Abstract
Cytochrome P450 1A2 (CYP1A2) is one of the most abundant and important drug metabolizing enzymes in human liver. However, little is known about the post-transcriptional regulation of CYP1A2, especially the mechanisms involving microRNAs (miRNAs). This study applied a systematic approach to investigate the post-transcriptional regulation of CYP1A2 by miRNAs. Candidate miRNAs targeting the 3′-untranslated region (3′-UTR) of CYP1A2 were screened in silico, resulting in the selection of sixty-two potential miRNAs for further analysis. The levels of two miRNAs, hsa-miR-132-5p and hsa-miR-221-5p, were inversely correlated with the expression of CYP1A2 mRNA transcripts in normal human liver tissue samples represented in The Cancer Genome Atlas (TCGA) dataset. The interactions between these miRNAs and cognate CYP1A2 mRNA sequences were evaluated using luciferase reporter gene studies and electrophoretic mobility shift assays, by which a direct interaction was confirmed involving hsa-miR-132-5p and a cognate binding site present in the CYP1A2 3′-UTR. Experiments by which hsa-miR-132-5p or random miRNA controls were introduced into HepG2, Huh-7 and HepaRG hepatic cell lines showed that only hsa-miR-132-5p suppressed the endogenous and lansoprazole-induced expression of CYP1A2, at biological activity, protein production, and mRNA transcript levels. Furthermore, 3-(4,5-di methylthiazol-2-yl)-2,5-diphenyltetrazolium bromide (MTT), and lactate dehydrogenase (LDH) assays showed that hsa-miR-132-5p attenuates CYP1A2-mediated, lansoprazole-enhanced, flutamide-induced hepatic cell toxicity. Results from multilayer experiments demonstrate that hsa-miR-132-5p suppresses the expression of CYP1A2 and that this suppression is able to decrease the extent of an adverse drug-drug interaction involving lansoprazole and flutamide.
Keywords: CYP1A2, hsa-miR-132-5p, Drug metabolizing enzymes, MicroRNA, Toxicity
1. Introduction
Endogenous or exogenous chemicals are activated or detoxified through two metabolic steps catalyzed by Phase I and Phase II liver enzymes. With a high concentration in the lipid bilayer of the endoplasmic reticulum of hepatocytes, the cytochrome P450 (CYP) enzyme family is primarily responsible for Phase I drug metabolism [1]. The CYP enzymes function as monoxygenases to catalyze the hydroxylation, dealkylation, dehalogenation, or other oxidative or reductive modifications of parent drugs [2,3]. Among liver CYP enzymes, CYP1A2, CYP2A6, CYP2B6, CYP2C8, CYP2C9, CYP2C19, CYP2D6, CYP2E1, and CYP3A4 are considered to be the most important isoenzymes pharmacologically, being involved in the metabolism of more than 90% of drugs [4]. CYP1A2 is highly expressed in human liver, accounting for approximately 13% of the total cytochrome P450 content [5], and CYP1A2 is responsible for metabolizing about 9% of clinically used drugs [6]. Besides drug metabolism, CYP1A2 also plays a major role in procarcinogen activation and drug-drug interactions [5,7].
The regulation of CYP1A2 expression is complicated. Similar to CYP1A1 and CYP1B1, CYP1A2 is primarily inducible through aromatic hydrocarbon receptor-mediated transactivation by ligand binding and nuclear translocation [8]. In addition, accumulating evidence indicates that CYP1A2 expression is influenced by genetic polymorphisms [9] and transcriptional factors [8,10]; recently it was been predicted that CYP1A2 may be subject to post-transcriptional regulation by microRNAs (miRNAs) [11].
MiRNAs are critically important in gene regulatory networks. They usually bind to their target sequences within the 3′-untranslated regions (3′-UTR) of mRNA molecules, which facilitates mRNA degradation or translational repression [11–13]. Some miRNAs have been found to interact directly with CYP450 mRNA transcripts. For example, CYP2C9 is regulated by miR-128 and miR-130b [14,15], CYP2C19 is modulated by miR-29a [16], CYP2E1 is suppressed by miR-378 [17], and CYP3A4 is regulated by miR-27a [18]. Analysis of the 3′-UTR of CYP1A2 suggests that it may be regulated by miRNAs [19–21]; however, a systematic/functional characterization has not been conducted.
CYP1A2 is inducible at both mRNA and protein levels by a variety of chemicals, including omeprazole, lansoprazole, 2,3,7,8-tetra chlorodibenzo-p-dioxin (TCDD), 3-methylcholanthrene, and rifampicin [22]. Omeprazole and lansoprazole are proton pump inhibitors (PPIs) that are sometimes co-administered with anticancer drugs to alleviate chemotherapy-induced nausea and vomiting [23]. As such, it is important to explore the role of CYP1A2-mediated drug-drug interactions involving drugs that are CYP1A2 substrates when combined with other drugs that may either induce or repress CYP1A2 expression in ways affecting drug safety or efficacy. The induction of CYP1A2 has been recognized as a possible reason for chemotherapy resistance [24]. Owing to its ability to convert drugs to toxic metabolites (e.g. the conversion of flutamide to liver toxicant 2-hydroxyflutamide [25]), the activity of CYP1A2 was found to mediate drug-induced adverse reactions, such as flutamide-induced liver toxicity [26]. However, the potential role of lansoprazole in a drug-drug interaction with flutamide is not well documented. If a specific miRNA is able to suppress the expression of CYP1A2, a novel miRNA-related mechanism may decrease the effect of CYP1A2-mediated drug-drug interactions involving a CYP1A2 substrate, such as flutamide, and a CYP1A2 inducer, such as lanzoprazole.
In this study, miRNAs that putatively regulate CYP1A2 were screened systematically using in silico analyses. Reporter gene assays and electrophoretic mobility shift assays demonstrated a specific and direct interaction between hsa-miR-132-5p and the CYP1A2 3′-UTR. The regulatory functions of hsa-miR-132-5p on endogenous and lansoprazole-induced CYP1A2 mRNA expression, protein production, and biological activity were detected in a terminally differentiated hepatic cell line and in hepatoma cells. Finally, cell proliferation and toxicity assays were applied to test the effect of the lansoprazole-flutamide interaction on CYP1A2-mediated cytotoxicity in hepatic cells.
2. Materials and methods
2.1. Cell culture
Human hepatic cancer cell lines (Huh-7 and HepG2) and human embryonic kidney (HEK) 293 T cells were obtained from the American Type Culture Collection (ATCC, Manassas, VA). These cell lines were cultured in Dulbecco’s minimum essential medium (DMEM, ATCC) supplemented with 10% fetal bovine serum (FBS, ATCC), 100 μg/mL streptomycin, and 100 IU/mL penicillin (ATCC). HepaRG cells were purchased from Life Technologies (Carlsbad, CA), seeded at 5 × 105 cells per well in 24-well plates, and maintained in William’s E medium supplemented with the Thaw, Plate, & General Purpose Medium Supplement (Life Technologies) for one day. Then the cells were incubated for additional 7 days in William’s E medium supplemented with Maintenance/Metabolism Medium Supplement. All cell lines were incubated in a humidified atmosphere containing 5% CO2 at 37 °C.
2.2. In silico analyses
The public database miRTar.human (http://mirtar.mbc.nctu.edu.tw/human/) was used to screen potential miRNA binding sites located in the 3′-UTR of CYP1A2. RNA hybrid, a RNA hybridization algorithm (http://bibiserv2.cebitec.unibielefeld.de/rnahybrid), was applied to calculate the minimum free energy of hybridization for potential miRNAs with their putative binding sites detected within the CYP1A2 mRNA sequence (NM_000761.4). Pearson’s correlation analysis (http://www.socscistatistics.com/tests/pearson/) was used to evaluate the relationships between CYP1A2 mRNA and the candidate miRNAs using The Cancer Genome Atlas (TCGA) dataset (http://cancergenome.nih.gov/), which includes miRNA levels and mRNA profiles of 49 cases of human non-tumor liver tissue samples.
2.3. Luciferase reporter gene assay
The pGL3-Control vector (Promega, Madison, WI) was modified by adding the Universal USER Cassette (New England Biolabs, Ipswich, MA), resulting in the pGL3-CU vector described in our previous study [14]. The pGL3-CU vector was digested with Xba I enzyme (New England Biolabs) and nicked with Nt. BbvCl (New England Biolabs). The core region of the CYP1A2 3′-UTR that harbors the putative binding sites for hsa-miR-132-5p and hsa-miR-221-5p was PCR amplified using the forward and reverse primers with extension oligonucleotides 5′-GGG AAA GU-3′ or 5′-GGA GAC AU-3′ on their 5′ ends as described in Table 1. The PCR products were digested with USER enzyme (New England Biolabs) and ligated into the linearized and nicked pGL3-CU vector according to the Universal USER Cassette protocol provided by the manufacturer. DNA sequence analysis was performed on the constructed plasmid, pGL3-CYP1A2, to confirm its authenticity.
Table 1.
Sequences of primers and oligonucleotides.
| Name | Sequence |
|---|---|
| For qRT-PCR | |
| CYP1A2 forward | 5′-GGG CAC TTC GAC CCT TAC AA-3′ |
| CYP1A2 reverse | 5′-GCA CAT GGC ACC AAT GAC G-3′ |
| GAPDH forward | 5′-GAA ATC CCA TCA CCA TCT TCC AGG-3′ |
| GAPDH reverse | 5′-GAG CCC CAG CCT TCT CCA TG-3′ |
| hsa-miR-132-5p forward | 5′-ACC GTG GCT TTC GAT TGT TAC T-3′ |
| hsa-miR-221-5p forward | 5′-ACC TGG CAT ACA ATG TAG ATT T-3′ |
| U6 forward | 5′-CTC GCT TCG GCA GCA CA-3′ |
| miRNA Universal reverse | 5′-AAC GCT TCA CGA ATT TGC GT-3′ |
| For luciferase reporter gene assay | |
| CYP1A2 3′-UTR forward | 5′-GGG AAA GU GAA GAC ACC ACC ATT CTG AGG-3 |
| CYP1A2 3′-UTR reverse | 5′-GGA GAC AU GGC AAA TCC ATA GAC ACA GAA A-3′ |
| hsa-miR-132-5p mutant | 5′-ACC GUA ACU UUC GAU UGU UAC U-3′ |
| hsa-miR-221-5p mutant | 5′-ACC UAA CAU ACA AUG UAG AUU U-3′ |
| For EMSA | |
| CYP1A2-miR-132-5p (6276–6297) | 5′-GCG AGU GGG GGC CAG CCA CGG G-3′ |
| CYP1A2-miR-132-5p (7547–7568) | 5′-CAU UAC AGG UGU GAG CCA CGG U-3′ |
| CYP1A2-miR-221-5p | 5′-GAG UCU UAC UCU GUU GCC AGG C-3′ |
| miRNA negative control | 5′-UCA CAA CCU CCU AGA AAG AGU AGA-3′ |
HepG2 and 293 T cells were plated at a density of 1 × 105 cells per well in a 96-well plate and allowed to attach for 24 h. Cells were transfected with the constructed luciferase reporter gene plasmids or the control luciferase plasmid (100 ng/well) plus pRL-SV40 renilla plasmid (Promega, 1 ng/well) using the Lipofectamine 2000 reagent (Life Technologies). Equal amounts (50 nM) of hsa-miR-132-5p mimic, inhibitor, or mutant; and hsa-miR-221-5p mimic, inhibitor, or mutant were co-transfected with the luciferase plasmids containing the CYP1A2 3′-UTR. The constructed luciferase plasmid pGL3-CYP1A2 was co-transfected with pRL-SV40 plasmid and miRNA negative control that served as a reference. Luciferase and renilla signals were determined 24 h after transfection using a Dual Luciferase Reporter Assay Kit (Promega). Five independent experiments were carried out.
