Abstract
Platelet endothelial cell adhesion molecule 1 (PECAM-1) is a cell adhesion protein involved in the regulation of cell adhesion and migration. Interestingly, several PECAM-1–deficient hematopoietic cells exhibit impaired chemotactic responses to stromal cell-derived factor 1 (SDF-1), a chemokine essential for B lymphopoiesis and bone marrow myelopoiesis. However, whether PECAM-1 is involved in SDF-1–regulated chemotaxis is unknown. We report here that SDF-1 induces tyrosine phosphorylation of PECAM-1 at its immunoreceptor tyrosine-based inhibition motifs in several hematopoietic cell lines via the Src family kinase Lyn, Bruton's tyrosine kinase, and JAK2 and that inhibition of these kinases reduced chemotaxis. Overexpression and knockdown of PECAM-1 enhanced and down-regulated, respectively, SDF-1–induced Gαi-dependent activation of the PI3K/Akt/mTORC1 pathway and small GTPase Rap1 in hematopoietic 32Dcl3 cells, and these changes in activation correlated with chemotaxis. Furthermore, pharmacological or genetic inhibition of the PI3K/Akt/mTORC1 pathway or Rap1, respectively, revealed that these pathways are independently activated and required for SDF-1–induced chemotaxis. When coexpressed in 293T cells, PECAM-1 physically associated with the SDF-1 receptor CXCR4. Moreover, PECAM-1 overexpression and knockdown reduced and enhanced SDF-1–induced endocytosis of CXCR4, respectively. Furthermore, when expressed in 32Dcl3 cells, an endocytosis-defective CXCR4 mutant, CXCR4-S324A/S325A, could activate the PI3K/Akt/mTORC1 pathway as well as Rap1 and induce chemotaxis in a manner similar to PECAM-1 overexpression. These findings suggest that PECAM-1 enhances SDF-1–induced chemotaxis by augmenting and prolonging activation of the PI3K/Akt/mTORC1 pathway and Rap1 and that PECAM-1, at least partly, exerts its activity by inhibiting SDF-1–induced internalization of CXCR4.
Keywords: C-X-C chemokine receptor type 4 (CXCR-4), cell migration, chemokine, myeloid cell, platelet endothelial cell adhesion molecule (PECAM-1), Ras-related protein 1 (Rap1), receptor internalization, signal transduction, tyrosine-protein kinase (tyrosine kinase), SDF-1
Introduction
Stromal cell-derived factor 1 (SDF-1),2 also known as CXCL12, is a chemokine constitutively expressed in the bone marrow stromal cells and has been demonstrated to be indispensable for B lymphopoiesis and bone marrow myelopoiesis by knock-out studies of this chemokine or its seven-transmembrane G-protein-coupled receptor CXCR4 in mice (1–3). In addition to its well established roles in chemotaxis and homing of hematopoietic cells to the bone marrow, SDF-1 may promote hematopoiesis synergistically with other cytokines by mainly enhancing survival of hematopoietic progenitors. CXCR4 mainly couples with the Gαi subunit of the heterotrimeric G protein complex, which dissociates from the receptor to inhibit adenylyl cyclase activity and triggers activation of the PI3K/Akt and MEK/ERK signaling pathways upon stimulation with SDF-1 (2). SDF-1 also activates the Rho family small GTPases, such as Rho, Rac, and Cdc42, and the Ras family small GTPases, such as Ras and Rap1 (4–8). It has also been reported that several tyrosine kinases, such as BTK, the Src family tyrosine kinase Lyn, and the JAK family tyrosine kinase JAK2, are coupled with CXCR4 and activated upon SDF-1 stimulation to mediate activation of intracellular signaling (2, 9–13). However, how these Gαi-dependent as well as presumably Gαi-independent signaling events are interconnected and collaboratively lead to SDF-1–induced migration of cells has remained to be clarified and may depend on cellular contexts. Upon ligand binding, CXCR4 is rapidly internalized by endocytosis, which is regulated by phosphorylation of serine residues in the C-terminal cytoplasmic tail and attenuates activation of intracellular signaling. Germinal mutations that truncate or alter the C-terminal region containing these serine residues cause a prolonged activation of CXCR4, leading to retention of myeloid cells to the bone marrow in warts, hypogammaglobulinemia, infection, and myelokathexis (WHIM) syndrome (2, 3). Recently, similar somatic mutations have been found in about 30% of patients with Waldenström's macroglobulinemia (14) and implicated in progression and therapy resistance of this low-grade lymphoma.
Platelet endothelial cell adhesion molecule 1 (PECAM-1) or CD31 is a 130-kDa glycoprotein member of the immunoglobulin superfamily of type I transmembrane cell adhesion molecules and is expressed on endothelial cells and various hematopoietic cells (15, 16). The cytoplasmic domain of PECAM-1 contains two immunoreceptor tyrosine-based inhibition motifs (ITIMs) that encompass Tyr663 and Tyr686, which become tyrosine-phosphorylated mainly by the Src family tyrosine kinases in response to various stimuli to recruit several SH2-domain containing signaling molecules, such as the protein-tyrosine phosphatase SHP2 and the Src family tyrosine kinases. Through binding with these and various other signaling molecules, PECAM-1 plays important roles in modulation of intracellular signaling mechanisms, including activation of the PI3K/Akt pathway and Rap1 (17–19), to regulate a variety of cellular events, such as cell adhesion, through integrin activation, chemotaxis, and apoptosis (15, 16). We have previously found that PECAM-1 is tyrosine-phosphorylated by the leukemogenic BCR/ABL fusion tyrosine kinase in hematopoietic cells, including primary Ph-positive acute lymphoblastic leukemia cells and transformed chronic myeloid leukemia cells, and implicated its roles in regulation of cell adhesion and apoptosis in response to the tyrosine kinase inhibitor imatinib (20). However, although PECAM-1 has been reported to be expressed also on various other types of leukemias, such as acute myeloid leukemia (AML) (21) and chronic lymphoid leukemia (22), its clinical relevance has remained controversial. The hematopoietic cytokine IL-3 has been shown to induce tyrosine phosphorylation of PECAM-1 in hematopoietic cells (23). Nevertheless, its significance in the signal transduction mechanisms by which IL-3 regulates proliferation and apoptosis of cells has remained unknown. Intriguingly, PECAM-1–deficient murine hematopoietic cells, including progenitor cells, megakaryocytes, and T-cells, showed an impairment in chemotactic response to SDF-1 (24, 25). However, whether and how PECAM-1 is involved in SDF-1/CXCR4–mediated signaling mechanisms regulating chemotaxis have yet to be elucidated.
In the present study, we show that PECAM-1 becomes tyrosine-phosphorylated on its ITIMs upon SDF-1 stimulation in various hematopoietic and leukemic cell lines. Furthermore, overexpression or knockdown of PECAM-1 in hematopoietic 32Dcl3 cells enhanced or down-regulated, respectively, SDF-1–induced activation of the PI3K/Akt/mTORC1 pathway and Rap1, which was required for and correlated with chemotaxis. Moreover, PECAM-1 was shown to associate physically with CXCR4 when coexpressed in 293T cells and was found to be colocalized with CXCR4 in 32Dcl3 cells. Furthermore, expression levels of PECAM-1 in 32Dcl3 cells negatively correlated with the SDF-1–induced down-regulation of the cell surface expression levels of CXCR4. Finally, an endocytosis-defective CXCR4 mutant, CXCR4-S324A/S325A (CXCR4-SA), mimicked PECAM-1 overexpression with its enhanced activities to activate the PI3K/Akt/mTORC1 pathway as well as Rap1 and to induce chemotaxis. These data suggest that PECAM-1 may enhance CXCR4-mediated chemotactic signaling pathways involving PI3K/Akt/mTORC1 and Rap1, at least partly, by inhibiting internalization of CXCR4.
Results
PECAM-1 becomes tyrosine-phosphorylated on ITIMs by SDF-1 stimulation in hematopoietic cells
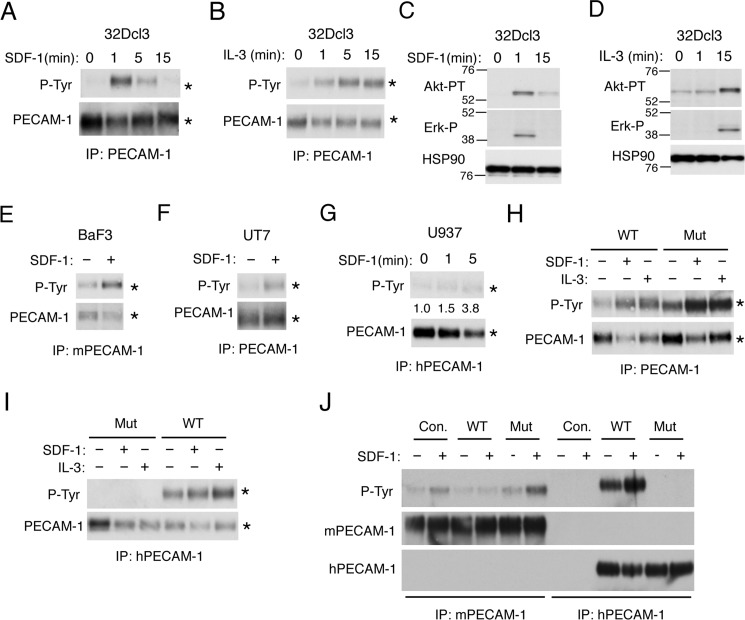
It has been reported that the hematopoietic cytokine IL-3 induces tyrosine phosphorylation of PECAM-1 in hematopoietic cells (23). However, whether chemokine SDF-1 also induces tyrosine phosphorylation of PECAM-1 is unknown. Thus, to address the possible involvement of PECAM-1 in SDF-1/CXCR4 signaling, we first examined whether SDF-1 induces tyrosine phosphorylation of PECAM-1. SDF-1 induced a transient tyrosine phosphorylation of PECAM-1 in murine hematopoietic 32Dcl3 cells, which was observed more rapidly than that induced by IL-3 and correlated with the time courses of activation of intracellular signaling induced by SDF-1 and IL-3 (Fig. 1, A–D). SDF-1–induced tyrosine phosphorylation of PECAM-1 was confirmed also in murine BaF3 cells and human leukemic cell lines UT7 and U937 (Fig. 1, E–G). These data suggest that PECAM-1 may be directly involved in SDF-1/CXCR4 signaling in hematopoietic cells.
Figure 1.
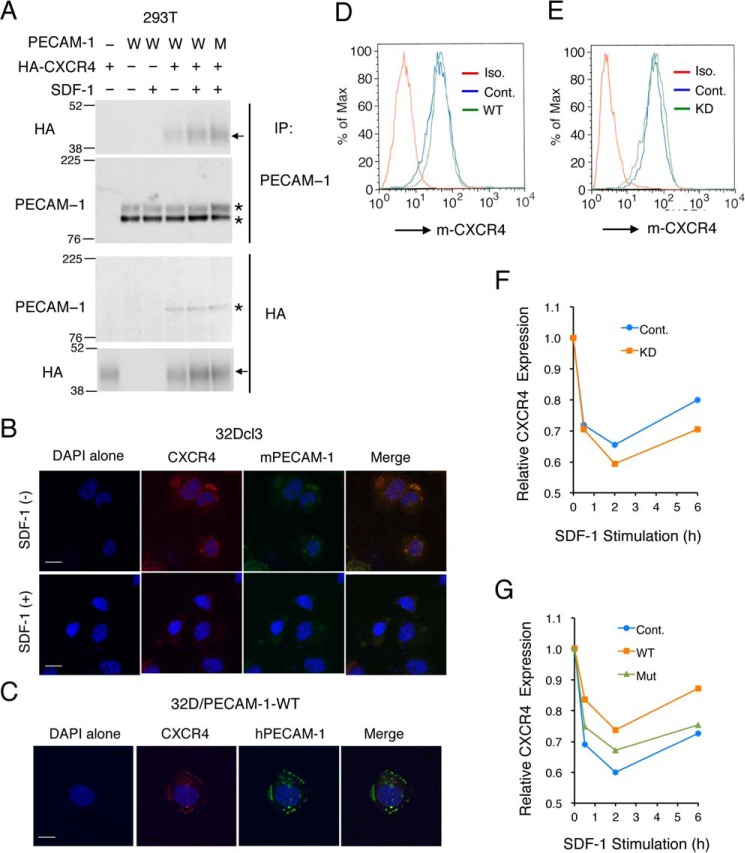
SDF-1 induces tyrosine phosphorylation of PECAM-1 on ITIMs in various hematopoietic cell lines. A and B, 32Dcl3 cells were starved of IL-3 for 4 h and left unstimulated or stimulated with 50 ng/ml SDF-1 or 10 ng/ml IL-3 for the indicated times. Cells were lysed and subjected to immunoprecipitation (IP) using antibody for PECAM-1 followed by immunoblot analysis with anti-phosphotyrosine (P-Tyr) and anti-PECAM-1 as indicated. Positions of PECAM-1 are indicated by asterisks. C and D, 32Dcl3 cells, starved of IL-3 for 12 h, were stimulated with 50 ng/ml SDF-1 or 10 ng/ml IL-3 for the indicated times. Cells were lysed and subjected to immunoblot analysis with antibodies against the indicated proteins. HSP90 was used as a loading control. Akt-PT, phospho-Thr308-Akt; Erk-P, phospho-Thr202/Tyr204-ERK. E, Baf3 cells, starved of IL-3 for 12 h, were left unstimulated or stimulated with 50 ng/ml SDF-1 for 1 min as indicated. Cell lysates were subjected to immunoprecipitation using antibody specific for murine PECAM-1 (mPECAM-1) and analyzed as in A. F, UT7 cells, starved of Epo for 4 h, were left unstimulated or stimulated with 50 ng/ml SDF-1 for 1min as indicated and analyzed. G, U937 cells were stimulated with 50 ng/ml SDF-1 for the indicated times and subjected to immunoprecipitation using antibody specific for human PECAM-1 (hPECAM-1) for immunoblot analysis. Relative levels of tyrosine-phosphorylated PECAM-1 were determined by densitometric analysis and are shown below the panel. H and I, 32Dcl3 cells overexpressing human PECAM-1-WT (WT) or human PECAM-1-ITIM(−) mutant (Mut) were starved of IL-3 for 4 h. Cells were stimulated with 50 ng/ml SDF-1 for 1 min, 10 ng/ml IL-3 for 5 min, or left unstimulated as indicated. Lysates were immunoprecipitated using antibody that reacts with both human and murine PECAM-1 (PECAM-1; SC-1506) or specific for human PECAM-1 (hPECAM-1; CS-3528) as indicated and analyzed using the antibodies indicated. J, 32Dcl3 cells overexpressing human PECAM-1-WT (WT) or human PECAM-1-ITIM(−) mutant (Mut) as well as vector control cells (Con.) were starved of IL-3 for 4 h. Cells were stimulated with 50 ng/ml SDF-1 for 1 min or left unstimulated as indicated. Lysates were immunoprecipitated using antibody that reacts specifically with murine PECAM-1 (mPECAM-1; SC-18916) or human PECAM-1 (hPECAM-1; CS-3528) as indicated and analyzed using the antibodies indicated. All the data shown are representative of experiments repeated at least three times with the exception of those shown in F, G, I, and J, which are representative of two repeated experiments.
