Abstract
The emergence of the plasmid-encoded colistin-resistance gene mcr-1 in Enterobacteriaceae represents a new threat to the treatment of infection in the clinical setting. A sensitive and rapid molecular method for detection of the mcr-1 gene in clinical isolates is needed to control the spread of this gene. In this study, we established a loop-mediated isothermal amplification (LAMP) assay for rapid detection of the mcr-1 gene. This assay was applied to cultured bacteria and spiked human stools. Real-time monitoring of turbidity and chromogenic visualization were used to assess the reaction results. The specificity and sensitivity of the primers in the LAMP reactions for detection of the mcr-1 gene were determined. All 20 clinically resistant isolates without the mcr-1 gene tested negative, indicating the high specificity of the LAMP primers. The sensitivity of LAMP, with a detection limit of 0.2 pg/μL DNA, was 10-fold greater than that of polymerase chain reaction (PCR). The assay was also conclusive when applied to human stools spiked with mcr-1-positive Escherichia coli. During clinical screening in a major hospital in Beijing, China, seven isolates were identified as positive from the 556 Enterobacteriaceae isolates. In conclusion, the LAMP assay we developed was useful for detection of the mcr-1 gene in the clinical setting.
Keywords: colistin, the mcr-1 gene, Enterobacteriaceae, LAMP, rapid detection
Introduction
Colistin is a polymyxin antibiotic that is considered as the final line of treatment for multidrug-resistant gram-negative bacteria (Li et al., 2006). In 2015, the first plasmid-mediated colistin-resistance gene, mcr-1, was identified in Enterobacteriaceae isolates of human and animal origin (Liu et al., 2016). Subsequently, the mcr-1 gene has been found in many countries around the world, as demonstrated by screening of early isolated strains and whole-genome sequencing of bacteria (Arcilla et al., 2016; Mulvey et al., 2016; Webb et al., 2016). Moreover, a report of a carbapenem-resistant Enterobacteriaceae (CRE) carrying the mcr-1 gene highlighted the importance of this new global public health issue (Yao et al., 2016).
Recently, various molecular methods, including conventional polymerase chain reaction (PCR) and real-time PCR, have been established for detection of the mcr-1 gene (Bontron et al., 2016; Liu et al., 2016). While these assays facilitate rapid, sensitive, and specific detection, they require specialized and expensive equipment. Therefore, another rapid and simple assay, termed loop-mediated isothermal amplification (LAMP), was developed to complement the existing PCR methods. LAMP is a nucleic acid detection technique that was first reported in 2000 and can be used to amplify DNA with Bst DNA polymerase under isothermal conditions, permitting autocycling strand displacement DNA synthesis (Notomi et al., 2000; Ushikubo, 2004). This method employs four to six primers recognizing a total of six to eight distinct sequences in the target DNA, giving the assay a high degree of specificity and amplification efficiency. LAMP is widely used for pathogen detection in infectious diseases (Parida et al., 2008).
Accordingly, in this study, we established and optimized a LAMP method for detection of the mcr-1 gene. The specificity and sensitivity of the LAMP reaction for detection of the mcr-1 gene were determined. In addition, the sensitivity of the LAMP assay for detection of the mcr-1 gene in fecal samples was also tested.
Materials and methods
Bacterial strains and preparation of templates
In total, 20 multidrug-resistant bacteria without the mcr-1 gene, as determined using the existing PCR method, were used as negative controls (Liu et al., 2016). Two isolates carrying the mcr-1 gene, Escherichia coli 12452 and E. coli 12536, were screened by our laboratory in 2016 and used as positive controls. The information on these strains is shown in Table 1. Genomic DNA from bacteria was extracted using a one-step microbial DNA extraction method based on Chelex-100 (de Lamballerie et al., 1992). In addition, in order to assess the sensitivity of the LAMP assay, genomic DNA was also extracted from E. coli 12452 using a Wizard genomic DNA purification kit (Promega, Madison, WI, USA) according to the manufacturer's instructions. Purified DNA was then prepared by serial 10-fold dilutions to give concentrations ranging from 20 ng/μL to 0.02 pg/μL (Liu et al., 2012).
Table 1.
