Abstract
Rubella is a viral infection that may cause fetal death or congenital defects, known as congenital rubella syndrome (CRS), during early pregnancy. The World Health Organization (WHO) recommends that countries assess the burden of rubella and CRS, including the determination of genotypes of circulating viruses. The goal of this study was to identify the genotypes of rubella viruses in the Democratic Republic of the Congo (DRC). Serum or throat swab samples were collected through the measles surveillance system. Sera that tested negative for measles IgM antibody were tested for rubella IgM antibody. Serum collected within 4 days of rash onset and throat swabs were screened by real-time RT-PCR for rubella virus RNA. For positive samples, an amplicon of the E1 glycoprotein gene was amplified by RT-PCR and sequenced. 11733 sera were tested for rubella IgM and 2816 (24%) were positive; 145 (5%) were tested for the presence of rubella RNA by real-time RT-PCR and 10 (7%) were positive. Seventeen throat swabs were analyzed by RT-PCR and three were positive. Sequences were obtained from eight of the positive samples. Phylogenetic analysis showed that the DRC rubella viruses belonged to genotypes 1B, 1E, 1G, and 2B. This report provides the first information on the genotypes of rubella virus circulating in the DRC. These data contribute to a better understanding of rubella burden and the dynamics of rubella virus circulation in Africa. Efforts to establish rubella surveillance in the DRC are needed to support rubella elimination in Africa.
Keywords: rubella, genotyping, DRC, Africa
INTRODUCTION
Rubella is a contagious infection caused by rubella virus that occurs usually in children and young adults. This illness has serious consequences in pregnant women causing a variety of birth defects known as congenital rubella syndrome (CRS) [Reef et al., 2011]. Worldwide, an estimated 110,000 babies are born with CRS every year [WHO, 2012]. Rubella virus is the sole member of the Rubivirus genus in the Togaviridae family. The genome of rubella virus is a single-stranded positive sense RNA of approximately 10 kb. The genome encodes two non-structural proteins (P90 and P150) and three virion proteins: the capsid protein and two envelope glycoproteins E2 and E1. Although rubella is serologically monotypic, sequence analyses of the E1 glycoprotein showed that distinct genetic variants of rubella virus exist. Rubella virus is classified into two clades; Clade 1 is divided in 10 genotypes (1a, 1B, 1C, 1D, 1E, 1F, 1G, 1H, 1I, and 1J) of which nine are recognized (uppercase letter) and one is provisional (lowercase letter). Clade 2 is divided in three genotypes (2A, 2B, and 2C) [WHO, 2013]. The World Health Organization (WHO) standard sequence window for rubella genotyping is a 739 nucleotide (nt) region (nts 8731–9469) within the E1 glycoprotein.
The WHO Measles and Rubella Laboratory Network has recommended the collection of rubella genotypic data to support the global control and elimination program [Centers-for-Disease-Control-and-Prevention, 2005].
The Democratic Republic of Congo (DRC) is a developing country with a population of 73 million (2012 estimate) living within an area of 2,345,000 km2 [CIA, 2012]. The national expanded program on immunization in the DRC includes the first dose of measles vaccine at 9 months of age. Rubella vaccination has not been introduced in the vaccination schedule in the DRC and rubella-specific surveillance is not in place. As a consequence, information about rubella and CRS in the DRC is very limited. The WHO recommends that countries without rubella vaccination program assess the burden of rubella and CRS before starting a routine vaccination program [WHO, 2011]. In 2004, the DRC established measles case-based surveillance following the WHO African Regional Office (WHO/AFRO) measles surveillance guidelines [WHO/AFRO, 2004]. The case definition for measles surveillance also captures some rubella cases. The surveillance program in support of measles elimination in the African region recommended that cases that tested negative or indeterminate for measles IgM antibody be tested for rubella IgM antibody [WHO/AFRO, 2004]. Virological surveillance data on rubella viruses are used to track progress towards the goal of rubella elimination, help with case classification, monitor transmission pathways, and identify interruption of transmission of endemic genotypes of the virus.