To determine the sites of tyrosine phosphorylation of PECAM-1 induced by SDF-1, we overexpressed human wild-type PECAM-1 (PECAM-1-WT) or the mutant PECAM-1 (PECAM-1-Mut) with Y663F and Y686F mutations introduced into both ITIMs (26, 27) in 32Dcl3 cells. When immunoprecipitated with an antibody reactive with both murine and human PECAM-1, PECAM-1 was found to be constitutively tyrosine-phosphorylated in cells overexpressing PECAM-1-Mut with increases in phosphorylation observed after stimulation with SDF-1 or IL-3 (Fig. 1H). However, when human PECAM-1-WT or PECAM-1-Mut was selectively immunoprecipitated with an antibody specific for human PECAM-1, it was revealed that PECAM-1-Mut was not tyrosine-phosphorylated with or without stimulation (Fig. 1I). In contrast, human PECAM-1-WT overexpressed in 32Dcl3 cells was rather heavily tyrosine-phosphorylated before stimulation with a modest increase in phosphorylation after stimulation with SDF-1 or IL-3. We next examined SDF-1–induced tyrosine phosphorylation of endogenous PECAM-1 in cells overexpressing human PECAM-1-WT or PECAM-1-Mut by immunoprecipitating murine PECAM-1 selectively using a species-specific antibody. Intriguingly, SDF-1–induced tyrosine phosphorylation was enhanced or diminished in cells overexpressing PECAM-1-Mut or PECAM-1-WT, respectively, in repeated experiments (Fig. 1J and data not shown). These data indicate that SDF-1 as well as IL-3 induces tyrosine phosphorylation of PECAM-1 exclusively in the ITIMs and that overexpression of PECAM-1-Mut enhances SDF-1–induced tyrosine phosphorylation on these motifs in endogenous PECAM-1.
Lyn, BTK, and JAK2 may be involved in tyrosine phosphorylation of the ITIMs in PECAM-1 and in migration of hematopoietic cells induced by SDF-1
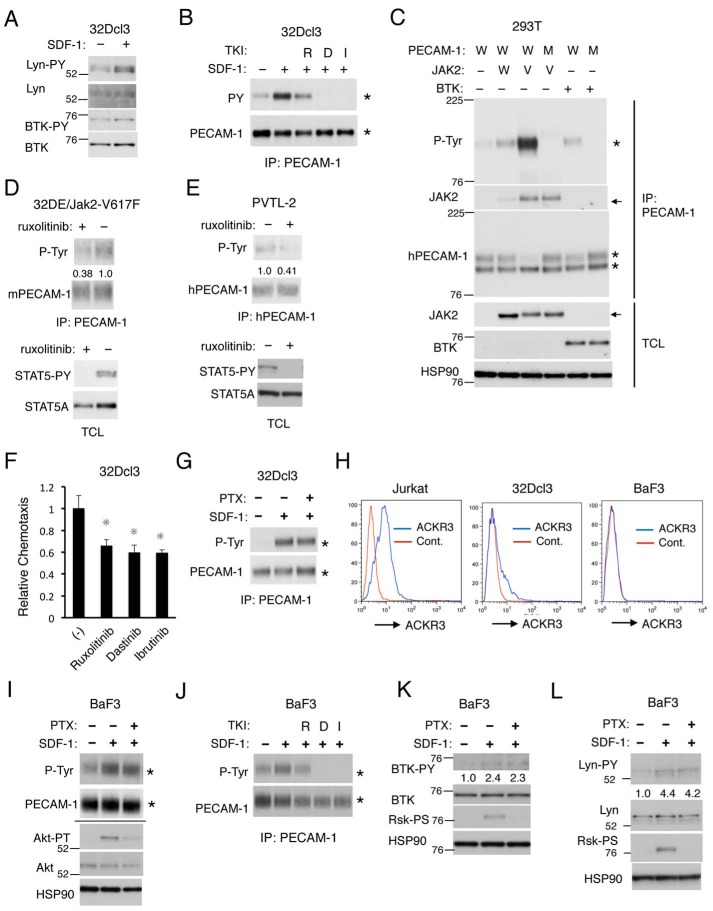
We next examined the tyrosine kinases involved in phosphorylation of PECAM-1 on the ITIMs upon stimulation by SDF-1 in hematopoietic cells. In this regard, previous studies have shown that the Src family tyrosine kinases, including Lyn, as well as BTK and JAK2 are involved in signaling from the SDF-1 receptor CXCR4 (2, 9–13). As shown in Fig. 2A, we confirmed that SDF-1 induced activation-specific tyrosine phosphorylation of Lyn and BTK in 32Dcl3 cells. Previous studies have repeatedly demonstrated that SDF-1 induces activation of JAK2 in hematopoietic cells (2, 9, 10, 13), although we failed to demonstrate its activation by SDF-1 stimulation using an activation-specific antibody, possibly because of its low sensitivity of detection. Thus, we examined the possible involvement of these kinases using their specific inhibitors. As shown in Fig. 2B, the Src family kinase inhibitor dasatinib or the BTK inhibitor ibrutinib abolished SDF-1–induced as well as basal tyrosine phosphorylation of PECAM-1 in 32Dcl3 cells. In contrast, the JAK family kinase inhibitor ruxolitinib partly inhibited SDF-1–induced tyrosine phosphorylation of PECAM-1 in these cells.
Figure 2.
SDF-1 induces tyrosine phosphorylation of PECAM-1 through Lyn, BTK, and JAK2 in a Gαi-independent manner. A, 32Dcl3 cells, starved of IL-3 for 4 h, were stimulated with 50 ng/ml SDF-1 for 1min or left unstimulated as indicated and lysed in radioimmune precipitation assay buffer. Lysates were subjected to immunoblot analysis using antibodies against the indicated proteins. Lyn-PY, phospho-Tyr396-Lyn; BTK-PY, phospho-Tyr223-BTK. B, 32Dcl3 cells, starved of IL-3 for 4 h, were left untreated or treated with 2 μm ruxolitinib (R), 20 nm dasatinib (D), or 5 μm ibrutinib (I) as indicated for 30 min. Cells were then stimulated for 1 min with 50 ng/ml SDF-1 or left unstimulated as indicated and analyzed. Positions of PECAM-1 are indicated by asterisks. C, 293T cells were transfected with plasmids coding for JAK2 wild type (W), JAK2-V617F (V), and BTK as well as PECAM-1-WT (W) or PECAM-1-Mut (M) as indicated. Anti-PECAM-1 immunoprecipitates and total cell lysates were subjected to immunoblot analysis using the indicated antibodies. HSP90 was used as a loading control. Arrows indicate positions of JAK2. Positions of molecular weight markers are also indicated. D, 32DE/JAK2-V617F cells were cultured for 6 h with or without 3 μm ruxolitinib. Cells were lysed and subjected to immunoprecipitation (IP) using antibody for PECAM-1 followed by immunoblotting with the indicated antibodies (upper panel). Total cell lysates were also subjected to immunoblot analysis with antibodies against the indicated proteins (lower panel). STAT5-PY, anti-phospho-Tyr694-STAT5. E, PVTL-2 cells were cultured with or without 2 μm ruxolitinib for 6 h (upper panel) or 1.5 μm ruxolitinib for 16 h (lower panel) and analyzed. F, 32Dcl3 cells were starved of IL-3 and cultured for 4 h in migration medium. Cells were then treated for 1 h with 1 μm ruxolitinib (R), 50 nm dasatinib (D), or 2 μm ibrutinib (I) or left untreated (−) as a control and subjected to chemotaxis assay with 100 ng/ml SDF-1. Relative chemotaxis was calculated by dividing the percentage of migrated cells pretreated with inhibitors by that of control cells. The data represent the mean of triplicates, and the asterisks indicate statistically significant (p < 0.05) differences compared with control by Student's t test. Error bars represent S.D. G, 32Dcl3 cells, starved of IL-3 for 4 h, were treated with 100 ng/ml PTX for 90 min or left untreated as indicated. Cells were then stimulated with 50 ng/ml SDF-1 for 1 min or left unstimulated as indicated and analyzed by immunoprecipitation with antibody for PECAM-1 (PECAM-1) and immunoblotting using the indicated antibodies. H, Jurkat, 32Dcl3, or BaF3 cells as indicated were starved in serum-free medium, and surface expression levels of ACKR3 were analyzed by flow cytometry. I, BaF3 cells, starved of IL-3 for 14 h, were treated with 100 ng/ml PTX for 60 min or left untreated as indicated. Cells were then stimulated with 50 ng/ml SDF-1 for 1 min or left unstimulated as indicated and analyzed. Anti-PECAM-1 immunoprecipitates and total cell lysates, shown above or below a horizontal line, respectively, were subjected to immunoblot analysis using the indicated antibodies. Akt-PT, phospho-Thr308-Akt. J, BaF3 cells, starved of IL-3 for 14 h, were left untreated or treated with 2 μm ruxolitinib (R), 20 nm dasatinib (D), or 5 μm ibrutinib (I) as indicated for 60 min. Cells were then stimulated for 1 min with 50 ng/ml SDF-1 or left unstimulated as indicated and analyzed. K, BaF3 cells, starved of IL-3 for 14 h, were treated with 100 ng/ml PTX for 60 min or left untreated as indicated. Cells were then stimulated with 50 ng/ml SDF-1 for 1 min or left unstimulated as indicated and analyzed. Relative levels of phosphorylation of BTK on Tyr223 were determined by densitometric analysis and are shown below the panel. Rsk-PS, phospho-Ser380-p90Rsk. L, BaF3 cells, starved of IL-3 for 3 h, were treated with 100 ng/ml PTX for 90 min or left untreated as indicated. Cells were then stimulated with 50 ng/ml SDF-1 for 1 min or left unstimulated as indicated and lysed in 1× Laemmli sample buffer to be analyzed. Relative levels of phosphorylation of Lyn on Tyr396 were determined by densitometric analysis and are shown below the panel. All the data shown are representative of experiments repeated at least three times with the exception of those shown in B, D, G, J, and K, which are representative of two repeated experiments. Cont., control; p-Tyr, phosphotyrosine; PY, phosphotyrosine; TKI, tyrosine kinase inhibitor; TCL, total cell lysate.
Previous studies have shown that Lyn and BTK phosphorylate PECAM-1 on the ITIMs (15, 28). We have also previously shown that Lyn phosphorylates PECAM-1-WT but not PECAM-1-Mut when coexpressed transiently in 293T cells (20). Thus, to confirm that BTK phosphorylates PECAM-1 in the ITIMs and to examine whether JAK2 similarly phosphorylates PECAM-1, we overexpressed BTK or JAK2 along with PECAM-1 in 293T cells. As expected, BTK induced tyrosine phosphorylation of PECAM-1-WT but not PECAM-1-Mut (Fig. 2C). JAK2 similarly induced tyrosine phosphorylation of PECAM-1-WT but not its mutant (Fig. 2C and data not shown). Interestingly, the constitutively activated JAK2-V617F mutant, implicated in pathogenesis of myeloproliferative neoplasms (29), physically associated with PECAM-1 much more strongly than wild-type JAK2 and induced a robust tyrosine phosphorylation of PECAM-1-WT but not PECAM-1-Mut. In accordance with this, PECAM-1 was constitutively tyrosine-phosphorylated along with a well established substrate of JAK2, STAT5, in 32D/EpoR cells expressing JAK2-V617F or in human leukemic PVTL-2 cells expressing this mutant, and this was inhibited by ruxolitinib (Fig. 2, D and E). Together, these results suggest that JAK2 as well as Lyn and BTK may be involved in tyrosine phosphorylation of PECAM-1 in the ITIMs in hematopoietic cells stimulated with SDF-1.