2.4. RNA electrophoretic mobility shift assays
All oligonucleotides and primers used in this study were obtained from Integrated DNA Technologies (Coraville, IA). The oligonucleotides for hsa-miR-132-5p were synthesized and 5′-labeled with cy5.5TM dye (dye-miR-132). The 2′-O-methyl-modified RNA oligonucleotides CYP1A2-Target-1 (nucleotide position 6276–6297) and CYP1A2-Target-2 (nucleotide position 7547–7568), corresponding to the hsa-miR-132-5p targeted sequences resident in 3′-UTR of CYP1A2, were 5′-labeled with IRDye®800 dye (IDT), dye-CYP1A2-Target-1 and dye-CYP1A2-Target-2. The unlabeled oligonucleotides, including the miRNA negative control (cold-NC) and hsa-miR-132-5p (cold-miR-132), were used in competition assays.
RNA electrophoretic mobility shift assays (EMSAs) were performed using LightShift Chemiluminescent RNA EMSA kits (Thermo Fisher Scientific, Waltham, MA) according to the manufacturer’s instructions. Experiments were performed independently at least five times. Briefly, 1× RNA EMSA binding buffer, 5% glycerol, 200 mM KCl, 100 mM MgCl2, and 200 nmol synthetic miRNA and/or cognate mRNA oligonucleotides were mixed in 20 μL reactions. Additional experiments were performed to determine if cellular factors would be recruited to miRNA/mRNA hybrid complexes. Cytoplasmic extracts from HepaRG cells were obtained using NE-PER Nuclear and Cytoplasmic extraction reagents (Thermo Fisher Scientific). Cytoplasmic extract (2 μg) and non-specific total RNA (1 μg, included in the RNA EMSA kit) were added to the basic reaction mixtures and incubated 20 min at room temperature to allow miRNA/RNA/protein complexes to form. Subsequently, antibodies against Ago1, Ago2, Ago3, or Ago4 (Abcam, Cambridge, MA) were used in the super-shift assays. In competition assays, unlabeled probes at 50-fold molar excesses were added to the reactions before the addition of dye-labeled probes. The reaction mixtures were separated by 12% PAGE electrophoresis at 4 °C and examined with an Odyssey CLx Infrared Imaging System (LI-COR Biosciences, Lincoln, NE).
2.5. Induction of CYP1A2 expression by lansoprazole
To induce CYP1A2 with lansoprazole (Sequoia Research Products Ltd, UK), Huh-7 cells were seeded at the density of 2 × 105 cells per well in 24-well plates. After incubation at 37 °C in a CO2 incubator for 24 h, the culture medium was exchanged with the complete medium containing lansoprazole at concentrations of 10 μM and 50 μM using five wells per treatment group. Treated cells were collected for CYP1A2 mRNA and protein assays after incubations for 0, 4, 12, and 24 h.
2.6. miRNA transfection
The hsa-miR-132-5p and hsa-miR-221-5p synthetic miRNA mimics, inhibitors, and negative control miRNA were purchased from Dharmacon GE (Lafayette, CO). MiRNA was transfected into Huh-7, HepG2, and HepaRG cells using Lipofectamine 2000 reagent (Life Technologies). After incubation at 37 °C in a CO2 incubator for 6 h, the transfection medium was exchanged with fresh complete medium. Cells were incubated further for 48 h and then harvested.
2.7. RNA extraction and quantitative real-time PCR analysis
Total RNA was extracted using miRNeasy Mini kits (Qiagen, Valencia, CA). cDNA for reverse-transcription polymerase chain reaction (RT-PCR) was synthesized with High-Capacity cDNA Reverse Transcription kits (Thermo Fisher Scientific). For the detection of mature miRNAs, small RNAs were reverse transcribed to cDNA using NCodeTM microRNA First-Strand cDNA Synthesis kits (Thermo Fisher Scientific). The expression levels of CYP1A2, endogenous control GAPDH, mature hsa-miR-132-5p, hsa-miR-221-5p, and endogenous control U6 were determined using an ABI Prism7900 Sequence Detection system (Thermo Fisher Scientific). Reactions were prepared according to the manufacturer’s instructions for QuantiFast SYBR® Green RT-PCR Kits (Life Technologies) using the primers listed in Table 1. The RNA expression levels for CYP1A2 mRNA or the miRNAs hsa-miR-132-5p and hsa-miR-221-5p were calculated relative to the expression of GAPDH or U6 small nuclear RNA, respectively.
2.8. Western blot assay
The cells were lysed in RIPA buffer (Thermo Scientific). Protein concentrations in lysate samples were quantified, separated by SDS-PAGE, transferred to 0.45 μm polyvinylidene fluoride (PVDF) membranes (Millipore), and incubated with the following primary antibodies diluted in Odyssey® Blocking Buffer (PBS, Cat No. 927–40003, LI-COR; Lincoln, NE): anti-CYP1A2 (mouse, 1:500; Cat No. sc-393783, Santa Cruz; Dallas, TX), anti-GAPDH (rabbit, 1:2000; Cat No. ab2634, Abcam; Cambridge, MA) at room temperature for 1 h. The membranes were then washed four times with PBST and incubated with the IRDye® 800CW Goat-anti-Mouse Antibody (1:5000; Cat No.926–32210, LI-COR; Lincoln, NE) or IRDye® 680RD Goat-anti-Rabbit Antibody (1:5000; Cat No.926–68071, LI-COR) diluted in Odyssey® Blocking Buffer. Quantitative analyses were performed using the Odyssey CLx Infrared Imaging System. The expression of GAPDH was used as the housekeeping gene control to confirm equal loading of the samples.
2.9. Measurement of CYP1A2 enzyme activity
CYP1A2 enzyme activity in HepG2 cells and Huh7 cells treated with lansoprazole, with or without hsa-miR-132-5p synthetic miRNA mimic, was measured using the substrate-specific P450-GloTM Assay (Promega, Madison, WI). To verify the specific inhibitory effects of the hsa-miR-132-5p mimic, CYP2B6 and CYP3A4 enzyme activities were also measured as negative controls. HepG2 cells and Huh7 cells were seeded at a density of 2 × 104 per well in 96-well plates. After being incubated overnight, synthetic hsa-miR-132-5p mimic together with negative control miRNA mimic were transfected into HepG2 cells and Huh7 cells using Lipofectamine 2000 reagent. After incubation at 37 °C in a CO2 incubator for 6 h, the transfection medium was exchanged with fresh medium containing lansoprazole (50 μM) and then incubated for a period of 12 h. The cells were washed with PBS twice and incubated with 50 μL of fresh culture medium containing the corresponding CYP1A2 substrate (Luciferin-1A2), CYP2B6 substrate (Luciferin-2B6), or CYP3A4 substrate (Luciferin-IPA) at 37 °C for 1 h. Then 25 μl of culture medium was transferred to a white 96-well plate together with 25 μL of luciferin detection reagent added in each well. The luminescence was measured with a BioTek Cytation 5 (BioTek, Winooski, VT) cell imaging multimode reader.
2.10. Cell viability assay
Cell viability was determined using the 3-(4,5-dimethylthiazol-2-yl)-2,5-diphenyltetrazolium bromide (MTT) (Sigma-Aldrich, St. Louis, MO) assay. HepG2 cells have been widely used for the detection of flutamide-dependent liver injury in previous studies [27–29]. In the present study, HepG2 cells were seeded at a density of 2 × 104 per well in 96-well plates. MiRNA transfection and lansoprazole induction were carried out as described above. After a period of 12 h incubation, flutamide (Sigma-Aldrich, St. Louis, MO) was dissolved in ethanol and added at a final concentration of 1 mM [29,30] to each well and incubated at 37 °C, 5% CO2 for 4 h. MTT labeling reagent (Sigma-Aldrich) was added to the cells, and the cells were incubated at 37 °C for 2 h. Dimethyl sulfoxide (DMSO, Sigma-Aldrich, St. Louis, MO) was then added, and the absorbance of stained cells was measured at 570 nm. Five experiments were performed independently.
2.11. LDH cytotoxicity assay
Flutamide-dependent cytotoxicity was detected using Pierce LDH Cytotoxicity Assay Kits (Thermo Scientific) according to the manufacturer’s instructions. HepG2 cells were treated in the same manner as described for the MTT-based cell viability assays. HepG2 cells without any treatment were seeded in triplicate and served separately as the spontaneous LDH activity controls and the maximum LDH activity controls. For the maximum LDH activity controls, 10 μL of Lysis Buffer (10×) was added to each well, and then incubated at 37 °C, 5% CO2 for 45 min. Aliquots (50 μL) of each sample medium were transferred to the corresponding wells of a fresh 96-well plate and mixed with 50 μL of Reaction Mixture. After incubation for 30 min at room temperature without light, 50 μL of Stop Solution was added to each sample well. The absorbance values at 490 nm and 680 nm were measured using a BioTek Cytation 5 cell imaging multimode reader. LDH activity was calculated by subtracting the 680 nm absorbance value from the 490 nm absorbance. The amount of LDH from each well was presented as the percentage of LDH released in cell culture media without treatment (control). Experiments were performed independently five times.
2.12. Statistical analyses
Data are presented as the mean ± standard error of the mean (SEM). All statistical analyses were performed with SPSS 17.0 software (IBM, Armonk, NY). Pearson’s correlation analysis was used to analyze the original data of human liver tissues to evaluate the correlations between the expression of CYP1A2 mRNA and the expression levels of hsa-miR-132-5p and hsa-miR-221-5p. Student’s unpaired t-test was used to analyze the difference of two independent groups, whereas one way analysis of variance (ANOVA) was used to determine the difference of more than two independent groups. Values of P < 0.05 were considered to be statistically significant.
