Abstract
Genome sequencing analysis has revealed at least 35 clusters of likely biosynthetic genes for secondary metabolites in Streptomyces ansochromogenes. Disruption of adpA encoding a global regulator (AdpA) resulted in the failure of nikkomycin production, whereas other antibacterial activities against Staphylococcus aureus, Bacillus cereus, and Bacillus subtilis were observed with the fermentation broth of ΔadpA but not with that of the wild-type strain. Transcriptional analysis showed that a cryptic gene cluster (pks7), which shows high identity with an oviedomycin biosynthetic gene cluster (ovm), was activated in ΔadpA. The corresponding product of pks7 was characterized as oviedomycin by MS and NMR spectroscopy. To understand the molecular mechanism of ovm activation, the roles of six regulatory genes situated in the ovm cluster were investigated. Among them, proteins encoded by co-transcribed genes ovmZ and ovmW are positive regulators of ovm. AdpA directly represses the transcription of ovmZ and ovmW. Co-overexpression of ovmZ and ovmW can relieve the repression of AdpA on ovm transcription and effectively activate oviedomycin biosynthesis. The promoter of ovmOI–ovmH is identified as the direct target of OvmZ and OvmW. This is the first report that AdpA can simultaneously activate nikkomycin biosynthesis but repress oviedomycin biosynthesis in one strain. Our findings provide an effective strategy that is able to activate cryptic secondary metabolite gene clusters by genetic manipulation of global regulatory genes.
Keywords: gene regulation, metabolism, natural product biosynthesis, polyketide, transcriptional coactivator, AdpA, Streptomyces ansochromogenes, cryptic gene clusters, global regulators, oviedomycin
Introduction
The crisis of antibiotic resistance has become an impending global problem, and it brings a significant threat to human health, so novel antibiotics are required to combat the evolving pathogens and new emerging diseases. Traditional large-scale screening of environmental microorganisms for bioactive molecules has been unable to meet the need (1). Therefore, it has become necessary to develop new technologies to discover novel bioactive compounds from diverse natural resources (2). Genome sequencing and in silico methods based on biological databases can rapidly pinpoint potential gene clusters for secondary metabolites, such as clustscan (3), CLUSEAN (4), SBSPKS toolbox (5), antiSMASH (6), etc. Thus, the productive capability of secondary metabolites in Streptomyces has been greatly underestimated. Cryptic secondary metabolite gene clusters have become a significant natural product reservoir, from which many more novel antibiotics could be discovered.
The expression of cryptic gene clusters is conditioned and can be activated by genetic manipulations of essential genes (7, 8). One of the approaches is to replace the native promoters of biosynthetic structural genes with constitutive promoters. For example, productions of the blue pigment indigoidine and two novel members of the polycyclic tetramate macrolactam family were activated by insertion of a strong and constitutive promoter in front of the transcriptional unit of structural genes of a small NRPS cluster and a hybrid type I PKS-NRPS cluster in Streptomyces albus J1074 (9). Another activation approach is to overexpress positive regulatory genes or to disrupt the negative regulatory genes situated in the cluster. For example, a giant type I modular PKS gene cluster of Streptomyces ambofaciens ATCC23877 spanning almost 150 kb was activated by overexpression of a LAL family regulatory gene, leading to the discovery of stambomycins, unusual glycosylated macrolides with unique chemical structures and promising antiproliferative activity against human cancer cell lines (10). Similarly, the production of chattamycins A and B in Streptomyces chattanoogensis was activated by overexpression of a pathway-specific activator gene chaI (11). Disruption of a putative repressor gene, pgaY, activated the production of UWM6 and rabelomycin in Streptomyces sp. PGA64 (12), and disruption of jadR2 activated jadomycin B production in Streptomyces venezuelae (13). Activation of cryptic gene clusters can also be achieved by genetic manipulation of global regulatory genes because they usually play a key role in controlling the check point for the onset of secondary metabolite biosynthesis. For example, disruption of dasR caused a significant increase in transcription of cpk genes in Streptomyces coelicolor (14). Two novel tylosin analogues were generated by disruption of wblA in Streptomyces ansochromogenes (15). Integration of a functional bldA gene activated the production of a polyeneoic acid amide in Streptomyces calvus (16). As one of the conserved global regulators in most of Streptomyces, AdpA can function as activator or repressor in terms of different target genes (17–19). In Streptomyces griseus, AdpA regulates more than 1000 genes, including those related to morphological development (20–22) and secondary metabolism, such as activator StrR for streptomycin (23) and GriR for grixazone biosynthesis (24). In Streptomyces lividans, the mutation of adpA affected the expression of more than 300 genes as revealed by transcriptomic analyses (25). Thus, the genetic manipulation of adpA could be an effective approach for the activation of cryptic gene clusters to gain novel antibiotics in Streptomyces. S. ansochromogenes is a natural producer of antifungal nikkomycin. Our previous results indicated that AdpA not only influenced morphological differentiation but also significantly affected nikkomycin production by directly controlling sanG, which encodes a pathway-specific transcriptional activator (26). antiSMASH analysis of S. ansochromogenes draft genome sequence indicated that 35 secondary metabolite gene clusters exist in the genome (27), of which only two corresponding secondary metabolites (nikkomycin and tylosin analogues) were identified. Therefore, it is expected to obtain more bioactive metabolites from the cryptic gene clusters by adpA manipulation.
In this work, we demonstrated that the cryptic oviedomycin biosynthetic gene cluster could be activated by disruption of adpA in S. ansochromogenes, and the activation mechanism was investigated. It is intriguing that the cluster-situated regulators (OvmZ and OvmW) are the targets of AdpA, and they collaboratively control oviedomycin biosynthesis.
Results
Activation of a cryptic oviedomycin biosynthetic gene cluster by disruption of adpA in S. ansochromogenes
A previous study (26) indicated that disruption of adpA in S. ansochromogenes (the wild-type strain) resulted in the failure of nikkomycin production. Due to the pleiotropic role of AdpA on target genes, it is interesting to know whether new bioactive compounds could be generated by adpA disruption. The fermentation broth of ΔadpA and the WT strain was subjected to bioassays against pathogenic bacteria. As a result, clear inhibition zones against Gram-positive bacteria (Staphylococcus aureus, Bacillus cereus, and Bacillus subtilis) were observed in the fermentation broth of ΔadpA (Fig. 1), which do not occur in the broth of WT strain. The results indicated that disruption of adpA led to the biosynthesis of other bioactive compound, whereas nikkomycin production was completely abolished. The bioactivity profiles of complemented strain (ΔadpAc) were restored to the same level as WT strain (Fig. 1), indicating that disruption of adpA is essential for both the production of new compound and the abolishment of nikkomycin biosynthesis.
Figure 1.
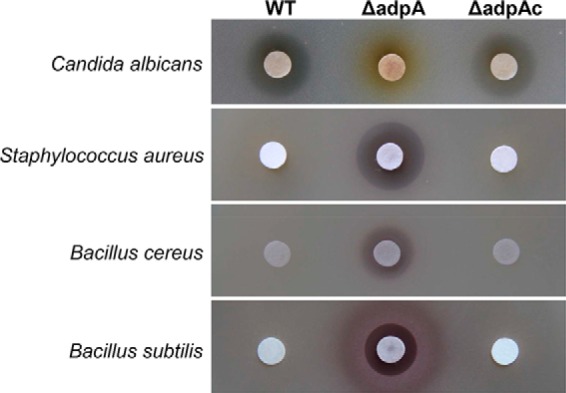
Bioassays of fermentation broth of wild-type strain and its derivatives. WT, S. ansochromogenes; ΔadpA, adpA disruption mutant; ΔadpAc, strain complemented by integrating a copy of adpA into the chromosome of ΔadpA. The strains were cultured in liquid MS for 4 days, and 200-μl supernatants from culture broths were assayed for bioactivity against C. albicans, S. aureus, B. cereus, and B. subtilis.
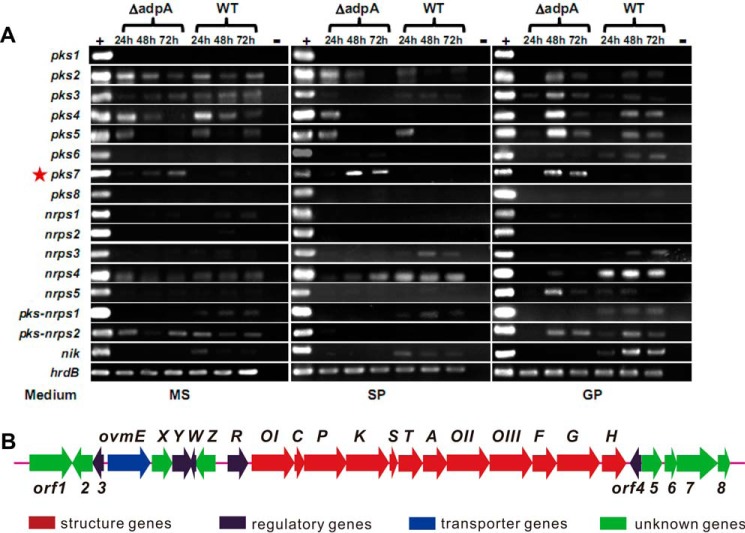
To verify the corresponding gene cluster(s) responsible for the newly emerged bioactive compound(s) in ΔadpA, we isolated the total RNA from both WT strain and ΔadpA grown in MS, SP, and GP media at different time courses, respectively. Transcription of the core genes (27) in each PKS and NRPS secondary metabolic gene cluster in S. ansochromogenes genome was analyzed by reverse transcription-PCR (RT-PCR). The gene clusters silent in WT strain but activated in ΔadpA are potentially responsible for the biosynthesis of the new bioactive compound. A core gene from pks7 in ΔadpA strain was transcribed in all detected media at each time course but not in WT strain (Fig. 2). Meanwhile, as we anticipated, the expression of nikkomycin biosynthetic gene cluster was impeded in ΔadpA (Fig. 2). These results indicated that pks7 was activated and may be responsible for the production of the bioactive metabolite in ΔadpA.
