Abstract
STAT5 proteins play a role in adipocyte development and function, but their specific functions are largely unknown. To this end, we used an unbiased MS-based approach to identify novel STAT5-interacting proteins. We observed that STAT5A bound the E1β and E2 subunits of the pyruvate dehydrogenase complex (PDC). Whereas STAT5A typically localizes to the cytosol or nucleus, PDC normally resides within the mitochondrial matrix where it converts pyruvate to acetyl-CoA. We employed affinity purification and immunoblotting to validate the interaction between STAT5A and PDC subunits in murine and human cultured adipocytes, as well as in adipose tissue. We found that multiple PDC subunits interact with hormone-activated STAT5A in a dose- and time-dependent manner that coincides with tyrosine phosphorylation of STAT5. Using subcellular fractionation and immunofluorescence microscopy, we observed that PDC-E2 is present within the adipocyte nucleus where it associates with STAT5A. Because STAT5A is a transcription factor, we used chromatin immunoprecipitation (ChIP) to assess PDC's ability to interact with STAT5 DNA-binding sites. These analyses revealed that PDC-E2 is bound to a STAT5-binding site in the promoter of the STAT5 target gene cytokine-inducible SH2-containing protein (cish). We have demonstrated a compelling interaction between STAT5A and PDC subunits in adipocytes under physiological conditions. There is previous evidence that PDC localizes to cancer cell nuclei where it plays a role in histone acetylation. On the basis of our ChIP data and these previous findings, we hypothesize that PDC may modulate STAT5's ability to regulate gene expression by controlling histone or STAT5 acetylation.
Keywords: acetyl coenzyme A (acetyl-CoA), adipocyte, growth hormone, pyruvate dehydrogenase complex (PDC), signal transducers and activators of transcription 1 (STAT1)
Introduction
The JAK/STAT (Janus kinase/signal transducer and activator of transcription)-signaling pathway is a ligand-activated signaling pathway that has known roles in the regulation of many cellular functions, including apoptosis, differentiation, immunological responses, and proliferation. A variety of hormones and cytokines that use the JAK/STAT pathway can modulate adipocyte differentiation and function. To date, of the seven known STATs (STAT1–4, STAT5A, STAT5B, and STAT6), STAT5 is the most characterized STAT protein involved in the regulation of adipogenesis and fat cell function. Although STAT5A and STAT5B are transcribed from separate genes, they are together referred to as STAT5 because they are highly homologous and share numerous overlapping functions. Studies from many laboratories have shown a role of STAT5 proteins, particularly STAT5A, in adipogenesis in vitro (1), and there is also evidence that STAT5 contributes to adipose tissue development in vivo (2, 3). In mature fat cells, however, the role of STAT5 is less clear. We know that STAT5 proteins are activated by growth hormone in adipocytes, suggesting a role in reducing lipid stores and regulating insulin action. Moreover, in the last decade, several STAT5 target genes have been identified in adipocytes, including fatty-acid synthase (4) and adiponectin (5). Although these studies support the notion that STAT5 proteins may be important in fat cell function, their primary function in adipocytes remains unknown.
To further understand the function of STAT5 proteins in adipocytes, we employed a non-biased mass spectrometry (MS)-based approach to identify novel STAT5-interacting proteins. To our surprise, two proteins that make up the pyruvate dehydrogenase complex (PDC)2 were found to interact with STAT5A proteins in 3T3-L1 adipocytes in a growth hormone (GH)-dependent manner. PDC is a multisubunit protein that is composed of pyruvate dehydrogenase (E1), dihydrolipoyl acetyltransferase (E2), dihydrolipoyl dehydrogenase (E3), and E3-binding protein (E3BP) subunits that concertedly convert pyruvate to acetyl-CoA, and whose expression was previously thought to be restricted to the mitochondrial matrix (6). When fully assembled, PDC is a megadalton complex composed of multiple copies of each subunit (7). The E1 component is further divided into E1α and E1β subunits that catalyze the oxidative decarboxylation and reductive acetylation of pyruvate in the presence of thiamine pyrophosphate. The E2 component then catalyzes the transfer of the acetyl group to coenzyme A (CoA) to form acetyl-CoA via electron transfer from the E2 lipoyl moiety to NAD+ by the E3 component (6). Originally known as protein X, the E3BP component binds to the E2 and E3 subunits to contribute to the complex's organization and assembly (8). Acetyl-CoA production via PDC is important in tissues that are active in fatty acid synthesis (9), including adipose tissue. We sought to validate the physical association between STAT5A and the E2 subunit of the pyruvate dehydrogenase complex (PDC-E2) in adipocytes under physiological conditions, and in all our studies, STAT5A was present in a complex with PDC following hormonal stimulation concomitantly with tyrosine phosphorylation.
Previous studies have demonstrated interactions between STAT3 and STAT5 with proteins that belong to the PDC complex (10–12). Studies performed in immune cells and lung cancer cells suggest that these STATs can translocate to the mitochondria and modulate PDC function (10, 12). Another study shows an interaction of PDC-E2 and STAT5 in the nucleus of mouse BaF3 pro-B cells (11). Our studies are the first to reveal that STAT5A can interact with PDC-E2 in adipocytes in a manner that highly correlates with STAT5 tyrosine phosphorylation. Moreover, our results demonstrate that this interaction occurs in human adipocytes as well as in subcutaneous and visceral mouse adipose tissue. In murine adipocytes, the nuclear location of PDC-E2 was verified by independent methods. Finally, our highly novel observations reveal that PDC-E2 is part of the GH-induced STAT5-binding complex in the promoter of the STAT5 target gene cytokine-inducible SH2-containing protein (cish). Notably, recent studies suggest that PDC can be present in the nucleus of several cancer cell lines where it generates acetyl-CoA in the nucleus to modulate histone acetylation (13). Based on our observations, we predict that association of PDC with STAT5A plays a role in the ability of STAT5 to regulate gene expression either by modulating STAT5 acetylation, histone acetylation, or the acetylation of other proteins present in the STAT5 DNA-binding complex. Although we demonstrated that PDC-E2 can bind to a STAT5-binding site, we predict that the PDC complex likely associates with other transcription factors where it can contribute to protein acetylation. Overall, our studies reveal that protein subunits comprising the acetyl-CoA-generating enzyme complex, PDC, can be present in adipocyte nuclei and be part of a STAT5 DNA-binding complex. These novel observations may shift our perspectives on the role of metabolic enzymes as well as their cellular locations and functions.
Results
To gain insight into the potential mechanisms involved in the ability of STAT5 to modulate fat cell function, we employed a non-biased MS-based approach to identify novel STAT5-interacting proteins. Briefly, STAT5A was immunoprecipitated from unstimulated and growth hormone-stimulated 3T3-L1 adipocytes. The immunoprecipitated proteins were separated by SDS-PAGE, and the gel was stained with Coomassie Blue to visualize protein bands for excision. Excised protein bands were subjected to in-gel tryptic digestion, and the resulting peptide fragments were analyzed and identified using liquid chromatography (LC)-MS/MS as described previously (14). The E1β and E2 subunits of the PDC were among the potential STAT5A-interacting proteins to emerge from our non-biased screening approach (supplemental Fig. S1). PDC-E2 was one of the few GH-regulated, STAT5A interaction partners that we discovered and the first association we sought to validate, as it had been previously reported in cultured immune cells (10, 11).
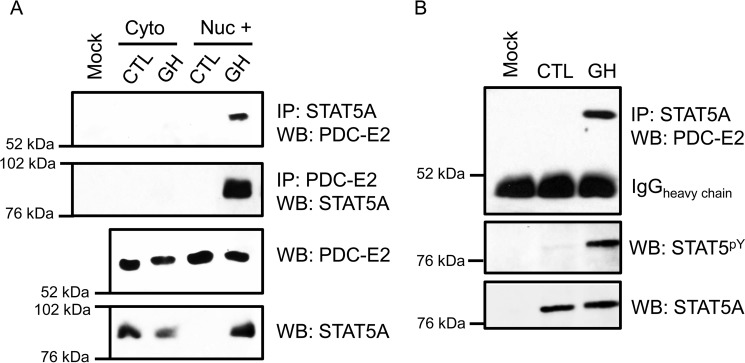
To determine whether STAT5A interacted with PDC-E2 under physiological conditions in adipocytes, we used an immunoprecipitation (IP) approach. As shown in Fig. 1, 3T3-L1 adipocytes were exposed to mouse GH (mGH) to induce STAT5 tyrosine phosphorylation and nuclear translocation and then fractionated into cytosolic and nuclear fractions. In Fig. 1A, IP with STAT5A (top panel) or IP with PDC-E2 (2nd panel) followed by Western blotting (WB) with PDC-E2 or STAT5A, respectively, clearly show an association between these two proteins, primarily within the nucleus-enriched (Nuc +) fraction under physiological conditions. We also performed this experiment with whole-cell extracts from human adipocytes (Fig. 1B) and again observed a GH-dependent interaction between STAT5A and PDC-E2.
