Abstract
Deregulation of the Wnt/β-catenin signaling pathway drives the development of colorectal cancer (CRC) but understanding of this pathway remains incomplete. Here we report that the damage-specific DNA-binding protein DDB2 is critical for β-catenin-mediated activation of RNF43, which restricts Wnt-signaling by removing Wnt receptors from the cell surface. Reduced expression of DDB2 and RNF43 was observed in human hyperplastic colonic foci. DDB2 recruited EZH2 and β-catenin at an upstream site in the Rnf43 gene, enabling functional interaction with distant TCF4/β-catenin binding sites in the intron of Rnf43. This novel activity of DDB2 was required for RNF43 function as a negative feedback regulator of Wnt-signaling. Mice genetically deficient in DDB2 exhibited increased susceptibility to colon tumor development in a manner associated with higher abundance of the Wnt receptor-expressing cells and greater activation of the downstream Wnt-pathway. Our results identify DDB2 as both a partner and regulator of Wnt-signaling with an important role in suppressing colon cancer development.
Keywords: Colon Cancer, Wnt/ β-catenin Signaling, DDB2, RNF43
Introduction
Colon cancer is driven by activating mutations in the Wnt/β-catenin signaling pathway. Over 80% of the colon cancer patients harbor mutations in APC that regulates the cellular level and activity of β-catenin (1,2). Moreover, there is evidence for activating mutations in β-catenin gene as well as inactivating mutation in the Axin gene (3). Despite these genetic lesions that cause cell-autonomous and intracellular stimulation of Wnt/β-catenin signaling, activation of the pathway through Frizzled (FZD) seven-transmembrane Wnt-receptors is also important in colon cancer. Blocking Wnt ligand secretion reduces β-catenin activity in colon cancer cells harboring APC truncating or β-catenin-stabilizing mutations (4,5). Blocking Wnt ligand secretion was also recently shown to be effective in treating colon cancer patient-derived xenografts (6). The availability of FZD receptors to bind the Wnt ligands is controlled by the so-called R-spondin/Lgr/RNF43 module (7), within which RNF43 is a ubiquitin ligase for the FZD receptor and R-spondin/Lgr block RNF43 inhibition of FZD (8,9). The frequent occurrence Rnf43 inactivating mutation (10) and R-spondin fusions (11) in colon cancer indicate the importance of the Wnt receptor activation for development and progression of the disease.
DDB2, encoded by the XPE gene, a p53-activated gene (12), participates in the early steps of nuclear excision repair (NER). DDB2 also possesses transcriptional regulatory activity. It represses expression of the antioxidant gene MnSOD (13). DDB2 also represses NEDD4L, an E3 ligase for Smad2/Smad3, in ovarian cancer cells to enhance TGF-β signaling (14). DDB2 also possesses transcriptional stimulatory activity. It binds to the promoter of the NFKBIA gene to activate its expression and regulate NF-kB activity, and that has been linked to regulation of invasiveness of breast cancer cells (15). That study also identified a DNA-element (TCCCCTTA) specifically recognized by DDB2 in the NFKBIA gene (14). The same DDB2-cognate element is found in NEDD4L that is repressed by DDB2. We observed that DDB2 stimulates expression of SEMA3A in colon cancer cells, and that involves an interaction with the XRCC5/6 proteins (16).
Previously, we showed that DDB2 plays a significant role in suppressing colon cancer metastasis. The protein level of DDB2 decreases in metastatic colon cancer, and that coincides also with loss of cell surface E-cadherin expression (17). Depletion of DDB2 in an orthotopic mouse model for colon cancer causes a strong increase in metastasis of the colon cancer cells to the liver. Reciprocally, expression of DDB2 suppresses metastatic potential of colon cancer cells harboring mutations in the Wnt-pathway (17). The metastasis suppressor function of DDB2, at least partly, correlates with suppression of epithelial to mesenchymal (EMT)-like changes of the colon cancer cells (17). For example, expression of DDB2 in SW620 cells, mesenchymal type, changes the morphology to epithelial type. Conversely, depletion of DDB2 in SW480, epithelial type, changes the morphology of those cells to mesenchymal-like. Here we provide evidence that DDB2 also plays a critical role in suppressing development of colon cancer at early stage by regulating Wnt-signaling.
Methods and Material
Plasmid, siRNA and Antibodies
pCDNA-V5-FZD5 was a kind gift from Dr. Hans Clevers. TCF4-DN in pLX303 was a gift from Dr. William Hahn (Addgene plasmid # 42592). Two distinct DsiRNA-DDB2, two DsiRNA-EZH2 and non-targeting DsiRNA-Control were purchased from IDT DNA pre-designed library. The sequences are listed in supplementary table S1. ON-TARGET plus PAF siRNA smartpool and non-targeting siRNA smartpool were purchased from Dharmacon. Antibodies used in this study include DDB2 (Western Blotting: 5416, Cell Signaling; Chromatin Immunoprecipitation: sc-25368, Santa Cruz; co-Immunoprecipitation: ab181136, Abcam), EZH2 (Western Blotting: 5246, Cell Signaling; Chromatin Immunoprecipitation: 39875, Active Motif), β-catenin (8480, Cell Signaling), RNF43 (orb140091, Biorbyt), EED (sc-28701, Santa Cruz), SUZ12 (3737, Cell Signaling), TCF4 (2569, Cell Signaling), V5 tag (R960-25, Invitrogen), T7 tag (69522, EMD Millipore), α-Tubulin (T9026, Sigma-Aldrich) and LRP5/6 (bs-2905R, BIOSS). HRP-conjugated secondary antibodies were purchased from BioRad.
Cell culture, Transfection and Stable Cell Lines
All cell lines are from ATCC. They were routinely authenticated based on morphology and growth characteristics. HCT116 cells and HT-29 cells were grown in DMEM medium containing 10% Fetal Bovine Serum and 1% Penicillin-Streptomycin at 37°C and 5% CO2. SW480 and SW620 were grown in RPMI medium containing 10% Fetal Bovine Serum and 1% Penicillin-Streptomycin at 37°C and 5% CO2. Cell transfection experiments were conducted using Lipofectamine2000 (Thermo Fisher Scientific) and following manufacturer’s recommendations. For transient expression of plasmids, cells were harvested 36hrs after transfection. For transient knockdown, cells were harvested 72h after siRNA transfection. Two distinct siRNAs targeting different regions of the gene were used to eliminate off-target effect. HCT116 cells stably expressing shRNA targeting DDB2 were described before (17). HCT116 cells stably expressing FLAG-HA-tagged DDB2 were established using a retroviral vector (16). And HCT116 cells stably expressing dn-TCF4 were established using a lentiviral vector and selected by Blasticidin.
CHIR99021, Deoxycholic Acid (DCA) and Recombinant Wnt3A Treatment
HCT 116 cells were treated with CHIR99021 (Stemgent) at 3.3µM for 16 hours in serum-free medium. HT-29 cells were treated with CHIR99021 at 6.7µM for 24 hours in serum-free medium. And HCT 116 cells and HT-29 cells were treated with DCA (Sigma-Aldrich) at 10µM for 2hours in serum-free medium. HT-29 cells were treated with recombinant human Wnt3A protein (R&D system) at 300ng/ml for 24 hours after 24 hours serum starvation.
Chromatin Immunoprecipitation
Chromatin Immunoprecipitation experiments were performed as described before (17). Anti-DDB2, anti-T7 tag, anti-EZH2, anti-β-Catenin and anti-TCF4 antibodies were used for immunoprecipitation. De-crosslinked chromatin was analyzed by qPCR using the primers described in supplementary table S2. Rnf43 Intron2-A, Intron2-B and c-Myc primers were described before (18). Axin2 Intron1 primers were purchased from Cell Signaling.
Gene Editing with CRISPR/Cas9 System
The deletion of target sequences in HT-29 cells was performed by the Genome Editing Core, University of Illinois at Chicago. Briefly, HT-29 cells were transfected with plasmids encoding Cas9 and guide RNAs (sgRNAs) that target the 132bps (Short Del) and 416bps (Long Del) fragments respectively. Clones were screened by PCR, and then confirmed by Sanger genomic DNA sequencing.
RNA Extraction and qPCR
Total RNA was extracted using TRIzol (Thermo Fisher Scientific) following manufacturer’s instructions. cDNA was synthesized using iScript cDNA Synthesis Kit (BioRad) and quantitative PCR was performed using iTaq universal SYBR Green supermix (BioRad) and a CFX96 system (BioRad). Oligonucleotides used as primers are described in supplementary table S3.