3. Results
3.1. Identification of potential miRNAs modulating CYP1A2
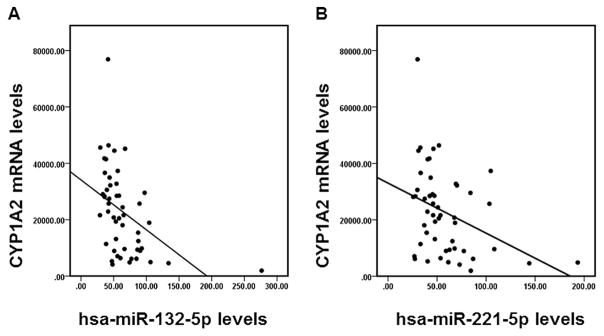
Sixty-two miRNAs were identified using the public database miRTar.human as candidate miRNAs binding to the 3′-UTR of CYP1A2, with the free energy of miRNA/mRNA hybridization less than −20 kcal/mol estimated by the RNAhybrid software (Table 2). To examine the possible correlations between the expression of candidate miRNAs and the expression of CYP1A2 mRNA in human tissue samples, we compared the expression profile of all 62 candidate miRNAs and the expression profile of CYP1A2 using the TCGA dataset. Pearson’s correlation analyses indicated that the expression of hsa-miR-132-5p and hsa-miR-221-5p had significant inverse associations (r = −0.449 for hsa-miR-132-5p and r = −0.367 for hsa-miR-221-5p, both P < 0.05) with the expression of CYP1A2 in non-tumor liver tissue samples (Fig. 1).
Table 2.
MicroRNAs targeting CYP1A2 3′-UTR predicted by miRTar.
| # | miRNAs | Position | Binding energy (kcal/mol)* |
|---|---|---|---|
| 1 | hsa-miR-1207-5p | 7647/7651 | −40.4/−38.4 |
| 2 | hsa-miR-939-5p | 7647 | −35.1 |
| 3 | hsa-miR-363-5p | 7199 | −33.8 |
| 4 | hsa-miR-3157-5p | 6752 | −33.6 |
| 5 | hsa-miR-762 | 7661/6278 | −33.5/−32.5 |
| 6 | hsa-miR-4322 | 7560 | −32.3 |
| 7 | hsa-miR-328-3p | 6270 | −32.2 |
| 8 | hsa-miR-3187-3p | 7120 | −32.1 |
| 9 | hsa-miR-1202 | 7530 | −31.6 |
| 10 | hsa-miR-3192-5p | 7350 | −31.5 |
| 11 | hsa-miR-149-3p | 7050 | −30.9 |
| 12 | hsa-miR-658 | 7042 | −30.7 |
| 13 | hsa-miR-671-5p | 6845 | −30.4 |
| 14 | hsa-miR-668-3p | 6470 | −30 |
| 15 | hsa-miR-128-3p | 7157 | −29.2 |
| 16 | hsa-miR-30b-3p | 6711/7048/7213/7349/7381 | −22.7/−29.2/−23/−27.7/−23.6 |
| 17 | hsa-miR-3116 | 7001 | −28.7 |
| 18 | hsa-miR-4286 | 6265 | −28.5 |
| 19 | hsa-miR-650 | 7348/7650 | −23.6/−28.3 |
| 20 | hsa-miR-4284 | 6872/7547 | −28.1/−24.1 |
| 21 | hsa-miR-143-3p | 7324 | −28 |
| 22 | hsa-miR-1287-5p | 6884 | −27.9 |
| 23 | hsa-miR-3178 | 7255 | −27.8 |
| 24 | hsa-miR-3174 | 6663/7031/7332 | −27.1/−26.8/−27.5 |
| 25 | hsa-miR-1322 | 7324 | −27.2 |
| 26 | hsa-miR-510-5p | 6467 | −27 |
| 27 | hsa-miR-3160-3p | 6659/7031/7332 | −26/−27/−25 |
| 28 | hsa-miR-4252 | 6647/7016/7317 | −24.9/−26.6/−26.8 |
| 29 | hsa-miR-132-5p | 6276/7547 | −24/−26.7 |
| 30 | hsa-miR-1909-3p | 7650 | −26.7 |
| 31 | hsa-miR-4265 | 7563 | −26.7 |
| 32 | hsa-miR-766-3p | 7006/7307 | −26.6/−20.1 |
| 33 | hsa-miR-4299 | 7660 | −26.6 |
| 34 | hsa-miR-3170 | 6820 | −26.1 |
| 35 | hsa-miR-4296 | 7566 | −25.8 |
| 36 | hsa-miR-1291 | 6343 | −25.6 |
| 37 | hsa-miR-25-5p | 7043 | −25.6 |
| 38 | hsa-miR-508-5p | 7007/7308 | −25.6/−21.3 |
| 39 | hsa-miR-542-3p | 6626 | −25.4 |
| 40 | hsa-miR-377-5p | 6670/7340 | −23.5/−24.6 |
| 41 | hsa-miR-4257 | 7342 | −24.6 |
| 42 | hsa-miR-485-5p | 6708/7077/7210 | −20.6/−24.5/−21.4 |
| 43 | hsa-miR-1228-3p | 6872/7541 | −24.3/−23.7 |
| 44 | hsa-miR-221-5p | 7000 | −24.1 |
| 45 | hsa-miR-138-5p | 7659 | −23.9 |
| 46 | hsa-miR-1827 | 7071/7351 | −23.8/−20.1 |
| 47 | hsa-miR-3176 | 6255 | −23.7 |
| 48 | hsa-miR-940 | 7071/7653 | −23.5/−22.5 |
| 49 | hsa-miR-490-3p | 6633 | −23.3 |
| 50 | hsa-miR-550a-5p | 7404 | −23.3 |
| 51 | hsa-miR-3155a | 6469 | −23.3 |
| 52 | hsa-miR-3199 | 7619 | −23.3 |
| 53 | hsa-miR-3128 | 6999 | −22.6 |
| 54 | hsa-miR-3125 | 7292 | −22.3 |
| 55 | hsa-miR-593-3p | 7146/7447 | −21.9/−21.9 |
| 56 | hsa-miR-122-3p | 6649 | −21.6 |
| 57 | hsa-miR-4316 | 7661 | −21.3 |
| 58 | hsa-miR-203a-3p | 7717 | −20.9 |
| 59 | hsa-miR-342-3p | 6870 | −20.9 |
| 60 | hsa-miR-625-5p | 7633 | −20.6 |
| 61 | hsa-miR-340-3p | 7446 | −20.3 |
| 62 | hsa-miR-4278 | 7639 | −20.3 |
Calculated by RNAhybrid software.
Fig. 1.
The correlation between the expression of CYP1A2 and the expression of miRNAs in non-tumor human liver samples. The correlation between the expression of CYP1A2 and the expression of hsa-miR-132-5p (A) or hsa-miR-221-5p (B) in non-tumor liver tissue samples based on data analysis derived from the TCGA dataset.
3.2. Hsa-miR-132-5p, but not hsa-miR-221-5p, suppressed CYP1A2 3′-UTR luciferase reporter gene activity
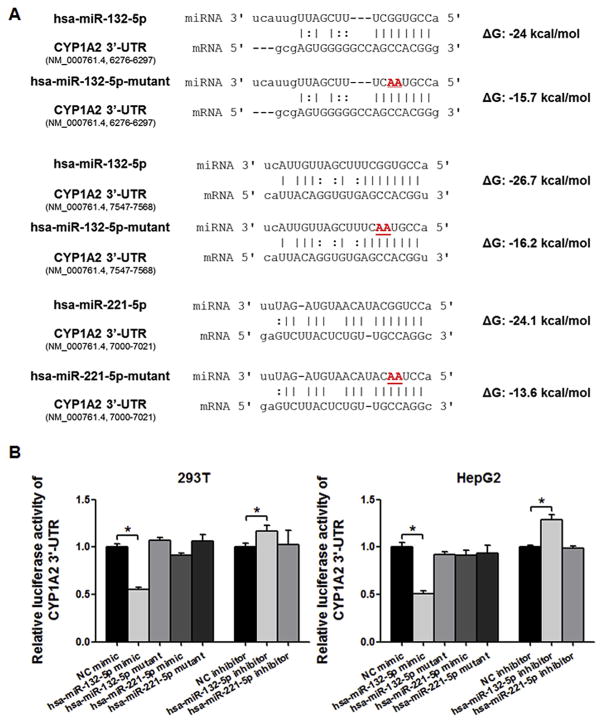
We explored the regulatory effects of hsa-miR-132-5p and hsa-miR-221-5p on the 3′-UTR of CYP1A2 using luciferase reporter assays and various miRNA mimics (Fig. 2A). As shown in Fig. 2B, the hsa-miR-132-5p mimic significantly decreased the luciferase activity produced by the reporter gene plasmid containing the 3′-UTR of CYP1A2 in 293T and HepG2 cells (55.3% and 50.8%, respectively, both P < 0.05). Notably, the inhibitory effect of hsa-miR-132-5p was abolished by an hsa-miR-132-5p inhibitor, which increased the luciferase activity in both cell lines (1.2-fold in 293T cells and 1.3-fold in HepG2 cells, both P < 0.05), compared with the negative control. However, neither the mimic nor the inhibitor of hsa-miR-221-5p changed significantly the luciferase activity in 293T or HepG2 cells. To confirm the specificity of hsa-miR-132-5p in suppressing the luciferase activity through binding to the CYP1A2 3′-UTR, we tested the inhibitory effect of the mutated version of the miRNA (Fig. 2A). Despite the presence of CYP1A2 3′-UTR, the hsa-miR-132-5p mutant failed to reduce luciferase activity (Fig. 2B). Collectively, these results demonstrated that hsa-miR-132-5p is a bona fide regulator of CYP1A2.
Fig. 2.
CYP1A2 3′-UTR luciferase reporter assays. (A) Sequences of wild type and mutant hsa-miR-132-5p and hsa-miR-221-5p, and targeted sequences of CYP1A2 3′-UTR are indicated. The free energies of hybridization between microRNA and targeted CYP1A2 3′-UTR predicted using RNAhybrid software are given. (B) hsa-miR-132-5p specifically regulates the luciferase activity produced by the reporter gene plasmid that contained the 3′-UTR of CYP1A2 both in 293 T and HepG2 cells, comparing to negative controls; whereas hsa-miR-221-5p does not show significant regulatory effects involving the CYP1A2 3′-UTR. Values are presented as the mean ± SEM from five independent experiments, *P < 0.05 versus NC mimic or NC inhibitor respectively.