To explore the roles of these kinases as well as the possible involvement of PECAM-1 phosphorylation in SDF-1–induced chemotactic signaling, we next examined the effects of ruxolitinib, dasatinib, and ibrutinib on migration of 32Dcl3 cells induced by SDF-1. Under the conditions where numbers of migrated cells were negligible in the absence of SDF-1 (data not shown), pretreatment of 32Dcl3 cells with these inhibitors significantly reduced numbers of migrated cells induced by SDF-1 (Fig. 2F). These results suggest that JAK2, Lyn, and BTK may be involved in SDF-1/CXCR4 signaling leading to chemotaxis and raise the possibility that PECAM-1 and its phosphorylation by these kinases may also be involved in the signaling mechanisms.
Finally, we examined whether these kinases were activated downstream of Gαi to induce tyrosine phosphorylation of PECAM-1. As shown in Fig. 2G, SDF-1–induced tyrosine phosphorylation of PECAM-1 was not inhibited by the Gαi-specific inhibitor pertussis toxin (PTX), thus indicating that it was not mediated through Gαi. To explore the possible involvement of atypical chemokine receptor 3 (ACKR3; previously named CXCR7), which binds to SDF-1 and is known to mediate signal in a G protein-independent manner or to modulate CXCR4 signaling (2) in the Gαi-independent induction of PECAM-1 phosphorylation, we examined the expression of ACKR3 in the cells used in the present study. In accordance with a previous report (30), ACKR3 was detected by flow cytometry at high levels in most of the T-cell acute lymphoblastic leukemia Jurkat cells (Fig. 2H). In contrast, ACKR3 could not be detected in BaF3 cells, in agreement with a previous report (31), whereas ACKR3 was detected at low levels in a portion of 32Dcl3 cells. Nevertheless, SDF-1 induced tyrosine phosphorylation of PECAM-1 in a Gαi-independent manner in BaF3 cells, and this was inhibited by the same tyrosine kinase inhibitors as in 32Dcl3 cells (Fig. 2, I and J). Furthermore, activation of BTK and Lyn induced by SDF-1 in BaF3 cells was not inhibited by PTX (Fig. 2, K and L). Together, these results suggest that SDF-1 induces tyrosine phosphorylation of PECAM-1 through CXCR4 but not ACKR3 by activating BTK, Lyn, and probably JAK2 in a Gαi-independent manner.
PECAM-1 may play a role in SDF-1–induced chemotaxis by enhancing activation of the PI3K/Akt/mTORC1 pathway
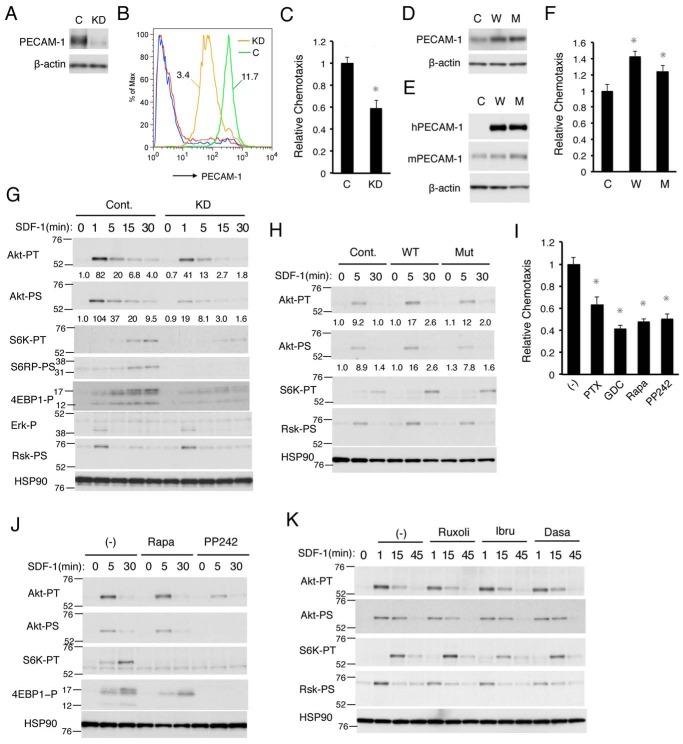
To explore further the possibility that PECAM-1 may play a role in SDF-1/CXCR4 signaling leading to chemotaxis, we next examined the effects of modulation of PECAM-1 expression on SDF-1–induced migration. As shown in Fig. 3, A–C, the expression level of PECAM-1 was remarkably decreased by retrovirus vector-mediated introduction of PECAM-1 shRNA into 32Dcl3 cells, which also reduced SDF-1–induced migration of these cells. In contrast, SDF-1–induced migration of 32Dcl3 cells was significantly enhanced by overexpression of human PECAM-1-WT and, to a lesser extent, by that of PECAM-1-Mut (Fig. 3, D–F). These results strongly support the idea that PECAM-1 may be involved in modulation of SDF-1/CXCR4 signaling leading to chemotaxis.
Figure 3.
PECAM-1 enhances SDF-1–induced chemotaxis mediated through activation of the PI3K/Akt/mTORC1 pathway. A, B, and C, 32Dcl3 cells infected with the retrovirus vector expressing PECAM-1 shRNA (KD) or luciferase shRNA as a control (C) were lysed and subjected to immunoblot analysis using antibodies against the indicated proteins in A, flow cytometric analysis for surface PECAM-1 expression in B, or chemotaxis assay with 100 ng/ml SDF-1 in C. β-Actin was used as a loading control. MFRs of PECAM-1 signals for vector control (green) and PECAM-1-knocked down cells (orange) are indicated, and red and blue lines represent negative controls for control and knockdown cells, respectively, in B. Relative chemotaxis was calculated by dividing the percentage of migrated cells by that of vector control cells. The data represent the mean of triplicates ±S.D., and the asterisks indicate statistically significant (p < 0.05) differences compared with control. D, E, and F, 32Dcl3 cells overexpressing human PECAM-1-WT (W) or human PECAM-1-Mut (M) as well as vector control cells (C) were lysed and analyzed by immunoblot analysis with antibodies against the indicated proteins in D and E or subjected to chemotaxis assay with 100 ng/ml SDF-1 in F. Error bars represent S.D. hPECAM-1, human PECAM-1; mPECAM-1, murine PECAM-1. G, 32Dcl3 cells expressing PECAM-1 shRNA (KD) or luciferase shRNA as control (Cont.) were starved of IL-3 for 12 h and stimulated with 5 ng/ml SDF-1 for the indicated times. Cell lysates were subjected to immunoblot analysis using antibodies against the indicated antibodies. Relative levels of phosphorylation were determined by densitometric analysis and are shown below panels. Positions of molecular weight markers are also indicated. Akt-PT, phospho-Thr308-Akt; Akt-PS, phospho-Ser473-Akt; S6K-PT, phospho-Thr389-p70S6K; S6RP-PS, phospho-Ser240/244-S6RP; 4EBP1-P, phospho-Thr37/46-4EBP1; Erk-P, phospho-Thr202/Tyr204-ERK; Rsk-PS, phospho-Ser380-p90Rsk. H, 32Dcl3 cells overexpressing human PECAM-1-WT (WT) or PECAM-1-Mut (Mut) as well as vector control cells (Cont.), starved of IL-3 for 4 h, were stimulated with 5 ng/ml SDF-1 for the indicated times and analyzed. I, 32Dcl3 cells were starved of IL-3 and cultured for 4 h in migration medium. Cells were then treated for 30 min with 100 ng/ml PTX, 1 μm GDC-0941 (GDC), 20 nm rapamycin (Rapa), or 1 μm PP242 or left untreated (−) as a control and subjected to chemotaxis assay with 100 ng/ml SDF-1. Relative chemotaxis was calculated by dividing the percentage of migrated cells for cells pretreated with inhibitors by that of control cells. Error bars represent S.D. J, 32Dcl3 cells were starved of IL-3 for 4 h and treated with 20 nm rapamycin (Rapa) or 1 μm PP242 for 30 min or left untreated as a control (−). Cells were then stimulated with 5 ng/ml SDF-1 for the indicated times and analyzed. K, 32Dcl3 cells were starved of IL-3 for 4 h and treated with 1 μm ruxolitinib (Ruxoli), 2 μm ibrutinib (Ibru), or 50 nm dasatinib (Dasa) for 1 h or left untreated as a control (−). Cells were then stimulated with 1 ng/ml SDF-1 for the indicated times and analyzed. All the data shown are representative of experiments repeated at least three times.
To explore which signaling pathways activated downstream of CXCR4 are involved in enhancement of chemotaxis by PECAM-1, we first examined the effects of PECAM-1 knockdown on activation of the PI3K/Akt/mTOR and MEK/ERK signaling pathways, which have been implicated in chemotactic signaling from CXCR4 (2, 3). As shown in Fig. 3G, SDF-1–induced activation-specific phosphorylation of Akt on Thr308 as well as Ser473 was slightly decreased and less sustained in 32Dcl3 cells when PECAM-1 was knocked down. In accordance with this, knockdown of PECAM-1 significantly inhibited SDF-1–induced activation of mTORC1 as judged by specific phosphorylation of its substrates (p70S6K and 4EBP-1) as well as its downstream signaling molecule (S6RP). In contrast, activation of the MEK/ERK pathway, as judged by activation-specific phosphorylation of ERK and its downstream signaling element p90Rsk, was not apparently affected by knockdown of PECAM-1. Consistent with these results, overexpression of PECAM-1-WT as well as PECAM-1-Mut enhanced SDF-1–induced activation of Akt and p70S6K (Fig. 3H). These data indicate that PECAM-1 may play a role in enhancement of the PI3K/Akt/mTORC1 pathway in these cells.
To examine whether the PI3K/Akt/mTORC1 pathway is involved in SDF-1–induced chemotaxis in 32Dcl3 cells, we next examined the effects of inhibitors for this pathway on chemotaxis. As shown in Fig. 3I, SDF-1–induced chemotaxis was inhibited by treatment with GDC-0941 (PI3K class IA inhibitor), rapamycin (mTORC1 inhibitor), and PP242 (mTORC1 and mTORC2 inhibitor) as well as PTX (Gαi inhibitor) in 32Dcl3 cells. We also confirmed that, under these conditions, rapamycin abolished SDF-1–induced activation of p70S6K without affecting activation of Akt and only slightly reducing the phosphorylation of 4EBP-1 in 32Dcl3 cells and that PP242 abolished phosphorylation of p70S6K and 4EBP-1 by mTORC1 as well as that of Akt on Ser473 by mTORC2 (Fig. 3J). Thus, rapamycin inhibited SDF-1–induced activation of mTORC1 and chemotaxis without affecting mTORC2 activation. Conversely, none of the inhibitors for tyrosine kinases involved in PECAM-1 phosphorylation significantly inhibited SDF-1–induced activation of the PI3K/Akt/mTORC1 pathway (Fig. 3K). Taken together, these results suggest that PECAM-1 may play a role in SDF-1–induced chemotaxis by enhancing activation of the PI3K/Akt/mTORC1 pathway independently of activation of the tyrosine kinases.
Rap1 but not Rac1 may mediate enhancement of SDF-1–induced chemotaxis by PECAM-1 independently of the PI3K/Akt pathway
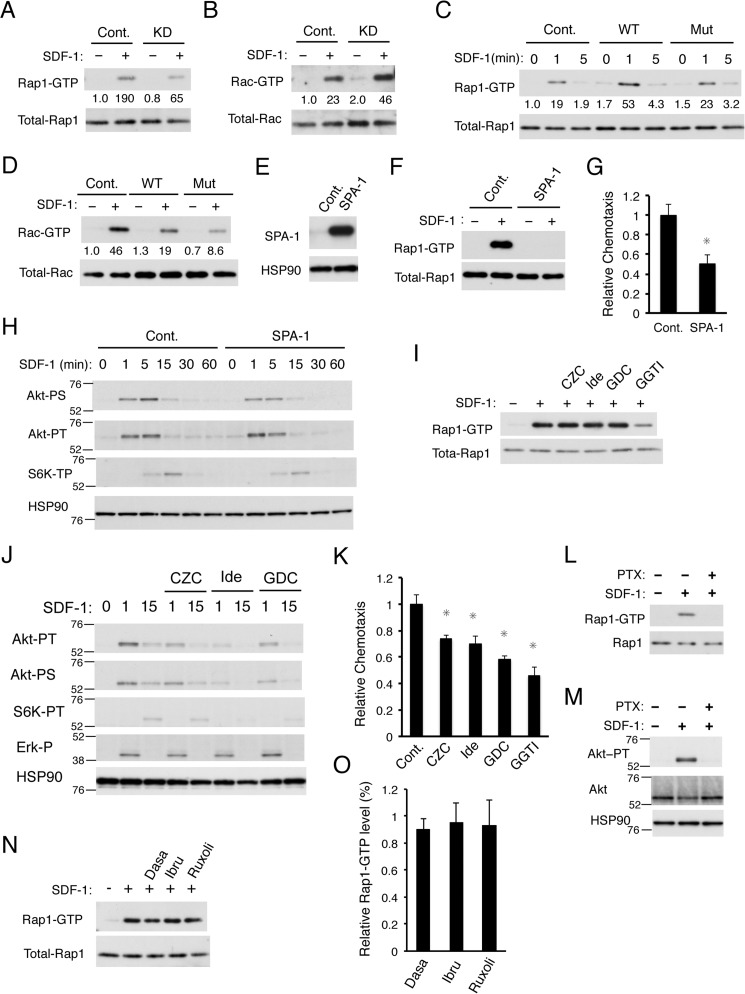
We have previously demonstrated that SDF-1 activates the Ras and Rho family GTPases Rap1 and Rac, respectively, which play important roles in regulation of chemotaxis, in 32Dcl3 cells (6, 7). Thus, to gain further insight into the mechanisms by which PECAM-1 enhances SDF-1–induced chemotaxis, we analyzed possible effects of PECAM-1 on SDF-1–induced activation of Rap1 and Rac. The affinity purification assays using specific probes revealed that knockdown of PECAM-1 reduced SDF-1–induced Rap1 activation in 32Dcl3 cells but enhanced the Rac activation (Fig. 4, A and B). Consistent with this, overexpression of PECAM-1-WT as well as PECAM-1-Mut enhanced or reduced SDF-1–induced activation of Rap1 or Rac, respectively (Fig. 4, C and D). Thus, the level of Rap1 activation induced by SDF-1 correlated with the expression level of PECAM-1 as well as SDF-1–induced chemotaxis, which suggests that Rap1 may be involved in enhancement of chemotactic response to SDF-1 by PECAM-1. In contrast, the level of SDF-1–induced Rac activation inversely correlated with that of PECAM-1 expression as well as chemotactic response.