Figure 1.
PDC-E2 interacts with STAT5A in GH-stimulated murine and human adipocytes. Fully differentiated 3T3-L1 adipocytes (A) or human adipocytes (B) were treated with 5 nm mGH (A) or hGH (B) for 15–20 min. Control (CTL) cells were untreated. A, monolayers were collected and subjected to subcellular fractionation that was optimized to separate cytoplasmic (cyto) and nuclear (nuc) compartments. The “Nuc +” fraction was free from cytoplasmic contamination and enriched for nuclei. Immunoprecipitation (IP) reactions utilized anti-STAT5A or anti-PDC-E2 antibodies and contained 300 μg of total protein per reaction. Interacting PDC-E2 (70 kDa) and STAT5A (95 kDa) proteins were detected by WB. Mock samples contained IP antibody but no extract. In the bottom two panels, the presence of STAT5A and PDC-E2 in the extracts used for immunoprecipitation was examined directly by Western blotting using 50 μg of total protein per lane. Experiments using 3T3-L1 adipocytes have been repeated more than three times on independent batches of cells. B, fully differentiated human adipocytes, derived from preadipocytes isolated from the visceral omental adipose depot of obese individuals, were purchased from Zenbio. Monolayers were collected, and whole-cell lysates were prepared. IP reactions and Western blotting were performed as described for A, except that 75 μg of total protein was used for each IP. The middle panel in B demonstrates the presence of activated STAT5 phosphorylated at tyrosine 694/699 (STAT5pY). The experiment with human adipocytes was performed one time.
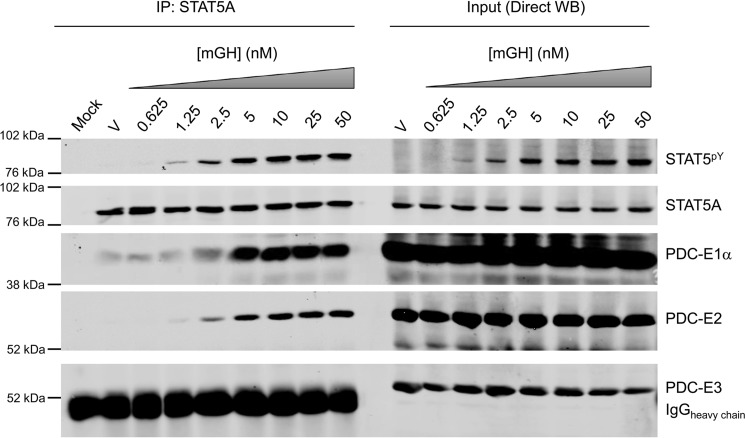
To assess the specificity of the STAT5A and PDC-E2 interaction, we examined its dose- and time-dependent regulation by STAT5 activators (Figs. 2 and 3). In Fig. 2, mature adipocytes were treated with a range of murine growth hormone (mGH) doses, and interactions between STAT5A and PDC subunits (E1, E2, and E3) were assessed. Whole-cell extracts were incubated with a STAT5A antibody, and associated proteins pulled down by immunoprecipitation were examined by Western blotting (left-hand side of Fig. 2). Direct Western blotting input controls are shown on the same gel in the right-hand side of Fig. 2. As expected and shown on the right side in Fig. 2, mGH exposure resulted in a dose-dependent tyrosine phosphorylation of STAT5 (STAT5pY). The left side of Fig. 2 also shows that mGH induced a dose-dependent association between STAT5A and multiple PDC subunits (E1α and E2 shown in Fig. 2 and supplemental Fig. S2 as well as E1β shown in supplemental Fig. S2). The band observed in the PDC-E3 blot is from the heavy chain of the immunoprecipitating antibody and masks any potential interaction with PDC-E3. Although we have not observed an interaction between STAT5A and the E3 subunit of PDC, our data cannot conclusively rule it out (Fig. 2 and data not shown).
Figure 2.
GH induces the interaction between STAT5A and multiple subunits of PDC in a dose-dependent manner. Fully differentiated 3T3-L1 adipocytes were treated with vehicle (V; 10 μm NaHCO3) or 0.625–50 nm mGH for 15 min. Monolayers were scraped into IP lysis buffer, and whole-cell lysates were prepared. IP experiments were performed using the anti-STAT5A antibody and 300 μg of lysate protein. The mock experiment contained IP antibody but no protein lysate. WB was used to examine the protein content of the immunoprecipitates (IP: STAT5A; left) and inputs (Direct WB; right). Molecular mass markers are shown to the left of each strip, and Western blotting targets are identified on the right. E1α (43 kDa), E2 (70 kDa), and E3 (55 kDa) are subunits of PDC. The expected molecular mass of STAT5A/STAT5pY is 95 kDa. This experiment was repeated at least two times on independent batches of adipocytes.
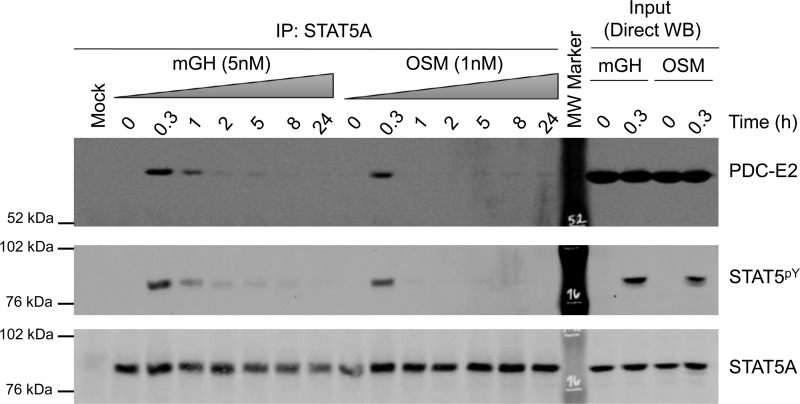
Figure 3.
GH and OSM induce the interaction between STAT5A and PDC-E2 in a time-dependent manner. Fully differentiated 3T3-L1 adipocytes were treated with mGH or OSM for the indicated times. Monolayers were scraped into IP lysis buffer, and whole-cell lysates were prepared. IP experiments were performed using the anti-STAT5A antibody and 300 μg of lysate protein. The mock experiment contained IP antibody but no lysate. Western blotting was used to examine the protein content of the immunoprecipitates (IP: STAT5A; left) and lysate inputs (Direct WB; right). This experiment was repeated three times on independent batches of adipocytes.
In the majority of our experiments, mock IPs containing all reagents except for cell lysates served as negative controls. Additionally, mock IPs comprised of an isotype control IgG and whole-cell lysates from vehicle- and mGH-stimulated 3T3-L1 adipocytes produced no bands at the molecular weight of the PDC-E2 and E1 subunits (data not shown).
We also assessed specificity by performing a time-course study with two known STAT5 activators, mGH and oncostatin M (OSM). As shown on the right in Fig. 3, an acute (0.3 h) treatment with GH or OSM resulted in STAT5 tyrosine phosphorylation without altering the levels of PDC-E2 or STAT5A expression. In addition, GH and OSM induced an association of STAT5A with PDC-E2 in a time-dependent manner that correlated with STAT5 tyrosine phosphorylation (shown on the left of Fig. 3).