Ubiquitination Assay
HT-29 cells were transfected with V5-tagged FZD5 vectors and His-tagged ubiquitin, as indicated, and either siRNA-Control or siRNA-DDB2. Also the parental, short del and long del HT-29 cells were used in the assay. The major procedure and material were described before (9). Briefly, the HT-29 cells were washed and collected in ice-cold PBS with N-ethylmaleimide (Sigma). Then the cells were lysed in lysis buffer consist of 10 mM Tris–HCl, 100 mM phosphate buffer, 6 M guanidinium pH 8.0, 20 mM imidazole, and 10 mM β-mercaptoethanol, followed by sonication. Then the lysates were centrifuged, and the supernatants were incubated with nickel-nitrilotriacetic acid–agarose beads (Qiagen) for 3 hours at room temperature. Beads were then washed with lysis buffer with 0.2% Triton X-100, followed by sequential washes with buffers 1, 2 and 3 (Buffer 1: 10 mM Tris–HCl pH 8.0, 8 M urea, 100 mM phosphate buffer pH 8.0, 10 mM β-mercaptoethanol and 0.2% Triton X-100; Buffer 2: 10 mM Tris–HCl pH 6.3, 8 M urea, 100 mM phosphate buffer pH 6.3, 10 mM β-mercaptoethanol and 0.2% Triton X-100; Buffer 3: 10 mM Tris–HCl pH 6.3, 8 M urea, 100 mM phosphate buffer pH 6.3, 10 mM β-mercaptoethanol and 0.1% Triton X-100). After washes, the His-tagged proteins were eluted in two beads volumes of elution buffer (10 mM Tris–HCl pH 7.0, 6.2 M urea, 100 mM phosphate buffer and 200 mM imidazole and 1x Laemmli buffer). For western blotting, anti-V5, anti-DDB2, anti-RNF43and anti-α-Tubulin antibodies were used.
Animals and Tissues
All mouse studies were performed following the guideline of the University of Illinois Animal Care Committee. The DDB2 whole body knock-out (DDB−/−) mouse line was described previously (19). In the AOM/DSS colon carcinogenesis model, 6 weeks old male DDB2+/+ and DDB2−/− mice were given a single I.P. injection of AOM (10 mg/kg). One week after AOM injection, animals received 2% DSS in their drinking water for 7 days. 12 DDB2+/+ mice and 14 DDB2−/− mice were sacrificed at 15 weeks. For immunohistochemistry, colorectal tissues were fixed in 10% buffered formalin and embedded in paraffin. RNA extracts were prepared from colorectal tissues using TRIzol and RNeasy Mini Kit (QIAGEN).
Immunohistochemistry
Immunohistochemistry of paraffin-embedded tissue samples was performed using Avidin/Biotin Blocking Kit, Vectastain ABC Kit and DAB Substrate Kit (Vector Laboratories) according to manufacturer’s instructions. DDB2 antibody was diluted 1:250 for human samples; RNF43 antibody was diluted 1:200 for mouse samples and 1:50 for human samples; LRP5/6 antibody was diluted 1:300 in 3% goat serum in TBS buffer. Nuclei were counter stained with hematoxylin. Negative controls were performed with normal rabbit IgG.
Human Colorectal specimen
All the human samples for tissue arrays were collected following University of Illinois guidelines and NIH policy on human sample studies. The tissue microarrays (TMA) were prepared by the Research Histology Core in UIC. The adequacy, quality, and structural integrity of the paraffin-embedded tissue blocks were assessed and verified prior to inclusion in the study. Colon resection cases provided sample tissue for construction of tissue arrays. The tissue arrays were designed and constructed based on prescribed practices. Hematoxylin and eosin-stained adjacent tissue sections were reviewed and used to identify areas of normal, hyperplastic, dysplastic and carcinomatous colonic tissue. Cores from archived normal colon specimens provided non-disease controls. Donor core tissues 0.6 mm diameter were obtained from corresponding regions in the donor paraffin block and embedded in a recipient block. A duplicate donor core tissue from each lesion was sampled across a patient’s colonic resection. The donor cores from all subjects were embedded in a tissue microarray (TMA) recipient block. Each of the tissue microarray cores was histologically examined to document and record tissue-sampling adequacy and diagnoses. All tissue samples were de-identified, containing no patient demographic information. Total 24 patients were analyzed by immunohistochemistry.
Statistical analyses
All statistical analyses were performed using GraphPad Prism 6 software. Unpaired, two-tailed Student t-tests and Mann-Whitney U-tests were used to analyze statistical significance. SD was indicated in the figure, and sample volume was indicated in the figure legends. Two-tailed, paired t-tests were used to analyze human TMA staining positivity. And the Spearman rank coefficient was used as a statistical measure of correlation between DDB2 expression and RNF43 expression in each patient. Unless otherwise indicated, all the experiments were performed at least three times.
Results
DDB2 activates expression of Wnt-receptor regulator Rnf43
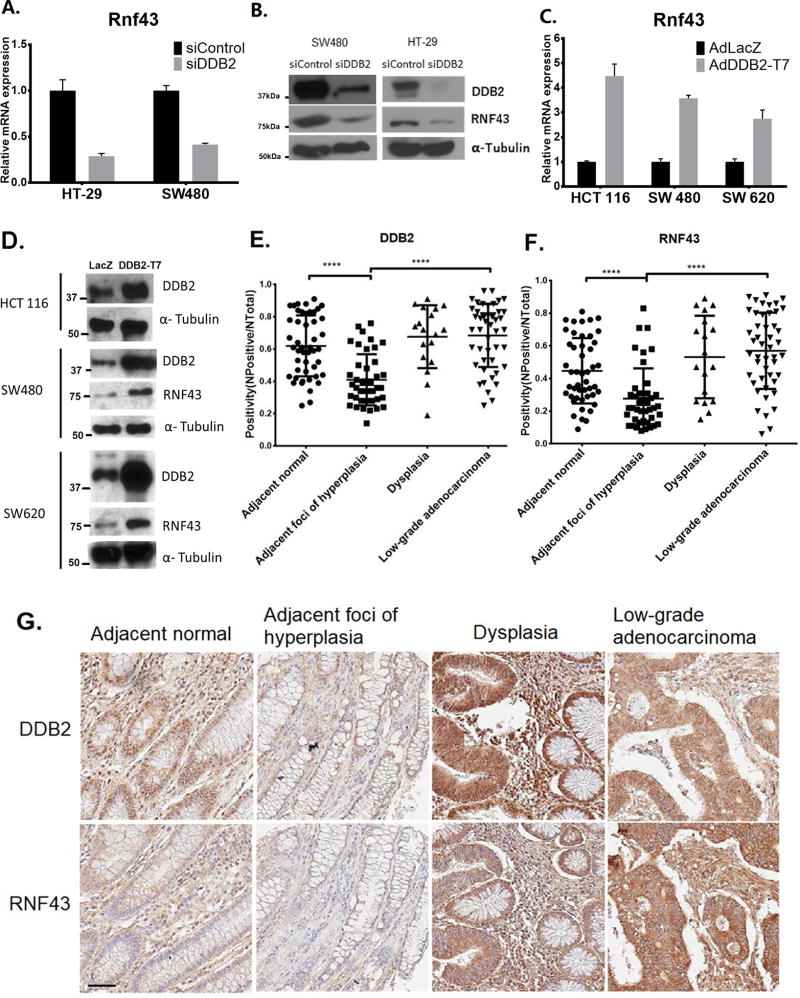
The transcriptional regulatory function of DDB2 plays significant tumor suppressor roles in colon cancer metastasis. We showed that depletion of DDB2 causes colon cancer cells to undergo epithelial to mesenchymal transition along with an increase in their metastatic potential (17). Since the Wnt/β-catenin pathway is important in the development and progression of colon cancer, we sought to investigate whether the regulatory mechanism of DDB2 interacts with the Wnt/β-catenin signaling pathway. We analyzed the Wnt/β-catenin pathway genes, and discovered a role of DDB2 in the activation of Rnf43 that regulates Wnt-signaling by removing the receptors from cell surface. We measured the mRNA expression level of Rnf43 with or without DDB2 shRNA or siRNA in several colon cancer cell lines. In SW480, HT-29 and HCT116 cells, silencing of DDB2 resulted in a loss of the Rnf43 transcript (Fig. 1A and supplemental Fig. S1A). HCT116 cells express a frame-shift mutant Rnf43-mRNA, and the mutant protein was not detected by the antibody (supplemental Fig. S1B). The protein levels of RNF43 in HT-29 and SW480 were reduced upon treatment with siRNA-DDB2 (Fig. 1B). Reciprocally, expression of DDB2 resulted in an increase in the level of Rnf43-mRNA. For example, HCT116, SW480 and SW620 cells were infected with adenovirus expressing DDB2 or LacZ. Quantitative RT-PCR assays of the RNAs from the infected samples showed that the Rnf43 mRNA level was increased in cells infected with adenovirus expressing DDB2 (Fig. 1C). Consistently, the protein level of RNF43 also increased in DDB2 overexpressing cells (Fig. 1D). These results show that DDB2 is required for expression of Rnf43 in colon cancer cells.
Figure 1. Expression of Rnf43 in colon cancer cells and in human colonic tissues correlates with DDB2.
(A) SW480 cells and HT-29 cells were transient transfected with siRNAs (siControl or siDDB2). Total RNA was analyzed using qRT-PCR for the mRNA level of Rnf43 N=3, Error bars indicate SD. (B) Total levels of DDB2, RNF43 and α-Tubulin (Loading Control) proteins were analyzed by Western Blotting. (C and D) The indicated cell lines were infected with adenovirus expressing either LacZ or T7 tagged DDB2 (T7-DDB2). qRT-PCR (C) was performed to analyze relative mRNA level of Rnf43expression (Normalized by gapdh). N=3, Error bars indicate SD. Western blotting (D) was performed to analyze the protein levels of DDB2, RNF43 and α-Tubulin. (E and F) Immunohistochemical staining of RNF43 and DDB2 in adjacent normal, adjacent foci of hyperplasia, dysplasia and low-grade adenocarcinoma samples were quantified as positivity (=Number of Positive/Number of Total). Positivity of DDB2 (E) and RNF43 (F) were plotted. N of Adjacent normal = 47, N of Adjacent foci of hyperplasia= 42, N of Dysplasia = 18, N of Low-grade adenocarcinoma = 47. **** P < 0.0001 (G) Representative images of DDB2 expression and RNF43 expression from same patient. Scale bar =50 µm.