Figure 4.
PECAM-1 enhances SDF-1–induced chemotaxis also through activation of Rap1 independently of the PI3K/Akt pathway. A and B, 32Dcl3 cells expressing PECAM-1 shRNA (KD) or luciferase shRNA as a control (Cont.) were starved of IL-3 for 4 h and stimulated for 1 min with 10 ng/ml SDF-1 or left unstimulated as indicated. Cells were lysed, and Rap1-GTP or Rac-GTP was affinity-purified from cell lysates with a GST-RalGDS-RBD or GST-PAK2-RBD fusion protein. Eluates from precipitates (upper panels) or total cell lysates (lower panels) were subsequently subjected to immunoblot analysis to estimate the expression levels of activated or total amount of these small GTPases, respectively, using antibodies against Rap1 and Rac. Relative levels of Rap1-GTP and Rac-GTP were determined by densitometric analysis and are shown below the panels. C and D, 32Dcl3 cells overexpressing human PECAM-1-WT (WT) or human PECAM-1-Mut (Mut) as well as vector control cells (Cont.) were starved of IL-3 for 4 h, stimulated with 10 ng/ml SDF-1 for the indicated times, and analyzed as described for A and B. E, 32Dcl3 cells overexpressing SPA-1 and vector control cells (Cont.) were lysed and subjected to immunoblot analysis with antibodies against the indicated proteins. F, 32Dcl3 cells overexpressing SPA-1 and vector-control cells (Cont.) were starved of IL-3 for 4 h. Cells were then simulated with 10 ng/ml SDF-1 for 1 min or left unstimulated as indicated and subjected to Rap1 activation assay. G, 32Dcl3 cells overexpressing SPA-1 and vector control cells (Cont.) were subjected to chemotaxis assay with 50 ng/ml SDF-1. The data represent the mean of triplicates, and the asterisks indicate statistically significant (p < 0.05) differences compared with control. Error bars represent S.D. H, 32Dcl3 cells overexpressing SPA-1 and vector control cells (Cont.), starved of IL-3 for 6 h, were stimulated with 1 ng/ml SDF-1 for the indicated times and subjected to immunoblot analysis using antibodies against the indicated proteins. Positions of molecular weight markers are also indicated. Akt-PT, phospho-Thr308-Akt; Akt-PS, phospho-Ser473-Akt; S6K-PT, phospho-Thr389-p70S6K. I, 32Dcl3 cells, starved of IL-3 for 4 h, were treated for 90 min with 3 μm CZC24832 (CZC), 3 μm idelalisib (Ide), 3 μm GDC-0941 (GDC), or 10 μm GGTI-298 (GGTI) as indicated or left untreated as a control. Cells were stimulated with 10 ng/ml SDF-1 for 1 min or left unstimulated as indicated and analyzed for Rap1 activation. J, 32Dcl3 cells were starved of IL-3 for 12 h and treated with 3 μm CZC24832 (CZC), 3 μm idelalisib (Ide), or 3 μm GDC-0941 (GDC) as indicated for 1 h or left untreated. Cells were then stimulated with 5 ng/ml SDF-1 for the indicated times and subjected to immunoblot analysis with antibodies against the indicated proteins. Akt-PT, phospho-Thr308-Akt; Akt-PS, phospho-Ser473-Akt; S6K-PT, phospho-Thr389-p70S6K; Erk-P, phospho-Thr202/Tyr204-ERK. K, 32Dcl3 cells were starved of IL-3 and cultured for 4 h in migration medium. Cells were then treated for 1 h with 3 μm CZC24832 (CZC), 3 μm idelalisib (Ide), 3 μm GDC-0941 (GDC), or 10 μm GGTI-298 (GGTI) or left untreated (Cont.) as a control and subjected to chemotaxis assay with 100 ng/ml SDF-1. Relative chemotaxis was calculated by dividing the percentage of migrated cells for cells pretreated with inhibitors by that of control cells. Error bars represent S.D. L and M, 32Dcl3 cells, starved of IL-3 for 4 h, were treated with 100 ng/ml PTX for 90 min or left untreated as indicated. Cells were then stimulated with 50 ng/ml SDF-1 for 1 min or left unstimulated as indicated and analyzed for activation of Rap1 in L and Akt in M. N and O, 32Dcl3 cells, starved of IL-3 for 4 h, were treated for 90 min with 50 nm dasatinib (Dasa), 2 μm ibrutinib (Ibru), or 1 μm ruxolitinib (Ruxoli) as indicated or left untreated. Cells were then stimulated with 10 ng/ml SDF-1 for 1 min or left unstimulated as indicated and analyzed for Rap1 activation. A representative result from three independent experiments is shown in N. The relative Rap1-GTP level was calculated by dividing the Rap1-GTP level of cells pretreated with the inhibitor with that of control cells, and the means from three independent experiments are plotted in O. Error bars represent S.D. All the data shown are representative of experiments repeated at least three times.
To confirm the role of Rap1 activation in SDF-1–induced chemotactic response and to examine the possible interaction between the activation mechanisms of Rap1 and the PI3K/Akt/mTORC1 pathway, we overexpressed SPA-1, a specific GTPase-activating protein for Rap1, to inhibit Rap1 activation in 32Dcl3 cells. As shown in Fig. 4, E and F, SPA-1 overexpression abolished SDF-1–induced Rap1 activation as expected. SPA-1 overexpression also reduced SDF-1–induced cell migration in 32Dcl3 cells (Fig. 4G). However, SPA-1 overexpression inhibited SDF-1–induced activation of neither Akt nor p70S6K significantly (Fig. 4H). However, none of the various PI3K inhibitors, including CZC-24832 (PI3Kγ inhibitor), idelalisib (PI3Kδ inhibitor), and GDC-0941 (PI3K class IA inhibitor) but only the Rap1 inhibitor GGTI-298 showed an inhibitory effect on SDF-1–induced Rap1 activation (Fig. 4I), whereas these PI3K inhibitors reduced SDF-1–induced activation of Akt and p70S6K to various extents (Fig. 4J). As expected, GGTI-298 as well as these PI3K inhibitors down-regulated SDF-1–induced chemotaxis of 32Dcl3 cells (Fig. 4K). It was also demonstrated that SDF-1–induced activation of Akt and Rap1 in 32Dcl3 cells (Fig. 4, L and M) as well as that of Akt and p90Rsk in Baf3 cells (Fig. 2, I, K, and L) was inhibited by the Gαi-specific inhibitor PTX, which is in contrast to SDF-1–induced tyrosine phosphorylation of PECAM-1 and activation of BTK or Lyn (Fig. 2, G, I, K, and L). However, none of the inhibitors for tyrosine kinases involved in PECAM-1 phosphorylation inhibited SDF-1–induced Rap1 activation (Fig. 4, N and O). Together, these data suggest that PECAM-1 enhancement of SDF-1–induced chemotaxis is mediated independently through activation of Rap1 and the PI3K/Akt pathway, which are activated through Gαi and independently of the tyrosine kinases activated by CXCR4.
PECAM-1 interacts with CXCR4 and prevents its internalization after SDF-1 stimulation
To explore further the mechanisms underlying PECAM-1–mediated enhancement of CXCR4 signaling, we next coexpressed PECAM-1 and HA-tagged CXCR4 in 293T cells to examine whether they physically interact with each other. Coimmunoprecipitation assays revealed that PECAM-1-WT as well as PECAM-1-Mut physically associated with CXCR4 in these cells independently of SDF-1 stimulation (Fig. 5A and data not shown). Although we could not detect the physical association of PECAM-1 with CXCR4 in 32Dcl3 cells using a similar coimmunoprecipitation assay, most likely due to its low sensitivity (negative data not shown), endogenous PECAM-1 as well as human PECAM-1 overexpressed in these cells was found to be colocalized with CXCR4 in these cells before treatment with SDF-1 by using immunofluorescence confocal microscopy (Fig. 5, B and C). These results raise the possibility that PECAM-1 may physically interact with CXCR4 in hematopoietic cells to enhance its downstream signaling leading to chemotaxis. Thus, we next examined whether PECAM-1 may modulate the cell surface expression level of CXCR4 in 32Dcl3 cells. As shown in Fig. 5, D and E, neither knockdown nor overexpression of PECAM-1 showed any effect on the expression level of CXCR4 in 32Dcl3 cells. However, knockdown of PECAM-1 enhanced the time-dependent down-regulation of CXCR4 cell surface expression after SDF-1 stimulation and retarded the recovery of its surface expression (Fig. 5F). In line with this, overexpression of PECAM-1-WT or, to a lesser extent, that of PECAM-1-Mut reduced the down-regulation of CXCR4 expression and accelerated its recovery (Fig. 5G). Together, these results suggest that PECAM-1 may physically interact with CXCR4 to retard SDF-1–induced endocytosis of CXCR4 or to enhance its recycling to the cell surface to increase its chemotactic signaling in hematopoietic cells.
Figure 5.
PECAM-1 interacts with CXCR4 on the cell surface to reduce its internalization after SDF-1 stimulation. A, 293T cells were transfected with plasmids encoding PECAM-1-WT (W), PECAM-1-Mut (M), and 3xHA-tagged CXCR4 as indicated. Cells were stimulated with 50 ng/ml SDF-1 for 1 min or left untreated as indicated before lysis. Anti-PECAM-1 or anti-HA immunoprecipitates (IP) as indicated were subjected to immunoblot analysis using the indicated antibodies. Positions of PECAM-1 are indicated by asterisks. Arrows indicate positions of HA-CXCR4. Positions of molecular weight markers are also indicated. B, 32Dcl3 cells, starved of IL-3 for 4 h, were treated with or without 50 ng/m SDF-1 for 5 min as indicated. Cells were then stained with anti-mouse CXCR4 antibody conjugated with PE (red) and anti-mouse PECAM-1 (mPECAM-1) antibody conjugated with APC (green) as well as with DAPI (blue) for nuclear staining. Representative confocal images of cells are shown. Scale bars correspond to 10 μm. C, 32Dcl3 cells overexpressing human PECAM-1-WT, starved of IL-3 for 4 h, were stained with anti-mouse CXCR4 (m-CXCR4) antibody conjugated with PE (red) and anti-human PECAM-1 (hPECAM-1) antibody conjugated with Alexa Fluor 647 (green) as well as with DAPI (blue) for nuclear staining. D, 32Dcl3 cells overexpressing human PECAM-1-WT (WT) or vector control cells (Cont.) were starved, and surface expression levels of CXCR4 were analyzed by flow cytometry. Iso., isotype control. E, 32Dcl3 cells expressing PECAM-1 shRNA (KD) or luciferase shRNA as a control (Cont.) were analyzed as described for D. F, 32Dcl3 cells expressing PECAM-1 shRNA (KD) or luciferase shRNA as a control (Cont.) were stimulated with 100 ng/ml SDF-1 for the indicated times, and surface expression levels of CXCR4 were analyzed by flow cytometry. Changes in CXCR4 expression levels as estimated by mean fluorescence intensity after SDF-1 stimulation are plotted. G, 32Dcl3 cells overexpressing human PECAM-1-WT (WT) or human PECAM-1-Mut (Mut) as well as vector control cells (Cont.) were stimulated as described for F. All the data shown are representative of experiments repeated at least three times.
Impairment of CXCR4 endocytosis enhances SDF-1–induced chemotaxis and activation of Rap1 and the PI3K/Akt/mTORC1 signaling pathway
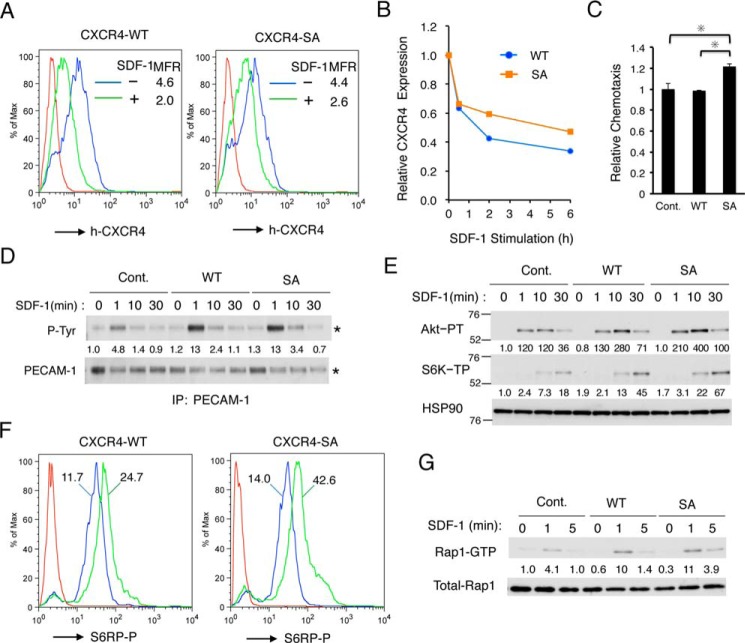
To explore the possible role impairment of CXCR4 endocytosis by PECAM-1 may play in enhancement of chemotactic signaling, we examined effects of the CXCR4-S324A/S325A mutation, which has been reported to impair its SDF-1–induced endocytosis (32), on CXCR4-mediated signaling in 32Dcl3 cells. We first confirmed that, when expressed in 32Dcl3 cells, the SDF-1–induced time-dependent down-regulation of human CXCR4-SA surface expression was retarded as compared with that of human CXCR4-WT (Fig. 6, A and B). When expressed at comparable levels in 32Dcl3 cells, human CXCR4-SA but not CXCR4-WT significantly enhanced SDF-1–induced chemotaxis (Fig. 6C). In accordance with the idea that PECAM-1 interacts with CXCR4, overexpression of human CXCR4-WT as well as CXCR4-SA enhanced SDF-1–induced tyrosine phosphorylation of PECAM-1 (Fig. 6D). Moreover, overexpression of CXCR4-WT or CXCR4-SA enhanced SDF-1–induced activation of the PI3K/Akt/mTORC1 pathway and Rap1; this effect was more prolonged in cells expressing CXCR4-SA than in cells expressing CXCR4-WT (Fig. 6, E–G). Together, these results support the idea that retention of CXCR4 on the cell surface by the SA mutation or PECAM-1 overexpression in cells stimulated with SDF-1 may enhance and prolong CXCR4-mediated activation of the PI3K/Akt/mTORC1 pathway and that of Rap1 to enhance chemotaxis.