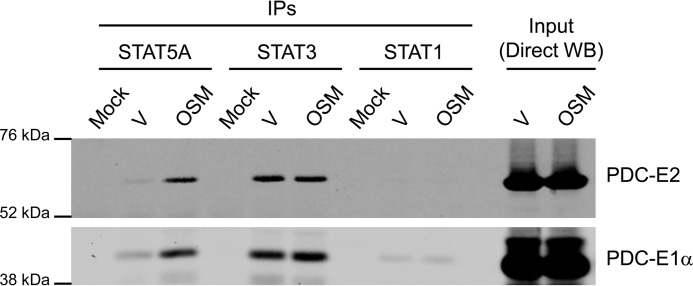
In addition to STAT5A, STAT1 and STAT3 are expressed in adipocytes and can be induced by OSM. To determine whether these other adipocyte-expressed STATs can associate with PDC-E2, we treated adipocytes for 20 min with OSM or vehicle and performed IP analysis with STAT1, STAT3, or STAT5A antibodies. As shown in Fig. 4, acute OSM stimulation induced an association between STAT5A and both E1α and E2 subunits of PDC, whereas STAT3 associated with these two PDC subunits in an OSM-independent manner. STAT1, however, did not significantly associate with either PDC subunit. We also induced STAT1 tyrosine phosphorylation with IFNγ and did not observe any association between STAT1 and PDC subunits under these conditions (data not shown).
Figure 4.
PDC subunits differentially interact with members of the STAT protein family. Fully differentiated 3T3-L1 adipocytes were treated with vehicle (V) (0.1% BSA/PBS) or 1 nm OSM for 20 min, a condition that activates STAT5, STAT3, and STAT1. Monolayers were scraped into IP lysis buffer, and whole-cell lysates were prepared. IP reactions were performed using antibodies targeted against the indicated STAT proteins and 300 μg of lysate protein. The mock experiment contained IP antibody but no lysate. Western blotting was used to examine the protein content of the immunoprecipitates (IPs; left) and lysate inputs (Direct WB; right). This experiment was repeated at least three times on independent batches of cells.
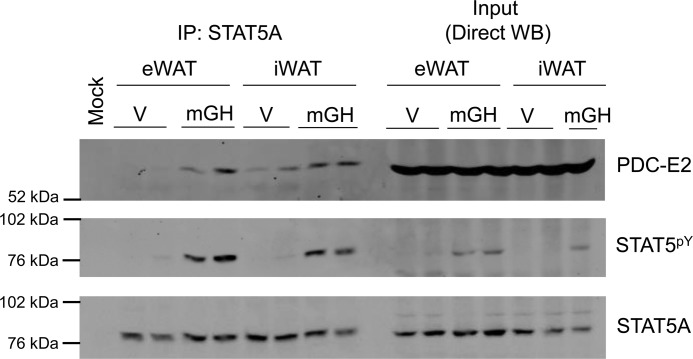
To determine whether the physical association between STAT5A and PDC-E2 could also occur in vivo, we injected mice with mGH (1.5 mg/kg) or vehicle 15 min prior to sacrifice. Epididymal white adipose tissue (eWAT) and inguinal adipose tissue (iWAT) depots were collected. Tissue homogenates were prepared and used for IP with the STAT5A antibody (Fig. 5, left) and for direct Western analysis of the IP input (Fig. 5, right). Mock contained IP antibody but no homogenate. As shown in Fig. 5, we observed a GH-induced interaction between STAT5A and PDC-E2 in both visceral (eWAT) and subcutaneous (iWAT) AT in vivo similar to our observations in cultured adipocytes (Figs. 1–4).
Figure 5.
STAT5A interacts with PDC-E2 in adipose tissue in vivo. Male C57BL/6J mice (12 weeks of age on a chow diet) were injected intraperitoneally with mGH (1.5 mg/kg) or vehicle (V: 0.1% BSA/PBS) 15 min prior to sacrifice. eWAT and iWAT white adipose tissue depots were collected in IP buffer and immediately placed on ice. eWAT and iWAT were homogenized using a Potter-Elvehjem tissue homogenizer and centrifuged. Floating lipid was removed, and the supernatants were used for IP and direct Western blotting experiments. IP experiments were performed using the anti-STAT5A antibody and 600 μg of tissue homogenate protein. The mock experiment contained IP antibody but no homogenate. Western blotting was used to examine the protein content of the immunoprecipitates (IP: STAT5A; left) and homogenate inputs (Direct WB; 150 μg of protein/lane; right). This experiment was performed one time, and two mice per condition were examined.
Several published studies have demonstrated the presence of STAT proteins in mitochondria (10, 15–19), prompting us to investigate whether a STAT5–PDC interaction could be detected in adipocyte mitochondria. Although we did visualize immunogold-labeled STAT5A localized within some mitochondria of 3T3-L1 adipocytes via transmission electron microscopy (data not shown), knocking down or activating STAT5A in adipocytes produced no significant alterations in mitochondrial gene expression or function (data not shown and supplemental Fig. S3). Based on a study in cancer cells that showed the PDC complex could be present in the nucleus (13), we employed immunofluorescence (IF) microscopy and subcellular fractionation to assess whether PDC could be present in the nucleus of fat cells. For IF microscopy, 3T3-L1 adipocytes were grown on coverslips and treated with mGH. IF was conducted using primary antibodies directed against PDC-E2 and STAT5pY. In Fig. 6, the green staining represents PDC-E2 (A) and the red staining represents tyrosine-phosphorylated STAT5 (B). Nuclei were counterstained with DAPI (Fig. 6, C and D, blue). Remarkably, we observed some PDC-E2 localized within the adipocyte nucleus as indicated by the cyan color inside the DAPI-stained nucleus (shown in the Merge panel, Fig. 6C). The yellow-orange color suggests co-localization of PDC-E2 and STAT5pY outside of the nucleus, and co-localization of PDC-E2 and STAT5pY within the adipocyte nucleus appears as white spots (Fig. 6, C and D).
Figure 6.
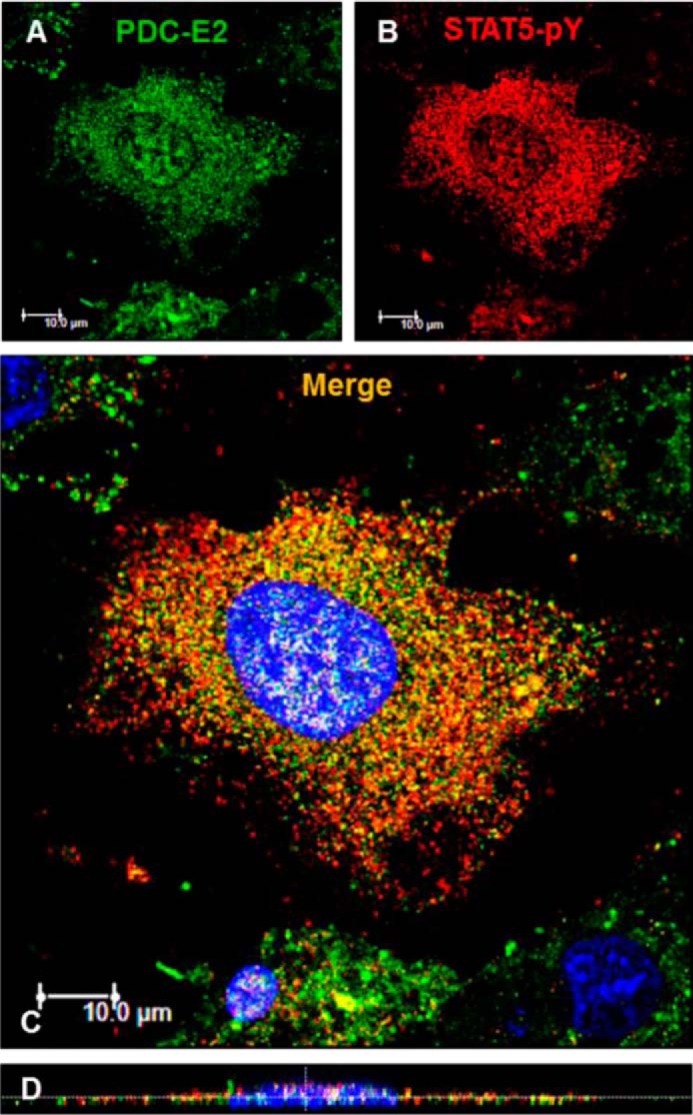
PDC-E2 interacts with STAT5pY in adipocyte nuclei. Mature 3T3-L1 adipocytes were seeded on coverslips, serum-deprived overnight, and then treated with 5 nm mGH for 20 min. After fixation, IF microscopy was conducted using primary antibodies directed against PDC-E2 (mouse primary) and STAT5pY (rabbit primary). A Dylight-488-conjugated anti-mouse secondary antibody (green; A) and an AlexaFluor 594-conjugated anti-rabbit secondary antibody (red; B) were used. Nuclei were counterstained with DAPI (4′,6-diamidino-2-phenylindole, a fluorescent stain that strongly binds DNA and stains the nucleus blue). Cell staining was imaged on a Leica TCS SP5 confocal laser-scanning fluorescence microscope using a ×63 oil immersion objective. The laser wavelengths were 405 nm (DAPI), 488 nm (green channel, 30% intensity; A), and 561 nm (red channel, 80% intensity; B). C, two-dimensional images from all three channels were merged. D, orthogonal sections were imaged using the z-stack feature (15 sections were scanned). This experiment was repeated at least two times on independent batches of cells.
