Abstract
Background:
Non-structural protein 4 (NSP4) is a critical protein for rotavirus (RV) replication and assembly. This protein has multiple domains and motifs that predispose its function and activity. NSP4 has a sequence divergence in human and animal RVs. Recently, 14 genotypes (E1-E14) of NSP4 have been identified, and E1 and E2 have been shown to be the most common genotypes in human.
Methods:
The gene and protein sequence of NSP4 in RV-positive samples were inspected with the aim of NSP4 genotyping and variation analysis in viroporin and other domains. P and G typings of RV samples were carried out by WHO primers using a semi-multiplex PCR method. Non-typeable RV samples were amplified by conserved primers and sequenced.
Results:
In viroporin and enterotoxin, conserved sequence was detected, and amino acids substitution with the same biochemical properties was found.
Conclusion:
Association of NSP4 genotype with P or G genotyping G1/G9 correlates with E1 genogroups. In electrophoretyping of RV, E2 genotype had a short pattern when compared to E1.
Keywords: NSP4, Rotavirus, Genotyping
INTRODUCTION
Rotavirus (RV) is a major pathogen causing severe viral diarrhea in young children and animals and is associated with sporadic outbreaks of diarrhea in elderly and immune-compromised patients[1-3]. RV is a genus of the Reoviridae family with 100 nm diameter and contains icosahedral nucleocapsid symmetry[4]. Mature viral particles are composed of triple-layered protein capsids that enclose dsRNA genome with 11 segments. This genome encodes six structural proteins and seven NSPs, and the role of these proteins in the pathogenesis of RV is more or less accepted[4,5].
Segment 10 of RV encodes a non-structural protein (NSP), NSP4, which is a multifunctional and complex protein[6]. On the other hand, NSP4 is the first described viral enterotoxin that can induce “dose-dependent” and “age-dependent” diarrhea without histological changes in mice[7]. Increasing evidence shows that this enterotoxin activates signal transduction pathway and increases intracellular calcium levels in cells by mobilizing calcium from endoplasmic reticulum that leads to chloride secretion[8]. Pathophysiological mechanism of diarrhea includes destruction of RV-infected enterocytes, malabsorption, and neurotoxic activity induced by NSP4 enterotoxin[9]. This protein is important for RV morphogenesis and pathogenesis and efficiently stimulates immune response and can be targeted for the development of an effective vaccine[5,10].
RV is genetically divided into 14 genogroups from E1-E14. NSP4 is a relatively small protein with 175 amino acids (aa)[11]. NSP4 has been indicated to be an ER-specific transmembrane glycoprotein and acts as an intracellular receptor that binds to double-layered particles via C-terminus sequence, thereby facilitating ER entrance and acquisition of outer coating to form triple-layered particles[12-14]. An active synthetic peptide, 114-135 residues of NSP4, induces age-dependent diarrhea in mice; however, anti-NSP4-specific antibodies reduce the symptoms of diarrhea by neutralizing the enterotoxin effects[7]. Other functional and structural domains in NSP4 sequence include a coiled-coil domain and two viral protein-binding domains[15]. NSP4 contains two N-glycosylation sites in the amino terminus that are important to functionality.
The main aim of this study was to determine the genotypic diversity among RV-positive specimens based on sequencing and phylogenetic analysis of the gene coding for NSP4. Overlapping domains in the amino acid sequence were also determined and explained. Moreover, we determined the gene constellation of NSP4 with P and G genotypes in our samples and also compared electrophoretypes with G or P genotyping.
MATERIALS AND METHODS
Samples
A total of 27 RV-positive samples were obtained from Virology Laboratory of Ahvaz Medical Science University (Ahvaz, Iran) from 2013 to 2015. Fecal samples were collected from children under five years old with diarrhea from Aboozar Children Hospital, Ahvaz. RV was detected in the samples using ELISA (Abbott Laboratories, Chicago, USA) and RT-PCR. The fecal samples were diluted to about 10% (wt/vol) in DMEM medium, and then an equal volume of trichlorotrifluoroethane (Freon) was added and clarified by a low speed centrifugation (~ 8000 ×g).
RT-PCR amplification and sequencing of the NSP4 Gene
RV dsRNA was extracted using guanidine/ phenol solution according to the manufacturer’s instructions (Sinaclon, Iran). Thereafter, RNA pellets were dissolved in 50 µl of diethyl pyrocarbonate-treated water and stored at -70 °C until use. NSP4 gene from group A RV was amplified by OneStep RT-PCR Kit (QIAGEN, Germany) with NSP4 F-6 5’-TTAAAAGTTCTGTTCCGAGAGAGCG-3’ and NSP4 R-6 5’-GTCACAYTAAGACCRTTCCTTCC AT-3’ primers.