Figure 2.
Transcriptional analysis of PKS and NRPS biosynthetic gene clusters in S. ansochromogenes and ΔadpA by RT-PCR. A, cDNA templates were synthesized from RNA of S. ansochromogenes cultivated in three different liquid media (MS, SP, and GP). The strain name is indicated at the top of the gel, and growth time is below the strain name. The media used are indicated at the bottom of the gels. S. ansochromogenes genomic DNA (+) and double-distilled H2O (−) were used as positive and negative controls for each primer pair. Each gene cluster is given at the left of the gels. B, genetic organization of pks7 cluster in S. ansochromogenes. The gene cluster contains 26 ORFs, and the gene ovmW was marked in this study.
Isolation, identification, and characterization of oviedomycin
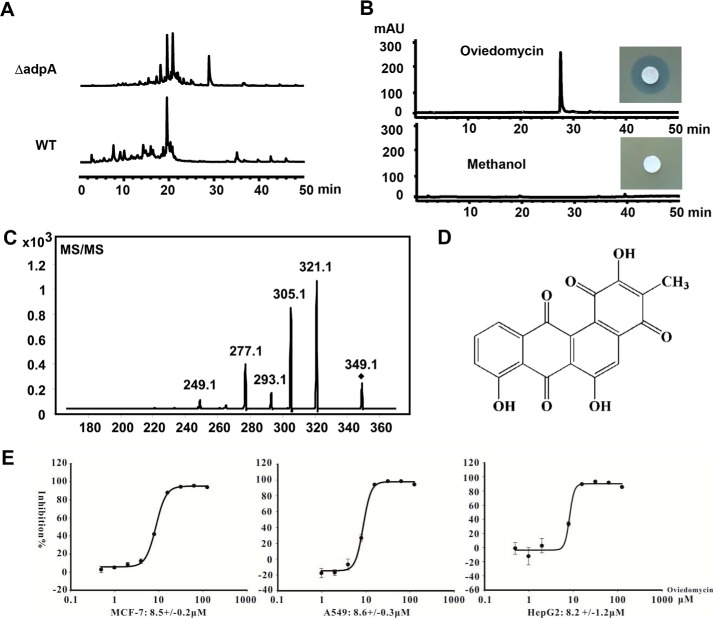
BLAST analysis revealed that pks7 contains 26 complete ORFs spanning 27 kb and shares more than 95% DNA sequence identities with the oviedomycin biosynthetic gene cluster in Streptomyces antibioticus ATCC 11891 (28). To verify if pks7 can also synthesize similar product in ΔadpA, the fermentation broth was analyzed by HPLC, and the active metabolite was isolated and characterized. On HPLC chromatograms, there was a distinct peak at a retention time of 28 min in the extract of ΔadpA, compared with that of WT strain (Fig. 3A). It exhibited inhibitory activity against S. aureus, whereas the HPLC solute methanol did not show inhibition zone (Fig. 3B). To determine the chemical structure of this compound, 3 liters of fermentation broth of ΔadpA in MS medium was harvested and extracted with chloroform. The organic phase was applied onto a Sephadex LH-20 column to remove most impurities, and finally the compound was separated and collected by semipreparative HPLC. The chemical structure of purified compound was determined by MS and NMR spectroscopy. A molecular ion peak at m/z− 349.0 was shown on the electrospray ionization-MS spectrum. MS/MS analysis revealed that most fragments were identical with those of the known angucyclinone oviedomycin (Fig. 3C) (29). Furthermore, 1H NMR and 13C NMR data were also consistent with those of oviedomycin (supplemental Fig. 1). Although the sugar side chain is not present in the structure as in other augucyclinone antibiotics, the purified oviedomycin showed good inhibitory activity against A549 (non-small-cell lung cancer), HepG2 (liver cancer) and MCF-7 (breast cancer) with IC50 < 20 μm (Fig. 3E). Thus, the disruption of adpA elicited the production of a compound potentially applicable as drug.
Figure 3.
Identification and bioassay of oviedomycin. A, HPLC analysis of oviedomycin production in WT strain and ΔadpA. B, HPLC analysis of purified oviedomycin and the bioassay against S. aureus. The solute methanol was used as the negative control. mAU, milli-absorbance unit. C, MS/MS analysis of oviedomycin. D, structure of oviedomycin. E, cytotoxic assay of oviedomycin. The cell lines used in the cytotoxin assay were MCF-7, A549, and HepG2. Error bars, S.D.
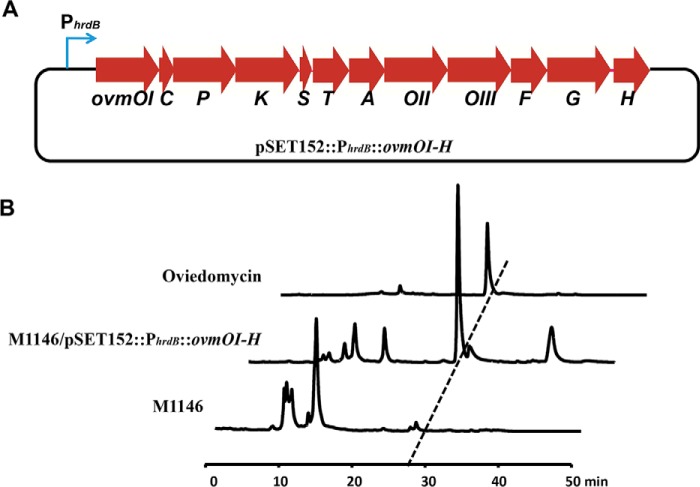
There are 12 genes (ovmOI–ovmH) assembling as one transcriptional unit responsible for oviedomycin biosynthesis in S. antibioticus ATCC 11891 (28). To verify whether pks7 is directly responsible for oviedomycin biosynthesis, the structural genes (ovmOI–ovmH) spanning >14-kb DNA fragments in this cluster were cloned into a Streptomyces integration vector pSET152 through Gibson assembly under the control of promoter PhrdB (Fig. 4A). The resulting construct was then introduced into S. coelicolor M1146 by conjugation for heterologous expression. As a result, oviedomycin was successfully produced in M1146 (Fig. 4B and supplemental Fig. 2), demonstrating that pks7 is undoubtedly responsible for oviedomycin biosynthesis in S. ansochromogenes, and then pks7 was designated as ovm.
Figure 4.
Heterologous expression of oviedomycin biosynthesis gene cluster. A, schematic representation of the plasmid used for heterologous expression of oviedomycin. B, heterologous expression of oviedomycin in M1146.
AdpA represses the expression of oviedomycin biosynthetic genes in S. ansochromogenes
To reveal the activation mechanism of ovm by disruption of adpA, the transcription of structural and regulatory genes in ovm were analyzed by quantitative real-time PCR (qRT-PCR)3 with the total RNA extracted from mycelia of WT strain and ΔadpA grown for 24 h.
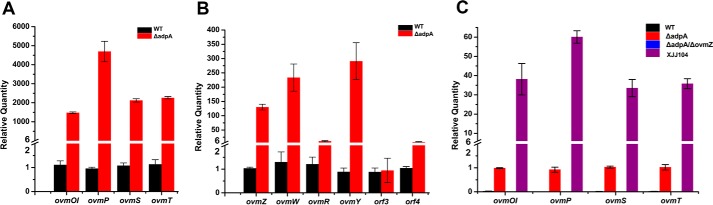
In ovm, structural genes, ovmOI–ovmH, are in the same transcriptional direction and probably constitute one operon. Among the structural genes, ovmC, ovmP, ovmK, ovmS, ovmT, and ovmA are responsible for the ring skeleton formation; ovmOI, ovmOII, and ovmOIII are oxygenases responsible for the introduction of those oxygens; and ovmF, ovmG, and ovmH are involved in polyketide metabolism (30). qRT-PCR showed that the transcription of four representative structural genes (ovmOI, ovmP, ovmS, and ovmT) from the operon (ovmOI-ovmH) in charge of oviedomycin biosynthesis increased 1000–4500-fold in ΔadpA compared with that in WT strain (Fig. 5A), indicating that AdpA plays a negative role in these genes' expression.
Figure 5.
Transcriptional analysis of oviedomycin biosynthetic genes by qRT-PCR. Total RNAs were isolated from mycelia of S. ansochromogenes and its derivatives after fermentation for 24 h. The constitutive hrdB transcript was used as an internal control. Error bars, S.D. A, transcriptional analysis of structural genes in WT strain and ΔadpA. B, transcriptional analysis of regulatory genes in WT strain and ΔadpA. C, transcriptional analysis of structural genes in WT strain, ΔadpA, XJJ104, and ΔadpA/ΔovmZ.
