Abstract
Among the features that distinguish type 1 innate lymphoid cells (ILC1s) from natural killer (NK) cells is a gene signature indicative of ‘imprinting’ by cytokines of the TGF-β family. We examined mice in which ILC1s and NK cells lacked SMAD4, a signal transducer that facilitates the canonical signaling pathway common to all cytokines of the TGF-β family. While SMAD4 deficiency did not affect ILC1 differentiation, NK cells unexpectedly acquired an ILC1-like gene signature and were unable to control tumor metastasis or viral infection. Mechanistically, SMAD4 restrained non-canonical TGF-β signaling mediated by the cytokine receptor TGF-βR1 in NK cells. NK cells from a SMAD4-deficient person affected by polyposis were also hyper-responsive to TGF-β. These results identify SMAD4 as a previously unknown regulator that restricts non-canonical TGF-β signaling in NK cells.
Natural Killer (NK) cells are a major innate source of interferon-γ (IFN-γ) during immune responses to virus-infected cells and tumor cells1. Additional subsets of innate lymphoid cells (ILCs) that produce IFN-γ have been identified and have been collectively designated ‘type 1 ILCs’ (ILC1s). However, the criteria used to distinguish ILC1s from NK cells and the relationship between the two cell types remain somewhat controversial. On the basis of ontogeny, mouse NK cells and ILC1s are thought to derive from developmentally distinct, restricted progenitor cells that branch from the shared common innate lymphoid progenitor, which gives rise to all ILCs2–7. Downstream of the common innate lymphoid progenitor, one branch leads to generation of the NK cell progenitor, which subsequently differentiates into mature NK cells, a path that requires the transcription factors T-bet and Eomes. Another branch includes progenitors with progressively restricted potential that culminate in the generation of T-bet-dependent and Eomes-independent ILC1s, as well as other ILC subsets that produce the cytokines IL-5 and IL-13 (ILC2s) or IL-17 and IL-22 (ILC3s)3–7. While committed ILC precursor cells capable of giving rise to all ILC populations in humans have been identified, whether human counterparts of mouse T-bet-dependent and Eomes-independent IFN-γ-producing ILC1s exist is matter of debate8–10.
Another criterion used to distinguish NK cells from ILC1s is based on mobility and location: NK cells recirculate throughout lymphoid organs, whereas ILC1s are found as resident cells in non-lymphoid tissues, such as the gut, liver, adipose tissue and salivary glands (SGs). In mice, tissue residency of ILC1s has been demonstrated by parabiosis11–13 and is possibly implemented by the constitutive expression of a specialized set of molecules, including the integrins CD49a (α1) and CD103 (αEβ7) and the activation marker CD69 (refs. 11–15). It has been shown that the development of tissue resident ILC1s is driven in part by the transcription factor Hobit16. Accordingly, Hobit-deficient mice lack liver CD49a+ ILC1s. Human counterparts of mouse tissue-resident ILC1s are present within the epithelium of the oral and intestinal mucosae14.
Finally, ILC1s and NK cells have been distinguished in mice on the basis of their transcriptome profiles3,17,18. In addition to Itga1 (which encodes CD49a), the ILC1 gene signature includes the expression of genes that encode the cytokine receptors IL-7R and IL-21R, the death-inducing molecule TRAIL and the AMP-degrading enzyme CD73, as well as germline transcripts Tcrg-V2 and Tcrg-V3 (which encode γ-chain variable regions of the T cell antigen receptor). The defining signature of NK cells includes the expression of genes that encode the integrin CD49b (α2) and inhibitory receptors for major histocompatibility complex class I, as well as Eomes, but essentially no expression of IL-7R. A subset of ILC1s in the SGs has been described that has the phenotype of both ILC1s and NK cells15. Like ILC1s, SG ILC1s express CD49a, CD103, TRAIL, CD73, IL-21R and Hobit. Similar to NK cells, SG ILC1s also express CD49b, T-bet and Eomes but lack IL-7R. However, in contrast to other ILC1s, SG ILC1s are weak producers of IFN-γ. These observations suggest that ILC1s might include a spectrum of cells with partially overlapping phenotypes and functions that vary depending on the tissue microenvironment.
Among various tissue factors, cytokines of the TGF-β (‘transforming growth factor-β’) family are emerging as crucial mediators in the differentiation of ILC1s. These cytokines transmit intracellular signals by forming a hetero-tetrameric receptor complex with two type I receptors and two type II receptors19. Type II receptors activate type I receptors, which are the main propagators of intracellular signals through recruitment and phosphorylation of the R-SMAD (receptor SMAD) transducers. Phosphorylated R-SMAD pair with SMAD4, which facilitates their translocation into the nucleus to initiate the transcription of hundreds of genes. Alternatively, the E3 ubiquitin ligase TRIM33 can compete with SMAD4 for binding to phosphorylated R-SMAD, and this complex mediates the transcription of a distinct set of genes20. Finally, cytokines of the TGF-β family can activate many other signal transducers, such as the kinases MAPK (‘mitogen activated protein kinase’) and PI3-K (‘phosphatidylinositol-3-OH kinase’) via non-canonical R-SMAD-independent pathways, some of which remain poorly defined21.
Published studies have shown that ablation of the type II TGF-β receptor TGFβR2 in SG ILC1s abolishes expression of CD49a, CD103, TRAIL and CD73 while enhancing both expression of Eomes and the ability to produce IFN-γ13. However, lack of TGFβR2 has little or no effect on gut or liver ILC1s, which suggests that other cytokines of the TGF-β family, such as bone-morphogenic proteins and activin proteins, might drive the differentiation of precursors into ILC1s in these tissues. Here we further pursued this hypothesis by generating mice in which both ILC1s and NK cells selectively lack SMAD4, which promotes canonical signaling of all cytokines of the TGF-β family. We expected SMAD4 deficiency to block ILC1 differentiation in all tissues. Moreover, given the reported inhibitory function of TGF-β in the development and function of NK cells22–24, we anticipated that a lack of SMAD4 in NK cells would enhance their function. In contrast, we found that SMAD4 deficiency did not visibly affect ILC1 differentiation but instead altered the phenotype of conventional NK cells; SMAD4-deficient NK cells had a gene signature that was ILC1-like and indicative of imprinting by TGF-β. SMAD4 deficiency affected the control of tumor metastasis and viral infection by NK cells by affecting both the function and proliferative capacity of these cells. SMAD4 served an essential function in curtailing the imprinting of NK cells by TGF-β by inhibiting non-canonical signaling through TGFβR1. Notably, NK cells from a SMAD4-deficient patient were also hyper-responsive to TGF-β. Our results indicate that SMAD4 unexpectedly functions as negative regulator of non-canonical TGF-β signaling in conventional NK cells.
RESULTS
SMAD4-deficient NK cells have ILC1-like characteristics
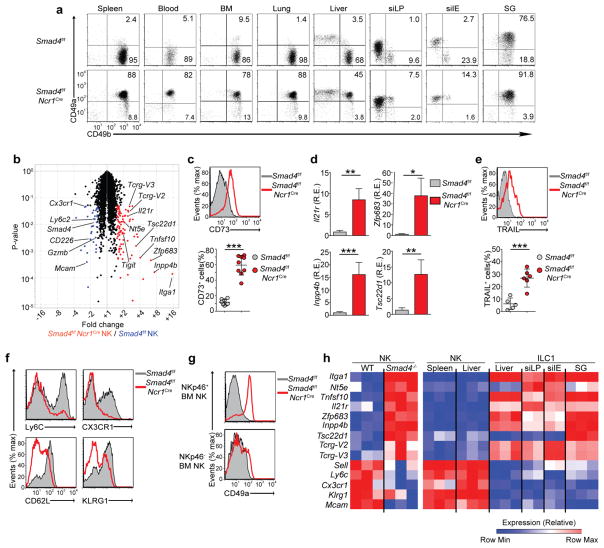
To assess the effect of cytokines of the TGF-β superfamily on ILC1 differentiation, we crossed mice carrying loxP-flanked Smad4 alleles (Smad4f/f)25 with mice in which sequence encoding ‘codon-improved’ Cre recombinase is knocked into the gene encoding the activating receptor NKp46 (Ncr1iCre)17. Smad4f/fNcr1iCre mice lacked SMAD4 in all cells expressing NKp46, which include NK cells, ILC1s and NKp46+ ILC3s, as well as RORγt+ ILC3s that have converted into ILC1s (also known as ‘ex-ILC3s’)17. We first assessed expression of CD49a and CD49b, which distinguish ILC1s from NK cells in multiple tissues. ILC1s from Smad4f/fNcr1iCre mice maintained high expression of CD49a in the liver, the lamina propria of the small intestine, the intraepithelium of the small intestine and the SGs, similar to Smad4f/f mice (Fig. 1a), which suggested that imprinting of all ILC1s by TGF-β was SMAD4 independent. Unexpectedly, CD49b+ NK cells from the spleen, blood, bone marrow (BM), lungs, liver and gut of Smad4f/fNcr1iCre mice expressed much more CD49a on the cell surface than did their counterparts from Smad4f/f mice (Fig. 1a), which suggested that SMAD4 deficiency induced the conversion of NK cells into ILC1-like cells.