Rnf43 is also a Wnt/β-catenin signaling target gene, and is activated by β-catenin (9). We sought to investigate whether the Wnt/β-catenin mediated activation of Rnf43 also involves DDB2. We employed CHIR99021, a GSK3 inhibitor, to activate β-catenin in colon cancer cells in the presence and absence of DDB2-depletion. HCT116 (supplemental Fig. S1C) and HT-29 cells (supplemental Fig. S1D) were treated with CHIR99021 for 16 hours and 24 hours respectively. Quantitative RT-PCR was performed on total RNAs isolated from the CHIR99021 or vehicle treated cells to measure the mRNA level of DDB2, Rnf43 and Axin2. Interestingly, CHIR99021 stimulated Rnf43 mRNA expression in control cells but not in the cells with silenced DDB2. However Axin2, as another Wnt/β-catenin target gene, is stimulated in both control cells and DDB2 silenced cells following CHIR99021 treatment. Moreover, deoxycholic acid (DCA) that was shown to activate downstream Wnt-signaling (20) exhibited similar effects on Rnf43 and Axin2 expression (supplemental Fig. S1E and F). These results suggest that DDB2 is required for the activation of Rnf43, but not Axin2, by Wnt/β-catenin signaling.
Reduced expression of RNF43 in human colonic hyperplasia correlates with reduced expression of DDB2
DDB2 is a p53-activated gene, and its expression in colon cancer is down regulated in high-grade metastatic colon cancer (17). However, deregulation of Wnt-signaling is one of the earliest events during colon cancer development. Moreover, in our mouse studies, we observed increased susceptibility of the DDB2-deficient mice in colon tumor development. Therefore, we analyzed human colonic specimens for expression of DDB2 and RNF43 at the early stages of tumor development. We analyzed tissue microarray samples at different stages from 24 patients who eventually developed adenocarcinoma. Immunohistochemical staining of the samples indicated down regulation of both DDB2 and RNF43 expression in the hyperplastic foci in a high proportion of the samples (Fig.1E–G). To examine whether RNF43 and DDB2 expressions are correlated in tissue microarray, we performed Spearman correlation analysis for DDB2 and RNF43 expression. Samples from each patient were analyzed separately, to eliminate various backgrounds among different patients (supplemental table S4). 16 out of 24 patients showed significantly correlated expressions of DDB2 and RNF43. The reduced expression of RNF43 in the early stage of foci development is interesting because often it is at that stage Wnt-signaling begins to get deregulated. We did not detect reductions in the low-grade adenocarcinoma samples. However, in Bittner Colon datasets (Oncomine), there were significant reductions of Rnf43 in the high-grade samples (supplemental Fig. S2) and that would be consistent with our previous observation on loss of DDB2 expression in high-grade samples (17).
Association of DDB2 with the upstream regulatory region in Rnf43 is critical for its activation by a GSK3-inhibitor
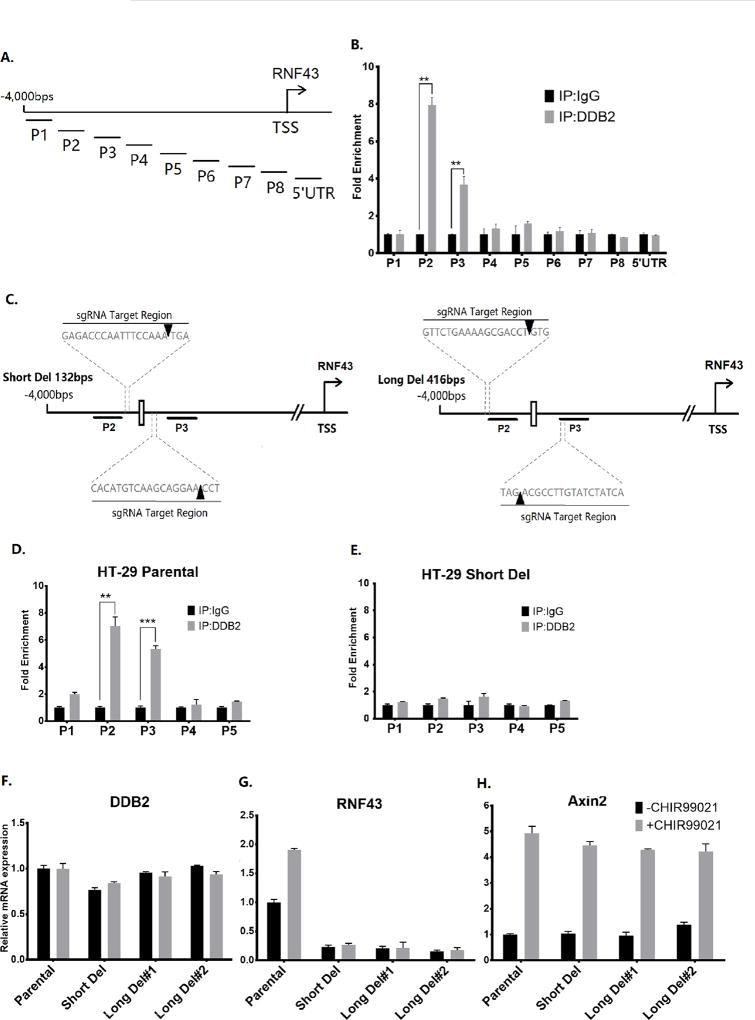
DDB2 associates with the promoter region of its target genes, including SOD2, NFKBIA and NEDD4L (13–15). To study whether DDB2 also associates with the Rnf43 promoter, Chromatin immunoprecipitation (ChIP) assays were conducted in HCT116 cells. Nine pairs of primers were designed to cover 4,000 bps upstream of Rnf43 transcription start site, and 500 bps in 5’UTR. The ChIP experiment results demonstrated interactions of DDB2 with two consecutive chromatin fragments, P2 and P3 (Figs. 2A and B). Interaction with the P2 region was confirmed also in cells expressing T7-tagged DDB2. Chromatin-IP with T7-antibody specifically showed interaction in cells expressing T7-DDB2, but not in cells expressing LacZ (supplemental Fig. S3). DDB2-antibody, on the other hand, showed interactions in both LacZ and T7-DDB2 expressing cells.
Figure 2. DDB2 binds to an upstream region in the Rnf43 gene and deletion of DDB2 binding region inhibits Rnf43 expression.
(A) The schematic representation of the human Rnf43upstream regulatory region. The thick truncated lines mark the regions covered by primer sets of interest. TSS: Transcription start site. (B) Chromatin-IP (ChIP) assays for binding of DDB2 with regulatory region upstream of the Rnf43gene. The ChIP assay was conducted to analyze the local enrichment of DDB2 across the upstream regulatory region and part of 5’ UTR of Rnf43gene in HCT 116 cells. The relative fold enrichment was quantified by normalization to input. And the fold enrichment of IgG is set as 1 for all primer sets. N = 5, Error bars indicate SD. **: P <0.01. (C) The schematic representation of the human Rnf43upstream regulatory region. The thick truncated lines mark the P2 and P3 regions. The white rectangle between P2 and P3 regions indicates the putative DDB2 binding element ‘TCCCCTAA’. Dash lines indicate location of the sequences targeted by sgRNAs used in CRISPR/Cas9 genome editing. The orange arrowheads indicate the precise region where the Cas9 Nuclease cuts the genomic sequence. (D and E) ChIP assay was performed to analyze the local enrichment of DDB2 in HT-29 Parental cells (D) and Short Del cells (E). The relative fold enrichment was quantified by normalization to input. And the fold enrichment of IgG is set as 1 for all primer sets. (F–H) Parental HT-29 cells, Short Del HT-29 cells and two clones of Long Del HT-29 cells were treated with DMSO or CHIR99021 (6.7µM, 24 hrs). The mRNA levels of DDB2 (F), Rnf43 (G) and Axin2 (H) were analyzed using qRT-PCR (Normalized by 18S rRNA). N = 3, Error bars indicate SD. **: P <0.01. ***: P <0.001
Interestingly, a putative DDB2 binding element TCCCCTAA, which is one nucleotide different from the ones found in NFKBIA (15) or NEDD4L (14), is located between the fragments P2 and P3. The available PAM motif in the region around that DDB2-cognate element and CRISPR/Cas9 genome editing technique allowed us to generate two HT-29 cell lines that harbor deletions of 132bp (between P2 and P3) and 416bp (partially overlapping with P2 and P3) (Figs. 2C). Both deletions removed the putative DDB2 binding element (clone selection data are included in supplemental Fig. S4). Loss of DDB2 interactions was confirmed by ChIP experiments. For example, ChIP experiment showed that DDB2 no longer associate with the P2 or P3 region in the 132bp-deleted HT-29 cells (Figs. 2D and E). Next, we analyzed CHIR99021-induced expression of Rnf43-mRNA in undeleted and deleted (both 132bp and 416bp) HT-29 cells. As expected from our DDB2 knockdown experiments, deletion of the DDB2-interaction site resulted in a loss of basal expression of Rnf43 in the HT-29 cells without impacting DDB2 expression (Figs. 2F, 2G, and Fig.S5). More interestingly, CHIR99021 treatment, which activated expression of Axin2, failed to activate Rnf43 expression in the deleted cells (Figs. 2G and H). That was observed in both 132bp and 416bp deleted HT-29 cells. These results indicate that DDB2 supports Wnt/β-catenin activation of the Rnf43 promoter by directly binding to the upstream region between P2 and P3.