Figure 6.
The S324A/S325A mutation in CXCR4 impairs endocytosis and prolongs SDF-1–induced activation of the PI3K/Akt/mTORC1 pathway and Rap1 to enhance chemotaxis. A, 32Dcl3 cells overexpressing human CXCR4-WT or human CXCR4-SA (SA) were cultured for 2 h with or without 500 ng/ml SDF-1 as indicated, and cell surface expression of human CXCR4 (h-CXCR4) was evaluated by flow cytometry. Red lines represent isotype controls. B, 32Dcl3 cells overexpressing human CXCR4 wild type (WT) or human CXCR4-SA (SA) were stimulated with 500 ng/ml SDF-1 for the indicated times, and surface expression levels of human CXCR4 were analyzed by flow cytometry. Changes in CXCR4 expression levels after SDF-1 stimulation are plotted. C, 32Dcl3 cells overexpressing CXCR4-WT (WT) or CXCR4-SA (SA) as well as vector control cells (Cont.) were subjected to chemotaxis assay with 50 ng/ml SDF-1. Relative chemotaxis was calculated by dividing the percentage of migrated cells by that of vector control cells. The data represent the mean of triplicates, and the asterisks indicate statistically significant (p < 0.05) differences compared with control. Error bars represent S.D. D, 32Dcl3 cells overexpressing CXCR4-WT (WT) or CXCR4-SA (SA) as well as vector control cells (Cont.) were starved of IL-3 for 4 h and then stimulated with 50 ng/ml SDF-1 for the indicated times. Cells were lysed and subjected to immunoprecipitation (IP) using antibody for PECAM-1 followed by immunoblot analysis with anti-phosphotyrosine (P-Tyr) and anti-PECAM-1. Relative levels of tyrosine-phosphorylated PECAM-1 were determined by densitometric analysis and are shown below the panel. E, 32Dcl3 cells overexpressing CXCR4-WT (WT) or CXCR4-SA (SA) as well as vector control cells (Cont.) were starved of IL-3 for 4 h and then stimulated with 5 ng/ml SDF-1 for the indicated times. Lysates were subjected to immunoblot analysis using antibodies against the indicated proteins. Relative levels of phosphorylated Akt or S6K are indicated. Positions of molecular weight markers are indicated. Akt-PT, phospho-Thr308-Akt; S6K-PT, phospho-Thr389-p70S6K. F, 32Dcl3 cells overexpressing CXCR4-WT (WT) or CXCR4-SA (SA) were starved of IL-3 for 12 h and then stimulated with 10 ng/ml SDF-1 for 30 min or left unstimulated. Cells were fixed, permeabilized, and analyzed by flow cytometry using anti-phospho-Ser240/244-S6RP antibody. Cells treated with or without SDF-1 are shown as a green or blue line, respectively, whereas a red line represents isotype control for the antibody used. MFR for each sample is indicated. S6RP-P, phospho-Ser240/244-S6RP. G, 32Dcl3 cells overexpressing CXCR4-WT (WT) and CXCR4-SA (SA) as well as vector control cells (Cont.) were starved of IL-3 for 4 h and then stimulated with 1 ng/ml SDF-1 for the indicated times. Cells were lysed and analyzed for Rap1 activation. Relative levels of Rap1-GTP were determined by densitometric analysis and are shown below the panel. All the data shown are representative of experiments repeated at least three times with the exception of those shown in D and F, which are representative of two repeated experiments.
Discussion
In this study, we have reported for the first time that SDF-1 induced tyrosine phosphorylation of PECAM-1 in the murine model hematopoietic cell lines 32Dcl3 and BaF3 as well as in other hematopoietic cell lines, including leukemic cells, such as UT7 and U937 (Fig. 1). The phosphorylation induced by SDF-1 was much more rapid and transient as compared with that induced by IL-3, which correlated with the difference in time course of signal activation by this chemokine and cytokine. We have further found that SDF-1 or IL-3 induces tyrosine phosphorylation of PECAM-1 exclusively in the ITIMs because human PECAM-1-Mut with Y663F and Y686F mutations introduced into these motifs did not show any tyrosine phosphorylation after stimulation (Fig. 1I). Intriguingly, however, SDF-1–induced as well as basal tyrosine phosphorylation of endogenous murine PECAM-1 was remarkably increased by overexpression of this mutant (Fig. 1, H and J). This might have been caused by SDF-1–induced as well as spontaneous dimerization of endogenous PECAM-1 with the overexpressed mutant because homodimerization of PECAM-1 induces its activation and phosphorylation in the ITIMs (15, 16). Thus, it is possible that the enhanced tyrosine phosphorylation of ITIMs in endogenous PECAM-1 might compensate for the lack of phosphorylation of this mutant to transduce chemotactic signals induced by SDF-1 in cells overexpressing this mutant. This possibility should be taken into consideration when we evaluate the functional significance of tyrosine phosphorylation of PECAM-1 in the ITIMs using cells overexpressing this mutant and needs to be addressed in future studies.
The present study further indicates that Lyn, BTK, and JAK2 should be involved in tyrosine phosphorylation of PECAM-1 in the ITIMs after SDF-1 stimulation. First, we confirmed activation of Lyn and BTK by SDF-1 stimulation using activation-specific antibodies in 32Dcl3 and BaF3 cells (Fig. 2, A, K, and L), which is in accordance with previous reports (11, 12). Although we could not demonstrate activation of JAK2 using a similar method, activation of JAK2 as well as JAK3 by SDF-1 stimulation has been reported previously in multiple studies (2, 9, 10, 13). Second, specific inhibitors for these kinases reduced the PECAM-1 phosphorylation induced by SDF-1 in 32Dcl3 and BaF3 cells (Fig. 2, B and J). Third, we demonstrated that Lyn, BTK, or JAK2 induces tyrosine phosphorylation of PECAM-1 in the ITIMs when coexpressed transiently in 293T cells in the present and previous studies (Fig. 2C) (20). In 32Dcl3 cells, dasatinib abolished tyrosine phosphorylation of PECAM-1, whereas ibrutinib and to a lesser degree ruxolitinib reduced the SDF-1–induced phosphorylation. Previously, Tourdot et al. (28) revealed that the Src family kinases, such as Lyn, phosphorylate Tyr686 in the C-terminal ITIM to enable phosphorylation of Tyr663 in the N-terminal ITIM by other kinases, including BTK, resulting in recruitment of the tandem SH2 domain-containing tyrosine phosphatase SHP2 in platelets. Thus, it is plausible that Lyn and BTK as well as JAK2 coordinately and sequentially phosphorylate the PECAM-1 ITIMs in SDF-1-stimulated hematopoietic cells. Interestingly, JAK2-V617F mutant, implicated in pathogenesis of myeloproliferative neoplasms, induced tyrosine phosphorylation of PECAM-1 in 293T and hematopoietic cells, including leukemic PVTL-2 cells (Fig. 2, C–E). Furthermore, JAK2-V617F physically associated with PECAM-1 much more strongly than wild-type JAK2 in 293T cells (Fig. 2C). We previously reported the tyrosine phosphorylation of PECAM-1 by BCR/ABL and its possible involvement in modulation of leukemogenic signaling from this aberrant tyrosine kinase (20). Future studies are warranted to elucidate a possible involvement of PECAM-1 in modulation of Jak2-V617F–mediated leukemogenic signaling.
In the present study, the expression levels of PECAM-1 correlated with SDF-1–induced migration, suggesting that PECAM-1 may play an enhancing role in regulation of SDF-1–induced chemotaxis. In agreement with this, PECAM-1–deficient murine hematopoietic cells, including progenitor cells, megakaryocytes, and T-cells, showed an impairment in chemotactic response to SDF-1 (24, 25). Furthermore, an anti-PECAM-1 monoclonal antibody reduced SDF-1–induced transendothelial migration of the human AML cell line HL-60 and primary AML cells (21). Nevertheless, it remains unknown how PECAM-1 modulates the CXCR4-mediated signaling mechanisms regulating chemotaxis. Although we have found that overexpression of PECMA-1-Mut also enhanced SDF-1–induced migration (Fig. 3F), overexpression of PECAM-1-Mut remarkably enhanced SDF-1–induced tyrosine phosphorylation of endogenous PECAM-1 (Fig. 1, H and J). Furthermore, inhibition of Lyn, BTK, or JAK2, responsible for tyrosine phosphorylation of the ITIMs, reduced SDF-1–induced chemotaxis (Fig. 2F). Thus, the significance of ITIM phosphorylation, which recruits SHP2 and other signaling molecules, in enhancement of chemotaxis remains to be clarified in future studies.
The levels of PECAM-1 expression and chemotaxis correlated with the SDF-1–induced activation of Akt and, more remarkably, mTORC1 but not that of ERK and p90Rsk (Fig. 3, G and H). Furthermore, chemotaxis was significantly inhibited by the mTORC1-specific inhibitor rapamycin among other inhibitors for the PI3K/mTOR pathway (Fig. 3I), which suggests that mTORC1 activated downstream of the PI3K/Akt pathway should play a critical role in enhancement of SDF-1–induced chemotaxis by PECAM-1. Previously, the effect of mTOR on cell migration has been attributed mainly to mTORC2 due to its ability to activate the Rho family small GTPases, such as Rac, and to control actin cytoskeleton organization (33–35). However, SDF-1–induced activation of Rac correlated inversely with the levels of PECAM-1 expression and chemotaxis as well as activation of the PI3K/Akt/mTORC1 pathway in the present study. In agreement with our results, previous studies have demonstrated that rapamycin or knockdown of the mTORC1 component Raptor inhibited SDF-1–induced chemotaxis of dendritic cells and the human cancer cell line HeLa (36, 37). Nevertheless, the exact mechanisms of mTORC1 enhancement of chemotaxis remain to be elucidated in future studies.
The levels of PECAM-1 expression and chemotaxis also correlated with SDF-1–induced activation of Rap1 but not Rac in 32Dcl3 cells (Fig. 4, A–D), thus suggesting that Rap1 may play a role in enhancement of chemotaxis by PECAM-1. It has previously been reported that SDF-1 induced activation of Rap1 in lymphocytes and that its inhibition by RapGAPII or SPA-1 inhibited chemotaxis, thus suggesting that Rap1 may transduce chemotactic signaling downstream of CXCR4 (4, 5). We also previously reported that SDF-1 induced activation of Rap1 as well as Rac in 32Dcl3 and BaF3 cells, although chemotaxis was significantly inhibited by transient expression of a dominant-negative mutant of Rac but not that of Rap1 (6, 7). In accordance with the previous report by Shimonaka et al. (5), however, SDF-1–induced chemotaxis was significantly reduced by abolition of Rap1 activation by SPA-1 (Fig. 4G). Furthermore, the Rap1 inhibitor GGTI-298 inhibited chemotaxis as well as Rap1 activation induced by SDF-1 (Fig. 4, I and K). Together, these results support the idea that Rap1 may play a role in enhancement of chemotaxis by PECAM-1.
As for the mechanisms for activation of Rap1 by CXCR4, we previously found that overexpression of the adaptor protein CrkL, which interacts with various signaling molecules, including guanine nucleotide exchange factors for the Ras and Rho family GTPases, enhanced SDF-1–induced activation of Rap1, Rac, and Ras as well as chemotaxis (7). However, we did not find any significant difference in CrkL expression levels in 32Dcl3 cells with decreased or increased PECAM-1 expression levels (negative data not shown). In this regard, it is interesting to note that Rap1 and Rac1 were reported to be antagonistically activated through competitive binding of the Crk family adaptor CrkII with C3G and DOCK180, two exchange factors for Rap1 and Rac, respectively (38). A similar mechanism may possibly explain the opposite effects of PECAM-1 on SDF-1–induced activation of Rap1 and Rac observed in the present study. Recently, Schmid et al. (39) reported that various chemoattractants, including SDF-1, activate β1 integrin through Rap1 activation mediated through PI3Kγ, a class IB isoform of PI3K, to induce chemotaxis of granulocytes and monocytes. However, although both Akt and Rap1 activation induced by SDF-1 was inhibited by the Gαi-specific inhibitor PTX (Fig. 4, L and M), neither PI3K inhibitors, including that for PI3Kγ, nor SPA-1 showed any significant effect on SDF-1–induced Rap1 or the PI3K/mTORC1 pathway activation, respectively (Fig. 4, H and I). Moreover, inhibition of tyrosine kinases responsible for induction of PECAM-1 phosphorylation in cells stimulated with SDF-1 did not significantly affect Rap1 activation (Fig. 4, N and O). Thus, PECAM-1 should enhance activation of Rap1 downstream of Gαi but independently of the PI3K pathway and without involving tyrosine phosphorylation of PECAM-1 or modulation of the expression level of CrkL.