Figure 1.
Altered phenotype of NK cells in Smad4f/fNcr1iCre mice. (a) Expression of CD49a and CD49b in NK1.1+CD3− cells from various tissues (above plots) of Smad4f/f and Smad4f/fNcr1iCre mice (left margin). Numbers in quadrants indicate percent cells in each throughout. (b) Microarray analysis of gene expression in Smad4f/fNcr1iCre splenic NK cells (n = 3 biological replicates) versus Smad4f/f NK cells (n = 3 replicates) (Smad4−/− NK/Smad4+/+ NK), plotted against P values (volcano plot); colors indicate transcripts significantly (P ≤ 0.05;Student’s t-test) upregulated (red) or downregulated (blue) by at least 1.5-fold in Smad4f/fNcr1iCre cells relative to their expression in Smad4f/f cells. (c) Surface expression of CD73 on Smad4f/f and Smad4f/fNcr1iCre splenic NK cells (key) (top), and quantification of results (bottom). (d) Expression of Il21r, Zfp683, Inpp4b, and Tsc22d1 mRNA in Smad4f/f and Smad4f/fNcr1iCre splenic NK cells (key); results are presented as relative expression (RE) to those of the control gene Gapdh. (e) Surface expression of TRAIL on Smad4f/f and Smad4f/fNcr1iCre splenic NK cells (key) after 48 h of culture with IL-2 (top), and quantification of results (bottom). (f) Surface expression of Ly6C, CX3CR1, CD62L and KLRG1 on Smad4f/f and Smad4f/fNcr1iCre splenic NK cells. (g) Surface expression of CD49a on NKp46− or NKp46+ NK cells from the BM of Smad4f/f and Smad4f/fNcr1iCre mice. (h) Expression of indicated genes (left margin) by Smad4f/f and Smad4f/fNcr1iCre splenic NK cells and by wild-type NK cells and ILC1s (on the website of www.immgen.org/databrowser/index.html the Immunological Genome Project). *P < 0.05, **P < 0.01 and ***P < 0.001 (unpaired Student’s t-test). Each symbol (c,e) represents an individual mouse; small horizontal lines indicate the mean (± s.d.). Data are representative of 3 experiments (a) or 1 experiment (b) or are pooled from at least three independent experiments with one to three mice per genotype in each (c–g; mean + s.d. d).
Further comparison of global gene expression in splenic NK cells from Smad4f/fNcr1iCre mice with that of such cells from Smad4f/f mice revealed that SMAD4-deficient NK cells expressed genes such as Itga1 (which encodes CD49a), Nt5e (which encodes CD73), Tnfsf10 (which encodes TRAIL), Il21r (which encodes IL-21 receptor), Zfp683 (which encodes Hobit) and Tsc22d1 (which encodes the transcription factor Tsc22d1), and Inpp4b (which encodes the phosphatase Inpp4b), whose expression is characteristic of ILC1s but not NK cells (Fig. 1b and Supplementary Fig. 1a). The expression of some ILC1 markers was confirmed by flow cytometry and RT-PCR analysis of freshly isolated SMAD4-deficient NK cells (Fig. 1c,d), but surface expression of TRAIL became apparent only after in vitro culture of SMAD4-deficient NK cells with IL-2 (Fig. 1e). The similarity of SMAD4-deficient NK cells to all ILC1s was further emphasized by the expression of germline Tcrg-V2 and Tcrg-V3 transcripts (Fig. 1b), which have been noted to be part of the ILC core signature18. SMAD4-deficient NK cells also had lower expression of some markers that are normally expressed on mature NK cells, such as Ly6C, CX3CR1, CD62L, KLRG1 and MCAM, than did Smad4f/f NK cells (Fig. 1b,f and Supplementary Fig. 1b); however they retained other features typical of NK cells, including expression of both Eomes and T-bet, along with lack of expression of IL-7R and CD69 (Supplementary Fig. 1c). Similar numbers of NKp46+ ILC3s were present in Smad4f/fNcr1iCre mice and Smad4f/f mice, and SMAD4-deficient NKp46+ ILC3s were functionally equivalent to their Smad4f/f counterparts (Supplementary Fig. 1d,e), which indicated that NKp46-driven deletion of SMAD4 affected mainly NK cells.
We sought to determine whether SMAD4 deficiency affected NK cell development in the BM. NK cells normally differentiate from immature NKp46− cells, with detectable expression of CD49a on the cell surface, into mature NKp46+CD49a− cells. While immature NK cells appeared to be similar in Smad4f/f mice and Smad4f/fNcr1iCre mice, mature SMAD4-deficient NK cells had substantial upregulation of CD49a in parallel with the deletion of Smad4 by the NKp46-driven iCre (Fig. 1g). A similar result was obtained for BM NK cells from Smad4f/fIl7rCre mice (Supplementary Fig. 1f), in which Smad4 is deleted in the common lymphoid progenitor cell. Overall, these data suggested that imprinting of SMAD4-deficient cells by TGF-β occurred at a late stage of NK cell development. We concluded that lack of SMAD4 biased conventional NK cells toward an ILC1-like phenotype, which most closely resembled that of SG ILC1s due to co-expression of markers of NK cells (Eomes and CD49b) and ILC1s (T-bet, CD49a and TSC22d1) (Fig. 1h).
SMAD4 deficiency impairs NK cell control of metastatic melanoma
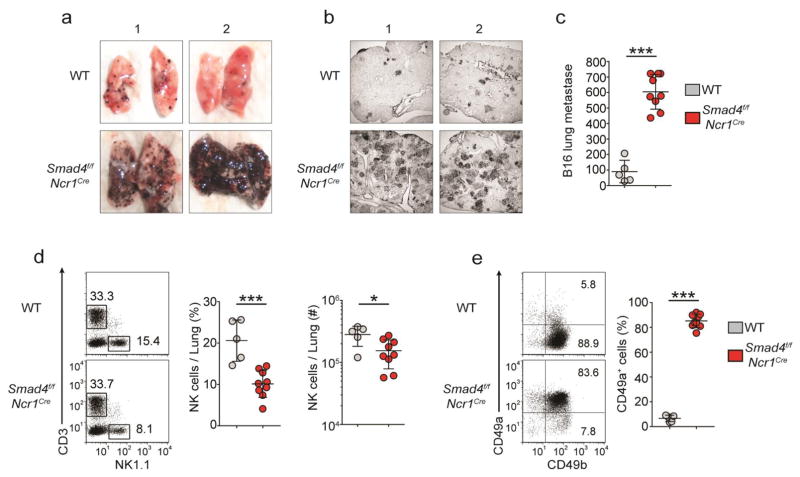
To assess the effect of SMAD4 deficiency on NK cell function in vivo, we chose the B16 melanoma model, since the containment of B16 lung metastases is highly dependent on NK cells through the production of IFN-γ and, to a lesser extent, the release of lytic granules26. We injected with 1 × 105 B16 cells intravenously into Smad4f/f and Smad4f/fNcr1iCre mice and assessed their lungs for pulmonary metastatic nodules after 15 d. Gross and microscopic examination of the lungs revealed that Smad4f/fNcr1iCre mice had a much greater burden of B16 nodules than did their control littermates (Fig. 2a,b). The lungs of Smad4f/fNcr1iCre mice had an average of 600 B16 foci per whole lung, whereas Smad4f/f mice had approximately sixfold fewer foci (Fig. 2c). Several Smad4f/fNcr1iCre mice had too many B16 lung metastases to count accurately and therefore were assigned scores of the highest definite value (700 foci) (Fig. 2c). These data demonstrated that Smad4f/fNcr1iCre mice limited B16 tumor metastasis much less effectively than did Smad4f/f mice.