In addition, there are six putative regulatory genes (orf3, ovmY, ovmW, ovmR, orf4, and ovmZ) in ovm. Orf3 shares 49% amino acid sequence identity with the ArsR family regulator Rv2642 from Mycobacterium tuberculosis H37Rv. OvmY shows 39% amino acid sequence identity with the TetR family regulator CprB from S. coelicolor. OvmR shares 61% amino acid sequence identity with the IclR family regulator WP_044364315 from Streptomyces natalensis. Orf3, OvmY, and OvmR contain a helix-turn-helix DNA-binding domain at the N terminus. Orf4 shares 62% amino acid sequence identity with the MarR family regulator SFJ27529 from Amycolatopsis sacchari and contains a helix-turn-helix domain at its interior. ovmW encoding a small protein with 63 amino acids was not annotated in S. antibioticus ATCC 11891, and OvmW shares 65% identity with MerR family regulator EpaH from Kitasatospora sp. HKI 714 (31). However, both OvmW and EpaH have no identity with MerR family proteins in the sequence of amino acids by alignment (see “Discussion”). There is another putative regulatory gene ovmZ adjacent to ovmW. Its encoding product shares 59% identity with ChaP from chattamycin biosynthetic gene cluster in S. chattanoogensis L10 (11), 43% identity with Aur1O from the auricin biosynthetic gene cluster in Streptomyces aureofaciens CCM 3239 (32), 40% identity with TylU from the tylosin biosynthetic gene cluster in Streptomyces fradiae T59235 (33), 38% identity with Med-ORF27 from the medermycin biosynthetic gene cluster in Streptomyces sp. AM-7161 (34), 37% identity with PgaK from the pga gene cluster in Streptomyces sp. PGA64 (35), and 34% identity with EpaI from the endophenazines biosynthesis gene cluster in Kitasatospora sp. HKI 714 (31). Despite the fact that OvmZ homologous proteins are widespread in Streptomyces, their functions have not been elucidated except for TylU, a regulator relating to tylosin production (33). It is noteworthy that most regulatory genes of this type situate in PKS biosynthetic gene clusters. Both ovmW and ovmZ were preliminarily predicted as regulatory genes. Their role in oviedomycin biosynthesis was assessed by qRT-PCR. The transcription of ovmY, ovmW, ovmR, orf4, and ovmZ was increased in ΔadpA compared with that in WT strain, and particularly the transcription of ovmZ, ovmW, and ovmY was increased >100-fold (Fig. 5B), whereas the transcription of orf3 was not affected. It was concluded that both structural and regulatory genes in ovm were up-regulated in ΔadpA, suggesting that AdpA plays a negative regulatory role in the transcription of cluster-situated regulator (CSR) genes except orf3. AdpA might regulate oviedomycin biosynthesis by controlling these CSR genes.
Coordinative regulation of OvmZ and OvmW on the oviedomycin biosynthetic gene cluster
To identify the roles of putative CSR genes on oviedomycin biosynthesis, disruptions of these genes were carried out via homologous recombination in both WT strain and ΔadpA. HPLC analysis of fermentation broth showed that oviedomycin production was not detected in ΔovmZ, ΔovmW, ΔovmR, ΔovmY, Δorf3, and Δorf4 strains as well as in WT strain (Fig. 6A), suggesting that none of the six regulatory genes plays the repressor role in oviedomycin biosynthesis. Moreover, oviedomycin produced in ΔadpA was completely abolished in the double mutants ΔadpA/ΔovmZ and ΔadpA/ΔovmW (Fig. 6B). A moderate decrease of oviedomycin production was detected in the double mutant ΔadpA/ΔovmR, whereas oviedomycin production in ΔadpA/ΔovmY, ΔadpA/Δorf3, and ΔadpA/Δorf4 strains was almost the same as in the ΔadpA strain (Fig. 6B). These results demonstrated that ovmZ and ovmW are essential for activation of oviedomycin biosynthetic gene cluster.
Figure 6.
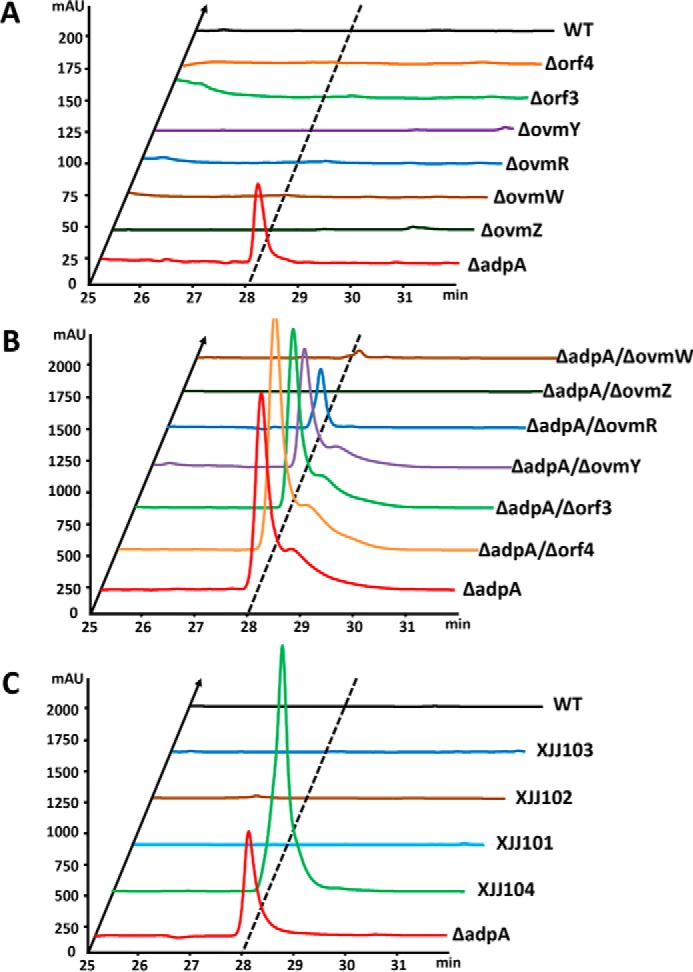
HPLC analysis of oviedomycin produced by S. ansochromogenes and its derivatives. A, the injection volume for ΔadpA is 10 μl, and the others are 100 μl. B, injection volume is 100 μl. C, injection volume is 50 μl.
To further clarify whether OvmZ, OvmW, and OvmR are positive regulators in oviedomycin biosynthesis, ovmZ, ovmW, or ovmR, respectively, was inserted into pKC1139 under the control of a strong constitutive expressed hrdB promoter (PhrdB) (36). The resulting recombinant plasmids were introduced into WT strain to generate XJJ101, XJJ102, and XJJ103, respectively. However, no oviedomycin was detected in these constructed strains, indicating that the overexpression of ovmZ, ovmW, or ovmR individually was not sufficient for activation of oviedomycin biosynthesis in WT strain. Because ovmZ and ovmW are adjacent and situated in the same operon in the cluster, we attempted to make co-overexpression of the two genes synchronously in WT strain by inserting them into pKC1139 as mentioned above. As anticipated, the resulting strain XJJ104 gave rise to much higher production of oviedomycin than ΔadpA (Fig. 6C).
Subsequently, the transcriptional analyses of structural genes (ovmOI, ovmP, ovmS, and ovmT) in WT, ΔadpA, ΔadpA/ΔovmZ, and XJJ104 strains were performed by qRT-PCR. The biosynthetic genes were not transcribed in ΔadpA/ΔovmZ as in WT strain, whereas the transcriptional levels were 30–60 times higher in XJJ104 than those in ΔadpA (Fig. 5C). Therefore, co-overexpression of ovmZ and ovmW can derepress oviedomycin biosynthesis in WT strain. OvmZ and OvmW proved to be positive regulators for oviedomycin biosynthesis, and the expression of structural genes is dependent on the co-expression of OvmZ and OvmW.
Cascade regulation of oviedomycin biosynthesis
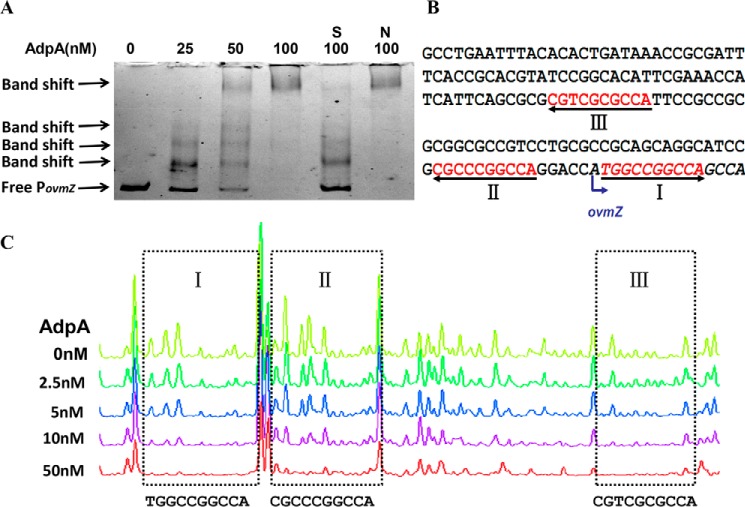
The above results showed that ovmZ and ovmW are required for oviedomycin biosynthesis, and global regulator AdpA exerts negative regulatory effect on oviedomycin biosynthesis by repressing the expression of ovmZW. To determine whether AdpA could directly regulate its target gene, AdpA-His6 was expressed in Escherichia coli BL21 (DE3) and purified through chromatography on nickel-nitrilotriacetic acid resin. EMSAs were performed by using AdpA-His6 with the promoter PovmZ as the probe. As anticipated, AdpA could form a complex with PovmZ in a concentration-dependent manner (Fig. 7A). EMSA suggested that AdpA inhibits oviedomycin biosynthesis in WT strain via directly repressing ovmZW transcription.
Figure 7.