DDB2 recruits β-catenin onto the upstream regulatory region in Rnf43 through an interaction with EZH2
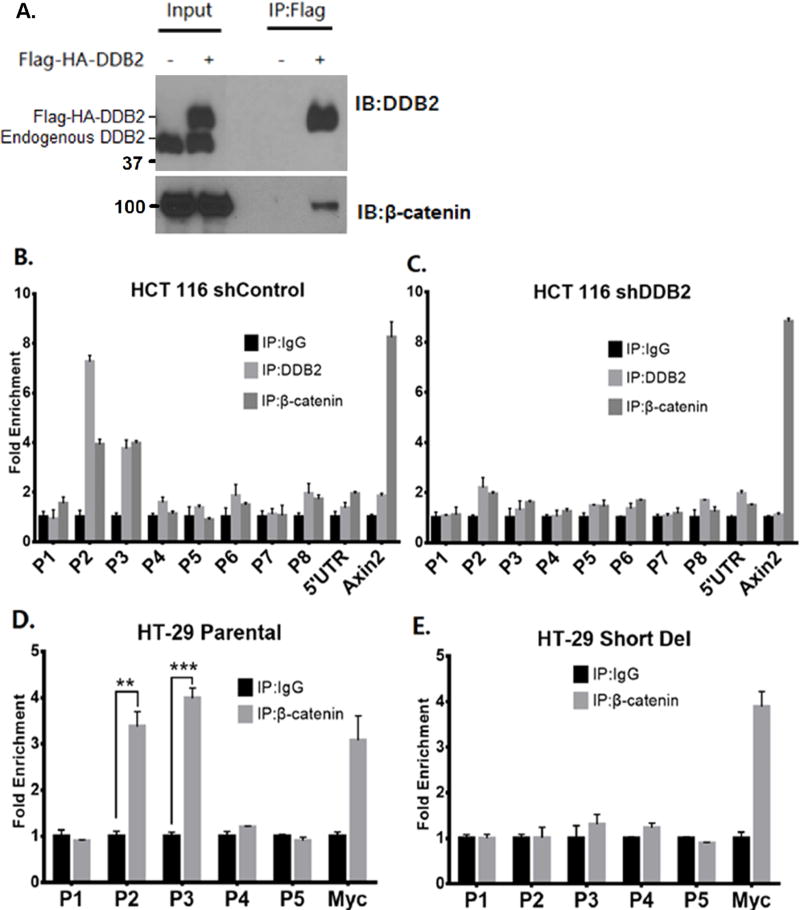
The 132bp region between P2 and P3 upstream chromatin fragments in the Rnf43 gene lacks any recognizable TCF-binding element. Moreover, a ChIP experiment failed to detect a TCF4 interaction in that upstream region (Supplemental Fig. S6). Interestingly, one study identified cis-regulatory elements bound by TCF4 in the introns of Rnf43 (18). We could confirm the interactions of TCF4 in those intronic regions by ChIP (Supplemental Fig. S6). However, those intronic cis-regulatory elements are insufficient to activate Rnf43, because the DDB2-depleted cells or cells lacking the 132bp upstream sequence failed to respond to GSK3–inhibitor or DCA. Therefore, we investigated the role of DDB2 in the mechanism by which Rnf43 responds to the GSK3–inhibitor CHIR99021, which functions by increasing nuclear β-catenin. We observed that DDB2 could associate with β-catenin. Immunoprecipitation of nuclear extracts from HCT116 cells that express Flag and HA tagged DDB2 with Flag-ab resulted in co-precipitation of β-catenin (Fig. 3A).
Figure 3. DDB2 recruits β-catenin onto Rnf43 upstream regulatory region.
(A) Nuclear extracts of HCT 116 cells expressing empty vector or Flag-HA-DDB2 were immunoprecipitated by ANTI-FLAG M2 affinity gel and analyzed by Western Blotting using DDB2 and total β-catenin antibodies. IP: Immunoprecipitation; IB: Immunoblotting. (B and C) ChIP assay was performed to analyze the local enrichment of DDB2 and β-catenin across the upstream regulatory region and part of 5’ UTR of Rnf43gene in HCT 116 cells expressing either shControl (B) or shDDB2 (C). The relative fold enrichment was quantified by normalization to input. And the fold enrichment of IgG is set as 1 for all primer sets. A primer set which amplifies the TCF/ β-catenin binding site on Axin2 intron1 (Cell Signaling) is used as positive control for β-catenin immunoprecipitation. (D and E) ChIP assay was performed to analyze the local enrichment of β-catenin from P1 to P5 regions of Rnf43gene in HT-29 Parental (D) and Short Del (E) cells. The relative fold enrichment was quantified by normalization to input. And the fold enrichment of IgG is set as 1 for all primer sets. A primer set which amplifies the TCF/ β-catenin binding site on c-Myc promoter is used as positive control. N = 3, Error bars indicate SD. **: P <0.01. ***: P <0.001
Next, we sought to determine whether β-catenin is recruited to the upstream region of Rnf43 by DDB2. A ChIP assay was performed in HCT 116 cells expressing control-shRNA or DDB2-shRNA. Clearly, there were enrichments of the P2 and P3 regions in the chromatin-IPs with antibodies against DDB2 and β-catenin when control-shRNA cells were used (Fig. 3B). However, in DDB2-shRNA cells, the bindings of both DDB2 and β-catenin were decreased in P2 and P3 regions (Fig. 3C), indicating that DDB2 recruits β-catenin onto the P2/P3 regions in the Rnf43 gene. A set of primers that amplifies the β-catenin binding region in the Axin2 promoter was used as a positive control, showing no effect of DDB2-depletion on β-catenin recruitment to the Axin2 promoter (Figs. 3B and C). Furthermore, a ChIP assay was performed in HT-29 parental cells and HT-29 cells harboring deletion in the upstream DDB2-binding region with β-catenin antibody. Consistent with the DDB2/β-catenin interaction, enrichments of the P2 and P3 regions with β-catenin antibody were observed in the parental HT-29 cells, but not in the DDB2-binding site deleted cells (Figs. 3D and E). Together, these results show that DDB2 recruits β-catenin onto the upstream P2/P3 regions in the Rnf43 gene.
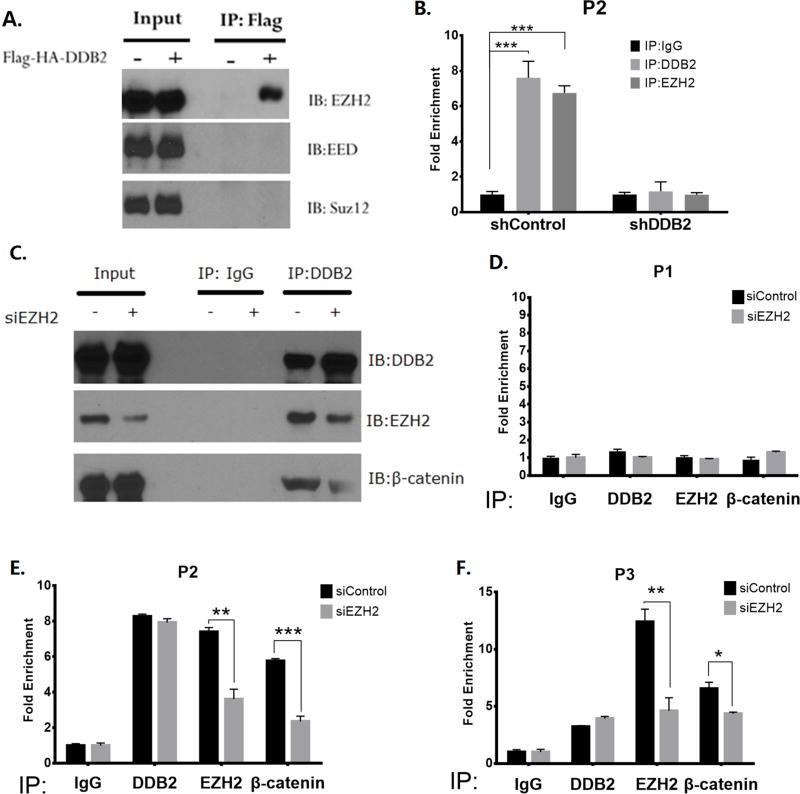
A previous study provided evidence for an interaction between β-catenin and EZH2, involving the PCNA-associated factor PAF (21). Moreover, a recent study demonstrated an interaction between DDB2 and EZH2 through the WD40 domain of DDB2 (14). Intriguingly, a publicly available ChIP-seq dataset (UCSC Genome Browser) indicated interaction of EZH2 with the upstream sequences in the Rnf43 gene around -2,900bp, which is the region between P2 and P3. Therefore, we tested whether EZH2 is a part of the DDB2 transcriptional complex. A co-immunoprecipitation experiment in HCT116 cells showed that DDB2 interacts with EZH2, but not with the other PRC2 components EED and Suz12 (Fig. 4A). Next, we conducted ChIP assay to test whether EZH2 is recruited by DDB2 onto the upstream region of Rnf43. The P2 region was tested in HCT116 cells with control-shRNA and DDB2-shRNA. As the shown in Fig 4B, the enrichments of both DDB2 and EZH2 decrease significantly upon depletion of DDB2, indicating that EZH2 is recruited by DDB2. Next, we considered the possibility that the interaction of DDB2 with β-catenin might be mediated by EZH2. As shown in Fig. 4C, indeed siRNA mediated knockdown of EZH2 resulted in a reduction of co-immunoprecipitation of β-catenin with DDB2. Moreover, depletion of EZH2 also resulted in a reduction of recruitment of β-catenin in the upstream regulatory region of Rnf43, between P2 and P3 (Figs. 4D–F), and inhibition of Rnf43 expression (supplemental Fig. S7). Together, these observations show that DDB2 recruits β-catenin onto the Rnf43 regulatory region requiring EZH2.