Figure 2.
SMAD4 is essential for NK cell–mediated anti-tumor immunity. (a,b) Images of whole lungs (a) and microscopy of tissue sections (12 μm thick) of a single lung lobe (b) from Smad4f/f and Smad4f/fNcr1iCre mice (two per genotype shown, 1,2 above images) 15 d after intravenous injection with 1 × 105 B16 cells. Original magnification (b), ×2.5. (c) Quantification of total black B16 foci on the surface of lungs of mice as in a. (d) Flow cytometry (left), frequency (middle) and absolute number (right) of lung NK cells from mice as in a,b. Numbers adjacent to outlined areas (left) indicate percent T cells (CD3+NK1.1−, top left) or NK cells (CD3−NK1.1+ cells, bottom right). (e) Expression of CD49a and CD49b in lung NK cells from mice as in a. Each symbol (c–e) represents an individual mouse; small horizontal lines indicate the mean (± s.d.). *P < 0.05 and ***P < 0.001 (unpaired Student’s t-test). Data are pooled from two independent experiments.
The overwhelming number of metastatic nodules present in Smad4f/fNcr1iCre mice mirrored the results of studies in which NK cells are absent in the lung27. Analysis of the frequency and number of NK cells revealed that these cells were present in the lungs of all mice at day 15 of B16 tumor metastatic model, although fewer were present in Smad4f/fNcr1iCre mice than in their Smad4f/f littermates (Fig. 2d). Moreover, Smad4f/fNcr1iCre NK cells had high expression of CD49a at day 15 (Fig. 2e), which demonstrated that these cells maintained their ILC1-like phenotype. We concluded that SMAD4 was essential for the control of B16 lung metastasis by NK cells.
NK cell effector functions require SMAD4
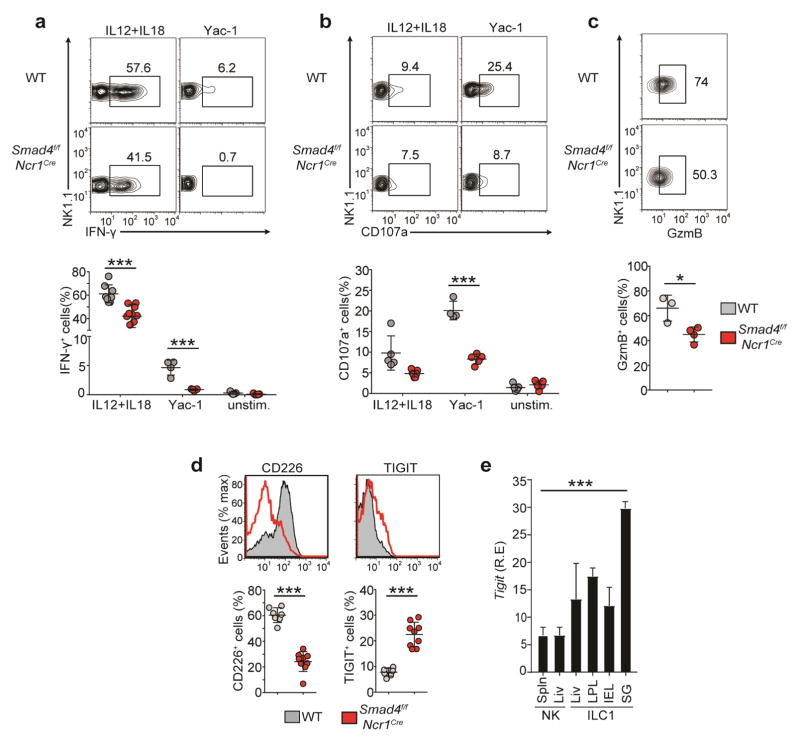
To gain insight into the mechanisms underlying the impaired control of tumor metastasis in vivo, we assessed the functional capacity of SMAD4-deficient NK cells in vitro. We found that SMAD4-deficient NK cells produced significantly less IFN-γ than did Smad4f/f NK cells in response to incubation with the cytokines IL-12 plus IL-18 or with YAC-1 mouse lymphoma cells, as target cells (Fig. 3a). SMAD4-deficient NK cells also had impaired lytic ability after either stimulation, manifested by less CD107a on the cell surface (Fig. 3b). The impaired lytic ability of SMAD4-deficient NK cells was consistent with a significant reduction in the expression of Gzmb mRNA (Fig. 1b) and its product, granzyme B (Fig. 3c). Thus, NK cells from Smad4f/fNcr1iCre mice are functionally deficient compared to their Smad4f/f counterparts.
Figure 3.
SMAD4-deficient NK cells have impaired functions. (a,b) Flow cytometry (top) and quantification (bottom) of intracellular IFN-γ (a) and surface CD107a (b) of splenic NK cells obtained from Smad4f/f and Smad4f/fNcr1iCre mice and left unstimulated (unstim.) or stimulated with IL-12 and IL-18 or 1 × 106 Yac-1 cells. Numbers adjacent to outlined areas (top) indicate percent NK1.1+IFN-γ+ cells (a) or NK1.1+CD107a+ cells (b). (c) Flow cytometry (top) and quantification (bottom) of granzyme B (GzmB) in splenic NK cells obtained from Smad4f/f and Smad4f/fNcr1iCre mice and stimulated for 48 h with IL-2. Numbers adjacent to outlined areas (top) indicate percent NK1.1+GzmB+ cells. (d) Flow cytometry of the ex vivo surface expression of CD226 (left) and TIGIT (right) on splenic NK cells from Smad4f/f and Smad4f/fNcr1iCre mice (top), and quantification of the results (bottom). (e) Expression of Tigit in NK cells and ILC1s (below plot) from the spleen (Spln), liver (Liv), SG, siLP, and siIE; [www.immgen.org/databrowser/index.html]. Each symbol (a–d) represents an individual mouse; small horizontal lines indicate the mean (± s.d.). *P < 0.05 and ***P < 0.001 (unpaired Student’s t-test). Data are pooled from at least two independent experiments (mean + s.d. in e).
Lack of SMAD4 alters expression of activating and inhibitory receptors
We also assessed the expression of activating and inhibitory receptors that control the recognition of tumor cells by NK cells. SMAD4-deficient NK cells had much lower expression of the activating receptor CD226 (DNAM-1), which promotes killing of tumor cells that express the ligands CD155 and CD112, including melanoma cells28,29, than that of Smad4f/f cells (Fig. 3d). Conversely, SMAD4-deficient NK cells had higher expression of the inhibitory receptor TIGIT (WUCAM), which also detects CD155 and CD112 (refs. 30–33), than that of Smad4f/f cells (Fig. 3d). We also noted that high TIGIT expression was a feature shared by all ILC1s (Fig. 3e), which further confirmed the ILC1-like phenotype of SMAD4-deficent NK cells. Thus, SMAD4-deficient NK cells recognized mainly melanoma cells and other tumors expressing CD155 and CD112 through the inhibitory receptor TIGIT rather than the activating receptor CD226. In summary, the greater abundance of B16 lung metastases observed in Smad4f/fNcr1iCre mice was probably due to multiple NK cell defects, including diminished functional capacity and unbalanced expression of TIGIT and CD226.