EMSA and DNase I footprinting assays. A, EMSA of His6-tagged AdpA protein with the probe PovmZ containing the promoter of ovmZ. Each lane contains 50 ng of DNA probes and 2 μg of poly(dI-dC). S, unlabeled specific probe (10-fold) was added; N, nonspecific probe (10-fold) was added. B, binding sequence (sites I, II, and III) of AdpA on the upstream region of ovmZ. The binding sites are shaded in red, and the translation start codons are indicated in italic type. C, DNase I footprinting assays of the His6-tagged AdpA protein binding to the probe PovmZ. Each line contains 200 ng of DNA probe incubated with increasing concentrations of AdpA protein (0, 2.5, 5, 10, and 50 nm). The binding sites are marked in the dashed rectangle, and the sequences are given below the dashed rectangle.
To determine the specific binding sites of AdpA on the upstream region of ovmZ, DNase I footprinting was performed to analyze the protected regions of FAM/HEX-labeled probe in the presence or absence of AdpA. Three AdpA protected binding sites (sites I, II, and III) spanning the translation start codons of ovmZ were clearly observed (Fig. 7, B and C). Site I is consistent with the consensus AdpA-binding sequence in S. griseus (37), and the other two sites showed some similarities to the consensus sequence in S. griseus and to that of the sanG promoter in S. ansochromogenes (26).
To confirm whether OvmZ and OvmW directly regulate the transcription of oviedomycin biosynthetic genes, the promoter region (PovmOI) of the first gene (ovmOI) of the co-transcribed structural gene operon in ovm was used as a probe. OvmZ and OvmW were expressed and purified from E. coli as described above. EMSAs were performed to show the binding activity of OvmZ and OvmW to the probe. However, no binding shift pattern was observed by using OvmZ-His6 or OvmW-His6 individually or both proteins together (supplemental Fig. 3). We suspected that OvmZ and OvmW might have no function when expressed in E. coli, or the binding conditions in vitro might be inappropriate so the gusA guided in vivo reporting system was taken into account. gusA encodes β-glucuronidase enzyme (GUS), which can hydrolyze 5-bromo-4-chloro-3-indolyl-β-d-glucoronide (X-Gluc) to form a blue precipitate, 5,5′-dibromo-4,4′-dichloro-indigo (38). An integrating gusA reporter plasmid, pIJ10500::PovmOI::gusA, was constructed and then introduced into S. coelicolor M1146 via conjugation to generate the XJJ105 strain. Subsequently, plasmids pKC1139::PhrdBZW, pKC1139::PhrdBZ, and pKC1139::PhrdBW were respectively introduced into M1146 to give XJJ106, XJJ107, and XJJ108 and also introduced into XJJ105 to give XJJ109, XJJ110, and XJJ111. After culture on an AS-1 plate containing X-Gluc for 3 days, the blue pigment only appeared in XJJ109, which carries both pKC1139::PhrdBZW and pIJ10500::PovmOI::gusA, whereas the other constructs had no obvious GUS activity (Fig. 8). This result confirmed that collaborated OvmZ-OvmW could directly activate PovmOI and in turn positively regulate the transcription of oviedomycin biosynthetic genes.
Figure 8.
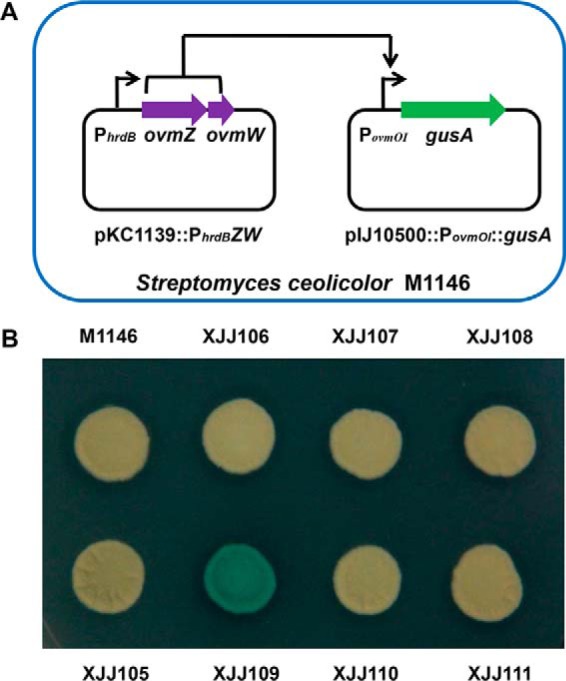
In vivo determination of the interaction between OvmZW and PovmOI. A, the plasmid pKC1139::PhrdBZW was used to express OvmZ and OvmW. The plasmid pIJ10500::PovmOI::gusA contains the promoter of ovmOI and the reporter gene gusA. They were introduced into M1146 to detect the activity of the ovmOI promoter. B, chromogenic assay on AS-1 plate containing the substrate X-Gluc. XJJ105 contains the plasmid pIJ10500::PovmOI::gusA; XJJ106 contains the plasmid pKC1139::PhrdBZW; XJJ107 contains the plasmid pKC1139::PhrdBZ; XJJ108 contains the plasmid pKC1139::PhrdBW; XJJ109 contains the plasmids pKC1139::PhrdBZW and pIJ10500::PovmOI::gusA; XJJ110 contains the plasmids pKC1139::PhrdBZ and pIJ10500::PovmOI::gusA; and XJJ111 contains the plasmids pKC1139::PhrdBW and pIJ10500::PovmOI::gusA.
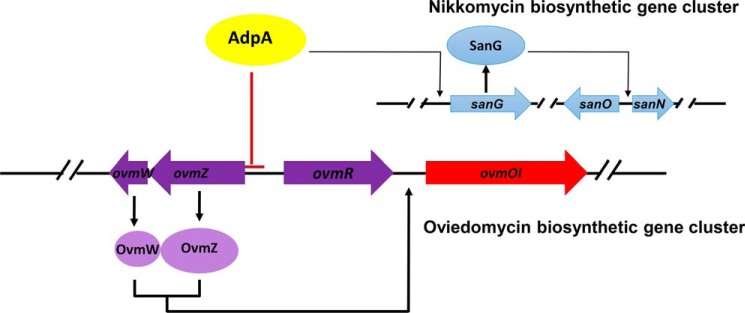
Taken together, the molecular mechanism activating a cryptic oviedomycin biosynthetic gene cluster in S. ansochromogenes was proposed (Fig. 9). In the wild-type strain, AdpA positively regulates nikkomycin production by activating the expression of the pathway-specific regulatory gene sanG. Meanwhile, it represses oviedomycin biosynthesis by repressing the positive regulatory genes ovmZ and ovmW. OvmZ and OvmW, as partner, coordinately regulate the transcription of structural genes in ovm. Co-overexpression of ovmZ and ovmW or disruption of adpA leads to the activation of ovm and, in turn, oviedomycin production.
Figure 9.
Model for the regulation of nikkomycin and oviedomycin biosynthesis by AdpA in S. ansochromogenes.
Discussion
Sequencing of Streptomyces genomes revealed the presence of a large number of secondary metabolic gene clusters for previously unsuspected products (39) and thus the potential to produce many more natural products than had previously been recognized. It has therefore become necessary to establish new methods to activate these “cryptic” pathways (7, 40, 41). Mining of genome sequences has revealed 35 gene clusters for secondary metabolite biosynthesis in S. ansochromogenes, a nikkomycin producer. Global regulatory genes, such as adpA, are also present in this strain. Although AdpA was proved to be an activator for nikkomycin production in our previous work (26), presumably it may play a repressor role for some other secondary metabolic gene clusters based on its pleiotropic roles on gene regulations and the vast target. Thus, disruption of global regulatory genes, including adpA, may be an effective strategy to activate cryptic gene clusters. In this work, disruption of adpA combined with transcription screening, bioassay, and compound characterization facilitated rapid localization of relevant gene clusters. Also, this is the first experimental evidence that AdpA positively and negatively regulates the expression of two antibiotic biosynthetic gene clusters (activating nikkomycin but repressing oviedomycin production) in the same strain. Manipulation of global regulatory genes can promote the discovery of novel bioactive compounds.
In elucidation of the regulatory mechanism of cryptic ovm gene cluster, two novel CSR genes encoding positive regulators OvmW and OvmZ were identified, which are essential for the activation of oviedomycin production. Interestingly, these two activators are dependent on each other and cooperatively function as partners. Constitutive expression of ovmZ and ovmW individually in WT strain could not activate oviedomycin biosynthesis, whereas co-overexpression of OvmZ and OvmW could relieve the repression of AdpA. OvmZ, consisting of 248 amino acids, shows no conserved domains in the database, and all of the orthologues are hypothetical proteins. OvmW comprises 63 amino acids, and its homologous gene is present but not annotated in the ovm gene cluster in S. antibioticus ATCC11891 (28). Although some of its orthologues are annotated as MerR family transcriptional regulators, alignment analysis of the OvmW amino acid sequence showed no homology to those of typical MerRs. Thus, this kind of small protein is also hypothetical. To date, six candidate orthologues of OvmZ (PgaK, Med-ORF27, ChaP, Aur1O, TylU, and EpaI) have been reported; however, their functions remain unclear. Most of the OvmZ orthologues are present in the type II PKS biosynthetic gene cluster, such as PgaK from the pga gene cluster (35), Med-ORF27 from the medermycin biosynthetic gene cluster (34), ChaP from the chattamycin biosynthetic gene cluster (11), Aur1O from the auricin biosynthetic gene cluster (32), and EpaI from the endophenazines biosynthesis gene cluster (31). Unlike OvmZ, there is no ovmW orthologue adjacent to them in the gene clusters except for epaI and tylU. epaH, an ovmW orthologue, is located downstream of epaI. In the type I PKS tylosin biosynthetic cluster, tylU, the so-called SARP “helper” gene, has homology with ovmZ, and there is another unannotated gene adjacent to tylU that is homologous to ovmW. In a previous report (33), tylU disruption caused a decrease of tylosin production, but the overexpression of tylU did not improve tylosin production in S. fradiae. Probably, the function of TylU was not elucidated clearly because the equivalent homologous gene of ovmW in tylosin gene cluster was not taken into consideration. Interestingly, a BLAST search against a database revealed that this kind of coupled regulatory gene is widespread in actinomycetes. Moreover, most of them are present in secondary metabolite biosynthetic gene clusters, as revealed by analyzing the flanking sequence of these coupled genes with antiSMASH (supplemental Table 1 and Fig. 4). Although the definite function and regulation mechanism remains a mystery, there is no doubt that they may play essential roles in different types of secondary metabolisms with an uncharacterized mechanism. It would be helpful to elucidate the mechanism of close cooperation of OvmZ and OvmW in the future.
