Abstract
It is well known that the reactive oxygen species NO can trigger cell death in plants and other organisms, but the underlying molecular mechanisms are not well understood. Here we provide evidence that NO may trigger cell death in tomato (Solanum lycopersicum) by inhibiting the activity of phosphoinositide-dependent kinase 1 (SlPDK1), a conserved negative regulator of cell death in yeasts, mammals, and plants, via S-nitrosylation. Biotin-switch assays indicated that SlPDK1 is a target of S-nitrosylation. Moreover, the kinase activity of SlPDK1 was inhibited by S-nitrosoglutathione in a concentration-dependent manner, indicating that SlPDK1 activity is abrogated by S-nitrosylation. The S-nitrosoglutathione–induced inhibition was reversible in the presence of a reducing agent but additively enhanced by hydrogen peroxide (H2O2). Our LC-MS/MS analyses further indicated that SlPDK1 is primarily S-nitrosylated on a cysteine residue at position 128 (Cys128), and substitution of Cys128 with serine completely abolished SlPDK1 kinase activity, suggesting that S-nitrosylation of Cys128 is responsible for SlPDK1 inhibition. In summary, our results establish a potential link between NO-triggered cell death and inhibition of the kinase activity of tomato PDK1.
Keywords: cell death, nitric oxide, nitrosylation, phosphorylation, protein kinase, phosphoinositide-dependent kinase
Introduction
NO potentiates the induction of hypersensitive cell death in soybean cells by reactive oxygen species (ROS)2 (1). However, the molecular mechanism of the NO-induced cell death remains an enigma. One potential mechanism is that the activity of proteins that control cell death may be altered by a posttranslational modification, S-nitrosylation. Functional groups containing a nitroso group attached to the sulfur atom of a thiol are called S-nitrosothiols (SNOs) (2). The addition of a nitroso group to a sulfur atom of a Cys residue of a protein is known as S-nitrosylation (3). S-nitrosylation is an enzyme-independent, posttranslational, and labile modification that can function as an on/off switch of protein activity (3–5). In cells, the extent of S-nitrosylation of proteins is largely determined by the level of S-nitrosoglutathione (GSNO), which serves as a stable and mobile NO pool and effectively transduces NO signaling (3, 4). S-nitrosoglutathione reductase (GSNOR) catalyzes GSNO reduction and thus regulates the cellular levels of both GSNO and protein S-nitrosylation (6). Loss of GSNOR leads to increased cellular levels of S-nitrosylated proteins (6, 7). Thousands of diverse classes of proteins, both in plants and in mammals, have been identified as targets of S-nitrosylation (8–12). In plants, proteins with diverse functions are S-nitrosylated at specific Cys residue(s), and their functions are either inhibited or enhanced by this modification (13–28).
3-Phosphoinositide-dependent protein kinase 1 (PDK1) and its downstream target, protein kinase B (PKB, also known as Akt), are central regulators of mammalian apoptosis (29–31). PKB is a member of the AGC family of protein kinases, which is activated by second messengers such as phospholipids and Ca2+ through PDK1. Mammalian PDK1 phosphorylates PKB to promote its function in suppressing programmed cell death (30–33). PKB negatively regulates apoptosis by phosphorylation and inactivation of pro-apoptotic factors such as BAD (Bcl-2-associated death) and activation of anti-apoptotic factors such as cAMP-response element-binding protein and IκB kinase (IKK) (30–32, 34). Deficiency of the PDK1 gene(s) in Drosophila (35), mice (36), yeast (37, 38), and tomato (39), respectively, results in lethality or severe apoptosis. PKB knock-out mice display spontaneous apoptosis in several different tissues (40). In tomato, the PKB/Akt homolog Adi3 (AvrPto-dependent Pto-interacting protein 3) physically interacts with and is phosphorylated by SlPDK1 (39). Silencing both SlPDK1 and Adi3 or treatment with a PDK1 inhibitor results in MAPKK kinase α–dependent cell death, indicating that Adi3 functions analogously to the mammalian PKB/Akt by negatively regulating cell death via PDK1 phosphorylation (39).
Yasukawa et al. (41) showed that NO donors induced S-nitrosylation and inactivation of PKB/Akt kinase activity in vitro and in vivo and that the mutant Akt1/PKB (224Cys→Ser) was resistant to S-nitrosylation by NO and its kinase inactivation (41). Although the NO and PDK1-PKB/Akt pathways are both key regulators of cell death, the link between these two pathways has not been firmly established in plants. Here we show that the kinase activity of tomato SlPDK1 was inhibited by GSNO in a concentration-dependent manner and that this inhibition was additively enhanced by H2O2. Interestingly, we determined that SlPDK1 was S-nitrosylated at Cys128. This Cys residue is critical for the function of SlPDK1 because a mutation of Cys128 to Ser completely abolished its kinase activity. Our results suggest that inhibition of SlPDK1 activity by S-nitrosylation at Cys128 might be a key molecular event underpinning NO-triggered cell death.
Results
Spontaneous cell death occurs in GSNOR1-silenced tomato plants
NO triggers cell death in soybean suspension cells (1). While investigating the role of GSNOR1 in disease resistance, we observed that spontaneous cell death frequently occurred in the leaves of GSNOR1-silenced tomato plants (Fig. 1), suggesting that GSNOR1 negatively regulates cell death. Increased cell death was also observed in GSNOR1 knock-out mice and Arabidopsis (19, 42) that had significantly higher levels of protein S-nitrosylation (7, 42). Our experiments, coupled with the previous studies, prompted us to postulate that the cell death observed in GSNOR1 knock-out (19) or knock-down plants (Fig. 1) was caused by S-nitrosylation of key negative regulator(s) of cell death.
Figure 1.

GSNOR1 is a negative regulator of cell death in tomato. Spontaneous cell death occurs on the leaves of tomato plants in which SlGSNOR1 expression was silenced by tobacco rattle virus (TRV)–induced gene silencing. Cell death is not observed in leaves of plants infected with the TRV2-0 empty vector control. Arrows indicate the areas with extensive cell death. The numbers below the figure indicate the number of plants displaying spontaneous cell death of 12 plants. This experiment was repeated three times with similar results.
Tomato SlPDK1 but not Adi3 is the target of S-nitrosylation
We took a candidate protein approach to search for negative regulators of cell death with functions potentially regulated by S-nitrosylation. We focused on proteins that were shown previously to be key negative regulators of cell death and S-nitrosylated. PKB rose to the top of our list because it is a conserved cell death regulator in yeasts (43), mammals (34), and plants (39), and it is also a target of S-nitrosylation in mice (41, 44). Most importantly, S-nitrosylation of PKB inhibits its kinase activity and, thus, its function (41). To test whether a plant PKB is a target of S-nitrosylation, a biotin-switch assay (45) was performed on tomato Adi3 fused to maltose-binding protein (MBP–Adi3). Adi3 is a homolog of PKB/Akt, which is a known negative regulator of tomato cell death (39). Adi3 was not S-nitrosylated because no band was detectable for MBP–Adi3 in the biotin-switch assay (Fig. 2A). Unexpectedly, we found that tomato SlPDK1, a protein kinase that acts upstream of Adi3, was S-nitrosylated (Fig. 2B). To confirm this result, the biotinylated MBP–SlPDK1 fusion protein was purified using neutravidin–agarose, and then a Western blot analysis was performed using an antibody against MBP. A band the size of MBP–SlPDK1 was detected (Fig. 2C), confirming that SlPDK1 is a target of S-nitrosylation. To test whether the SlPDK1 is S-nitrosylated in planta, the FLAG–SlPDK1 fusion protein was transiently expressed in NbGSNOR1-silenced Nicotiana benthamiana leaves via Agrobacterium infiltration. The purpose of silencing NbGSNOR1 was to create a high cellular SNO level to facilitate the detection of S-nitrosylation. NbGSNOR1 was silenced using TRV-induced gene silencing (VIGS) (46). 2 days after infiltration, total protein was extracted from the infiltrated leaf area and biotinylated using the biotin-switch–based approach. The biotinylated proteins were then purified using neutravidin–agarose beads followed by Western blotting using an antibody against FLAG. The transiently expressed FLAG–SlPDK1 was indeed S-nitrosylated in planta, further confirming that SlPDK1 is a target of S-nitrosylation (Fig. 2D). It has been reported previously that VIGS of SlPDK1 in tomato or application of a PDK1 inhibitor results in the early death of tomato seedlings and suspension cells, respectively (39). Therefore, SlPDK1 satisfies the two criteria we set for the candidate protein: that it negatively regulates cell death and is S-nitrosylated.