G and P genotyping
Genotyping of samples was performed according to the WHO Manual of RV detection and characterization methods with minimal modification at genotyping protocols. Semi-multiplex PCR was employed for the amplification of VP7 and VP4 genes. aBT1 G1, aCT2 G2, G3-Aust, aDT4 G4, aAT8 G8, and G9, as forward primers (Asian type), and End9, as a reverse primer, for VP7 amplification were used for genotyping by semi-multiplex PCR. VP4 gene amplification of different genotypes were performed by con3, as well as 2T-1 P[4], 1T-1 P[8], and 3T-1 P[6] forward and reverse primers, respectively. The cDNA was constructed with RevertAid First Strand cDNA Synthesis Kit (Life Technologies, USA) and gene specific consensus End 9 and con3 primers (1 µM final concentration). Typically, the first-round PCR was performed in a 50-µl volume containing 1 × PCR buffer (75 mM Tris/HCl [pH 8.8], 2 mM MgCl2, 200 µM dNTP mix, the appropriate primer mixture (500 nm each), 1 U Taq polymerase, and 5 µl cDNA. PCR reaction was run for 35 cycles of 94 °C for 45 s, 50 °C for 30 s, and 72 °C for 1 min, with a final extension step of 72 °C for 10 min.
RNA electrophoretyping
SDS-PAGE was performed with slab gels using Laemmli’s method[16]. After electrophoresis, gels were fixed and stained simultaneously in a solution containing 5% ethanol, 1% nitric acid, and 0.1% AgNO3 for 5 min; thereafter, the solution was discarded. The gels were rinsed three times with distilled water for 10 s and then developed with a solution of 1.3% NaOH, 0.5% Na2CO3, and 0.4% HCOH (30%) for 1 to 2 min until the appearance of dark-stained bands on the yellow background[17]. Development was stopped with a solution containing 5% ethanol and 1% nitric acid for 1 min, and the stopping solution was then discarded[18,19].
RESULTS
NSP4 coding sequence analysis
Totally, 26 high-quality sequences were obtained from 27 samples of NSP4 coding sequence gene. The sequences were submitted to the GenBank with accession numbers KT148598-KT148623. Based on the RV Classification Working Group, NSP4 has 14 genotypes[20,21]. RotaC2.0 was utilized, as a regularly updated online automatic web application, for RV NSP4 genotyping. The detected genotypes included E1 (20 cases, ~77%) and E2 (6 cases, ~23%). NetNGlyc 1.0 Server was employed for the prediction of N-glycosylation sites. Position 8 of NSP4 protein sequence revealed N-glycosylation site patterns Asn, Tyr, Thr in E1 and E2 genotype but at position 18 in E1 and E2 genotype indicated patterns Asn, Asp, Thr and Asn, Ser, Thr, respectively.
In the double-layered particle-binding site (161-175) of E1 and E2 genotypes, non-charged and charged aa (169 and 174) substitutions (S, K) were frequently detected, respectively (Fig. 1). In position 169 of E1 and E2 genotypes, amino acids serine and lysine were frequently detected. However, in some of the positions (162,164, 165, 166, 168, 170, and 173) of double-layered particle-binding region, conserved aa was detected. Diarrhea inducing motifs 114 to 135 in E1 and E2 genotypes in our samples had variations in some positions. Position 131 in NSP4 of non-avian RVs is histidine or tyrosine.
Fig.1.
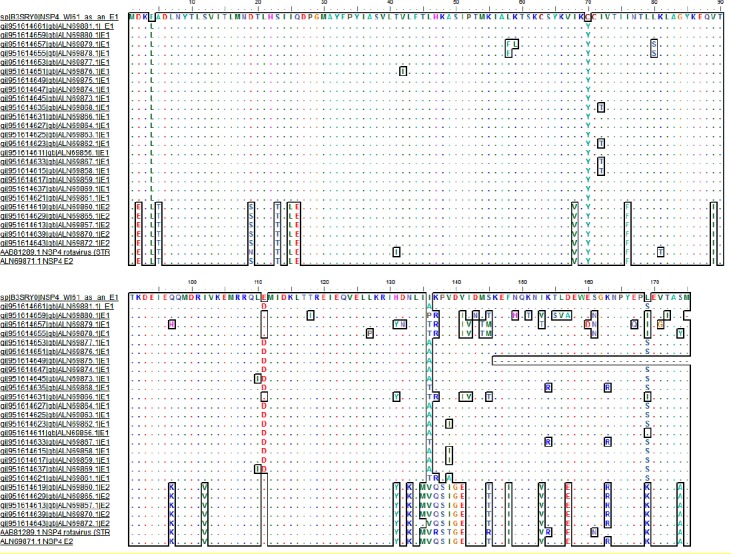
Multiple sequence alignments (MSAS) of protein NSP4 human rotavirus group A by accession number. For MSAS, muscle algorithm was applied, and E1 and E2 of NSP4 genogroups were compared to WI61 and RV5 (Accession numbers: ABV53305.1 and AAB81289.1), respectively.
