Abstract
Sendai virus (SeV), which causes respiratory diseases in rodents, possesses the C protein that blocks the signal transduction of interferon (IFN), thereby escaping from host innate immunity. We previously demonstrated by using protein crystallography that two molecules of Y3 (the C-terminal half of the C protein) can bind to the homodimer of the N-terminal domain of STAT1 (STAT1ND), elucidating the mechanism of inhibition of IFN-γ signal transduction. SeV C protein also blocks the signal transduction of IFN-α/β by inhibiting the phosphorylation of STAT1 and STAT2, although the mechanism for the inhibition is unclear. Therefore, we sought to elucidate the mechanism of inhibition of the IFN signal transduction via STAT1 and STAT2. Small angle X-ray scattering analysis indicated that STAT1ND associates with the N-terminal domain of STAT2 (STAT2ND) with the help of a Gly-rich linker. We generated a linker-less recombinant protein possessing a STAT1ND:STAT2ND heterodimeric structure via an artificial disulfide bond. Analytical size-exclusion chromatography and surface plasmon resonance revealed that one molecule of Y3 can associate with a linker-less recombinant protein. We propose that one molecule of C protein associates with the STAT1:STAT2 heterodimer, inducing a conformational change to an antiparallel form, which is easily dephosphorylated. This suggests that association of C protein with the STAT1ND:STAT2ND heterodimer is an important factor to block the IFN-α/β signal transduction.
Keywords: innate immunity, interferon, negative-strand RNA virus, signal transduction, small-angle X-ray scattering (SAXS), surface plasmon resonance (SPR), paramyxovirus
Introduction
Sendai virus (SeV),2 which is known as a virus causing acute respiratory infection in mice, belongs to the genus Respirovirus of the family Paramyxoviridae and has a negative-sense, single-stranded RNA genome. SeV has two accessory proteins, V and C, that are not essential for viral multiplication but have a role in increasing viral growth and pathogenicity (1–5). V protein is produced using a V mRNA transcribed from the P gene, into which one G residue is inserted at the editing point, and it suppresses induction of IFN-β by interaction with a virus RNA sensor protein, melanoma differentiation-associated gene 5, and its downstream transcription regulator, interferon regulatory factor-3 (6, 7). On the other hand, C proteins comprise a nested set of four independently initiated and carboxyl-coterminal proteins C (amino acids (aa) 1 to 204), Y1 (aa 24 to 204), Y2 (aa 30 to 204), and C′ (with an 11-aa addition to the N terminus of C) (Fig. 1A), and are translated from P and V mRNAs in a different reading frame from that of P and V proteins. Among C proteins species, C is mainly expressed in the infected cells (reviewed in Refs. 8 and 9).
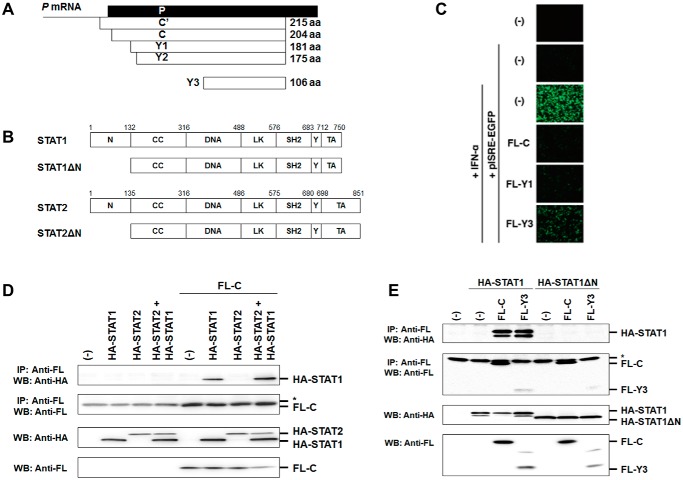
Figure 1.
Generation of N-terminal domain-deleted mutants of STAT1 and STAT2 and functional analysis of C proteins. A, schematic diagram of constructs of C′, C, Y1, Y2, and Y3. B, linear representation of the domains in human STAT1, STAT2, and their N-terminal domain-deleted mutants (STAT1ΔN and STAT2ΔN). N, N-terminal domain; CC, coiled-coil domain; DNA, DNA-binding domain; LK, linker domain; SH2, SH2 domain; Y, phosphorylated tyrosine residue; TA, transcription activation domain. C, to estimate the strength of the response to IFN-α, subconfluent 293T cells were transfected with pISRE-EGFP and an expression plasmid for FL-C, FL-Y1, or FL-Y3, and IFN-α (20 units/ml) was added to the culture medium at 6 h after transfection. At 24 h post-transfection, photographs were taken under an immunofluorescent microscope. D, HeLa cells were transfected with an expression vector for HA-STAT1, HA-STAT2, or FL-C. A portion of the cell lysates prepared at 24 h post-transfection were mixed as indicated and immunoprecipitated (IP) with an anti-FLAG antibody (Anti-FL) together with protein G-Sepharose. The immunoprecipitates were separated by SDS-PAGE followed by Western blot analysis (WB) using an anti-HA antibody (Anti-HA) and anti-FL antibody. A part of the cell lysates was used to confirm protein expression using anti-HA and anti-FL antibodies. E, HeLa cells were transfected with an expression vector for HA-STAT1 or HA-STAT1ΔN together with an expression vector for FL-C or FL-Y3. At 24 h post-transfection, the cell lysate was immunoprecipitated with anti-FL antibody and analyzed by Western blotting using anti-HA and anti-FL antibodies. *, a light chain of IgG.
C protein regulates viral RNA synthesis to reduce IFN-inducing RNA species (10–12) as well as controlling viral genome polarity (13, 14). In addition, C protein facilitates the budding of matured viral particles (12–19). Therefore, SeV can effectively produce particles at the late stage after infection when C protein is accumulated. Furthermore, C protein blocks the signal transduction of IFN-α/β by inhibiting phosphorylation of Tyr701 in transcription factor STAT1 and phosphorylation of Tyr690 in STAT2, in which inhibition of the Tyr phosphorylation of STAT2 is pivotal in inhibition of the signal transduction (20–22). On the other hand, binding of C protein to STAT1 seems to be important to block the signal transduction of IFN-α/β (23, 24).
STAT1 and STAT2 have a common domain structure, in which an N-terminal domain is linked to a core fragment, including a DNA-binding domain, SH2 domain, and tyrosine residue targeted for phosphorylation, via a linker peptide, and immediately followed by a transactivation domain (25) (Fig. 1B). The N-terminal domain of STAT1 is responsible for dimerization in its non-phosphorylated form. Because of the flexibility between the N-terminal domain and core fragment, STAT1 and STAT2 have a diversity of structures. Non-phosphorylated STAT1 generates a homodimer in a parallel form in which two SH2 domains are oriented in the same direction or in an antiparallel form in which two SH2 domains are oriented in opposite directions (26–28). After stimulation by IFN-γ, STAT1 is converted into a DNA-binding form in which phosphorylated Tyr701 interacts with the SH2 domain in the other subunit, together with the disappearance of interaction between the two N-terminal domains (29, 30).
C protein blocks the signal transduction of IFN-γ (31, 32). The C-terminal half of C protein (aa 99–204, named Y3; Fig. 1A) associates with the N-terminal domain of STAT1 (STAT1ND). In a previous report, we proposed the inhibition mechanism of STAT1 activation by C protein on the basis of the structure of the Y3–STAT1ND complex determined by X-ray crystallography (33). That is, with the association of one molecule of C protein, the STAT1 homodimer is induced to take the antiparallel form, in which STAT1 is easily dephosphorylated on Tyr701. On the other hand, with the association of two molecules of C protein, the STAT1 homodimer takes the parallel form, in which STAT1 is resistant to dephosphorylation. However, C protein strengthens the binding of two STAT1NDs and forms high-molecular-weight complexes with phosphorylated STAT1, preventing STAT1 from binding to a γ-activated sequence. On the other hand, it remains unclear how C protein inhibits IFN-α/β-induced phosphorylation of both STAT1 and STAT2.
In this study, we found that the inhibition of IFN-α-induced phosphorylation of STAT1 and STAT2 by C protein is partly explained by the action of the Y3 domain that can bind to STAT1ND. Small angle X-ray scattering (SAXS) and mutational analyses demonstrated the inhibition mechanism of STAT2 phosphorylation by the Y3 domain of C protein in a STAT1-dependent manner. It was also revealed that C protein inhibits phosphorylation of STAT2 in both STAT1-dependent and -independent manners.
Results
Association of C protein with STAT1
We first confirmed inhibition of signal transduction in cells transiently expressing Y3. 293T cells were transfected with the reporter plasmid pISRE-EGFP together with a plasmid for expression of the FL-C protein and its deletion mutants, FL-Y1 and FL-Y3 (Fig. 1A). IFN-α was added to the cell culture medium at 6 h post-transfection, and the cells were observed by fluorescent microscopy at 24 h post-transfection (Fig. 1C). IFN-α-induced fluorescence from pISRE-EGFP was restricted by co-expression of FL-C and FL-Y1. Fluorescence was also inhibited by FL-Y3 with a slightly reduced efficiency (Fig. 1C). Binding capacities of C protein with STAT1 and STAT2 were analyzed by an immunoprecipitation method using mixtures of cell lysates prepared from HeLa cells that express HA-STAT1, HA-STAT2, or FL-C. Immunoprecipitation of the mixtures with an anti-FLAG tag antibody and subsequent Western blot analysis showed that C protein associates with STAT1 but not with STAT2 (Fig. 1D). Although C protein may interact with STAT2 via the association with STAT1, an interaction between C protein and STAT2 was not observed even in the presence of a large amount of STAT1.