This article summarizes the results of rubella testing obtained through the measles case-based surveillance system. The epidemiology of laboratory-confirmed rubella cases and genotyping of rubella virus strains identified in the DRC from 2004 to 2013 are described.
MATERIALS AND METHODS
Sample Collection
All samples were collected in the DRC as part of routine measles surveillance from 2004 to 2013. Serum samples were collected from patients who met the measles case definition, using the WHO/AFRO guideline for measles surveillance [WHO/AFRO, 2004]. Throat swabs samples were collected when possible from the same patients. A suspected measles case was defined as any person with generalized maculopapular rash and fever plus one of the following: cough, coryza or conjunctivitis, or any person in whom a clinician suspects measles.
Enzyme-Linked Immune-Sorbent Assay (ELISA)
All samples were sent to the national reference laboratory at the Institut National de Recherches Biomedical (INRB) for laboratory testing. All samples were first screened for measles IgM antibody by ELISA using a commercial kit, Enzygnost® anti-measles virus IgM (Siemens, Marburg, Germany). Samples that were determined to be measles IgM negative or indeterminate were tested for rubella IgM antibody using the Enzygnost® anti-rubella virus IgM kit (Siemens). Patients with rubella IgM positive samples were classified as laboratory confirmed rubella cases.
Laboratory Molecular Testing of Rubella Samples
Screening of rubella IgM positive serum and throat swabs from cases that were also IgM positive by serology, collected between 2006 and 2013 was undertaken using the RT-PCR technique to detect rubella virus RNA as described in Namuwulya et al. [2014]. Briefly, the end point RT-PCR was performed using the Qiagen One-Step RT-PCR kit (Qiagen, Valencia, CA) and one forward and two reverse primers that amplify a 185-bp region in the rubella E1 protein coding region. All RT-PCR positive samples and all samples positive for rubella IgM by ELISA tests were sent to the Centers for Disease Control and Prevention (CDC) in Atlanta, USA, for sequencing and genotyping. At the CDC, blood derived samples were used for further testing if they were collected less than four days from symptom onset, or if the date of collection was unknown, since previous work showed that blood samples collected later than three days after onset were unlikely to be positive [Zheng et al., 2013].
RNA Extraction and FTA Cards
Serum and throat samples, 200 μl of each, were either directly added on 1/4 inch punches from an FTA card (Capitol Scientific, Austin, TX) for transportation purposes or RNA was first extracted from 140 μl of the serum/throat swab specimens using the QiaAmp Viral RNA Mini Kit (Qiagen, Valencia, CA) according to the manufacturer’s instructions. Thirty microliters of RNA was then added on 1/4 inch punches from an FTA card and at the CDC re-extracted from FTA card using the QiaAmp Viral Mini Kit according to the manufacturer’s instructions with the following modifications. The punches were placed in 1.5 μl microfuge tubes containing 600 μl of the kit lysis buffer, the recommended amount of carrier RNA, and 150 μl of PBS. The tubes were vortexed, incubated at room temperature for 10 min, and spun at 13000 rpm for 2 min. After removal of 700 μl to a new tube, the rest of the extraction was carried out according to the manufacturer’s instructions, with a final elution volume of 60 μl. For laboratory negative controls, RNA was extracted from 140 μl of transport media for the throat swabs and blank FTA card punches for the blood/serum spots.
Real-Time RT-PCR and Nested-Set Amplifications
A real-time RT-PCR assay, which was used to detect the presence of rubella virus RNA in serum (145) and throat swab(17) samples using the Invitrogen Superscript III Platinum qRT-PCR kit (Invitrogen, Carlsbad, CA), amplified a 185 nucleotide (nt) fragment of the E1 coding region as previously described [Abernathy et al., 2009]. The assay was modified by the addition of a second reverse primer (RV12-2: CCACGAGCCGCGAACAGTCG), which enhanced detection of Clade 2 rubella viruses by changing three of the nucleotides in the primer sequence to match the Clade 2 consensus sequence for the region. All throat swab RNAs were tested, but RNAs from blood spots were screened by real-time RT-PCR only if they were collected within three days of the onset of symptoms. RNA samples that were positive by real-time RT-PCR were used as templates in one or more of four nested RT-PCR assays using specific primers pairs (Table I). The nested RT-PCR assays were designed to amplify regions of the RV E1 coding region which encompassed the 739-nt region recommended by WHO for rubella genotyping. Samples with low Cts (30–34) on real time PCR analyses were sequenced using templates amplified by the 739-nt nested RT-PCR. Those with higher Cts (>35), required a pair of nested sets of primers as follows: (i) the 601-nt set and the 138-nt Clade 1 set for the 1B and 1G sequence and (ii) the 601-nt and 138-nt Clade 2 set for the 2B sequences.