NK cell population expansion during infection requires SMAD4
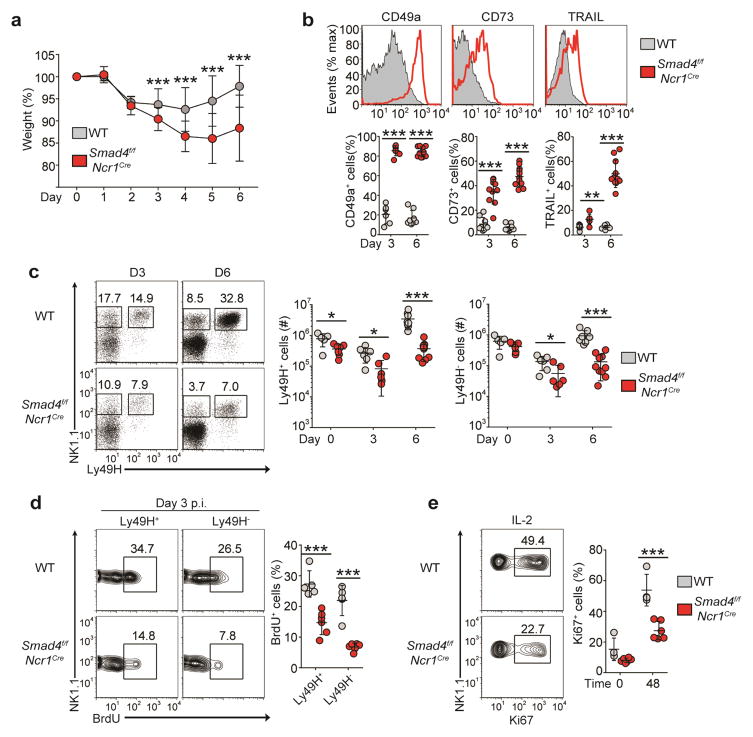
In C57BL/6 mice, NK cells contain infection with mouse cytomegalovirus (MCMV) through the activating receptor Ly49H, which detects the MCMV-encoded m157 protein on infected cells; this triggers the activation, population expansion and effector functions of NK cells34. To determine whether SMAD4 deficiency affected the anti-MCMV response, we injected 1 × 105 plaque-forming units of MCMV into Smad4f/f and Smad4f/fNcr1iCre mice and monitored them, from day 0 through day 6 after infection, for weight loss, which reflects the severity of viral infection. Smad4f/fNcr1iCre mice lost significantly more weight than did Smad4f/f mice on days 3–6 after infection (Fig. 4a), indicative of an impaired capacity to control MCMV infection. During the course of infection, SMAD4-deficient NK cells maintained or had enhanced expression of ILC1 markers (Fig. 4b).
Figure 4.
SMAD4 is necessary for the anti-viral function of NK cells. (a) Weight of Smad4f/f and Smad4f/fNcr1iCre mice at various times (horizontal axis) after infection with MCMV (5 × 104 plaque-forming units), presented relative to weight before infection. (b) Flow cytometry (top) and quantification (bottom) of the ex vivo surface expression of ILC1 markers on splenic NK cells from mice as in a at day 6 (top) or day 3 or 6 (bottom) after infection with MCMV. (c) Flow cytometry of splenic NK cells (gated on CD3−CD19−) from mice as in a at day 3 or 6 after infection with MCMV (left), and absolute number of Ly49H+ (middle) and Ly49H− (right) splenic NK cells. Numbers adjacent to outlined areas indicate percent NK1.1+Ly49H− cells (left) or NK1.1+Ly49H+ cells (right). (d) Flow cytometry of the incorporation of BrdU by Ly49H+ or Ly49H− splenic NK cells from mice as in a at day 3 after infection with MCMV (left), and quantification of BrdU+ Ly49H+ or Ly49H− cells (right). Numbers adjacent to outlined areas (left) indicate percent NK1.1+BrdU+ (proliferating) cells. (e) Flow cytometry (left) and quantification (right) of Ki67 expression by splenic NK cells from uninfected mice assessed after 48 h of culture with IL-2 (48) or immediately after isolation without further culture (0). Numbers adjacent to outlined areas (left) indicate percent NK1.1+Ki67+ (proliferating) cells. Each symbol (b–e) represents an individual mouse; small horizontal lines indicate the mean (± s.d.). *P < 0.05, **P < 0.01 and ***P < 0.001 (unpaired Student’s t-test). Data are pooled from four (a–d; mean ± s.d. in a) or three (e) independent experiments with at least two mice per genotype per time point in each.
The NK cell responses of mice were further assessed at 3 d and 6 d after infection. Naive Smad4f/f and Smad4f/fNcr1iCre mice had similar numbers of splenic Ly49H+ cells (Fig. 4c and Supplementary Fig. 2a). At day 3 after infection, differences in the frequency and number of Ly49H+ and Ly49H− NK cells were observed (Fig. 4c). The considerable population expansion of Ly49H+ NK cells evident in Smad4f/f mice at day 6 after infection was much lower in Smad4f/fNcr1iCre mice (Fig. 4c). Similarly, fewer Ly49H− NK cells, which undergo more limited population expansion during infection, were present in Smad4f/fNcr1iCre mice than in Smad4f/f mice at day 6 after infection (Fig. 4c). These results indicated that SMAD4-deficient NK cells were defective in the ability to proliferate and/or survive during infection with MCMV.
To directly assess NK cell proliferation, we infected Smad4f/f and Smad4f/fNcr1iCre mice with MCMV and injected the thymidine analog bromodeoxyuridine (BrdU) intraperitoneally into the mice at day 3 after infection, then assessed incorporation of BrdU into NK cells 4 h later. Considerable incorporation of BrdU by NK cells, particularly the Ly49H+ population, was obvious in Smad4f/f mice, whereas much less BrdU was taken up by both Ly49H+ and Ly49H− SMAD4-deficient NK cells (Fig. 4d). We also used expression of the proliferation marker Ki67 to assess NK cell proliferation in response to cytokine stimulation in vitro. SMAD4-deficient NK cells had significantly lower expression of Ki67 than that of Smad4f/f NK cells (Fig. 4e). NK cell viability during MCMV infection and in vitro culture was similar in Smad4f/f mice and Smad4f/fNcr1iCre mice (Supplementary Fig. 2b,c). These data demonstrated that SMAD4 was critical for the population expansion of NK cells during viral infection. The failure of Ly49H+ NK cell populations to expand adequately in Smad4f/fNcr1iCre mice during MCMV infection contributed to the enhanced morbidity of these mice.
SMAD4 restricts the imprinting of NK cells by TGF-β independently of TGFβR2
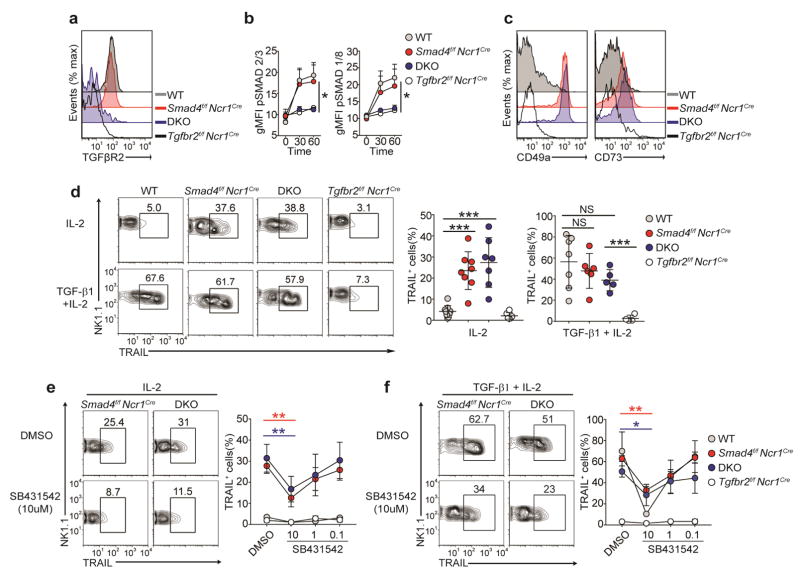
We hypothesized that the SMAD4-deficient NK cells were hyper-responsive to TGF-β. In support of our hypothesis, we found that Smad4f/f and SMAD4-deficient NK cells upregulated expression of the ILC1 markers CD49a, CD73 and TRAIL when cultured in vitro with TGF-β but not when cultured with other cytokines of this family (BMP4 and activin A) (Supplementary Fig. 3). Since a published study showed that imprinting of SG ILC1s by TGF-β requires TGFβR2 (ref. 13), we next investigated whether imprinting of SMAD4-deficient NK cells by TGF-β also depended on TGFβR2. To test that hypothesis, we generated mice that were doubly deficient in SMAD4 and TGFβR2 (Smad4f/fTgfbr2f/fNcr1iCre; called ‘DKO’ here). We confirmed that NK cells from DKO mice no longer expressed TGFβR2 ex vivo or after in vitro culture and that they were unable to engage in canonical TGF-β signaling, which requires phosphorylation of R-SMAD (Fig. 5a,b and Supplementary Fig. 4a,b). However, NK cells from DKO mice maintained high expression of CD49a and CD73 ex vivo (Fig. 5c) and upregulated their expression of TRAIL when cultured in vitro with IL-2 (Fig. 5d). DKO NK cells further increased their expression of TRAIL in response to TGF-β plus IL-2 (Fig. 5d), which demonstrated that SMAD4-deficient NK cells were responsive to TGF-β even in the absence of TGFβR2. Extending that conclusion was the finding that while TGFβR2 deficiency alone affected the imprinting of SG ILC1s by TGF-β, concomitant lack of SMAD4 and TGFβR2 restored the expression of CD49a in SG ILC1s (Supplementary Fig. 4c). Thus, the TGF-β responsiveness of SMAD4-deficient NK cells and ILC1s was not dependent on TGFβR2.