Figure 2.
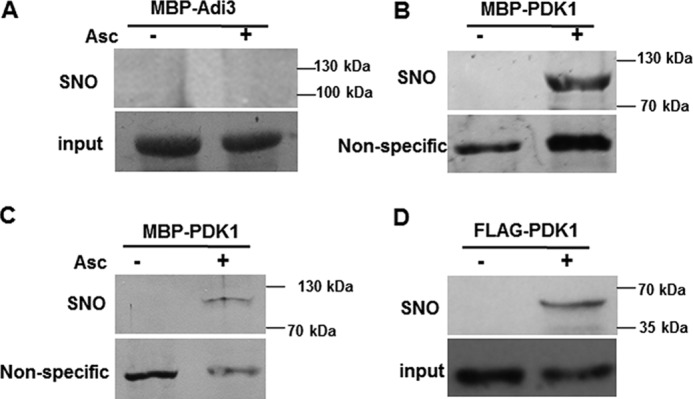
SlPDK1 is a target of S-nitrosylation. A and B, MBP–Adi3 (A) and MBP-SlPDK1 (B) were purified from bacterial extracts and treated with 1 mm GSNO. S-nitrosylation was detected by biotin-switch assay using a biotin antibody. Asc, ascorbate. C, biotinylated MBP–SlPDK1 protein was purified with neutravidin–agarose, separated by SDS-PAGE, and detected by immunoblot assay with an anti-MBP antibody. Coomassie Blue was used to stain MBP–Adi3 protein for loading control in A (input), and the nonspecific bands detected in immunoblots were used as controls in B and C. D, the FLAG–SlPDK1 fusion protein transiently expressed in the leaves of NbGSNOR1-silenced N. benthamiana plant is S-nitrosylated in vivo. Equal loading was verified with FLAG antibody against the input protein extracts. These experiments were repeated three times with similar results.
GSNO inhibits tomato SlPDK1 kinase activity in vitro
Because SlPDK1 is S-nitrosylated, we reasoned that its kinase activity could be altered in the presence of GSNO. To test this possibility, we performed kinase assays in the presence and absence of GSNO using MBP–SlPDK1 purified from Escherichia coli and a kinase-deficient version of Adi3 (Adi3K337Q) as substrate. Adi3K337Q loses most autophosphorylation activity, but it is trans-phosphorylated by PDK1 (39). Trans-phosphorylation of Adi3K337Q by SlPDK1 was inhibited by GSNO in a concentration-dependent manner (Fig. 3, left), suggesting that S-nitrosylation of SlPDK1 negatively regulates its kinase activity.
Figure 3.
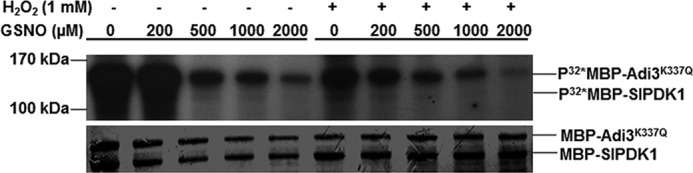
GSNO and H2O2 additively inhibit the kinase activity of MBP–SlPDK1 in vitro. A, the trans-phosphorylation activity of MBP–SlPDK1 fusion protein on MBP–Adi3K337Q, a kinase-dead version of Adi3, was inhibited in vitro by GSNO in a concentration-dependent manner, and this inhibition was additively enhanced by 1 mm H2O2. Equal loading was confirmed by the Coomassie Blue staining; The experiment was repeated three times with similar results.
Inhibition of SlPDK1 activity is reversible by reducing agent but enhanced additively by H2O2
Because of the labile nature of S-nitrosylation, a reducing agent, such as DTT, can reverse this modification. When DTT was added to the kinase assay buffer, it clearly reversed the inhibition of SlPDK1 kinase activity by GSNO (Fig. 4A). This result suggests that GSNO affects the kinase activity of SlPDK1 through a reversible thiol modification (47).
Figure 4.
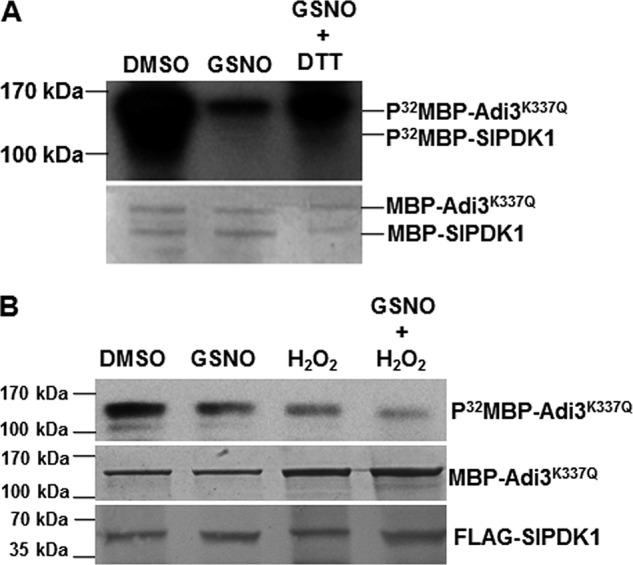
Inhibition of SlPDK1 kinase activity is reversible by reducing agent in vitro but enhanced additively by H2O2 when transiently expressed in N. benthamiana leaves. A, DTT partially reversed the inhibitory effect of GSNO on trans-phosphorylation of MBP–Adi3 K337Q by MBP–SlPDK1 in vitro. The Coomassie Blue–stained gel is shown for loading control. B, the kinase activity of SlPDK1 transiently expressed in the leaves of N. benthamiana plants was additively inhibited by 2 mm GSNO and 1 mm H2O2. 35S::FLAG-SlPDK1 was transiently expressed in N. benthamiana via agroinfiltration. The kinase activity of the cell extracts from the infiltrated area was determined using purified kinase-dead MBP-Adi3K337Q as a substrate. Equal loading for MBP–Adi3 K337Q was shown by Coomassie Blue staining, and equal input for FLAG–SlPDK1 was shown by Western blot analysis using FLAG antibody. The experiments were repeated three with similar results.
NO and H2O2 have been shown to synergistically induce cell death in soybean suspension cells (1), and they also act synergistically in inhibiting PKB activity in mammalian cells (41). To test whether H2O2 and GSNO can synergistically inhibit the kinase activity of SlPDK1, we performed the kinase assay in the presence of H2O2 alone or in the presence of both H2O2 and GSNO. H2O2 alone inhibited the kinase activity of SlPDK1 (Fig. 3, first lane on the right; compare the H2O2+/GSNO− lane with the H2O2−/GSNO− lane). However, the combination of H2O2 and GSNO had a stronger inhibitory effect on SlPDK1 kinase activity than either treatment alone (Fig. 3). These observations correlate with the facts that H2O2 enhances the level of NO donor-induced S-nitrosylation of PKB (41) and that NO promotes ROS-induced cell death (1, 48).