To confirm that C protein associates with STAT1ND, FL-C or FL-Y3 was transfected with HA-STAT1 or HA-STAT1ΔN. Western blot analysis showed that C protein and Y3 could not associate with STAT1ΔN, unlike the full-length STAT1, indicating that STAT1ND is important for association with C protein (Fig. 1E).
Inhibition of STAT1 phosphorylation by C protein
To investigate whether association between C protein and STAT1 is essential for the inhibition of IFN-α-induced STAT1 phosphorylation, HA-STAT1 was expressed with FL-C or FL-Y3 in STAT1-null U3A cells. Because the expression level of FL-C was higher than that of FL-Y3 when the same amount of each plasmid was applied, the amount of the FL-C plasmid was reduced to adjust the expression level of FL-C to that of FL-Y3. At 30 min after stimulation with IFN-α, which was added at 24 h after transfection, the cell lysate was prepared. Western blot analysis showed that phosphorylation of HA-STAT1 was almost completely inhibited in the presence of FL-C protein (Fig. 2, A and C). HA-STAT1ΔN was also phosphorylated at Tyr701 in the presence of IFN-α, indicating that STAT1ND is not essential for the phosphorylation (Fig. 2A). Interestingly, STAT1ΔN was phosphorylated at a low level in the absence of IFN-α. FL-C protein also significantly inhibited the phosphorylation of HA-STAT1ΔN (Fig. 2, A and C). On the other hand, as shown previously (33), FL-Y3 inhibited the IFN-α-induced phosphorylation of HA-STAT1 more weakly than did FL-C protein, and FL-Y3 much more weakly inhibited the phosphorylation of HA-STAT1ΔN (Fig. 2, B and C).
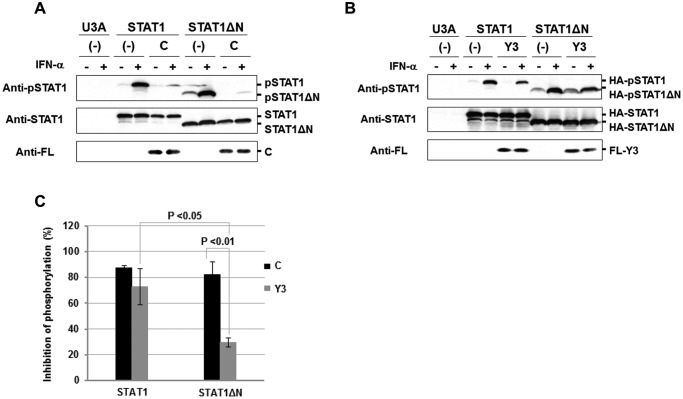
Figure 2.
Inhibition of IFN-α-induced tyrosine phosphorylation of STAT1 in the presence of C protein. U3A cells were transfected with an expression vector for HA-STAT1 or HA-STAT1ΔN together with an expression vector for FL-C (A) or FL-Y3 (B). At 0.5 h after stimulation with IFN-α (1,000 units/ml), proteins in the cell extracts were separated by SDS-PAGE for Western blot analysis with an anti-STAT1 antibody (anti-STAT1), an anti-Tyr701-phosphorylated STAT1 antibody (anti-pSTAT1) and anti-FL. C, the rate of phosphorylation inhibition was determined on the basis of averaged signal intensities of HA-pSTAT1 in the presence or absence of FL-C (A) or FL-Y3 (B), which was calculated from three independent experiments. Intensities of bands were measured using ImageJ version 1.47, and signal intensity of HA-STAT1 was used as an internal standard. An error bar indicates standard deviation. p value was calculated on the basis of Welch's test.
These results indicate that the action of C protein for inhibition of STAT1 phosphorylation cannot be explained solely by its ability to associate with STAT1ND. The N-terminal region (aa 1–97) of C protein seems to be important for the STAT1ND-independent inhibition of STAT1 phosphorylation. The localization of C protein may be important for the inhibition, because the N-terminal 1–23–aa region has been reported to cause membrane association (34). However, STAT1ND is likely to be essential for inhibition of STAT1 phosphorylation by the C-terminal half of C protein, Y3.
Inhibition of STAT2 phosphorylation by C protein
To investigate the effects of C protein on STAT2 phosphorylation, FL-STAT2 or FL-STAT2ΔN were expressed with FL-C or FL-Y3 in STAT2-null U6A cells. In this case, the amount of each plasmid was also adjusted so that the expression level of FL-C became equivalent to that of FL-Y3. After stimulation with IFN-α for 1 h, FL-C protein was also found to considerably inhibit the phosphorylation of FL-STAT2ΔN as well as that of full-length FL-STAT2 (Fig. 3, A and D). On the other hand, FL-Y3 inhibited the phosphorylation of FL-STAT2 at a lower rate than FL-C protein did, and the ratio of FL-pSTAT2ΔN versus FL-STAT2ΔN indicates that FL-Y3 inhibited the FL-STAT2ΔN phosphorylation moderately (Fig. 3, B and D).
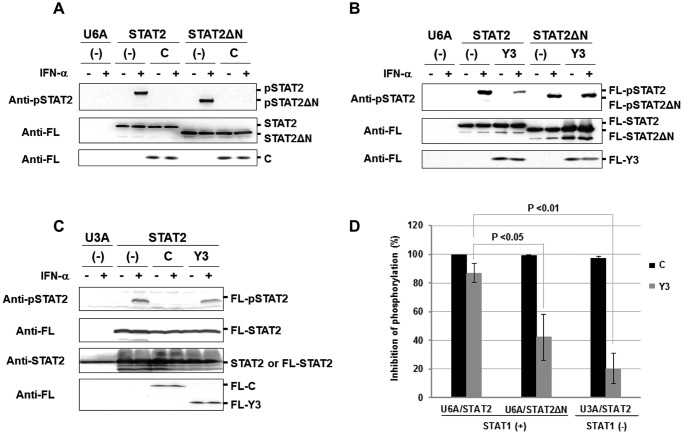
Figure 3.
Inhibition of IFN-α-induced phosphorylation of STAT2 in the presence of C protein. U6A cells in a 35-mm dish were transfected with an expression vector for FL-STAT2 (1 μg) or FL-STAT2ΔN (1 μg) together with an expression vector for FL-C (0.4 μg) (A) or FL-Y3 (1 μg) (B). At 24 h post-transfection, the cells were stimulated with IFN-α (1,000 units/ml) for 1 h. C, U3A cells were transfected with an expression vector for FL-STAT2 together with an expression vector for FL-C or FL-Y3. At 24 h post-transfection, the cells were stimulated with IFN-α (1,000 units/ml) for 1 h. Proteins in the cell extracts were separated by SDS-PAGE for Western blot analysis using an anti-Tyr690-phosphorylated STAT2 antibody (Anti-pSTAT2) and anti-FL. D, the rate of phosphorylation inhibition was determined on the basis of averaged signal intensity of FL-STAT2 in A–C, which was calculated from three independent experiments. Signal intensity of FL-STAT2 was used as an internal standard.
The phosphorylation state of endogenous STAT2 was also analyzed using STAT1-null U3A cells transiently expressing FL-C or FL-Y3 after stimulation with IFN-α. However, the analysis was difficult probably because of the low expression level of STAT2 or the low sensitivity of the antibody against phosphorylated STAT2. To increase the intracellular expression level of STAT2, U3A cells were transfected with an expression vector for FL-STAT2 together with an expression vector for FL-C or FL-Y3. The results showed that FL-C protein significantly inhibited the phosphorylation of FL-STAT2 in the absence of STAT1 as it did in the presence of STAT1 (Fig. 3, C and D). On the other hand, FL-Y3 hardly inhibited the phosphorylation of FL-STAT2 in the absence of STAT1.
These results indicate that C protein inhibits the IFN-α-induced phosphorylation of STAT2 in at least two different ways, STAT1-dependent and STAT1-independent. It is likely that Y3 predominantly inhibits the phosphorylation in a STAT1-dependent way. STAT2ND seems to be important for the STAT1-dependent inhibition of STAT2 phosphorylation by C protein.
Association of STAT1ND with STAT2ND
Considering that STAT2ND displays a high sequence homology to STAT1ND (identity = 46.9%), STAT1ND may interact with STAT2ND to generate a heterodimer, similar to the STAT1ND homodimer. At first, we tried to detect interaction between STAT1ND and STAT2ND by using ProS2-STAT1ND and STAT2ND separately purified from Escherichia coli cell lysate (supplemental Fig. S1). Although size-exclusion chromatography indicated that the homodimerization of ProS2-STAT1ND was not impaired, interaction between STAT1ND and STAT2ND was not detected, probably due to weakness of the association under a general condition. Thus, we generated a STAT1ND:STAT2ND fusion protein linked by a Gly- and Ser-rich linker-peptide consisting of 40 amino acids (Fig. 4A).