Viroporin domain of NSP4 sequence, which extends between 47 to 92 amino acids, contains two subdomains: pentalysine and amphipathic[8]. Pentalysine in our sequence contained lysine at positions 55, 59, 62, 66, and 69 as mentioned previously[8] (Fig. 1). Moreover, cysteine residues located at positions 63 and 71 of viroporin domain are conserved and play vital roles in disulfide bond
formation and NSP4 oligomerization[8]. In many protein sequences, sites of the E2 and E1 genotypes (2, 111, 157, and 163), replacement of acidic or basic aa has been noticed, but in some of the positions (26, 97, 133, 141, and 169), polar amino acid was substituted with non-polar aa (Fig. 1).
NSP4, P, and G typing and gene constellation
Genotyping analysis of RV in 27 samples showed the following result: G1 in 7 cases (25.9%), G2 in 8 cases (29.7%), G3 in 2 cases (7.4%), and G9 in 10 cases (37%). The amplicon sizes of G1, G2, G3, G4, and G9 genotypes were 749, 652, 813, 584, and 305 bp, respectively (Fig. 2b). The G8 genotype was not detected in the samples, but mixed infections were found in three samples, two G1/G2 and one G2/G9. The emerging G9 genotype occurred in several countries with a frequency between 2.2 and 11.1%; however, it showed a high frequency in Turkey (40.5%), where it was the most common genotype[2].
Fig. 2.
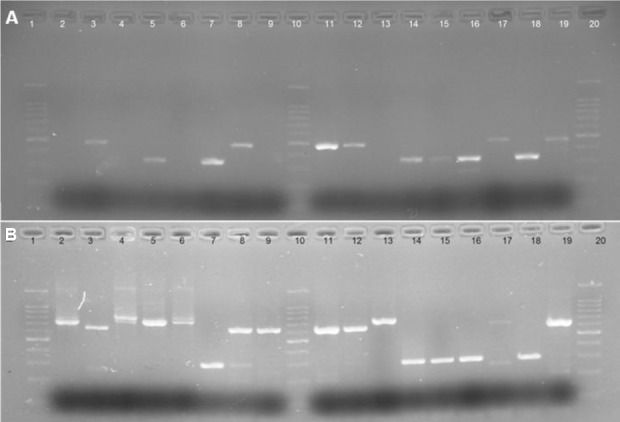
PCR pattern of G and P type human rotavirus. PCR amplicon of rotavirus typing (A) Representative agarose gel electrophoresis of PCR products of semi-multiplex P typing. Lanes 1, 10, and 20, 100 bp ladder; lane 18, RV4 (P8) representing positive control; lanes 3, 8, 11, 12, 17, and 19, P[4] genotype; lanes 5, 7, 14, 15, and 16, P[8] genotype; lane 2, negative control. Lanes 4, 9, and 13, negative results. (B) Representative agarose gel electrophoresis of PCR products of semi-multiplex G typing. Lanes 1, 10, and 20, 100 bp ladder; lane 2, RV4 (G1) representing positive control; lanes 4, 5, and 6, G1 genotype (749 bp); lanes 3, 8, 9, 11, 12, and 19, G2 genotype (652 bp); lanes 7, 14, 15, and 18, G9 (305 bp); lanes 17, 16, and 13, negative results.
The most frequent P types were P[8] in 17 (62.9%) samples, P[4] in 8 (29%), and P[6] in 1 (3%). Amplicon sizes of P[4], P[8], P[6], and P[9] genotypes were 484, 346, 260, and 392 bp, respectively; P[9] genotype was not identified (Fig. 2a). In all G/P samples, the constellation of G1/G9 and G2 was associated with P[8] and P[4], respectively. G3P [6] was identified in one of the samples, and in all samples, constellation of G1, 9/P[8]/E1 was shown. Besides, G2P[4] genotype was associated with E2 genotype but G3 with E1 genotype.