Figure 4. EZH2 mediates DDB2/β-catenin interaction on Rnf43 upstream regulatory region.
(A) Nuclear extracts of HCT 116 cells expressing empty vector or Flag-HA-DDB2 were immunoprecipitated by ANTI-FLAG M2 affinity gel and analyzed by Western Blotting using EZH2, EED and SUZ12 antibodies. IP: Immunoprecipitation; IB: Immunoblotting. (B) ChIP assay was conducted to analyze the local enrichment of DDB2 and EZH2 at P2 region in HCT 116 cells expressing shControl or shDDB2. The relative fold enrichment was quantified by normalization to input, and the fold enrichment of IgG is set as 1. N = 3, Error bars indicate SD. ***: P <0.001. (C) Nuclear extracts of HCT 116 cells transfected with siRNA-Control or siRNA-EZH2 were immunoprecipitated by normal Rabbit IgG or DDB2 antibodies. The results were analyzed by Western Blotting using DDB2, EZH2 and β-catenin antibodies. IP: Immunoprecipitation; IB: Immunoblotting. (D–F) EZH2 mediates DDB2- β-catenin interaction on P2 and P3 regions. The ChIP assays were conducted to analyze the enrichment of DDB2, EZH2 and β-catenin at P1(D), P2(E) and P3(F) regions in HCT 116 cells transfected with siRNA-Control or siRNA-EZH2. The relative fold enrichment was quantified by normalization to input, and the fold enrichment of IgG is set as 1. N = 3, Error bars indicate SD. *: P <0.05,**: P <0.01,***: P <0.001
Evidence for interaction between the DDB2-binding and TCF4-binding chromatins in the Rnf43 gene
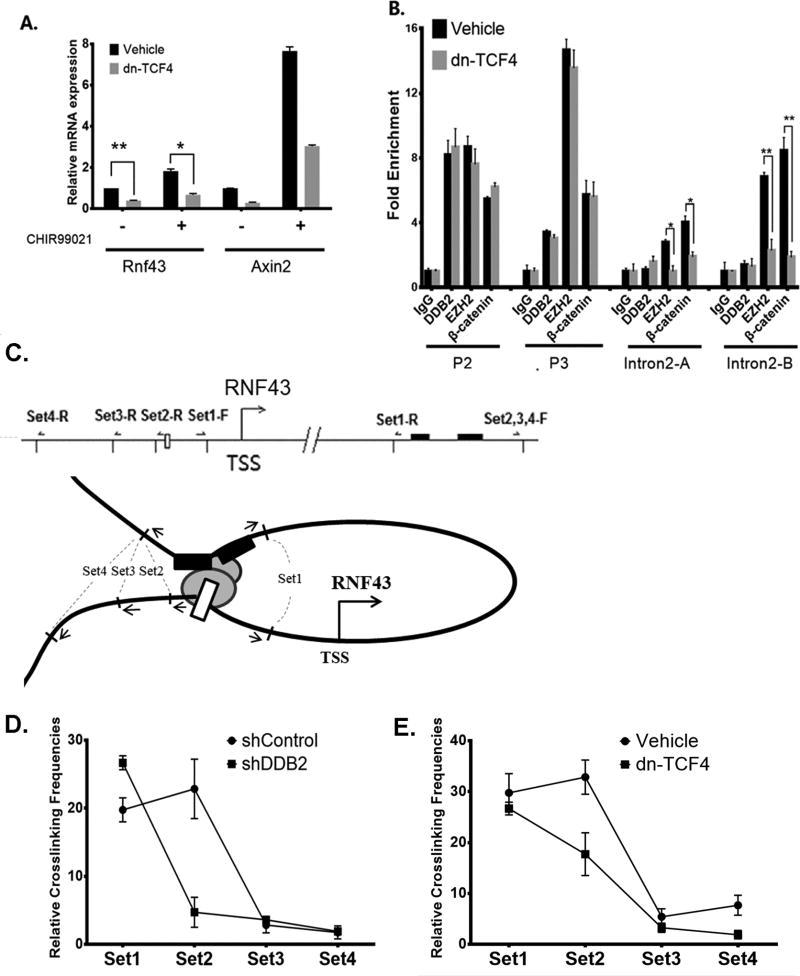
Previous studies identified two TCF4/β-catenin binding sites within intron 2 of human Rnf43 gene (18). As pointed out above, we confirmed binding of TCF4 and β-catenin at those two sites (supplemental Fig. S6). We sought to investigate whether TCF4 is important for GSK3-inhibitor induced expression of Rnf43. A dominant-repressive form of TCF4 (dn-TCF4) was expressed in HCT116 cells. Expression of dnTCF4 inhibited expression of Rnf43 (Fig. 5A), supporting the notion that TCF4 is required for expression of Rnf43. Also, we conducted ChIP assays in the presence or absence of dnTCF4 expression. As shown in Fig. 5B, expression of dnTCF4 had very little impact on bindings of β-catenin and EZH2 onto the upstream P2/P3 region. The bindings of β-catenin as well as EZH2 in the intron2, on the other hand, were significantly inhibited by dnTCF4. Inhibition of Rnf43 expression and inhibition of β-catenin binding by dn-TCF4 imply that the TCF4-sites in the introns are important for expression. Since the DDB2-binding is critical for expression of Rnf43, we considered the possibility of a functional interaction between the DDB2-binding chromatin and the TCF4-binding chromatin at intron 2 of the Rnf43 gene. We carried out chromosome conformation capture (3C) assays to investigate an interaction between the two regions in Rnf43 chromatin in the presence and absence of DDB2, following a strategy described before (22). Briefly, HCT116 cells expressing Control-shRNA or DDB2-shRNA were crosslinked by formaldehyde to link the chromatin segments that are in close spatial proximity. Chromatins were then digested with BglII, followed by ligation with DNA ligase. These steps link distal DNA sequences that are brought together because of chromatin interactions for analyses by PCR. The interactions were analyzed using 4 sets of PCR primers as depicted schematically in Fig 5C (also see supplemental table S5). In those experiments, we observed clear evidence for interactions with primer sets 1 and 2 (Fig. 5D). Moreover, the interaction with the primer set 2 was dependent upon the presence of DDB2. These results demonstrate a DDB2-dependent interaction between the upstream chromatin region near DDB2-binding site and the intron region in Rnf43 gene. To further investigate the involvement of the TCF-sites in the introns for the interaction, we carried out the 3C experiments with cells expressing dominant negative TCF4 (dnTCF4). Expression of dnTCF4 caused a significant reduction in the interaction observed with the primer set 2 (Fig. 5E). These results confirm involvement of TCF-factors, bound to the TCF-elements in the intron, in chromatin interaction with DDB2 bound to the upstream P2/P3 region in the Rnf43 gene.
Figure 5. Chromatin looping structure between DDB2-binding and TCF4-binding regions in the Rnf43 gene.
(A) The expression of dominant negative TCF4 decreased Rnf43expression. HCT 116 cells stably expressing empty vector or dominant-negative TCF4 (dn-TCF4) were treated with DMSO or CHIR99021 (3.3µM, 16hrs). Relative mRNA levels of Rnf43and Axin2 were analyzed by qRT-PCR. N = 3, Error bars indicate SD. (B) Expression of dominant negative TCF4 reduced EZH2 and β-catenin interaction with TCF4 binding regions. The ChIP assay was performed in HCT 116 cells stably expressing empty vector or dn-TCF4. The enrichment of DDB2, EZH2 and β-catenin was analyzed at P2, P3 as well as at the TCF4 binding sites in the intron regions of Rnf43gene. The relative fold enrichment was quantified by normalization to input, and the fold enrichment of IgG is set as 1. N = 5, Error bars indicate SD. *: P <0.05,**: P <0.01. (C) A schematic diagram showing the location of primer and probe sets designed for the Chromatin Conformation Capture (3C) assay. The white rectangle indicates the putative DDB2 binding element at upstream regulatory region in the Rnf43gene. The black bars mark two binding sites of TCF4 in intron2 of the Rnf43gene. The black truncated lines across the Rnf43gene indicate the sites that are recognized by BglII restriction endonuclease. The arrows mark the primers for each of the primer sets. Four sets of primers are designed to detect physical interactions between restriction fragments. (D and E) Expression of DDB2-shRNA (D) or expression of a dominant negative mutant of TCF4 (E) decreases relative crosslinking frequency at Set2. The 3C assays were conducted in HCT 116 cells expressing shControl vs. shDDB2 (D); or control vector vs. dn-TCF expression vector (E). Four sets of primers and probes were analyzed using Taqman qPCR. A serial dilution of the control template was used for standard curve; and the relative crosslinking frequencies were normalized by gapdh loading control. N=4, Error bars indicate SD.