3.3. Hsa-miR-132-5p interacted directly with CYP1A2 3′-UTR mRNA
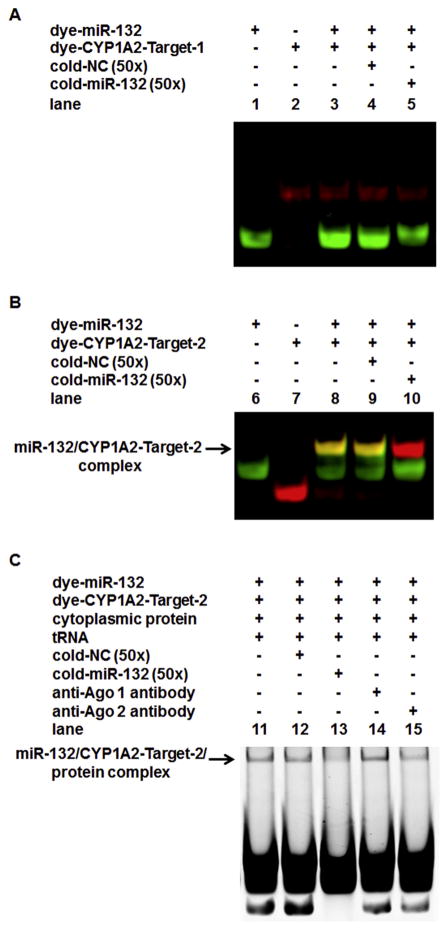
RNA EMSAs were performed to test whether hsa-miR-132-5p interacts directly with its target mRNA sequences in the 3′-UTR of CYP1A2. Two targeted sequences predicted within the CYP1A2 3′-UTR mRNA, CYP1A2-Target-1 (located at the nucleotide position 6276–6297 of the CYP1A2 gene) and CYP1A2-Target-2 (located at the nucleotide position 7547–7568 of the gene), were analyzed in the experiment. Dye-miR-132 (in green fluorescence) interacted with dye-CYP1A2-Target-2 (in red fluorescence) to form a stable miRNA/mRNA complex (Fig. 3B, lane 8, the yellow band, indicated by an arrow), whereas dye-miR-132 did not form a miRNA/mRNA complex with CYP1A2-Target-1 (Fig. 3A, lane 3). In competition assays, when unlabeled hsa-miR-132-5p (cold-miR-132, 50×) was added to the mixture, it appeared to consume almost the total amount of dye-CYP1A2-Target-2 present, resulting in a complex comprised of the excess unlabeled hsa-miR-132 with the dye-CYP1A2-Target-2 oligo, resulting in a red fluorescent band lacking the green fluorescence associated with dye-miR-132 (Fig. 3B, lane 10, indicated by an arrow). The complete absence of a yellow complex, indicating the presence of both dye-labeled oligonucleotides with electrophoretic migration coincident with that of the miRNA/CYP1A2-Target 2 RNA complex, is consistent with nearly total depletion of the specific complex detected in lane 8 by competition with the unlabeled oligonucleotide. In contrast, a similar effect was not observed when the miRNA negative control was applied (cold-NC, 50×, Fig. 3B, lane 9), confirming that the successful competition observed in lane 10 was sequence-specific. Adding HepaRG cytoplasmic extract to the hsa-miR-132-5p/CYP1A2-Target-2 mixture resulted in a shifted band with reduced electrophoretic mobility, indicating that a new complex was formed by miRNA-mRNA-protein interactions (Fig. 3C, lane 11, indicated by an arrow). The new band representing the hsa-miR-132-5p/CYP1A2-Target-2/protein complex was weakened by adding excess cold-miR-132 probes (Fig. 3C, lane 13). When the intensity of this new band in lane 13 was compared to that of the band in lane 11 representing the dye-hsa-miR-132-5p/dye-CYP1 A2-Target-2/protein complex, it was found that the intensity of the band in lane 13 was decreased to 68.7% (P < 0.05), indicating that the complex formed by the interaction among hsa-miR-132-5p, CYP1A2-Target-2 and proteins was also sequence-specific. Furthermore, addition of the anti-Ago 2 antibody to the mixture of hsa-miR-132-5p/CYP1A2-Target-2/pro tein reduced the intensity of the complex to 76.7% (P < 0.05, lane 15 versus lane 11), whereas addition of the anti-Ago 1 antibody to the mixture (lane 14) did not change the intensity of the hsa-miR-132-5p/CYP1A2-Target-2/protein complex, suggesting that the protein Ago-2 was involved in the miRNA-mRNA-protein interaction specifically, although the super-shift band was unable to be detected (it was beyond detection possibly due to the technical limitation). Therefore, these results exhibited the direct and specific interaction between hsa-miR-132-5p and its cognate target within the transcript of CYP1A2 3′-UTR.
Fig. 3.
RNA EMSAs for detecting the interaction between hsa-miR-132-5p and CYP1A2 3′-UTR mRNA. (A) Lane 1 to 5 showed no interactions between hsa-miR-132-5p (dye labeled miR-132, dye-miR-132, was indicated by green fluorescence) and its first target at the CYP1A2 3′-UTR mRNA (dye-labeled target-1, dye-CYP1A2-Target-1, located at nucleotide position 6276–6297 of the gene, indicated by red fluorescence). (B) Lane 6 to 10 demonstrate the specific interaction between hsa-miR-132-5p (dye-miR132, indicated by green fluorescence) and its second target at the CYP1A2 3′-UTR mRNA (dye-labeled target-2, dye-CYP1A2-Target-2, located at nucleotide position 7547–7568 of the gene, indicated in red fluorescence). A stable dye-miR-132/dye-CYP1A2-Target-2 complex in yellow fluorescence was formed, as shown in lane 8 indicated by an arrow. In competition assays, the excessive amount of unlabeled hsa-miR-132-5p (cold-miR-132, 50×) interacted with almost the total amount of dye-CYP1A2-Target-2, which competitively inhibited the interaction between the labeled dye-miR-132 and dye-CYP1A2-Target-2, thus the band in red fluorescence appeared in lane 10 (which represented the cold-miR-132/dye-CYP1A2-Target-2 complex, and was indicated by an arrow, whereas the excessive amount of unlabeled miRNA negative control (cold-NC, 50×) was applied, no such a band was observed in lane 9. (C) When the HepaRG cytoplasmic extract was added to the mixture of hsa-miR-132-5p and CYP1A2-Target-2, a new band with reduced electrophoretic mobility appeared in lane 11 (indicated by an arrow), suggesting that an hsa-miR-132-5p/CYP1A2-Target-2/protein complex was formed. As shown in lane 13, the hsa-miR-132-5p/ CYP1A2-Target-2/protein complex was weakened by adding an excessive amount of cold-miR-132 probes. The intensity of the band was decreased to 68.7 ± 1.9% (n = 5, P < 0.05) in comparison to that of in lane 12 (cold-NC, 50×). When the anti-Ago 2 antibody was further added to the mixture (lane 15), the band density of hsa-miR-132-5p/CYP1A2 mRNA/protein complex was reduced to 76.7 ± 2.6% (n = 5, P < 0.05, lane 15 versus lane 11). The addition of the anti-Ago 1 antibody to the mixture (lane 14) did not change the intensity of the hsa-miR-132-5p/CYP1A2-Target-2/protein complex. (For interpretation of the references to colour in this figure legend, the reader is referred to the web version of this article.)
3.4. Hsa-miR-132-5p regulates CYP1A2 mRNA and protein expression
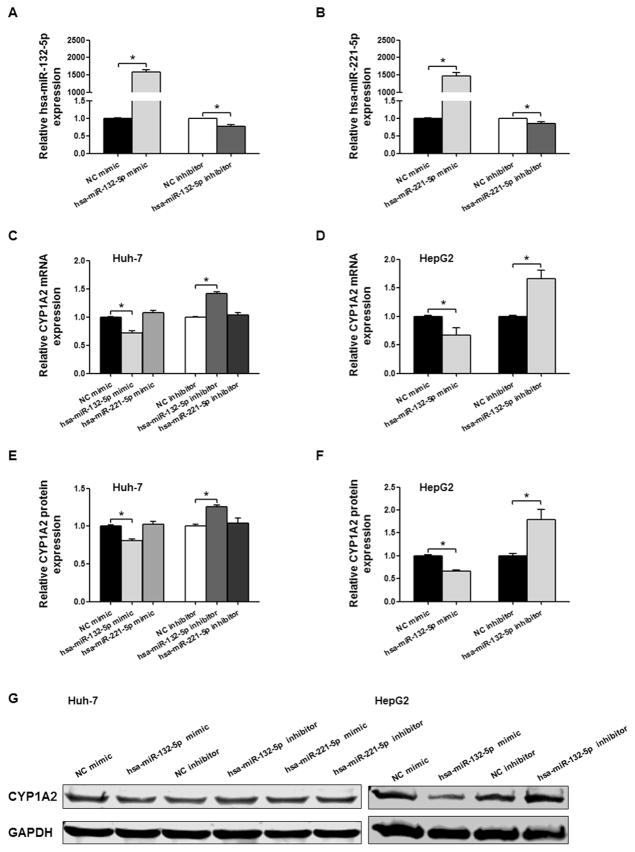
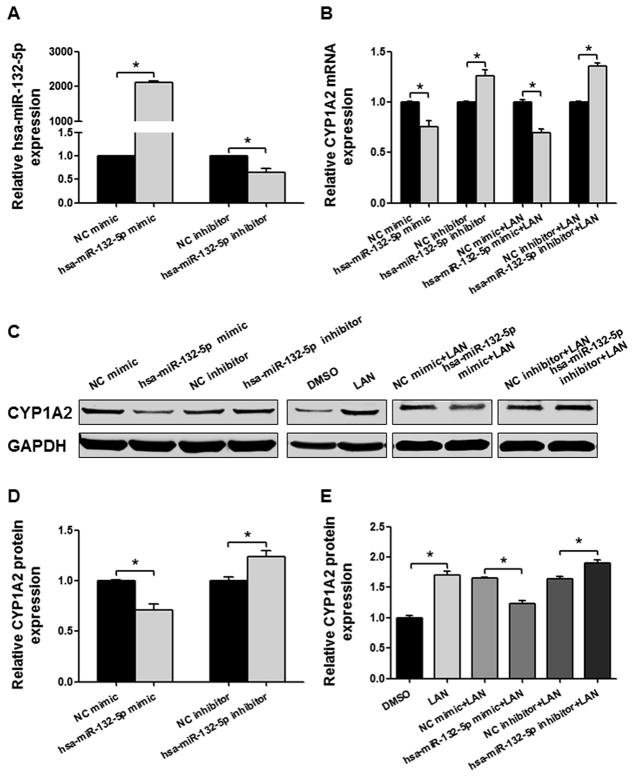
To address the regulatory role of hsa-miR-132-5p, we tested the effects of the hsa-miR-132-5p mimic and an antisense hsa-miR-132-5p inhibitor on the expression of CYP1A2 in Huh-7 human hepatic cancer cells. Transfection efficiencies for the mimic and the inhibitor were confirmed by measuring hsa-miR-132-5p and hsa-miR-221-5p expression levels in Huh-7 cells (Fig. 4A and B). Transfection of the hsa-miR-132-5p mimic suppressed the expression of CYP1A2 mRNA to 71.5%, compared with the negative control; whereas the hsa-miR-132-5p inhibitor displayed a “sponge” effect and elevated the expression of CYP1A2 mRNA by 1.4-fold (Fig. 4C, P < 0.05). In agreement with the observed changes in mRNA expression, reduced levels of CYP1A2 protein were detected by Western blot in the hsa-miR-132-5p mimic-treatment group (80.7%, P < 0.05), and increased CYP1A2 levels were found in the hsa-miR-132-5p inhibitor-treatment group compared with the control group (1.3-fold, P < 0.05, Fig. 4E and G). We also explored the effect of hsa-miR-221-5p on the expression of CYP1A2. Under the same conditions, hsa-miR-221-5p did not influence the expression of CYP1A2 mRNA transcripts (Fig. 4C) or CYP1A2 protein (Fig. 4E and G). The regulatory effects of hsa-miR-132-5p mimic and miRNA inhibitor on endogenous CYP1A2 expression were also confirmed in HepG2 cells. CYP1A2 mRNA and protein levels were decreased to 66.6% and 66.4% of those in control after hsa-miR-132-5p mimic transfection, and were increased to 1.7- and 1.8-fold of those in control upon hsa-miR-132-5p inhibition, respectively (Fig. 4D, F and G, all P < 0.05).