Phylogenetic tree
Phylogenetic analysis confirmed the results obtained by sequencing analysis for NSP4 genotyping assay (Fig. 3). Multiple sequence alignments were conducted by muscle algorithm using MEGA 6.0 software[22]. E1 and E2 were observed in separated clades and were also compared with some of the reference sequences in the GenBank.
Fig. 3.
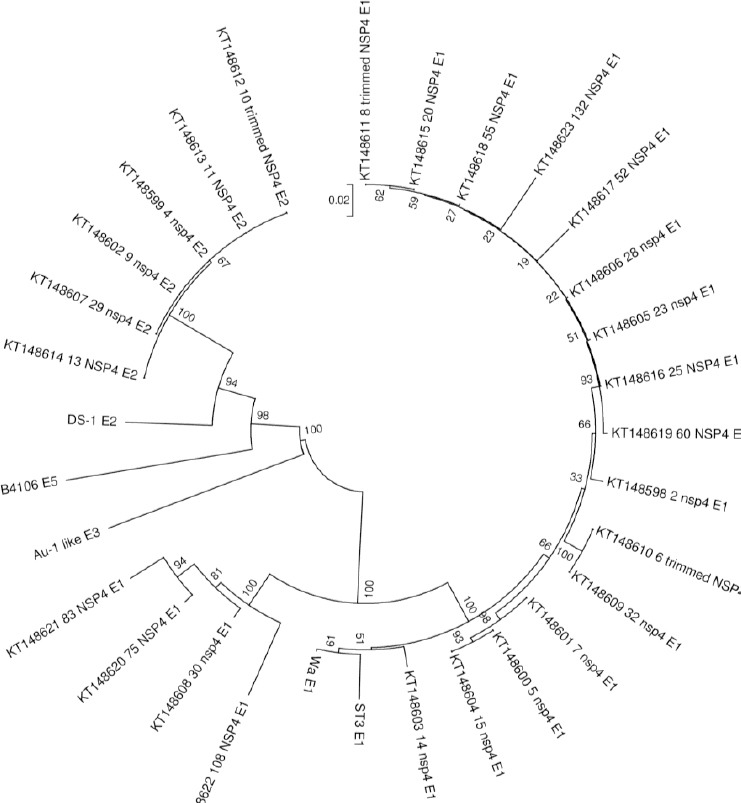
Phylogenetic tree of nucleotide sequences (nt 21-675, strain WA) of human RV strains genotyped and characterized in this study. Reference strains are named, and sample strains are indicated by their GenBank accession numbers. Bar show 0.02 substitutions per nucleotide. The most commonly detected RV genotypes in Iran were G4P[8] and G3P[6] genotypes alongside RV3 and McN13 prototype strains. The tree was inferred utilizing the nucleotide neighbor-joining algorithm with 200 bootstrapping replication of MEGA software.
SDS-PAGE analysis of samples
SDS-PAGE was carried out to represent the electrophoretic pattern of genotypes G and P and also to compare their patterns together. In the combination of G1/G9P[8], a similar pattern was detected (Fig. 4). Electrophoretic movement of VP4 and VP7 segments in these genotypes has been shown in the same position. The short electrophoretic pattern of G2/P4 genotype (samples 4, 6, 7, and 8) with a totally different pattern from G1/G9P[8] was presented. Moreover, in 10 segments of G2P[4] corresponding to E2 genotype a different pattern was presented.
Fig. 4.
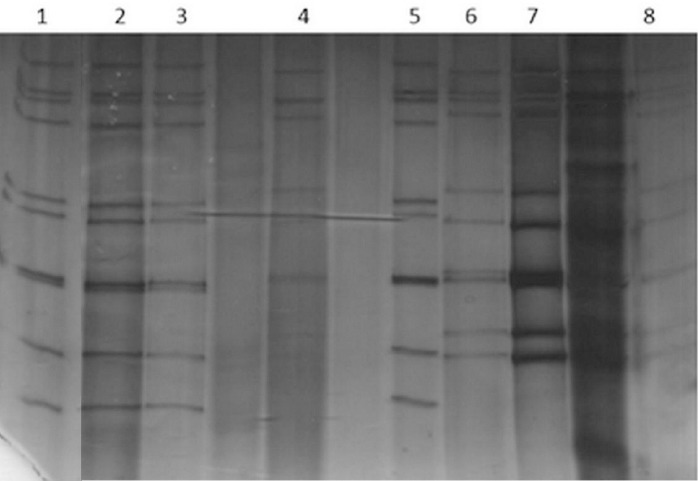
Electrophoretyping of RV from different genotypes. Lanes 1 and 5, G1P[8]; lanes 2 and 3, G1P[8]; lanes 4, 6, 7, and 8, G2P[4]. The short electrophoretic pattern in G2P[4] E2 genotype has been shown.