SlPDK1 kinase activity is additively inhibited by GSNO and H2O2 when transiently expressed in the leaves of N. benthamiana
To explore whether GSNO and H2O2 can additively inhibit SlPDK1 activity in plant, we tested the effect of GSNO and H2O2 on the FLAG–SlPDK1 transiently expressed in N. benthamiana leaves via Agrobacterium infiltration. We purified MBP–Adi3K337Q from E. coli and used it as a phosphorylation substrate for transiently expressed FLAG–SlPDK1 present in extracts from N. benthamiana leaves. The phosphorylation reaction (purified MBP–Adi3K337Q + leaf protein extracts) was performed either in the presence of 2 mm GSNO or 1 mm H2O2 individually or both. Consistent with the in vitro assays (Fig. 3), GSNO or H2O2 individually inhibited the activity of MBP–SlPDK1, and when they were added in combination, MBP–SlPDK1 activity was further inhibited (Fig. 4B). These results support the conclusion that SlPDK1 activity is additively inhibited by both GSNO and H2O2 when it was expressed in planta.
Tomato SlPDK1 is S-nitrosylated at Cys128 and Cys466
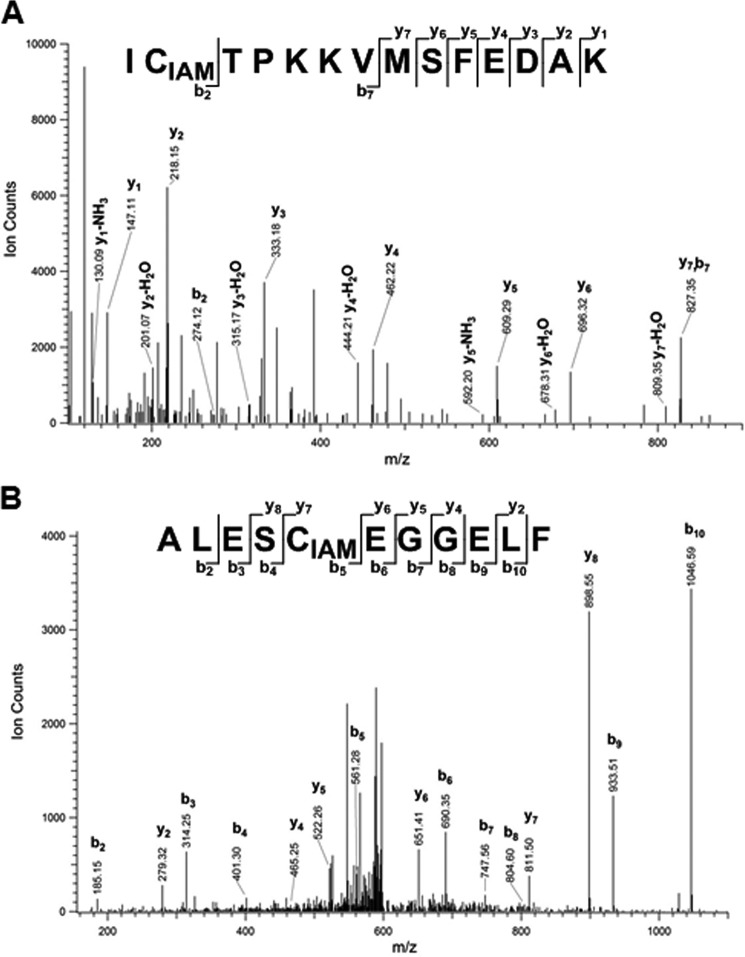
LC-MS/MS approaches based on the biotin-switch assay have been widely used for the identification of S-nitrosylated Cys sites within proteins (49–51). In our experiment, a differential alkylation strategy modified from a method published previously (52) was applied to the purified wild-type MBP–SlPDK1 fusion protein. S-nitrosylation of SlPDK1 was chemically induced by reaction with GSNO. SlPDK1 was subsequently alkylated with N-ethylmaleimide (NEM) to block free thiols, and after alkylation, the SNO groups were reduced and labeled with iodoacetamide (IAM). In this approach, the Cys residues labeled by IAM are identified as target sites of S-nitrosylation. Following alkylation, the protein sample was enzymatically digested and analyzed by LC-MS/MS. When trypsin was used for protein digestion, we identified peptides containing two of the four Cys residues in SlPDK1, Cys214 and Cys466. Of the two detected Cys residues, only Cys466 (ICTPKKVMSFEDAK), but not Cys214, was determined to be S-nitrosylated (Fig. 5A).
Figure 5.
MS/MS identification of S-nitrosylated Cys-containing peptides after differential alkylation. A, representative annotated MS/MS spectrum of IAM-labeled Cys466-containing peptide with ascorbate/CuCl treatment. Trypsin was used for protein digestion. B, representative annotated MS/MS spectrum of IAM-labeled Cys128-containing peptide with ascorbate/CuCl treatment. Asp-N was used for protein digestion. The IAM-modified peptides were not observed in the absence of ascorbate. The peptide sequence is shown at the top of each spectrum with the annotation of the identified fragment ions. For clarity, only major identified peaks are annotated.
To identify peptides containing Cys128 or Cys244, we used a different protease, Asp-N, for protein digestion based on an in silico digestion prediction (53–54). With this approach, a peptide that contained Cys128 with an IAM modification was identified, indicating that Cys128 (ALESCEGGELF) is a redox-sensitive site susceptible to SNO modification (Fig. 5B). In sum, these results indicated that the Cys128 and Cys466 residues of SlPDK1 are S-nitrosylated.
Cys128 is critical for the kinase activity of SlPDK1
To identify the Cys residues that are important for the kinase activity of SlPDK1, phosphorylation assays were performed for the WT as well as the four Cys→Ser mutant MBP fusion proteins. Mutations at Cys214 and Cys466 had no effect on the kinase activity of MBP–SlPDK1 in the absence of GSNO (Fig. 6). However, MBP–SlPDK1 kinase activity was completely abolished by the Cys128 mutation, whereas some residual activity was observed for the Cys244 mutant (Fig. 6). These results indicate that Cys128 and Cys244, but not Cys214 and Cys466, are critical for the kinase activity of SlPDK1. As expected, the kinase activities of the WT MBP–SlPDK1 and the 214Cys→Ser and 466Cys→Ser mutants were severely inhibited by GSNO (Fig. 6), indicating that these two residues are not responsible for the inhibition. The residual kinase activity of the 244Cys→Ser mutant was sensitive to GSNO (Fig. 6), suggesting that an additional Cys residue could be responsible for the kinase inhibition, and this is most likely Cys128. Therefore, it is highly possible that S-nitrosylation at Cys128 is responsible for the inhibition of SlPDK1 kinase activity. Thus, our results established a potential link between NO-triggered cell death and the inhibition of SlPDK1 kinase activity by S-nitrosylation at Cys128.
Figure 6.