TABLE I.
Primer Sequences for Four Nested RT-PCR Assays
| Primer name | Primer sequence PCR | product size | Sequence region and clade specificity |
|---|---|---|---|
| RV8633F-1st round | AGCGACGCGGCCTGCTGGGG | 945 | 739-nt common to clade 1 and 2 |
| RV9577R-1st round | CGCCCAGGTCTGCCGGGTCTC | ||
| RV8669F-2nd round | GTGATGAGCGTGTTCGCCCTT | 873 | |
| RV9541R-2nd round | GTGTGTGCCATACACCACGCC | ||
| RV11 (8812F)-1st round | CAACACGCCGCACGGACAAC | 766 | 601-nt fragment, common to clade 1 and 2 |
| RV9577R-1rst round | CGCCCAGGTCTGCCGGGTCTC | ||
| RV8823F-2nd round | ACGGACAACTCGAGGTCC | 727 | |
| RV9541R-2nd round | GTGTGTGCCATACACCACGCC | ||
| RV8699F-1st round | GTGATGAGCGTGTTCGCCCTT | 328 | 138-nt fragment specific to clade 1 |
| RV12 (8996R)-1st round | CCACAAGCCGCGAGCAGTCA | ||
| RV8691-2nd rounda | CTAGCTACGTCCAGCACCCT | 271 | |
| RV8961R-2nd round | CAAACCGGGGAGGCCCA | ||
| RV8669F-2B-1st round | GTGATGAGCGTGTTCGCCCT | 328 | 138-nt fragment specific to clade 2 |
| RV12-2 (8996R)-1st round | CCACGAGCCGCGAACAGTCG | ||
| RV8691F-2B-2nd round | CTAGCTACGTCCAGCACCC | 271 | |
| RV8961R-2nd round | CAAACCGGGGAGGCCCA |
All nested RT-PCR assays were performed with the Superscript III One-Step Platinum Taq HiFi kit (Invitrogen Carlsbad, CA) modified by the addition of betaine to a final concentration of 1M (Sigma, St. Louis, MO) and using 2.5 μl of extracted RNA as the first round template in a total reaction volume of 25 μl. Cycling conditions for round one consisted of 30 min at 55°C, 2 min at 94°C and 40 cycles of 10 sec at 94°C, 15 sec at 55°C, and 1 min at 68 °C. For the second round, 1 μl of the first round PCR was transferred to the second round reaction tube and the 30 min at 55°C RT phase was eliminated. Negative and positive controls were carried through both rounds and strict segregation of master mix preparation and template addition was followed.
Sequencing and Phylogenetic Analysis
DNAs from nested RT-PCR positive reactions were purified using the Charge Switch PCR Clean-Up kit (Invitrogen Carlsbad, CA). The 739 nt sequences of the standard window used for genotyping RVs were determined bi-directionally using Applied Biosystems Prism BigDye Terminator Cycle Sequencing Ready Reaction Kit (Applied Biosystems, Foster City, CA) and a 3130 DNA sequencer (Applied Biosystems). Assembly of sequences was performed with Sequencher V5.2.3 (Gene Codes, Ann Arbor, MI). Genotype assignments were made using an alignment of sequences from the DRC and the 32 WHO reference virus sequences [WHO, 2013] using the neighbor joining algorithm of the MEGA6 program [Tamura et al., 2013]. Rubella sequences from the neighboring countries of Sudan [Omer et al., 2010], Uganda [Namuwulya et al., 2014], and Tanzania [Centers-for-Disease-Control-and-Prevention, 2013] were also used in this analysis. Rubella sequences from this study were deposited on GenBank under accession numbers KU218397-KU218404.