DISCUSSION
The aim of this study was the analysis of nucleotide and amino acid sequences of NSP4 in RV-positive samples. In this report, the samples were genotyped based on G or P genotyping orresponding with VP7 and VP4 gene segments, and gene constellation of VP7, VP4, and NSP4 was also compared.
Since NSP4 is the most important and multi-functional protein of RV[6], analysis of this gene and its protein in clinical samples may be significant.
However, RV genome is segmented, and evolution of each segment can be analyzed for G or P typing[21,23]. As a result, genotyping of NSP4 gene in the samples was carried out using RotaC2.0 web server, and E1 and E2 genotypes were identified. From 27 sequences samples, E1 (20 cases, ~77%) and E2 (6 cases, ~23%) were obtained. In general, E1 and E2 genotypes are dominant E genotypes in human RV samples[15,24]. Therefore, it could be assumed that the diversity of NSP4 genes among human RV strains is restricted to two of the five NSP4 genotypes described. This protein comprises multiple domains, and each domain has a specific function. Viroporin is one of the important domains of NSP4 enterotoxin protein, which comprises two motifs: pentalysine and amphipathic[8,25]. The presence of a conserved pentalysine domain among the NSP4 protein sequence has been demonstrated, and the alteration of this domain in clinical samples is probably associated with variation in the pathogenesis of virus.
Asparagine residues have been demonstrated to be glycosylated at positions 8 and 18[26]. In general, in N-linked glycosylation, Asparagine residues are located in a specific pattern sequence in the primary structure (Asn-X-Ser or Asn-X-Thr or in rare instances Asn-X-Cys). Among the E1 and E2 genotypes, the amino acid residue 8 in NSP4 protein was found in Asn-Tyr-Thr glycosylation pattern, while the amino acid number 18 was consistently observed in As-Asp-Thr pattern for E1 genotype and As-Ser-Thr pattern for E2 genotypes. Previously, experimental and sequence analysis has been suggested that all gene reassortants in RV are not random[27-30]; the existence of RV genogroups is the strongest evidence in favor of this argument. Moreover, protein-protein interactions in RV particles play an important role in the evolution of these viruses[27]. There is a strong association between P and G typing with E typing in RV samples.
Gene association G1, 9/P[8]/E1, and G2P[4]/E2 were detected, and no inter-genogroup reassorting containing VP7, VP4, and NSP4 occurred in these conventional strains.
Based on previous epidemiological studies from Iran[31-42], there is a rare report on G9 genotype; however, in this study, G9 genotype (~37%) was detected in Khuzestan geographical region. Presently, G9 genotype has become the fifth most common genotype identified in humans[43]. In the samples used in our study, genotype G9 was associated with P[8], a genotype that is responsible for the majority of infections in the world. However, other VP4 genotypes such as P[4], P[6], P[11], and P[19] are also reported with G9. One of the major limitations of RV genotyping with WHO protocols is that G or P genotyping primers do not cover all samples with similar genotypes. Therefore, designing the conserved primers for non-typeable G and P genotypes are inevitable. In this study, we designed some primers to amplify the conserved region of VP4 and VP7 genes. Subsequently, the non-typeable RV samples were subjected to PCR amplification by those primers, and the PCR products were sequenced by Sanger sequencing method.
ACKNOWLEDGMENTS
The present study is a part of a grant (No. 92150) supported by Health Research Institute, Infectious and Tropical Diseases Research Center, Ahvaz Jundishapur University of Medical Sciences, Ahvaz, Iran.
Footnotes
CONFLICT OF INTEREST. None declared.
