Abstract
TWIST1, an epithelial-mesenchymal transition (EMT) transcription factor, is critical for oncogene-driven non-small cell lung cancer (NSCLC) tumorigenesis. Given the potential of TWIST1 as a therapeutic target, a chemical-bioinformatic approach using connectivity mapping (CMAP) analysis was used to identify TWIST1 inhibitors. Characterization of the top ranked candidates from the unbiased screen revealed that harmine, a harmala alkaloid, inhibited multiple TWIST1 functions including single-cell dissemination, suppression of normal branching in 3D epithelial culture, and proliferation of oncogene driver-defined NSCLC cells. Harmine treatment phenocopied genetic loss of TWIST1 by inducing oncogene-induced senescence or apoptosis. Mechanistic investigation revealed that harmine targeted the TWIST1 pathway through its promotion of TWIST1 protein degradation. As dimerization is critical for TWIST1 function and stability, the effect of harmine on specific TWIST1 dimers was examined. TWIST1 and its dimer partners, the E2A proteins, which were found to be required for TWIST1-mediated functions, regulated the stability of the other heterodimeric partner post-translationally. Harmine preferentially promoted degradation of the TWIST1-E2A heterodimer compared to the TWIST-TWIST1 homodimer and targeting the TWIST1-E2A heterodimer was required for harmine cytotoxicity. Finally, harmine had activity in both transgenic and patient-derived xenograft (PDX) mouse models of KRAS mutant NSCLC. These studies identified harmine as a first-in-class TWIST1 inhibitor with marked anti-tumor activity in oncogene-driven NSCLC including EGFR mutant, KRAS mutant and MET altered NSCLC.
Keywords: KRAS, TWIST1 inhibitor, lung cancer, EMT, E2A
INTRODUCTION
As the leading cause of cancer deaths in the U.S. and worldwide, lung cancer remains a major public health problem. Recent advances in classifying non-small cell lung cancer (NSCLC) into molecularly-defined subgroups that respond to targeted therapies have shifted the treatment paradigm in NSCLC from standard chemotherapy to a personalized therapeutic approach. While initial response rates to these targeted therapies are high, resistance is all but inevitable (1–3). Additionally, patients with the most frequent molecular driver, mutant KRAS, lack effective targeted therapies (4). New strategies are needed to effectively and durably target oncogene-driven NSCLC.
Epithelial-mesenchymal transition (EMT) is a reversible biological process that allows for the transdifferentiation of epithelial cells to adopt a mesenchymal phenotype, resulting in an increase in motility and a loss of epithelial polarity (5). EMT-transcription factors (EMT-TF) such as the TWIST, SNAIL and ZEB proteins are drivers of the EMT transcriptional program. Expression of EMT-TFs and subsequent induction of EMT is associated with invasion, dissemination, metastasis, suppression of oncogene-induced senescence (OIS) and apoptosis, and promotion of a cancer stem cell phenotype (5,6). Additionally, induction of EMT and expression of EMT-TF have also been implicated in resistance to chemotherapy, radiation and targeted therapies (2,7,8). Developing molecules that can effectively inhibit the activity of EMT-TFs would have significant therapeutic implications given the diverse role of EMT-TF in both tumorigenesis, metastasis and therapeutic resistance.
We previously demonstrated that TWIST1 is required for tumorigenesis in NSCLC characterized by defined oncogenic drivers including KRAS mutant, EGFR mutant and MET amplified/mutant tumors (9,10). We have also demonstrated that Twist1 cooperates with mutant Kras to induce lung adenocarcinoma in vivo and that suppression of Twist1 expression can lead to OIS and oncogene-induced apoptosis (OIA) (9,10). TWIST1 has been shown to promote tumorigenesis in breast and prostate carcinomas through induction of EMT, invasion, and metastasis as well as suppression of OIS and apoptosis (11–17). While TWIST1 has been implicated in tumorigenesis through its ability to promote EMT and metastasis, we have demonstrated that TWIST1 in oncogene-driven NSCLC functions to primarily suppress OIS and OIA (9,10). Taken together, data from these previous studies suggests that pharmacological inhibition of TWIST1 may be a valuable therapeutic strategy across multiple solid tumors. In the current study, we identified and characterized harmine as the first pharmacologic inhibitor of TWIST1 with significant anti-tumor activity in oncogene-driven lung cancer.
Materials and Methods
Cell Lines and Reagents
All human non-small cell lung cancer cell lines, (A549, H460, H358, H23, H727, H23, Calu-1, Calu-6, PC-9, H1975, H3255, Hcc827, H1650, H1437, H596, H1648, and H1993) and embryonic kidney cell line HEK 293T were obtained from the American Type Culture Collection (ATCC) and grown in media as recommended by ATCC. Cell lines were authenticated using a short tandem repeat (STR) DNA profiling from the cell bank from which they were acquired. Cell lines were authenticated when initially acquired and every six months using a commercial vendor. Cell lines were tested for mycoplasma every six months using MycoAlert Detection Kit (Lonza). Harmine (286044-1G) and cyclohexamide (C4859) was purchased from Sigma-Aldrich and Q-VD-oPH (A1901) was purchased from ApexBio Technology.
Cell viability assays
For all harmine experiments, NSCLC cell lines were seeded in 96 wells at appropriate cell density based on their optimal growth rates. Following a 24 hour incubation, cells were treated with harmine for 24, 48, 72 hrs. For all E2A knockdown and Twist1-E2A harmine rescue experiments, NSCLC cell lines were seeded in 96 wells at appropriate cell density and were infected with lentivirus for 24 hours. Following 24 hours of infection, lentivirus was replaced with normal growth media or media with harmine. Cell viability was assessed at 24 and 48 hours following harmine treatment or at Day 4, 5 and 6. Cell viability was assessed using CellTiter96® Aqueous One Solution Cell Proliferation Assay kit (Promega) or CellTiter-Glo (Promega) according to manufacturer’s protocol. For all viability experiments, experimental treatment groups were performed in quadruplet and experiments were performed at least twice to ensure consistent results. All viability data was normalized to its appropriate non-treated control. IC50 values were calculated using Prism V6 software.
SA-β-galactosidase staining
SA-β-galactosidase staining was performed as previously described utilizing the Senescence β-Galactosidase Staining Kit (Catalogue #9860) from Cell Signaling (10).
Colony formation assay
On day 4 or 6 after infection with the indicated shRNA lentiviruses, cells were plated in 12-well plates at a density of 5,000–10,000 cells/well. On day 12, the cells were stained with crystal violet (0.5% in 95% ethanol) as previously described (10). For all colony forming experiments, experimental treatment groups were performed in triplicate and experiments were repeated at least twice to ensure consistent results.
Quantitative RT-PCR
Total RNA was isolated from cells using the QIAprep RNeasy Kit (Qiagen) according to manufacturer’s protocol. Using 1μg of RNA, cDNA was generated using the High-Capacity cDNA Reverse Transcription Kit (Applied Biosystems). 34 ng equivalents of cDNA were applied for amplification of the transcript described below using an Applied Biosystems StepOne RT-PCR system (Perkin Elmer Applied Biosystems) for 40 cycles using the PowerUp™ SYBR® Green Master Mix (Perkin Elmer Applied Biosystems) or TaqMan® Universal PCR Master Mix (Perkin Elmer Applied Biosystems) according to manufacturer’s protocol. Taqman was used for determine baseline TWIST1 and TCF3 mRNA levels in Fig. 3B, while SYBR Green was used in Fig. 4E. Following amplification, data was analyzed using the Applied Biosystems StepOne Real-Time PCR Software (Perkin Elmer Applied Biosystems). For each sample, the RNA levels of genes of interest were standardized using a housekeeping gene (18s) within that sample. The level of gene of interest was normalized to the expression of that gene from the appropriate comparator sample. Primer list is available in Supplementary Table 2–3.
Immunoblot analysis
After treatment with harmine or infection with lentivirus, cells were lysed and protein was quantified, prepared, and western blots were performed as previously described (10). Supplementary Table 4 contains details of primary antibodies used. The antigen-antibody complexes were visualized by chemiluminescence (ECL and ECL-plus reagent by GE Healthcare).
Connectivity MAP analysis
A gene signature after silencing of TWIST1 was generated based on our previous published data (10) and used to query against publically available gene expression profiles for a panel of drugs (http://www.broad.mit.edu/cmap). Connectivity analysis was performed as previously described (19).
3D organoid assay
Generation of the 3D organoid system, activation of TWIST1, and subsequent analysis of dissemination and branching was performed as previously described (20).
Lentiviral shRNA and cDNA Overexpression Experiments
293T cells were seeded (4 × 106 cells) in T-25 flasks and lentiviral particles were generated using a four-plasmid system and infected as according to the TRC Library Production and Performance Protocols, RNAi Consortium, Broad Institute (21), and as previously described (10). A full list of constructs used is available in Supplemental Tables 5–7. The sequence of these constructs and any primers used are available upon request.
Luciferase promoter reporter assay
Luciferase promoter reporter assays were performed as previously described (18). Cell extracts were prepared 48 hours after transfection in passive lysis buffer, and the reporter activity was measured using the Dual-Luciferase Reporter Assay System (Promega).