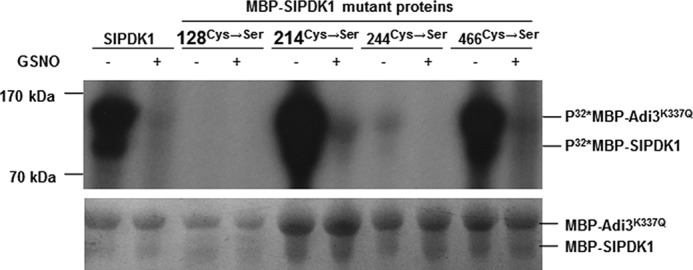
Cys128 and Cys244 are critical for the kinase activity of MBP–SlPDK1. The four Cys residues (Cys128, Cys214, Cys244, and Cys466) within SlPDK1 were mutated to Ser individually by site-directed mutagenesis to create four substituted versions of MBP–SlPDK1. The kinase assay was performed for the WT MBP–SlPDK1 and these four mutant fusion proteins using MBP–Adi3K337Q as substrate in the absence (−) or presence (+) of 2 mm GSNO. Equal loading was shown by Coomassie Blue staining. This experiment was repeated three times with similar results.
Discussion
Loss of GSNOR1 results in spontaneous cell death in different plant species
It has been reported that loss of GSNOR1 results in a substantial increase in the level of protein S-nitrosylation and increased cell death in response to pathogen infections both in mammals and plants (7, 19, 42). Unexpectedly, we observed a spontaneous cell death phenotype in SlGSNOR1-silenced plants even in the absence of pathogen infection (Fig. 1), raising the possibility that S-nitrosylation acts as a regulator of cell death. Identification of such a negative regulator(s) with a function that is inhibited by S-nitrosylation will enable the molecular mechanism by which NO induces cell death to be unraveled.
SlPDK1 activity is inhibited by both NO and H2O2 in an S-nitrosylation–dependent manner
Because H2O2 and NO operate together in the cell death signaling cascades (for review, see Ref. 55), identification of factors that interact with both H2O2 and NO could allow the discovery of the point of overlap between the two pathways (4, and 55). In an attempt to identify such factors, we focused on Adi3, a homolog of Akt/PBK in mammals, because it is not only a negative regulator both in tomato and mouse (39, 41), but it is also a target of S-nitrosylation in the mouse (39). In addition, S-nitrosylation of mouse PKB/Akt at Cys224 inactivates its function in vitro and in vivo, and H2O2 has a synergistic effect on the inhibition of Adi3 kinase activity (41). In contrast, we found that, unlike the mouse PKB/Akt, tomato Adi3 was not modified by S-nitrosylation (Fig. 2A). This result was not totally unexpected because Cys224 is not conserved in tomato Adi3. Surprisingly, we found that SlPDK1, which functions upstream of Adi3, was S-nitrosylated at Cys128 and Cys466 (Figs. 2, B–D, and 5). These two Cys residues are not conserved in the mammalian PDK1 homolog, which explains why mammalian PDK1 has not been identified as a target of S-nitrosylation. Similar to what has been observed for PKB/Akt in mice (41), GSNO inhibited the SlPDK1 kinase activity in a concentration-dependent manner (Figs. 3 and 4B), and this inhibition was enhanced in the presence of H2O2 (Figs. 3 and 4B) and reversed by addition of the reducing agent DTT (Fig. 4A). The strong additive effect of NO and H2O2 on plant cell death (1) as well as on the inhibition of the SlPDK1 kinase activity (Figs. 3 and 4B) implied that the cell death observed in the SlPDK1-silenced plants might be a consequence of the inhibition of the SlPDK1 kinase activity and that the PDK1 in plants might be one of the long-sought factors targeted by both NO and H2O2 to trigger cell death (1, 48). This statement is supported by the fact that silencing SlPDK1 leads to a lethal phenotype (39).
As Cys128 but not Cys466 was critical for SlPDK1 kinase activity (Fig. 6), and Cys128 was the major site of S-nitrosylation (Figs. 5B and 6), it is most likely that inhibition of the kinase activity of SlPDK1 by S-nitrosylation at Cys128 is responsible for the cell death observed in the SlGSNOR1-silenced tomato plants (Fig. 1). One might argue that abolishing the SlPDK1 activity by the 128Cys→Ser substitution could be the result of disrupting the overall structure of SlPDK1. However, it is also possible that S-nitrosylation at Cys128 could inhibit the kinase activity of SlPDK1 through altering its structure. Together, these results suggest that NO targets different components in the conserved PI3K–PDK1–PKB/Akt signaling pathway to regulate cell death in mice and tomato, demonstrating that distinct differences exist between plants and mammals in the control of NO-induced cell death.
Hu et al. (12) recently analyzed the flanking sequences of 1195 S-nitrosylated Cys residues identified in Arabidopsis proteins and found that EXC was a predominant putative consensus sequence. Interestingly, the two amino acid residues preceding Cys128 are Glu and Ser, which perfectly matches the consensus EXC sequence and provides additional support for the theory that Cys128 is the major site of S-nitrosylation in SlPDK1. The other three Cys residues within SlPDK1 do not match this consensus site. Additionally, our result validates the consensus sequence identified in Arabidopsis by Hu et al. (12) and suggests that this consensus sequence is conserved in different plant species.
NO and H2O2 target multiple proteins in the cell death pathway
Given the nonspecific nature of S-nitrosylation, it is possible that multiple proteins that play both positive and negative roles in cell death are simultaneously modified by S-nitrosylation. The final fate of the cells is determined by many factors, including local cellular NO concentration, the subcellular localization of these cell death regulators, and the sensitivity of different cell death regulators to S-nitrosylation (35, 56, 57). The co-existence of multiple positive and negative regulators of cell death that are regulated by S-nitrosylation enables cells to fine-tune cell death signaling more precisely and with more flexibility.
In mammals, S-nitrosylation of GAPDH triggers binding to Siah1 and mediates the nuclear translocation of GAPDH (58). In the nucleus, GAPDH stabilizes Siah1, enhancing its E3 ubiquitin ligase activity, which ultimately leads to cell death (58). In plants, GAPDH is a target of S-nitrosylation (8, 22). These observations raise the possibility that a similar pathway is also operating in plants and that GAPDH could be another key factor in NO-induced cell death. Interestingly, Han et al. (59) recently showed that the cytosolic GAPDH in N. benthamiana plants (GAPCs) negatively regulates autophagy through interacting with autophagy-related protein 3 (ATG3). Silencing of GAPCs enhances N gene-mediated cell death and plant resistance against both incompatible and compatible pathogens. It appears that the GAPDH in plants functions differently from its counterpart in mammals.
The RBOHD subunit of the Arabidopsis NAPDH oxidase complex is S-nitrosylated at Cys890, and this modification abolishes its ability to synthesize ROS (19). As a result, when the Cys890 mutant version of AtRBOHD was transformed into the Arabidopsis rbohd mutant, the transgenic plants underwent increased cell death compared with those transformed with a WT AtRBOHD after being challenged with an avirulent bacterial strain (19). On the other hand, instead of inhibiting the function of a protein, S-nitrosylation of the Arabidopsis cytosolic APX1 at Cys32 enhances its H2O2 scavenging activity, resulting in increased resistance to oxidative stress (25). The 32Cys→Ser mutation reduces the activity of APX1 and abolishes its GSNO-enhanced activity (25). As a result, induction of flg22-induced expression of resistance-related marker genes in the apx1 mutant was rescued by the WT APX1-FLAG but not APX1C32S-FLAG transgene (25). In mice, the 224Cys→Ser mutation of Akt1/PKB does not inhibit its kinase activity, and thus, the mutant Akt1/PKB was resistant to S-nitrosylation and kinase inactivation (41). However, in our case, the 128Cys→Ser mutation completely abolished the kinase activity of SlPDK1 (Fig. 6), and, therefore, it is not possible to create a mutant version of SlPDK1 in which the kinase activity is still maintained but no longer inhibited by S-nitrosylation. As a result, this prevents us from generating a transgenic line that could be resistant to NO-induced cell death.