DDB2 is required for RNF43-mediated ubiquitination of the Wnt-receptor FZD5
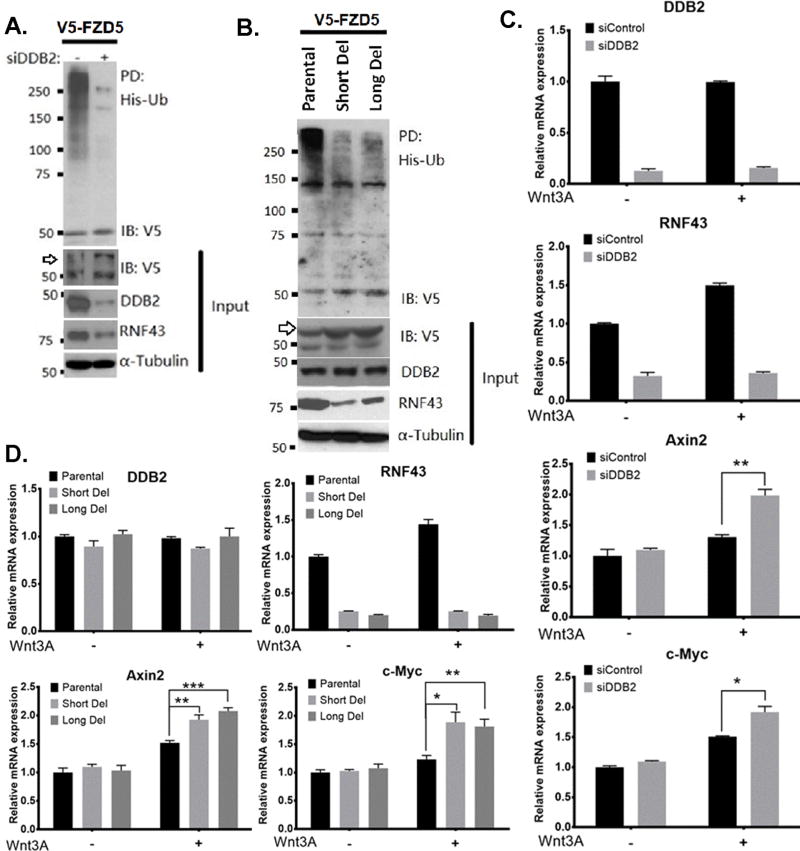
RNF43 is an E3 ubiquitin ligase for the Wnt receptor Frizzled and LRP5/6, and plays a negative regulatory role that attenuates Wnt-signaling (7). Since DDB2 is required for RNF43 expression, we investigated whether DDB2 deficiency leads to inhibition of Wnt-receptor ubiquitination. We analyzed ubiquitination of V5-tagged FZD5 (V5-FZD5) in HT-29 cells that co-express His-tagged Ub along with control-siRNA or DDB2-siRNA. His-tagged-ubiquitinated proteins were collected using Ni-agarose beads and the bound proteins were assayed for the presence of FZD5 using V5-ab. The HT-29 cells express RNF43 at high levels, and poly-ubiquitinated FZD5 can be detected easily. Interestingly, the DDB2-depleted cells exhibited a strong inhibition of the FZD5 poly-ubiquitination as well as an increased level of the mature form of V5-FZD5 (Fig. 6A). Next, we compared the HT-29 cells with those harboring deletions in the Rnf43 upstream region. Lack of DDB2 binding reduced protein expression of RNF43 in the deleted cells. Those cells also exhibited greatly reduced poly-ubiquitination of FZD5 compared to the parental HT-29 cells (Fig. 6B). These results show that the interaction of DDB2 with the upstream region of Rnf43 is critical for ubiquitination of the Wnt-receptors.
Figure 6. DDB2 is required for RNF43-mediated FZD5 ubiquitination and regulation of the Wnt target genes in colon cancer cells.
(A) DDB2 deficiency leads to inhibition of FZD5 ubiquitination. HT-29 cells were transfected with V5–FZD5, His-ubiquitin and siRNA-Control or siRNA-DDB2, as indicated. His-Ubiquitinated proteins were collected by nickel-nitrilotriacetic acid–agarose beads and the presence of ubiquitinated V5-FZD5 is detected by V5 antibody. (B) Lack of DDB2 bound region in Rnf43gene results in inhibition of FZD5 ubiquitination. HT-29 Parental, Short Del and Long Del cells were transfected with V5–FZD5 and His-ubiquitin. And the assay was performed as described above. IP, Immunopull-down; IB: Immunoblotting; Ub, ubiquitin. Arrows indicate mature, glycosylated V5-conjugated FZD5. (C) HT-29 cells transiently expressing siControl or siDDB2 were treated with vehicle or human recombinant Wnt3A protein (300ng/ml, 24hrs). Relative mRNA levels of DDB2, Rnf43, Axin2 and c-Myc were analyzed using qRT-PCR (Normalized by 18S rRNA). N=3. All error bars indicate SD. (D) Lack of DDB2 binding region in Rnf43gene increases the expression of Wnt/β-catenin pathway target genes upon Wnt3a treatment. HT-29 Parental cells, Short Del cells and Long Del cells were treated with vehicle or human recombinant Wnt3A protein (300ng/ml, 24hrs). Relative mRNA levels of DDB2, Rnf43, Axin2 and c-Myc were analyzed using qRT-PCR (Normalized by 18S rRNA). N=3. All error bars indicate SD. *: P <0.05, **: P <0.01, ***: P <0.001
A recent study indicated that the RNF43 protein could sequester TCF4 in the nuclear periphery (23). We sought to investigate whether expression of DDB2 would induce perinuclear accumulation of TCF4. We expressed mVenus tagged DDB2 (green fluorescence) in SW620 cells and subjected the cells for immunostaining for TCF4 (red fluorescence). As shown in supplemental Fig. S8A, DDB2 expressing cells exhibited re-localization of TCF4. Moreover, when we compared HT-29 cells with HT-29 cells lacking the DDB2-site in the Rnf43 promoter, there were cells in the parental line exhibiting perinuclear staining for TCF4, but that was absent in the DDB2-site deleted line (Supplemental Fig. S8B).
Based on these, we predicted that cells lacking binding of DDB2 to the Rnf43 regulatory region or cells with reduced expression of DDB2 would exhibit enhanced response to Wnt-ligands. To investigate that possibility, we compared HT-29 cells lacking DDB2-binding with the parental cells for response to exogenous Wnt3a. Following treatments with vehicle or Wnt3a, total RNA from the cells were assayed for expression of the Wnt response genes. In cells lacking DDB2-binding, Wnt3a had only a marginal effect on the expression of Rnf43. On the other hand, expression of Axin2 and c-Myc were greater in cells lacking DDB2-interaction when treated with Wnt3a (Fig. 6C). Similar observations were made in cells in which the level of DDB2 was diminished by siRNA expression. HT-29 cells expressing DDB2-siRNA exhibited a greater response to Wnt3a induced expression of the Wnt-target genes (Fig. 6D). These results confirm a role of DDB2 in the feed back regulation of Wnt-signaling through RNF43.