Transgenic mice
All mice were housed in pathogen free facilities and all experimental procedures were approved by the Institutional Animal Care and Use Committee of The Johns Hopkins University.
Inducible Twist1/KrasG12D transgenic mice in the FVB/N inbred background were of the genotype: CCSP-rtTA/tetO-KrasG12D/Twist1-tetO-luc (CRT). All the mice were weaned 3–4 weeks of age and then placed on doxycycline at 4–6 weeks of age. The CRT mice treated had similar levels of tumor burden per CT. Micro-CT imaging and quantification of tumor burden was performed as previously described (9).
For in vivo experiments, harmine was dissolved in normal saline by heating and sonication. The mice received 10 mg/kg harmine or saline via intraperitoneal injection daily, 5 days a week for 3 weeks.
For the 3D epithelial cell culture experiments, mammary organoids were isolated from the previously described, CMV::rtTA;TRE-Twist1 mice (9,20).
Histology and immunohistochemistry
Immunohistochemistry and Histology were performed as previously described (22). Primary antibodies were used at the following dilutions: Ki-67 at 1:2000 and cleaved caspase 3 at 1:500.
Statistics
Student’s t-test was performed where indicated. For dissemination experiments (Figure 1C–J), data did not follow a normal distribution, therefore non-parametric comparison testing was performed (Kruskal-Wallis). For PDX experiment, a Student t-test was performed on the finalized tumor volumes between the harmine and vehicle treated groups.
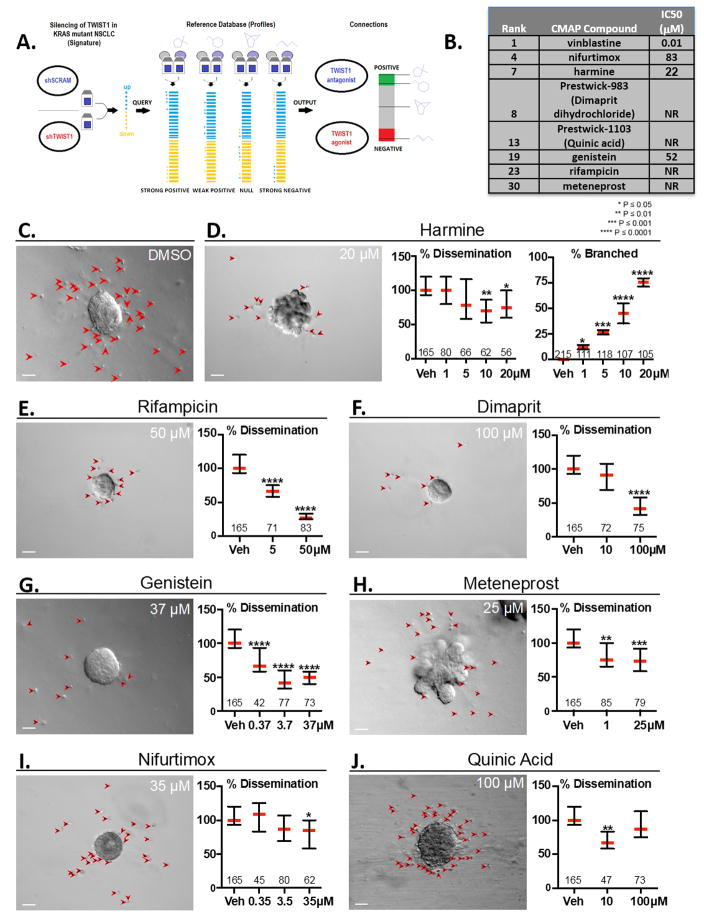
Figure 1. Connectivity MAP (CMAP) analysis identifies compounds which inhibit NSCLC proliferation, TWIST1-dependent dissemination and TWIST1-suppression of organoid branching.
(A) CMAP Analysis Schematic. A gene signature for TWIST1 knockdown was utilized to query the Connectivity Map (Broad Institute). (B) Chart depicting eight of the top 30 ranked compounds which were selected for further analysis. IC50 was determined in two KRAS mutant NSCLC cell lines (A549 and H460) at 72 hours using MTS assays. (C) TWIST1 induction with doxycycline results in profound dissemination of cells and prevents branching (DMSO, upper left panel). Primary Twist1 inducible breast epithelial cells are implanted as organoids in 3D culture. Red arrowheads indicate disseminated cells. (D–J) Candidate compounds inhibit Twist1-induced 3D dissemination and/or restore FGF2-induced branching of primary breast epithelial cells in vitro. Values within the graph indicate the number of organoids quantified per treatment condition. Dissemination data is normalized to the median of each experimental replicate. Error bars, 95% Confidence Intervals. Treatment with small molecules listed at the indicated doses induced statistically significant (Kruskal-Wallis test, P<.05) reductions in dissemination. (D) Branching data is presented as mean of each experimental replicate. See Supplementary Fig. 1A–C for representative images of unbranched vs. branched organoids. Error bars, ±SD. Treatment with harmine at the indicated doses induced statistically significant (One-way ANOVA, P<.05) increases in branching. Scale bar, 50μm.
RESULTS
Identification of novel TWIST1 inhibitors with a Connectivity MAP analysis
Transcription factors, such as TWIST1, have long been considered a potential therapeutic target for cancer, given their essential role in modulating transcriptional networks that drive tumorigenesis. Despite their potential as drug targets, effective small molecule therapies that inhibit transcription factor function remain elusive (23). Therefore, the identification of inhibitors of oncogenic EMT transcription factors would be a significant advance in cancer therapeutics and potentially lead to therapies which would not only inhibit tumor growth but metastatic potential as well. Connectivity mapping (CMAP) is a tool that compares gene signature changes associated with biological manipulation or disease to corresponding changes produced by potential drugs (19). In order to identify potential TWIST1 targeting chemical compounds, we utilized our previously published TWIST1 knockdown expression profile (10) as the query signature for CMAP analysis. CMAP analysis generated a rank list of the 6100 compounds based on statistical correlation between changes in global gene expression signature induced by drug versus that induced by TWIST1 knockdown (Fig. 1A, Supplementary Table 1). We selected eight of the top thirty ranked compounds based on a review of the literature suggesting that these compounds inhibited pathways important in tumorigenesis or anti-cancer activity. These compounds were evaluated for their respective cytotoxic activity in vitro in KRAS mutant NSCLC cell lines (Fig. 1A). Of these compounds, only four demonstrated anti-proliferative effects in NSCLC (Figure 1B). To identify inhibitors of TWIST1 that suppress TWIST1-dependent processes that are independent of proliferation such as single-cell dissemination, we utilized our previously published 3D organoid assay with primary breast epithelial cells derived from our doxycycline inducible CMV:rtTA;TRE-Twist1 mouse model (9,20). We have previously shown that expression of Twist1 inhibits normal branching morphogenesis and leads to rapid and widespread dissemination of primary breast epithelial cells from the 3D organoid (Fig. 1C) which can be reversed by removing doxycycline from the medium (20). We therefore examined whether selected candidate compounds could inhibit the Twist1-dependent dissemination in this model. Remarkably, all of seven compounds tested resulted in a significant decrease in Twist1-mediated dissemination (Fig 1D–J) compared to the vehicle control (Fig. 1C). In this 3D organoid system, FGF2 treatment induces normal branching morphogenesis of primary breast epithelial cells (Supplementary Fig. 1A–C) (20). We next examined the ability of these compounds to restore FGF2-induced branching which we have previously shown is inhibited by Twist1 expression in this model (20). Two compounds, meteneprost and the harmala alkaloid, harmine were able to restore FGF2-induced branching in a dose dependent manner (Figure 1D, 1H) compared to the vehicle control (Fig. 1C). Interestingly, three harmine analogues were in the top 80 compounds identified in the CMAP analysis, with two of the compounds being in the top 40 (Supplementary Table 1). Previous studies have reported that harmine and its analogues have marked anti-tumor activity (24–26). While two compounds screened, harmine and meteneprost, inhibited TWIST1-dependent single epithelial cell dissemination and TWIST1-suppresion of epithelial organoid branching (Figure 1D, 1H), harmine was the only compound screened that inhibited growth in lung cancer cell lines as well as the aforementioned TWIST1-dependent functions in our mammary 3D organoid system. Given these factors, we decided to further characterize the biological activities of harmine as a TWIST1 inhibitor.