Experimental procedures
Plant materials
Tomato (S. lycopersicum) and N. benthamiana were used in our experiments. The plants were grown in a growth room at 20 °C during the dark and 22 °C during the light, with a photoperiod of 16 h.
Virus-induced gene silencing
A 461-bp cDNA fragment of the tomato SlGSNOR1 ORF (NM_001251867) and a 415-bp fragment of NbGSNOR1 were amplified, respectively, via RT-PCR from total RNA extracted from the respective plants. The amplified fragments were cloned into the Gateway entry vector pENTR/D (Invitrogen) and subsequently recombined into the pTRV2-attR2-attR1 vector via LR reaction (46). For the VIGS assay, pTRV1, pTRV2, TRV2-SlGSNOR1, and pTRV2-NbGSNOR1 were introduced into Agrobacterium strain GV3101. Agrobacterium containing the binary vectors were grown and infiltrated into leaves as described previously (46, and 60). The primers used for PCR amplification were as follows: SlGSNOR1-F, caccTGCCTTCTTGGATGTGGTGTTC; SlGSNOR1-L, TTGAAACCAC CAAAAGCAGT TCC; NbGSNOR1-F, caccTGCCTTCTTGGATGTGGTGTTC; and NbGSNOR1-R, TTGAAACCACCAAAAGCWGTTCC.
pTRV1 and pTRV2-SlGSNOR1 or pTRV1 and pTRV2-NbGSNOR1 were co-inoculated into 3-week-old tomato and N. benthamiana seedlings, respectively, by Agrobacterium-mediated infiltration as described previously (46, 60), and the phenotypes were assayed 2 to 3 weeks later. Each silencing experiment was carried out a minimum of three times with at least six plants.
In vitro site-directed mutagenesis and protein expression
Site-directed mutagenesis was carried out using the GeneTailor site-directed mutagenesis system (Invitrogen). The primers used in the mutagenesis were as follows: SlPDK1-F, ATGTTGGCATTGGTAGGGG; SlPDK1-R, TCACCGGTTCTGGAGAGCT; SlPDK1-Mu1-F, TGTACATGGCACTTGAGTCTGCTGAAGGTGGAGA; SlPDK1-Mu1-R, AGACTCAAGTGCCATGTACAGTGAAAAAGTG; SlPDK1-Mu2-F, CTGCATCAGATGACAAAGCCGCTACTTTTGTGG; SlPDK1-Mu2-R, GGCTTTGTCATCTGATGCAGCATTTGGAAG; SlPDK1-Mu3-F, ATGATCTTTGGGCACTTGGCGCCACATTGTATCA; SlPDK1-Mu3-R, GCCAAGTGCCCAAAGATCATTTCCAAAAGT; SlPDK1-Mu4-F, GCCCTTCACAGTTCAAGATTGCTACACCAAAGAA; and SlPDK1-Mu4-R, AATCTTGAACTGTGAAGGGCTTGTGACTTG. The bold and underlined letters indicate the mutations generated.
Constructs for protein expression were transformed into E. coli strain BL21 Star (DE3) (Invitrogen) and grown overnight in 2 ml of LB at 37 °C. A 200-μl aliquot of the culture was added to 10 ml of LB, grown at 37 °C to A600 = 0.5, and protein expression was induced with 100 μm isopropyl 1-thio-β-d-galactopyranoside for 3 h at 28 °C. Cells were harvested by centrifugation and lysed by sonication in 1 ml of extraction buffer (50 mm Tris (pH 8.0), 50 mm NaCl, 5 mm EDTA, 0.1% Triton X-100, and general protease inhibitors (Sigma)), and debris was pelleted by centrifugation. Proteins were purified from the supernatants by adding amylose resin (New England Biolabs and FLAG M2–agarose (Sigma), incubating at 4 °C for 30 min with rotation, pelleting the resin by centrifugation, and washing three times with extraction buffer.
Kinase assays
Phosphorylation assays were performed as described previously (39). Briefly, the MBP translational fusion proteins were immobilized on 50 μl of amylose beads, washed three times with kinase buffer without DTT and ATP, and then incubated with or without GSNO of different concentrations at 4 °C for 1 h before the in vitro kinase assay was performed. After the incubation, the beads were washed once with kinase buffer without ATP and DTT and then incubated with 5 μCi of [γ-32P]ATP in 50 μl of kinase buffer. The Adi3/Pdk1 kinase buffer contained 10 mm Tris (pH 7.5), 150 mm NaCl, 10 mm MgCl2, and 20 μm ATP. Reactions were carried out at room temperature for 30 min and analyzed by 10% SDS-PAGE with phosphorimaging (Molecular Dynamics) visualization. For cross-phosphorylation assays, MBP translational fusion proteins were purified separately on 50 μl of amylose resin, and proteins left on the resins were mixed and used in a kinase assay, with the buffer of the enzyme carrying out the phosphorylation. Kinase assays were repeated a minimum of three times.
For the kinase assay in planta, SlPDK1 was cloned into pEarleyGate 202 to generate 35SPro::FLAG-SlPDK1 and was subsequently transformed into Agrobacterium strain GV2260. The Agrobacterium solution carrying 35SPro::FLAG-SlPDK1 was infiltrated into N. benthamiana leaves, and the protein extracts in the infiltrated areas were used for in planta kinase assay with purified MBP-Adi3K337Q from E. coli as substrate.
Biotin labeling of S-nitrosylated proteins and purification of biotinylated proteins
The Cys residue of a protein can be biotinylated in the presence of biotin (covalently attaching biotin to a protein). The biotin-switch assay has been developed to identify S-nitrosylated proteins (45) and consists of three principle steps: blocking of free cysteine thiols (non-S-nitrosylated thiols) by S-methylthiolation with methyl methanethiosulfonate (MMTS), removing the NO group from the Cys residues with ascorbate, and labeling of thiols by S-biotinylation with biotin-HPDP, a reactive mixed disulfide of biotin. The S-nitrosylated protein can be purified using streptavidin or avidin beads, which bind biotin with an extremely high affinity and specificity. The degree of S-nitrosylation reflected by biotinylation can be determined by Western blotting with a biotin antibody, a protein-specific antibody, or an antibody for a tag.
Arabidopsis cells were frozen in liquid nitrogen and ground to a fine powder. Proteins were extracted by adding 12 ml of HEN buffer (25 mm HEPES (pH 7.7), 1 mm EDTA, and 0.1 mm neocuproine) to 10 g of cell powder. Cell debris was removed by centrifugation (20,000 × g, 10 min at 4 °C), and protein concentration was determined according to Bradford with bovine serum albumin as standard.
The in vitro S-nitrosylation and subsequent biotinylation of S-nitrosylated proteins were performed as described in Ref. 45 with minor modifications (8). After treating the supernatant with GSNO for 20 min at room temperature, the proteins were incubated with 20 mm MMTS and 2.5% SDS at 50 °C for 20 min with frequent vortexing for blocking non-nitrosylated free Cys residues. Residual MMTS was removed by precipitation with 2 volumes of −20 °C acetone, and the proteins were resuspended in 0.1 ml of HENS buffer (HEN buffer containing 1% SDS)/1 mg of protein. Biotinylation was achieved by adding 2 mm biotin–HPDP and 1 mm ascorbate and incubation at room temperature for 1 h.
After removing biotin-HPDP, the precipitated proteins were resuspended in 0.1 ml of HENS buffer/1 mg of protein and 2 volumes of neutralization buffer (20 mm HEPES (pH 7.7), 100 mm NaCl, 1 mm EDTA, and 0.5% Triton X-100). A total of 15 μl of neutravidin–agarose/1 mg of protein was added and incubated for 1 h at room temperature. The matrix was washed extensively with 20 volumes of washing buffer (600 mm NaCl in neutralization buffer), and bound proteins were eluted with 100 mm β-mercaptoethanol in neutralization buffer (8).