RESULTS
Epidemiology of Rubella Infection in the DRC
From 2004 to 2013, 12,255 serum samples collected through the measles surveillance system were found negative or indeterminate for measles infection after measles IgM testing in the laboratory in the DRC. Of these serum samples, 11,733 were analyzed for the presence of rubella IgM and 522 were not analyzed. Of these 11,733 samples, 2,816 (24%) were positive for rubella IgM, 8,447 were negative, and 470 had indeterminate results for rubella IgM (Table II). The number of rubella IgM positive cases increased from 73 in 2004 to 766 in 2013. The percentage of rubella IgM positive cases varied from 13% in 2005 to 46% in 2013 (Table II).
TABLE II.
Number of Specimens Tested for Rubella IgM Antibodies and Results, DRC, 2004–2013
| Year | Measles IgM negative and indeterminate | Specimens not tested for rubella IgM | Specimens tested for rubella IgM | Number of rubella IgM positive (%) | Number rubella IgM negative | Indeterminate results |
|---|---|---|---|---|---|---|
| 2004 | 362 | 4 | 358 | 73 (20) | 280 | 5 |
| 2005 | 915 | 4 | 911 | 118 (13) | 763 | 30 |
| 2006 | 1,672 | 14 | 1,658 | 256 (15) | 1,323 | 79 |
| 2007 | 1,522 | 8 | 1,514 | 266 (18) | 1,174 | 74 |
| 2008 | 953 | 2 | 951 | 244 (26) | 666 | 41 |
| 2009 | 728 | 5 | 723 | 110 (15) | 587 | 26 |
| 2010 | 1,190 | 2 | 1,188 | 186 (16) | 947 | 55 |
| 2011 | 1,387 | 73 | 1,314 | 320 (24) | 936 | 58 |
| 2012 | 1,866 | 404 | 1,462 | 477 (33) | 934 | 51 |
| 2013 | 1,660 | 6 | 1,654 | 766 (46) | 837 | 51 |
| Total | 12,255 | 522 | 11,733 | 2,816 (24) | 8,447 | 470 |
Geographic and Age Distribution of Rubella Infection
Geographic origin was identified for all but two samples. Rubella cases were identified from all eleven provinces of the country with the highest positivity rate found in Bandudu province (43%) followed by Bas-Congo (41%), Kinshasa (33%), and Nord-Kivu (25%) (Table III). Of the 2,816 confirmed rubella case-patients, 2,794 (99%) had information on age. The age distribution of confirmed rubella cases ranged from 1 month to 49 years. Mean age was 21.6 years and median age was 24.5 years. The majority of rubella cases was concentrated in the group of children aged 5–9 years (43%) followed by age 1–4 years (31%) and 10–14 years (16%). Five percent (5%) were 15 years of age or older and 4% were less than 1 year (Fig. 1). Among rubella positive cases, gender information was available for 2,789 (99%) of which 1,409 (50.5%) were male and 1,380 (49.5%) were female. Eighty-one rubella cases (3%) were among women of reproductive age (15–49 years).
TABLE III.
Distribution of Laboratory-Confirmed Rubella Cases by Provinces, DRC, 2004–2013
| Province | Measles IgM negative and indeterminate | Specimen tested for rubella IgM | Rubella IgM-positive cases | Rubella positivity percentage (%) |
|---|---|---|---|---|
| Bas-Congo | 689 | 634 | 257 | 41 |
| Bandundu | 648 | 608 | 264 | 43 |
| Equateur | 1,120 | 1,092 | 271 | 25 |
| Katanga | 3,000 | 2,902 | 589 | 20 |
| Kinshasa | 1,544 | 1,403 | 457 | 33 |
| Kassai Occidental | 780 | 743 | 164 | 22 |
| Kasai Oriental | 963 | 946 | 124 | 13 |
| Maniema | 419 | 398 | 55 | 14 |
| Nord Kivu | 603 | 585 | 148 | 25 |
| Province Orientale | 2,033 | 1,973 | 380 | 19 |
| Sud Kivu | 454 | 447 | 106 | 24 |
| Unknown origin | 2 | 2 | 1 | 50 |
| Total | 12,255 | 11,733 | 2,816 | 24 |
Fig. 1.