Harmine inhibits growth in oncogene driver defined NSCLC cell lines and phenocopies loss of TWIST1
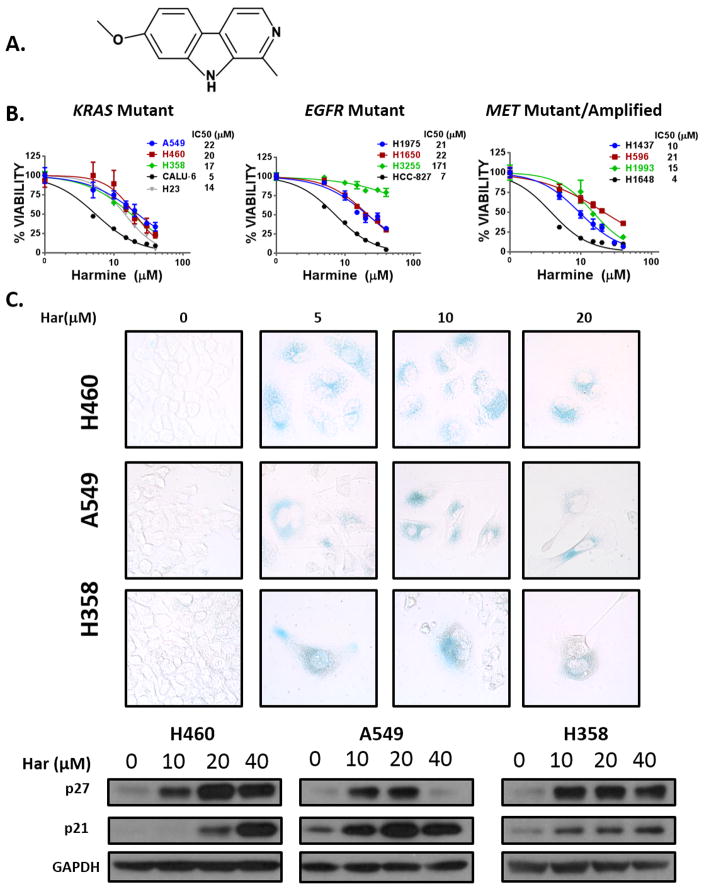
We first characterized the growth inhibitory effects of harmine (Fig. 2A) across a panel of oncogene driver defined NSCLC lines which we had previously reported as dependent on TWIST1 expression (10). Similar to genetic silencing of TWIST1, we found harmine treatment to be cytotoxic across the panel of oncogene driver defined NSCLC lines (KRAS mutant, MET amplified/mutant, and EGFR mutant) (Fig. 2B). While harmine was more potent in cell lines with high TWIST1 levels (Fig. 2B), harmine also had activity in cell lines with low TWIST1 expression (Supplementary Fig. 2) (27). Additionally, harmine had activity in NSCLC cell lines with primarily epithelial or mesenchymal phenotypes, suggesting that the activity of harmine is independent of EMT status (27–29). We had previously demonstrated that inhibition of TWIST1 leads to oncogene-induced senescence (OIS) in KRAS mutant NSCLC lines (9). Similar to silencing of TWIST1, treatment of the KRAS mutant NSCLC lines A549, H460, and H358 with harmine-induced changes characteristic of OIS, including positive Senescence-Associated Beta-galactosidase (SA-β-Gal) staining and induction of p21 and p27 (Fig. 2C). We also found that harmine treatment induced OIS in NSCLC cell lines with EGFR and MET mutations (Supplementary Fig. 3A–B). We have previously shown that TWIST1 is required for suppression of OIA in a subset of lung cancer cells including Calu-6 and H23 (10). Consistent with a TWIST1-suppressive effect, after harmine treatment, we observed a significant induction of apoptosis, in a dose dependent manner in both lines (Supplementary Fig. 4A). The growth inhibition by harmine in these cells was dependent on apoptosis as co-treatment with the pan caspase inhibitor, Q-VD-oPH, prevented the cytotoxic effects of harmine (Supplementary Fig. 4B). Furthermore, BCL-2 overexpression partially prevented harmine-induced apoptosis, suggesting that the intrinsic apoptotic pathway is required for the growth inhibitory effects of harmine in these lines (Supplementary Fig. 4C).
Figure 2. Harmine inhibits growth through the induction of oncogene-induced senescence (OIS) in oncogene driver defined NSCLC cell lines.
(A) Chemical structure of the harmala alkaloid, harmine. (B) MTS assays demonstrating growth inhibition in the indicated KRAS mutant, EGFR mutant, and MET mutant/amplified NSCLC cells following harmine treatment at 72 hours. Data represent mean ±SD (n=4 technical replicates). (C) UPPER: Senescence-Associated Beta-galactosidase (SA-β-Gal) staining demonstrating that harmine (Har) treatment leads to oncogene-induced senescence (OIS) in KRAS mutant NSCLC cell lines. Cells were treated at the indicated doses for 72 hours and stained 7 days following treatment. Images were obtained with bright field objective at 40× magnification. LOWER: Western blot demonstrating a marked increase in p21 and p27 expression 48 hours after harmine treatment at the indicated doses.
Harmine treatment results in TWIST1 protein degradation
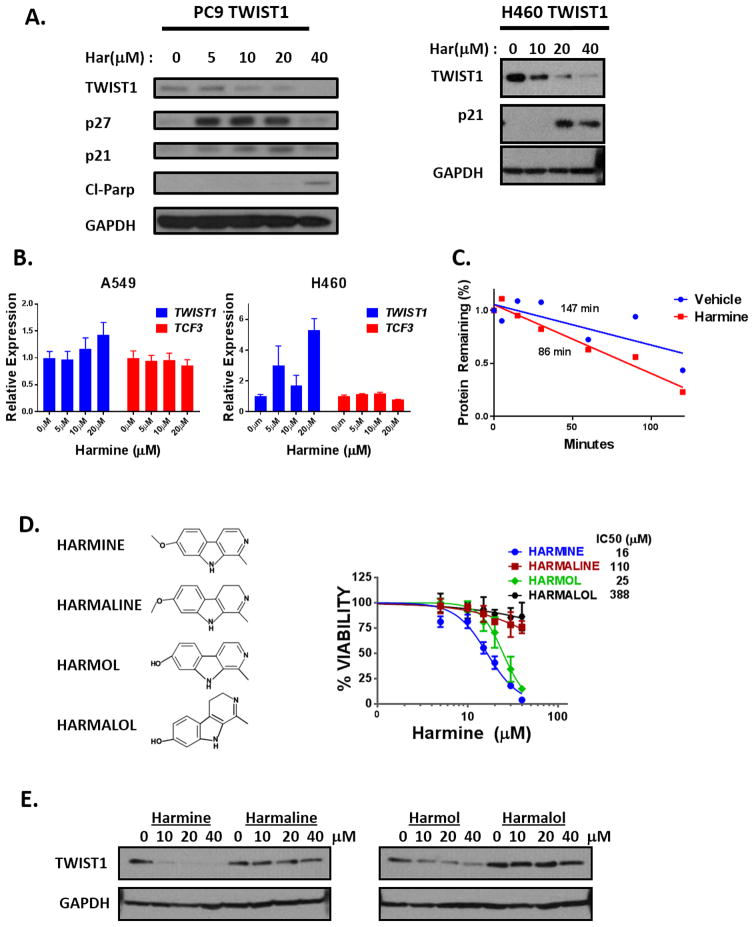
Previous studies have demonstrated that modulation of TWIST1 protein stability is a critical regulatory mechanism of TWIST1 function (30–33). To determine whether harmine directly targets TWIST1 or the TWIST1 pathway, we first examined the effect of harmine on TWIST1 protein stability. Harmine treatment reduced the levels of TWIST1 protein in a dose and time dependent manner as shown by western blotting and this was accompanied by a reciprocal induction of p21 (Fig. 3A, Supplementary Fig. 5A), a known TWIST1 repressed transcriptional target gene (34,35). The effect of harmine on TWIST1 expression appears to occur through a post-translational mechanism as harmine treatment did not decrease TWIST1 mRNA levels (Fig. 3B) but did decrease the half-life of the TWIST1 protein (Fig. 3C, Supplementary Fig. 5B).
Figure 3. Harmine treatment leads to TWIST1 protein degradation.
(A) LEFT: Representative western blots demonstrating 72 hours of harmine treatment promotes TWIST1 degradation and subsequent oncogene-induced senescence (p27 and/or p21) and apoptosis (Cl-Parp) in a dose-dependent manner in the EGFR mutant NSCLC cell line, PC-9. RIGHT: Western blot demonstrating reduction of exogenous TWIST1 protein expression as well as induction of p21 after 72 hours of harmine treatment in the KRAS mutant NSCLC cell line, H460. (B) Quantitative RT-PCR analysis of TWIST1 and TCF3 transcripts following 72 hours of harmine treatment in A549 and H460 cells which failed to detect a decrease in mRNA levels of TWIST1 or TCF3. All harmine treatment groups are normalized to untreated group. Data represent mean ±SD (n=3 technical replicates). (C) Harmine treatment decreases the half-life of TWIST1 protein. Protein concentration was quantified using densitometry and protein half-lives were determined using linear regression. (D) LEFT: Chemical structures of Harmala Alkaloid compounds identified as potential TWIST1 inhibitors from CMAP analysis. RIGHT: MTS assays demonstrating growth inhibition of a H460 TWIST1 NSCLC cell line in a dose dependent manner following treatment with indicated harmala alkaloids at 72 hours. Data represent mean ±SD (n=4 technical replicates). (E) Western blot demonstrating reduction of exogenous TWIST1 protein expression by harmine and harmol in H460 TWIST1 overexpressing cells 72 hours after treatment with the indicated doses.