Because genetic manipulations of other regulatory genes in the cluster have no effect on oviedomycin biosynthesis, it seems that OvmZ and OvmW are the direct effectors and dominate the transcription of the structural genes. Gene disruption of ovmZ and ovmW in ΔadpA and their overexpression in WT strain revealed that they are positive regulatory genes. However, EMSAs showed that the OvmZ and OvmW expressed in E. coli failed to bind to their target PovmOI, suggesting that the additional factors or ligands may be required for their activity or for proper protein folding. To verify this assumption, in vivo genetic construction with gusA reporter gene was performed, indicating that the ovmOI promoter is the direct target of co-expressed OvmZ and OvmW. Unfortunately, we failed to detect OvmZ and OvmW proteins by using Western blot hybridization and electrospray ionization-MS detection of the whole proteins of cell lysates in ΔadpA or even in the wild-type strain containing the plasmid for overexpression of OvmZ and OvmW, suggesting that the fast degradation of expressed protein or posttranslational modification might take place in Streptomyces. This could be a serious obstacle in resolving the regulation mechanisms for this kind of regulator. Development of adequate techniques to overcome this issue will be helpful.
Angucycline antibiotics have become the largest group of type II PKS-engineered products because they possess diverse chemical structures and potent biological activities against a number of pathogenic bacteria as well as cancer cell lines (42). Intriguingly, most angucycline antibiotic gene clusters are usually cryptic or silent, and their activation mechanisms still remain unknown (13, 43, 44). For example, jadomycin, as one of the intensively studied angucycline members, is produced under stress conditions, such as heat shock, phage infection, and ethanol treatment. But there is no ovmZ orthologous within the gene cluster. pga cluster is also a cryptic angucycline cluster, and genetic manipulations of all CSR genes could not activate its expression (35). A functionally unknown ovmZ homologous gene, pgaK, has not been genetically manipulated. It would not be surprising that PgaK could not work alone because a partner protein gene is absent in the cluster. Another cryptic angucycline cluster is the chattamycin biosynthetic gene cluster. Although there is ovmZ orthologous in this cluster, the direct effector of regulation seems to be another positive regulator, ChaI, because the transcription of the cluster could be activated by constitutive expression of ChaI (11). Thus, it would be more complicated to activate cryptic gene clusters for angucycline antibiotics by manipulating the CSR genes. In our study, the biosynthesis of an angucycline antibiotic, oviedomycin, was activated by disruption of adpA. This study indicated that genetic manipulation of the global regulatory genes for activating cryptic gene clusters could be an effective approach.
Experimental procedures
Strains, plasmids, and growth conditions
Bacterial strains and plasmids used in this study are listed in Table 1, and primers are listed in supplemental Table 2. S. ansochromogenes (the wild-type strain), S. coelicolor M1146, and their derivative strains were grown on mannitol soya flour medium agar or in yeast extract-malt extract (YEME) liquid medium at 28 °C (45). Mannitol soya flour liquid medium was used as the fermentation medium for oviedomycin production. E. coli JM109, C41 (DE3), ET12567/pUZ8002, and their derivative strains were grown at 37 °C in LB medium containing necessary antibiotics for propagating plasmids. The final concentrations of antibiotics used were as follows: 100 μg/ml ampicillin, 100 μg/ml apramycin, 100 μg/ml kanamycin, and 12.5 μg/ml chloramphenicol in LB for E. coli; 25 μg/ml nalidixic acid in mannitol soya flour medium for selection of Streptomyces exconjugants; 10 μg/ml apramycin and 10 μg/ml kanamycin in YEME and mannitol soya flour medium for S. ansochromogenes; and 50 μg/ml apramycin in YEME and mannitol soya flour medium for S. coelicolor M1146.
Table 1.
Strains and plasmids used in this study
| Name | Description | Sources |
|---|---|---|
| Strains | ||
| S. ansochromogenes | Wild-type strain | Ref. 49 |
| ΔadpA | adpA disruption mutant | This study |
| ΔovmZ | ovmZ disruption mutant | This study |
| ΔovmW | ovmW disruption mutant | This study |
| ΔovmR | ovmR disruption mutant | This study |
| Δorf3 | orf3 disruption mutant | This study |
| Δorf4 | orf4 disruption mutant | This study |
| ΔadpA/ΔovmZ | adpA and ovmZ disruption mutant | This study |
| ΔadpA/ΔovmW | adpA and ovmW disruption mutant | This study |
| ΔadpA/ΔovmR | adpA and ovmR disruption mutant | This study |
| ΔadpA/ Δorf3 | adpA and orf3 disruption mutant | This study |
| ΔadpA/Δorf4 | adpA and orf4 disruption mutant | This study |
| XJJ101 | ovmZ constitutive expression strain | This study |
| XJJ102 | ovmW constitutive expression strain | This study |
| XJJ103 | ovmR constitutive expression strain | This study |
| XJJ104 | ovmZ and ovmW constitutive expression strain | This study |
| S. coelicolor | ||
| M1146 | Δact Δred Δcpk Δcda | Ref. 50 |
| XJJ105 | M1146 derivative containing ovmOI promoter-gusA fusion plasmid | This study |
| XJJ106 | M1146 derivative containing ovmZW overexpression plasmid | This study |
| XJJ107 | M1146 derivative containing ovmZ overexpression plasmid | This study |
| XJJ108 | M1146 derivative containing ovmW overexpression plasmid | This study |
| XJJ109 | M1146 derivative containing ovmOI promoter-gusA fusion plasmid and ovmZW overexpression plasmid | This study |
| XJJ110 | M1146 derivative containing ovmOI promoter-gusA fusion plasmid and ovmZ overexpression plasmid | This study |
| XJJ111 | M1146 derivative containing ovmOI promoter-gusA fusion plasmid and ovmW overexpression plasmid | This study |
| E. coli | ||
| JM109 | F,′ proA+B+, lacIq, Δ(lacZ)M15/Δ(lac-proAB), gyrA96, recA1, relA1, endA1, hsdR17 | Ref. 51 |
| ET12567/pUZ8002 | recE, dam, dcm, hsdS, CmR, StrR, TetR, KmR | Ref. 52 |
| C41(DE3) | F−, ompT, gal dcm hsd SB (rB−, mB−) (DE3) | Lucigen |
| S. aureus CGMCC1.89 | Indicator strain for bioassays | CGMCCa |
| B. cereus CGMCC1.1626 | Indicator strain for bioassays | CGMCC |
| B. subtilis CGMCC1.1630 | Indicator strain for bioassays | CGMCC |
| C. albicans CGMCC2.4159 | Indicator strain for bioassays | CGMCC |
| Plasmids | ||
| pKC1139 | E. coli-Streptomyces shuttle plasmid contains a Streptomyces temperature-sensitive origin of replication | Ref. 53 |
| pIJ10500 | HygR, a derivative of pMS82 containing ΦBT1 integrase gene | Ref. 54 |
| pET23b::adpA | pET23b derivative with adpA coding region | Ref. 26 |
| pAD4 | Plasmid used for disruption of adpA | Ref. 26 |
| pAD5 | pSET152 derivative with intact adpA and its native promoter | Ref. 26 |
| pUZ101 | Plasmid used for construction of pKC1139::ZUD | This study |
| pZP101 | Plasmid used as an amplification template for probe PovmZ | This study |
| pKC1139::ovmZUD | Plasmid used for disruption of ovmZ | This study |
| pKC1139::ovmWUD | Plasmid used for disruption of ovmW | This study |
| pKC1139::ovmRUD | Plasmid used for disruption of ovmR | This study |
| pKC1139::orf3UD | Plasmid used for disruption of orf3 | This study |
| pKC1139::orf4UD | Plasmid used for disruption of orf4 | This study |
| pKC1139:: PhrdBZ | pKC1139 derivative with intact ovmZ and hrdB promoter | This study |
| pKC1139:: PhrdBW | pKC1139 derivative with intact ovmW and hrdB promoter | This study |
| pKC1139::PhrdBZW | pKC1139 derivative with intact ovmZW and hrdB promoter | This study |
| pKC1139::PhrdBR | pKC1139 derivative with intact ovmR and hrdB promoter | This study |
| pIJ10500::PovmOI::gusA | pIJ10500 derivative with intact gusA and ovmOI promoter | This study |
a CGMCC, China General Microbiological Culture Collection.