Figure 5.
Imprinting of SMAD4-deficient NK cells by TGF-β is independent of TGFβR2. (a) Surface expression of TGFβR2 on splenic NK cells obtained from Smad4f/f, Smad4f/fNcr1iCre, Smad4f/fTgfbr2f/fNcr1iCre (DKO) and Tgfbr2f/fNcr1iCre mice (key) after culture for 48 h with IL-2. (b) Quantification of intracellular phosphorylated SMAD2 and SMAD3 (p-SMAD2/3) and phosphorylated SMAD1 and SMAD8 (p-SMAD1/8) in splenic NK cells obtained from mice as in a (key) and stimulated for 30 or 60 min (horizontal axis) with TGF-β1 and IL-2, assessed by flow cytometry and presented as geometric mean fluorescence intensity (gMFI). (c) Surface expression of CD49a (left) and CD73 (right) on splenic NK cells from mice as in a (key). (d) Flow cytometry (left) and quantification (right) of TRAIL expression on splenic NK cells of genotypes as in a (above plots (left) or key (middle and right)) after 48 h of culture with IL-2 or TGF-β1 plus IL-2. (e,f) Flow cytometry (left) and quantification (right) of TRAIL in splenic NK cells of indicated genotypes (above plots (left) or key (right)) after 48 h of culture with IL-2 (e) or TGF-β1 plus IL-2 (f) in the presence of the vehicle DMSO or the TGFβR1 inhibitor SB431542 (at a concentration of 10 μM (left) or various concentrations (horizontal axis, right)). Numbers above outlined areas (d–f) indicate percent NK1.1+TRAIL+ cells. Each symbol (d) represents an individual mouse; small horizontal lines indicate the mean (± s.d.). *P < 0.05, **P < 0.01 and ***P < 0.001 (unpaired Student’s t-test). Data are representative of 4 experiments (a) or 5 experiments (c) or are pooled from at least two independent experiments with at least one mouse per genotype in each (b,d–f; mean ± s.d. in e,f).
SMAD4 inhibits a non-canonical TGFβR1-dependent pathway
We sought to determine whether deletion of receptors for the TGF-β family that transmit canonical signals through SMAD4 would be sufficient to elicit the paradoxical imprinting by TGF-β observed for SMAD4-deficient NK cells. We noticed that splenic NK cells expressed mainly type II receptors for BMPs (such as BMPR2) and TGFβR2 (Supplementary Fig. 4d). However, NK cells deficient in either BMPR2 or TGFβR2 seemed to be similar to Smad4f/f NK cells (Supplementary Fig. 4e), which suggested that abolishing BMPR2- or TGFβR2-mediated canonical signaling was not sufficient to elicit paradoxical imprinting by TGF-β equivalent to that seen for SMAD4-deficient NK cells.
We finally sought to determine whether the imprinting of SMAD4-deficient splenic NK cells by TGF-β depended on TGFβR1. To test this, we measured TRAIL expression in SMAD4-deficient and DKO NK cells cultured in vitro with or without a small-molecule antagonist specific for TGFβR1 (SB431542). TRAIL was chosen because it is not detected ex vivo but becomes apparent after in vitro culture, especially in the presence of TGF-β. Therefore, TRAIL can be used as a sensitive ‘read-out’ of NK cell responsiveness to TGF-β. While TRAIL was expressed on the surface of cultured SMAD4-deficient and DKO NK cells and its expression was substantially upregulated by TGF-β, inhibition of TGFβR1 resulted in a dose-dependent diminution in the surface expression of TRAIL on both SMAD4-deficient NK cells and DKO NK cells (Fig. 5e,f). No difference in cell survival was observed during this culture (Supplementary Fig. 4f), which demonstrated that the effect of SB431542 was specific to TGFβR1 signaling. We concluded that SMAD4 restricted the imprinting of NK cells by TGF-β in a TGFβR2-independent, TGFβR1-dependent manner.
Human SMAD4-deficiency amplifies NK cell responsiveness to TGF-β
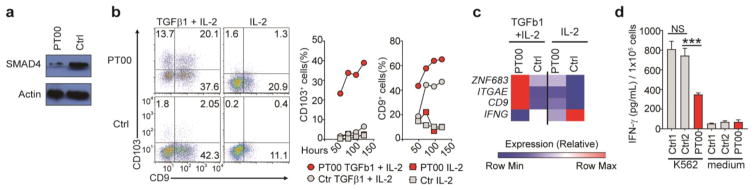
Germline mutations in the gene encoding human SMAD4 are responsible for some cases of juvenile polyposis syndrome, an autosomal dominant disease characterized by the development of intestinal polyps and an increased risk of gastrointestinal cancer35–37. Thus, we sought to determine whether our finding that SMAD4 restrained the imprinting of murine NK cells by TGF-β was applicable to human NK cells. We obtained a blood sample from a patient (PT00) affected by polyposis, who had a 25–base pair deletion in the sequence encoding the MH2 domain of SMAD4. Expression of SMAD4 protein was much lower in NK cells from PT00 than in control NK cells from healthy donors (Fig. 6a), which confirmed that this mutation resulted in haploinsufficiency for SMAD4. We assessed the expression of CD103 and CD9, two markers that have been shown to reflect the imprinting of human ILC1s by TGF-β14, as well as IFN-γ production and expression of mRNA encoding Hobit. NK cells freshly isolated from the blood of PT00 did not express CD103 or CD9 (data not shown), possibly due to the remaining SMAD4 protein. However, after culture with TGF-β, NK cells from PT00 upregulated their surface expression of CD103 and CD9 much more than did NK cells from healthy donors (Fig. 6b,c). Moreover, NK cells obtained from PT00 and exposed to TGF-β also showed increased expression of mRNA encoding Hobit (Fig. 6c) and less production of IFN-γ after co-culture with K562 target cells (Fig. 6d) than did control NK cells from healthy donors. Together these data indicated that SMAD4 modulated the sensitivity of human NK cells to TGF-β. An excessive NK cell response to TGF-β in patients with loss-of-function mutations in the gene encoding SMAD4 might contribute to their susceptibility to colon cancer.
Figure 6.
Human NK cells with a SMAD4 mutation are hyper-responsive to TGF-β. (a) Immunoblot analysis of total SMAD4 and actin (loading control) in human NK cells from a SMAD4-deficient patient (PT00) and a healthy control donor (Ctrl1). (b) Flow cytometry (left) and quantification (right) of CD103 and CD9 in NK cells from donors as in a and cultured with TGF-β1 plus IL-2 or with IL-2 alone. (c) Expression of ZNF683 (encoding Hobit), ITGAE (encoding CD103), CD9 (encoding CD9) and IFNG (encoding IFN-γ) (left margin) in NK cells from donors as in a and cultured overnight with TGF-β1 and IL-2 or with IL-2 alone. (d) Secretion of IFN-γ by NK cells from donors as in a (two healthy donors: Ctrl1 and Ctrl2) cultured overnight alone (Medium) or together with K562 target cells. ***P < 0.001 (unpaired Student’s t-test). Data are representative of two (a), QQ (c) or at least three (b,d) independent experiments (mean + s.d. in d).
DISCUSSION
SMAD4 is a known positive regulator of TGF-β canonical signaling that facilitates the translocation of R-SMAD into the nucleus for the initiation of transcription. Given that TGF-β imprints ILC1s13,14 and impedes the development and function of NK cells22,23, we anticipated that SMAD4 deficiency in these cells would promote the conversion of ILC1s into NK-like cells and enhance the function of differentiated NK cells. Paradoxically, ILC1s remained intact, while NK cells converted into ILC1-like cells with impaired anti-viral and anti-tumor function. Despite having a defect of TGF-β canonical signaling, SMAD4-deficient NK cells were hyper-responsive to TGFβR1-mediated signals. Thus, our study has demonstrated that SMAD4 is also a negative regulator of a non-canonical TGF-β signaling pathway in NK cells.