In addition to harmine, several other harmala alkaloid compounds were identified in our CMAP analysis. To determine if particular structural features of harmala alkaloids were important for the induction of TWIST1 degradation, we selected three related compounds that differed from harmine in key structural positions (Fig. 3D, left panel). We were interested in both the 7-methoxy structural moiety which has previously been shown to effect cytotoxicity and neurotoxicity and the saturation level of the pyridine ring, which has been demonstrated to affect the biological activity of beta-carbolines (24–26,36). Of note, harmine and harmaline have a methoxy group, whereas harmol and harmalol have hydroxyl groups at the position R7. Conversely, harmine and harmol contain pyridine rings with three versus two double bonds found in harmaline and harmalol. The compounds (harmine and harmol) which contained pyridine rings with three double bonds exhibited the most cytotoxicity suggesting that this feature was critical for tumor growth inhibition and TWIST1 degradation (Fig. 3D–E). However, the presence of a methoxy or hydroxyl group, at the position R7 did not correlate with increased cytotoxicity and the ability to degrade TWIST1. Notably, the relatively potency of these compound directly correlated with their ability to lead to TWIST1 degradation (Fig. 3D–E).
TWIST1 and the E2A proteins reciprocally stabilize each other and harmine leads to degradation of both components of this dimer
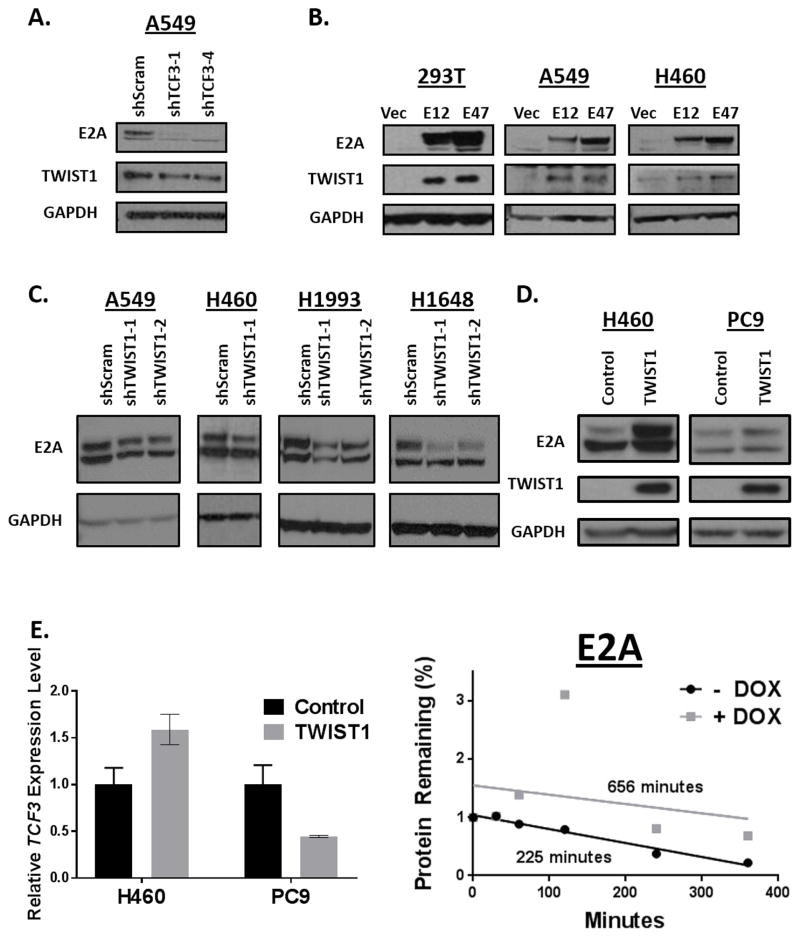
The TWIST1 protein forms both homo- and hetero-dimers and its functions are dependent on its respective dimer partner (37). We and others have previously shown that heterodimerization with the proteins encoded by the TCF3 gene, here after referred to as the E2A proteins (E12 and E47), are critical for TWIST1 function in tumorigenesis and EMT (18,38,39) and potentially TWIST1 protein stability in osteoblasts (31). Therefore, we decided to examine the role of the E2A proteins in regulating both TWIST1 protein stability in NSCLC and response to harmine. To examine the effect of modulating the levels of TWIST1 and E2A proteins on the stability of the dimer partner, we knocked down and overexpressed each of the proteins and examined the effect on the protein level of its dimer partner. Silencing of TCF3 in the KRAS mutant NSCLC cell line A549 induced modest downregulation of TWIST1 (Fig. 4A) and conversely overexpression of the E12 or E47 induced robust upregulation of TWIST1 (Fig. 4B). In both KRAS mutant (A549, H460), MET-amplified (H1648, H1993) NSCLC cell lines, silencing of TWIST1 induced downregulation of E2A proteins (Fig. 4C). Conversely, overexpression of TWIST1 in the same KRAS mutant lines and an EGFR mutant NSCLC cell line (PC9) increased E2A protein expression (Fig. 4D). To explore the mechanism of TWIST1 regulation of E2A, we analyzed TCF3 mRNA levels following TWIST1 overexpression. We found that H460 and PC9 cell lines TWIST1 overexpression does not result in a marked increase in TCF3 RNA levels, and in fact decreases TCF3 mRNA levels in PC9 cells, suggesting that it is unlikely that TWIST1 regulates E2A transcriptionally. Additionally, we found that TWIST1 overexpression in multiple cell lines leads to an increase in E2A protein half-life (Fig. 4E, Supplementary Fig. 5C–D). These results suggest that TWIST1 regulates expression of the E2A proteins post-translationally.
Figure 4. The E2A proteins and TWIST1 reciprocally stabilize each other.
(A) Silencing of TCF3 leads to downregulation of TWIST1 in the KRAS mutant NSCLC cell line, A549. Cells were infected with the indicated shRNAs and were harvested for western blot analysis 96 hours following infection. (B) Overexpression of the E2A proteins, E12 or E47 induces upregulation of TWIST1 in 293T cells as well as in KRAS mutant NSCLC cells (A549, H460). 293T cells were harvested 72 hours following transfection. Experiments in A549 and H460 were performed in stable E12 or E47 overexpressing cell lines. (C) Silencing of TWIST1 induces downregulation of E2A proteins in KRAS mutant (A549, H460) as well as in MET amplified (H1648, H1993) NSCLC cell lines. Cells were infected with the indicated shRNAs and were harvested for western blot analysis 96 hours following infection. (D) Overexpression of TWIST1 induces the E2A proteins in KRAS mutant NSCLC cells (H460) and EGFR mutant NSCLC cells (PC-9). H460 cells were harvested once stable cell lines were established. PC9 TRE3G-TWIST1 cells were harvested following 500 ng/ml treatment of doxycycline for 24 hours. (E) LEFT: TWIST1 induction of E2A proteins is not accompanied by a robust upregulation of TCF3 mRNA in KRAS mutant (H460) and EGFR mutant NSCLC cells (PC-9). All TWIST1 overexpressing groups are normalized to untreated group (n=3 technical replicates). Data represent mean ±SD. RIGHT: TWIST1 overexpression increases the half-life of E2A proteins. Protein concentration was quantified using densitometry and protein half-lives were determined using linear regression.
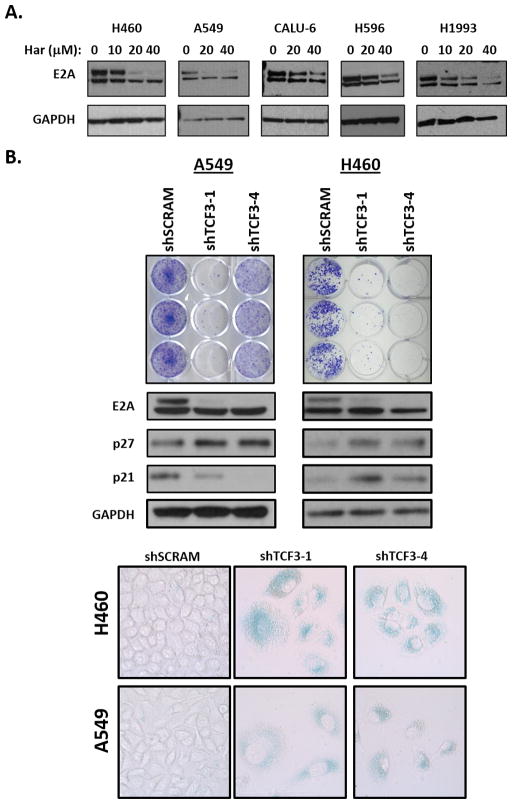
Given the ability of the E2A proteins and TWIST1 to reciprocally stabilize each other, we examined the effect of harmine on the E2A proteins. Remarkably, treatment with harmine resulted in a dose-dependent decrease of E2A protein expression in both KRAS mutant and MET-mutant/amplified NSCLC cell lines (Fig. 5A). Additionally, this decrease in E2A protein appears to be post-transcriptional as harmine treatment does not decrease TCF3 RNA levels (Fig. 3B). To further characterize the potential role of the E2A proteins in TWIST1-mediated lung tumorigenesis, we silenced TCF3 in KRAS mutant NSCLC cell lines. Silencing of TCF3 also resulted in OIS, phenocopying silencing of TWIST1. Similar to our previously published results (9,10), both growth inhibition in colony formation assays and OIS as evidenced by increased SA-β-Gal staining and p21/p27 levels were observed with silencing of TCF3 (Fig. 5B).