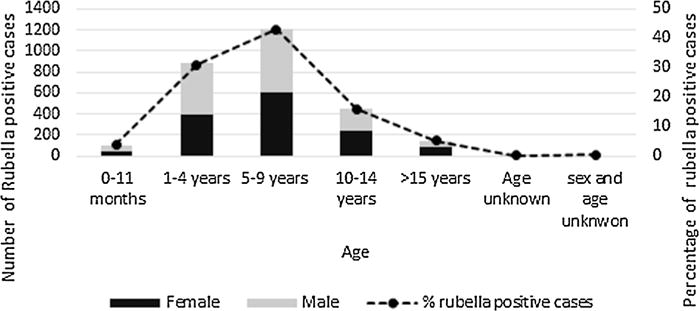
Age and sex distribution of rubella-confirmed cases in DRC, 2004–2013.
Rubella Virus Genotypes and Their Geographic Locations
To characterize the virus, RNA samples were first analyzed by real-time PCR to identify the viral RNA. There were no samples available for PCR testing between 2004 and 2005. Of 162 samples (17 were from throat swabs and 145 were from rubella IgM positive serum) analyzed by real-time PCR, 13 samples including 10 serum and three throat swabs samples were positive. Of those, the 739nt window of the E1 glycoprotein gene could be amplified for eight samples and the PCR products were sequenced. Phylogenetic analysis showed that the DRC rubella viruses belonged to genotypes 1B, 1E, 1G (1 sample each), and 2B (5 samples) (Fig. 2). Genotype 1E was detected in 2007, genotype 1B in 2008, genotype 1G in 2011, and genotype 2B in 2012 and 2013.
Fig. 2.
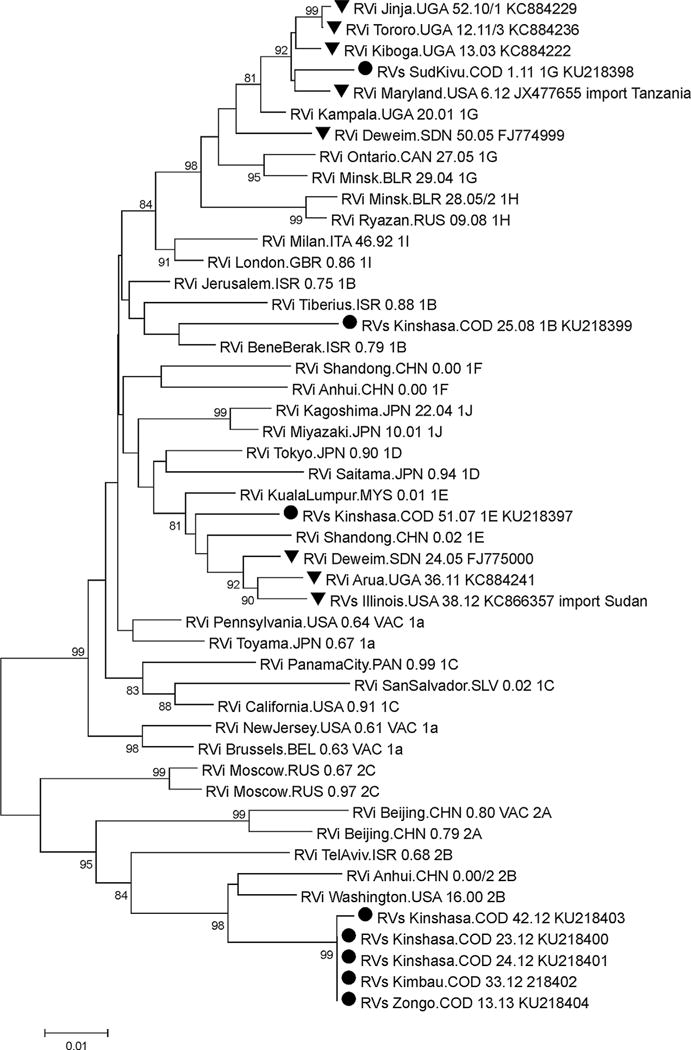
Phylogenetic analysis of rubella virus strains found in the DRC (black circles). Rubella virus strains from neighboring countries are also indicated (black triangles). Remaining sequences are WHO reference sequences. Significant bootstrap values (>80%) are indicated.