Fig. 4.
Detection of hsa-miR-132-5p and hsa-miR-221-5p regulatory effects on CYP1A2 expression in Huh-7 cells and HepG2 cells. Relative hsa-miR-132-5p (A) and hsa-miR-221-5p expression (B) in Huh-7 cells transfected with microRNA mimics or inhibitors. U6 was used as a loading control. (C) Relative CYP1A2 mRNA expression in Huh-7 cells when transfected with hsa-miR-132-5p or hsa-miR-221-5p mimics or inhibitors, and GAPDH mRNA expression was used as an internal control. (D) Relative CYP1A2 mRNA expression in HepG2 cells when transfected with hsa-miR-132-5p mimics or inhibitors, and GAPDH mRNA expression was used as an internal control. (E) Densitometry analysis shows the protein expression of CYP1A2 in Huh-7 cells transfected with hsa-miR-132-5p or hsa-miR-221-5p mimics or inhibitors, and GAPDH was used as loading control. (F) Densitometry analysis shows the protein expression of CYP1A2 in HepG2 cells transfected with hsa-miR-132-5p mimics or inhibitors, and GAPDH was used as loading control. (G) Representative Western blot analysis shows the protein expression of CYP1A2 in Huh-7 cells and HepG2 cells transfected with hsa-miR-132-5p or hsa-miR-221-5p mimics or inhibitors. The bar graph represents the densitometry analysis of CYP1A2 expression from five independent experiments and intensities of bands were normalized to the amount of GAPDH. All values are represented as the mean ± SEM from five independent experiments, * P < 0.05 versus NC mimic or NC inhibitor respectively.
3.5. Hsa-miR-132-5p suppressed endogenous and lansoprazole-induced CYP1A2 expression in HepaRG cells
The terminally differentiated hepatic cell line HepaRG that expresses drug metabolizing enzymes and transporters (DMETs) at levels similar to those of primary hepatocytes was selected to test the regulatory effects of hsa-miR-132-5p on the expression of the endogenous CYP1A2 gene. As shown in Fig. 5A, hsa-miR-132-5p levels in HepaRG cells were elevated by transfection with the miRNA mimic and decreased by transfection with the inhibitor. Similar to the observation in Huh-7 cells, transfection with hsa-miR-132-5p resulted in a statistically significant suppression of the endogenous CYP1A2 level. CYP1A2 mRNA and protein levels were decreased to 75.5% and 71.1% of those in controls by the transfection with the hsa-miR-132-5p mimic, and were increased to about 1.3- and 1.2-fold by transfection with the hsa-miR-132-5p inhibitor, respectively (Fig. 5B–D, all P < 0.05).
Fig. 5.
Hsa-miR-132-5p regulates endogenous and lansoprazole-induced exogenous CYP1A2 expression in HepaRG cells. (A) Relative hsa-miR-132-5p expression when transfected with microRNA mimics or inhibitors. U6 was used as a loading control. (B) Relative CYP1A2 mRNA expression when transfected with hsa-miR-132-5p mimics and inhibitors with or without lansoprazole, and GAPDH mRNA expression was used as an internal control. (C) Representative Western blot analysis shows the CYP1A2 expression transfected with hsa-miR-132-5p mimics and inhibitors with or without lansoprazole, and GAPDH was used as a loading control. (D, E) Densitometry analysis of CYP1A2 expression from five independent experiments and the intensities of bands were normalized to the amount of GAPDH. All values are represented as the mean ± SEM from five independent experiments (*, P < 0.05).
Lansoprazole is a well-recognized CYP1A2 inducer in hepatic cells [31] and a similar inductive effect on the expression of CYP1A2 was observed in HepaRG cells (Fig. 5C and E). Transfection with hsa-miR-132-5p suppressed lansoprazole-induced CYP1A2 expression. In the presence of lansoprazole, the mRNA and protein levels of CYP1A2 were decreased to 69.3% and 75.0% of those in the controls after hsa-miR-132-5p mimic transfection, respectively; whereas transfection with the hsa-miR-132-5p inhibitor increased the mRNA and protein levels of CYP1A2 to 1.4- and 1.2-fold of those in the controls, respectively (Fig. 5B, C and E, all P < 0.05).
3.6. Hsa-miR-132-5p suppressed endogenous and lansoprazole-induced CYP1A2 expression in Huh-7 and HepG2 cells
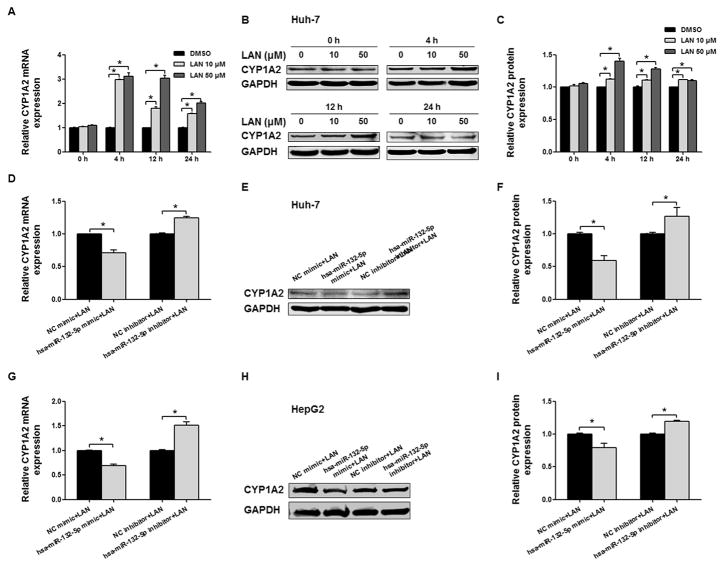
A previous study reported that CYP1A2 could be induced in the hepatic cancer cell line HepG2 by 50 μM lansoprazole [31]. As shown in Fig. 6A–C, the administration of lansoprazole to Huh-7 cells at concentrations of 10 and 50 μM for 4, 12, or 24 h significantly elevated the levels of both CYP1A2 mRNA and protein above their levels at the initial time point. The observed induction of CYP1A2 by 50 μM lansoprazole was most prominent at 4 and 12 h, which was comparable to the reported results in HepG2 cells. Consistent with its inhibitory effect on the expression of endogenous CYP1A2 in Huh-7 cells (Fig. 4), hsa-miR-132-5p also inhibited lansoprazole-induced expression of CYP1A2 in Huh-7 cells. The hsa-miR-132-5p mimic attenuated the elevation of CYP1A2 expression in the presence of lansoprazole at both the mRNA level (70.1% of that in control, P < 0.05) and the protein level (59.4% of that in control, P < 0.05), whereas inhibition of hsa-miR-132-5p by its inhibitor further increased the expression of CYP1A2 (1.2-and 1.3-fold of that in control, for mRNA and protein levels, respectively; both P < 0.05) (Fig. 6D–F).
Fig. 6.
Hsa-miR-132-5p regulates endogenous and lansoprazole-induced exogenous CYP1A2 expression in Huh-7 and HepG2 cells. (A) Relative CYP1A2 mRNA expression in Huh-7 cell when induced by lansoprazole (10 and 50 μM, dissolved in DMSO) at different time points (0, 4, 12, and 24 h), and GAPDH mRNA expression was used as an internal control. (B) Representative Western blot analysis shows the CYP1A2 expression in Huh-7 cells induced by lansoprazole (10 and 50 μM) at different time points (0, 4, 12, and 24 h), and GAPDH was used as loading control. (C) The bar graphs represent the densitometry analysis of CYP1A2 protein expression from five independent experiments (mean ± SEM) and the intensities of bands were normalized to the amount of GAPDH (*, P < 0.05). (D, E, F) Induction of CYP1A2 by lansoprazole was modulated by hsa-miR-132-5p in both mRNA and protein levels in Huh-7 cells, and GAPDH was used as an internal control. (G, H, I) Lansoprazole-induced exogenous CYP1A2 expression in HepG2 cells could be regulated by hsa-miR-132-5p, and GAPDH was used as an internal control. Error bars represent ± SEM (n = 5); *, P < 0.05.
Similar regulatory effects of hsa-miR-132-5p on lansoprazole-induced CYP1A2 expression were observed in HepG2 cells. The elevated CYP1A2 expression in the presence of lansoprazole was inhibited by hsa-miR-132-5p significantly (69.1% and 79.2% of those control for mRNA and protein levels after the transfection of mimic, respectively; 1.5- and 1.2-fold of those in control for mRNA and protein levels after the transfection of inhibitor, respectively; Fig. 6G–I, all P < 0.05).