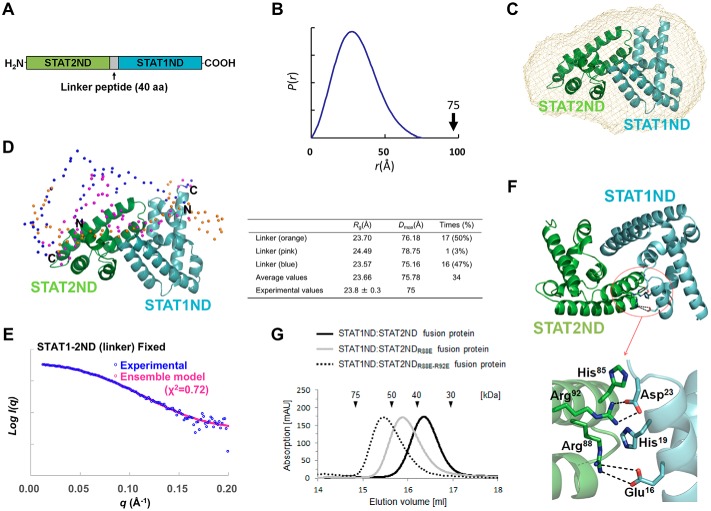
Figure 4.
Interaction between STAT1ND and STAT2ND. A, schematic diagram of the STAT1ND:STAT2ND fusion protein. B, distance distribution function of the fusion protein obtained by an SAXS experiment. C, heterodimer model of STAT1ND (cyan) and STAT2ND (green), shown as ribbon representation, is imposed on a low-resolution bead model calculated using DAMMIF by fitting to the experimental scattering curve shown in B. The STAT1ND:STAT2ND heterodimer model was created by replacing one subunit in the STAT1ND homodimer (PDB code 3WWT) with a STAT2ND homology model generated using SWISS-MODEL server (EXPASY) and the “align” command in PyMOL (58). D, an ensemble model generated by EOM analysis using atomic coordinates of the STAT1ND:STAT2ND heterodimer model. Cα atoms in the linker peptide are shown in sphere representation. STAT1ND and STAT2ND are colored in blue and orange, respectively, and their N and C termini are marked. Structural parameters obtained by EOM analysis are shown in the table. E, experimental scattering curve of the fusion protein and theoretical scattering curve of the ensemble model are colored in blue and magenta, respectively. F, the STAT1ND:STAT2ND heterodimer model is shown in ribbon representation. Carbon atoms in STAT1ND and STAT2ND are colored in cyan and green, respectively. The region enclosed with a red circle is enlarged below. Glu16, Asp23, and His19 from STAT1ND and His85, Arg88, and Arg92 from STAT2ND are shown in a stick model. The dotted lines represent possible hydrogen bonds. G, analytical size-exclusion chromatograms of STAT1ND:STAT2ND (black line), STAT1ND:STAT2NDR88E (gray line), or STAT1ND:STAT2NDR88E-R92E fusion protein (dotted line) are shown. All of the structural drawings were made using PyMOL.
The distance distribution function P(r) of the fusion protein obtained by SAXS analysis showed a single peak with no tailing. The maximum dimension (Dmax) of particles was estimated to be 75 Å (Fig. 4B). Dmax of the STAT1ND:STAT2ND heterodimer model (72 Å), which was created on the basis of the crystal structure of the STAT1ND homodimer (Fig. 4C), is in good agreement with the estimated value. The structure of the fusion protein is thought to be compact like that of the STAT1ND:STAT2ND heterodimer model, whereas the linker peptide between STAT1ND and STAT2ND may be flexible. A dummy atom model, reconstructed from experimental SAXS data by using the program DAMMIF, also showed good agreement with the STAT1ND:STAT2ND heterodimer model (Fig. 4C). No electron density corresponding to the linker peptide was observed in the dummy atom model, suggesting that the linker peptide fluctuates in a solution.
Based on the ensemble optimization method (EOM), an ensemble structure of the STAT1ND:STAT2ND fusion protein was made by using an experimental scattering curve and atomic coordinates of STAT1ND and STAT2ND connected by a flexible linker (supplemental Fig. S2). Although each structure obtained by the EOM was dissimilar to that of the STAT1ND homodimer, all of the structures were compact. Next, we fixed the relative orientation between STAT1ND and STAT2ND as in the STAT1ND:STAT2ND heterodimer model and rebuilt the ensemble model. The resultant ensemble structure exhibited good agreement with the experimental scattering curve (Fig. 4, D and E). Heterodimerization of STAT1ND and STAT2ND is likely to occur in a manner similar to that for the homodimerization of STAT1ND.
In the models, Arg88 and Arg92 of STAT2ND interact with Glu16 and Asp23 of STAT1ND, respectively (Fig. 4F). To validate the heterodimeric model, Arg88 and Arg92 of STAT2ND were each replaced by Glu and the mutated fusion proteins were analyzed by size-exclusion chromatography. The estimated molecular masses of STAT1ND:STAT2NDR88E fusion protein (49.7 kDa) and STAT1ND:STAT2NDR88E-R92E fusion protein (61.1 kDa) were larger than that of the wild-type fusion protein (37.5 kDa) (Fig. 4G), suggesting that the whole structure of the mutants was not compact.
Inhibition of IFN-α signal transduction in the presence of STAT2 variants
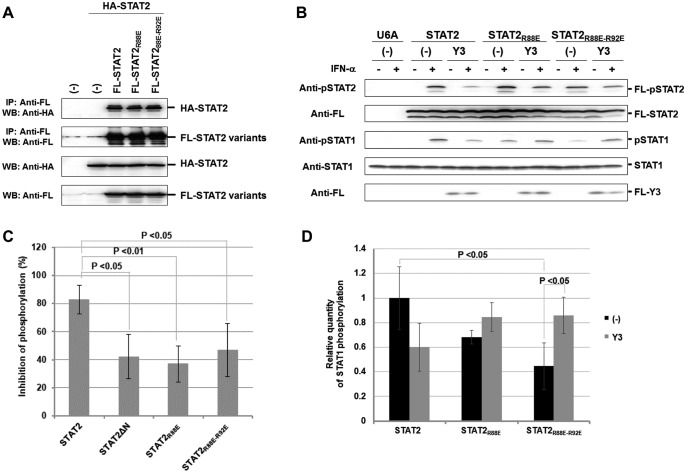
Co-immunoprecipitation experiments showed that R88E and R92E mutations in FL-STAT2 have no influence on the interaction with wild-type HA-STAT2 (Fig. 5A), indicating that homodimerization of STAT2 is not impaired by the mutations. To examine whether interaction between STAT1ND and STAT2ND is related to the inhibition of STAT2 phosphorylation by Y3, FL-STAT2 or the R88E mutant (FL-STAT2R88E) or R88E-R92E mutant (FL-STAT2R88E-R92E) were transiently expressed with FL-Y3 in U6A cells. The amount of the FL-Y3 plasmid co-transfected with FL-STAT2R88E or FL-STAT2R88E-R92E was adjusted so that expression levels of FL-Y3 became equivalent. At 1 h after stimulation with IFN-α, inhibition rates of phosphorylation in FL-STAT2R88E or FL-STAT2R88E-R92E by FL-Y3 were significantly decreased (Fig. 5, B and C). The inhibition rates were comparable with that in FL-STAT2ΔN (Fig. 5C, data from Fig. 3D). These results suggest that interaction between STAT1ND and STAT2ND is necessary for the STAT1-dependent inhibition of STAT2 phosphorylation by C protein. On the other hand, although endogenous STAT1 was not phosphorylated in response to stimulation with IFN-α in U6A cells, it was phosphorylated when FL-STAT2 or one of the mutants was transiently expressed (Fig. 5, B and D). In the case of FL-STAT2R88E or FL-STAT2R88E-R92E-expressed cells, the phosphorylation level of STAT1 was decreased to about half. An interaction between STAT1ND and STAT2ND may promote the IFN-α-induced phosphorylation of STAT1. Additionally, phosphorylated STAT1 was accumulated by Y3 in FL-STAT2R88E or FL-STAT2R88E-R92E-expressed cells. Long-term IFN-α stimulation causes accumulation of phosphorylated STAT1 in C protein-expressing cells (21). Loss of the interaction between STAT1ND and STAT2ND may facilitate the accumulation of phosphorylated STAT1 by C protein.
Figure 5.