In vitro S-nitrosylation of PDK1 and differential alkylation of Cys residues
The purified MBP–PDK1 was reconstituted in 250 mm HEPES (pH 7.7), 1 mm EDTA, 0.1 mm neocuproine, and 0.1% SDS and treated with 10 mm DTT for 30 min at 37 °C to release all free thiols on the protein. DTT was subsequently removed by using a 0.5-μm Amicon filter with 10-kDa molecular weight cutoff (EMD Millipore, Billerica, MA). To induce S-nitrosylation, the protein solution was divided into two aliquots and treated with and without GSNO, respectively, in 1% DMSO at 37 °C for 30 min in the dark. After this step, unmodified cysteine residues were blocked with 50 mm NEM at 55 °C for 30 min in the dark in 2% SDS, and the excess NEM was removed by cold acetone precipitation. Protein pellets were subsequently dissolved in 50 mm HEPES (pH 7.7) with 0.5% SDS. S-nitrosylated cysteine residues were selectively reduced by adding 5 mm sodium ascorbate and 5 μm CuCl. 40 mm IAM was added to label the nascent free thiols in the dark at 37 °C for 1 h. The protein samples were then purified and concentrated in 50 mm Tris-HCl (pH 7.5) by using an Amicon filter. Protein digestion was performed at 37 °C overnight by adding trypsin or Asp-N with an enzyme-to-protein ratio 1:200. After digestion, 0.1% TFA was added to stop the reaction. Peptide samples were cleaned up with Omix C18 Ziptip (Agilent) and put in 0.1% formic acid prior to LC-MS/MS.
LC-MS/MS and data analysis
LC-MS/MS analysis was performed using the nano-Aquity UPLC system (Waters Corp., Milford, MA) with a homemade 75-μm inner diameter × 70 cm reverse-phase capillary column packed with 3 μm C18 resin (Phenomenex, Torrance, CA). The LC system was operated at a constant flow rate of 300 nl/min over a 100-min gradient. Mobile phases A and B were 0.1% (v/v) formic acid in water and 0.1% (v/v) formic acid in ACN, respectively. The gradient started with 100% of mobile phase A to 60% of mobile phase B. MS analysis was performed on an LTQ-Orbitrap Velos mass spectrometer (Thermo Scientific, San Jose, CA) coupled with an electrospray ionization interface using a homemade 150-μm outer diameter × 20-μm inner diameter chemically etched electrospray emitter. The heated capillary temperature and spray voltage were 350 °C and 2.2 kV, respectively. Full MS spectra were recorded at a resolution of 60 K over a range of m/z 300–2000 with an automated gain control value of 1 × 106. The most abundant 10 parent ions were selected for MS/MS using collision-induced dissociation with a normalized collision energy setting of 35. Precursor ion activation was performed with an isolation width of 2.5 Da, a minimal intensity of 1000 counts, and an activation time of 0.1 s. A dynamic exclusion time of 60 s was used.
LC-MS/MS raw data were converted into data files using Bioworks Cluster 3.2 (Thermo Fisher Scientific, Cambridge, MA), and an MSGF plus algorithm (v9979, released in March 2014) was used to search MS/MS spectra against a sequence database containing MBP and PDK1. The key search parameters used were 20-ppm tolerance for precursor ion masses, 0.5-Da tolerance for fragment ions, partial tryptic search with up to two missed cleavages, dynamic oxidation of methionine (15.9949 Da), dynamic NEM modification of Cys (125.0477 Da), and dynamic IAM modification of Cys (+57.0215 Da). Peptides were identified from database search results applying the following criteria: MSGF SpecE value <10−9, Q value <0.01 (false discovery rate <1%), and mass measurement error <10 ppm (±5 ppm). IAM-modified Cys residues were identified as redox-sensitive sites susceptible to S-nitrosylation.
Author contributions
J.-Z. L. designed the experiments. J.-Z. L., M. N., Z. L., and W.-L. Q. performed the experiments shown in Figs. 1–4 and 6. W.-J. Q. and J. D. designed and J. D. carried out the experiments shown in Fig. 5. J.-Z. L., J. D., S. A. W., and W.-J. Q. analyzed the data. J.-Z. L. wrote the manuscript with input from J. D., S. A. W., and W.-J. Q.
Acknowledgments
We thank Prof. Gregory Martin and Dr. Timothy P Devarenne for providing the MBP–SlPDK1, MBP–Adi3, and MBP–Adi3K337Q constructs. We also thank Prof. Meihao Sun for providing technical support with protein purification. The mass spectrometry analyses were performed in the Environmental Molecular Sciences Laboratory, a Department of Energy national scientific user facility located at Pacific Northwest National Laboratory that is operated by the Battelle Memorial Institute for the Department of Energy under Contract DE-AC05-76RL0 1830.
This work was supported by National Natural Science Foundation of China Grants 31371401 and 31571423 (to J.-Z. L.), a Department of Energy early career research award (to W.-J. Q.), and National Science Foundation Plant Genome Research Program 0820642 and Department of Agriculture National Institute of Food and Agriculture (NIFA) Hatch Project 3808 (to S. A. W.). The authors declare that they have no conflicts of interest with the contents of this article.
- ROS
- reactive oxygen species
- SNO
- S-nitrosothiol
- GSNO
- S-nitrosoglutathione
- GSNOR
- S-nitrosoglutathione reductase
- PKB
- protein kinase B
- MBP
- maltose-binding protein
- VIGS
- virus-induced gene silencing
- TRV
- tobacco rattle virus
- NEM
- N-ethylmaleimide
- IAM
- iodoacetamide
- MMTS
- methyl methanethiosulfonate
- RBOHD
- respiratory burst oxidase homolog D
- HPDP
- hexyl-3′-(2′-pyridyldithio)-propionamide.