3.7. Hsa-miR-132-5p inhibits the lansoprazole-induced CYP1A2 enzyme activity and attenuates CYP1A2-mediated flutamide-induced cytotoxicity
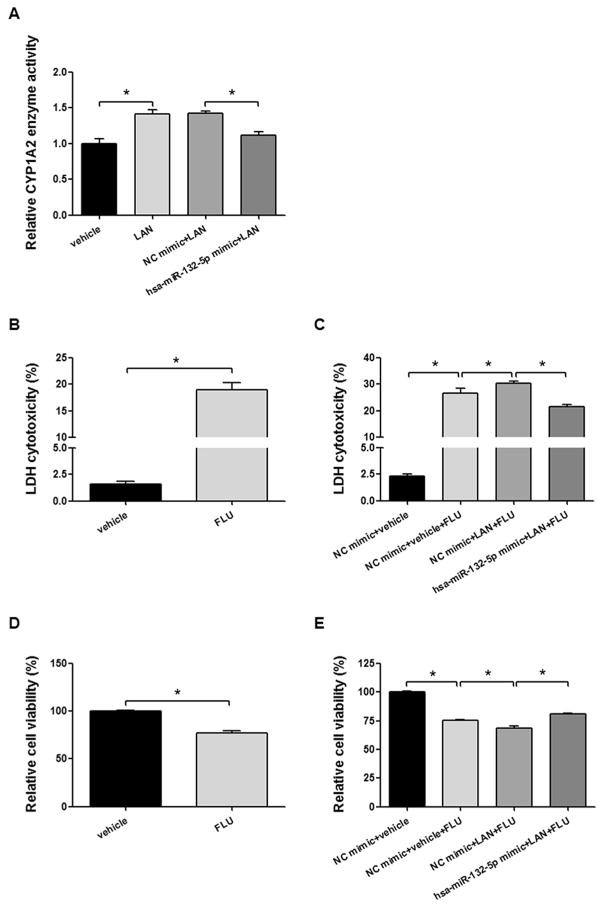
Lansoprazole induced CYP1A2 enzyme activity by 1.4-fold in HepG2 cells. Lansoprazole-induced CYP1A2 activity was decreased to 78.5% of its induced level by transfection with the hsa-miR-132-5p mimic (Fig. 7A, all P < 0.05). However, the hsa-miR-132-5p mimic did not inhibit CYP2B6 and CYP3A4 enzyme activity (data not shown). Similar results were also observed in Huh-7 cells (data not shown).
Fig. 7.
Hsa-miR-132-5p attenuates the lansoprazole-induced CYP1A2 enzyme activity and CYP1A2-mediated flutamide-dependent cytotoxicity in HepG2 cells. (A) HepG2 cells were treated by lansoprazole (LAN, 50 μM, dissolved in DMSO) for 12 h. LAN increased CYP1A2 enzyme activity to 1.4-fold. Hsa-miR-132-5p mimic decreased the LAN-induced CYP1A2 enzyme activity to 78.5%. (B, D) HepG2 cells were treated with flutamide (FLU, 1 mM, dissolved in ethanol) for 4 h. Cytotoxicity by LDH assays (B) and cell viability by MTT assays (D) were detected. (C, E) HepG2 cells were transfected with hsa-miR-132-5p mimic or negative control mimic (NC mimic), then treated by LAN (50 μM) for 12 h following FLU (1 mM) treatment for 4 h. Then cytotoxicity by LDH assays (C) and cell viability by MTT assays (E) were detected. Error bars represent ± SEM (n = 5); *, P < 0.05.
Since lansoprazole induces the expression and activity of CYP1A2 in hepatic cells, we explored whether it can influence the cytotoxic effect of flutamide. Hepatic cellular toxicity of flutamide was assessed using LDH and MTT assays in HepG2 cells. As shown in Fig. 7B, flutamide treatment resulted in 19.0% cytotoxicity in HepG2 cells using the LDH release assay, compared to 1.6% cytotoxicity associated with the vehicle control (Fig. 7B, P < 0.05), which is consistent with the cell viability results measured by the MTT assay, with 76.8% cells survival (in terms of 23.2% cytotoxicity) following flutamide treatment, compared to the vehicle control (Fig. 7D, P < 0.05). To explore whether hsa-miR-132-5p can modulate the cellular response generated by the co-treatment of lansoprazole and flutamide, we transfected the hsa-miR-132-5p mimic into HepG2 cells prior to co-treatment with lansoprazole and flutamide. The hsa-miR-132-5p mimic attenuated the effect of CYP1A2-mediated flutamide-dependent cytotoxicity. As shown by LDH assays in Fig. 7C, the cytotoxicity of flutamide was 26.6% in the group treated with negative control (NC) mimic + vehicle + flutamide, compared to the 2.3% in the group treated with NC mimic + vehicle (P < 0.05). The cytotoxicity observed in the group exposed to the combination of NC mimic + lansoprazole + flutamide was increased to 30.4% (P < 0.05), indicating that lansoprazole enhanced flutamide-dependent cell toxicity. However, the hsa-miR-132-5p mimic attenuated the lansoprazole-enhanced and flutamide-dependent cytotoxicity to 21.4% (P < 0.05), in the group treated with the hsa-miR-132-5p mimic + lansoprazole + flutamide. In the cell viability evaluation using MTT assays, similar results were observed. Briefly, transfection with the hsa-miR-132-5p mimic attenuated lansoprazole-mediated, flutamide-dependent cell death (P < 0.05, Fig. 7E). These observations indicate that hsa-miR-132-5p is a potential mediator participating in the effects of a drug-drug interaction between lansoprazole and flutamide by suppressing CYP1A2 activity.
4. Discussion
MiRNAs are key post-transcriptional regulators of gene expression and, thus, are involved in many biological and pathological processes, including metabolism, cell proliferation and death, and tumorigenesis [32]. Hsa-miR-132-5p has been reported to be associated with certain types of human malignancies. It plays a tumor suppressive role in several human malignancies, including HCC [33], prostate cancer [34], osteosarcoma [6], non-small cell lung cancer [35], and ovarian cancer [36]. By contrast, it was shown to be an oncogene in gastric cancer [37], colorectal carcinoma [38], pancreatic cancer [39], hemangioma [40], and chronic lymphocytic leukemia [41]. And its role in breast cancer is controversial, either promoting or suppressing tumorigenesis [40,42].
Much evidence indicates that miRNAs play an important role for inter-individual variability in the expression of pharmacologically important DMETs [11,43–47]. DMET-targeting miRNAs affect drug metabolism and efficacy; in turn, drugs influence the expression of miRNAs [12,21]. In the current study, we investigated the role of miRNAs in regulating an important drug metabolizing enzyme, CYP1A2. We first screened candidate miRNAs by in silico analysis and found 62 miRNAs (Table 2) that might potentially interact with the 3′-UTR of CYP1A2. Then, two miRNAs (hsa-miR-132-5p and hsa-miR-221-5p) were selected from 62 miRNAs for wet-lab validation, based on statistically significant inverse correlations between their expression levels and the expression levels of CYP1A2 mRNA in a group of non-tumor liver tissues. Finally, of the two miRNAs, only hsa-miR-132-5p was proved to exert an inhibitory effect on the expression of CYP1A2 at the transcript, protein and enzyme activity levels.
RNA EMSAs were performed to determine whether hsa-miR-132-5p was able to interact with the 3′-UTR of CYP1A2 directly. It was predicted that hsa-miR-132-5p could have two potential targeting sites in the 3′-UTR of CYP1A2. As to binding strength, the first binding site (CYP1A2-Target-1) at the nucleotide position 6276–6297 of the transcript was −24 kcal/mol; and the second binding site (CYP1A2-Target-2) at the nucleotide position 7547–7568 of the CYP1A2 3′-UTR was -26.7 kcal/mol. The results from EMSAs echoed the in silico calculations because only hsa-miR-132-5p interactions at the second binding site in the CYP1A2 3′-UTR could be detected under our experimental conditions. Upon adding the HepaRG cytoplasmic extracts, a shifted band was observed, indicating the formation of a miRNA/RNA/protein complex. The Argonaute protein family members, including Ago-1, Ago-2, Ago-3, and Ago-4, bind miRNAs to constitute key components of RNA-induced silencing complexes (RISCs) which function to suppress the expression of targeted mRNA transcripts. In our study, the miRNA/RNA/protein complex we observed was weakened when the Ago-2 antibody was added; in contrast, addition of Ago-1, -3, and -4 antibodies showed no effects on the density of the complex (data of Ago 3 and 4 not shown), suggesting that the effect of Ago-2 is specific. Although a super-shift band produced by the addition of the Ago-2 antibody was beyond detection due to technical limitations, EMSAs suggested that Ago-2 may be involved in the modulation of CYP1A2 expression.
A series of miRNA transfection assays using the terminally differentiated hepatic cell line HepaRG and hepatoma cell lines Huh-7 and HepG2 demonstrated that hsa-miR-132-5p suppressed the expression of CYP1A2 mRNA and protein significantly. The suppressive effect on the expression of CYP1A2 was not a byproduct of the inhibition of hepatic cell proliferation by hsa-miR-132-5p because cell viability was not affected after the transfection of hsa-miR-132-5p mimic into the hepatic cell lines Huh-7 and HepG2 (data not shown). Also, the expression and biological activity levels of other CYP enzymes, such as CYP2B6 and CYP3A4, were not affected by the transfection of hsa-miR-132-5p into these hepatic cell lines under the same conditions (data not shown).
Lansoprazole is a well-recognized inducer of CYP1A2, as demonstrated in the human hepatoma cell line HepG2 [31,48]. In our study, CYP1A2 could also be induced by lansoprazole in terminally differentiated HepaRG cells and in Huh-7 human hepatoma cells in a concentration-dependent manner. According to our results in Huh-7 cells, the strongest induction was observed at 4 and 12 h, and more obvious effects appeared in cells treated with 50 μM lansoprazole than those exposed to 10 μM lansoprazole. Further, lansoprazole-induced CYP1A2 expression and biological activity could be suppressed by hsa-miR-132-5p in HepaRG, Huh-7, and HepG2 cells.