The effect of that regulatory function of SMAD4 is relevant to people with inherited defects in SMAD4, which might hamper the ability of NK cells to effectively control tumor cells and might contribute, at least in part, to their susceptibility to polyposis and colon cancer. Consistent with that, NK cells obtained from a SMAD4-haploinsufficient person had higher expression of ILC1 surface markers and less production of IFN-γ after exposure to TGF-β than did NK cells from healthy donors. It is likely that SMAD4 deficiency in humans also affects T cell responses via a similar mechanism. In mice, selective deletion of SMAD4 in T cells results in the spontaneous formation of intestinal tumors38,39 and defective anti-tumor T cell responses40. Moreover, while TGFβR2 deficiency impairs the generation of regulatory T cells, concomitant deletion of SMAD4 in T cells offsets TGFβR2 deficiency and leads to the partial recovery of regulatory T cells in vivo40. Various human genetic mutations can affect SMAD4 (ref. 35); some might cause haplo-insufficiency, while others might have dominant negative effects. Future studies should establish whether a dysregulated non-canonical TGF-β pathway is engendered by all mutations in the gene encoding SMAD4 or only a portion of them.
The phenotype of TGF-β-imprinted SMAD4-deficient NK cells was notably similar to that previously reported for tissue resident ILC1s, such as SG, intestinal and liver ILC1s. However, in the context of those studies, our results indicate that imprinting by TGF-β occurs through distinct modalities in different cell types. In SMAD4-deficient NK cells, TGF-β acts through a TGFβR1-dependent, TGFβR2-independent non-canonical pathway. In SG ILC1s, TGF-β acts through a non-canonical pathway that is TGFβR2 dependent and SMAD4 independent13. In other cells, such as those in the liver and gut, imprinting by TGF-β might occur through pathways that require various other members of the TGF-β or TGF-β receptor superfamilies. It will be important to define these variables in the development of the as-yet- poorly understood ILC1s, such as liver ILC1s. Future studies will also need to determine how SMAD4 restrains non-canonical TGFβR1 signaling. Among the signal transducers potentially involved in the regulation of non-canonical pathways, it is worth considering TRIM33, an E3 ligase linked to the degradation of TGFβR1 as well as to competing with SMAD4 for binding R-SMAD receptors and inducing their translocation to the nucleus20,41. The absence of SMAD4 might enhance the translocation of TRIM33–R-SMAD receptors complexes to the nucleus while diminishing the TRIM33-mediated control of TGFβR1 in the cytosol.
Our results have identified Hobit as a key transcription factor integral to the TGF-β signature of ILC1s. Hobit is a transcription factor related to the transcription factor Blimp that shapes various characteristics of tissue-resident memory T cells and liver ILC1s, particularly their ability to acquire tissue-residency features16. Our results have demonstrated that TGF-β signaling induces Hobit and suggest that it might be a central node in the network responsible for the ILC1 functional program. Future studies should determine whether Hobit induces the expression of molecules with regulatory functions, such as CD73 and TRAIL, in addition to molecules involved in tissue residency.
ILC1s can promote anti-tumor responses in an IL-15-rich environment, which stimulates cytotoxicity42. Our study has suggested that in TGF-β-rich tumors, ILC1s might in fact be ineffective or facilitate tumor growth because of their reduced capacity for IFN-γ production and increased expression of regulatory molecules. Accordingly, another paper has shown that ILC1s found in tumors undergo strong imprinting by TGF-β and are incapable of effective anti-tumor responses43. Thus, the effect of ILC1s in tumors might depend on the tumor environment. We found that one common feature of ILC1s and ILC1-like cells was the expression of TIGIT, an inhibitory receptor specific for CD115 and CD112 that attenuates lymphocyte activation. Checkpoint-blockade strategies that target TIGIT might effectively activate ILC1s increasing anti-tumor innate immune responses.
METHODS
Mice
Smad4f/f (Smad4tm2.1Cxd/J) mice were obtained from the Jackson Laboratory and were designed with loxP sites flanking exon 8 of Smad4 (corresponding to the MH2 domain of the protein). Tgfbr2f/f (B6;129-Tgfbr2tm1Karl/J) mice were obtained from the Jackson Laboratory. Bmpr2f/f mice were provided by M. Rabinovitch. Il7rCre mice were provided by H.-R. Rodewald44. Ncr1iCre mice, in which sequence encoding ‘codon-improved’ iCre recombinase is inserted into the 3′ untranslated region of Ncr1 (which encodes NKp46)17, were bred to Smad4f/f, Tgfbr2f/f and Bmpr2f/f mice to generate Smad4f/fNcr1iCre, Tgfbr2f/fNcr1iCre, Smad4f/fTgfbr2f/fNcr1iCre and Bmpr2f/fNcr1iCre mice. All mice were bred at a pathogen-free facility at Washington University. Age- and sex- matched animals were used throughout the experiments (littermates for most). All animal experiments were conducted per the USA Public Health Service Policy of Humane Care and Use of Laboratory Animals. All protocols were approved by the Institutional Animal Care and Use Committee (School of Medicine, Washington University, St. Louis, MO).
Antibodies and Flow cytometry
Fluorochrome- and biotin-conjugated antibodies to mouse NK1.1 (PK136), CD3 (145-2C11), CD19 (1D3), CD45 (30-F11), CD49b (HMa2), CD73 (TY/11.8), TRAIL (N2B2), Ly49H (3D10), Ki67 (SolA15), Ly6C (HK1.4), CD5 (53-7.2), GzmB (GB11), KLRG1 (2F1), Eomes (Dan11mag), T-bet (eBIO 4B10), RORct (AFKJS-9), IL-22 (1H8PWSR), and IFN-γ (xma1.2) were purchased from eBioscience. Fluorochrome- and biotin-conjugated antibodies to mouse CD49a (Ha31/8), CD69 (H1.2F3) CD62L (MEL-14), TIGIT (1G9), Thy1.2 (53-2.1), CD107a (1D4B), pSMAD2/3 (072-670), pSMAD1/8 (N6-1233), and human CD103 (Ber-ACT8) were purchased from BD Biosciences. Biotin-conjugated TGFβR2 (Cat # BAF532) and human CD9 (209306) were purchased from R&D Systems. Fluorochrome-conjugated antibodies to mouse CX3CR1 (SAO11F11) and CCR6 (29-2L17) was purchased from Biolegend. Streptavidin-APC, Streptavidin-PerCP/Cy5.5, and Streptavidin-PE-Cy7 were purchased from Invitrogen, Biolegend, and eBioscience, respectively. The isolation of splenic, lung, hepatic and intestinal lymphocytes was done as previously described15. Stained cells were analyzed on a FACSCanto using Diva software or on a FACSCalibur using CellQuest software. For transcription-factor detection, cells were first stained for surface markers, followed by fixation and permeabilization with the Foxp3 staining buffer set (eBioscience). Cells were then resuspended in 1× permwash solution and stained with antibodies.
Stimulation
Splenocytes were made into single-cell suspensions, followed by the addition of addition of IL-12 (10 ng/ml, Peprotech) and IL-18 (100 ng/ml, MBLI) or 1 × 106 Yac-1 cells. After 1 h, brefeldin A (10ug/ml, Sigma) was added to each well and stimulation was allowed to continue for a total of 6 h. Cells were then washed, surface stained and fixed using IC fixation buffer (eBioscience). Fixed cells were permeabilized with 1× permwash buffer and stained for intracellular cytokines. For detection of phosphorylated SMAD proteins, splenocytes were made into single-cell suspensions that were incubated in serum-free medium for 1 h at 37 °C. TGF-β1 (10 ng) and IL-2 were then added to cultures, and after 30 or 60 min, cells were fixed, permeabilized, and stained according to the manufacturer’s (BD Phosflow Cell Signaling) instructions. For ILC3 stimulation, lymphocytes were isolated from the small intestine lamina propria and cultured with 10 ng/mL IL-23 (R&D) for 3.5 hours. Brefeldin A was added for the last three hours.