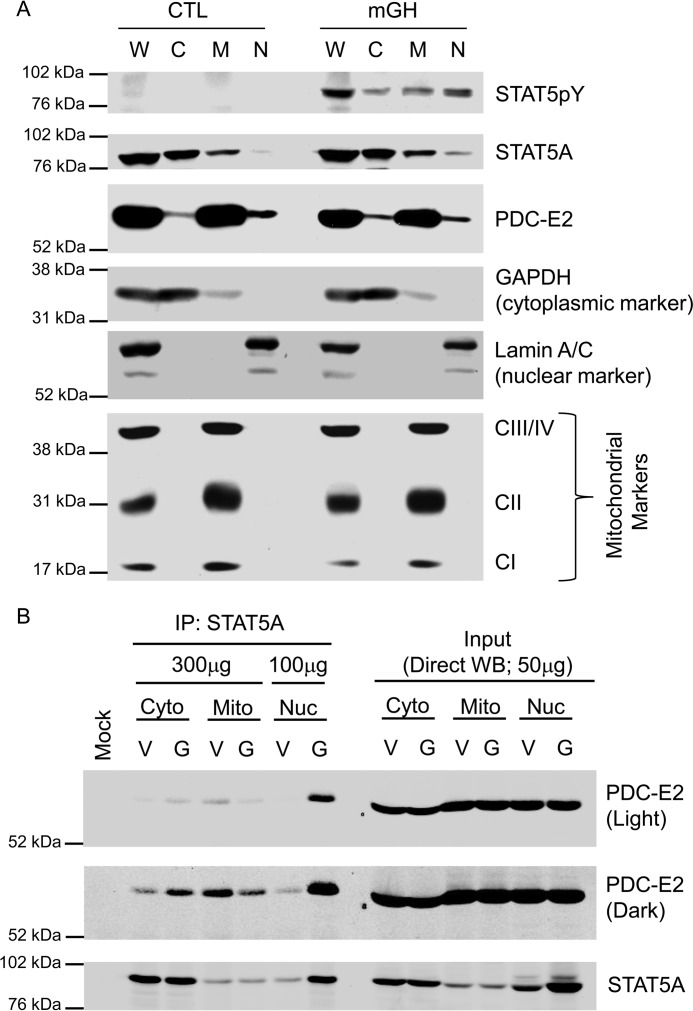
The co-localization of PDC-E2 and STAT5pY shown by confocal microscopy falls short of demonstrating whether these two proteins physically associate in the adipocyte nucleus. To confirm the presence of PDC-E2 in the adipocyte nuclei, we fractionated whole-cell extracts to obtain cytosolic, mitochondrial, and nuclear extracts. As shown in Fig. 7A, we observed STAT5pY only in GH-treated cells. We also observed that STAT5 is enriched in the cytosol, whereas PDC-E2 is enriched in the mitochondria. However, PDC-E2 was also present in the nuclei, and STAT5A was present in mitochondria. Analysis of GAPDH confirmed that there was no cytosolic contamination of the nuclear fractions, and the pattern of Lamin expression confirmed that the mitochondrial fraction was not contaminated by nuclear protein. Also, the expression of three mitochondrial markers confirmed that the nuclear fraction was not contaminated with mitochondrial proteins. To assess whether PDC-E2 and STAT5A can physically associate in the nucleus, we performed IP and Western blot analysis on cellular fractions. As shown in Fig. 7B, the expression of PDC-E2 does not change in any fraction following GH treatment. Direct Western blot analysis of the input samples also confirmed the expected increase in STAT5A in the nucleus following GH stimulation. Notably, the interaction between STAT5A and PDC-E2 occurred primarily in the nucleus.
Figure 7.
PDC-E2 is present in adipocyte nuclei where it associates with activated STAT5A. Mature 3T3-L1 adipocytes were serum-deprived overnight and then treated with 5 nm mGH (G) or vehicle (V) for 20 min. Control (CTL) plates were untreated. A, monolayers were detached using trypsin and then subjected to subcellular fractionation using a cytoplasmic, nuclear, and mitochondrial fractionation kit. Cytoplasmic (C), mitochondrial (M), and nuclear (N) fractions from each treatment group were analyzed by immunoblotting. A whole-cell lysate (W) of each treatment group was also prepared separately and analyzed alongside the fractions. CI (20 kDa), CII (30 kDa), and CIII/IV (40/48 kDa) indicate subunits of the electron transport chain complexes that were immunoblotted as mitochondrial marker proteins. GAPDH (37 kDa) and Lamin A/C (62/69 kDa) were used as cytoplasmic and nuclear markers, respectively. B, monolayers were scraped into nuclear homogenization buffer and subjected to subcellular fractionation using differential centrifugation. The cytoplasmic (Cyto), mitochondrial (Mito), and nuclear (Nuc) extracts from each treatment group were subjected to immunoprecipitation using an anti-STAT5A antibody. The mock experiment contained IP antibody but no extract. The amount of total protein used in each IP is indicated in the figure. Western blotting was used to examine the protein content of the immunoprecipitates (IP: STAT5A; left) and subcellular fraction inputs (Direct WB; 50 μg of protein/lane; right). Fractionation results were replicated at least three times on independent batches of adipocytes.
cish, or cis, is a well-established STAT5 target gene in several cells, including adipocytes. Expression of cish is acutely induced by activated STAT5 binding to its promoter, and the CISH protein acts a negative regulator to shut down JAK-STAT signaling (20–22). As shown in Fig. 8, we performed ChIP analysis to examine protein binding to an established STAT-binding site in the cish promoter. As expected, GH treatment resulted in a significant increase in STAT5 binding to the cish promoter. Notably, we also observed PDC-E2 binding to this same site in a growth hormone-dependent manner. Because the magnitude of binding was lower than we observed for STAT5, we also examined the binding of a known STAT5 transcriptional coactivator, CREB-binding protein (CBP). We observed that this coactivator bound the cish promoter at levels very similar to what we observed for PDC-E2.
Figure 8.
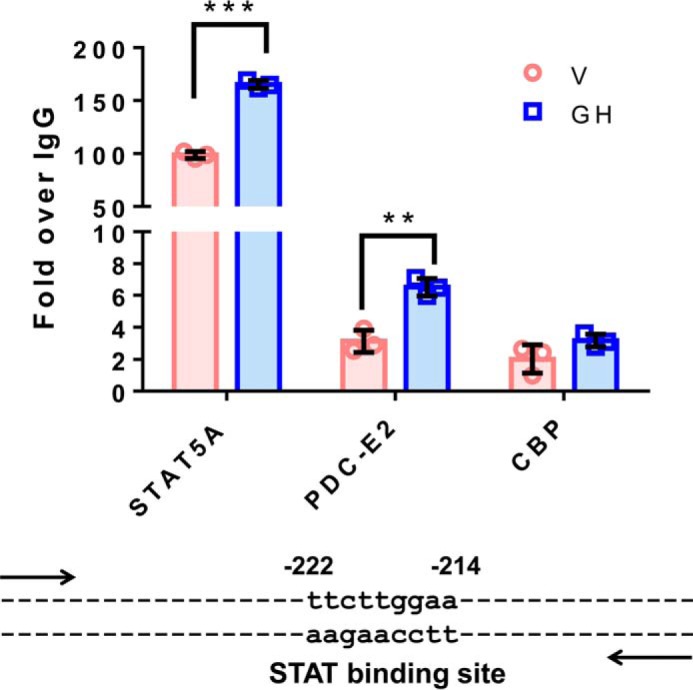
PDC-E2 binds to a STAT5 DNA-binding site within the cish promoter. Fully differentiated 3T3-L1 adipocytes were serum-deprived overnight and then treated with vehicle (V; NaHCO3) or 5 nm mGH for 20 min. Chromatin was cross-linked using DSG and 1% formaldehyde, and nuclear extracts were subjected to ChIP using anti-STAT5A, CBP, or PDC-E2 antibodies. The cish promoter DNA bound to each immunoprecipitated protein was quantified by qPCR, and percent input was calculated. Non-immune isotype-specific IgGs were used as negative controls, and all data were normalized against IgG binding and represented as fold over IgG. For each antibody, GH treatments (blue bars) were compared with vehicle (pink bars), and statistical significance was determined using the Student's t test and assigned as **, p < 0.005, and ***, p ≤ 0.001. These results have been replicated well over three times using independent batches of adipocytes. The cish gene is a well-known STAT5 target gene, and the STAT-binding site that we examined within the cish promoter is shown below the graph. The numbers above the binding site identify the location of the STAT-binding site relative to the cish transcription start site.