The PPI lansoprazole is a commonly used drug in clinical practice to inhibit the production of stomach acid in patients. Often, it is co-administered with other drugs; therefore, it is meaningful to investigate drug-drug interaction effects involving lansoprazole [49–52]. For example, to relieve the gastrointestinal side effects (nausea and vomiting) during the treatment of advanced prostate cancer patients with flutamide (its common side effects include nausea and vomiting), lansoprazole might be co-administered. However, the interaction between lansoprazole and flutamide should be considered since a severe adverse reaction could occur. Flutamide is an androgen receptor antagonist that is primarily prescribed to treat advanced prostate cancer in men and to treat acne, hirsutism, and alopecia in women with polycystic ovary syndrome [53]. Despite the efficacy of its anti-androgen effects, flutamide-dependent hepatotoxicity represents a significant health concern, since the incidence of the adverse reaction could be as high as 9% in patients [54], and the severity of flutamide-dependent hepatotoxicity could lead to liver transplantation or death [55]. As a result, the US Food and Drug Administration issued a Black Box Warning to alert doctors and patients that “flutamide can cause liver failure.” Although the mechanism of action for flutamide-dependent hepatotoxicity largely remains unclear, several metabolites, such as 2-hydroxylflutamide [27] and 4-nitro-3-(trifluoromethyl)pheny lamine or FLU-1 [56], have been reported to associate with metabolite-triggered hepatotoxicity. We think that the mechanisms of flutamide-induced hepatotoxicity are complicated and multiple toxic metabolites are possibly involved in the process. In human hepatocytes, the major metabolite 2-hydroxyflutamide at least partially contributes to flutamide-induced hepatotoxicity. As a mitochondrial toxicant, upon binding to several key proteins in the mitochondrial electron transport chain, 2-hydroxyflutamide can inhibit the enzymatic activities of NADH ubiquinone oxidoreductase, succinate dehydrogenase, and ATP synthase, thus decreasing cellular ATP content, weakening the mitochondrial respiratory capacity, and increasing the level of superoxide, finally resulting in cell apoptosis [27,57]. Importantly, the catalytic activity of CYP1A2 is primarily responsible for converting flutamide to the toxic metabolite 2-hydroxyflutamide [25]. In the current study, the observed cellular toxicity of flutamide in HepG2 cells correlated inversely with hsa-miR-132-5p suppression of CYP1A2 activity. When increased CYP1A2 expression was induced by lansoprazole, flutamide toxicity also increased. However, transfection with hsa-miR-132-5p resulted in suppression of CYP1A2 enzymatic activity and decreased flutamide toxicity, demonstrating that hsa-miR-132-5p attenuated CYP1A2-mediated, lansoprazole-induced, flutamide-dependent hepatic cell toxicity. These results may provide insights on the role of miRNAs in preventing or treating CYP1A2-mediated adverse drug reactions.
In conclusion, using integrative approaches, including in silico and in vitro methods, we demonstrated that hsa-miR-132-5p directly interacts with the CYP1A2 3′-UTR to suppress its expression, which may play an important role in drug-drug interactions involving CYP1A2.
Acknowledgments
This study was supported and funded by the National Center for Toxicological Research (NCTR), U.S. Food and Drug Administration with additional support to Y.C. from the National Natural Science Foundation of China (Grant No. 81672408). Y.C., L.Z., Y.W., and Z. R. were supported by the appointment to the Postgraduate Research Program at the NCTR administered by the Oak Ridge Institute for Science Education through an interagency agreement between the U.S. Department of Energy and the U.S. Food and Drug Administration (FDA).
Abbreviations
- CYP
cytochrome P450
- miRNAs
microRNAs
- DMETs
drug metabolizing enzymes and transporters
- Ago
argonaute RISC catalytic component
- RISC
RNA-induced silencing complex
- FBS
fetal bovine serum
- EMSA
electrophoretic mobility shift assay
- HCC
hepatocellular carcinoma
- PPI
proton pump inhibitor
Footnotes
Conflict of interest
The authors declare no conflict of interest associated with this manuscript. The information in these materials is not a formal dissemination of information by the U.S. Food and Drug Administration.
Disclaimer
The information in these materials is not a formal dissemination of the U.S. Food and Drug Administration.
Author contributions
Y.C., D.Y., and B.N. proposed and designed the study. Y.C. finished the first draft of the manuscript. Y.C., L.Z., Y.W., J.Z., and B. K. conducted experiments. C.Y., L.Z., W.H.T., B.K., S.C., Z.R., L.G. N. M., F.Q., K.H., D.L., D.Y., W.T., and B.N. analyzed data and revised the manuscript.
References
- 1.Anzenbacher P, Anzenbacherova E. Cytochromes P450 and metabolism of xenobiotics. Cell Mol Life Sci. 2001;58:737–747. doi: 10.1007/PL00000897. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Heller F. Genetics/genomics and drug effects. Acta Clin Belg. 2013;68:77–80. doi: 10.2143/ACB.3210. [DOI] [PubMed] [Google Scholar]
- 3.Lin J, Schyschka L, Muhl-Benninghaus R, Neumann J, Hao L, Nussler N, et al. Comparative analysis of phase I and II enzyme activities in 5 hepatic cell lines identifies Huh-7 and HCC-T cells with the highest potential to study drug metabolism. Arch Toxicol. 2012;86:87–95. doi: 10.1007/s00204-011-0733-y. [DOI] [PubMed] [Google Scholar]
- 4.Masek V, Anzenbacherova E, Etrych T, Strohalm J, Ulbrich K, Anzenbacher P. Interaction of N-(2-hydroxypropyl)methacrylamide copolymer-doxorubicin conjugates with human liver microsomal cytochromes P450: comparison with free doxorubicin. Drug Metab Dispos. 2011;39:1704–1710. doi: 10.1124/dmd.110.037986. [DOI] [PubMed] [Google Scholar]
- 5.Wang B, Zhou SF. Synthetic and natural compounds that interact with human cytochrome P450 1A2 and implications in drug development. Curr Med Chem. 2009;16:4066–4218. doi: 10.2174/092986709789378198. [DOI] [PubMed] [Google Scholar]
- 6.Yang J, Gao T, Tang J, Cai H, Lin L, Fu S. Loss of microRNA-132 predicts poor prognosis in patients with primary osteosarcoma. Mol Cell Biochem. 2013;381:9–15. doi: 10.1007/s11010-013-1677-8. [DOI] [PubMed] [Google Scholar]
- 7.Simonsson M, Veerla S, Markkula A, Rose C, Ingvar C, Jernstrom H. CYP1A2–a novel genetic marker for early aromatase inhibitor response in the treatment of breast cancer patients. BMC Cancer. 2016;16:256. doi: 10.1186/s12885-016-2284-3. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Zhou SF, Yang LP, Zhou ZW, Liu YH, Chan E. Insights into the substrate specificity, inhibitors, regulation, and polymorphisms and the clinical impact of human cytochrome P450 1A2. AAPS J. 2009;11:481–494. doi: 10.1208/s12248-009-9127-y. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Ghotbi R, Christensen M, Roh HK, Ingelman-Sundberg M, Aklillu E, Bertilsson L. Comparisons of CYP1A2 genetic polymorphisms, enzyme activity and the genotype-phenotype relationship in Swedes and Koreans. Eur J Clin Pharmacol. 2007;63:537–546. doi: 10.1007/s00228-007-0288-2. [DOI] [PubMed] [Google Scholar]
- 10.Quattrochi LC, Vu T, Tukey RH. The human CYP1A2 gene and induction by 3-methylcholanthrene. A region of DNA that supports AH-receptor binding and promoter-specific induction. J Biol Chem. 1994;269:6949–6954. [PubMed] [Google Scholar]
- 11.Swart M, Dandara C. Genetic variation in the 3’-UTR of CYP1A2, CYP2B6, CYP2D6, CYP3A4, NR1I2, and UGT2B7: potential effects on regulation by microRNA and pharmacogenomics relevance. Front Genet. 2014;5:167. doi: 10.3389/fgene.2014.00167. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Koturbash I, Tolleson WH, Guo L, Yu D, Chen S, Hong H, et al. microRNAs as pharmacogenomic biomarkers for drug efficacy and drug safety assessment. Biomark Med. 2015;9:1153–1176. doi: 10.2217/bmm.15.89. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Guo H, Ingolia NT, Weissman JS, Bartel DP. Mammalian microRNAs predominantly act to decrease target mRNA levels. Nature. 2010;466:835–840. doi: 10.1038/nature09267. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Yu D, Green B, Marrone A, Guo Y, Kadlubar S, Lin D, et al. Suppression of CYP2C9 by microRNA hsa-miR-128-3p in human liver cells and association with hepatocellular carcinoma. Sci Rep. 2015;5:8534. doi: 10.1038/srep08534. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Rieger JK, Reutter S, Hofmann U, Schwab M, Zanger UM. Inflammation-associated microRNA-130b down-regulates cytochrome P450 activities and directly targets CYP2C9. Drug Metab Dispos. 2015;43:884–888. doi: 10.1124/dmd.114.062844. [DOI] [PubMed] [Google Scholar]
- 16.Yu D, Green B, Tolleson WH, Jin Y, Mei N, Guo Y, et al. MicroRNA hsa-miR-29a-3p modulates CYP2C19 in human liver cells. Biochem Pharmacol. 2015;98:215–223. doi: 10.1016/j.bcp.2015.08.094. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Mohri T, Nakajima M, Fukami T, Takamiya M, Aoki Y, Yokoi T. Human CYP2E1 is regulated by miR-378. Biochem Pharmacol. 2010;79:1045–1052. doi: 10.1016/j.bcp.2009.11.015. [DOI] [PubMed] [Google Scholar]
- 18.Pan YZ, Gao W, Yu AM. MicroRNAs regulate CYP3A4 expression via direct and indirect targeting. Drug Metab Dispos. 2009;37:2112–2117. doi: 10.1124/dmd.109.027680. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Ingelman-Sundberg M, Sim SC, Gomez A, Rodriguez-Antona C. Influence of cytochrome P450 polymorphisms on drug therapies: pharmacogenetic, pharmacoepigenetic and clinical aspects. Pharmacol Ther. 2007;116:496–526. doi: 10.1016/j.pharmthera.2007.09.004. [DOI] [PubMed] [Google Scholar]