REFERENCES
- 1.Anderson EJ, Weber SG. Rotavirus infection in adults. The lancet infectious diseases. 2004;4(2):91–99. doi: 10.1016/S1473-3099(04)00928-4. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Durmaz R, Kalaycioglu AT, Acar S, Bakkaloglu Z, Karagoz A, Korukluoglu G, Ertek M, Torunoglu MA. Prevalence of rotavirus genotypes in children younger than 5 years of age before the introduction of a universal rotavirus vaccination program:report of rotavirus surveillance in Turkey. PloS one. 2014;9(12):e113674. doi: 10.1371/journal.pone.0113674. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Desselberger U. Rotaviruses. Virus research. 2014;190:75–96. doi: 10.1016/j.virusres.2014.06.016. [DOI] [PubMed] [Google Scholar]
- 4.Estes M, greenberg H. In: Fields virology. 6 ed. Knipe DM, Howley PM, editors. Philadelphia: Wolters Kluwer Health/Lippincott Williams & Wilkins; 2013. pp. 1347–1401. [Google Scholar]
- 5.Trask SD, McDonald SM, Patton JT. Structural insights into the coupling of virion assembly and rotavirus replication. Nature reviews microbiology. 2012;10(3):165–177. doi: 10.1038/nrmicro2673. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Ball JM, Mitchell DM, Gibbons TF, Parr RD. Rotavirus NSP4:a multifunctional viral enterotoxin. Viral immunol. 2005;18(1):27–40. doi: 10.1089/vim.2005.18.27. [DOI] [PubMed] [Google Scholar]
- 7.Ball JM, Tian P, Zeng CQ, Morris AP, Estes MK. Age-dependent diarrhea induced by a rotaviral nonstructural glycoprotein. Science. 1996;272(5258):101–104. doi: 10.1126/science.272.5258.101. [DOI] [PubMed] [Google Scholar]
- 8.Hyser JM, Collinson-Pautz MR, Utama B, Estes MK. Rotavirus disrupts calcium homeostasis by NSP4 viroporin activity. mBio. 2010;1(5):e00265–10. doi: 10.1128/mBio.00265-10. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Hagbom M, Sharma S, Lundgren O, Svensson L. Towards a human rotavirus disease model. Ccurrent opinion in virology. 2012;2(4):408–418. doi: 10.1016/j.coviro.2012.05.006. [DOI] [PubMed] [Google Scholar]
- 10.Kavanagh OV, Ajami NJ, Cheng E, Ciarlet M, Guerrero RA, Zeng CQ, Crawford SE, Estes MK. Rotavirus enterotoxin NSP4 has mucosal adjuvant properties. Vaccine. 2010;28(18):3106–3111. doi: 10.1016/j.vaccine.2010.02.063. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Chang KO, Kim YJ, Saif LJ. Comparisons of nucleotide and deduced amino acid sequences of NSP4 genes of virulent and attenuated pairs of group A and C rotaviruses. Virus genes. 1999;18(3):229–233. doi: 10.1023/a:1008068218966. [DOI] [PubMed] [Google Scholar]
- 12.Lopez T, Camacho M, Zayas M, Najera R, Sanchez R, Arias CF, Lopez S. Silencing the morphogenesis of rotavirus. Journal of virology. 2005;79(1):184–192. doi: 10.1128/JVI.79.1.184-192.2005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Maruri-Avidal L, Lopez S, Arias CF. Endoplasmic reticulum chaperones are involved in the morphogenesis of rotavirus infectious particles. Journal of virology. 2008;82(11):5368–5380. doi: 10.1128/JVI.02751-07. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Patton JT, Silvestri LS, Tortorici MA, Vasquez-Del Carpio R, Taraporewala ZF. Rotavirus genome replication and morphogenesis:role of the viroplasm. Current topics in microbiology and immunology. 2006;309:169–187. doi: 10.1007/3-540-30773-7_6. [DOI] [PubMed] [Google Scholar]
- 15.Araujo IT, Heinemann MB, Mascarenhas JD, Assis RM, Fialho AM, Leite JP. Molecular analysis of the NSP4 and VP6 genes of rotavirus strains recovered from hospitalized children in Rio de Janeiro Brazil. Journal of medical microbiology. 2007;56(Pt 6):854–859. doi: 10.1099/jmm.0.46787-0. [DOI] [PubMed] [Google Scholar]
- 16.Westermeier R. Electrophoresis in Practice:A Guide to Methods and Applications of DNA and Protein Separations. USA: Wiley; 2005. [Google Scholar]