Discussion
Because STAT5 has a prominent role in adipocyte development and function, we sought to identify novel STAT5-interacting proteins in fat cells to further understand the role of this signaling molecule and transcription factor. To our surprise, two mitochondrial proteins belonging to the pyruvate dehydrogenase complex were identified from our unbiased screening method. Notably, both of these proteins, PDC-E1 and PDC-E2, interacted with STAT5A in a GH-regulated manner, suggesting that STAT5 only interacts with the PDC complex when it is activated by tyrosine phosphorylation. Our results clearly demonstrate that STAT5A and the E2 subunit of PDC (PDC-E2) associate in murine and human adipocytes under physiological conditions following GH stimulation (Fig. 1). We demonstrated the specificity of this interaction by showing dose and time effects concurrent with GH stimulation and STAT5 activation (Figs. 2 and 3 and supplemental Fig. S2). In addition, we observed that other STAT5 activators could also induce the physical association of STAT5A and PDC-E2 in adipocytes (Figs. 3 and 4), and we showed that STAT5A interacts with the E1α and E1β subunits of PDC as well (Figs. 2 and 4 and supplemental Fig. S2). Notably, we detected the interaction of STAT5A and PDC-E2 in both subcutaneous and visceral adipose tissue from GH-injected mice (Fig. 5). The results of these studies led to nearly 2 years of negative data as we focused on the potential function of a PDC-E2/STAT5A association in the mitochondria. Although we could confirm that STAT5A was present in the mitochondria by subcellular fractionation (Fig. 7, A and B) and by transmission electron microscopy (data not shown), we were unable to substantially perturb mitochondrial function with either GH stimulation or knockdown of STAT5A in adipocytes (supplemental Fig. S3). However, we remained confident in our observations demonstrating the association of STAT5A and PDC-E2 in adipocytes in vitro and in vivo (Figs. 1–5).
Independent studies also support an interaction of STAT5 and PDC proteins in non-adipocyte cells as a previous study had shown that STAT5 proteins can associate with PDC-E2 in a leukemic T cell line (10). These studies also demonstrated translocation of tyrosine-phosphorylated STAT5 into mitochondria and STAT5 binding to the D-loop regulatory region of mitochondrial DNA in vitro (10). Another group demonstrated that PDC can translocate from the mitochondria to the nucleus in cancer cells (13). Based on all these observations and our own, we looked for PDC in the nucleus of adipocytes. Using a variety of methods, including IF microscopy, cell fractionation, and Western blotting analysis, we confirmed that PDC complex proteins are present in the nucleus and mitochondria of cultured adipocytes (Figs. 6 and 7). Although we observed that the association of STAT5A and PDC-E2 only occurs when STAT5 is tyrosine-phosphorylated (Figs. 1–5 and 7), we have never observed a translocation of PDC-E2 to the nucleus nor a translocation of STAT5 to the mitochondria (Fig. 7B, right side of the figure). Based on our observations, some of the PDC complex appears to be present in the nucleus under basal conditions and can associate with STAT5A (Figs. 7 and 8).
Similar to our studies with GH in adipocytes, the association of either STAT5A or STAT5B with PDC-E2 in BaF3 pro-B cells was IL-3-dependent and correlated with STAT5 tyrosine phosphorylation (11). These authors also suggest that PDC-E2 may act as a coactivator of STAT5 based on data showing that PDC-E2 overexpression in BaF3 cells increased the expression of socs3 (suppressor of cytokine signaling 3) (11), a well-characterized STAT target gene. In addition to STAT5, we observed that STAT3 could also associate with PDC-E1α and E2 in adipocytes under stimulated and unstimulated conditions (Fig. 4). However, we did not evaluate the subcellular location of this interaction. There is published evidence that STAT3 can be present in the mitochondria (15, 17, 19, 23, 24) or can translocate to the mitochondria to regulate electron transport activity (18, 19, 24). A recent paper (12) has shown that in the mitochondria of A549 lung cancer cells, STAT3 associates with the PDC-E1 subunit to enhance the conversion of pyruvate to acetyl-CoA and to elevate the mitochondrial membrane potential, which promotes ATP synthesis. Although we cannot rule out a role of STAT3 in the mitochondrial function of PDC in adipocytes, our results strongly suggest that the association of PDC complex proteins with STAT5A in adipocytes is likely due to the interaction of these proteins in the nucleus.
As indicated above, recent studies indicate that PDC can be present in the nucleus to generate acetyl-CoA as a substrate in histone acetylation (13). Of note, there is already evidence that other acetyl-CoA-generating enzymes can be present in the nucleus (25) and that the acetyl-CoA-generating enzyme, ATP-citrate lyase, can link cellular metabolism to histone acetylation (26). The effects of a high-fat diet (HFD) on acyl-CoAs and histone acetylation have recently been investigated in mouse white adipose tissue (WAT). Mice consuming a HFD have reduced levels of acetyl-CoA in WAT that are associated with decreased histone acetylation (27). In addition, genetic deletion of ATP-citrate lyase (acly) in cultured adipocytes reduced both acetyl-CoA and histone acetylation levels (27). Collectively, these studies suggest an important role for acetyl-CoA-generating enzymes in the nucleus. Although it is unclear whether the PDC complex makes a substantial contribution to histone or protein acetylation in the nucleus, our studies suggest that, like ATP-citrate lyase, the PDC complex might also affect acetyl-CoA levels in the nucleus of adipocytes as it does in cancer cells (13). Several proteins that contain mitochondrion-targeting sequences also reside in the nucleus (28). Also, the evidence that there are multiple acetyl-CoA-generating enzymes in the nucleus suggests that this process is regulated, dynamic, and likely to be important (25).
In summary, an unbiased MS-based approach identified PDC-E1β and E2 as proteins that could associate with STAT5. Although studies in BaF3 cells had previously shown an interaction between STAT5 proteins and PDC-E2 (10, 11), our studies are the first observation that STAT5A can interact with PDC-E2 in adipocytes in a manner that highly correlates with STAT5 tyrosine phosphorylation (Figs. 1–5). Moreover, this interaction occurs in human adipocytes (Fig. 1), as well as in subcutaneous and visceral mouse adipose tissue (Fig. 5). The interaction of STAT3 with subunits of the PDC complex is distinct from that of STAT5 in that it is not coincident with STAT3 activation by tyrosine phosphorylation (Fig. 4). The subcellular localization of PDC-E2 was verified by independent methods, and we demonstrated that PDC-E2 is present in the adipocyte nucleus where it interacts with activated STAT5A (Figs. 6 and 7). Finally, our highly novel observations reveal that PDC-E2 is part of the GH-induced STAT5-binding complex that regulates the promoter of the cish gene (Fig. 8).
Future studies will be complex because knockdown of PDC proteins and the resulting disruption of its mitochondrial activity would greatly impact cellular function. Our immunoprecipitation results clearly demonstrate an inducible interaction between STAT5 and multiple subunits of PDC (E2, E1α, and E1β). We also show that PDC-E2 localizes within the adipocyte nucleus where it associates with STAT5 and can bind to STAT5 DNA-binding sites. Previously published studies demonstrate that the entire PDC complex localizes within cancer cell nuclei where it generates acetyl-CoA to modulate histone acetylation (13). These findings together with our data suggest that STAT5 likely interacts with the entire PDC complex, which may provide a local pool of acetyl-CoA to regulate gene expression by modulating either STAT5 acetylation, histone acetylation, or the acetylation of other proteins present in the STAT5 DNA-binding complex. Studies are ongoing to test this hypothesis. Moreover, our data do not reveal whether or not there is a direct physical interaction between STAT5 and any of the PDC subunits. Further studies are needed to identify specific interaction surfaces between STAT5 and one or more of the PDC subunits.