Geographic location of rubella strains showed that three different genotypes were found in Kinshasa province; genotype 1B (RVs Kinshasa.COD 25.08), genotype 1E (RVs Kinshasa.COD 51.07), and genotype 2B (RVs Kinshasa.COD 42.12, Rvs Kinshasa. COD 23.12, and Rvs Kinshasa.COD 24.12). Genotype 2B was also found in Banbundu (RVs Kimbau.COD 33.12) and Equateur (RVs Zongo.COD.13.13) provinces, while genotype 1G was found in Sud Kivu (RVs SudKivu.COD 1.11) province (Fig. 3).
Fig. 3.
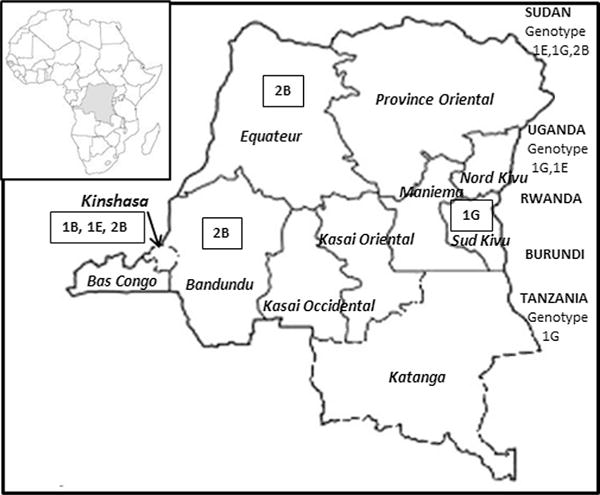
Geographical location of rubella virus genotypes (in rectangles) found in the Democratic Republic of the Congo. Rubella genotypes found in neighboring countries are also indicated under the country’s name.
DISCUSSION
This report provides the first information on the genotypes of rubella viruses found in the DRC. As of 2014, rubella vaccine was not introduced in the national immunization program and there was no rubella surveillance system in the DRC. The percentage of rubella confirmed cases in the DRC (24%) was similar to the rubella positivity rate (25%) previously found in African region during 2002–2009 [Goodson et al., 2011]. Both rates were derived from ELISA testing of measles surveillance samples that tested measles IgM negative. The majority of reported rubella cases during 2004–2013 occurred in persons older than 5 years (64%). These findings are similar to the results reported from a study of rubella cases in Uganda in 2003–2012 [Namuwulya et al., 2014]. Three percent (3%) of rubella cases occurred among women older than 15 years which corresponds to child bearing age in Africa, and therefore represents a risk for congenital rubella syndrome (CRS). This is slightly lower than the data recently reported on rubella in the African region where 5% of confirmed rubella cases were found among women of reproductive age [Goodson et al., 2011]. It is noteworthy that this report is based only on samples collected for measles surveillance and thus may not represent the complete picture of rubella in the DRC.