Bacterial strains used in this study.
| No. | Species | Source |
|---|---|---|
| 1 | Escherichia coli 12452 | Clinical isolate |
| 2 | Escherichia coli 12536 | Clinical isolate |
| 3 | Escherichia coli 12266 | Clinical isolate |
| 4 | Escherichia coli 12278 | Clinical isolate |
| 5 | Klebsiella pneumonia 12146 | Clinical isolate |
| 6 | Klebsiella pneumonia 12163 | Clinical isolate |
| 7 | Acinetobacter baumannii 12223 | Clinical isolate |
| 8 | Acinetobacter baumannii 12250 | Clinical isolate |
| 9 | Stenotrophomonas maltophilia 12285 | Clinical isolate |
| 10 | Stenotrophomonas maltophilia 12455 | Clinical isolate |
| 11 | Pseudomonas aeruginosa 12343 | Clinical isolate |
| 12 | Pseudomonas aeruginosa 12352 | Clinical isolate |
| 13 | Enterobacter cloacae 12105 | Clinical isolate |
| 14 | Enterobacter cloacae 12211 | Clinical isolate |
| 15 | Proteus mirabilis 12116 | Clinical isolate |
| 16 | Proteus mirabilis 12407 | Clinical isolate |
| 17 | Serratia marcescens 12208 | Clinical isolate |
| 18 | Serratia marcescens 12330 | Clinical isolate |
| 19 | Salmonella typhimurium 12306 | Clinical isolate |
| 20 | Salmonella typhimurium 12161 | Clinical isolate |
| 21 | Shigella flexneri 12221 | Clinical isolate |
| 22 | Shigella flexneri 12169 | Clinical isolate |
Primer design
The sequence of the mcr-1 gene was downloaded from the GenBank database (accession number LC114018.1), and the gene-specific primers were then designed using the Primer Explorer (version 5) software (http://primerexplorer.jp/lampv5e/index.html). First, the forward inner primer (FIP), backward inner primer (BIP), outer forward primer (F3), and outer backward primer (B3) were designed. Then, additional loop primers (LF and LB) were designed to accelerate the amplification reaction (Notomi et al., 2000). All primers were synthesized by Sangon Biotech Co., Ltd. (Shanghai, China).
Spiked stools and DNA extraction
Fifty milligrams of fresh feces from healthy individuals was suspended in 200 μL distilled water by vigorous shaking for 5 min. An amount of 108 bacteria (corresponding to 0.2 OD600) or dilution was resuspended in 50 μL of 0.85% NaCl and spiked in 200 μL of human fecal suspension (Bontron et al., 2016). The total DNA was extracted from spiked stool samples with a QIAamp DNA Stool Mini Kit (Qiagen GmbH, Hilden, Germany) according to the manufacturer's instructions. DNA was recovered in 100 μL distilled water, and the LAMP assay was performed with 2 μL DNA template.
LAMP reaction
The LAMP assay was performed in 25 μL reaction mixtures containing the following components: 20 mM Tris-HCl (pH 8.8), 10 mM KCl, 10 mM (NH4)2SO4, 8 mM MgSO4, 0.8 M betaine, 0.1% Tween-20, 1.4 mM of each dNTP, and 8 U Bst DNA polymerase. The amounts of primers needed for one reaction were 40 pmol for FIP and BIP, 20 pmol for LF and LB, and 5 pmol for F3 and B3. Finally, an appropriate amount of genomic DNA template was added to the reaction tube (Liu et al., 2012). The mixture was covered with 25 μL wax and incubated for 60 min in a dry bath incubator with a controlled temperature. Temperature was optimized by testing at 60–67°C.
LAMP assay amplification products were detected using two different methods: turbidimetry and visual detection. For monitoring of turbidity, real-time amplification by the LAMP assay was monitored using a Loopamp real-time turbidimeter (LA320-C; Eiken Chemical Co. Ltd., Tokyo, Japan). For visual detection, manganese ion (Mn2+) and calcein, a fluorescent metal indicator, were added to the reaction mixture, which appeared orange because calcein was quenched by Mn2+. For a positive reaction, Mn2+ formed a magnesium pyrophosphate precipitate (an insoluble salt) during amplification, and the color of the reaction system changed from orange to green. For a negative reaction, there was no color change because amplification did not occur (Mori et al., 2001).