Although we demonstrated that PDC-E2 could bind to one STAT5-binding site, we predict that the PDC complex also associates with other transcription factors where it can contribute to protein acetylation. It will be interesting to determine what proportion of STAT5-binding sites contains PDC as part of the DNA-binding complex. Overall, our studies demonstrate that components of the PDC complex, in particular PDC-E2, can be present in adipocyte nuclei and be part of a DNA-binding complex in the cish promoter. These novel observations may shift our perspectives on the role of metabolic enzymes as well as their cellular locations and functions.
Experimental procedures
Cell culture
Murine 3T3-L1 preadipocytes were grown to 2 days after confluence in Dulbecco's modified Eagle's media (DMEM; from Sigma) with 10% bovine calf serum. The preadipocytes were induced to differentiate using a standard protocol and MDI induction mixture containing 3-isobutylmethylxanthine, dexamethasone, insulin and 10% fetal bovine serum (FBS) in DMEM (29). HyClone calf and fetal bovine sera were purchased from Thermo Fisher Scientific or GE Healthcare. The medium was changed every 48–72 h during growth and differentiation. Cells were serum-deprived by changing the medium to DMEM containing 0.3% bovine serum albumin (BSA; catalog no. A6003, Sigma) for 16–24 h before treatment with mGH, OSM, or appropriate vehicle. Recombinant murine GH (catalog no. CYT-540) was purchased from ProSpec (East Brunswick, NJ) or Dr. A. F. Parlow at the National Hormone and Peptide Program (NHPP; Torrance, CA). GH from either source yielded similar results. Recombinant OSM (catalog no. 495-MO-025/CF) was from R&D Systems (Minneapolis, MN). For experiments in human adipocytes, human omental adipocytes from subjects with a body mass index (BMI) greater than 30 (Item no. OA-1012-3) were purchased in 12-well plates from Zen-Bio (Research Triangle Park, NC). Human adipocytes were maintained as instructed by Zen-Bio in Omental Adipocyte Medium Human (catalog no. OM-AM) upon receipt in our laboratory until use within 3 weeks. Prior to treatment with hGH, the cells were serum-deprived in Omental Basal Medium (catalog no. OM-BM) for 16–24 h. HGH/HumanKine® (catalog no. H5916) was from Sigma.
Preparation of whole-cell extracts
Cell monolayers were rinsed once with phosphate-buffered saline (PBS) and then scraped into non-denaturing IP buffer that contained 10 mm Tris (pH 7.4), 150 mm NaCl, 1 mm EGTA, 1 mm EDTA, 1% Triton X-100, 0.5% IGEPAL CA-630, protease inhibitors (1 mm phenylmethylsulfonyl fluoride, 1 μm pepstatin, 50 trypsin inhibitory milliunits of aprotinin, 10 μm leupeptin, 1 mm 1,10-phenanthroline), and phosphatase inhibitor (0.2 mm sodium vanadate). The cell suspension was incubated on ice for at least 30 min or subjected to a freeze/thaw cycle at −80 °C and then passed through a 20-gauge needle three times. The whole-cell extract was clarified by centrifugation at 13,000 × g for 10 min at 4 °C.
Subcellular fractionation
Cell monolayers from ten 10-cm culture plates were scraped into NHB buffer composed of 20 mm Tris (pH 7.4), 10 mm NaCl, 3 mm MgCl2 and the same protease and phosphatase inhibitors used for IP buffer. After adding 0.15% IGEPAL CA-630 (from 10% stock) to the cell suspension, it was homogenized on ice using 10–16 strokes in a Dounce homogenizer. The extract was centrifuged at 2400–3500 rpm in a Beckman GS-6KR centrifuge with a swinging bucket rotor. Subsequently, the supernatant containing cytosol and mitochondria was centrifuged again at 18,000 × g for 30 min at 4 °C to pellet mitochondria, which was then resuspended in IP buffer, whereas the supernatant containing cytosol was transferred to a fresh tube. The nuclear pellet from the first centrifugation was washed once with half of the initial volume of NHB buffer and re-centrifuged at 4 °C for 5 min at 540 rpm in the Beckman GS-6KR centrifuge. The supernatant was discarded, and the nuclear pellet was resuspended in IP buffer and incubated on ice for 0.5–1 h. To break open the nuclei, the nuclear extract was passed through a 20-gauge needle four times and clarified by centrifuging at 13,000 × g for 10 min at 4 °C. To separate cytosol, mitochondria, and nuclear fractions, we also utilized the Abcam cell fractionation kit (catalog no. ab109719, Cambridge, MA) according to the manufacturer's instructions with the following modification for cell type. We fractionated confluent, mature 3T3-L1 adipocytes grown and differentiated on 10-cm plates. Two to five plates per treatment group were used, and the cells were detached using 1.5 ml of 0.25% trypsin/EDTA per plate. Trypsin was inactivated with 4 ml of DMEM containing 10% calf serum per plate prior to collecting the cells by centrifugation. Manufacturer's instructions were followed for the remainder of the protocol.
Preparation of adipose tissue extracts from mice
Chow-fed 12-week-old C57BL6/J male mice from a breeding colony at Pennington Biomedical Research Center were administered either a vehicle composed of 0.1% BSA in PBS or 1.5 mg/kg mGH in a volume of 240–300 μl via intraperitoneal injection. At the time of injection, the mice weighed 24–30 g. Fifteen minutes after injection, mice were then euthanized by carbon dioxide asphyxiation and cervical dislocation. Epididymal and inguinal (lymph nodes removed) white adipose tissue (WAT) depots were immediately excised and placed in IP buffer (with 2× concentration of protease and phosphatase inhibitors) on ice. This experiment was approved by the Institutional Animal Care and Use Committee at Pennington Biomedical Research Center. The WAT depots were homogenized on ice using a Potter-Elvehjem tissue homogenizer and centrifuged at 10,000 × g for 10 min at 4 °C. The floating lipid layer was removed, and the remaining whole-tissue extract was used for immunoprecipitation and immunoblotting.
Measurement of protein concentration
Protein content of cell and tissue extracts was quantified using the bicinchoninic acid (BCA) assay kit from Sigma (catalog no. BCA1).
Gel electrophoresis and immunoblotting
Samples were separated on 7.5 or 10% SDS-polyacrylamide gels (acrylamide, catalog no. EC-890, from National Diagnostics, Atlanta, GA) and transferred to nitrocellulose membranes (catalog no. 162-0115, Bio-Rad) as described previously (30). Results were visualized with horseradish peroxidase-conjugated secondary antibodies (Jackson ImmunoResearch) and enhanced chemiluminescence (Pierce/Thermo Fisher Scientific) (30) or IRDye-conjugated secondary antibodies (Licor Biosciences, Lincoln, NE) and scanned with the Odyssey infrared scanner (Licor Biosciences) (31).
Immunoprecipitation
Cell or tissue extracts (300–600 μg of total protein) were incubated with 4–5 μg of immunoprecipitating antibody overnight on a mini-tube rotator at 4 °C. Protein A-conjugated agarose (IPA300 protein A resin; catalog no. 10-2003-02) from Repligen or protein A/G plus-agarose (catalog no. sc-2003) beads from Santa Cruz Biotechnology (Dallas, TX) were added to the antibody/epitope mixture, and the conjugation reaction proceeded for an additional 1–3 h at 4 °C with rotation. Following conjugation to the bead resin, the beads were pelleted by centrifugation at 13,000 rpm for 1–3 min at 4 °C. The supernatant was removed by aspiration, and the beads were washed three times with ice-cold 1× IP buffer. Between each wash, the beads were pelleted by centrifugation at 10,000 rpm for 1–3 min at 4 °C, and the supernatant was removed by aspiration. After the final wash, the IP antibody and immunoprecipitated proteins were eluted from the bead resin into 2× SDS loading buffer (LB) by boiling the samples for 10 min at 95–100 °C. Samples were flicked multiple times during the heat step to ensure efficient elution. The samples were briefly centrifuged, and the supernatants were analyzed by SDS-PAGE and immunoblotting. A mock sample containing no IP antibody, no sample, or a non-immune isotype control antibody was used as a negative control for each IP experiment.