Inhibition of the phosphorylation of STAT2 with decreased affinity to STAT1ND in the presence of Y3. A, HeLa cells were transfected with an expression vector for FL-STAT2, FL-STAT2R88E, or FL-STAT2R88E-R92E together with an expression vector for wild-type HA-STAT2. At 24 h post-transfection, the cell lysate was immunoprecipitated with anti-FL antibody and analyzed by Western blotting using anti-HA and anti-FL antibodies. B, U6A cells were transfected with an expression vector for FL-STAT2, FL-STAT2R88E, or FL-STAT2R88E-R92E together with an expression vector for FL-Y3. At 24 h post-transfection, the cells were stimulated with IFN-α (1,000 units/ml) for 1 h (B). Proteins in the cell extracts were separated by SDS-PAGE for Western blot analysis using anti-pSTAT2, anti-FL, anti-STAT1, and anti-phosphorylated STAT1 antibodies. C, the rate of phosphorylation inhibition was determined on the basis of averaged signal intensity of FL-pSTAT2 in B, which was calculated from three independent experiments. Signal intensity of FL-STAT2 was used as an internal standard. D, relative quantities of phosphorylated STAT1 were determined on the basis of the average signal intensity of pSTAT1 in B, which was calculated from three independent experiments. Signal intensity of STAT1 was used as an internal standard.
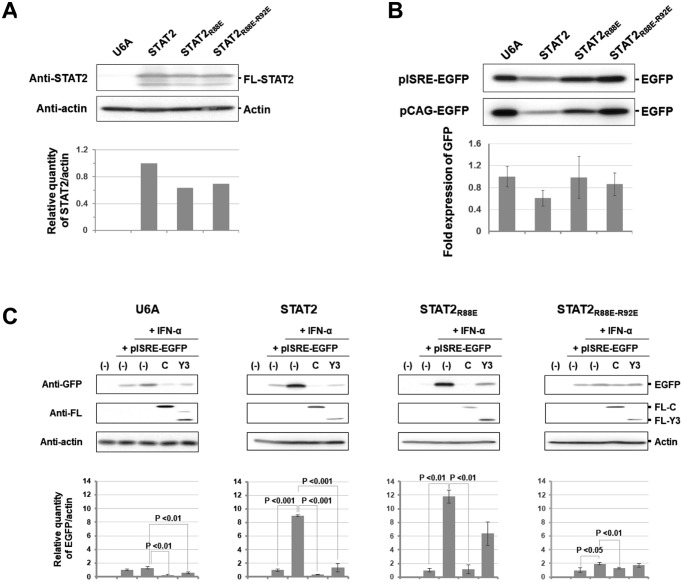
To investigate the strength of signal transduction, U6A cells constitutively expressing wild-type FL-STAT2 (U6A-STAT2WT cells), the R88E mutant (U6A-STAT2R88E cells), or the R88E-R92E mutant (U6A-STAT2R88E-R92E cells) were established. Western blot analysis showed that expression levels of wild-type FL-STAT2 and the mutants in U6A cells were comparable (Fig. 6A). After each cell line had been transfected with pISRE-EGFP or pCAG-EGFP, the cell lysate was prepared for Western blot analysis to detect the expression of EGFP. The basal expression levels of EGFP under control of the ISRE promoter were different in U6A cells. However, it was confirmed that the expression level of EGFP under control of the ISRE promoter was correlated to that under control of the actin promoter. The difference in the basal expression levels of EGFP seems to be mainly due to the transfection efficiency of the cells (Fig. 6B).
Figure 6.
Response to IFN-α of U6A cells constitutively expressing STAT2 mutants. A, quantification analysis of constitutively expressed wild-type or a mutant of FL-STAT2 in U6A cells. The cell lysate was analyzed by Western blotting using an anti-STAT2 antibody (anti-STAT2) and anti-actin antibody (anti-actin). B, comparison of ISRE reporter activities at the basal level in U6A cells constitutively expressing wild-type or a mutant of FL-STAT2. To estimate the strength of ISRE reporter activities in the absence of IFN-α stimulation, subconfluent U6A cells constitutively expressing FL-STAT2 or a mutant were transfected with pISRE-EGFP. At 24 h after transfection, the cell lysate was analyzed by Western blotting using an anti-GFP antibody (anti-GFP). At the same time, U6A cells were transfected with pCAG-EGFP, and the cell lysate was used as an external standard. C, EGFP reporter assay for signal transduction of IFN-α using U6A cells. To estimate the strength of the response to IFN-α, subconfluent U6A cells and cells constitutively expressing FL-STAT2 or a mutant were transfected with pISRE-EGFP and an expression plasmid for FL-C or FL-Y3, and IFN-α (2,000 units/ml) was added at 16 h post-transfection. After 8 h, the cell lysate was analyzed by Western blotting using anti-GFP (anti-GFP), anti-actin, and anti-FL antibodies. Quantified intensity of EGFP calculated from three independent experiments is shown in the graph. An error bar indicates standard deviation. Signal intensity of actin was used as an internal standard in this figure.
Each cell line was transfected with pISRE-EGFP along with the expression vector of FL-C or FL-Y3 to investigate the response after addition of IFN-α (Fig. 6C). Although U6A cells were unresponsive to IFN-α, FL-C protein seemed to inhibit the basal activity level of the ISRE promoter. As in U6A cells, a specific response after stimulation with IFN-α was weak in U6A-STAT2R88E-R92E cells, suggesting that the innate immunity system is disordered by the mutation. On the other hand, IFN-α induced EGFP expression in U6A-STAT2WT and U6A-STAT2R88E cells. FL-C protein was found to inhibit the signal transduction of IFN-α in U6A cells expressing FL-STAT2 or its mutants. On the other hand, the rate of inhibition of signal transduction by FL-Y3 was decreased in U6A-STAT2R88E and U6A-STAT2R88E-R92E cells compared with that in U6A-STAT2WT cells, indicating that interaction between STAT1ND and STAT2ND is important for Y3 to block the IFN-α signal transduction.
Importance of interaction between STAT1ND and STAT2ND in SeV-mediated inhibition of signal transduction
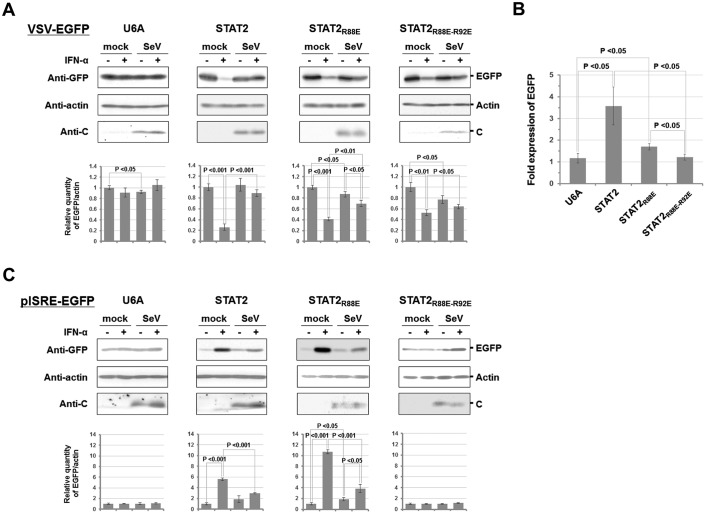
To determine whether interaction between STAT1ND and STAT2ND is important for SeV to escape from host innate immunity, SeV-infected U6A cells or the derivatives were superinfected with recombinant vesicular stomatitis virus expressing enhanced GFP (rVSV-EGFP) in the absence or presence of IFN-α. After incubation for 10 h, EGFP expression was evaluated by Western blot analysis using an anti-GFP antibody (Fig. 7A). The expression level of EGFP in U6A cells, showing VSV replication, was almost unchanged by the addition of IFN-α and/or by SeV infection probably because of STAT2 deficiency. On the other hand, VSV replication in mock-infected U6A cells in which FL-STAT2 or its mutants had been introduced was reduced in the presence of IFN-α, although the rates of reduction in VSV replication in U6A-STAT2R88E and U6A-STAT2R88E-R92E cells were lower than in U6A-STAT2 cells. The innate immunity system seems to be partially impaired by mutations in STAT2. The decrease in the response to IFN-α was more pronounced in U6A-STAT2R88E-R92E cells than in U6A-STAT2R88E cells.
Figure 7.
Antiviral action of IFN-α in U6A cells expressing STAT2 mutants after SeV infection. A, U6A cells constitutively expressing FL-STAT2 or the mutant were infected with SeV. At 1 h post-infection, the cells were incubated with serum-free DMEM for 4 h at 37 °C, followed by the addition of IFN-α (2,000 units/ml) to the culture medium. After an additional incubation for 9 h, the cells were superinfected with rVSV-EGFP. After further incubation for 10 h, cell lysates were prepared to estimate the expression levels of rVSV-EGFP-derived EGFP and SeV C protein by Western blot analysis. B, ratio of EGFP expression in IFN-α-treated SeV-infected cells to that in mock-infected cells. C, to monitor the signal transduction of IFN-α in SeV-infected cells, subconfluent U6A cells were transfected with pISRE-EGFP. After 10 h, the cells were infected with SeV as mentioned above. The cells were incubated with serum-free DMEM for 4 h at 37 °C, followed by the addition of IFN-α (2,000 units/ml) to the culture medium. After an additional incubation for 9 h, the cell lysate was analyzed by Western blotting. Relative densities of EGFP in A and C are shown as a bar graph, in which density of EGFP in the IFN-α-untreated mock-infected cells was normalized to 1.