- 20.Ramamoorthy A, Skaar TC. In silico identification of microRNAs predicted to regulate the drug metabolizing cytochrome P450 genes. Drug Metab Lett. 2011;5:126–131. doi: 10.2174/187231211795305258. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Mishra PJ. The miRNA-drug resistance connection: a new era of personalized medicine using noncoding RNA begins. Pharmacogenomics. 2012;13:1321–1324. doi: 10.2217/pgs.12.121. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Diaz D, Fabre I, Daujat M, Saint Aubert B, Bories P, Michel H, et al. Omeprazole is an aryl hydrocarbon-like inducer of human hepatic cytochrome P450. Gastroenterology. 1990;99:737–747. doi: 10.1016/0016-5085(90)90963-2. [DOI] [PubMed] [Google Scholar]
- 23.Ettinger DS, Armstrong DK, Barbour S, Berger MJ, Bierman PJ, Bradbury B, et al. Antiemesis. J Natl Compr Can Net. 2012;10:456–485. doi: 10.6004/jnccn.2012.0047. [DOI] [PubMed] [Google Scholar]
- 24.AbuHammad S, Zihlif M. Gene expression alterations in doxorubicin resistant MCF7 breast cancer cell line. Genomics. 2013;101:213–220. doi: 10.1016/j.ygeno.2012.11.009. [DOI] [PubMed] [Google Scholar]
- 25.Shet MS, McPhaul M, Fisher CW, Stallings NR, Estabrook RW. Metabolism of the antiandrogenic drug (Flutamide) by human CYP1A2. Drug Metab Dispos: Biol Fate Chem. 1997;25:1298–1303. [PubMed] [Google Scholar]
- 26.Gomez JL, Dupont A, Cusan L, Tremblay M, Suburu R, Lemay M, et al. Incidence of liver toxicity associated with the use of flutamide in prostate cancer patients. Am J Med. 1992;92:465–470. doi: 10.1016/0002-9343(92)90741-s. [DOI] [PubMed] [Google Scholar]
- 27.Ball AL, Kamalian L, Alfirevic A, Lyon JJ, Chadwick AE. Identification of the additional mitochondrial liabilities of 2-hydroxyflutamide when compared with its parent compound, flutamide in HepG2 cells. Toxicol Sci: Off J Soc Toxicol. 2016;153:341–351. doi: 10.1093/toxsci/kfw126. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Choucha Snouber L, Bunescu A, Naudot M, Legallais C, Brochot C, Dumas ME, et al. Metabolomics-on-a-chip of hepatotoxicity induced by anticancer drug flutamide and Its active metabolite hydroxyflutamide using HepG2/C3a microfluidic biochips. Toxicol Sci: Off J Soc Toxicol. 2013;132:8–20. doi: 10.1093/toxsci/kfs230. [DOI] [PubMed] [Google Scholar]
- 29.Tolosa L, Gomez-Lechon MJ, Perez-Cataldo G, Castell JV, Donato MT. HepG2 cells simultaneously expressing five P450 enzymes for the screening of hepatotoxicity: identification of bioactivable drugs and the potential mechanism of toxicity involved. Arch Toxicol. 2013;87:1115–1127. doi: 10.1007/s00204-013-1012-x. [DOI] [PubMed] [Google Scholar]
- 30.Fau D, Eugene D, Berson A, Letteron P, Fromenty B, Fisch C, et al. Toxicity of the antiandrogen flutamide in isolated rat hepatocytes. J Pharmacol Exp Ther. 1994;269:954–962. [PubMed] [Google Scholar]
- 31.Krusekopf S, Roots I, Hildebrandt AG, Kleeberg U. Time-dependent transcriptional induction of CYP1A1, CYP1A2 and CYP1B1 mRNAs by H+/K+-ATPase inhibitors and other xenobiotics. Xenobiotica. 2003;33:107–118. doi: 10.1080/0049825021000023978. [DOI] [PubMed] [Google Scholar]
- 32.Filipowicz W, Bhattacharyya SN, Sonenberg N. Mechanisms of post-transcriptional regulation by microRNAs: are the answers in sight? Nat Rev Genet. 2008;9:102–114. doi: 10.1038/nrg2290. [DOI] [PubMed] [Google Scholar]
- 33.Liu K, Li X, Cao Y, Ge Y, Wang J, Shi B. MiR-132 inhibits cell proliferation, invasion and migration of hepatocellular carcinoma by targeting PIK3R3. Int J Oncol. 2015;47:1585–1593. doi: 10.3892/ijo.2015.3112. [DOI] [PubMed] [Google Scholar]
- 34.Formosa A, Lena AM, Markert EK, Cortelli S, Miano R, Mauriello A, et al. DNA methylation silences miR-132 in prostate cancer. Oncogene. 2013;32:127–134. doi: 10.1038/onc.2012.14. [DOI] [PubMed] [Google Scholar]
- 35.Liu X, Yan S, Pei C, Cui Y. Decreased microRNA-132 and its function in human non-small cell lung cancer. Mol Med Rep. 2015;11:3601–3608. doi: 10.3892/mmr.2015.3222. [DOI] [PubMed] [Google Scholar]
- 36.Chung YW, Bae HS, Song JY, Lee JK, Lee NW, Kim T, et al. Detection of microRNA as novel biomarkers of epithelial ovarian cancer from the serum of ovarian cancer patients. Int J Gynecol Cancer. 2013;23:673–679. doi: 10.1097/IGC.0b013e31828c166d. [DOI] [PubMed] [Google Scholar]
- 37.Gao FY, Liu QY, Yuan L, Xuan SY. Upregulation of microRNA-132 in gastric cancer promotes cell proliferation via retinoblastoma 1 targeting. Mol Med Rep. 2015;12:7005–7010. doi: 10.3892/mmr.2015.4253. [DOI] [PubMed] [Google Scholar]
- 38.Schetter AJ, Leung SY, Sohn JJ, Zanetti KA, Bowman ED, Yanaihara N, et al. MicroRNA expression profiles associated with prognosis and therapeutic outcome in colon adenocarcinoma. JAMA. 2008;299:425–436. doi: 10.1001/jama.299.4.425. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Park JK, Henry JC, Jiang J, Esau C, Gusev Y, Lerner MR, et al. miR-132 and miR-212 are increased in pancreatic cancer and target the retinoblastoma tumor suppressor. Biochem Biophys Res Commun. 2011;406:518–523. doi: 10.1016/j.bbrc.2011.02.065. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Anand S, Majeti BK, Acevedo LM, Murphy EA, Mukthavaram R, Scheppke L, et al. MicroRNA-132-mediated loss of p120RasGAP activates the endothelium to facilitate pathological angiogenesis. Nature medicine. 2010;16:909–914. doi: 10.1038/nm.2186. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Calin GA, Liu CG, Sevignani C, Ferracin M, Felli N, Dumitru CD, et al. MicroRNA profiling reveals distinct signatures in B cell chronic lymphocytic leukemias. Proc Natl Acad Sci USA. 2004;101:11755–11760. doi: 10.1073/pnas.0404432101. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Li S, Meng H, Zhou F, Zhai L, Zhang L, Gu F, et al. MicroRNA-132 is frequently down-regulated in ductal carcinoma in situ (DCIS) of breast and acts as a tumor suppressor by inhibiting cell proliferation. Pathol Res Pract. 2013;209:179–183. doi: 10.1016/j.prp.2012.12.002. [DOI] [PubMed] [Google Scholar]
- 43.Tsuchiya Y, Nakajima M, Takagi S, Taniya T, Yokoi T. MicroRNA regulates the expression of human cytochrome P450 1B1. Cancer Res. 2006;66:9090–9098. doi: 10.1158/0008-5472.CAN-06-1403. [DOI] [PubMed] [Google Scholar]
- 44.Yu D, Tolleson WH, Knox B, Jin Y, Guo L, Guo Y, et al. Modulation of ALDH5A1 and SLC22A7 by microRNA hsa-miR-29a-3p in human liver cells. Biochem Pharmacol. 2015;98:671–680. doi: 10.1016/j.bcp.2015.09.020. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Jin Y, Yu D, Tolleson WH, Knox B, Wang Y, Chen S, et al. MicroRNA hsa-miR-25-3p suppresses the expression and drug induction of CYP2B6 in human hepatocytes. Biochem Pharmacol. 2016;113:88–96. doi: 10.1016/j.bcp.2016.06.007. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Wang Y, Yu D, Tolleson WH, Yu LR, Green B, Zeng L, et al. A systematic evaluation of microRNAs in regulating human hepatic CYP2E1. Biochem Pharmacol. 2017 doi: 10.1016/j.bcp.2017.04.020. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.Zeng L, Chen Y, Wang Y, Yu LR, Knox B, Chen J, et al. MicroRNA hsa-miR-370-3p suppresses the expression and induction of CYP2D6 by facilitating mRNA degradation. Biochem Pharmacol. 2017 doi: 10.1016/j.bcp.2017.05.018. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48.Preissner S, Kroll K, Dunkel M, Senger C, Goldsobel G, Kuzman D, et al. SuperCYP: a comprehensive database on Cytochrome P450 enzymes including a tool for analysis of CYP-drug interactions. Nucleic Acids Res. 2010;38:D237–D243. doi: 10.1093/nar/gkp970. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49.Dilger K, Zheng Z, Klotz U. Lack of drug interaction between omeprazole, lansoprazole, pantoprazole and theophylline. Br J Clin Pharmacol. 1999;48:438–444. doi: 10.1046/j.1365-2125.1999.00043.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50.Granneman GR, Karol MD, Locke CS, Cavanaugh JH. Pharmacokinetic interaction between lansoprazole and theophylline. Ther Drug Monit. 1995;17:460–464. doi: 10.1097/00007691-199510000-00005. [DOI] [PubMed] [Google Scholar]
- 51.Wedemeyer RS, Blume H. Pharmacokinetic drug interaction profiles of proton pump inhibitors: an update. Drug Saf. 2014;37:201–211. doi: 10.1007/s40264-014-0144-0. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 52.Vakily M, Lee RD, Wu J, Gunawardhana L, Mulford D. Drug interaction studies with dexlansoprazole modified release (TAK-390MR), a proton pump inhibitor with a dual delayed-release formulation: results of four randomized, double-blind, crossover, placebo-controlled, single-centre studies. Clin Drug Investig. 2009;29:35–50. doi: 10.2165/0044011-200929010-00004. [DOI] [PubMed] [Google Scholar]
- 53.Osculati A, Castiglioni C. Fatal liver complications with flutamide. Lancet. 2006;367:1140–1141. doi: 10.1016/S0140-6736(06)68498-5. [DOI] [PubMed] [Google Scholar]
- 54.Cetin M, Demirci D, Unal A, Altinbas M, Guven M, Unluhizarci K. Frequency of flutamide induced hepatotoxicity in patients with prostate carcinoma. Hum Exp Toxicol. 1999;18:137–140. doi: 10.1177/096032719901800301. [DOI] [PubMed] [Google Scholar]
- 55.Garcia Cortes M, Andrade RJ, Lucena MI, Sanchez Martinez H, Fernandez MC, Ferrer T, et al. Flutamide-induced hepatotoxicity: report of a case series. Rev Esp Enferm Dig. 2001;93:423–432. [PubMed] [Google Scholar]
- 56.Matsuzaki Y, Nagai D, Ichimura E, Goda R, Tomura A, Doi M, et al. Metabolism and hepatic toxicity of flutamide in cytochrome P450 1A2 knockout SV129 mice. J Gastroenterol. 2006;41:231–239. doi: 10.1007/s00535-005-1749-y. [DOI] [PubMed] [Google Scholar]
- 57.Coe KJ, Jia Y, Ho HK, Rademacher P, Bammler TK, Beyer RP, et al. Comparison of the cytotoxicity of the nitroaromatic drug flutamide to its cyano analogue in the hepatocyte cell line TAMH: evidence for complex I inhibition and mitochondrial dysfunction using toxicogenomic screening. Chem Res Toxicol. 2007;20:1277–1290. doi: 10.1021/tx7001349. [DOI] [PMC free article] [PubMed] [Google Scholar]