Antibodies
Anti-STAT5A (L-20; sc-1081; rabbit polyclonal), PDC-E2 (C-9; sc-271534; mouse monoclonal), Lamin A/C (N-18; sc-6215; goat polyclonal), CBP (C-20; sc-583; rabbit polyclonal), and STAT3 (C-20; sc-482; rabbit polyclonal) antibodies were purchased from Santa Cruz Biotechnology. For ChIP applications, the TransCruz formulation of the anti-STAT5A and CBP antibodies was used. Anti-STAT1 rabbit polyclonal IgG (catalog no. 9172) was from Cell Signaling Technology. Anti-PDC-E1α (8D10E3; ab110334; mouse monoclonal) and GAPDH (ab9485; rabbit polyclonal) antibodies, and the MitoProfile® Total OXPHOS Rodent WB antibody mixture (ab110413; mouse monoclonal) were from Abcam. We used both mouse monoclonal (clone 8-5-2; 05-495) and rabbit polyclonal (07-586) anti-phospho-STAT5 A/B (Tyr-694/699) antibodies from Millipore to detect tyrosine-phosphorylated STAT5 (STAT5pY). Anti-PDC-E3 (DLD; PA5-27367; rabbit polyclonal) IgG and the non-immune isotype control mouse (catalog no. 31878) and rabbit (catalog no. PA5-23094) IgGs were purchased from Thermo Fisher Scientific.
IF microscopy
Mature 3T3-L1 adipocytes were seeded on coverslips, serum-deprived overnight in 0.3% BSA/DMEM, and then treated with 5 nm mGH for 20 min. After fixation with 3.7% paraformaldehyde (Electron Microscopy Services, Hatfield, PA; catalog no. 15710) and blocking with 10% BSA, IF was conducted using primary antibodies directed against PDC-E2 (mouse) and STAT5-pY (rabbit). A Dylight 488-conjugated anti-mouse antibody (catalog no. 35502) and an AlexaFluor 594-conjugated anti-rabbit antibody (catalog no. A-11037) from Thermo Fisher Scientific were used as secondary antibodies. IF microscopy using secondary antibody only and non-immune mouse- or rabbit-isotype control primary antibodies were performed to confirm the absence of non-specific binding. Nuclei were counterstained with DAPI (Thermo Fisher Scientific; catalog no. 62248). Cell staining was imaged on a Leica TCS SP5 confocal laser-scanning fluorescence microscope using a ×63 oil immersion objective. The lasers for wavelengths 405 nm (DAPI) and 488 nm (green channel) were set at 30% intensity and 80% for 561 nm (red channel). Two-dimensional images from all three channels were merged. Orthogonal sections were imaged using the z-stack feature (15 sections were scanned).
ChIP
Mature 3T3-L1 adipocytes (7–15 days post-MDI) were serum-deprived in 0.3% BSA/DMEM for 18–24 h prior to treatment with vehicle or mGH for 20 min. Seven to nine 10-cm plates per treatment group were used (∼4 × 107 cells). ChIP experiments were performed using the SimpleChIP® kit (Magnetic Beads) from Cell Signaling Technology (catalog no. 9003S; Danvers, MA) according to the manufacturer's instructions with a few modifications. Briefly, to cross-link cofactors and direct DNA-binding proteins to DNA, cells were incubated with 2 mm disuccinimidyl glutarate (DSG; catalog no. 20593) from Thermo Fisher Scientific for 30 min at room temperature, followed by incubation with 1% formaldehyde (catalog no. 252549) from Sigma for 10 min at room temperature. The cross-linking reaction was quenched by the addition of glycine provided in the kit. For adherent adipocytes, the medium was aspirated, and plates were rinsed two times with ice-cold PBS. Cells from each plate were scraped into 1 ml of ice-cold PBS + PIC (protease inhibitor mixture provided with the kit) and for each treatment combined in a 15-ml conical tube and centrifuged as directed. Nuclei preparation, chromatin digestion, and ChIP assays were performed as indicated in the manufacturer's instructions. For each ChIP, 15–30 μg of digested, cross-linked chromatin were incubated with 2 μg of specific antibody (rabbit anti-STAT5A, mouse anti-PDC-E2, and rabbit anti-CBP) or non-immune IgG control antibody (rabbit IgG or mouse IgG). Input and immunoprecipitated DNA were quantitated using the real-time quantitative PCR (qPCR) method in a total volume of 10 μl (2 μl of DNA and 8 μl of reaction master mix) using an Applied Biosystems 7900HT Fast Real-Time PCR system. We used the qPCR amplification program specified in the kit instructions with the exception that the initial denaturation step was performed for 10 min at 95 °C. The primer sequences used to generate the amplicon (−278/−167) containing the STAT5-binding site within the cish promoter were 5′-CGTCCAGCGATACGATTGGT-3′ (forward) and 5′-CAGGCGTCTAGTGCTTTGGA-3′ (reverse). To confirm specificity of binding to the STAT5 DNA-binding site, the following primer sequences were used to target a section within the cish-coding region (+3992/+4058) that lacks the STAT DNA-binding consensus sequence, 5′-TACCCCTTCCAACTCTGACTGAGC-3′ (forward) and 5′-TTCCCTCCAGGATGTGACTGTG-3′ (reverse). Amplification efficiency for each reaction was determined using a standard curve prepared from serial dilutions of the 2% input chromatin DNA. ChIP efficiencies were calculated using the percent input method and the following equation: percent input = 2% × 2(C(T) 2% input sample − C(T) IP sample), where C(T) = threshold cycle of the PCR.
Statistical analyses
Data for ChIP analyses were plotted as mean ± S.D., and t tests were performed to assess statistical significance. Differences with p < 0.05 were considered statistically significant.
Author contributions
A. J. R. and J. M. S. designed the experiments. A. J. R., H. H., and J. M. S. conducted the experiments. A. J. R. and H. H. analyzed the results. J. M. S. and A. J. R. wrote the majority of the manuscript, and H. H. edited the paper.
Supplementary Material
Acknowledgments
We thank Anik Boudreau for technical support and manuscript editing. We are also grateful to Drs. Randy Mynatt, David Burk, and Rob Noland for sharing resources and equipment and for providing technical advice that helped us assess the association of PDC with STAT5A in vitro and in vivo. This project used Genomics and Cell Biology and Bioimaging core facilities that are supported in part by COBRE (NIH8 1P30GM118430-01) and NORC (NIH 1P30-DK072476) center grants from the National Institutes of Health.
This work was supported by National Institutes of Health Grant R01DK052968-17 (to J. M. S.). The authors declare that they have no conflicts of interest with the contents of this article. The content is solely the responsibility of the authors and does not necessarily represent the official views of the National Institutes of Health.
This article contains supplemental Figs. S1–S3 and Experimental procedures.
- PDC
- pyruvate dehydrogenase complex
- mGH
- murine growth hormone
- hGH
- human growth hormone
- OSM
- oncostatin M
- IP
- immunoprecipitation
- IF
- immunofluorescence
- DSG
- disuccinimidyl glutarate
- eWAT
- epididymal white adipose tissue
- iWAT
- inguinal white adipose tissue
- cish
- cytokine-inducible SH2-containing protein
- CBP
- CREB-binding protein
- HFD
- high-fat diet
- CREB
- cAMP-response element-binding protein
- SH
- Src homology
- WB
- Western blotting
- BMI
- body mass index
- qPCR
- quantitative PCR.