VSV replication was increased in SeV-infected U6A-STAT2 cells compared with that in mock-infected U6A-STAT2 cells in the presence of IFN-α, indicating that SeV inhibits the signal transduction. However, the rate of increase in VSV replication caused by SeV infection was lower in U6A-STAT2R88E and U6A-STAT2R88E-R92E cells than in U6A-STAT2 cells (Fig. 7A).
To compare the anti-IFN-α actions of SeV among the established U6A cells, the ratio of EGFP expression in IFN-α-treated SeV-infected cells to that in IFN-α-treated mock control cells was calculated. As shown in Fig. 7B, the ratio in U6A-STAT2R88E cells was lower than that in U6A-STAT2 cells. In addition, the ratio in U6A-STAT2R88E-R92E cells was decreased to about 1, indicating that the anti-IFN-α effect of SeV had almost disappeared.
Western blot analysis using U6A cells transfected with pISRE-EGFP, a reporter plasmid for ISRE promoter activity, showed that the expression level of EGFP in U6A cells was almost unchanged by the addition of IFN-α and/or by SeV infection (Fig. 7C). Similar results were obtained when U6A-STAT2R88E-R92E cells were used, suggesting that R88E and R92E double mutations severely inhibited ISRE signal transduction. In the case of U6A-STAT2 and U6A-STAT2R88E cells, ISRE signal transduction was responsive to IFN-α, and SeV infection moderately decreased EGFP expression, suggesting that SeV weakened the antivirus action of the added IFN-α (Fig. 7C). The ISRE promoter was strongly driven by IFN-α in U6A-STAT2R88E cells and it was more strongly driven than in U6A-STAT2 cells, but the reason is unknown. In summary, interaction between STAT1ND and STAT2ND seems to be important for the regulation of IFN-α-induced innate immunity, and SeV efficiently inhibits IFN-α signal transduction in the presence of STAT1ND–STAT2ND interaction.
Interactions among Y3, STAT1ND, and STAT2ND
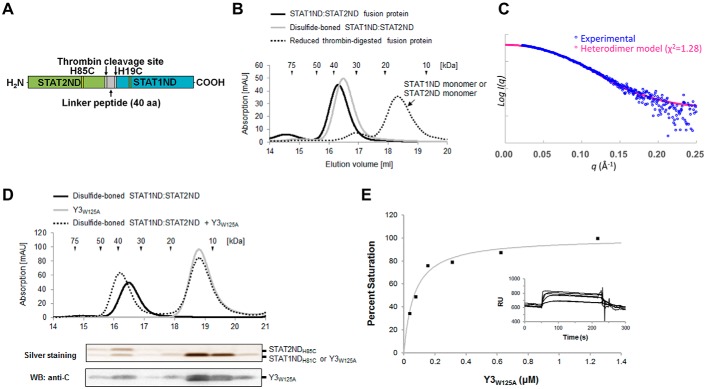
When STAT1ND forms a homodimer, His19 in one subunit is close to His81 in the other subunit. To generate a linker-less recombinant protein possessing a STAT1ND:STAT2ND heterodimeric structure, a STAT1ND mutant, in which His19 was replaced by Cys, was linked to a STAT2ND mutant, in which His85 (corresponding to His81 in STAT1ND) was replaced by Cys, via a linker peptide harboring two thrombin-recognition sites (Fig. 8A). After purification, the fusion protein was incubated in the presence of oxidant cystine to facilitate the formation of a disulfide bond, followed by digestion with thrombin to remove the linker peptide. Analytical size-exclusion chromatography showed that the estimated molecular mass of the resulting protein was 39 kDa in the presence of 0.5 mm cystine, which is close to the calculated molecular mass (33 kDa) based on the sequence. However, the estimated molecular mass in the presence of a reductant (1 mm dithiothreitol) was 18 kDa, which is close to the calculated molecular masses of STAT1ND (16 kDa) and STAT2ND (18 kDa) (Fig. 8B). Although STAT1ND and STAT2ND at high concentrations are known to form a homodimer, it is unlikely that the conditions used in the experiment were sufficient for formation of a homodimer. Furthermore, the experimental scattering curve of the resulting protein, which was obtained by SAXS measurement, exhibited good agreement with the calculated scattering curve of the STAT1ND:STAT2ND heterodimer model (Fig. 8C). These results indicate that the resulting protein takes a heterodimeric structure similar to the STAT1ND homodimeric structure.
Figure 8.
Binding between Y3 and STAT1ND:STAT2ND heterodimer. A, schematic diagram of the fusion protein to generate a disulfide-bonded STAT1ND:STAT2ND heterodimer. B, analytical size-exclusion chromatograms of the fusion protein (black line) and the disulfide-bonded STAT1ND:STAT2ND heterodimer in the presence (dotted line) or absence (gray line) of 1 mm dithiothreitol. C, experimental scattering curve of the disulfide-bonded STAT1ND:STAT2ND heterodimer in the presence of 0.5 mm cystine and theoretical scattering curve of STAT1ND:STAT2ND heterodimer are colored in blue and magenta, respectively. D, analytical size-exclusion chromatograms of Y3W125A (gray line) and disulfide-bonded STAT1ND:STAT2ND heterodimer in the presence (dotted line) or absence (black line) of Y3W125A. Proteins in fractions eluted from a mixture of the disulfide-bonded STAT1ND:STAT2ND heterodimer and 10-fold excess of Y3W125A were separated by SDS-PAGE, followed by silver staining and Western blotting using an anti-C antibody. E, binding of Y3W125A to the disulfide-bonded STAT1ND:STAT2ND heterodimer immobilized on the sensor chip. The maximal increase in RU indicates that one molecule of Y3W125A binds to the STAT1ND:STAT2ND heterodimer. The graph shows the binding ratio against the concentration of Y3W125A. Each point in the graph corresponds to experimental data, whereas the curve is theoretically drawn. The inset shows the change in sensorgrams after injecting Y3W125A. RU just before immobilization of the disulfide-bonded STAT1ND:STAT2ND heterodimer was set to 0.
We found that replacement of the Trp125 residue with Ala in Y3, which is located on a molecular surface distant from the STAT1ND-binding site (supplemental Fig. S3), facilitated in vitro analysis by increasing the solubility of Y3. We thus used Y3W125A instead of Y3 in the following experiments. To examine whether Y3 binds to the disulfide-bonded STAT1ND:STAT2ND heterodimer, the heterodimer was fractionated by analytical size-exclusion chromatography in the presence of Y3W125A, and analyzed by SDS-PAGE (Fig. 8D). The results of silver staining and Western blot analyses using an anti-C protein antibody showed that Y3W125A was co-eluted with the disulfide-bonded STAT1ND:STAT2ND heterodimer. To elucidate the stoichiometry of Y3 to the disulfide-bonded STAT1ND:STAT2ND heterodimer by SAXS analysis, a fully Y3-bound heterodimer was prepared from a mixture of the heterodimer with an excess of Y3 by using size-exclusion chromatography. However, errors of the experimental molecular weight and radius of gyration (Rg) of the complex measured by SAXS analysis are too large to distinguish one Y3-bound and two Y3-bound types (supplemental Table S1).
We also performed binding analysis by a surface plasmon resonance (SPR) experiment to determine the stoichiometry between Y3W125A and the disulfide-bonded STAT1ND:STAT2ND heterodimer. Because the STAT1ND:STAT2ND heterodimer was immobilized on an NTA sensor chip via the histidine tag, the Y3W125A mutant without the histidine tag should be used as an analyte. The analyte was prepared by digestion of ProS-Y3W125A, in which the histidine-tagged ProS2 was fused at the N terminus, with thrombin. Increased responsive unit (RU) values caused by the association of Y3W125A with the disulfide-bonded STAT1ND:STAT2ND heterodimer were obtained by injecting different concentrations of Y3W125A. Affinity analysis showed that our experimental data fitted well to a 1:1 binding mode, indicating that one molecule of Y3W125A binds to the disulfide-bonded STAT1ND:STAT2ND heterodimer (Fig. 8E). The dissociation constant (Kd) of the heterodimer for Y3W125A was determined to be 0.065 ± 0.032 μm. Y3W125A may have a stronger affinity for the STAT1ND:STAT2ND heterodimer than for the STAT1ND homodimer (the Kd1 and Kd2 values for the STAT1ND homodimer were determined to be 0.68 ± 0.17 and 0.49 ± 0.08 μm, respectively) (33), although the methods used to estimate the dissociation constant were different. In addition, stoichiometry in the fully Y3-bound disulfide-bonded STAT1ND:STAT2ND heterodimer was analyzed on the basis of the intensity of band signals of STAT2NDH85C (supplemental Fig. S4A) and Y3 (supplemental Fig. S4B). According to the standard curves to determine concentrations of STAT2NDH85C and Y3, the prepared complex contained almost equimolar concentrations of STAT2NDH85C and Y3 (supplemental Fig. S4C), supporting the results obtained by the SPR experiments.