PCR assay
PCR was performed to compare assay sensitivities for detection of the mcr-1 gene using the previously reported CLR5-F and CLR5-R primers (Liu et al., 2016). PCR tubes were set up using 12.5 μL PCR MasterMix reagents (Tiangen Biotech Co., Ltd., Beijing, China), 1 μM CLR5-F and CLR5-R primers (0.4 μM), and 1 μL DNA template in a final volume of 25 μL. The thermal cycler parameters were 3 min at 95°C; 30 cycles of 30 s at 95°C, 30 s at 55°C, and 30 s at 72°C; followed by 5 min at 72°C. The PCR-amplified products were analyzed by 1% agarose gel electrophoresis and stained with GelRed nucleic acid gel stain (Biotium, Hayward, CA, USA). Images were documented using a Bio-Rad Gel Doc EQ imaging system (Bio-Rad, Hercules, CA, USA).
Results
Ethics statement
All volunteers provided written, informed consent to participate in this study, which was reviewed and approved by the ethics committee of the Academy of Military Medical Sciences, China. All experiments were performed in accordance with relevant guidelines and regulations.
Primer screening
Five sets of primers for detection of the mcr-1 gene were designed. According to the primer parameters and variants of mcr-1 gene mutations (Thanh et al., 2016), we excluded the fourth and fifth sets of primers and evaluated the other three sets of primers. Under the same reaction conditions, three turbidity curves were observed, indicating that all three primers had the ability to amplify the target fragment. Through this analysis, we found that the third set of primers allowed for completion of the amplification in the shortest time (Figure 1). Primers in the third primer set (Table 2) were chosen as the final primers for mcr-1 gene detection by LAMP. For more information on the five sets of primers, see the Supplementary Materials.
Figure 1.
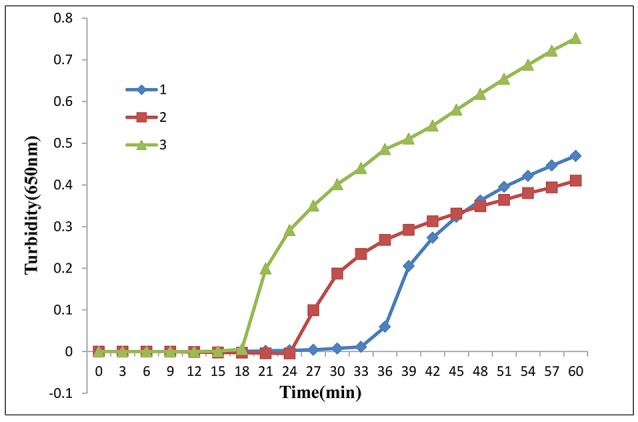
LAMP assay primer screening. Three sets of primers amplified the target gene under the same conditions. Turbidity was monitored by a Loopamp real-time turbidimeter at 650 nm every 6 s.
Table 2.
Primers used for amplification of the mcr-1 gene.
| Primer | Sequence (5′-3′) | Location |
|---|---|---|
| 3-F3 | ACAAGCAACCAAGCCTGAT | 681–699 |
| 3-B3 | GCGCCCAGATAGCTGAAC | 876–893 |
| 3-FIP | GAAGCTGACATGATCGGCGCGCGTAAGCCACGCCTAGTG | 745–765, 703–720 |
| 3-BIP | TTCCCACAGCTTGCCAAGATCGGCACAGAATACGCCGTCG | 787–808, 851–868 |
| 3-LB | AGCAATGTCACATCGTGCG | 826–844 |
Optimization of temperature for the lamp assay
Various reaction temperatures ranging from 60 to 67°C were employed at 1°C intervals to optimize the amplification. As shown in Figure 2, LAMP assay amplification occurred in the range of 60–67°C, and shorter reaction times were observed at 65, 66, and 67°C compared with those at other temperatures. We chose 66°C as the optimum temperature due to the higher peak and stronger enzyme activity in the LAMP assay (the optimum temperature for enzymatic activity of the Bst DNA polymerase was 65°C). Finally, we chose 66°C as the reaction temperature for later experiments.
Figure 2.