References
- 1. Richard A. J., and Stephens J. M. (2011) Emerging roles of JAK-STAT signaling pathways in adipocytes. Trends Endocrinol. Metab. 22, 325–332 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2. Stewart W. C., Pearcy L. A., Floyd Z. E., and Stephens J. M. (2011) STAT5A expression in Swiss 3T3 cells promotes adipogenesis in vivo in an athymic mice model system. Obesity 19, 1731–1734 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3. Teglund S., McKay C., Schuetz E., van Deursen J. M., Stravopodis D., Wang D., Brown M., Bodner S., Grosveld G., and Ihle J. N. (1998) Stat5a and Stat5b proteins have essential and nonessential, or redundant, roles in cytokine responses. Cell 93, 841–850 [DOI] [PubMed] [Google Scholar]
- 4. Hogan J. C., and Stephens J. M. (2005) The regulation of fatty acid synthase by STAT5A. Diabetes 54, 1968–1975 [DOI] [PubMed] [Google Scholar]
- 5. White U. A., Maier J., Zhao P., Richard A. J., and Stephens J. M. (2016) The modulation of adiponectin by STAT5-activating hormones. Am. J. Physiol. Endocrinol. Metab. 310, E129–E136 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6. Patel M. S., and Roche T. E. (1990) Molecular biology and biochemistry of pyruvate dehydrogenase complexes. FASEB J. 4, 3224–3233 [DOI] [PubMed] [Google Scholar]
- 7. Zhou Z. H., Liao W., Cheng R. H., Lawson J. E., McCarthy D. B., Reed L. J., and Stoops J. K. (2001) Direct evidence for the size and conformational variability of the pyruvate dehydrogenase complex revealed by three-dimensional electron microscopy. The “breathing” core and its functional relationship to protein dynamics. J. Biol. Chem. 276, 21704–21713 [DOI] [PubMed] [Google Scholar]
- 8. Hiromasa Y., Fujisawa T., Aso Y., and Roche T. E. (2004) Organization of the cores of the mammalian pyruvate dehydrogenase complex formed by E2 and E2 plus the E3-binding protein and their capacities to bind the E1 and E3 components. J. Biol. Chem. 279, 6921–6933 [DOI] [PubMed] [Google Scholar]
- 9. Sugden M. C., and Holness M. J. (2003) Recent advances in mechanisms regulating glucose oxidation at the level of the pyruvate dehydrogenase complex by PDKs. Am. J. Physiol. Endocrinol. Metab. 284, E855–E862 [DOI] [PubMed] [Google Scholar]
- 10. Chueh F. Y., Leong K. F., and Yu C. L. (2010) Mitochondrial translocation of signal transducer and activator of transcription 5 (STAT5) in leukemic T cells and cytokine-stimulated cells. Biochem. Biophys. Res. Commun. 402, 778–783 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11. Chueh F. Y., Leong K. F., Cronk R. J., Venkitachalam S., Pabich S., and Yu C. L. (2011) Nuclear localization of pyruvate dehydrogenase complex-E2 (PDC-E2), a mitochondrial enzyme, and its role in signal transducer and activator of transcription 5 (STAT5)-dependent gene transcription. Cell. Signal. 23, 1170–1178 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12. Xu Y. S., Liang J. J., Wang Y., Zhao X. J., Xu L., Xu Y. Y., Zou Q. C., Zhang J. M., Tu C. E., Cui Y. G., Sun W. H., Huang C., Yang J. H., and Chin Y. E. (2016) STAT3 undergoes acetylation-dependent mitochondrial translocation to regulate pyruvate metabolism. Sci. Rep. 6, 39517. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13. Sutendra G., Kinnaird A., Dromparis P., Paulin R., Stenson T. H., Haromy A., Hashimoto K., Zhang N., Flaim E., and Michelakis E. D. (2014) A nuclear pyruvate dehydrogenase complex is important for the generation of acetyl-CoA and histone acetylation. Cell 158, 84–97 [DOI] [PubMed] [Google Scholar]
- 14. Kheterpal I., Coleman L., Ku G., Wang Z. Q., Ribnicky D., and Cefalu W. T. (2010) Regulation of insulin action by an extract of Artemisia dracunculus L. in primary human skeletal muscle culture: a proteomics approach. Phytother. Res. 24, 1278–1284 [DOI] [PubMed] [Google Scholar]
- 15. Boengler K., Hilfiker-Kleiner D., Heusch G., and Schulz R. (2010) Inhibition of permeability transition pore opening by mitochondrial STAT3 and its role in myocardial ischemia/reperfusion. Basic Res. Cardiol. 105, 771–785 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16. Khan R., Lee J. E., Yang Y. M., Liang F. X., and Sehgal P. B. (2013) Live-cell imaging of the association of STAT6-GFP with mitochondria. PLoS ONE 8, e55426. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17. Szczepanek K., Chen Q., Larner A. C., and Lesnefsky E. J. (2012) Cytoprotection by the modulation of mitochondrial electron transport chain: the emerging role of mitochondrial STAT3. Mitochondrion 12, 180–189 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18. Tammineni P., Anugula C., Mohammed F., Anjaneyulu M., Larner A. C., and Sepuri N. B. (2013) The import of the transcription factor STAT3 into mitochondria depends on GRIM-19, a component of the electron transport chain. J. Biol. Chem. 288, 4723–4732 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19. Wegrzyn J., Potla R., Chwae Y. J., Sepuri N. B., Zhang Q., Koeck T., Derecka M., Szczepanek K., Szelag M., Gornicka A., Moh A., Moghaddas S., Chen Q., Bobbili S., Cichy J., et al. (2009) Function of mitochondrial Stat3 in cellular respiration. Science 323, 793–797 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20. Davey H. W., McLachlan M. J., Wilkins R. J., Hilton D. J., and Adams T. E. (1999) STAT5b mediates the GH-induced expression of SOCS-2 and SOCS-3 mRNA in the liver. Mol. Cell. Endocrinol. 158, 111–116 [DOI] [PubMed] [Google Scholar]
- 21. Ling C., and Billig H. (2001) PRL receptor-mediated effects in female mouse adipocytes: PRL induces suppressors of cytokine signaling expression and suppresses insulin-induced leptin production in adipocytes in vitro. Endocrinology 142, 4880–4890 [DOI] [PubMed] [Google Scholar]
- 22. Matsumoto A., Masuhara M., Mitsui K., Yokouchi M., Ohtsubo M., Misawa H., Miyajima A., and Yoshimura A. (1997) CIS, a cytokine inducible SH2 protein, is a target of the JAK-STAT5 pathway and modulates STAT5 activation. Blood 89, 3148–3154 [PubMed] [Google Scholar]
- 23. Gough D. J., Corlett A., Schlessinger K., Wegrzyn J., Larner A. C., and Levy D. E. (2009) Mitochondrial STAT3 supports Ras-dependent oncogenic transformation. Science 324, 1713–1716 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24. Szczepanek K., Chen Q., Derecka M., Salloum F. N., Zhang Q., Szelag M., Cichy J., Kukreja R. C., Dulak J., Lesnefsky E. J., and Larner A. C. (2011) Mitochondrial-targeted signal transducer and activator of transcription 3 (STAT3) protects against ischemia-induced changes in the electron transport chain and the generation of reactive oxygen species. J. Biol. Chem. 286, 29610–29620 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25. Choudhary C., Weinert B. T., Nishida Y., Verdin E., and Mann M. (2014) The growing landscape of lysine acetylation links metabolism and cell signalling. Nat. Rev. Mol. Cell Biol. 15, 536–550 [DOI] [PubMed] [Google Scholar]
- 26. Wellen K. E., Hatzivassiliou G., Sachdeva U. M., Bui T. V., Cross J. R., and Thompson C. B. (2009) ATP-citrate lyase links cellular metabolism to histone acetylation. Science 324, 1076–1080 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27. Carrer A., Parris J. L., Trefely S., Henry R. A., Montgomery D. C., Torres A., Viola J. M., Kuo Y. M., Blair I. A., Meier J. L., Andrews A. J., Snyder N. W., and Wellen K. E. (2017) Impact of a high-fat diet on tissue acyl-CoA and histone acetylation levels. J. Biol. Chem. 292, 3312–3322 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28. Monaghan R. M., and Whitmarsh A. J. (2015) Mitochondrial proteins moonlighting in the nucleus. Trends Biochem. Sci. 40, 728–735 [DOI] [PubMed] [Google Scholar]
- 29. Richard A. J., Amini Z. J., Ribnicky D. M., and Stephens J. M. (2012) St. John's Wort inhibits insulin signaling in murine and human adipocytes. Biochim. Biophys. Acta 1822, 557–563 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30. Richard A. J., Amini-Vaughan Z., Ribnicky D. M., and Stephens J. M. (2013) Naringenin inhibits adipogenesis and reduces insulin sensitivity and adiponectin expression in adipocytes. Evid. Based Complement Alternat. Med. 2013, 549750. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31. Fuller S., Richard A. J., Ribnicky D. M., Beyl R., Mynatt R., and Stephens J. M. (2014) St. John's Wort has metabolically favorable effects on adipocytes in vivo. Evid. Based Complement Alternat. Med. 2014, 862575. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.