Discussion
We previously reported that the association of one molecule of C protein with the STAT1ND homodimer could cause reduction of phosphorylated STAT1 molecules, whereas the association of two molecules of C protein causes reduction of intact STAT1 molecules by complex formation, thereby inhibiting IFN-γ signaling (33). C protein is also known to inhibit IFN-α-induced phosphorylation of STAT1 and STAT2 molecules, which is thought to be important for escaping from host innate immunity.
Because C protein is unable to directly interact with STAT2, the mechanism by which STAT2 phosphorylation is inhibited has remained elusive. STAT1ND, which has a strong affinity toward C protein, is expected to interact with the N-terminal domain of STAT2 (STAT2ND) on the basis of the sequence similarity. However, no interactions between N-terminal domains of hetero-STAT proteins have been detected by using the yeast two-hybrid system (35). Thus, the N-terminal domain of STATs is believed to specifically form a homodimer, not a heterodimer. On the other hand, it was recently shown by equilibration sedimentation analysis with proteins prepared from a Baculovirus expression system that STAT1ND can bind to STAT2ND with significantly strong affinity (36). Although our gel-filtration results did not support the strong affinity of STAT1ND with STAT2ND, STAT1ND might have the potential to form a heterodimer with STAT2ND.
Gly-rich linkers have been proven to be useful to gain structural insights for weak or transient interactions by creating a covalent link between the structural elements (37). For example, an interaction between P and NP proteins from measles virus in the family Paramyxoviridae was determined by using X-ray and NMR techniques with the help of a Gly-rich linker peptide (38). In this study, we demonstrated that STAT1ND can associate with STAT2ND with the help of a Gly-rich linker or artificial disulfide bond. The use of a Gly-rich linker was also shown to be helpful for detecting the possible heterodimerization between proteins that each easily forms a stable homodimer.
By using STAT2 mutants with decreased affinity with STAT1ND, the interaction between STAT1ND and STAT2ND was found to be important for Y3 to inhibit IFN-α signaling. Additionally, one molecule of Y3 was suggested to associate with the heterodimeric structure of STAT1ND and STAT2ND (Fig. 8), in contrast to previous results showing that two molecules of Y3 associate with the homodimeric structure of STAT1ND. Although two Y3-binding sites are formed at the dimer interface in the STAT1 homodimer, Y3 mainly interacts with one of the two STAT1ND subunits. Therefore, a Y3 molecule is likely to be accommodated into one of the two possible Y3-binding sites in the STAT1ND:STAT2ND heterodimer, where Y3 mainly interacts with STAT1ND.
It was still unclear how the formation of a tertiary complex by STAT1, STAT2, and C proteins leads to the inhibition of STAT2 phosphorylation. However, the STAT1:STAT2 heterodimer is assumed to be dephosphorylated in the same manner as the STAT1 homodimer. We propose the following scenario for C protein–mediated inhibition of STAT2 phosphorylation. One molecule of C protein associates with the dimeric structure formed between STAT1ND and STAT2ND. Thus, the C protein-bound STAT1:STAT2 heterodimer is induced to take an antiparallel form, which is easily dephosphorylated.
Seven STAT proteins (STAT1–4, -5a, -5b, and -6) have been identified in humans. These proteins are crucial to transduce the signaling of numerous ligands into the nucleus, including cytokines, growth factors, and hormones. Dysregulated activation of STAT signaling is involved in chronic inflammation and cancer, including blood malignancies and solid tumors (39, 40). Recently, it was reported that the loss of interaction between N-terminal domains of STAT1 causes a collapse of bacterial resistance in mice (41). In addition, the loss of interaction between N-terminal domains of STAT3 has been suggested to be involved in inflammatory hepatocellular adenoma in humans (42). In U6A-STAT2R88E-R92E cells, specific activity of the ISRE promoter induced by IFN-α was significantly impaired (Figs. 6 and 7A), and the antiviral effect of IFN-α was also reduced (Fig. 7, A and C). Detailed characterization of the cells, especially in terms of the relationship with disease, will be awaited.
The current study showed that C protein has a STAT1-dependent mechanism for inhibition of STAT2 phosphorylation. On the other hand, the results also showed that C protein can inhibit the phosphorylation of STAT2 through a different mechanism in which interaction between STAT1ND and STAT2ND is unnecessary. IFN-α-induced phosphorylation of Tyk2 has been reported to be inhibited in SeV-infected cells (43), although the mechanism is unclear. C protein may inhibit the phosphorylation of Tyk2, leading to the inhibition of phosphorylation of STAT1 and STAT2 by Tyk2. A study to understand the alternative mechanism is now in progress.
Experimental procedures
Cells, antibodies, and viruses
293T cells, which are human embryonic kidney cells expressing simian virus 40 T antigen, and HeLa cells (CCL-2; human cervical cancer-derived cells) were purchased from RIKEN Cell Bank and ATCC, respectively. U3A cells (STAT1-null cells) and U6A cells (STAT2-null cells), of which the parent cells are 2fTGH cells (human fibrosarcoma cells), were purchased from DS Pharma Biochemical (Osaka, Japan). These cells were propagated in Dulbecco's modified minimum essential medium (DMEM) supplemented with 10% fetal calf serum (Biological Industries, Kibbutz, Israel) and 100 units/ml of penicillin G, 100 μg/ml of streptomycin (Invitrogen). To maintain U3A and U6A cell lines constitutively expressing proteins by using the pKS336 vector, the medium was additionally supplemented with 10 μg/ml of blasticidin S (Wako Pure Chemical Industries, Osaka, Japan). Mouse monoclonal antibodies against HA tag (G036; Abcam), FLAG tag (M2; Sigma), STAT1 (610185; BD Transduction Laboratories), and actin (MAB1501; Chemicon International), rabbit monoclonal antibodies against Tyr701-phosphorylated STAT1 (sc-7988-R; Santa Cruz Biotechnology) and Tyr690-phosphorylated STAT2 (sc-21689-R; Santa Cruz Biotechnology), and rabbit polyclonal antibodies against green fluorescent protein (GFP) (sc-8334; Santa Cruz Biotechnology) and STAT2 (C-20, sc-476; Santa Cruz Biotechnology) were used according to the manufacturer's instructions. Anti-SeV C rabbit antiserum was generated by using purified Y3 protein as an antigen. rVSV-EGFP was described previously (12). Wild-type SeV derived from a cDNA of the Z strain was propagated in embryonated chicken eggs (44), and infectivity was measured using an immunofluorescent infectious focus assay and expressed as cell infectious units/ml (45).
Plasmid construction
The cDNA fragments encoding N terminally HA-tagged STAT1 lacking the N-terminal domain (HA-STAT1ΔN; aa 132–750), N terminally HA-tagged STAT2 (HA-STAT2), N terminally FLAG-tagged STAT2 (FL-STAT2; aa 1–851), N terminally FLAG-tagged STAT2 lacking the N-terminal domain (FL-STAT2ΔN; aa 135–851), and EGFP were prepared by using PCR and subcloned into the pCAGGS vector under control of the chicken β-actin promoter (46) to generate pCAG-HA-STAT1ΔN, pCAG-HA-STAT2, pCAG-FL-STAT2, pCAG-FL-STAT2ΔN, and pCAG-EGFP, respectively. For the expression of R88E and R88E-R92E mutants of FL-STAT2 (FL-STAT2R88E and FL-STAT2R88E-R92E), mutations were introduced into the pCAG-FL-STAT2 using a KOD-Plus Mutagenesis Kit (TOYOBO). For preparation of cell lines with constitutive protein expression, the cDNA fragments of FL-STAT2 and its mutants were subcloned into the pKS336 vector that had the human elongation factor promoter and blasticidin S-resistant gene (47). pCAG-HA-STAT1, an expression plasmid for N terminally HA-tagged STAT1 (HA-STAT1; aa 1–750), and pCAG-FL-C and pCAG-FL-Y3, expression plasmids for N terminally FLAG-tagged SeV C (FL-C; aa 1–204), and Y3 (FL-Y3; aa 99–204), respectively, were described previously (33). pISRE-EGFP, which possesses an ISRE element upstream of the egfp gene, was described previously (48).