Figure 5. Harmine leads to degradation of the E2A proteins which are required for suppression of OIS.
(A) Western blot demonstrating dose-dependent downregulation of E2A proteins in KRAS mutant (H460, A549 and Calu-6) and MET mutant/amplified (H596 and H1993) NSCLC cell lines following 72 hours of harmine treatment. (B) UPPER: shRNA silencing of TCF3 in KRAS mutant NSCLC cell lines (A549 and H460) leads to growth inhibition as demonstrated in triplicates of crystal violet staining. MIDDLE/LOWER: shRNA silencing of TCF3 induces OIS as shown by western blot demonstrating a marked increase in p21 and/or p27 expression and positive SA-β-Gal staining (LOWER). For western blotting, cells were infected with the indicated shRNAs for 96 hours and harvested. For SA-β-Gal staining cells were infected with the indicated shRNAs and stained for 10 days following infection. Images were obtained with bright field objective at 40× magnification.
As discussed above, TWIST1 is required for suppression of OIA in a subset of lung cancer cells and we wanted to examine whether we could observe a similar phenotype after silencing of TCF3 (10). Genetic silencing of TCF3 in the KRAS mutant cell lines, Calu-6 and H23, resulted in significant growth inhibition and a corresponding increase in apoptosis (Supplementary Fig. 6A–C) phenocopying our previous studies with silencing of TWIST1 (10) and studies above with harmine (Supplementary Fig. 4). Furthermore, we demonstrated that the growth inhibition after silencing of TCF3 was dependent on apoptosis as pre-treatment with the pan caspase inhibitor, Q-VD-oPh rescued cell viability (Supplementary Fig. 6C).
The TWIST1/E2A heterodimer is critical for TWIST1 function and therapeutic response to harmine
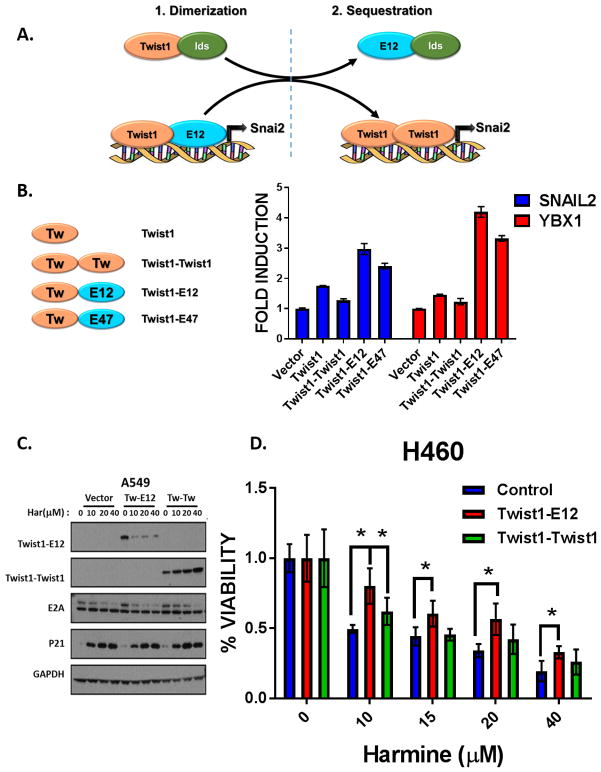
The exact mechanism of how TWIST1 and E2A interact to promote lung tumorigenesis is unknown; however we propose two potential mechanisms for the cooperation between TWIST1 and the E2A proteins, E12 or E47, to promote their neoplastic phenotypes. In the first scenario (dimerization), the TWIST1-E12 heterodimer is directly required for the transcriptional activity of TWIST1 and induces tumorigenesis (Fig. 6A). The second scenario (sequestration) suggests that E12/E47-mediated sequestration of Ids (Inhibitor of DNA-binding proteins) leads to TWIST1 transcriptional activity through allowing increased TWIST1 homodimer formation (Fig. 6A). In order to determine which mechanism is important for TWIST1 transcriptional activity, we first performed a luciferase assay utilizing the promoters of SNAI2 and YBX1, known transcriptional targets of TWIST1 (40,41). As expected, increased induction of SNAI2 and YBX1 promoter activity was seen in cells transiently overexpressing Twist1. To determine the transcriptional function of the TWIST1 homo- and heterodimeric proteins, we expressed Twist1 tethered Twist1, Twist1 tethered E12 and Twist1 tethered E47 fusion proteins in these luciferase assays (42,43). The Twist1-E12 and Twist1-E47 had significantly increased transcriptional activity compared to the Twist1-Twist1 homodimer or Twist1 alone (Fig. 6B). Since the Twist1-E2A heterodimers appeared to be the most potent inducer of transcription and harmine led to degradation of both TWIST1 and the E2A proteins we examined the relative effect of harmine on stability of the TWIST1 homo and heterodimers. We observed that harmine was most effective against the Twist1-E12 heterodimer following harmine treatment (Fig. 6C). Further supporting a role for the TWIST1 heterodimer in TWIST1 function and therapeutic response, overexpression of TWIST1 or the E2A proteins, E12 and E47, were able to rescue the harmine-induced growth inhibition in the KRAS mutant NSCLC cell lines A549 and H460 (Supplementary Fig. 7A–C). Furthermore, overexpression of the Twist1-E12 heterodimer similarly lead to a rescue of harmine-induced cytotoxicity, while overexpression of the Twist1-Twist1 heterodimer failed to lead to such rescue at most doses of harmine (Fig. 6D). Together these data support the model that the E2A proteins are necessary for TWIST1 functions and that degradation of TWIST1-E2A heterodimer is critical for harmine-induced cytotoxicity.
Figure 6. The TWIST/E2A heterodimer is critical for TWIST1 function and therapeutic response to harmine.
(A) Proposed model of potential mechanism(s) of TWIST1/E2A cooperation in tumorigenesis. (B) Luciferase assay showing increased induction of SNAI2 and YBX1 promoter activity in cells transiently overexpressing either Twist1 alone, Twist1-Twist1 homodimer, or Twist1-E12 heterodimer compared to the reporter activity with the vector alone (fold induction = 1) after 48 hours. All luciferase values were normalized to the corresponding renilla luciferase value in each well. Differences were statistically different for each loci for Twist1-E12 or Twist1-E47 versus Twist1 or Twist1-Twist1, p < 0.0005, 2-tailed Student’s t-test. (C) Western blot demonstrating preferential downregulation of the Twist1-E12 heterodimer in A549 KRAS mutant cell line following 48 hour treatment with harmine at the indicated doses. (D) Representative Cell-Titer Glo assay demonstrating that overexpression of Twist1-E12 heterodimer rescues harmine-induced growth inhibition in a KRAS mutant NSCLC cell line (H460), while the Twist1-Twist1 heterodimer fails to prevent harmine-induced cytotoxicity. Data represent mean ±SD (n=4 technical replicates). *, p<.05, 2-tailed Student’s t-test.
Harmine has in vivo activity in both transgenic and patient-derived xenograft mouse models of KRAS mutant lung cancer
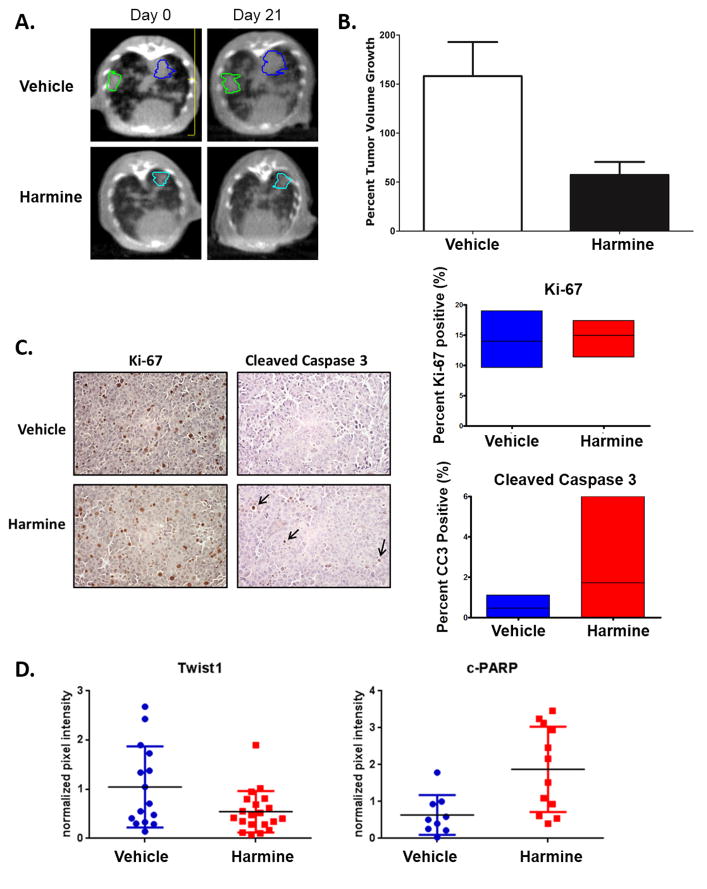
Having observed in vitro activity in KRAS mutant NSCLC cell lines, we wanted to determine whether harmine would have in vivo efficacy in a KrasG12D/Twist1 mouse model of autochthonous lung adenocarcinoma. We treated the CCSP-rtTA/tetO-KrasG12D/Twist1-tetO7-luc (CRT) mice, which overexpress mutant Kras and Twist1 predominantly in the type II cells of the mouse lung and form lung adenocarcinoma by 15 weeks (9), with harmine for 3 weeks and measured index lung tumor volumes in mice at baseline and weekly with serial micro-computed tomography (microCT). Micro-CT images, comparing tumor volume at baseline and at the end of treatment, revealed that treatment with harmine decreased the tumor volume growth rate (Fig. 7A). Of note, the anti-tumor activity of harmine in our Kras/Twist1 transgenic was similar to the tumor stasis seen in the same mice following genetic suppression of Twist1 expression as we have previously published (9) Additionally, harmine significantly inhibited tumor growth in a patient-derived xenograft (PDX) mouse model of KRAS mutant lung cancer (Supplementary Fig. 8A). Treatment of the animals with harmine resulted in no observable toxicity and body weight was unaffected (data not shown).