References
- 1. Delledonne M., Xia Y., Dixon R. A., and Lamb C. (1998) Nitric oxide functions as a signal in plant disease resistance. Nature 394, 585–588 [DOI] [PubMed] [Google Scholar]
- 2. Zhang Y., and Hogg N. (2005) S-nitrosothiols: cellular formation and Transport. Free Radic. Biol. Med. 38, 831–838 [DOI] [PubMed] [Google Scholar]
- 3. Stamler J. S. (1994) Redox signaling: nitrosylation and related target interactions of nitric oxide. Cell 78, 931–936 [DOI] [PubMed] [Google Scholar]
- 4. Grennan A. K. (2007) Protein S-nitrosylation: potential targets and roles in signal transduction. Plant Physiol. 144, 1237–1239 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5. Hess D. T., and Stamler J. S. (2012) Regulation by S-nitrosylation of protein post-translational modification. J. Biol. Chem. 287, 4411–4418 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6. Liu L., Hausladen A., Zeng M., Que L., Heitman J., and Stamler J. S. (2001) A metabolic enzyme for S-nitrosothiol conserved from bacteria to humans. Nature 410, 490–494 [DOI] [PubMed] [Google Scholar]
- 7. Feechan A., Kwon E., Yun B. W., Wang Y., Pallas J. A., and Loake G. J. (2005) A central role for S-nitrosothiols in plant disease resistance. Proc. Natl. Acad. Sci. U.S.A. 102, 8054–8059 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8. Lindermayr C., Saalbach G., and Durner J. (2005) Proteomic identification of S-nitrosylated proteins in Arabidopsis. Plant Physiol. 137, 921–930 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9. Sell S., Lindermayr C., and Durner J. (2008) Identification of S-nitrosylated proteins in plants. Methods Enzymol. 440, 283–293 [DOI] [PubMed] [Google Scholar]
- 10. Lee T. Y., Chen Y. J., Lu C. T., Ching W. C., Teng Y. C., Huang H. D., and Chen Y. J. (2012) dbSNO: a database of cysteine S-nitrosylation. Bioinformatics 28, 2293–2295 [DOI] [PubMed] [Google Scholar]
- 11. Zhang X., Huang B., Zhang L., Zhang Y., Zhao Y., Guo X., Qiao X., and Chen C. (2012) SNObase, a database for S-nitrosation modification. Protein Cell 3, 929–933 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12. Hu J., Huang X., Chen L., Sun X., Lu C., Zhang L., Wang Y., and Zuo J. (2015) Site-specific nitrosoproteomic identification of endogenously S-nitrosylated proteins in Arabidopsis. Plant Physiol. 167, 1731–1746 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13. Belenghi B., Romero-Puertas M. C., Vercammen D., Brackenier A., Inzé D., Delledonne M., and Van Breusegem F. (2007) Metacaspase activity of Arabidopsis thaliana is regulated by S-nitrosylation of a critical cysteine residue. J. Biol. Chem. 282, 1352–1358 [DOI] [PubMed] [Google Scholar]
- 14. Romero-Puertas M. C., Laxa M., Mattè A., Zaninotto F., Finkemeier I., Jones A. M., Perazzolli M., Vandelle E., Dietz K. J., and Delledonne M. (2007) S-nitrosylation of peroxiredoxin II promotes peroxynitrite-mediated tyrosine nitration. Plant Cell 19, 4120–4130 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15. Tada Y., Spoel S. H., Pajerowska-Mukhtar K., Mou Z., Song J., Wang C., Zuo J., and Dong X. (2008) Plant immunity requires conformational changes [corrected] of NPR1 via S-nitrosylation and thioredoxins. Science 321, 952–956 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16. Wang Y. Q., Feechan A., Yun B. W., Shafiei R., Hofmann A., Taylor P., Xue P., Yang F. Q., Xie Z. S., and Pallas J. A., Chu C. C., Loake G. J. (2009) S-nitrosylation of AtSABP3 antagonizes the expression of plant immunity. J. Biol. Chem. 284, 2131–2137 [DOI] [PubMed] [Google Scholar]
- 17. Chen R., Sun S., Wang C., Li Y., Liang Y., An F., Li C., Dong H., Yang X., Zhang J., and Zuo J. (2009) The Arabidopsis PARAQUAT RESISTANT2 gene encodes an S-nitrosoglutathione reductase that is a key regulator of cell death. Cell Res. 19, 1377–1387 [DOI] [PubMed] [Google Scholar]
- 18. Lindermayr C., Sell S., Müller B., Leister D., and Durner J. (2010) Redox regulation of the NPR1-TGA1 system of Arabidopsis thaliana by nitric oxide. Plant Cell 22, 2894–2907 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19. Yun B. W., Feechan A., Yin M., Saidi N. B., Le Bihan T., Yu M., Moore J. W., Kang J. G., Kwon E., Spoel S. H., Pallas J. A., and Loake G. J. (2011) S-nitrosylation of NADPH oxidase regulates cell death in plant immunity. Nature 478, 264–268 [DOI] [PubMed] [Google Scholar]
- 20. Keyster M., Klein A., Egbichi I., Jacobs A., and Ludidi N. (2011) Nitric oxide increases the enzymatic activity of three ascorbate peroxidase isoforms in soybean root nodules. Plant Signal Behav. 6, 956–961 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21. Terrile M. C., París R., Calderón-Villalobos L. I., Iglesias M. J., Lamattina L., Estelle M., and Casalongué C. A. (2012) Nitric oxide influences auxin signaling through S-nitrosylation of the Arabidopsis TRANSPORT INHIBITOR RESPONSE 1 auxin receptor. Plant J. 70, 492–500 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22. Lin A., Wang Y., Tang J., Xue P., Li C., Liu L., Hu B., Yang F., Loake G. J., and Chu C. (2012) Nitric oxide and protein S-nitrosylation are integral to hydrogen peroxide-induced leaf cell death in rice. Plant Physiol. 158, 451–464 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23. de Pinto M. C., Locato V., Sgobba A., Romero-Puertas Mdel C., Gadaleta C., Delledonne M., and De Gara L. (2013) S-nitrosylation of ascorbate peroxidase is part of programmed cell death signaling in tobacco Bright Yellow-2 cells. Plant Physiol. 163, 1766–1775 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24. Begara-Morales J. C., Sánchez-Calvo B., Chaki M., Valderrama R., Mata-Pérez C., López-Jaramillo J., Padilla M. N., Carreras A., Corpas F. J., and Barroso J. B. (2014) Dual regulation of cytosolic ascorbate peroxidase (APX) by tyrosine nitration and S-nitrosylation. J. Exp. Bot. 65, 527–538 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25. Yang H., Mu J., Chen L., Feng J., Hu J., Li L., Zhou J. M., and Zuo J. (2015) S-nitrosylation positively regulates ascorbate peroxidase activity during plant stress responses. Plant Physiol. 167, 1604–1615 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26. Albertos P., Romero-Puertas M. C., Tatematsu K., Mateos I., Sánchez-Vicente I., Nambara E., and Lorenzo O. (2015) S-nitrosylation triggers ABI5 degradation to promote seed germination and seedling growth. Nat. Commun. 6, 8669. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27. Wang P., Du Y., Hou Y. J., Zhao Y., Hsu C. C., Yuan F., Zhu X., Tao W. A., Song C. P., and Zhu J. K. (2015) Nitric oxide negatively regulates abscisic acid signaling in guard cells by S-nitrosylation of OST1. Proc. Natl. Acad. Sci. U.S.A. 112, 613–618 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28. Mengel A., Ageeva A., Georgii E., Bernhardt J., and Wu K., Durner J., and Lindermayr C. (2017) Nitric oxide modulates histone acetylation at stress genes by inhibition of histone deacetylases. Plant Physiol. 173, 1434–1452 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29. Downward J. (1998) Mechanisms and consequences of activation of protein kinase B/Akt. Curr. Opin. Cell Biol. 10, 262–267 [DOI] [PubMed] [Google Scholar]
- 30. Vivanco I., and Sawyers C. L. (2002) The phosphatidylinositol 3-kinase-AKT pathway in human cancer. Nat. Rev. Cancer 2, 489–501 [DOI] [PubMed] [Google Scholar]