For bacterial expression, cDNA fragments encoding Y3 and STAT1ND were cloned into the NdeI-XhoI sites of the original or the modified pCold ProS2 vector (33). For expression of the W125A mutant of Y3 (Y3W125A), mutagenesis was performed using a KOD-Plus Mutagenesis Kit. For the expression of a fusion protein of STAT1ND with STAT2ND via a linker peptide (STAT1ND:STAT2ND fusion protein), the DNA fragment encoding STAT1ND was cloned into the NdeI-XhoI sites of the original pCold ProS2 vector to generate pCold ProS2-STAT1ND. Then the DNA fragment encoding the N-terminal domain of STAT2 (STAT2ND; aa 1–129) was inserted just after the His-tag sequence of pCold ProS2-STAT1ND using an In-Fusion HD Cloning Kit (Clontech). The resulting plasmid was amplified by PCR with a forward primer, 5′-GGAATTCCATATGTCTCAGTGGTACGAACTTCAG-3′ (underline indicating an EcoRI site), and reverse primer 1, 5′-GGAATTCAGAACCACCGCCACCGCTCCCACCGCCGCCAGAACCGCCACCTCCTGAGCTTTGTTCCAATTGGG-3′ (underline indicating an EcoRI site). The PCR product was digested with EcoRI, followed by self-ligation to generate an expression plasmid for the fusion protein of STAT1ND with STAT2ND via a short linker peptide. To extend the length of the linker peptide, the resulting plasmid was amplified by PCR with the aforementioned forward primer and reverse primer 2, 5′-GGAATTCGGTACCGCCTCCGCCGGAACCACCACCGCCAGAACCACCGCCACC-3′ (underline indicating an EcoRI site), followed by digestion with EcoRI and self-ligation. The process was repeated using the forward primer and reverse primer 3, 5′-GGAATTCTGATCCTCCACCTCCTGATCCTCCGCCTCCGGTACCGCCTCC-3′ (underline indicating an EcoRI site), generating an expression plasmid for STAT1ND fused with STAT2ND via a Gly- and Ser-rich linker peptide of 40 aa in length. For expression of the STAT1ND:STAT2NDR88E and STAT1ND:STAT2NDR88E-R92E fusion proteins, which have mutation in the STAT2ND sequence of the STAT1ND:STAT2ND fusion protein, mutagenesis was performed using a KOD-Plus Mutagenesis Kit. For constructing a recombinant protein with a disulfide-bonded heterodimeric structure of STAT1ND and STAT2ND (disulfide-bonded STAT1ND:STAT2ND heterodimer), the cDNA fragment encoding the H19C mutant of STAT1ND (STAT1NDH19C), of which the amino terminus was linked to the carboxyl terminus of the H85C mutant of STAT2ND (STAT1NDH85C) via the linker peptide NH2-LELVPRGSSSGGGGSGGGGSGGGGSGGGGSGGGGTGGGGSGGGGSEFLVPRGSGS-COOH (underline indicating a thrombin-recognition site), was synthesized by Integrated DNA Technologies (Coralville, IA) and cloned into the NdeI-XhoI sites of the modified pCold ProS2 vector.
Co-immunoprecipitation and Western blotting
HeLa cells in a 12-well plate were transfected with the indicated plasmids using FuGENE HD reagent (Promega). After 24 h, the cells were solubilized in 0.25 ml of Nonidet P-40 lysis buffer (1% Nonidet P-40, 25 mm Tris-HCl (pH 7.6), 150 mm NaCl, 1 mm EDTA, 5% glycerol and Complete Mini Protease Inhibitor Mixture (Roche Diagnostics)). Cell lysates were immunoprecipitated with an anti-FLAG antibody together with protein G-Sepharose (GE Healthcare). The immunoprecipitates were washed three times with cell lysis buffer and once with wash II buffer (50 mm Tris-HCl (pH 7.4), 50 mm NaCl, and 5 mm EDTA) and then analyzed by Western blotting using an anti-HA or anti-FLAG antibody after separation by SDS-PAGE. Protein bands were detected by using horseradish peroxidase (HRP)-conjugated anti-mouse IgG antibody and Luminata Forte Western HRP Substrate (Millipore), followed by analysis using a LumiCube imaging analyzer (Liponics, Tokyo, Japan). A part of the cell lysates was also processed for SDS-PAGE and Western blotting to confirm protein expression.
Reporter assay
For IFN-α/β signal transduction, subconfluent 293T cells in a 35-mm dish were transfected with pISRE-EGFP (0.5 μg) and one of the C expression plasmids (0.5 μg) using FuGENE HD reagent, and IFN-α (20 units/ml; Mochida Pharmaceutical, Tokyo, Japan) was added to the culture medium at 6 h post-transfection. At 24 h, photographs were taken under an immunofluorescent microscope (TE2000-S; Nikon). When using U6A cells constitutively expressing FL-STAT2, FL-STAT2R88E, or FL-STAT2R88E-R92E, subconfluent cells in a 24-well plate were transfected with pISRE-EGFP (1 μg) together with pCAG-FL-C (0.4 μg) or pCAG-FL-Y3 (1 μg) using FuGENE HD reagent. The amount of pCAG-FL-C was adjusted so that the expression level of FL-C became equivalent to that of FL-Y3. At 16 h after transfection, the culture medium was replaced by a fresh medium supplemented with IFN-α (2,000 units/ml). At 24 h, cells were observed under a fluorescent microscope, and cell lysates were subsequently prepared for Western blot analysis using an anti-GFP antibody and an anti-actin antibody. Protein bands were visualized as described above. Band signals were quantified using ImageJ version 1.47 (National Institutes of Health, Bethesda, MD). Band intensities of actin were used for calibration among samples. Standard deviations were calculated from the data of at least three experiments.
Protein preparations
Expression of wild-type Y3, Y3W125A, or ProS2-Y3W125A was carried out in E. coli BL21(DE3) CodonPlus RIL (Novagen) at 15 °C for 24 h by induction with 0.2 mm isopropyl 1-thio-β-d-galactopyranoside, and expression of the STAT1ND:STAT2ND fusion protein was carried out in E. coli BL21(DE3)pLysS (Novagen) in a similar manner. All proteins were N terminally histidine-tagged and purified by nickel affinity chromatography using His-Bind Resin (Novagen) according to the supplier's instruction manual. For preparation of the disulfide-bonded STAT1ND:STAT2ND heterodimer, the STAT1NDH19C:STAT2NDH85C fusion protein was digested by thrombin using a Thrombin Cleavage Capture Kit (Novagen) according to the supplier's instruction manual in the presence of 0.5 mm cystine and was separated from the linker peptide by using nickel affinity chromatography. For preparation of the histidine tag-less Y3W125A, ProS2-Y3W125A was digested by thrombin, and the histidine-tagged ProS2 was removed by using His-Bind Resin. For preparation of the complex of disulfide-bonded STAT1ND:STAT2ND heterodimer with Y3 or Y3W125A, a large-scale supernatant from Y3- or Y3W125A-expressing E. coli cell lysate was mixed with the disulfide-bonded STAT1ND:STAT2ND heterodimer and separated from free Y3 or Y3W125A by gel filtration chromatography equilibrated with the above buffer containing 0.5 mm cystine instead of 1 mm dithiothreitol.
Protein concentrations of Y3, Y3W125A, ProS2-Y3W125A, histidine tag-less Y3W125A, STAT1ND:STAT2ND fusion protein, and disulfide-bonded STAT1ND:STAT2ND heterodimer were determined by measuring the absorbance at 280 nm using the molar extinction coefficients 22,460, 16,960, 25,900, 16,960, 44,920, and 37,840 m−1 cm−1, respectively.
Size-exclusion chromatogram analysis
A portion of the protein solution was injected into HHa Superdex 200 10/300 GL HPLC column (GE Healthcare) equilibrated with 0.1 m Tris-HCl buffer (pH 8.0), containing 100 mm NaCl at room temperature at a flow rate of 0.7 ml/min. The column was calibrated with a Gel Filtration Calibration Kit (GE Healthcare).
Small angle X-ray scattering analysis
All of the SAXS measurements were carried out at 4 °C. SAXS measurement of the STAT1ND:STAT2ND fusion protein was performed with a BioSAXS1000 system mounted on a MicroMax007HF X-ray generator (Rigaku, Tokyo, Japan). Samples in a buffer containing 20 mm Tris-HCl (pH 7.6), 0.1 m NaCl, 5% (v/v) glycerol, 1 mm EDTA, and 1 mm DTT were used for SAXS measurement. A PILATUS100K detector (DECTRIS, Baden-Dättwil, Switzerland) at a distance of 484 mm from the sample was used to measure scattering intensities. The X-ray wavelength was 1.5418 Å. One-dimensional scattering data I(q) as a function of q (q = 4πsinθ/λ, where 2θ is the scattering angle and λ is the wavelength) were obtained through radial averaging by using the program SAXSlab (Rigaku). SAXS measurements of the thrombin-digested STAT1ND:STAT2ND fusion protein and its Y3-bound form were performed on the BL15A2 beamline at the Photon Factory (Tsukuba, Japan). To remove aggregated proteins, samples were purified by Superdex 200 10/300 GL size-exclusion chromatography (GE Healthcare) before SAXS measurements. Samples in a buffer containing 20 mm Tris-HCl (pH 7.6), 0.2 m NaCl, 1 mm EDTA, and 0.5 mm cystine were used. A PILATUS2M detector (DECTRIS) at a distance of 1603 mm from the sample was used. The X-ray wavelength was 1.2184 Å. Radial averaging of scattering intensity was performed by the program SAnglar (49). Because no inter-particle interference was detected at several protein concentrations, I(q) data from different protein concentrations were merged and then used for structural analysis. SAXS data are summarized in supplemental Fig. S5 and Table S1. All data were processed by using software applications embedded in the ATSAS package (50). The radius of gyration Rg and forward scattering intensity I(0) were estimated from the Guinier plot of I(q) in the smaller angle region of qRg < 1.3. The distance distribution function P(r) was calculated by the program GNOM (51). The maximum particle dimension Dmax was estimated from the P(r) function as the distance r for which P(r) = 0 (52). The molecular weight of the sample was estimated by comparing I(0)/c (where c is the protein concentration) of the sample to that of standard protein: ovalbumin, BSA, lysozyme, RNase A, and ubiquitin. Dummy atom models were produced by the program DAMMIF (53). Ten independently calculated, low-resolution dummy atom models were averaged by the program DAMAVER (54). Superposition of the molecular envelopes and model structures was performed by the program SUPCOMB (55). An ensemble model of the STAT1ND:STAT2ND fusion protein was built by the program EOM version 1 (56). Theoretical I(q) from the model structure was calculated by the program CRYSOL (57).