DDB2 deficiency increases abundance of Wnt-receptors and enhances Wnt-pathway in vivo in a mouse model for colon cancer
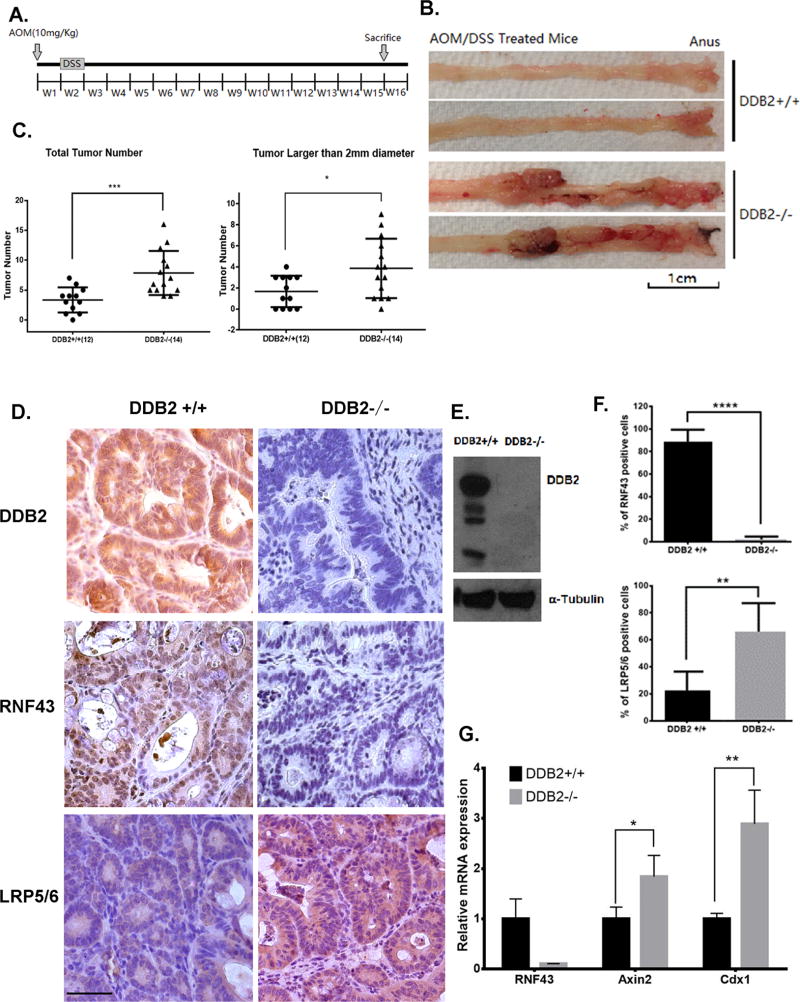
Because Wnt-signaling is a major driver for colon cancer, we investigated whether DDB2 deficiency promotes colorectal tumor development. To investigate the effects of DDB2 on endogenous colorectal cancer, we subjected the male wild-type and DDB2 −/− mice to Azoxymethane (AOM)/Dextran Sulfate (DSS) chemical carcinogenesis protocol (Fig. 7A). When compared 15 weeks after AOM injection, the DDB2−/− mice developed many more and much larger size tumors compared to the wild-type mice (Figs. 7B and C). Whole-exome sequencing identified activating mutation in the Ctnnb1 gene, which encodes β-catenin, in all tumor samples of both genotypes (supplemental Fig. S9A). There was no significant difference in the profile of total variants between the wild-type samples and the DDB2−/− samples (supplemental Figs. S9B and C). Also, there was no consistent mutation of the major mutational colon cancer driver genes, other than that of Ctnnb1 (supplemental Figs. S9D–F). The whole-exome sequencing results suggest that activating mutation in the Ctnnb1 gene is the major driver of colon tumor development in AOM/DSS mouse model.
Figure 7. DDB2 deficiency in mouse colon tumors inhibits RNF43 expression and increases abundance of Wnt receptors.
(A) Schematic of the AOM/DSS protocol. Mice were injected with AOM one week prior to beginning the 7-day treatment of DSS. AOM/DSS treated mice were sacrificed 15 weeks after AOM injection. (B) Representative images of mouse colorectum. At 15 weeks, male DDB2+/+ and DDB2−/− mice were sacrificed and the entire colorectal tissues were excised and whole mounts were examined from proximal to distal ends using light microscopy. Increased numbers of nodular and polyploid colonic tumors were observed in the colorectum of the DDB2−/− mice. (C) DDB2 −/− mice developed greater number of tumor nodules than wild-type mice. Quantification of total tumor nodules and the tumors larger than 2-mm diameter observed in 12 DDB2 +/+ mice and 14 DDB2−/− mice. (D) Colorectal tumors from DDB2−/− mice showed significantly decreased level of RNF43 expression and up-regulated level of LRP5/6. Immunohistochemical staining for DDB2, RNF43 and LRP5/6 were performed on the colorectal tissue sections from DDB2+/+ and DDB2−/− mice and images were taken under 40x objective. (Scale bar=50 µm). (E)Western blotting shows the knockout of DDB2 in colon tumor samples. (F) Quantification of the percentage of RNF43 positive and LRP5/6 positive cells. N=15. (G) Upregulated expression of Wnt target genes in DDB2−/− tumors. RNA was extracted from colorectal tumors isolated from DDB2+/+ mice and DDB2−/− mice, and the relative mRNA levels of Rnf43, Axin2 and Cdx1 were examined by qPCR. N=3. All error bars indicate SD. *: P <0.05, **: P <0.01, ***: P <0.001
Next, we investigated whether the increased susceptibility of tumor development is related to deficiency in RNF43 and increased Wnt signaling. We performed immunohistochemical staining of the tumor tissue sections for RNF43, as well as for LRP5/6 that are co-receptors of Wnt ligands. As shown in Figs. 7D–F, the tumor sections in the DDB2−/− mice are significantly deficient in RNF43. Consistent with that, compared to wild-type mice, the protein levels of LRP5/6 were significantly higher in the tumor sections derived from the DDB2−/− mice (Fig. 7D–F). We also examined the mRNA expression of Wnt target genes Axin2 and Cdx1 in these tumor samples. Both Axin2 and Cdx1 expression were increased in the tumor tissues derived from the DDB2−/− mice (Fig. 7G). These results suggest that DDB2 deficiency promotes colorectal tumor progression via hyper-activation of Wnt signaling.
Furthermore, we investigated if DDB2 deficiency regulates RNF43 and Wnt signaling in normal coloretum. Immunohistochemical staining was performed on normal colon tissue sections to examine the protein expression of RNF43 in the wild-type and DDB2−/− mice. RNF43 was barely detected in the colon crypts of DDB2−/− mice (Supplemental Figs. S10A and B). Additionally, there were increases in the mRNA level of Axin2 and Cdx1 in DDB2−/− mice colorectum (Supplemental Figs. S10C and D). Together, these data suggested that DDB2 is a key activator of RNF43, which negatively regulates Wnt/β-catenin signaling and suppresses colorectal tumor progression.
Discussion
The results presented here are significant in several ways. First, we show that DDB2 is critical for expression of Rnf43, a regulator of Wnt-signaling. It binds to an upstream regulatory region of Rnf43 and recruits EZH2 as well as β-catenin. Moreover, DDB2 mediates an interaction of that upstream regulatory region with the previously identified TCF4-binding region to activate expression of Rnf43. The observations also demonstrate a novel interplay between DDB2 and Wnt-signaling in which DDB2 functions both as a partner as well as a regulator of Wnt-signaling. Furthermore, we provided in vivo evidence linking DDB2-mediated activation of Rnf43 to inhibition of colon cancer.
In canonical Wnt-signaling, upon ligand-induced activation of the pathway, β-catenin is stabilized and accumulates in the nucleus to associate with the TCF/LEF family of transcription factors and activate expression of the TCF/LEF target genes (24,25). A study with over-expression of a dominant negative TCF4 indicated that all β-catenin/target gene interactions involve TCF/LEF factors (26). It is noteworthy that the study with dominant negative TCF4 focused mainly on genes with ChIP signals between −2.5 to +2.5 kb. DDB2-dependent recruitment of β-catenin at the −2.9 kb region of Rnf43 in our study was not inhibited by dominant negative TCF4 (Fig. 5B). DDB2 recruits β-catenin through an interaction with EZH2, which was previously shown to associate with β-catenin and activate Wnt-target genes (21). Our results also show that TCF4 and DDB2 both are important for expression of Rnf43. Moreover, a functional interaction exists between the DDB2-binding and TCF4-binding chromatins in the Rnf43 gene in colon cancer cells. Therefore, it is possible that the TCF4 bound β-catenin and the DDB2 bound EZH2 mediate an interaction that is critical for expression of Rnf43 (supplemental Fig. S11).
The observation that DDB2 is critical for the Wnt-pathway activation of Rnf43 is significant because Rnf43 is a tumor suppressor that regulates Wnt-signaling by regulating the Wnt-receptors through poly-ubiquitnation (7). Colon cancer cells secrete high-levels of Wnt-ligands, which bind to the Wnt-receptors to enhance the signaling pathway even in the presence of activating mutations in the APC gene (5). Inhibition of that pathway through inhibition of Wnt-ligand secretion reduces expression of the Wnt-target genes, resulting in a loss of tumorigenicity in colon cancer cells harboring activating mutations in the Wnt-pathway (5). That is consistent also with our studies comparing colon tumor development in DDB2−/− and DDB2+/+ mice. It was shown that AOM induces mutations in K-ras and β-catenin that alters both Ras pathway and Wnt pathway in rat model (27). However, in our C57BL/6 mouse model, AOM/DSS carcinogenesis protocol only generated activating mutations in the ctnnb1 gene in tumors derived from both strains (Supplemental Figs. S9D–F), which makes it a perfect model to study the regulation of Wnt pathway in colon tumors. In that regard it is interesting that the DDB2−/− mice developed greater number and larger size tumors compared to the DDB2+/+ mice. The increased susceptibility coincided with a loss of Rnf43 expression and increased levels of the Wnt-receptors LRP5/6. Moreover, the Wnt-target genes were expressed at higher levels in the DDB2−/− tumors. In human colon cancer cells, we observed that cells lacking DDB2 expression or cells lacking DDB2-binding in the Rnf43 gene are also deficient in poly-ubiquitination of the Wnt-receptor FZD5, and that those cells exhibit greater response to exogenous Wnt-ligand. In addition, we observed a strong correlation between the expression of RNF43 and DDB2 in human colonic tissues, especially in hyperplasia. These observations from human samples strongly suggest a role of DDB2/RNF43 axis in the early steps involved in the pathogenesis of colon cancer.