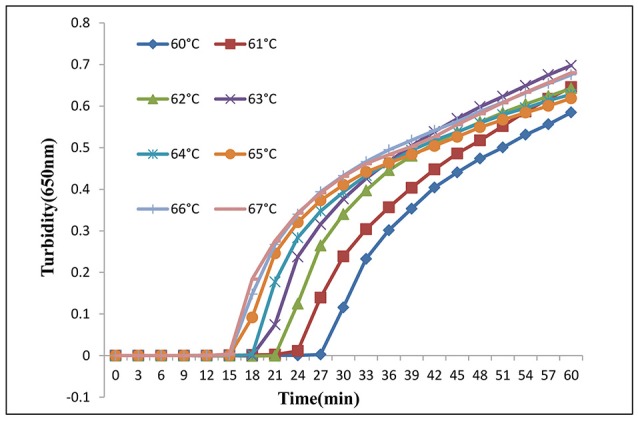
LAMP assay temperature optimization. Different temperatures (60–67°C at 1°C intervals) were tested and 66°C was chosen as the optimal temperature for LAMP amplification.
Specificity of the lamp assay for the mcr-1 gene
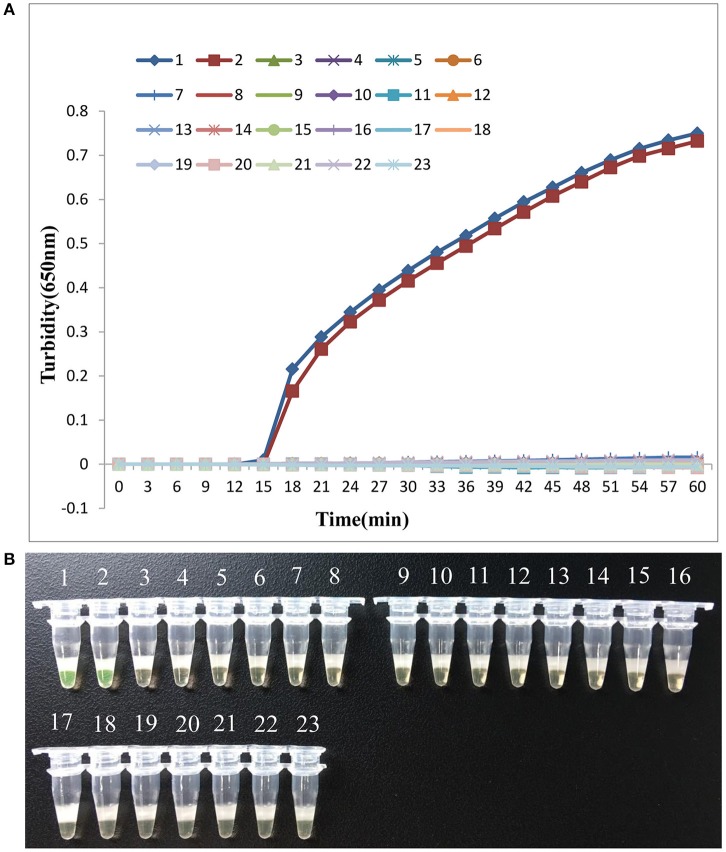
Next, we analyzed the specificity of LAMP detection of the mcr-1 gene. As shown in Figure 3, only when E. coli 12452 and E. coli 12536 were used as the templates, both turbidity and visual detection appeared positive. Importantly, 20 multidrug-resistant bacteria without the mcr-1 gene, including the blank control, tested negative, indicating that the LAMP assay was highly specific for the mcr-1 gene.
Figure 3.
LAMP assay specificity. Specificity of the LAMP method for detecting the mcr-1 gene by real-time turbidimeter (A) or visual detection based on a fluorescent metal indicator (B). Amplification was performed at 66°C for 60 min. 1, E. coli 12452; 2, E. coli 12536; 3, E. coli 12266; 4, E. coli 12278; 5, K. pneumonia 12146; 6, K. pneumonia 12163; 7, Acinetobacter baumannii 12223; 8, A. baumannii 12250; 9, Stenotrophomonas maltophilia 12285; 10, S. maltophilia 12455; 11, Pseudomonas aeruginosa 12343; 12, P. aeruginosa 12352; 13, Enterobacter cloacae 12105; 14, E. cloacae 12211; 15, Proteus mirabilis 12116; 16, P. mirabilis 12407; 17, Serratia marcescens 12208; 18, S. marcescens 12330; 19, Salmonella typhimurium 12306; 20, S. typhimurium 12161; 21, Shigella flexneri 12221; 22, Shigella flexneri 12169; 23, negative control (double-distilled water).