- 17.Qu L, Li X, Wu G, Yang N. Efficient and sensitive method of DNA silver staining in polyacrylamide gels. Electrophoresis. 2005;26(1):99–101. doi: 10.1002/elps.200406177. [DOI] [PubMed] [Google Scholar]
- 18.An ZW, Xie LL, Cheng H, Zhou Y, Zhang Q, He XG, Huang HS. A silver staining procedure for nucleic acids in polyacrylamide gels without fixation and pretreatment. Analytical biochemistry. 2009;391(1):77–79. doi: 10.1016/j.ab.2009.04.036. [DOI] [PubMed] [Google Scholar]
- 19.Byun SO, Fang Q, Zhou H, Hickford JG. An effective method for silver-staining DNA in large numbers of polyacrylamide gels. Analytical biochemistry. 2009;385(1):174–175. doi: 10.1016/j.ab.2008.10.024. [DOI] [PubMed] [Google Scholar]
- 20.Matthijnssens J, Ciarlet M, McDonald SM, Attoui H, Banyai K, Brister JR, Buesa J, Esona MD, Estes MK, Gentsch JR, Iturriza-Gomara M, Johne R, Kirkwood CD, Martella V, Mertens PP, Nakagomi O, Parreno V, Rahman M, Ruggeri FM, Saif LJ, Santos N, Steyer A, Taniguchi K, Patton JT, Desselberger U, Van Ranst M. Uniformity of rotavirus strain nomenclature proposed by the Rotavirus Classification Working Group (RCWG) Archives of virology. 2011;156(8):1397–1413. doi: 10.1007/s00705-011-1006-z. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Matthijnssens J, Ciarlet M, Rahman M, Attoui H, Banyai K, Estes MK, Gentsch JR, Iturriza-Gomara M, Kirkwood CD, Martella V, Mertens PP, Nakagomi O, Patton JT, Ruggeri FM, Saif LJ, Santos N, Steyer A, Taniguchi K, Desselberger U, Van Ranst M. Recommendations for the classification of group A rotaviruses using all 11 genomic RNA segments. Archives of virology. 2008;153(8):1621–1629. doi: 10.1007/s00705-008-0155-1. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Tamura K, Stecher G, Peterson D, Filipski A, Kumar S. MEGA6:Molecular Evolutionary Genetics Analysis version 6.0. Molecular biology and evolution. 2013;30(12):2725–2729. doi: 10.1093/molbev/mst197. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Maes P, Matthijnssens J, Rahman M, Van Ranst M. RotaC:a web-based tool for the complete genome classification of group A rotaviruses. BMC microbiology. 2009;9:238. doi: 10.1186/1471-2180-9-238. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Ben Hadj Fredj M, Zeller M, Fodha I, Heylen E, Chouikha A, Van Ranst M, Matthijnssens J, Trabelsi A. Molecular characterization of the NSP4 gene of human group A rotavirus strains circulating in Tunisia from 2006 to 2008. Infection, genetics and evolution. 2012;12(5):997–1004. doi: 10.1016/j.meegid.2012.02.011. [DOI] [PubMed] [Google Scholar]
- 25.Browne EP, Bellamy AR, Taylor JA. Membrane-destabilizing activity of rotavirus NSP4 is mediated by a membrane-proximal amphipathic domain. The Journal of general virology. 2000;81(Pt 8):1955–1959. doi: 10.1099/0022-1317-81-8-1955. [DOI] [PubMed] [Google Scholar]
- 26.Bergmann CC, Maass D, Poruchynsky MS, Atkinson PH, Bellamy AR. Topology of the non-structural rotavirus receptor glycoprotein NS28 in the rough endoplasmic reticulum. The EMBO journal. 1989;8(6):1695–1703. doi: 10.1002/j.1460-2075.1989.tb03561.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Heiman EM, McDonald SM, Barro M, Taraporewala ZF, Bar-Magen T, Patton JT. Group A human rotavirus genomics:evidence that gene constellations are influenced by viral protein interactions. Journal of virology. 2008;82(22):11106–11116. doi: 10.1128/JVI.01402-08. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Cunliffe NA, Bresee JS, Gentsch JR, Glass RI, Hart CA. The expanding diversity of rotaviruses. Lancet. 2002;359(9307):640–642. doi: 10.1016/S0140-6736(02)07781-4. [DOI] [PubMed] [Google Scholar]
- 29.Iturriza-Gomara M, Anderton E, Kang G, Gallimore C, Phillips W, Desselberger U, Gray J. Evidence for genetic linkage between the gene segments encoding NSP4 and VP6 proteins in common and reassortant human rotavirus strains. Journal of clinical microbiology. 2003;41(8):3566–3573. doi: 10.1128/JCM.41.8.3566-3573.2003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Nakagomi O, Nakagomi T. Genomic relationships among rotaviruses recovered from various animal species as revealed by RNA-RNA hybridization assays. Research in veterinary science. 2002;73(3):207–214. doi: 10.1016/s0034-5288(02)00097-8. [DOI] [PubMed] [Google Scholar]