Gene disruptions
To construct an adpA disruption mutant (ΔadpA) in S. ansochromogenes and its derivatives, the plasmid pAD4 (26) was first introduced into E. coli ET12567/pUZ8002 by transformation and then introduced into S. ansochromogenes by conjugal transfer. The transformants resistant to kanamycin (Kanr) but sensitive to apramycin (Aprs) were selected as the double crossover disrupted strains and confirmed by PCR with primer pair ad1F/ad2R and using their genomic DNA as the template. For complementation analysis, the plasmid pAD5 (26) was introduced into ΔadpA by conjugal transfer from ET12567/pUZ8002, and the complemented strain was confirmed by PCR with primer pair ad1F/ad2R. To construct recombinant plasmid for ovmZ disruption, the 1468-bp DNA fragment corresponding to the upstream region of ovmZ was amplified by PCR with primer pair UZF/UZR. The PCR product was purified by gel purification (Qiagen) and digested with HindIII and XbaI and then inserted into the HindIII/XbaI site of pKC1139 to generate pUZ101. The 1441-bp DNA fragment corresponding to the downstream region of ovmZ was amplified by PCR with primer pair DZF/DZR, followed by digestion with HindIII and EcoRI after purification, and then inserted into the HindIII/EcoRI site of pUZ101 to generate pKC1139::ovmZUD. The resulting plasmid was introduced into S. ansochromogenes by conjugal transfer from ET12567/pUZ8002. The transformants conferring apramycin resistance were obtained, and then the colonies, which had lost the apramycin resistance after growing for 4 days at 42 °C, were selected as the double crossover disrupted mutant of ovmZ. This mutant strain (ΔovmZ) was confirmed by PCR using primer pair UZF/DZR and its chromosome DNA as the template to generate a 2909-bp fragment. For the double disruptions of adpA and ovmZ, the plasmid pAD4 was introduced into ΔovmZ by conjugal transfer from ET12567/pUZ8002. The double mutant strain (ΔadpA/ΔovmZ) conferring kanamycin resistance (Kanr) but sensitive to apramycin (Aprs) was selected and further confirmed by PCR as described above. The construction of single disruption strains of ovmW, ovmR, ovmY, orf3, and orf4 and the double disruption strains of these genes together with adpA were performed as mentioned above.
Construction for constitutive overexpression of regulatory genes
To construct constitutive overexpression plasmids for regulatory genes, Streptomyces multicopy number plasmid pKC1139 was used to clone the regulatory genes under the control of the hrdB promoter (PhrdB). The ovmZ coding region was amplified from the genomic DNA of S. ansochromogenes with primer pair ovmZF/ovmZR, and the hrdB promoter was amplified from the genomic DNA of S. coelicolor M145 using primer pair hrdBpF/hrdBpR. The primer hrdBpR was phosphorylated with T4 polynucleotide kinase (New England Biolabs) before PCR amplification to facilitate subsequent ligation reactions. The PCR product of the ovmZ coding region was digested with EcoRI, and the PCR product of the hrdB promoter was digested with XbaI. After purification from gel, two fragments were ligated together with XbaI/EcoRI-digested plasmid pKC1139 to generate pKC1139::PhrdBZ. The plasmids pKC1139::PhrdBW, pKC1139::PhrdBZW, and pKC1139::PhrdBR were constructed in a similar way. The primer pairs used in the constructions are listed in supplemental Table 1. The resulting plasmids pKC1139::PhrdBZ, pKC1139::PhrdBW, pKC1139::PhrdBZW, and pKC1139::PhrdBR, respectively, were introduced into S. ansochromogenes to obtain XJJ101, XJJ102, XJJ103, and XJJ104.
Heterologous expression of ovm in S. coelicolor M1146
The plasmid for heterologous expression of ovm was generated by Gibson assembly. The ovm cluster containing 12 structural genes (ovmOI–ovmH) was separately amplified by PCR to split into three DNA fragments (F1 with 5005 bp, F2 with 5000 bp, and F3 with 4418 bp) using three sets of primer pairs (F1F/F2R, F2F/F2R, and F2F/F2R) and S. ansochromogenes genomic DNA as the template. The hrdB promoter was amplified with the primer pair hrdBpF′/hrdBpR′ from genomic DNA of S. coelicolor M145. It contains a 30-nt sequence at the 5′-end of PhrdB overlapped with the one end of pSET152 digested with EcoRV and a 36-nt sequence at the 3′-end of PhrdB overlapped with the 5′-end of F1 fragment. F1, F2, and F3 fragments contain a 30-nt sequence overlapped with each other as the genetic organization order of the ovm cluster, and the F3 fragment has a 30-nt sequence at its 3′-end overlapped with the other end of pSET152 as mentioned above. 15 μl of Gibson assembly mix, ∼180 ng of purified ovm fragments, and ∼20 ng of PhrdB fragment were added in a final volume of 20 μl of reaction mixture. The reactions were performed at 50 °C for 1 h (46). Then the assembled DNA was transformed into the E. coli JM109 competent cells. The resulting plasmid, pSET152::PhrdB::ovmOI-H, was further transferred into S. coelicolor M1146 by conjugation from ET12567/pUZ8002. The regenerated recombinant strain was used for heterologous expression of oviedomycin.
Isolation, purification, and characterization of oviedomycin
ΔadpA was cultivated in a 5-liter fermentor containing 3 liters of mannitol soya flour liquid medium for 3 days at 28 °C and 200 rpm. The cultures were filtered through gauze, extracted with chloroform, dried in vacuo, and then redissolved in a small amount of methanol. Sephadex LH-20 column was applied for further purification. Active fractions were collected by semipreparative HPLC equipped with ZORBAX SB-C18 reverse-phase column (9.4 × 250 mm, 5 μm; Agilent) at 3 ml/min using a linear gradient elution. The elution solution was acetonitrile from 5 to 100% in water with 0.1% trifluoroacetic for 40 min followed by 100% acetonitrile with 0.1% trifluoroacetic for 10 min. MS analysis was performed on an LTQ Orbitrap hybrid mass spectrometer (Thermo Fisher Scientific) equipped with a Dionex Ultimate 3000 nano-low system and a nano-electrospray ion source. NMR spectra were recorded on a 500-MHZ Bruker spectrometer using DMSO-d6 as the solvent. Normal detection of oviedomycin was performed by HPLC with a ZORBAX SB-C18 reverse phase column (4.6 × 250 mm, 5 μm; Agilent). Samples were eluted at 1 ml/min with the same linear gradient as mentioned above.
Expression and purification of AdpA, OvmZ, and OvmW
The plasmid pET23b::adpA used for AdpA expression was constructed previously by Pan et al. (26). To construct recombinant plasmids for OvmZ and OvmW expression, the coding regions of ovmZ and ovmW were amplified from genomic DNA of S. ansochromogenes by PCR using primer pairs ovmZ1F/ovmZ1R and ovmW1F/ovmW1R. The PCR products were digested with NdeI and XhoI and then inserted into the NdeI/XhoI site of pET23b to generate plasmids pET23b::ovmZ and pET23b::ovmW. To construct the plasmid for the co-overexpression of OvmZ-OvmW, the ovmZ and ovmW coding regions with His6 label at the 3′-end were amplified from genomic DNA of S. ansochromogenes by PCR using primer pairs ovmZ1F/ovmZ2R and ovmW2F/ovmW1R. The PCR product of ovmZ was digested with NdeI/BbsI, and the PCR product of ovmW was digested with BbsI/XhoI. The purified fragments were ligated together with NdeI/XhoI linear plasmid pET23b to generate pET23b::ovmZW. The resulting plasmids were introduced into E. coli C41 (DE3) for the expression of OvmZ and OvmW.
Electrophoretic mobility shift assays and DNase I footprinting
The EMSAs were performed as described previously (47). The upstream regions (probes PovmZ and PovmOI) of ovmZ and ovmOI were amplified from genomic DNA of S. ansochromogenes by PCR using primer pairs ovmZpF/ovmZpR and ovmOIpF/ovmOIpR, respectively. The probe PovmZ was inserted into the EcoRV site of pBluescript KS+ to generate the plasmid pZP101, and then it was used as the template for PCR amplification using primers FAM-M13F/HEX-M13R to prepare fluorescently labeled probe PovmZ. The probes PovmZ and PovmOI were subsequently incubated with various concentrations of the protein individually at 25 °C for 30 min in 20 μl of reaction mixture. For competition assays, the unlabeled probe (specific competitor) or the nonspecific probe (a DNA fragment of the internal sequence of ovmOI amplified by PCR with primer pair OIF/OIR), respectively, was added to the binding reaction mixtures. After incubation, the samples were loaded on 4% (w/v) native polyacrylamide gels for electrophoresis in 4 °C. The fluorescently labeled DNA was detected by Tanon-5200 Multi. The unlabeled probe DNA in the gel was stained with SYBR Gold nucleic acid gel stain for 30 min and photographed under UV transillumination using Quantity One.
DNase I footprinting assays were performed as described previously (47). The probes (200 ng) and different amounts of AdpA were added to a final reaction volume of 50 μl and incubated at 25 °C for 30 min. DNase I (Promega) digestions were carried out for 50 s at 25 °C and stopped by adding 10 μl of EGTA. The purified samples were added to 9.5 μl of HiDi formamide and 0.5 μl of GeneScan-LIZ500 size standard, and the mixture was then analyzed with a 3730XL DNA analyzer. Subsequently, the results were analyzed with the software GeneMarker version 2.2.