Sensitivity of the lamp assay for the mcr-1 gene
To determine the sensitivity of primers in LAMP detection of the mcr-1 gene, purified genomic DNA was extracted from E. coli 12452 and subjected to serial 10-fold dilutions. PCR was also carried out with the same amount of purified DNA template. As shown in Figure 4, the six reaction curves of real-time turbidity indicated that six serial 10-fold dilutions of the template were detected (20 ng/μL to 0.2 pg/μL), consistent with results of the visual method (positive reactions turned green, whereas negative reactions remained orange). The detection limit of the LAMP assay for the mcr-1 gene was 0.2 pg/μL, which was 10 times more sensitive than conventional PCR.
Figure 4.
LAMP assay sensitivity. Comparative sensitivities of the LAMP assay (A) and (B) and traditional PCR (C) for detection of the mcr-1 gene. 1–7:10-fold serial dilution of genomic DNA extracted from E. coli 12452; 8: negative control (double-distilled water). The expected PCR product size was 309 bp.
Evaluation of the lamp assay in spiked stool samples
Because the mcr-1 gene is often detected in human and cattle feces, LAMP assays were applied to human stools spiked with different amounts of mcr-1-positive bacteria. PCR was also carried out as a control. The results showed that the detection limit of the LAMP assay for human stools was 103 bacteria, which was also 10 times more sensitive than conventional PCR. Thus, LAMP assays can be efficiently applied to detection of the mcr-1 gene in fecal samples.
Detection of the mcr-1 gene in clinical isolates
A LAMP-based surveillance method for detection of the mcr-1 gene in clinical isolates was conducted in a local hospital in Beijing, China. In total, 556 multidrug-resistant Enterobacteriaceae were collected in 2016 from patients with suspected infections; these bacteria had been screened for the mcr-1 gene by PCR using the above primers (Liu et al., 2016).
Subsequently, the results of LAMP assays showed that seven mcr-1-positive isolates, including six E. coli and one Klebsiella pneumoniae, were detected from 556 bacteria. These results were consistent with the results of the PCR assay. Finally, these mcr-1-positive isolates were validated again by PCR-based sequencing. Thus, these findings demonstrated that LAMP assays were well suited for large-scale clinical screening.
Discussion
In Europe and the United States of America, clinical infections caused by multidrug-resistant or pan-resistant Acinetobacter baumannii and Pseudomonas aeruginosa are often treated with topical polymyxins. These drugs are also used to treat severe infections caused by CRE combined with other types of antibiotics (Falagas et al., 2010). However, the use of antibiotics in agriculture and the nonstandard use of antibiotics in the clinic have accelerated the emergence of drug-resistant bacteria. In the second half of 2015, the first plasmid-mediated colistin-resistance gene, mcr-1, was reported in China, attracting the attention of scientists and physicians worldwide (Liu et al., 2016). By the second half of 2016, mcr-1-positive isolates were found in more than 30 countries around the world (Al-Tawfiq et al., 2017). The mcr-1 gene was found in bacteria isolated from various food animals, the environment (river water), various types of meats and vegetables, and infected patients and asymptomatic human carriers, including international travelers (Bernasconi et al., 2016; Hu et al., 2016; Liu et al., 2016; Zurfuh et al., 2016). The mcr-1 gene has been found in Enterobacteriaceae, mostly E. coli and K. pneumonia (Quan et al., 2017). The positive rate of animal mcr-1-positive isolates is much higher than that of clinical isolates (Liu et al., 2016). Therefore, mcr-1-positive strains can spread in hospitals, causing outbreaks and epidemics of nosocomial infections. Thus, CRE carrying the mcr-1 gene will undoubtedly become a major global public health issue (Falgenhauer et al., 2016; Yao et al., 2016).