Cell culture
Splenic NK cells were either enriched by magnetic sorting (CD49b microbeads; Miltenyi Biotec) or single-cell suspensions of whole spleen were used. Cells were cultured in complete medium with IL-2 (1,000 U/ml; Roche) and/or TGF-β1 (10 ng; Peprotech), BMP4 (10 ng; Peprotech) or activin A (10 ng; Peprotech) for 48 h. SB431542 (Sigma) was solubilized in DMSO and added at the beginning of culture.
Tumor model and MCMV infection
1 × 105 B16-F10 melanoma cells in HBBS were injected into the tail vein of mice. At day 15 after injection, tumor foci were quantified in the lungs with the use of a dissecting microscope. A lobe of the lung was also flash-frozen for tissue sections. For MCMV infection, mice were given MCMV (5 × 104 plaque-forming units) intraperitoneally, and their weight was monitored every day for 6 d. At day 3, mice were given intraperitoneal injection of 2 mg BrdU and, 4 h later, incorporation was measured using the BrdU Flow Kit (BD). Annexin V surface expression was measured using the Annexin V apoptosis detection kit (BD).
qRT-PCR
For gene-expression analysis by qRT-PCR, CD49b+ splenic NK cells were enriched by magnetic sorting (Miltenyi Biotec), were purified by flow cytometry (BD FACSAria II) (NK1.1+CD49b+CD3−CD19−) and were frozen in RLT buffer (Qiagen). RNA was extracted with an RNeasy Mini kit (Qiagen). cDNA was synthesized from RNA with Superscript III first-strand synthesis system for RT-PCR (Invitrogen). cDNA expression was analyzed by quantitative PCR using iTaq Universal SYBR Green Supermix (Bio-Rad Laboratories) and a StepOnePlus system (Applied Biosystems). The expression of target genes was calculated and normalized to the expression of the control gene Gapdh using the 2−ΔΔCT method.
Primer Pairs
Primer Pairs for qRT-PCR Primers were used as follows:
Gapdh forward-ACGGCAAATTCAACGGCACAGTCA;
Gapdh reverse-TGGGGGCATCGGCAGAAGG;
Il21r forward- GGCTGCCTTACTCCTGCTG;
Il21r reverse-TCATCTTGCCAGGTGAGACTG;
Zfp683 forward-CTCAGCCACTTGCAGACTCA
Zfp683 reverse-CTGTCGGTGGAGGCTTTGTA
Inpp4b forward- AGAACCTCAGATGGTGGCAAA
Inpp4b reverse- CCCGCTCAGACTTTCTGGTG
Tsc22d1 forward- CCAGTGGCGATGGATCTAGGA
Tsc22d1 reverse- CTTGCACCAGAGCTATTGTCA
Human samples
The study was performed in accordance with the protocols approved by the institutional review board (School of Medicine, Washington University, St. Louis, MO). All subjects provided written informed consent. Blood was collected from a patient (PT00) with polyposis and a SMAD4 mutation (c1351_1375del25) or healthy donors. CD56+ cells were enriched from the blood by magnetic cell sorting (Miltenyi Biotec), purified by flow cytometry (BD FACSAria II) (CD56+CD3−CD19−) and underwent population expansion in vitro as previously described45. NK cells were cultured with K562 at a ratio of 1:4 overnight. IFN-γ amounts were then measured in the supernatant by cytometric bead array (BD).
Immunoblot analysis
Samples were lysed sequentially in NP-40 lysis buffer (0.5% NP-40, 2 mM EDTA, 150 mM NaCl, pH 7.4), spun at 13,000 g for 10 minutes to pellet nuclei. Lysis buffers also contained aprotinin, EDTA, leupeptin, PMSF, sodium orthovanadate, and phosphatase inhibitor (cocktail 3 from Sigma-Aldrich). Samples were denatured and reduced by addition of 4X LDS sample buffer, 10% β-mercaptoethanol, and heating to 95° C for 10 minutes (Invitrogen). Samples were run on an 8% TRIS/SDS polyacrylamide gel with a 4% stacker and transferred to nitrocellulose. Blots were blocked for 1 hour at room temperature in 5% non-fat dry milk in PBS + 0.05% Tween 20. Anti-human SMAD4 and pan-actin were used as primary antibodies (Cell Signaling Technologies 9515P and 8685P). Secondary was anti-rabbit-HRP (Southern Biotech, 4010-05). Chemiluminescent reagents from Thermo Scientific were used to visualize bands (West Pico (34080) and West Femto (34095)).
Microarray and data analysis
RNA was obtained ex vivo from mouse splenic NK cells (NK1.1+CD49b+CD3−CD19−) purified by flow cytometry (BD FACSAria II) or from human NK cells after overnight culture with TGF-β1 (2 ng) and/or IL-2. Total RNA was isolated (RNeasy micro kit, QIAGEN), amplified, and hybridized to the Affymetrix Mouse Gene (v.1.0) or Affymetrix Human Gene (v.1.0) ST arrays. Data were analyzed with GenePattern software (Broad Institute) as previously described18.
Statistical analysis
Statistical analyses were performed using Prism 5.0 (GraphPad Software) or Multiplot for volcano plots. All data were analyzed with an unpaired Student’s t-test. A P value of ≤0.05 was considered significant.
A Life Sciences Reporting Summary for this paper is available online.
Data availability
The data that support the findings of this study are available from the corresponding author upon request. The accession codes are GSE100246 and GSE100247.
Supplementary Material
Acknowledgments
We thank the Genome Technology Access Center in the Department of Genetics at Washington University School of Medicine for help with genomic analysis; H.-R. Rodewald (DKZF, Heidelberg, Germany) for Il7rCre mice; M. Rabinovitch (Stanford University, USA) for Bmpr2f/f mice. Supported by the US National Institutes of Health (UO1 AI095542, RO1 DE025884, and RO1 DK103039 to M. Colonna; RO1 CA176695 to M. Cella; and 5T32CA009547-30 to V.S.C.), Crohn’s & Colitis Foundation of America (274415) and Cancer Research Institute (Fellowship to J.K.B).
Footnotes
AUTHOR CONTRIBUTIONS
V.S.C., T.K.U., L.C-B., J.K.B, Q.W., S.G. and M. Cella performed experiments and analyzed the data; V.S.C., M.L.R, and M. Cella performed in silico analysis; A.J.W. provided materials and clinical expertise; V.S.C., M. Cella and M. Colonna designed the experiments; and V.S.C., S.G., Cella and M. Colonna wrote the manuscript.
COMPETING FINANCIAL INTERESTS
The authors declare no competing financial interests.