Figure 7. Harmine has activity in a KrasG12D/Twist1 mouse model of autochthonous lung adenocarcinoma.
(A) Representative microCT images of autochthonous lung tumors showing decreased lung tumor growth 3 weeks following treatment with harmine versus vehicle in the CCSP-rtTA/tetO-KrasG12D/Twist1-tetO7-luc (CRT) mice. Contoured lung tumors represent index lesions followed serially for tumor volume quantification. (B) Lung tumor volumes were quantified at baseline and weekly with serial micro-computed tomography (microCT) in the same CRT mice. Micro- CT images were reviewed by a board certified radiation oncologist on multiple index tumors in a blinded fashion (n=18 tumors for vehicle from 6 mice and n=16 from 5 mice for harmine). Difference was statistically different using a Mann-Whitney test, P=0.0025. Volumes were normalized to the starting volume, t= 0 before harmine treatment, and percent tumor volume growth was then calculated by (normalized tumor vol. × 100%) − 100%. (C) LEFT: Treatment with harmine results in similar proliferation levels as measured by Ki-67 staining, but increased apoptosis as measured by cleaved caspase 3 staining indicated by black arrows. RIGHT: Quantification and comparison of Ki-67 staining (n=12 tumors for vehicle and n=15 for harmine) (p= 0.3621) and cleaved caspase 3 IHC (n=12 tumors for vehicle and n=16 for harmine) (p= 0.0453). (D) Quantification of Twist1 and cleaved PARP protein levels in vehicle and harmine treated animals at 3 weeks. Proteins were normalized to luciferase protein levels to control for possible differences in tumor burden. Differences were statistically significant (using Student’s T-Test) for Twist1 (n=15 tumors for vehicle and n=20 tumors for harmine), P < 0.04 and c-PARP (n=9 tumors for vehicle and n=12 tumors for harmine), P < 0.005 respectively.
We then examined the potential mechanisms of growth inhibition after harmine treatment in vivo. We first examined whether a decrease in proliferation was responsible for the observed growth inhibition, however, we observed no significant difference in proliferation rate as measured by Ki-67 staining after harmine treatment (Fig. 7B). We next examined whether increased apoptosis contributed to the growth inhibitory effects of harmine and we did observe increased apoptosis as measured by cleaved caspase 3 and cleaved PAPP with harmine (Fig. 7B–C; Supplementary Fig. 8B). Most notably, harmine treatment led to marked decrease in Twist1 protein in the mouse lung tumors (Fig. 7C, Supplementary Fig. 8B). Thus, harmine has cytotoxic effects in vivo on Kras mutant, Twist1 overexpressing lung adenocarcinoma, which are accompanied by Twist1 degradation.
DISCUSSION
Using a Connectivity MAP chemical-bioinformatic analysis, we identified multiple compounds that recapitulated the genetic signature of TWIST1 knockdown. We subsequently assayed seven of the top ranked candidates from this screen for their ability to inhibit multiple TWIST1-mediated in vitro phenotypes that included single-cell dissemination and suppression of FGF2-dependent branching of mammary epithelial cells in a 3D organoid system. All seven compounds tested were able to inhibit TWIST1-mediated dissemination, while the harmala alkaloid compound, harmine, was also able to restore FGF2-dependent mammary epithelial cell branching. We chose to further characterize harmine and demonstrated that it had significant growth inhibitory activity in multiple oncogene-driven NSCLC cell lines. We demonstrated that harmine not only leads to TWIST1 degradation but also phenocopied the loss of TWIST1 by inducing either OIS or OIA in previously defined subsets of NSCLC cell lines. Remarkably, in our mouse model of Kras mutant, Twist1 overexpressing lung cancer, harmine had significant anti-tumor activity, no overt toxicity, and led to decreased expression of Twist1 protein in vivo.
These studies also found that TWIST1 and its binding partners, the E2A proteins reciprocally regulate the stability of each other. Additionally, we demonstrated that genetic silencing of TCF3 in part phenocopies the loss of TWIST1, with loss of TCF3 expression reactivating latent senescence and apoptotic programs. Our data also suggests that the TWIST1-E2A heterodimer, rather than the TWIST1-TWIST1 homodimer, is critical for the transcriptional activation of TWIST1 target genes important for tumorigenesis. Interestingly, the TWIST1-E12 heterodimer has been previously shown to be critical for the ability of TWIST1 to cooperate with RAS to promote mammary tumorigenesis and suppress senescence (38). Additionally, the metastatic ability of TWIST1 in prostate cancer cells requires the ability of TWIST1 to heterodimerize with the E2A proteins (18). Our data demonstrated that harmine preferentially induces degradation of the TWIST1-E2A heterodimer rather than the TWIST1-TWIST1 heterodimer and degradation of the heterodimer is required for the cytotoxic effects of harmine. Future studies will be aimed at determining the mechanism(s) by which harmine leads to TWIST1 degradation, specifically the TWIST1-E2A heterodimer.
While harmine was not associated with overt toxicity in our in vivo model, harmine has been found to have potentially dose-limiting neurotoxicity in humans (25,44). Previous structure-activity studies have determined that harmine derivatives with substituents at position-2 and -9 can modulate the cytotoxic effects of harmine, while the addition of a bulky substituent at the -7 position can ameliorate the neurotoxicity associated with harmine (24–26,44). While micromolar range doses were required for harmine-mediated cytotoxicity in vitro, it should be noted that we were able to achieve doses in vivo that both inhibited tumor growth and promoted TWIST1 degradation without noticeable side effects. Current efforts in our laboratory are ongoing to identify harmine analogues or related compounds that allow for more potent inhibition of TWIST1 transcriptional activity without associated neurotoxicity.
In summary, we identified harmine as a first-in-class inhibitor of TWIST1 with broad cytotoxic activity in the three major classes of oncogene-driven NSCLC, EGFR-mutant, KRAS mutant, and c-MET-amplified/mutant. Given that we have previously established the requirement of TWIST1 for tumorigenesis in oncogene-driven lung cancer with these genetic backgrounds (10), using harmine derivatives may be a viable therapeutic option to treat oncogene-driven NSCLC both in the treatment-naïve and acquired resistance setting. Additionally, as TWIST1 is rarely expressed post-natally (45,46), pharmacological inhibition of TWIST1 may be associated with minimal side-effects. TWIST1 has been implicated in oncogenesis, EMT, metastasis, therapeutic resistance, and tumor stem cell maintenance, across multiple solid tumors including head and neck, lung, breast, and prostate cancers (18,47–51). The use of harmine and potential analogues has far-reaching therapeutic implications given the diverse roles of TWIST1 in tumorigenesis, metastasis and therapeutic response. For this reason, we are currently screening harmine derivatives that allow for more potent, specific inhibition of TWIST1 which we can bring to the clinic.
Supplementary Material
IMPLICATIONS.
TWIST1 is required for oncogene-driven NSCLC tumorigenesis and EMT, thus harmine and its analogues/derivatives represent a novel therapeutic strategy to treat oncogene-driven NSCLC as well as other solid tumor malignancies.
