Society. 2013;172(3):962–74. doi: 10.1016/j.jconrel.2013.09.015. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Deng ZJ, Morton SW, Ben-Akiva E, Dreaden EC, Shopsowitz KE, Hammond PT. Layer-by-layer nanoparticles for systemic codelivery of an anticancer drug and siRNA for potential triple-negative breast cancer treatment. ACS nano. 2013;7(11):9571–84. doi: 10.1021/nn4047925. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Dreaden EC, Kong YW, Morton SW, Correa S, Choi KY, Shopsowitz KE, et al. Tumor-Targeted Synergistic Blockade of MAPK and PI3K from a Layer-by-Layer Nanoparticle. Clinical cancer research : an official journal of the American Association for Cancer Research. 2015;21(19):4410–9. doi: 10.1158/1078-0432.CCR-15-0013. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Dreaden EC, Morton SW, Shopsowitz KE, Choi JH, Deng ZJ, Cho NJ, et al. Bimodal tumor-targeting from microenvironment responsive hyaluronan layer-by-layer (LbL) nanoparticles. ACS nano. 2014;8(8):8374–82. doi: 10.1021/nn502861t. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Morton SW, Shah NJ, Quadir MA, Deng ZJ, Poon Z, Hammond PT. Osteotropic therapy via targeted layer-by-layer nanoparticles. Advanced healthcare materials. 2014;3(6):867–75. doi: 10.1002/adhm.201300465. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Poon Z, Chang D, Zhao X, Hammond PT. Layer-by-layer nanoparticles with a pH-sheddable layer for in vivo targeting of tumor hypoxia. ACS nano. 2011;5(6):4284–92. doi: 10.1021/nn200876f. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Roh YH, Lee JB, Shopsowitz KE, Dreaden EC, Morton SW, Poon Z, et al. Layer-by-layer assembled antisense DNA microsponge particles for efficient delivery of cancer therapeutics. ACS nano. 2014;8(10):9767–80. doi: 10.1021/nn502596b. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Elbakry A, Wurster EC, Zaky A, Liebl R, Schindler E, Bauer-Kreisel P, et al. Layer-by-layer coated gold nanoparticles: size-dependent delivery of DNA into cells. Small. 2012;8(24):3847–56. doi: 10.1002/smll.201201112. [DOI] [PubMed] [Google Scholar]
- 42.Elbakry A, Zaky A, Liebl R, Rachel R, Goepferich A, Breunig M. Layer-by-layer assembled gold nanoparticles for siRNA delivery. Nano letters. 2009;9(5):2059–64. doi: 10.1021/nl9003865. [DOI] [PubMed] [Google Scholar]
- 43.Wurster EC, Elbakry A, Gopferich A, Breunig M. Layer-by-layer assembled gold nanoparticles for the delivery of nucleic acids. Methods in molecular biology. 2013;948:171–82. doi: 10.1007/978-1-62703-140-0_12. [DOI] [PubMed] [Google Scholar]
- 44.http://microview.sourceforge.net/MicroView-Reference-Guide.html.
- 45.Santiago Correa KYC, Dreaden Erik C, Renggli Kasper, Shi Aria, Gu Li, Shopsowitz Kevin E, Quadir Mohiuddin A, Ben-Akiva Elana, Hammond Paula T. Highly Scalable, Closed-Loop Synthesis of Drug-Loaded, Layer-by-Layer Nanoparticles. Advanced Functional Materials. 2015 doi: 10.1002/adfm.201504385. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Dreaden EC, Kong YW, Morton SW, Correa S, Choi KY, Shopsowitz KE, et al. Tumor-Targeted Synergistic Blockade of MAPK and PI3K from a Layer-by-Layer Nanoparticle. Clin Cancer Res. 2015;21(19):4410–9. doi: 10.1158/1078-0432.ccr-15-0013. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.Whitehead KA, Langer R, Anderson DG. Knocking down barriers: advances in siRNA delivery. Nature reviews Drug discovery. 2009;8(2):129–38. doi: 10.1038/nrd2742. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48.Farhat FS, Temraz S, Kattan J, Ibrahim K, Bitar N, Haddad N, et al. A phase II study of lipoplatin (liposomal cisplatin)/vinorelbine combination in HER-2/neu-negative metastatic breast cancer. Clinical breast cancer. 2011;11(6):384–9. doi: 10.1016/j.clbc.2011.08.005. [DOI] [PubMed] [Google Scholar]
- 49.Mylonakis N, Athanasiou A, Ziras N, Angel J, Rapti A, Lampaki S, et al. Phase II study of liposomal cisplatin (Lipoplatin) plus gemcitabine versus cisplatin plus gemcitabine as first line treatment in inoperable (stage IIIB/IV) non-small cell lung cancer. Lung cancer. 2010;68(2):240–7. doi: 10.1016/j.lungcan.2009.06.017. [DOI] [PubMed] [Google Scholar]
- 50.Ravaioli A, Papi M, Pasquini E, Marangolo M, Rudnas B, Fantini M, et al. Lipoplatin monotherapy: A phase II trial of second-line treatment of metastatic non-small-cell lung cancer. Journal of chemotherapy. 2009;21(1):86–90. doi: 10.1179/joc.2009.21.1.86. [DOI] [PubMed] [Google Scholar]
- 51.Stathopoulos GP, Boulikas T. Lipoplatin formulation review article. Journal of drug delivery. 2012;2012:581363. doi: 10.1155/2012/581363. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.