The present study has revealed that PECAM-1 physically interacts with CXCR4 when coexpressed in 293T cells (Fig. 5A). Recently, dela Paz et al. (40) have reported that PECAM-1 binds CXCR4 indirectly through interaction with heparan sulfate proteoglycans, which bind to heparin-binding sites in the PECAM-1 extracellular domain, in endothelial cells as well as in HEK293 cells. However, although heparin, a competitive inhibitor of heparan sulfate-protein interactions, inhibited activation of SDF-1–induced CXCR4 signaling in accordance with a previous study (41), it did not affect the physical association between PECAM-1 and CXCR4 in 293T cells in the present study (negative data not shown). PECAM-1-Mut as well as PECAM-1-WT associated with CXCR4 in 293T, thus suggesting that the ITIMs should not be directly involved in binding with CXCR4 (Fig. 5A). Although we could confirm that CXCR4 and PECAM-1 were colocalized in 32Dcl3 cells by confocal microscopy (Fig. 5, B and C), we could not demonstrate the physical interaction by the coimmunoprecipitation assay in these cells, most likely because of the relatively low expression levels (negative data not shown). Thus, further studies are required to confirm and elucidate the precise mechanisms of physical association of PECAM-1 with CXCR4 in hematopoietic cells.
The PECAM-1 expression levels negatively or positively correlated with the rate of decrease or recovery, respectively, of the surface CXCR4 levels after SDF-1 stimulation in 32Dcl3 cells (Fig. 5, F and G). Thus, it is plausible that PECAM-1 may physically interact with CXCR4 to reduce down-regulation of its surface expression level after SDF-1 stimulation to enhance chemotactic signaling. Consistent with this hypothesis, the endocytosis-defective CXCR4-SA mutant showed increased abilities to activate the PI3K/Akt/mTORC1 pathway as well as Rap1, leading to enhanced chemotaxis in a manner similar to overexpression of PECAM-1 (Fig. 6). Intriguingly, similar germ line mutations in the C terminus of CXCR4 that block receptor internalization after SDF-1 stimulation result in persistent CXCR4 activation, leading to WHIM syndrome (3). Furthermore, somatic mutations of CXCR4 in the C terminus identical or similar to those found in WHIM syndrome have been recently found in about 30% of patients with Waldenström's macroglobulinemia (14). One of the most common WHIM-like CXCR4 mutations in this lymphoma, S338X, has been confirmed to show decreased CXCR4 down-regulation, leading to enhanced and sustained signaling, including Akt activation, and to contribute to disease progression and in the acquisition of resistance to various therapeutic agents (42, 43). Thus, it is tempting to speculate that PECAM-1 expressed in malignant hematopoietic cells may modulate disease progression or confer therapy resistance on these cells in the SDF-1–rich bone marrow environment, which warrants investigation in future studies.
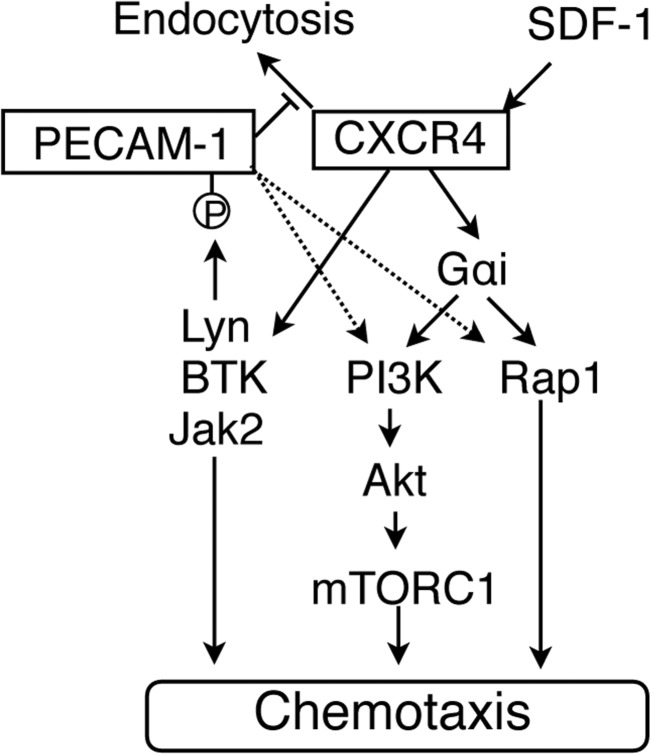
Molecular mechanisms underlying the enhancing effect of PECAM-1 on CXCR4-mediated chemotactic signaling are schematically and hypothetically depicted in Fig. 7. SDF-1 induces tyrosine phosphorylation of PECAM-1 in hematopoietic cells coordinately and sequentially by Lyn, BTK, and JAK2 in a Gαi-independent manner. Although overexpression of PECAM-1-Mut defective in phosphorylation enhanced SDF-1–induced chemotaxis similarly to that of PECAM-1-WT, it remarkably enhanced SDF-1–induced tyrosine phosphorylation of endogenous PECAM-1. Thus, the significance of tyrosine phosphorylation of ITIMs in PECAM-1 on chemotaxis remains to be elucidated in future studies. However, activation of these tyrosine kinases should play a role in CXCR4-mediated chemotactic signaling through mechanisms not involving the PI3K/Akt pathway or Rap1 because inhibition of these kinases reduced chemotaxis without significantly affecting activation of these signaling events. PECAM-1 increases chemotaxis by enhancing and prolonging Gαi-mediated activation of the PI3K/Akt/mTORC1 pathway and Rap1 by inhibiting internalization of CXCR4 and possibly accelerating its recycling back to the cell surface. However, the possibility that PECAM-1 may more directly enhance activation of the PI3K/Akt pathway and Rap1, similarly to when PECAM-1 is activated by cross-linking (17–19), needs to be addressed in future studies. It should also be noted that activation of the MEK/ERK pathway and Rac may also mediate chemotactic signaling from CXCR4 as we reported previously in the same hematopoietic cell model system (6, 7). Nevertheless, it was found not to be involved in enhancement of CXCR4 signaling by PECAM-1. Finally, future studies are warranted to address the possibility that PECAM-1 may also enhance CXCR4-mediated signaling, regulating other cellular effects, such as cell proliferation and survival, in normal as well as malignant hematopoietic cells similarly to the mutations in CXCR4 found in WHIM syndrome and Waldenström's macroglobulinemia.
Figure 7.
A schematic model for the molecular mechanisms underlying the enhancing effect of PECAM-1 on CXCR4-mediated chemotactic signaling.
Experimental procedures
Cells and reagents
Murine IL-3-dependent cell lines, 32Dcl3 and BaF3, and a clone of 32Dcl3 expressing the Epo receptor, 32D/EpoR, have been described previously (6, 44) and were maintained in RPMI 1640 medium supplemented with 10% FCS and 5 units/ml recombinant murine IL-3 or 1 unit/ml human recombinant Epo. A human leukemic cell line expressing the endogenous EpoR, UT7 (45), was kindly provided by Dr. N. Komatsu and maintained in RPMI 1640 medium with 10% FCS and 1 unit/ml Epo. The human AML cell line U937 was obtained from the Riken cell bank (Ibaraki, Japan) and maintained in RPMI 1640 medium with 10% FCS. The T-cell acute lymphoblastic leukemia cell line Jurkat was also maintained in RPMI 1640 medium with 10% FCS. A leukemic cell line expressing the constitutively activated JAK2-V617F mutant, PVTL-2, was established from a patient with AML transformed from polycythemia vera and maintained in RPMI 1640 medium with 10% FCS.3 For starvation from cytokines and SDF-1, cells were cultured for the indicated times in ASF104 medium (Ajinomoto, Tokyo, Japan) without FCS. PLAT-A, an amphotropic virus-packaging cell line, and 293T, a human embryonic kidney cell line, were kindly provided by Dr. T. Kitamura and Dr. S. Yamaoka, respectively, and maintained in DMEM supplemented with 10% FCS.
Recombinant murine SDF-1 and IL-3 were purchased from R&D Systems (Abington, UK) and Peprotech (Rocky Hill, NJ), respectively. Recombinant human Epo was kindly provided by Chugai Pharmaceutical Co. Ltd. (Tokyo, Japan). Dasatinib was purchased from Toronto Research Chemicals Inc. (Toronto, Canada). The BTK inhibitor ibrutinib and the Rap1 inhibitor GGTI-298 were purchased from Cayman Chemical (Ann Arbor, MI). The JAK family kinase inhibitors ruxolitinib and JAKI-1 were purchased from LC Laboratories (Woburn, MA) and Calbiochem, respectively. GDC-0941 was purchased from Active Biochemicals Co., Ltd. (Wanchai, Hong Kong). Idelalisib and CZC24832 were purchased from AdooQ BioScience (Irvine, CA). PTX was purchased from List Biochemical Laboratories (Campbell, CA). Rapamycin was purchased from Cell Signaling Technology (Beverly, MA). PP242 was purchased from Wako (Osaka, Japan).
Antibodies against PECAM-1 (SC-1506 and SC-18916), Lyn (SC-15), Rap1 (SC-65), STAT5A (SC-1081), and HSP90 (SC-13119) were purchased from Santa Cruz Biotechnology (Santa Cruz, CA). An anti-phosphotyrosine monoclonal antibody (4G10; 05-321) and an antibody against Rac (05-389) were purchased from Millipore (Billerica, MA). Antibodies against human PECAM-1 (CS-3528), mouse PECAM-1 (CS-77699), phospho-Tyr223-BTK (CS-5082), phospho-Thr202/Tyr204-ERK (CS-9106), phospho-Ser380-p90Rsk (CS-9335), phospho-Thr37/46-4EBP1 (CS-2855), phospho-Thr389-p70S6K (CS-9234), phospho-Ser240/244-S6RP (CS-2215), Akt (CS-2920), phospho-Thr308-Akt (CS-9275), phospho-Ser473-Akt (CS-9271), phospho-Tyr694-STAT5 (CS-9359), and HA (CS-3724) were purchased from Cell Signaling Technology. Antibody against β-actin (A1978) was purchased from Sigma. Antibody against phospho-Tyr396-Lyn was purchased from Epitomics Inc. (Burlingame, CA). Antibody against SPA-1 was kindly provided by Dr. M. Hattori (46).
Expression plasmids
The retrovirus vectors pMXs-IG and pMXs-puro were kindly provided by Dr. T. Kitamura. Expression plasmids for human PECAM-1 and its mutant with Y663F and Y686F mutations in the ITIMs, PECAM-1-ITIM(−), in pcDNA3.0 vector were kindly provided by Dr. D. Newman (26, 27). The coding regions for wild-type and ITIM(−) mutant of PECAM-1 were subcloned from these plasmids into retroviral vectors pMXs-IG (EcoRI) and pMXs-puro (EcoRI) using the restriction enzyme in parentheses. Retroviral pMLP shRNA-mir expression vector (transOMIC Technologies, Huntsville, AL) was used to express shRNAs designed to target PECAM-1 as well as luciferase as a control. The target sequence for murine PECAM-1 (NM_001032378) was 5′-AAGCCGAAGTTAGAGTTCTCCT-3′ and for luciferase was 5′-AACCGGCTGAAGAGCCTGATCA-3′. Retrovirus expression plasmids for JAK2 and the JAK2-V617F mutant (pRev-TRE-JAK2, pRev-TRE-JAK2-V617F, and pRx-JAK2-V617F) were described previously (47, 48). A lentivirus expression vector for JAK2-V617F (Pcw107-JAK2-V617F-V5) was a gift from David Sabatini and Kris Wood (Addgene plasmid number 64610) (49). The lentiviral packaging plasmid psPAX2 and the envelope-expressing plasmid pMD2.G were gifts from Didier Trono (Addgene plasmid numbers12260 and 12259, respectively). An expression plasmid of BTK (pWZL-Neo-Myr-Flag-BTK) was a gift from William Hahn and Jean Zhao (Addgene plasmid number 20432) (50). The coding region for BTK was subcloned between the BamHI and XhoI sites of pMXs-puro. An expression plasmid for SPA-1 in pSRα-myc vector was kindly provided by Dr. M. Hattori (51). The coding region for SPA-1 was subcloned into pMXs-puro (BamHI) using the restriction enzyme in parentheses. An expression vector for 3xHA-tagged CXCR4 in pcDNA3.1 was obtained from cDNA Resource Center (Bloomsburg, PA). Expression plasmids for CXCR4 wild type and its endocytosis-impaired mutant with serines in the C-terminal region changed to alanine (S324A/S325A) in pcDNA3.0 vector were kindly provided by Dr. A. Marchese (32). For construction of pMXs-puro-CXCR4-wild type and pMXs-puro-CXCR4-SA, the region coding for CXCR4 wild type or CXCR4-SA was excised from these plasmids by digestion with both EcoRI and XbaI and subcloned between the EcoRI and XhoI sites of pMXs-puro by blunting the XbaI and XhoI sites.
Transfection and infection
For transient expression in 293T cells, cells were transfected with the indicated plasmids using Lipofectamine reagent (Invitrogen) according to the manufacturer's instructions. Cells were harvested 48 h after transfection for immunoprecipitation and immunoblotting.