Acknowledgments
Financial Support: This work was supported by the following funding sources: Dr. Burns have received research funding for this project from an American Lung Association Award (LCD 257864), V Foundation Scholar Award, Sidney Kimmel Foundation (SKF-15-099) and a Doris Duke Charitable Foundation Clinical Scientist Award (2015097) and a UPCI LUNG SPORE CDA P50CA090440. This project used the UPCI Animal, Flow Cytometry and Biostatistics Facilities that are supported in part by award P30CA047904. Zachary Yochum received support from the Howard Hughes Medical Institute (HHMI) Medical Fellowship, T32GM008421-22 and a 1F30CA213765-01. PT Tran was supported by the Nesbitt Family; Uniting Against Lung Cancer Foundation; Movember-Prostate Cancer Foundation; Commonwealth Foundation; American Lung Association (LCD-339465); Sidney Kimmel Foundation (SKF-13-021); DoD (W81XWH-13-1-0182); ACS (122688-RSG-12-196-01-TBG); NIH/NCI (R01CA166348). AJ Ewald was supported by the American Cancer Society (RSG-12-141-01–CSM to AJE), the National Institutes of Health (NIGMS 3T32GM007309 to NMN), and the Pink Agenda & Breast Cancer Research Foundation to AJE.
The authors thank James G. Herman, MD, Laura Stabile, PhD, and Frank P. Vendetti PhD at the University of Pittsburgh for the discussion, advice regarding this work and supply of resources when applicable. In addition, we would like to acknowledge the helpful comments and suggestions provided by the members of the Rudin, Hann, Burns and Tran laboratories.
Footnotes
Disclosure of Potential Conflicts of Interest
No potential conflicts of interest were disclosed.
Author’s Contributions
Conception and design: Z.A.Y., J.C., L.M., C.R.M., P.T. and T.F.B
Development of methodology: Z.A.Y., J.C., L.M., N.M.N., A.J.E., P.T.T. and T.F.B
Acquisition of data (provided animals, acquired and managed patients, provided facilities, etc.): Z.A.Y., J.C., L.M., N.M.N., M.A., S.C., S.K., S.C.C., J.E. and T.F.B
Analysis and interpretation of data (e.g., statistical analysis, biostatistics, computational analysis): Z.A.Y., J.C., L.M., N.M.N., A.J.E., P.T.T. and T.F.B.
Writing, review, and/or revision of the manuscript: Z.A.Y., J.C., A.J.E. C.M.R., P.T.T. and T.F.B
Administrative, technical, or material support (i.e., reporting or organizing data, constructing databases): Z.A.Y., J.C., and T.F.B
Study supervision: P.T. and T.F.B
References
- 1.Katayama R, Shaw AT, Khan TM, Mino-Kenudson M, Solomon BJ, Halmos B, et al. Mechanisms of acquired crizotinib resistance in ALK-rearranged lung Cancers. Sci Transl Med. 2012;4(120):120ra117. doi: 10.1126/scitranslmed.3003316. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Sequist LV, Waltman BA, Dias-Santagata D, Digumarthy S, Turke AB, Fidias P, et al. Genotypic and Histological Evolution of Lung Cancers Acquiring Resistance to EGFR Inhibitors. Sci Transl Med. 2011;3(75):75ra26. doi: 10.1126/scitranslmed.3002003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Yu HA, Riely GJ, Lovly CM. Therapeutic strategies utilized in the setting of acquired resistance to EGFR tyrosine kinase inhibitors. Clin Cancer Res. 2014;20(23):5898–5907. doi: 10.1158/1078-0432.CCR-13-2437. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Cox AD, Fesik SW, Kimmelman AC, Luo J, Der CJ. Drugging the undruggable RAS: Mission possible? Nat Rev Drug Discov. 2014;13(11):828–851. doi: 10.1038/nrd4389. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Thiery JP, Acloque H, Huang RY, Nieto MA. Epithelial-mesenchymal transitions in development and disease. Cell. 2009;139(5):871–890. doi: 10.1016/j.cell.2009.11.007. [DOI] [PubMed] [Google Scholar]
- 6.Puisieux A, Brabletz T, Caramel J. Oncogenic roles of EMT-inducing transcription factors. Nat Cell Biol. 2014;16(6):488–494. doi: 10.1038/ncb2976. [DOI] [PubMed] [Google Scholar]
- 7.Davis FM, Stewart TA, Thompson EW, Monteith GR. Targeting EMT in cancer: opportunities for pharmacological intervention. Trends Pharmacol Sci. 2014;35(9):479–488. doi: 10.1016/j.tips.2014.06.006. [DOI] [PubMed] [Google Scholar]
- 8.Ansieau S, Collin G, Hill L. EMT or EMT-Promoting Transcription Factors, Where to Focus the Light? Front Oncol. 2014;4:353. doi: 10.3389/fonc.2014.00353. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Tran PT, Shroff EH, Burns TF, Thiyagarajan S, Das ST, Zabuawala T, et al. Twist1 suppresses senescence programs and thereby accelerates and maintains mutant Kras-induced lung tumorigenesis. PLoS Genet. 2012;8(5):e1002650. doi: 10.1371/journal.pgen.1002650. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Burns TF, Dobromilskaya I, Murphy SC, Gajula RP, Thiyagarajan S, Chatley SN, et al. Inhibition of TWIST1 leads to activation of oncogene-induced senescence in oncogene-driven non-small cell lung cancer. Mol Cancer Res. 2013;11(4):329–338. doi: 10.1158/1541-7786.MCR-12-0456. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Ansieau S, Bastid J, Doreau A, Morel AP, Bouchet BP, Thomas C, et al. Induction of EMT by twist proteins as a collateral effect of tumor-promoting inactivation of premature senescence. Cancer cell. 2008;14(1):79–89. doi: 10.1016/j.ccr.2008.06.005. [DOI] [PubMed] [Google Scholar]
- 12.Cheng GZ, Chan J, Wang Q, Zhang W, Sun CD, Wang LH. Twist transcriptionally up-regulates AKT2 in breast cancer cells leading to increased migration, invasion, and resistance to paclitaxel. Cancer Res. 2007;67(5):1979–1987. doi: 10.1158/0008-5472.CAN-06-1479. [DOI] [PubMed] [Google Scholar]
- 13.Cheng GZZW, Sun M, Wang Q, Coppola D, Mansour M, et al. Twist is transcriptionally induced by activation of STAT3 and mediates STAT3 oncogenic function. J Biol Chem. 2008;(283):14665–14673. doi: 10.1074/jbc.M707429200. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Pham CG, Bubici C, Zazzeroni F, Knabb JR, Papa S, Kuntzen C, et al. Upregulation of Twist-1 by NF-kappaB blocks cytotoxicity induced by chemotherapeutic drugs. Molecular and cellular biology. 2007;27(11):3920–3935. doi: 10.1128/MCB.01219-06. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Yang J, Mani SA, Donaher JL, Ramaswamy S, Itzykson RA, Come C, et al. Twist, a master regulator of morphogenesis, plays an essential role in tumor metastasis. Cell. 2004;117(7):927–939. doi: 10.1016/j.cell.2004.06.006. [DOI] [PubMed] [Google Scholar]
- 16.Yang MH, Wu MZ, Chiou SH, Chen PM, Chang SY, Liu CJ, et al. Direct regulation of TWIST by HIF-1alpha promotes metastasis. Nat Cell Biol. 2008;10(3):295–305. doi: 10.1038/ncb1691. [DOI] [PubMed] [Google Scholar]
- 17.Zhuo WL, Wang Y, Zhuo XL, Zhang YS, Chen ZT. Short interfering RNA directed against TWIST, a novel zinc finger transcription factor, increases A549 cell sensitivity to cisplatin via MAPK/mitochondrial pathway. Biochemical and biophysical research communications. 2008;369(4):1098–1102. doi: 10.1016/j.bbrc.2008.02.143. [DOI] [PubMed] [Google Scholar]
- 18.Gajula RP, Chettiar ST, Williams RD, Nugent K, Kato Y, Wang H, et al. Structure-function studies of the bHLH phosphorylation domain of TWIST1 in prostate cancer cells. Neoplasia. 2015;17(1):16–31. doi: 10.1016/j.neo.2014.10.009. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Lamb JCE, Peck D, Modell JW, Blat IC, Wrobel MJ, et al. The Connectivity Map: using gene-expression signatures to connect small molecules, genes, and disease. Science. 2006;313(5795):1929–1935. doi: 10.1126/science.1132939. [DOI] [PubMed] [Google Scholar]
- 20.Shamir ER, Pappalardo E, Jorgens DM, Coutinho K, Tsai WT, Aziz K, et al. Twist1-induced dissemination preserves epithelial identity and requires E-cadherin. J Cell Biol. 2014;204(5):839–856. doi: 10.1083/jcb.201306088. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Moffat J, Grueneberg DA, Yang X, Kim SY, Kloepfer AM, Hinkle G, et al. A lentiviral RNAi library for human and mouse genes applied to an arrayed viral high-content screen. Cell. 2006;124(6):1283–1298. doi: 10.1016/j.cell.2006.01.040. [DOI] [PubMed] [Google Scholar]