Detection of drug resistance genes is critical for the prevention of bacterial resistance. Conventional and real-time PCR methods have been established for detection of the mcr-1 gene (Bontron et al., 2016; Liu et al., 2016). However, these methods require expensive equipment. Thus, to complement existing methods, we established a simple and efficient LAMP method. The LAMP assay was found to be highly specific for the mcr-1 gene, and the detection limit of the LAMP assay for the mcr-1 gene was 0.2 pg/μL, which was 10 times more sensitive than that of conventional PCR. In addition, the LAMP assay could be efficiently applied to detection of the mcr-1 gene in fecal samples.
Conclusion
In this study, we established a LAMP detection method for the mcr-1 gene; this method was found to be a rapid, specific, sensitive, and cost-effective tool for the identification of resistance genes. We anticipate that this method may be used routinely in the screening of clinically resistant bacteria. To the best of our knowledge, this is the first detailed report on the application of LAMP for detection of the mcr-1 gene.
Author contributions
LH and WL designed experiments; DZ, SH, and HL carried out experiments; ZY, YS, and XH analyzed experimental results; QZ and YW collected clinical isolates and related information; DZ wrote the manuscript.
Conflict of interest statement
The authors declare that the research was conducted in the absence of any commercial or financial relationships that could be construed as a potential conflict of interest.
Footnotes
Funding. This work received support from the Beijing Natural Science Foundation (grant number 7164289) and the National Natural Science Foundation of China (grant number 81373077).
Supplementary material
The Supplementary Material for this article can be found online at: https://www.frontiersin.org/articles/10.3389/fmicb.2017.02356/full#supplementary-material
References
- Al-Tawfiq J. A., Laxminarayan R., Mendelson M. (2017). How should we respond to the emergence of plasmid-mediated colistin resistance in humans and animals? Int. J. Infect. Dis. 54, 77–84. 10.1016/j.ijid.2016.11.415 [DOI] [PubMed] [Google Scholar]
- Arcilla M. S., Van Hattem J. M., Matamoros S., Melles D. C., Penders J., De Jong M. D., et al. (2016). Dissemination of the mcr-1 colistin resistance gene. Lancet Infect. Dis. 16, 147–149. 10.1016/S1473-3099(15)00541-1 [DOI] [PubMed] [Google Scholar]
- Bernasconi O. J., Kuenzli E., Pires J., Tinguely R., Carattoli A., Hatz C., et al. (2016). Travelers can import colistin-resistant enterobacteriaceae, including those possessing the plasmid-mediated mcr-1 gene. Antimicrob. Agents Chemother. 60, 5080–5084. 10.1128/AAC.00731-16 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bontron S., Poirel L., Nordmann P. (2016). Real-time PCR for detection of plasmid-mediated polymyxin resistance (mcr-1) from cultured bacteria and stools. J. Antimicrob. Chemother. 71, 2318–2320. 10.1093/jac/dkw139 [DOI] [PubMed] [Google Scholar]
- de Lamballerie X., Zandotti C., Vignoli C., Bollet C., de Micco P. (1992). A one-step microbial DNA extraction method using “Chelex 100” suitable for gene amplification. Res. Microbiol. 143, 785–790. 10.1016/0923-2508(92)90107-Y [DOI] [PubMed] [Google Scholar]
- Falagas M. E., Rafailidis P. I., Matthaiou D. K. (2010). Resistance to polymyxins: mechanisms, frequency and treatment options. Drug Resist. Updat. 13, 132–138. 10.1016/j.drup.2010.05.002 [DOI] [PubMed] [Google Scholar]
- Falgenhauer L., Waezsada S. E., Yao Y., Imirzalioglu C., Kasbohrer A., Roesler U., et al. (2016). Colistin resistance gene mcr-1 in extended-spectrum β-lactamase-producing and carbapenemase-producing Gram-negative bacteria in Germany. Lancet Infect. Dis. 16, 282–283. 10.1016/S1473-3099(16)00009-8 [DOI] [PubMed] [Google Scholar]