- 31.Modaress S, Rahbarimanesh AA, Edalat R, Sohrabi A, Modarres S, Gomari H, Motamedirad M, Sayari AA. Human rotavirus genotypes detection among hospitalized children, a study in Tehran, Iran. Archives of Iranian medicine. 2011;14(1):39–45. [PubMed] [Google Scholar]
- 32.Modarres S, Rahbarimanesh AA, Karimi M, Modarres S, Motamedi-Rad M, Sohrabi A, Nasiri-Oskoii N. Electrophoretic RNA genomic profiles of rotavirus strains prevailing among hospitalized children with acute gastroenteritis in tehran, iran. Archives of Iranian medicine. 2008;11(5):526–531. [PubMed] [Google Scholar]
- 33.Farahtaj F, Gallimore CI, Iturriza-Gomara M, Taremi M, Zali MR, Edalatkhah H, Fayaz A, Gray JJ. Rotavirus VP7 VP4 and VP6 genotypes co-circulating in Tehran Iran between 2003 and 2004. Epidemiology and infection. 2007;135(5):834–838. doi: 10.1017/S0950268806007485. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Zarnani AH, Modarres SH, Jadali F, Sabahi F, Moazzeni SM, Vazirian F. Role of rotaviruses in children with acute diarrhea in Tehran, Iran. Journal of clinical virology. 2004;29(3):189–193. doi: 10.1016/S1386-6532(03)00123-9. [DOI] [PubMed] [Google Scholar]
- 35.Samarbafzadeh A, Tehrani EM, Makvandi M, Taremi M. Epidemiological aspects of rotavirus infection in Ahwaz, Iran. Journal of health, population, and nutrition. 2005;23(3):245–249. [PubMed] [Google Scholar]
- 36.Eesteghamati A, Gouya M, Keshtkar A, Najafi L, Zali MR, Sanaei M, Yaghini F, El Mohamady H, Patel M, Klena JD, Teleb N. Sentinel hospital-based surveillance of rotavirus diarrhea in iran. The journal of infectious diseases. 2009;200(Suppl 1):S244–S247. doi: 10.1086/605050. [DOI] [PubMed] [Google Scholar]
- 37.Khalili B, Cuevas LE, Reisi N, Dove W, Cunliffe NA, Hart CA. Epidemiology of rotavirus diarrhoea in Iranian children. Journal of medical virology. 2004;73(2):309–312. doi: 10.1002/jmv.20092. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Shoja Z, Tagliamonte M, Jalilvand S, Ziaee AA, Mokhtari-Azad T, Hamkar R, Shahmahmoodi S, Rezaei F, Tornesello M, Buonaguro FM, Nategh R, Buonaguro L. Molecular characterization analysis of the outer protein layer (VP7) from human rotavirus A genotype G1 isolate identified in Iran:implications for vaccine development. New microbiologica. 2012;35(4):415–427. [PubMed] [Google Scholar]
- 39.Mayameii A, Shapouri MR, Ghorbanpour M, Hajikolaei MR, Keyvanfar H. Molecular G typing of bovine rotaviruses in Iran. Pakistan journal of biological sciences. 2007;10(19):3466–3469. doi: 10.3923/pjbs.2007.3466.3469. [DOI] [PubMed] [Google Scholar]
- 40.Ghorashi Z, Behbahan AG, Oskouei SA. Rotavirus enteric infection in children of northwest Iran. The pediatric infectious disease journal. 2011;30(7):616–618. doi: 10.1097/INF.0b013e31820a45cb. [DOI] [PubMed] [Google Scholar]
- 41.Shoja Z, Jalilvand S, Mokhtari-Azad T, Nategh R. Epidemiology of cocirculating human rotaviruses in Iran. The pediatric infectious disease journal. 2013;32(4):e178–e181. doi: 10.1097/INF.0b013e31827ee392. [DOI] [PubMed] [Google Scholar]
- 42.Afshar A, Tadayon RA. Rotavirus in diarrhoeic calves in Iran. Veterinary record. 1979;105(17):400. doi: 10.1136/vr.105.17.400. [DOI] [PubMed] [Google Scholar]
- 43.Doro R, Laszlo B, Martella V, Leshem E, Gentsch J, Parashar U, Banyai K. Review of global rotavirus strain prevalence data from six years post vaccine licensure surveillance:is there evidence of strain selection from vaccine pressure? Infection, genetics and evolution. 2014;28:446–461. doi: 10.1016/j.meegid.2014.08.017. [DOI] [PMC free article] [PubMed] [Google Scholar]