References
- 1.Marcus A, et al. Recognition of tumors by the innate immune system and natural killer cells. Adv Immunol. 2014;122:91–128. doi: 10.1016/B978-0-12-800267-4.00003-1. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Yu X, et al. The basic leucine zipper transcription factor NFIL3 directs the development of a common innate lymphoid cell precursor. eLife. 2014;3:1–20. doi: 10.7554/eLife.04406. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Klose CSN, et al. Differentiation of type 1 ILCs from a common progenitor to all helper-like innate lymphoid cell lineages. Cell. 2014;157:340–356. doi: 10.1016/j.cell.2014.03.030. [DOI] [PubMed] [Google Scholar]
- 4.Constantinides MG, McDonald BD, Verhoef PA, Bendelac A. A committed precursor to innate lymphoid cells. Nature. 2014;508:397–401. doi: 10.1038/nature13047. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Daussy C, et al. T-bet and Eomes instruct the development of two distinct natural killer cell lineages in the liver and in the bone marrow. J Exp Med. 2014;211:563–577. doi: 10.1084/jem.20131560. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Constantinides MG, et al. PLZF expression maps the early stages of ILC1 lineage development. Proc Natl Acad Sci USA. 2015;112:5123–5128. doi: 10.1073/pnas.1423244112. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Geiger TL, Sun JC. Development and maturation of natural killer cells. Curr Opin Immunol. 2016;39:82–89. doi: 10.1016/j.coi.2016.01.007. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Bernink JH, et al. Human type 1 innate lymphoid cells accumulate in inflamed mucosal tissues. Nat Immunol. 2013;14:221–229. doi: 10.1038/ni.2534. [DOI] [PubMed] [Google Scholar]
- 9.Simoni Y, et al. Human Innate lymphoid cell subsets possess tissue-type based heterogeneity in phenotype and frequency. Immunity. 2017;46:148–161. doi: 10.1016/j.immuni.2016.11.005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Lim AI, et al. Systemic human ILC precursors provide a substrate for tissue ILC differentiation. Cell. 2017;168:1086–1100e1010. doi: 10.1016/j.cell.2017.02.021. [DOI] [PubMed] [Google Scholar]
- 11.Sojka DK, et al. Tissue-resident natural killer (NK) cells are cell lineages distinct from thymic and conventional splenic NK cells. eLife. 2014;3:e01659. doi: 10.7554/eLife.01659. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Gasteiger G, Fan X, Dikiy S, Lee SY, Rudensky AY. Tissue residency of innate lymphoid cells in lymphoid and nonlymphoid organs. Science. 2015;350:981–985. doi: 10.1126/science.aac9593. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Cortez VS, et al. Transforming growth factor-β signaling guides the differentiation of innate lymphoid cells in salivary glands. Immunity. 2016;44:1127–1139. doi: 10.1016/j.immuni.2016.03.007. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Fuchs A, et al. Intraepithelial type 1 innate lymphoid cells are a unique subset of IL-12- and IL-15-responsive IFN-γ-producing cells. Immunity. 2013;38:769–781. doi: 10.1016/j.immuni.2013.02.010. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Cortez VS, Fuchs A, Cella M, Gilfillan S, Colonna M. Cutting edge: salivary gland NK cells develop independently of Nfil3 in steady-state. J Immunol. 2014;192:4487–4491. doi: 10.4049/jimmunol.1303469. [DOI] [PubMed] [Google Scholar]
- 16.Mackay LK, et al. Hobit and Blimp1 instruct a universal transcriptional program of tissue residency in lymphocytes. Science. 2016;352:459–463. doi: 10.1126/science.aad2035. [DOI] [PubMed] [Google Scholar]
- 17.Narni-Mancinelli E, et al. Fate mapping analysis of lymphoid cells expressing the NKp46 cell surface receptor. Proc Natl Acad Sci USA. 2011;108:18324–18329. doi: 10.1073/pnas.1112064108. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Robinette ML, et al. Transcriptional programs define molecular characteristics of innate lymphoid cell classes and subsets. Nat Immunol. 2015;16:306–317. doi: 10.1038/ni.3094. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Massagué J. TGFβ signalling in context. Nat Rev Mol Cell Biol. 2012;13:616–630. doi: 10.1038/nrm3434. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.He W, et al. Hematopoiesis controlled by distinct TIF1γ and Smad4 branches of the TGFβ pathway. Cell. 2006;125:929–941. doi: 10.1016/j.cell.2006.03.045. [DOI] [PubMed] [Google Scholar]
- 21.Zhang YE. Non-Smad signaling pathways of the TGF-β family. CSH Perspec Biol. 2017:9. doi: 10.1101/cshperspect.a022129. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Laouar Y, Sutterwala FS, Gorelik L, Flavell RA. Transforming growth factor-β controls T helper type 1 cell development through regulation of natural killer cell interferon-γ. Nat Immunol. 2005;6:600–607. doi: 10.1038/ni1197. [DOI] [PubMed] [Google Scholar]
- 23.Marcoe JP, et al. TGF-β is responsible for NK cell immaturity during ontogeny and increased susceptibility to infection during mouse infancy. Nat Immunol. 2012;13:843–850. doi: 10.1038/ni.2388. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Viel S, et al. TGF-β inhibits the activation and functions of NK cells by repressing the mTOR pathway. Sci Signal. 2016;9:ra19. doi: 10.1126/scisignal.aad1884. [DOI] [PubMed] [Google Scholar]
- 25.Yang X, Li C, Herrera PL, Deng CX. Generation of Smad4/Dpc4 conditional knockout mice. Genesis. 2002;32:80–81. doi: 10.1002/gene.10029. [DOI] [PubMed] [Google Scholar]
- 26.Takeda K, et al. IFN-γ production by lung NK cells is critical for the natural resistance to pulmonary metastasis of B16 melanoma in mice. J Leukoc Biol. 2011;90:777–785. doi: 10.1189/jlb.0411208. [DOI] [PubMed] [Google Scholar]
- 27.van Helden MJ, et al. Terminal NK cell maturation is controlled by concerted actions of T-bet and Zeb2 and is essential for melanoma rejection. J Exp Med. 2015;212:2015–2025. doi: 10.1084/jem.20150809. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Lakshmikanth T, et al. NCRs and DNAM-1 mediate NK cell recognition and lysis of human and mouse melanoma cell lines in vitro and in vivo. J Clin Invest. 2009;119:1251–1263. doi: 10.1172/JCI36022. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Gilfillan S, et al. DNAM-1 promotes activation of cytotoxic lymphocytes by nonprofessional antigen-presenting cells and tumors. J Exp Med. 2008;205:2965–2973. doi: 10.1084/jem.20081752. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Johnston RJ, et al. The immunoreceptor TIGIT regulates antitumor and antiviral CD8+ T cell effector function. Cancer Cell. 2014;26:923–937. doi: 10.1016/j.ccell.2014.10.018. [DOI] [PubMed] [Google Scholar]
- 31.Chauvin JM, et al. TIGIT and PD-1 impair tumor antigen-specific CD8+ T cells in melanoma patients. J Clin Invest. 2015;125:2046–2058. doi: 10.1172/JCI80445. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Stanietsky N, et al. The interaction of TIGIT with PVR and PVRL2 inhibits human NK cell cytotoxicity. Proc Natl Acad Sci USA. 2009;106:17858–17863. doi: 10.1073/pnas.0903474106. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Boles KS, et al. A novel molecular interaction for the adhesion of follicular CD4 T cells to follicular DC. Eur J Immunol. 2009;39:695–703. doi: 10.1002/eji.200839116. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Vidal SM, Lanier LL. NK cell recognition of mouse cytomegalovirus-infected cells. Curr Top Microbiol Immunol. 2006;298:183–206. doi: 10.1007/3-540-27743-9_10. [DOI] [PubMed] [Google Scholar]
- 35.Miyaki M, Kuroki T. Role of Smad4 (DPC4) inactivation in human cancer. Biochem Biophys Res Commun. 2003;306:799–804. doi: 10.1016/s0006-291x(03)01066-0. [DOI] [PubMed] [Google Scholar]
- 36.Larsen Haidle J, Howe JR. Juvenile Polyposis Syndrome. GeneReviews. 1993:1–28. [Google Scholar]
- 37.Howe JR, et al. Mutations in the SMAD4/DPC4 gene in juvenile polyposis. Science. 1998;280:1086–1088. doi: 10.1126/science.280.5366.1086. [DOI] [PubMed] [Google Scholar]
- 38.Kim BG, et al. Smad4 signalling in T cells is required for suppression of gastrointestinal cancer. Nature. 2006;441:1015–1019. doi: 10.1038/nature04846. [DOI] [PubMed] [Google Scholar]
- 39.Li MO, Flavell RA. TGF-β: a master of all T cell trades. Cell. 2008;134:392–404. doi: 10.1016/j.cell.2008.07.025. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Gu AD, et al. A critical role for transcription factor Smad4 in T cell function that is independent of transforming growth factor β receptor signaling. Immunity. 2015;42:68–79. doi: 10.1016/j.immuni.2014.12.019. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Huang F, Chen YG. Regulation of TGF-β receptor activity. Cell Biosci. 2012;2:9. doi: 10.1186/2045-3701-2-9. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Dadi S, et al. Cancer immunosurveillance by tissue-resident innate lymphoid cells and innate-like T cells. Cell. 2016;164:365–377. doi: 10.1016/j.cell.2016.01.002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Gao Y, et al. Tumor immunoevasion by the conversion of effector NK cells into type 1 innate lymphoid cells. Nat Immunol. 2017;18:XXX–XXX. doi: 10.1038/ni.3800. [DOI] [PubMed] [Google Scholar]
- 44.Schlenner SM, et al. Fate mapping reveals separate origins of T cells and myeloid lineages in the thymus. Immunity. 2010;32:426–436. doi: 10.1016/j.immuni.2010.03.005. [DOI] [PubMed] [Google Scholar]
- 45.Cella M, Colonna M. Cloning human natural killer cells. Methods Mol Biol. 2000;121:1–4. doi: 10.1385/1-59259-044-6:1. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
Data Availability Statement
The data that support the findings of this study are available from the corresponding author upon request. The accession codes are GSE100246 and GSE100247.