Construction of plasmids and generation of strains for gusA reporter system
The promoter of ovmOI was amplified from genomic DNA of S. ansochromogenes by PCR using primer pair PovmOIF/PovmOIR. The gusA coding region was amplified from pGUS (48) with primers gusAF/R. The primer gusAF was phosphorylated with T4 polynucleotide kinase before PCR amplification to facilitate subsequent ligation reactions. The PCR products of gusA coding region and the ovmOI promoter were digested with NotI and XbaI, respectively, and were subsequently ligated together with XbaI/NotI-digested pIJ8660 plasmid to generate pIJ8660::PovmOI::gusA. The fragment tfd-PovmOI-gusA was then amplified from the plasmid pIJ8660::PovmOI::gusA by PCR with primer pair tfdF/gusA2R. The PCR product was digested with XhoI/NotI and then inserted into the XhoI/NotI site of pIJ10500 to generate the plasmid pIJ10500::PovmOI::gusA. The resulting plasmid was transformed into S. coelicolor M1146 to obtain XJJ105. Three plasmids (pKC1139::PhrdBZW, pKC1139::PhrdBZ, and pKC1139::PhrdBW) were introduced into M1146, respectively, to obtain XJJ106, XJJ107, and XJJ108, and also the three plasmids were introduced into XJJ105 to obtain XJJ109, XJJ110, and XJJ111, respectively. The strains were cultured on AS-1 plates with X-Gluc at a final concentration of 40 μm at 30 °C for 3 days to allow the blue color formation.
RNA isolation, RT-PCR, and qRT-PCR
Total RNA was isolated from S. ansochromogenes and its derivatives, and RT-PCR and qRT-PCR were performed as described previously (27, 44).
Bioassays of oviedomycin
To detect the biological activity against S. aureus, B. cereus, and B. subtilis, 1-ml overnight cultures of indicator strains were well dispersed in 100 ml of predissolved LB agar and poured into 15-cm plates, respectively. Oxford cups were placed on the plates, and fermentation extracts were then added into the Oxford cups. After incubation for 12 h at 37 °C, the inhibition zones were observed and assessed. Bioassays against Candida albicans were carried out by a disk diffusion method as described previously (15). Bioassays for anticancer activity were performed with 3-(4,5-dimethyl-2-thiazolyl)-2,5-diphenyl-2H-tetrazolium bromide (see supplemental material).
Author contributions
J. X., Y. T., and H. T. designed the study. J. X. performed most of the experiments, analyzed the primary data, and wrote the draft of the manuscript. J. Zhang) participated in the bioassay and structure analysis of oviedomycin. J. Zhuo assisted with strain fermentation. Y. L. participated in the protein purification and analysis. Y. T. revised the manuscript and wrote partial contents. H. T. supervised the whole research work and revised the manuscript. All authors reviewed the results and approved the final version of the manuscript.
Supplementary Material
Acknowledgments
We thank Drs. Guomin Ai and Jinwei Ren (Institute of Microbiology, Chinese Academy of Sciences, Beijing, China) for assistance with MS and NMR spectroscopy and Dr. Mervyn J. Bibb (Department of Molecular Microbiology, John Innes Centre, Norwich, UK) for kindly providing the plasmid pGUS.
This work was supported by Ministry of Science and Technology of China Grants 2015CB150600 and 2013CB734001 and National Natural Science Foundation of China Grants 31370097 and 31571281. The authors declare that they have no conflicts of interest with the contents of this article.
This article contains supplemental Tables 1 and 2 and Figs. 1–4.
- qRT-PCR
- quantitative real-time PCR
- CSR
- cluster-situated regulator
- GUS
- β-glucuronidase enzyme
- X-Gluc
- 5-bromo-4-chloro-3-indolyl-β-d-glucoronide
- YEME
- yeast extract-malt extract
- nt
- nucleotide.
References
- 1. Baltz R. H. (2006) Marcel Faber Roundtable: is our antibiotic pipeline unproductive because of starvation, constipation or lack of inspiration? J. Ind. Microbiol. Biotechnol. 33, 507–513 [DOI] [PubMed] [Google Scholar]
- 2. van Wezel G. P., McKenzie N. L., and Nodwell J. R. (2009) Chapter 5. Applying the genetics of secondary metabolism in model actinomycetes to the discovery of new antibiotics. Methods Enzymol. 458, 117–141 [DOI] [PubMed] [Google Scholar]
- 3. Starcevic A., Zucko J., Simunkovic J., Long P. F., Cullum J., and Hranueli D. (2008) ClustScan: an integrated program package for the semi-automatic annotation of modular biosynthetic gene clusters and in silico prediction of novel chemical structures. Nucleic Acids Res. 36, 6882–6892 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4. Weber T., Rausch C., Lopez P., Hoof I., Gaykova V., Huson D. H., and Wohlleben W. (2009) CLUSEAN: a computer-based framework for the automated analysis of bacterial secondary metabolite biosynthetic gene clusters. J. Biotechnol. 140, 13–17 [DOI] [PubMed] [Google Scholar]
- 5. Anand S., Prasad M. V., Yadav G., Kumar N., Shehara J., Ansari M. Z., and Mohanty D. (2010) SBSPKS: structure based sequence analysis of polyketide synthases. Nucleic Acids Res. 38, W487–W496 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6. Medema M. H., Blin K., Cimermancic P., de Jager V., Zakrzewski P., Fischbach M. A., Weber T., Takano E., and Breitling R. (2011) antiSMASH: rapid identification, annotation and analysis of secondary metabolite biosynthesis gene clusters in bacterial and fungal genome sequences. Nucleic Acids Res. 39, W339–W346 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7. Liu G., Chater K. F., Chandra G., Niu G., and Tan H. (2013) Molecular regulation of antibiotic biosynthesis in Streptomyces. Microbiol. Mol. Biol. Rev. 77, 112–143 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8. Kealey C., Creaven C. A., Murphy C. D., and Brady C. B. (2017) New approaches to antibiotic discovery. Biotechnol. Lett. 39, 805–817 [DOI] [PubMed] [Google Scholar]
- 9. Olano C., García I., González A., Rodriguez M., Rozas D., Rubio J., Sánchez-Hidalgo M., Braña A. F., Méndez C., and Salas J. A. (2014) Activation and identification of five clusters for secondary metabolites in Streptomyces albus J1074. Microb. Biotechnol. 7, 242–256 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10. Laureti L., Song L., Huang S., Corre C., Leblond P., Challis G. L., and Aigle B. (2011) Identification of a bioactive 51-membered macrolide complex by activation of a silent polyketide synthase in Streptomyces ambofaciens. Proc. Natl. Acad. Sci. U.S.A. 108, 6258–6263 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11. Zhou Z., Xu Q., Bu Q., Guo Y., Liu S., Liu Y., Du Y., and Li Y. (2015) Genome mining-directed activation of a silent angucycline biosynthetic gene cluster in Streptomyces chattanoogensis. Chembiochem 16, 496–502 [DOI] [PubMed] [Google Scholar]
- 12. Metsä-Ketelä M., Ylihonko K., and Mäntsälä P. (2004) Partial activation of a silent angucycline-type gene cluster from a rubromycin β producing Streptomyces sp. PGA64. J. Antibiot. 57, 502–510 [DOI] [PubMed] [Google Scholar]
- 13. Xu G., Wang J., Wang L., Tian X., Yang H., Fan K., Yang K., and Tan H. (2010) “Pseudo” γ-butyrolactone receptors respond to antibiotic signals to coordinate antibiotic biosynthesis. J. Biol. Chem. 285, 27440–27448 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14. Nazari B., Kobayashi M., Saito A., Hassaninasab A., Miyashita K., and Fujii T. (2013) Chitin-induced gene expression in secondary metabolic pathways of Streptomyces coelicolor A3(2) grown in soil. Appl. Environ. Microbiol. 79, 707–713 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15. Lu C., Liao G., Zhang J., and Tan H. (2015) Identification of novel tylosin analogues generated by a wblA disruption mutant of Streptomyces ansochromogenes. Microb. Cell. Fact. 14, 173–184 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16. Kalan L., Gessner A., Thaker M. N., Waglechner N., Zhu X., Szawiola A., Bechthold A., Wright G. D., and Zechel D. L. (2013) A cryptic polyene biosynthetic gene cluster in Streptomyces calvus is expressed upon complementation with a functional bldA gene. Chem. Biol. 20, 1214–1224 [DOI] [PubMed] [Google Scholar]
- 17. Gallegos M. T., Schleif R., Bairoch A., Hofmann K., and Ramos J. L. (1997) Arac/XylS family of transcriptional regulators. Microbiol. Mol. Biol. Rev. 61, 393–410 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18. Tobes R., and Ramos J. L. (2002) AraC-XylS database: a family of positive transcriptional regulators in bacteria. Nucleic Acids Res. 30, 318–321 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19. Akanuma G., Hara H., Ohnishi Y., and Horinouchi S. (2009) Dynamic changes in the extracellular proteome caused by absence of a pleiotropic regulator AdpA in Streptomyces griseus. Mol. Microbiol. 73, 898–912 [DOI] [PubMed] [Google Scholar]