- Hu Y., Liu F., Lin I. Y., Gao G. F., Zhu B. (2016). Dissemination of the mcr-1 colistin resistance gene. Lancet Infect. Dis. 16, 146–147. 10.1016/S1473-3099(15)00533-2 [DOI] [PubMed] [Google Scholar]
- Li J., Nation R. L., Turnidge J. D., Milne R. W., Coulthard K., Rayner C. R., et al. (2006). Colistin: the re-emerging antibiotic for multidrug-resistant Gram-negative bacterial infections. Lancet Infect. Dis. 6, 589–601. 10.1016/S1473-3099(06)70580-1 [DOI] [PubMed] [Google Scholar]
- Liu W., Zou D., Li Y., Wang X., He X., Wei X., et al. (2012). Sensitive and rapid detection of the New Delhi Metallo-Beta-Lactamase gene by loop-mediated isothermal amplification. J. Clin. Microbiol. 50, 1580–1585. 10.1128/JCM.06647-11 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Liu Y. Y., Wang Y., Walsh T. R., Yi L. X., Zhang R., Spencer J., et al. (2016). Emergence of plasmid-mediated colistin resistance mechanism MCR-1 in animals and human beings in China: a microbiological and molecular biological study. Lancet Infect. Dis. 16, 161–168. 10.1016/S1473-3099(15)00424-7 [DOI] [PubMed] [Google Scholar]
- Mori Y., Nagamine K., Tomita N., Notomi T. (2001). Detection of loop-mediated isothermal amplification reaction by turbidity derived from magnesium pyrophosphate formation. Biochem. Biophys. Res. Commun. 289, 150–154. 10.1006/bbrc.2001.5921 [DOI] [PubMed] [Google Scholar]
- Mulvey M. R., Mataseje L. F., Robertson J., Nash J., Boerlin P., Toye B., et al. (2016). Dissemination of the mcr-1 colistin resistance gene. Lancet Infect. Dis. 16, 147–149. 10.1016/S1473-3099(16)00067-0 [DOI] [PubMed] [Google Scholar]
- Notomi T., Okayama H., Masubuchi H., Yonekawa T., Watanabe K., Amino N., et al. (2000). Loop-mediated isothermal amplification of DNA. Nucleic Acids Res. 28:E63. 10.1093/nar/28.12.e63 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Parida M., Sannarangaiah S., Dash P. K., Rao P. V., Morita K. (2008). Loop mediated isothermal amplification (LAMP): a new generation of innovative gene amplification technique; perspectives in clinical diagnosis of infectious diseases. Rev. Med. Virol. 18, 407–421. 10.1002/rmv.593 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Quan J., Li X., Chen Y., Jiang Y., Zhou Z., Zhang H., et al. (2017). Prevalence of mcr-1 in Escherichia coli and Klebsiella pneumoniae recovered from bloodstream infections in China: a multicentre longitudinal study. Lancet Infect. Dis. 17, 400–410. 10.1016/S1473-3099(16)30528-X [DOI] [PubMed] [Google Scholar]
- Pham Thanh D., Thanh Tuyen H., Nguyen Thi Nguyen T., Chung The H., Wick R. R., Thwaites G. E., et al. (2016). Inducible colistin resistance via a disrupted plasmid-borne mcr-1 gene in a 2008 Vietnamese Shigella sonnei isolate. J. Antimicrob. Chemother. 71, 2314–2317. 10.1093/jac/dkw173 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ushikubo H. (2004). Principle of LAMP method–a simple and rapid gene amplification method. Uirusu 54, 107–112. 10.2222/jsv.54.107 [DOI] [PubMed] [Google Scholar]
- Webb H. E., Granier S. A., Marault M., Millemann Y., Den Bakker H. C., Nightingale K. K., et al. (2016). Dissemination of the mcr-1 colistin resistance gene. Lancet Infect. Dis. 16, 144–145. 10.1016/S1473-3099(15)00538-1 [DOI] [PubMed] [Google Scholar]
- Yao X., Doi Y., Zeng L., Lv L., Liu J. H. (2016). Carbapenem-resistant and colistin-resistant Escherichia coli co-producing NDM-9 and MCR-1. Lancet Infect. Dis. 16, 288–289. 10.1016/S1473-3099(16)00057-8 [DOI] [PubMed] [Google Scholar]
- Zurfuh K., Poirel L., Nordmann P., Nuesch-Inderbinen M., Hachler H., Stephan R. (2016). Occurrence of the plasmid-borne mcr-1 colistin resistance gene in extended-spectrum-β-lactamase-producing Enterobacteriaceae in river water and imported vegetable samples in Switzerland. Antimicrob. Agents Chemother. 60, 2594–2595. 10.1128/AAC.00066-16 [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.