- 31. Duronio V. (2008) The life of a cell: apoptosis regulation by the PI3K/PKB pathway. Biochem. J. 415, 333–344 [DOI] [PubMed] [Google Scholar]
- 32. Fresno Vara J. A., Casado E., de Castro J., Cejas P., Belda-Iniesta C., and González-Barón M. (2004) PI3K/Akt signalling pathway and cancer. Cancer Treat. Rev. 30, 193–204 [DOI] [PubMed] [Google Scholar]
- 33. Zhu J., Huang J. W., Tseng P. H., Yang Y. T., Fowble J., Shiau C. W., Shaw Y. J., Kulp S. K., and Chen C. S. (2004) From the cyclooxygenase-2 inhibitor celecoxib to a novel class of 3-phosphoinositide-dependent protein kinase-1 inhibitors. Cancer Res. 64, 4309–4318 [DOI] [PubMed] [Google Scholar]
- 34. Desai S., Pillai P., Win-Piazza H., and Acevedo-Duncan M. (2011) PKC-ι promotes glioblastoma cell survival by phosphorylating and inhibiting BAD through a phosphatidylinositol 3-kinase pathway. Biochim. Biophys. Acta 1813, 1190–1197 [DOI] [PubMed] [Google Scholar]
- 35. Choi B. M., Pae H. O., Jang S. I., Kim Y. M., and Chung H. T. (2002) Nitric oxide as a pro-apoptotic as well as anti-apoptotic modulator. J. Biochem. Mol. Biol. 35, 116–126 [DOI] [PubMed] [Google Scholar]
- 36. Venigalla R. K., McGuire V. A., Clarke R., Patterson-Kane J. C., Najafov A., Toth R., McCarthy P. C., Simeons F., Stojanovski L., and Arthur J. S. (2013) PDK1 regulates VDJ recombination, cell-cycle exit and survival during B-cell development. EMBO J. 32, 1008–1022 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37. Casamayor A., Torrance P. D., Kobayashi T., Thorner J., and Alessi D. R. (1999) Functional counterparts of mammalian protein kinases PDK1 and SGK in budding yeast. Curr. Biol. 9, 186–197 [DOI] [PubMed] [Google Scholar]
- 38. Deak M., Casamayor A., Currie R. A., Downes C. P., and Alessi D. R. (1999) Characterisation of a plant 3-phosphoinositide-dependent protein kinase-1 homologue which contains a pleckstrin homology domain. FEBS Lett. 451, 220–226 [DOI] [PubMed] [Google Scholar]
- 39. Devarenne T. P., Ekengren S. K., Pedley K. F., and Martin G. B. (2006) Adi3 is a Pdk1-interacting AGC kinase that negatively regulates plant cell death. EMBO J. 25, 255–265 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40. Chen W. S., Xu P. Z., Gottlob K., Chen M. L., Sokol K., Shiyanova T., Roninson I., Weng W., Suzuki R., Tobe K., Kadowaki T., and Hay N. (2001) Growth retardation and increased apoptosis in mice with homozygous disruption of the Akt1 gene. Genes Dev. 15, 2203–2208 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41. Yasukawa T., Tokunaga E., Ota H., Sugita H., Martyn J. A., and Kaneki M. (2005) S-nitrosylation-dependent inactivation of Akt/protein kinase B in insulin resistance. J. Biol. Chem. 280, 7511–7518 [DOI] [PubMed] [Google Scholar]
- 42. Liu L., Yan Y., Zeng M., Zhang J., Hanes M. A., Ahearn G., McMahon T. J., Dickfeld T., Marshall H. E., Que L. G., and Stamler J. S. (2004) Essential roles of S-nitrosothiols in vascular homeostasis and endotoxic shock. Cell 116, 617–628 [DOI] [PubMed] [Google Scholar]
- 43. Jeon B. W., Kim K. T., Chang S. I., and Kim H. Y. (2002) Phosphoinositide 3-OH kinase/protein kinase B inhibits apoptotic cell death induced by reactive oxygen species in Saccharomyces cerevisiae. J. Biochem. 131, 693–699 [DOI] [PubMed] [Google Scholar]
- 44. Lu X. M., Lu M., Tompkins R. G., and Fischman A. J. (2005) Site-specific detection of S-nitrosylated PKB α/Akt1 from rat soleus muscle using CapLC-Q-TOF(micro) mass spectrometry. J. Mass Spectrom. 40, 1140–1148 [DOI] [PubMed] [Google Scholar]
- 45. Jaffrey S. R., and Snyder S. H. (2001) The biotin switch method for the detection of S-nitrosylated proteins. Sci STKE 2001, pl1. [DOI] [PubMed] [Google Scholar]
- 46. Liu Y., Schiff M., and Dinesh-Kumar S. P. (2002) Virus-induced gene silencing in tomato. Plant J. 31, 777–786 [DOI] [PubMed] [Google Scholar]
- 47. Astier J., and Lindermayr C. (2012) Nitric oxide-dependent posttranslational modification in plants: an update. Int. J. Mol. Sci. 13, 15193–15208 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48. Durner J., Wendehenne D., and Klessig D. F. (1998) Defense gene induction in tobacco by nitric oxide, cyclic GMP, and cyclic ADP-ribose. Proc. Natl. Acad. Sci. U.S.A. 95, 10328–10333 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49. Hao G., Derakhshan B., Shi L., Campagne F., and Gross S. S. (2006) SNOSID, a proteomic method for identification of cysteine S-nitrosylation sites in complex protein mixtures. Proc. Natl. Acad. Sci. U.S.A. 103, 1012–1017 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50. Derakhshan B., Wille P. C., and Gross S. S. (2007) Unbiased identification of cysteine S-nitrosylation sites on proteins. Nat. Protoc. 2, 1685–1691 [DOI] [PubMed] [Google Scholar]
- 51. Duan J., Gaffrey M. J., and Qian W. J. (2017) Quantitative proteomic characterization of redox-dependent post-translational modifications on protein cysteines. Mol. Biosyst. 13, 816–829 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 52. Liu M., Hou J., Huang L., Huang X., Heibeck T. H., Zhao R., Pasa-Tolic L., Smith R. D., Li Y., Fu K., Zhang Z., Hinrichs S. H., and Ding S. J. (2010) Site-specific proteomics approach for study protein S-nitrosylation. Anal. Chem. 82, 7160–7168 [DOI] [PubMed] [Google Scholar]
- 53. Choudhary G., Wu S. L., Shieh P., and Hancock W. S. (2003) Multiple enzymatic digestion for enhanced sequence coverage of proteins in complex proteomic mixtures using capillary LC with ion trap MS/MS. J. Proteome Res. 2, 59–67 [DOI] [PubMed] [Google Scholar]
- 54. Swaney D. L., Wenger C. D., and Coon J. J. (2010) Value of using multiple proteases for large-scale mass spectrometry-based proteomics. J. Proteome Res. 9, 1323–1329 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 55. Zaninotto F., La Camera S., Polverari A., and Delledonne M. (2006) Cross talk between reactive nitrogen and oxygen species during the hypersensitive disease resistance response. Plant Physiol. 141, 379–383 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 56. Mannick J. B. (2007) Regulation of apoptosis by protein S-nitrosylation. Amino Acids 32, 523–526 [DOI] [PubMed] [Google Scholar]
- 57. Li H., Wan A., Xu G., and Ye D. (2013) Small changes huge impact: the role of thioredoxin 1 in the regulation of apoptosis by S-nitrosylation. Acta Biochim. Biophys. Sin. 45, 153–161 [DOI] [PubMed] [Google Scholar]
- 58. Hara M. R., Agrawal N., Kim S. F., Cascio M. B., Fujimuro M., Ozeki Y., Takahashi M., Cheah J. H., Tankou S. K., Hester L. D., Ferris C. D., Hayward S. D., Snyder S. H., and Sawa A. (2005) S-nitrosylated GAPDH initiates apoptotic cell death by nuclear translocation following Siah1 binding. Nat. Cell Biol. 7, 665–674 [DOI] [PubMed] [Google Scholar]
- 59. Han S., Wang Y., Zheng X., Jia Q., Zhao J., Bai F., Hong Y., and Liu Y. (2015) Cytoplastic glyceraldehyde-3-phosphate dehydrogenases interact with ATG3 to negatively regulate autophagy and immunity in Nicotiana benthamiana. Plant Cell 27, 1316–1331 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 60. Liu J. Z., Blancaflor E. B., and Nelson R. S. (2005) The tobacco mosaic virus 126-kilodalton protein, a constituent of the virus replication complex, alone or within the complex aligns with and traffics along microfilaments. Plant Physiol. 138, 1853–1865 [DOI] [PMC free article] [PubMed] [Google Scholar]