Genomic characterization was performed on eight samples by amplification of a 739 window of the E1 region as recommended by WHO guidelines [WHO, 2013]. Phylogenetic analysis identified rubella viruses of distinct genotypes circulating in the DRC between 2007 and 2013: 1B, 1E, 1G, and 2B. Genotype 1B was found in 2008. This is the first report of this virus in the Central African region. This may be because only very few countries in Africa have studied the molecular biology of rubella viruses circulating in their country. Genotype 1E, found in the DRC in 2007 was previously identified in Sudan in 2005 [Omer et al., 2010] and more recently in Uganda in 2011 [Namuwulya et al., 2014]. Sequence analysis showed that the sequence of the DRC strain 1E differs from the 1E strain from Sudan and Uganda sequences by 3%. Genotype 1G was found in the DRC in 2011. This genotype is known to be prevalent in central Africa [WHO, 2007; Abernathy et al., 2011]. It was found in Uganda in 2003, 2011–2012 [Namuwulya et al., 2014], in Sudan in 2005 [Omer et al., 2010] and more recently imported into the USA from Tanzania in 2012 [Centers-for-Disease-Control-and-Prevention, 2013]. The genotype 1G sequence from the DRC differs from the Tanzanian strain by 1% and from Uganda’s sequences by 2%. It differs from the Sudan 2005 sequence by 3%. The 739nt of E1 gene sequence was identical between four of the genotype 2B viruses and one virus, RVs/Kinshasa.COD/42.12, varied by two nucleotides. This genotype was found in Sudan in 2006 [Omer et al., 2010]. Sequence comparison of the 224 nucleotides available in GenBank for Sudan 2006 with the DRC genotype 2B sequences showed that they differ by 4%.
This study has some limitations. The low percentage of rubella RNA positive samples (8%) may be because that the analysis were performed on samples collected for the purpose of measles surveillance with unknown storage histories. The majority of the samples were derived from blood which has been shown to have a low positivity rate compared to respiratory samples even on days immediately after symptom onset. The low amount of rubella RNA present in blood derived samples likely also explains why the 739nt of the E1 glycoprotein gene could be amplified from only eight samples [Abernathy et al., 2009; Zheng et al., 2013]. The origin of rubella viruses could not be traced due to the absence of a rubella surveillance system in the DRC. It is therefore not possible to say if these rubella strains are endemic or imported.
This report provides base line information on rubella epidemiology and genetic information on rubella strains circulating in the DRC. These data will help track endemic viruses and monitor the evolution of rubella virus once the rubella vaccine is introduced in the country. Furthermore, these data will contribute to a better understanding of the dynamics of rubella virus circulation in Africa. Efforts to establish rubella surveillance in the DRC and strengthen virologic surveillance of rubella viruses are needed in order to provide support to the rubella control in Africa.
Acknowledgments
We want to thank the WHO DRC office for logistical help in the country. We would like to thank INRB personnel; Yvonne Lay Mowele, Seraphine Wanzambi, Naomie Mitonga, and Frida Nkawa, for technical assistance. We are grateful to Albert Mbule, WHO DRC office, and Jean-Claude Changa-Changa, INRB, for their help on data management. We are grateful to Olen Kew, CDC Atlanta and Paul Rota, CDC Atlanta for guidance, helpful discussions, and constructive criticisms leading to the completion of this work.
Grant sponsor: The Bill and Melinda Gates Foundation through the SURVAC Project; Grant sponsor: The World Health Organization; Grant sponsor: The U.S. Centers for Diseases Control and Prevention; Grant sponsor: The CDC Foundation; Grant sponsor: The Ministry of Health of the Democratic Republic of the Congo
Footnotes
DISCLAIMER
The findings and conclusions in this report are those of the author(s) and do not necessarily represent the official position of the Centers for Disease Control and Prevention. Names of specific vendors, manufacturers, or products are included for public health and informational purposes; inclusion does not imply endorsement of the vendors, manufacturers, or products by the Centers for Disease Control and Prevention or the US Department of Health and Human Services.