- 22.Tran PT, Bendapudi PK, Lin HJ, Choi P, Koh S, Chen J, et al. Survival and Death Signals Can Predict Tumor Response to Therapy After Oncogene Inactivation. Science Translational Medicine. 2011;3(103):103ra199–103ra199. doi: 10.1126/scitranslmed.3002018. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Hagenbuchner JAM. Targeting transcription factors by small compounds--Current strategies and future implications. Biochem Pharmacol. 2016;107:1–13. doi: 10.1016/j.bcp.2015.12.006. [DOI] [PubMed] [Google Scholar]
- 24.Cao R, Fan W, Guo L, Ma Q, Zhang G, Li J, et al. Synthesis and structure-activity relationships of harmine derivatives as potential antitumor agents. Eur J Med Chem. 2013;60:135–143. doi: 10.1016/j.ejmech.2012.11.045. [DOI] [PubMed] [Google Scholar]
- 25.Chen Q, Chao R, Chen H, Hou X, Yan H, Zhou S, et al. Antitumor and neurotoxic effects of novel harmine derivatives and structure-activity relationship analysis. Int J Cancer. 2005;114(5):675–682. doi: 10.1002/ijc.20703. [DOI] [PubMed] [Google Scholar]
- 26.Ishida J, Wang HK, Bastow KF, Hu CQ, Lee KH. Antitumor agents 201. Cytotoxicity of harmine and beta-carboline analogs. Bioorg Med Chem Lett. 1999;9(23):3319–3324. doi: 10.1016/s0960-894x(99)00598-3. [DOI] [PubMed] [Google Scholar]
- 27.Pallier K, Cessot A, Cote JF, Just PA, Cazes A, Fabre E, et al. TWIST1 a new determinant of epithelial to mesenchymal transition in EGFR mutated lung adenocarcinoma. PLoS One. 2012;7(1):e29954. doi: 10.1371/journal.pone.0029954. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Kitai H, Ebi H, Tomida S, Floros KV, Kotani H, Adachi Y, et al. Epithelial-to-Mesenchymal Transition Defines Feedback Activation of Receptor Tyrosine Kinase Signaling Induced by MEK Inhibition in KRAS-Mutant Lung Cancer. Cancer Discov. 2016;6(7):754–769. doi: 10.1158/2159-8290.CD-15-1377. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Matsubara D, Ishikawa S, Oguni S, Aburatani H, Fukayama M, Niki T. Molecular predictors of sensitivity to the MET inhibitor PHA665752 in lung carcinoma cells. J Thorac Oncol. 2010;5(9):1317–1324. doi: 10.1097/JTO.0b013e3181e2a409. [DOI] [PubMed] [Google Scholar]
- 30.Demontis S, Rigo C, Piccinin S, Mizzau M, Sonego M, Fabris M, et al. Twist is substrate for caspase cleavage and proteasome-mediated degradation. Cell Death Differ. 2006;13(2):335–345. doi: 10.1038/sj.cdd.4401744. [DOI] [PubMed] [Google Scholar]
- 31.Hayashi M, Nimura K, Kashiwagi K, Harada T, Takaoka K, Kato H, et al. Comparative roles of Twist-1 and Id1 in transcriptional regulation by BMP signaling. J Cell Sci. 2007;120(Pt 8):1350–1357. doi: 10.1242/jcs.000067. [DOI] [PubMed] [Google Scholar]
- 32.Su YW, Xie TX, Sano D, Myers JN. IL-6 stabilizes Twist and enhances tumor cell motility in head and neck cancer cells through activation of casein kinase 2. PLoS One. 2011;6(4):e19412. doi: 10.1371/journal.pone.0019412. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Weiss MB, Abel EV, Mayberry MM, Basile KJ, Berger AC, Aplin AE. TWIST1 Is an ERK1/2 Effector That Promotes Invasion and Regulates MMP-1 Expression in Human Melanoma Cells. Cancer research. 2012;72(24):6382–6392. doi: 10.1158/0008-5472.CAN-12-1033. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Shiota M, Izumi H, Onitsuka T, Miyamoto N, Kashiwagi E, Kidani A, et al. Twist and p53 reciprocally regulate target genes via direct interaction. Oncogene. 2008;27(42):5543–5553. doi: 10.1038/onc.2008.176. [DOI] [PubMed] [Google Scholar]
- 35.Takahashi E, Funato N, Higashihori N, Hata Y, Gridley T, Nakamura M. Snail regulates p21(WAF/CIP1) expression in cooperation with E2A and Twist. Biochemical and biophysical research communications. 2004;325(4):1136–1144. doi: 10.1016/j.bbrc.2004.10.148. [DOI] [PubMed] [Google Scholar]
- 36.Cao R, Chen Q, Hou X, Chen H, Guan H, Ma Y, et al. Synthesis, acute toxicities, and antitumor effects of novel 9-substituted beta-carboline derivatives. Bioorg Med Chem. 2004;12(17):4613–4623. doi: 10.1016/j.bmc.2004.06.038. [DOI] [PubMed] [Google Scholar]
- 37.Franco HL, Casasnovas J, Rodriguez-Medina JR, Cadilla CL. Redundant or separate entities?--roles of Twist1 and Twist2 as molecular switches during gene transcription. Nucleic Acids Res. 2011;39(4):1177–1186. doi: 10.1093/nar/gkq890. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Jacqueroud L, Bouard C, Richard G, Payen L, Devouassoux-Shisheboran M, Spicer DB, et al. The Heterodimeric TWIST1-E12 Complex Drives the Oncogenic Potential of TWIST1 in Human Mammary Epithelial Cells. Neoplasia. 2016;18(5):317–327. doi: 10.1016/j.neo.2016.03.007. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Chang AT, Liu Y, Ayyanathan K, Benner C, Jiang Y, Prokop JW, et al. An evolutionarily conserved DNA architecture determines target specificity of the TWIST family bHLH transcription factors. Genes Dev. 2015;29(6):603–616. doi: 10.1101/gad.242842.114. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Casas E, Kim J, Bendesky A, Ohno-Machado L, Wolfe CJ, Yang J. Snail2 is an essential mediator of Twist1-induced epithelial mesenchymal transition and metastasis. Cancer research. 2011;71(1):245–254. doi: 10.1158/0008-5472.CAN-10-2330. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Shiota M, Izumi H, Onitsuka T, Miyamoto N, Kashiwagi E, Kidani A, et al. Twist promotes tumor cell growth through YB-1 expression. Cancer research. 2008;68(1):98–105. doi: 10.1158/0008-5472.CAN-07-2981. [DOI] [PubMed] [Google Scholar]
- 42.Castanon I, Von Stetina S, Kass J, Baylies MK. Dimerization partners determine the activity of the Twist bHLH protein during Drosophila mesoderm development. Development (Cambridge, England) 2001;128(16):3145–3159. doi: 10.1242/dev.128.16.3145. [DOI] [PubMed] [Google Scholar]
- 43.Firulli BA, Redick BA, Conway SJ, Firulli AB. Mutations within helix I of Twist1 result in distinct limb defects and variation of DNA binding affinities. J Biol Chem. 2007;282(37):27536–27546. doi: 10.1074/jbc.M702613200. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Zhang XF, Sun RQ, Jia YF, Chen Q, Tu RF, Li KK, et al. Synthesis and mechanisms of action of novel harmine derivatives as potential antitumor agents. Sci Rep. 2016;6:33204. doi: 10.1038/srep33204. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Chen YT, Akinwunmi PO, Deng JM, Tam OH, Behringer RR. Generation of a Twist1 conditional null allele in the mouse. Genesis. 2007;45(9):588–592. doi: 10.1002/dvg.20332. [DOI] [PubMed] [Google Scholar]
- 46.Pan DFM, Lopes A, Wang YX. Twist-1 is a PPARdelta-inducible, negative-feedback regulator of PGC-1alpha in brown fat metabolism. Cell. 2009;137:73–86. doi: 10.1016/j.cell.2009.01.051. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.Xue G, Restuccia DF, Lan Q, Hynx D, Dirnhofer S, Hess D, et al. Akt/PKB-mediated phosphorylation of Twist1 promotes tumor metastasis via mediating cross-talk between PI3K/Akt and TGF-beta signaling axes. Cancer Discov. 2012;2(3):248–259. doi: 10.1158/2159-8290.CD-11-0270. [DOI] [PubMed] [Google Scholar]
- 48.Jin HO, Hong SE, Woo SH, Lee JH, Choe TB, Kim EK, et al. Silencing of Twist1 sensitizes NSCLC cells to cisplatin via AMPK-activated mTOR inhibition. Cell Death Dis. 2012;3:e319. doi: 10.1038/cddis.2012.63. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49.Ansieau S, Morel AP, Hinkal G, Bastid J, Puisieux A. TWISTing an embryonic transcription factor into an oncoprotein. Oncogene. 2010;29(22):3173–3184. doi: 10.1038/onc.2010.92. [DOI] [PubMed] [Google Scholar]
- 50.Xu Y, Qin L, Sun T, Wu H, He T, Yang Z, et al. Twist1 promotes breast cancer invasion and metastasis by silencing Foxa1 expression. Oncogene. 2016 doi: 10.1038/onc.2016.286. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 51.Yang WH, Su YH, Hsu WH, Wang CC, Arbiser JL, Yang MH. Imipramine blue halts head and neck cancer invasion through promoting F-box and leucine-rich repeat protein 14-mediated Twist1 degradation. Oncogene. 2016;35(18):2287–2298. doi: 10.1038/onc.2015.291. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.