To obtain 32Dcl3 cells overexpressing human PECAM-1 or PECAM-1-ITIM(−), cells were infected with the recombinant retrovirus obtained from PLAT-A transfected with pMXs-IG or pMXs-puro constructed to express human PECAM-1 or PECAM-1-ITIM(−) as described previously (52). Cells were sorted for GFP expression with flow cytometry when using pMXs-IG vector or selected with medium containing 1 μg/ml puromycin when using pMXs-puro vector. Overexpression of PECAM-1 or its mutant was confirmed by immunoblotting.
To knock down PECAM-1 in 32Dcl3 cells, the cells were infected with the recombinant retrovirus obtained from PLAT-A transfected with pMLP-shRNA-PECAM-1 or pMLP-shRNA-luciferase as a control. Infected cells were selected in medium containing 1 μg/ml puromycin followed by sorting for GFP-expressing cells by flow cytometry. Knockdown of PECAM-1 expression was confirmed by immunoblotting and flow cytometry.
32D/EpoR cells expressing JAK2-V617F were obtained by infection with pRx-JAK2-V617F followed by selection with neomycin and withdrawal from Epo. 32Dcl3 cells overexpressing SPA-1, CXCR4-WT, or CXCR4-SA and vector control cells were obtained by infection with pMXs-puro-SPA-1, pMX-puro-CXCR4-WT, pMXs-puro-CXCR4-SA, or pMXs-puro, respectively, followed by selection with 1 μg/ml puromycin. Expression of SPA-1 was confirmed by immunoblotting, and expression of CXCR4-WT and its mutant was confirmed by flow cytometry.
Immunoprecipitation and immunoblotting
For immunoprecipitation experiments, cells were lysed in a lysis buffer containing 1% Triton X-100, 50 mm Tris-HCl (pH 7.5), 100 mm NaCl, 5 mm EDTA, 50 mm NaF, 40 mm β-glycerophosphate, 1 mm sodium orthovanadate, 1 mm phenylmethylsulfonyl fluoride, and 10 μg/ml each of aprotinin and leupeptin. For detection of the activated form of Lyn or BTK, cells were lysed, respectively, in 1× Laemmli sample buffer and sheared by several passes through a 27-gauge needle or in radioimmune precipitation assay buffer containing 1% Nonidet P-40, 20 mm Tris-HCl (pH 7.4), 75 mm NaCl, 0.1% sodium dodecyl sulfate, and 0.5% deoxycholic acid supplemented with phosphatase and protease inhibitors as described above and clarified by centrifugation. Cell lysates were subjected to immunoprecipitation and immunoblotting as described previously (53). For immunoblot analysis of total cell lysates, samples were prepared by mixing an aliquot of cell lysates with an equal volume of 2× Laemmli sample buffer and heating at 100 °C for 5min. All the data shown are representative of experiments repeated at least three times unless indicated otherwise.
Small GTPase activation assay
Small GTPase activation in cells was examined by using activation-specific probes for Rap1 (GST-RalGDS-RBD) and Rac (GST-PAK2-RBD) as described previously (54, 55). In brief, cells were lysed and incubated with the fusion proteins precoupled to glutathione-agarose beads. GTP-bound small GTPases were eluted from the beads and analyzed by immunoblotting. All the data shown are representative of experiments repeated at least three times.
Chemotaxis assay
Chemotaxis assays were performed in 24-well plates using 5-μm porosity Transwell inserts (Costar, Kennebunk, ME). Cells were washed once and incubated in RPMI 1640 medium containing 0.5% heat-inactivated bovine serum albumin (migration medium) for 4 h at 37 °C. Cells were then resuspended at 2 × 106/ml in migration medium, and 100 μl of cells was added to the insert (upper chamber), and 600 μl of migration buffer with or without SDF-1 was added to the lower chamber. After 4-h incubation, the number of viable cells migrated to the lower chamber was counted by the trypan blue dye exclusion method. For assessment of the effect of various inhibitors on chemotaxis, cells were preincubated with the indicated concentrations of inhibitors for 30 min before addition to the insert. The data represent the mean of triplicates ±S.D. An unpaired two-tailed Student's t test was used to calculate differences between means; differences were considered significant when p < 0.05. All the data shown are representative of experiments repeated at least three times.
Flow cytometric analyses of PECAM-1, CXCR4, ACKR3, and phospho-Ser240/244-S6RP
For flow cytometric analyses of the cell surface expression of mouse PECAM-1, cells were incubated on ice for 45 min with APC-rat anti-mouse CD31 (102510, Biolegend, San Diego, CA) or without antibody as a control. After washing, the samples were analyzed by flow cytometry. For analyses of the cell surface expression of murine CXCR4, cells were incubated with PE-rat anti-mouse CD184 (BD551966, BD Biosciences) or APC-rat anti-mouse CD184 (BD558644, BD Biosciences) and PE-rat IgG2b κ isotype control (BD555848, BD Biosciences) or APC-rat IgG2b κ isotype control (17-4031, eBioscience, Santa Clara, CA) on ice for 45 min. For analyses of the cell surface expression of human CXCR4, cells were incubated with PE-mouse anti-human CD184 (BD557145, BD Biosciences) or PE-mouse IgG2a κ isotype control (BD556653, BD Biosciences) on ice for 45 min. For analyses of the cell surface expression of ACKR3, cells were incubated with rabbit-anti-ACKR3 (51024-1-AP, Proteintech), which reacts with both human and murine ACKR3, or rabbit-IgG (SC-2027, Santa Cruz Biotechnology) as a control on ice for 45 min and then with PE-conjugated goat anti-rabbit IgG (heavy + light) (4052-09, Southern Biotech) for 45 min.
For analyses of phospho-Ser240/244-S6RP, cells were fixed and permeabilized using a Cytofix/Cytoperm kit (BD554714, BD Biosciences) according to the manufacturer's instructions. Fixed cells were incubated on ice for 60 min with either anti-phospho-Ser240/244-S6RP (CS-5364, Cell Signaling Technology) or anti-rabbit monoclonal antibody (CS-3900, Cell Signaling Technology) as an isotype control and then with PE-conjugated goat anti-rabbit IgG (heavy + light) for 30 min in the dark. After washing, the samples were analyzed by flow cytometry. The data were analyzed using FlowJo software (Tree Star, Inc., Ashland, OR). Mean fluorescence ratio (MFR) was calculated by dividing mean fluorescence intensity of each sample with that of isotype control. All the data shown are representative of experiments repeated at least three times unless indicated otherwise.
Confocal immunofluorescence microscopy
Cells were incubated with PE-anti-mouse CXCR4 and APC-anti-mouse CD31 or Alexa Fluor 647-anti-human CD31 (BD561654, BD Biosciences) on ice for 45 min. After washing, the cytospin samples were prepared and mounted with Prolong® Gold antifade reagent with DAPI (Thermo Fisher Scientific) and visualized using a laser-scanning confocal microscope (FluoView FV10i, Olympus, Tokyo, Japan) equipped with a 60× oil immersion objective lens. All the data shown are representative of experiments repeated three times.
Author contributions
Y. U. and O. M. contributed to project conception and the design of experiments. Y. U. performed most of the experiments and analyzed the results. H. A., K. O., S. I., A. N., G. O., and T. K. also performed experiments and analyzed results. Y. U. and O. M. wrote the paper with contributions from all of the other coauthors.
Acknowledgments
We thank Drs. D. Newman, A. Marchese, N. Komatsu, T. Kitamura, S. Yamaoka, M. Hattori, W. Hahn, J. Zhao, D. Sabatini, K. Wood, and D. Trono for the generous gifts of experimental materials.
This work was supported by Ministry of Education, Culture, Sports, Science and Technology of Japan Grants 15K09467, 24591384, and 26461416. The authors declare that they have no conflicts of interest with the contents of this article.
S. Ishida, H. Akiyama, Y. Umezawa, K. Okada, A. Nogami, G. Oshikawa, T. Nagao, and O. Miura, manuscript in preparation.
- SDF-1
- stromal cell-derived factor-1
- WHIM
- warts, hypogammaglobulinemia, infection, and myelokathexis
- PECAM-1
- platelet endothelial cell adhesion molecule-1
- ITIM
- immunoreceptor tyrosine-based inhibition motif
- AML
- acute myeloid leukemia
- PTX
- pertussis toxin
- CXCR4-SA
- CXCR4-S324A/S325A
- Epo
- erythropoietin
- EpoR
- Epo receptor
- MFR
- mean fluorescence ratio
- BTK
- Bruton's tyrosine kinase
- SH2
- Src homology 2
- Mut
- mutant
- ACKR3
- atypical chemokine receptor 3
- mTOR
- mechanistic target of rapamycin
- CrkL
- Crk-like
- GAP
- GTPase-activating protein
- GDS
- guanine nucleotide dissociation stimulator
- RBD
- Rap-binding domain
- APC
- allophycocyanin
- PE
- phycoerythrin.
References
- 1. Nagasawa T. (2015) CXCL12/SDF-1 and CXCR4. Front. Immunol. 6, 301. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2. Pawig L., Klasen C., Weber C., Bernhagen J., and Noels H. (2015) Diversity and inter-connections in the CXCR4 chemokine receptor/ligand family: molecular perspectives. Front. Immunol. 6, 429. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3. Pozzobon T., Goldoni G., Viola A., and Molon B. (2016) CXCR4 signaling in health and disease. Immunol. Lett. 177, 6–15 [DOI] [PubMed] [Google Scholar]
- 4. McLeod S. J., Li A. H., Lee R. L., Burgess A. E., and Gold M. R. (2002) The Rap GTPases regulate B cell migration toward the chemokine stromal cell-derived factor-1 (CXCL12): potential role for Rap2 in promoting B cell migration. J. Immunol. 169, 1365–1371 [DOI] [PubMed] [Google Scholar]
- 5. Shimonaka M., Katagiri K., Nakayama T., Fujita N., Tsuruo T., Yoshie O., and Kinashi T. (2003) Rap1 translates chemokine signals to integrin activation, cell polarization, and motility across vascular endothelium under flow. J. Cell Biol. 161, 417–427 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6. Arai A., Jin A., Yan W., Mizuchi D., Yamamoto K., Nanki T., and Miura O. (2005) SDF-1 synergistically enhances IL-3-induced activation of the Raf-1/MEK/Erk signaling pathway through activation of Rac and its effector Pak kinases to promote hematopoiesis and chemotaxis. Cell. Signal. 17, 497–506 [DOI] [PubMed] [Google Scholar]
- 7. Arai A., Aoki M., Weihua Y., Jin A., and Miura O. (2006) CrkL plays a role in SDF-1-induced activation of the Raf-1/MEK/Erk pathway through Ras and Rac to mediate chemotactic signaling in hematopoietic cells. Cell. Signal. 18, 2162–2171 [DOI] [PubMed] [Google Scholar]
- 8. Sahin A. O., and Buitenhuis M. (2012) Molecular mechanisms underlying adhesion and migration of hematopoietic stem cells. Cell Adh. Migr. 6, 39–48 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9. Vila-Coro A. J., Rodríguez-Frade J. M., Martín De Ana A., Moreno-Ortíz M. C., Martínez-A C., and Mellado M. (1999) The chemokine SDF-1alpha triggers CXCR4 receptor dimerization and activates the JAK/STAT pathway. FASEB J. 13, 1699–1710 [PubMed] [Google Scholar]
- 10. Zhang X. F., Wang J. F., Matczak E., Proper J. A., and Groopman J. E. (2001) Janus kinase 2 is involved in stromal cell-derived factor-1α-induced tyrosine phosphorylation of focal adhesion proteins and migration of hematopoietic progenitor cells. Blood 97, 3342–3348 [DOI] [PubMed] [Google Scholar]
- 11. Nakata Y., Tomkowicz B., Gewirtz A. M., and Ptasznik A. (2006) Integrin inhibition through Lyn-dependent cross talk from CXCR4 chemokine receptors in normal human CD34+ marrow cells. Blood 107, 4234–4239 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12. de Gorter D. J., Beuling E. A., Kersseboom R., Middendorp S., van Gils J. M., Hendriks R. W., Pals S. T., and Spaargaren M. (2007) Bruton's tyrosine kinase and phospholipase Cγ2 mediate chemokine-controlled B cell migration and homing. Immunity 26, 93–104 [DOI] [PubMed] [Google Scholar]
- 13. Montresor A., Bolomini-Vittori M., Toffali L., Rossi B., Constantin G., and Laudanna C. (2013) JAK tyrosine kinases promote hierarchical activation of Rho and Rap modules of integrin activation. J. Cell Biol. 203, 1003–1019 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14. Hunter Z. R., Xu L., Yang G., Zhou Y., Liu X., Cao Y., Manning R. J., Tripsas C., Patterson C. J., Sheehy P., and Treon S. P. (2014) The genomic landscape of Waldenstrom macroglobulinemia is characterized by highly recurring MYD88 and WHIM-like CXCR4 mutations, and small somatic deletions associated with B-cell lymphomagenesis. Blood 123, 1637–1646 [DOI] [PubMed] [Google Scholar]
- 15. Newman P. J., and Newman D. K. (2003) Signal transduction pathways mediated by PECAM-1: new roles for an old molecule in platelet and vascular cell biology. Arterioscler. Thromb. Vasc. Biol. 23, 953–964 [DOI] [PubMed] [Google Scholar]