The observations that DDB2 is required for Wnt-activated expression of Rnf43 identify DDB2 as a new component in the Wnt/β-catenin signaling pathway in colon cancer cells. The deregulation of the Wnt-signaling pathway in colon cancer is poorly understood because, in addition to activating pro-cancer genes, this pathway also activates expression of several tumor suppressor genes, including Rnf43. The observations that DDB2 is required for expression of Rnf43 and that DDB2 expression is diminished in high-grade colon cancer offer new insights into the mechanisms of Wnt-pathway deregulation that drive aggressive progression of colon cancer. The observations that DDB2 interacts with β-catenin to activate Rnf43 expression also suggest that there might be other Wnt-target tumor suppressor genes that are activated in a manner similar to Rnf43.
Supplementary Material
Acknowledgments
Authors thank Ms. Maureen Regan of the Genome Editing Core, University of Illinois at Chicago; Ms. Wenjun Bie from Dr. Angela Tyner’s lab and Mr. Xinyu Chen from Dr. Nissim Hay’s lab, University of Illinois at Chicago.
Financial support: P. Raychaudhuri is supported by grants from NIH-NCI (CA156164, CA175380, CA177655), and by a merit grant from the Veteran’s Administration (BX 000131). S. Bagchi is supported by the NCI grant CA156164. B.J. Merrill is supported by grant from NIH (HD081534).
Footnotes
Author Contributions
SH designed and carried out the experiments, analyzed data and contributed in writing the paper. DF generated several reagents and protocols for the experiments and performed WES analysis. BM helped designing the experiments and participated in writing the paper. SB made the initial observation on DDB2 regulation of Rnf43 and participated in designing the experiments. PR participated in designing the experiments, analyzing the data and writing the paper.
References
- 1.Brannon AR, Vakiani E, Sylvester BE, Scott SN, McDermott G, Shah RH, et al. Comparative sequencing analysis reveals high genomic concordance between matched primary and metastatic colorectal cancer lesions. Genome Biol. 2014;15(8):454. doi: 10.1186/s13059-014-0454-7. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Rubinfeld B, Albert I, Porfiri E, Munemitsu S, Polakis P. Loss of beta-catenin regulation by the APC tumor suppressor protein correlates with loss of structure due to common somatic mutations of the gene. Cancer Research. 1997;57(20):4624–30. [PubMed] [Google Scholar]
- 3.Liu WG, Dong XY, Mai M, Seelan RS, Taniguchi K, Krishnadath KK, et al. Mutations in AXIN2 cause colorectal cancer with defective mismatch repair by activating beta-catenin/TCF signalling (vol 26, pg 146, 2000) Nat Genet. 2000;26(4):501-01. doi: 10.1038/79859. [DOI] [PubMed] [Google Scholar]
- 4.Jung YS, Jun S, Lee SH, Sharma A, Park JI. Wnt2 complements Wnt/beta-catenin signaling in colorectal cancer. Oncotarget. 2015;6(35):37257–68. doi: 10.18632/oncotarget.6133. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Voloshanenko O, Erdmann G, Dubash TD, Augustin I, Metzig M, Moffa G, et al. Wnt secretion is required to maintain high levels of Wnt activity in colon cancer cells. Nat Commun. 2013;4 doi: 10.1038/ncomms3610. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Madan B, Ke Z, Harmston N, Ho SY, Frois AO, Alam J, et al. Wnt addiction of genetically defined cancers reversed by PORCN inhibition. Oncogene. 2016;35(17):2197–207. doi: 10.1038/onc.2015.280. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.de Lau W, Peng WC, Gros P, Clevers H. The R-spondin/Lgr5/Rnf43 module: regulator of Wnt signal strength. Gene Dev. 2014;28(4):305–16. doi: 10.1101/gad.235473.113. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Zebisch M, Xu Y, Krastev C, MacDonald BT, Chen MR, Gilbert RJC, et al. Structural and molecular basis of ZNRF3/RNF43 transmembrane ubiquitin ligase inhibition by the Wnt agonist R-spondin. Nat Commun. 2013;4 doi: 10.1038/ncomms3787. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Koo BK, Spit M, Jordens I, Low TY, Stange DE, van de Wetering M, et al. Tumour suppressor RNF43 is a stem-cell E3 ligase that induces endocytosis of Wnt receptors. Nature. 2012;488(7413):665–9. doi: 10.1038/nature11308. [DOI] [PubMed] [Google Scholar]
- 10.Giannakis M, Hodis E, Mu XJ, Yamauchi M, Rosenbluh J, Cibulskis K, et al. RNF43 is frequently mutated in colorectal and endometrial cancers. Nat Genet. 2014;46(12):1264–66. doi: 10.1038/ng.3127. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Seshagiri S, Stawiski EW, Durinck S, Modrusan Z, Storm EE, Conboy CB, et al. Recurrent R-spondin fusions in colon cancer. Nature. 2012;488(7413) doi: 10.1038/nature11282. 660-+ [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Hwang BJ, Ford JM, Hanawalt PC, Chu G. Expression of the p48 xeroderma pigmentosum gene is p53-dependent and is involved in global genomic repair. Proc Natl Acad Sci U S A. 1999;96(2):424–8. doi: 10.1073/pnas.96.2.424. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Minig V, Kattan Z, van Beeumen J, Brunner E, Becuwe P. Identification of DDB2 protein as a transcriptional regulator of constitutive SOD2 gene expression in human breast cancer cells. J Biol Chem. 2009;284(21):14165–76. doi: 10.1074/jbc.M808208200. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Zhao R, Cui T, Han C, Zhang X, He J, Srivastava AK, et al. DDB2 modulates TGF-beta signal transduction in human ovarian cancer cells by downregulating NEDD4L. Nucleic Acids Res. 2015 doi: 10.1093/nar/gkv667. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Ennen M, Klotz R, Touche N, Pinel S, Barbieux C, Besancenot V, et al. DDB2: A Novel Regulator of NF-kappa B and Breast Tumor Invasion. Cancer Research. 2013;73(16):5040–52. doi: 10.1158/0008-5472.CAN-12-3655. [DOI] [PubMed] [Google Scholar]
- 16.Fantini D, Huang S, Asara JM, Bagchi S, Raychaudhuri P. Chromatin association of XRCC5/6 in the absence of DNA damage depends on the XPE gene product DDB2. Mol Biol Cell. 2017;28(1):192–200. doi: 10.1091/mbc.E16-08-0573. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Roy N, Bommi PV, Bhat UG, Bhattacharjee S, Elangovan I, Li J, et al. DDB2 suppresses epithelial-to-mesenchymal transition in colon cancer. Cancer research. 2013;73(12):3771–82. doi: 10.1158/0008-5472.CAN-12-4069. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Takahashi N, Yamaguchi K, Ikenoue T, Fujii T, Furukawa Y. Identification of Two Wnt-Responsive Elements in the Intron of RING Finger Protein 43 (RNF43) Gene. Plos One. 2014;9(1) doi: 10.1371/journal.pone.0086582. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Yoon T, Chakrabortty A, Franks R, Valli T, Kiyokawa H, Raychaudhuri P. Tumor-prone phenotype of the DDB2-deficient mice. Oncogene. 2005;24(3):469–78. doi: 10.1038/sj.onc.1208211. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Pai R, Tarnawski AS, Tran T. Deoxycholic acid activates beta-catenin signaling pathway and increases colon cell cancer growth and invasiveness. Mol Biol Cell. 2004;15(5):2156–63. doi: 10.1091/mbc.E03-12-0894. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Jung HY, Jun S, Lee M, Kim HC, Wang X, Ji H, et al. PAF and EZH2 induce Wnt/beta-catenin signaling hyperactivation. Mol Cell. 2013;52(2):193–205. doi: 10.1016/j.molcel.2013.08.028. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Dekker J, Marti-Renom MA, Mirny LA. Exploring the three-dimensional organization of genomes: interpreting chromatin interaction data. Nature Reviews Genetics. 2013;14(6):390–403. doi: 10.1038/nrg3454. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Loregger A, Grandl M, Mejias-Luque R, Allgauer M, Degenhart K, Haselmann V, et al. The E3 ligase RNF43 inhibits Wnt signaling downstream of mutated beta-catenin by sequestering TCF4 to the nuclear membrane. Sci Signal. 2015;8(393) doi: 10.1126/scisignal.aac6757. [DOI] [PubMed] [Google Scholar]
- 24.Cadigan KM, Waterman ML. TCF/LEFs and Wnt Signaling in the Nucleus. Csh Perspect Biol. 2012;4(11) doi: 10.1101/cshperspect.a007906. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Korswagen HC, Clevers HC. Activation and repression of Wingless/Wnt target genes by the TCF/LEF-1 family of transcription factors. Cold Spring Harb Sym. 1999;64:141–47. doi: 10.1101/sqb.1999.64.141. [DOI] [PubMed] [Google Scholar]
- 26.Schuijers J, Mokry M, Hatzis P, Cuppen E, Clevers H. Wnt-induced transcriptional activation is exclusively mediated by TCF/LEF. Embo Journal. 2014;33(2):146–56. doi: 10.1002/embj.201385358. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Takahashi M, Wakabayashi K. Gene mutations and altered gene expression in azoxymethane-induced colon carcinogenesis in rodents. Cancer Sci. 2004;95(6):475–80. doi: 10.1111/j.1349-7006.2004.tb03235.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.