References
- Abernathy E, Cabezas C, Sun H, Zheng Q, Chen MH, Castillo-Solorzano C, Ortiz AC, Osores F, Oliveira L, Whittembury A, Andrus JK, Helfand RF, Icenogle J. Confirmation of rubella within 4 days of rash onset: Comparison of rubella virus RNA detection in oral fluid with immunoglobulin M detection in serum or oral fluid. J Clin Microbiol. 2009;47:182–188. doi: 10.1128/JCM.01231-08. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Abernathy ES, Hubschen JM, Muller CP, Jin L, Brown D, Komase K, Mori Y, Xu W, Zhu Z, Siqueira MM, Shulga S, Tikhonova N, Pattamadilok S, Incomserb P, Smit SB, Akoua-Koffi C, Bwogi J, Lim WW, Woo GK, Triki H, Jee Y, Mulders MN, de Filippis AM, Ahmed H, Ramamurty N, Featherstone D, Icenogle JP. Status of global virologic surveillance for rubella viruses. J Infect Dis. 2011;204:S524–S532. doi: 10.1093/infdis/jir099. [DOI] [PubMed] [Google Scholar]
- Centers-for-Disease-Control-and-Prevention. Global measles and rubella laboratory network, January 2004–June 2005. MMWR Morb Mortal Wkly Rep. 2005;54:1100–1104. [PubMed] [Google Scholar]
- Centers-for-Disease-Control-and-Prevention. Three cases of congenital rubella syndrome in the postelimination era–Maryland, Alabama, and Illinois, 2012. MMWR Morb Mortal Wkly Rep. 2013;62:226–229. [PMC free article] [PubMed] [Google Scholar]
- CIA. Africa: The Democratic Republic of the Congo 2012 [Google Scholar]
- Cooray S, Warrener L, Jin L. Improved RT-PCR for diagnosis and epidemiological surveillance of rubella. J Clin Virol. 2006;35:73–80. doi: 10.1016/j.jcv.2004.12.020. [DOI] [PubMed] [Google Scholar]
- Goodson JL, Masresha B, Dosseh A, Byabamazima C, Nshimirimana D, Cochi S, Reef S. Rubella epidemiology in Africa in the prevaccine era, 2002–2009. J Infect Dis. 2011;204:S215–225. doi: 10.1093/infdis/jir108. [DOI] [PubMed] [Google Scholar]
- Namuwulya P, Abernathy E, Bukenya H, Bwogi J, Tushabe P, Birungi M, Seguya R, Kabaliisa T, Alibu VP, Kayondo JK, Rivailler P, Icenogle J, Bakamutumaho B. Phylogenetic analysis of rubella viruses identified in Uganda, 2003–2012. J Med Virol. 2014;86:2107–2113. doi: 10.1002/jmv.23935. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Omer A, Abdel Rahim EH, Ali EE, Jin L. Primary investigation of 31 infants with suspected congenital rubella syndrome in Sudan. Clin Microbiol Infect. 2010;16:678–682. doi: 10.1111/j.1469-0691.2009.02966.x. [DOI] [PubMed] [Google Scholar]
- Reef SE, Strebel P, Dabbagh A, Gacic-Dobo M, Cochi S. Progress toward control of rubella and prevention of congenital rubella syndrome–worldwide, 2009. J Infect Dis. 2011;204:S24–27. doi: 10.1093/infdis/jir155. [DOI] [PubMed] [Google Scholar]
- Tamura K, Stecher G, Peterson D, Filipski A, Kumar S. MEGA6: Molecular evolutionary genetics analysis version 6.0. Mol Biol Evol. 2013;30:2725–2729. doi: 10.1093/molbev/mst197. [DOI] [PMC free article] [PubMed] [Google Scholar]
- WHO. Update standard nomenclature for wild type rubella virus. Wkly Epidemiol Rec. 2007;82:216–222. [PubMed] [Google Scholar]
- WHO. Rubella vaccines: WHO position paper. Wkly Epidemiol Rec. 2011;86:301–316. [PubMed] [Google Scholar]
- WHO. Rubella. 2012 Fact sheet 367. [Google Scholar]
- WHO. Rubella virus nomenclature update. Wkly Epidemiol Rec. 2013;88:337–348. [PubMed] [Google Scholar]
- WHO/AFRO. Guidelines for measles surveillance. World Health Organization Regional Ofice for Africa; 2004. pp. 1–37. [Google Scholar]
- Zheng Q, Abernathy ES, Sun H, Zhu Z, de Filippis A, Akoua-Koffi C, Ahmed H, Morris-Glasgow V, Quist-Therson M, Icenogle JP. Genotyping of rubella virus RNA in sera and dried blood spots collected during routine surveillance and in archival sera. J Virol Methods. 2013;187:284–287. doi: 10.1016/j.jviromet.2012.11.023. [DOI] [PubMed] [Google Scholar]