- 16. Privratsky J. R., and Newman P. J. (2014) PECAM-1: regulator of endothelial junctional integrity. Cell Tissue Res. 355, 607–619 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17. Pellegatta F., Chierchia S. L., and Zocchi M. R. (1998) Functional association of platelet endothelial cell adhesion molecule-1 and phosphoinositide 3-kinase in human neutrophils. J. Biol. Chem. 273, 27768–27771 [DOI] [PubMed] [Google Scholar]
- 18. Reedquist K. A., Ross E., Koop E. A., Wolthuis R. M., Zwartkruis F. J., van Kooyk Y., Salmon M., Buckley C. D., and Bos J. L. (2000) The small GTPase, Rap1, mediates CD31-induced integrin adhesion. J. Cell Biol. 148, 1151–1158 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19. Ferrero E., Belloni D., Contini P., Foglieni C., Ferrero M. E., Fabbri M., Poggi A., and Zocchi M. R. (2003) Transendothelial migration leads to protection from starvation-induced apoptosis in CD34+CD14+ circulating precursors: evidence for PECAM-1 involvement through Akt/PKB activation. Blood 101, 186–193 [DOI] [PubMed] [Google Scholar]
- 20. Wu N., Kurosu T., Oshikawa G., Nagao T., and Miura O. (2013) PECAM-1 is involved in BCR/ABL signaling and may downregulate imatinib-induced apoptosis of Philadelphia chromosome-positive leukemia cells. Int. J. Oncol. 42, 419–428 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21. Gallay N., Anani L., Lopez A., Colombat P., Binet C., Domenech J., Weksler B. B., Malavasi F., and Herault O. (2007) The role of platelet/endothelial cell adhesion molecule 1 (CD31) and CD38 antigens in marrow microenvironmental retention of acute myelogenous leukemia cells. Cancer Res. 67, 8624–8632 [DOI] [PubMed] [Google Scholar]
- 22. Tonino S. H., Spijker R., Luijks D. M., van Oers M. H., and Kater A. P. (2008) No convincing evidence for a role of CD31-CD38 interactions in the pathogenesis of chronic lymphocytic leukemia. Blood 112, 840–843 [DOI] [PubMed] [Google Scholar]
- 23. Wheadon H., Edmead C., and Welham M. J. (2003) Regulation of interleukin-3-induced substrate phosphorylation and cell survival by SHP-2 (Src-homology protein tyrosine phosphatase 2). Biochem. J. 376, 147–157 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24. Dhanjal T. S., Pendaries C., Ross E. A., Larson M. K., Protty M. B., Buckley C. D., and Watson S. P. (2007) A novel role for PECAM-1 in megakaryocytokinesis and recovery of platelet counts in thrombocytopenic mice. Blood 109, 4237–4244 [DOI] [PubMed] [Google Scholar]
- 25. Ross E. A., Freeman S., Zhao Y., Dhanjal T. S., Ross E. J., Lax S., Ahmed Z., Hou T. Z., Kalia N., Egginton S., Nash G., Watson S. P., Frampton J., and Buckley C. D. (2008) A novel role for PECAM-1 (CD31) in regulating haematopoietic progenitor cell compartmentalization between the peripheral blood and bone marrow. PLoS One 3, e2338. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26. Jackson D. E., Kupcho K. R., and Newman P. J. (1997) Characterization of phosphotyrosine binding motifs in the cytoplasmic domain of platelet/endothelial cell adhesion molecule-1 (PECAM-1) that are required for the cellular association and activation of the protein-tyrosine phosphatase, SHP-2. J. Biol. Chem. 272, 24868–24875 [DOI] [PubMed] [Google Scholar]
- 27. Newman D. K., Hamilton C., and Newman P. J. (2001) Inhibition of antigen-receptor signaling by platelet endothelial cell adhesion molecule-1 (CD31) requires functional ITIMs, SHP-2, and p56(lck). Blood 97, 2351–2357 [DOI] [PubMed] [Google Scholar]
- 28. Tourdot B. E., Brenner M. K., Keough K. C., Holyst T., Newman P. J., and Newman D. K. (2013) Immunoreceptor tyrosine-based inhibitory motif (ITIM)-mediated inhibitory signaling is regulated by sequential phosphorylation mediated by distinct nonreceptor tyrosine kinases: a case study involving PECAM-1. Biochemistry 52, 2597–2608 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29. Levine R. L., Pardanani A., Tefferi A., and Gilliland D. G. (2007) Role of JAK2 in the pathogenesis and therapy of myeloproliferative disorders. Nat. Rev. Cancer 7, 673–683 [DOI] [PubMed] [Google Scholar]
- 30. Melo R. C. C., Longhini A. L., Bigarella C. L., Baratti M. O., Traina F., Favaro P., de Melo Campos P., and Saad S. T. (2014) CXCR7 is highly expressed in acute lymphoblastic leukemia and potentiates CXCR4 response to CXCL12. PLoS One 9, e85926. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31. Onishi C., Mori-Kimachi S., Hirade T., Abe M., Taketani T., Suzumiya J., Sugimoto T., Yamaguchi S., Kapur R., and Fukuda S. (2014) Internal tandem duplication mutations in FLT3 gene augment chemotaxis to Cxcl12 protein by blocking the down-regulation of the Rho-associated kinase via the Cxcl12/Cxcr4 signaling axis. J. Biol. Chem. 289, 31053–31065 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32. Marchese A., and Benovic J. L. (2001) Agonist-promoted ubiquitination of the G protein-coupled receptor CXCR4 mediates lysosomal sorting. J. Biol. Chem. 276, 45509–45512 [DOI] [PubMed] [Google Scholar]
- 33. Jacinto E., Loewith R., Schmidt A., Lin S., Rüegg M. A., Hall A., and Hall M. N. (2004) Mammalian TOR complex 2 controls the actin cytoskeleton and is rapamycin insensitive. Nat. Cell Biol. 6, 1122–1128 [DOI] [PubMed] [Google Scholar]
- 34. Hernández-Negrete I., Carretero-Ortega J., Rosenfeldt H., Hernández-García R., Calderón-Salinas J. V., Reyes-Cruz G., Gutkind J. S., and Vázquez-Prado J. (2007) P-Rex1 links mammalian target of rapamycin signaling to Rac activation and cell migration. J. Biol. Chem. 282, 23708–23715 [DOI] [PubMed] [Google Scholar]
- 35. Liu L., Das S., Losert W., and Parent C. A. (2010) mTORC2 regulates neutrophil chemotaxis in a cAMP- and RhoA-dependent fashion. Dev. Cell 19, 845–857 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36. Delgado-Martín C., Escribano C., Pablos J. L., Riol-Blanco L., and Rodríguez-Fernández J. L. (2011) Chemokine CXCL12 uses CXCR4 and a signaling core formed by bifunctional Akt, extracellular signal-regulated kinase (ERK)1/2, and mammalian target of rapamycin complex 1 (mTORC1) proteins to control chemotaxis and survival simultaneously in mature dendritic cells. J. Biol. Chem. 286, 37222–37236 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37. Dillenburg-Pilla P., Patel V., Mikelis C. M., Zárate-Bladés C. R., Doçi C. L., Amornphimoltham P., Wang Z., Martin D., Leelahavanichkul K., Dorsam R. T., Masedunskas A., Weigert R., Molinolo A. A., and Gutkind J. S. (2015) SDF-1/CXCL12 induces directional cell migration and spontaneous metastasis via a CXCR4/Gαi/mTORC1 axis. FASEB J. 29, 1056–1068 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38. Vallés A. M., Beuvin M., and Boyer B. (2004) Activation of Rac1 by paxillin-Crk-DOCK180 signaling complex is antagonized by Rap1 in migrating NBT-II cells. J. Biol. Chem. 279, 44490–44496 [DOI] [PubMed] [Google Scholar]
- 39. Schmid M. C., Franco I., Kang S. W., Hirsch E., Quilliam L. A., and Varner J. A. (2013) PI3-kinase γ promotes Rap1a-mediated activation of myeloid cell integrin α4β1, leading to tumor inflammation and growth. PLoS One 8, e60226. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40. dela Paz N. G., Melchior B., Shayo F. Y., and Frangos J. A. (2014) Heparan sulfates mediate the interaction between platelet endothelial cell adhesion molecule-1 (PECAM-1) and the Gαq/11 subunits of heterotrimeric G proteins. J. Biol. Chem. 289, 7413–7424 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41. Seeger F. H., Rasper T., Fischer A., Muhly-Reinholz M., Hergenreider E., Leistner D. M., Sommer K., Manavski Y., Henschler R., Chavakis E., Assmus B., Zeiher A. M., and Dimmeler S. (2012) Heparin disrupts the CXCR4/SDF-1 axis and impairs the functional capacity of bone marrow-derived mononuclear cells used for cardiovascular repair. Circ. Res. 111, 854–862 [DOI] [PubMed] [Google Scholar]
- 42. Roccaro A. M., Sacco A., Jimenez C., Maiso P., Moschetta M., Mishima Y., Aljawai Y., Sahin I., Kuhne M., Cardarelli P., Cohen L., San Miguel J. F., Garcia-Sanz R., and Ghobrial I. M. (2014) C1013G/CXCR4 acts as a driver mutation of tumor progression and modulator of drug resistance in lymphoplasmacytic lymphoma. Blood 123, 4120–4131 [DOI] [PubMed] [Google Scholar]
- 43. Cao Y., Hunter Z. R., Liu X., Xu L., Yang G., Chen J., Patterson C. J., Tsakmaklis N., Kanan S., Rodig S., Castillo J. J., and Treon S. P. (2015) The WHIM-like CXCR4(S338X) somatic mutation activates AKT and ERK, and promotes resistance to ibrutinib and other agents used in the treatment of Waldenström's macroglobulinemia. Leukemia 29, 169–176 [DOI] [PubMed] [Google Scholar]
- 44. Miura O., D'Andrea A., Kabat D., and Ihle J. N. (1991) Induction of tyrosine phosphorylation by the erythropoietin receptor correlates with mitogenesis. Mol. Cell. Biol. 11, 4895–4902 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45. Komatsu N., Nakauchi H., Miwa A., Ishihara T., Eguchi M., Moroi M., Okada M., Sato Y., Wada H., Yawata Y., Suda T., and Miura Y. (1991) Establishment and characterization of a human leukemic cell line with megakaryocytic features: dependency on granulocyte-macrophage colony-stimulating factor, interleukin 3, or erythropoietin for growth and survival. Cancer Res. 51, 341–348 [PubMed] [Google Scholar]
- 46. Jin A., Kurosu T., Tsuji K., Mizuchi D., Arai A., Fujita H., Hattori M., Minato N., and Miura O. (2006) BCR/ABL and IL-3 activate Rap1 to stimulate the B-Raf/MEK/Erk and Akt signaling pathways and to regulate proliferation, apoptosis, and adhesion. Oncogene 25, 4332–4340 [DOI] [PubMed] [Google Scholar]
- 47. Nagao T., Oshikawa G., Wu N., Kurosu T., and Miura O. (2011) DNA damage stress and inhibition of Jak2-V617F cause its degradation and synergistically induce apoptosis through activation of GSK3β. PLoS One 6, e27397. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48. Nagao T., Kurosu T., Umezawa Y., Nogami A., Oshikawa G., Tohda S., Yamamoto M., and Miura O. (2014) Proliferation and survival signaling from both Jak2-V617F and Lyn involving GSK3 and mTOR/p70S6K/4EBP1 in PVTL-1 cell line newly established from acute myeloid leukemia transformed from polycythemia vera. PLoS One 9, e84746. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49. Martz C. A., Ottina K. A., Singleton K. R., Jasper J. S., Wardell S. E., Peraza-Penton A., Anderson G. R., Winter P. S., Wang T., Alley H. M., Kwong L. N., Cooper Z. A., Tetzlaff M., Chen P. L., Rathmell J. C., et al. (2014) Systematic identification of signaling pathways with potential to confer anticancer drug resistance. Sci. Signal. 7, ra121. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50. Boehm J. S., Zhao J. J., Yao J., Kim S. Y., Firestein R., Dunn I. F., Sjostrom S. K., Garraway L. A., Weremowicz S., Richardson A. L., Greulich H., Stewart C. J., Mulvey L. A., Shen R. R., Ambrogio L., et al. (2007) Integrative genomic approaches identify IKBKE as a breast cancer oncogene. Cell 129, 1065–1079 [DOI] [PubMed] [Google Scholar]
- 51. Tsukamoto N., Hattori M., Yang H., Bos J. L., and Minato N. (1999) Rap1 GTPase-activating protein SPA-1 negatively regulates cell adhesion. J. Biol. Chem. 274, 18463–18469 [DOI] [PubMed] [Google Scholar]
- 52. Oshikawa G., Nagao T., Wu N., Kurosu T., and Miura O. (2011) c-Cbl and Cbl-b ligases mediate 17-allylaminodemethoxygeldanamycin-induced degradation of autophosphorylated Flt3 kinase with internal tandem duplication through the ubiquitin proteasome pathway. J. Biol. Chem. 286, 30263–30273 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 53. Miura O., Cleveland J. L., and Ihle J. N. (1993) Inactivation of erythropoietin receptor function by point mutations in a region having homology with other cytokine receptors. Mol. Cell. Biol. 13, 1788–1795 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 54. Arai A., Nosaka Y., Kanda E., Yamamoto K., Miyasaka N., and Miura O. (2001) Rap1 is activated by erythropoietin or interleukin-3 and is involved in regulation of β1 integrin-mediated hematopoietic cell adhesion. J. Biol. Chem. 276, 10453–10462 [DOI] [PubMed] [Google Scholar]
- 55. Nosaka Y., Arai A., Kanda E., Akasaki T., Sumimoto H., Miyasaka N., and Miura O. (2001) Rac is activated by tumor necrosis factor α and is involved in activation of ERK. Biochem. Biophys. Res. Commun. 285, 675–679 [DOI] [PubMed] [Google Scholar]