- 20. Yamazaki H., Ohnishi Y., and Horinouchi S. (2000) An A-factor-dependent extracytoplasmic function sigma factor (sigma (AdsA)) that is essential for morphological development in Streptomyces griseus. J. Bacteriol. 182, 4596–4605 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21. Yamazaki H., Ohnishi Y., and Horinouchi S. (2003) Transcriptional switch on of ssgA by A-factor, which is essential for spore septum formation in Streptomyces griseus. J. Bacteriol. 185, 1273–1283 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22. Kato J. Y., Suzuki A., Yamazaki H., Ohnishi Y., and Horinouchi S. (2002) Control by A-factor of a metalloendopeptidase gene involved in aerial mycelium formation in Streptomyces griseus. J. Bacteriol. 184, 6016–6025 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23. Ohnishi Y., Kameyama S., Onaka H., and Horinouchi S. (1999) The A-factor regulatory cascade leading to streptomycin biosynthesis in Streptomyces griseus: identification of a target gene of the A-factor receptor. Mol. Microbiol. 34, 102–111 [DOI] [PubMed] [Google Scholar]
- 24. Higashi T., Iwasaki Y., Ohnishi Y., and Horinouchi S. (2007) A-factor and phosphate depletion signals are transmitted to the grixazone biosynthesis genes via the pathway-specific transcriptional activator GriR. J. Bacteriol. 189, 3515–3524 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25. Guyet A., Benaroudj N., Proux C., Gominet M., Coppée J. Y., and Mazodier P. (2014) Identified members of the Streptomyces lividans AdpA regulon involved in differentiation and secondary metabolism. BMC Microbiol. 14, 81–93 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26. Pan Y., Liu G., Yang H., Tian Y., and Tan H. (2009) The pleiotropic regulator AdpA-L directly controls the pathway-specific activator of nikkomycin biosynthesis in Streptomyces ansochromogenes. Mol. Microbiol. 72, 710–723 [DOI] [PubMed] [Google Scholar]
- 27. Zhong X., Tian Y., Niu G., and Tan H. (2013) Assembly and features of secondary metabolite biosynthetic gene clusters in Streptomyces ansochromogenes. Sci. China Life. Sci. 56, 609–618 [DOI] [PubMed] [Google Scholar]
- 28. Lombó F., Braña A. F., Salas J. A., and Méndez C. (2004) Genetic organization of the biosynthetic gene cluster for the antitumor angucycline oviedomycin in Streptomyces antibioticus ATCC 11891. Chembiochem 5, 1181–1187 [DOI] [PubMed] [Google Scholar]
- 29. Méndez C., Künzel E., Lipata F., Lombó F., Cotham W., Walla M., Bearden D. W., Braña A. F., Salas J. A., and Rohr J. (2002) Oviedomycin, an unusual angucyclinone encoded by genes of the oleandomycin-producer Streptomyces antibioticus ATCC11891. J. Nat. Prod. 65, 779–782 [DOI] [PubMed] [Google Scholar]
- 30. Lombó F., Abdelfattah M. S., Braña A. F., Salas J. A., Rohr J., and Méndez C. (2009) Elucidation of oxygenation steps during oviedomycin biosynthesis and generation of derivatives with increased antitumor activity. Chembiochem 10, 296–303 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31. Heine D., Martin K., and Hertweck C. (2014) Genomics-guided discovery of endophenazines from Kitasatospora sp. HKI 714. J. Nat. Prod. 77, 1083–1087 [DOI] [PubMed] [Google Scholar]
- 32. Novakova R., Bistakova J., Homerova D., Rezuchova B., and Kormanec J. (2002) Cloning and characterization of a polyketide synthase gene cluster involved in biosynthesis of a proposed angucycline-like polyketide auricin in Streptomyces aureofaciens CCM 3239. Gene 297, 197–208 [DOI] [PubMed] [Google Scholar]
- 33. Bate N., Bignell D. R., and Cundliffe E. (2006) Regulation of tylosin biosynthesis involving “SARP-helper” activity. Mol. Microbiol. 62, 148–156 [DOI] [PubMed] [Google Scholar]
- 34. Ichinose K., Ozawa M., Itou K., Kunieda K., and Ebizuka Y. (2003) Cloning, sequencing and heterologous expression of the medermycin biosynthetic gene cluster of Streptomyces sp. AM-7161: towards comparative analysis of the benzoisochromanequinone gene clusters. Microbiology 149, 1633–1645 [DOI] [PubMed] [Google Scholar]
- 35. Guo F., Xiang S., Li L., Wang B., Rajasärkkä J., Gröndahl-Yli-Hannuksela K., Ai G., Metsä-Ketelä M., and Yang K. (2015) Targeted activation of silent natural product biosynthesis pathways by reporter-guided mutant selection. Metab. Eng. 28, 134–142 [DOI] [PubMed] [Google Scholar]
- 36. Du D., Zhu Y., Wei J., Tian Y., Niu G., and Tan H. (2013) Improvement of gougerotin and nikkomycin production by engineering their biosynthetic gene clusters. Appl. Microbiol. Biotechnol. 97, 6383–6396 [DOI] [PubMed] [Google Scholar]
- 37. Yamazaki H., Tomono A., Ohnishi Y., and Horinouchi S. (2004) DNA-binding specificity of AdpA, a transcriptional activator in the A-factor regulatory cascade in Streptomyces griseus. Mol. Microbiol. 53, 555–572 [DOI] [PubMed] [Google Scholar]
- 38. Myronovskyi M., Welle E., Fedorenko V., and Luzhetskyy A. (2011) Beta-glucuronidase as a sensitive and versatile reporter in actinomycetes. Appl. Environ. Microbiol. 77, 5370–5383 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39. Bentley S. D., Chater K. F., Cerdeño-Tárraga A. M., Challis G. L., Thomson N. R., James K. D., Harris D. E., Quail M. A., Kieser H., Harper D., Bateman A., Brown S., Chandra G., Chen C. W., Collins M., et al. (2002) Complete genome sequence of the model actinomycete Streptomyces coelicolor A3(2). Nature 417, 141–147 [DOI] [PubMed] [Google Scholar]
- 40. Li Y., and Tan H. (2017) Biosynthesis and molecular regulation of secondary metabolites in microorganisms. Sci. China Life Sci. 60, 935–938 [DOI] [PubMed] [Google Scholar]
- 41. Niu G., Chater K. F., Tian Y., Zhang J., and Tan H. (2016) Specialised metabolites regulating antibiotic biosynthesis in Streptomyces spp. FEMS Microbiol. Rev. 40, 554–573 [DOI] [PubMed] [Google Scholar]
- 42. Kharel M. K., Pahari P., Shepherd M. D., Tibrewal N., Nybo S. E., Shaaban K. A., and Rohr J. (2012) Angucyclines: biosynthesis, mode-of-action, new natural products, and synthesis. Nat. Prod. Rep. 29, 264–325 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43. Wang L., Tian X., Wang J., Yang H., Fan K., Xu G., Yang K., and Tan H. (2009) Autoregulation of antibiotic biosynthesis by binding of the end product to an atypical response regulator. Proc. Natl. Acad. Sci. U.S.A. 106, 8617–8622 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44. Zou Z., Du D., Zhang Y., Zhang J., Niu G., and Tan H. (2014) A γ-butyrolactone-sensing activator/repressor, JadR3, controls a regulatory mini-network for jadomycin biosynthesis. Mol. Microbiol. 94, 490–505 [DOI] [PubMed] [Google Scholar]
- 45. Kieser T., Bibb M. J., Buttner M. J., Chater K. F., and Hopwood D. A. (2000) Practical Streptomyces Genetics, pp. 406–412, John Innes Foundation, Norwich, UK [Google Scholar]
- 46. Gibson D. G., Young L., Chuang R. Y., Venter J. C., Hutchison C. A. 3rd, and Smith H. O. (2009) Enzymatic assembly of DNA molecules up to several hundred kilobases. Nat. Methods 6, 343–345 [DOI] [PubMed] [Google Scholar]
- 47. Li Y., Li J., Tian Z., Xu Y., Zhang J., Liu W., and Tan H. (2016) Coordinative modulation of chlorothricin biosynthesis by binding of the glycosylated intermediates and end product to a responsive regulator ChlF1. J. Biol. Chem. 291, 5406–5417 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48. Sherwood E. J., and Bibb M. J. (2013) The antibiotic planosporicin coordinates its own production in the actinomycete Planomonospora alba. Proc. Natl. Acad. Sci. U.S.A. 110, E2500–E2509 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49. Zeng H., Tan H., and Li J. (2002) Cloning and function of sanQ: a gene involved in nikkomycin biosynthesis of Streptomyces ansochromogenes. Curr. Microbiol. 45, 175–179 [DOI] [PubMed] [Google Scholar]
- 50. Gomez-Escribano J. P., and Bibb M. J. (2011) Engineering Streptomyces coelicolor for heterologous expression of secondary metabolite gene clusters. Microb. Biotechnol. 4, 207–215 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 51. Sambrook J., and Russell D. W. (2001) Molecular Cloning: A Laboratory Manual, 3rd Ed., Appendix A3.7, Cold Spring Harbor Laboratory Press, NY [Google Scholar]
- 52. Paget M. S., Chamberlin L., Atrih A., Foster S. J., and Buttner M. J. (1999) Evidence that the extracytoplasmic function σ factor σE is required for normal cell wall structure in Streptomyces coelicolor A3(2). J. Bacteriol. 181, 204–211 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 53. Bierman M., Logan R., O'Brien K., Seno E. T., Rao R. N., and Schoner B. E. (1992) Plasmid cloning vectors for the conjugal transfer of DNA from Escherichia coli to Streptomyces spp. Gene 116, 43–49 [DOI] [PubMed] [Google Scholar]
- 54. Pullan S. T., Chandra G., Bibb M. J., and Merrick M. (2011) Genome-wide analysis of the role of GlnR in Streptomyces venezuelae provides new insights into global nitrogen regulation in actinomycetes. BMC Genomics 12, 175. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.