IFN-α treatment of SeV-infected cells
U6A cells in 12-well plates were infected with SeV at a multiplicity of infection of 15. After 1-h incubation at 37 °C, inocula were removed and supplemented with serum-free DMEM at 37 °C for 4 h, followed by the addition of IFN-α (2,000 units/ml). After an additional 10-h incubation, the cells were superinfected with rVSV-EGFP at a multiplicity of infection of 5 and incubated at 37 °C for 9 h. GFP expression was observed using an immunofluorescent microscope. The cells were then lysed in SDS sample buffer, and proteins were analyzed by SDS-PAGE followed by Western blotting using anti-GFP and anti-C protein antibodies. Protein bands were visualized and analyzed as described above.
Surface plasmon resonance
SPR experiments were performed at 25 °C using a Biacore X system (Biacore). For the interaction analysis, proteins were prepared in a buffer containing 10 mm HEPES (pH 7.4), 150 mm NaCl, and 50 μm EDTA and 0.01% Tween 20. All experiments were performed using an NTA sensor chip (GE Healthcare) and the same buffer at a flow rate of 10 μl/min. For each cycle, the chip was activated by injecting 50 μl of 0.5 mm NiCl2 over the flow cell, and the purified disulfide-bonded STAT1ND:STAT2ND heterodimer was immobilized. As a result, a RU of 630 ± 50 was achieved. Different concentrations of histidine tag-less Y3W125A (ranging from 0.04 to 20 μm) were flowed in the cell for 180 s. The affinity analysis was done using the binding responses recorded at 120 s after injection of each concentration of Y3W125A. RU values obtained by the injection of each concentration of Y3W125A using an unimmobilized sensor chip were used as a reference. Each experiment was done in duplicate with similar results.
Author contributions
K. O. conducted most of the experiments, analyzed the results, and wrote the first draft of the paper. T. O., Y. M., and M. S. conducted experiments of the SAXS analysis and molecular modeling. T. I. conducted experiments using SeV and rVSV-EGFP with K. O., and T. S. wrote the paper with K. O.
Supplementary Material
Acknowledgments
We are grateful to the beam-line staff at the Photon Factory BL15A2 for assistance with data collection and to Taeko Akiyoshi for technical assistance. We are also grateful to the staff of the Analysis Center of Life Science, Hiroshima University for the use of their facilities.
This work was supported by grants from the INAMORI Foundation (to K. O.), GlaxoSmithKline Japan (to K. O.), the Takeda Science Foundation (to K. O.), Japan Society for the Promotion of Science Grants 15K151430 and 16H051975 (to T. S. and K. O.), and the Japan Agency for Medical Research and Development (to T. S.). The authors declare that they have no conflicts of interest with the contents of this article.
This article contains supplemental Table S1 and Figs. S1–S5.
- SeV
- Sendai virus
- EGFP
- enhanced green fluorescent protein
- SAXS
- small angle X-ray scattering
- STAT1
- signal transducer and activator of transcription 1
- STAT2
- signal transducer and activator of transcription 2
- VSV
- vesicular stomatitis virus
- EOM
- ensemble optimization method
- RU
- response unit
- aa
- amino acid(s)
- SH2
- Src homology domain 2.
References
- 1. Kato A., Kiyotani K., Kubota T., Yoshida T., Tashiro M., and Nagai Y. (2007) Importance of the anti-interferon capacity of Sendai virus C protein for pathogenicity in mice. J. Virol. 81, 3264–3271 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2. Kato A., Kiyotani K., Sakai Y., Yoshida T., Shioda T., and Nagai Y. (1997) Importance of the cysteine-rich carboxyl-terminal half of V protein for Sendai virus pathogenesis. J. Virol. 71, 7266–7272 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3. Huang C., Kiyotani K., Fujii Y., Fukuhara N., Kato A., Nagai Y., Yoshida T., and Sakaguchi T. (2000) Involvement of the zinc-binding capacity of Sendai virus V protein in viral pathogenesis. J. Virol. 74, 7834–7841 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4. Kato A., Kiyotani K., Sakai Y., Yoshida T., and Nagai Y. (1997) The paramyxovirus, Sendai virus, V protein encodes a luxury function required for viral pathogenesis. EMBO J. 16, 578–587 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5. Gotoh B., Takeuchi K., Komatsu T., Yokoo J., Kimura Y., Kurotani A., Kato A., and Nagai Y. (1999) Knockout of the Sendai virus C gene eliminates the viral ability to prevent the interferon-α/β-mediated responses. FEBS Lett. 459, 205–210 [DOI] [PubMed] [Google Scholar]
- 6. Sakaguchi T., Irie T., Kuwayama M., Ueno T., Yoshida A., and Kawabata R. (2011) Analysis of interaction of Sendai virus V protein and melanoma differentiation-associated gene 5. Microbiol. Immunol. 55, 760–767 [DOI] [PubMed] [Google Scholar]
- 7. Irie T., Kiyotani K., Igarashi T., Yoshida A., and Sakaguchi T. (2012) Inhibition of interferon regulatory factor 3 activation by paramyxovirus V protein. J. Virol. 86, 7136–7145 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8. Nagai Y., Takakura A., Irie T., Yonemitsu Y., and Gotoh B. (2011) Sendai virus: evolution from mouse pathogen to a state-of-the-art tool in virus research and biotechnology. in The Biology of Paramyxoviruses (Samal S. K., ed) pp. 115–173, Caister Academic Press, Norfolk, UK [Google Scholar]
- 9. Lamb R. A., and Parks G. D. (2013) Paramyxoviridae: the viruses and their replication. in Fields Virology (Knipe D. M., and Howley P. M., eds) 6th Ed., pp. 957–995, Lippincott Williams & Wilkins, Philadelphia [Google Scholar]
- 10. Komatsu T., Takeuchi K., Yokoo J., and Gotoh B. (2004) C and V proteins of Sendai virus target signaling pathways leading to IRF-3 activation for the negative regulation of interferon-β production. Virology 325, 137–148 [DOI] [PubMed] [Google Scholar]
- 11. Takeuchi K., Komatsu T., Kitagawa Y., Sada K., and Gotoh B. (2008) Sendai virus C protein plays a role in restricting PKR activation by limiting the generation of intracellular double-stranded RNA. J. Virol. 82, 10102–10110 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12. Irie T., Nagata N., Igarashi T., Okamoto I., and Sakaguchi T. (2010) Conserved charged amino acids within Sendai virus C protein play multiple roles in the evasion of innate immune responses. PLoS ONE 5, e10719. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13. Irie T., Nagata N., Yoshida T., and Sakaguchi T. (2008) Paramyxovirus Sendai virus C proteins are essential for maintenance of negative-sense RNA genome in virus particles. Virology 374, 495–505 [DOI] [PubMed] [Google Scholar]
- 14. Irie T., Okamoto I., Yoshida A., Nagai Y., and Sakaguchi T. (2014) Sendai virus C proteins regulate viral genome and antigenome synthesis to dictate the negative genome polarity. J. Virol. 88, 690–698 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15. Irie T., Inoue M., and Sakaguchi T. (2010) Significance of the YLDL motif in the M protein and Alix/AIP1 for Sendai virus budding in the context of virus infection. Virology 405, 334–341 [DOI] [PubMed] [Google Scholar]
- 16. Irie T., Nagata N., Yoshida T., and Sakaguchi T. (2008) Recruitment of Alix/AIP1 to the plasma membrane by Sendai virus C protein facilitates budding of virus-like particles. Virology 371, 108–120 [DOI] [PubMed] [Google Scholar]
- 17. Irie T., Shimazu Y., Yoshida T., and Sakaguchi T. (2007) The YLDL sequence within Sendai virus M protein is critical for budding of virus-like particles and interacts with Alix/AIP1 independently of C protein. J. Virol. 81, 2263–2273 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18. Sakaguchi T., Kato A., Sugahara F., Shimazu Y., Inoue M., Kiyotani K., Nagai Y., and Yoshida T. (2005) AIP1/Alix is a binding partner of Sendai virus C protein and facilitates virus budding. J. Virol. 79, 8933–8941 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19. Sugahara F., Uchiyama T., Watanabe H., Shimazu Y., Kuwayama M., Fujii Y., Kiyotani K., Adachi A., Kohno N., Yoshida T., and Sakaguchi T. (2004) Paramyxovirus Sendai virus-like particle formation by expression of multiple viral proteins and acceleration of its release by C protein. Virology 325, 1–10 [DOI] [PubMed] [Google Scholar]
- 20. Garcin D., Latorre P., and Kolakofsky D. (1999) Sendai virus C proteins counteract the interferon-mediated induction of an antiviral state. J. Virol. 73, 6559–6565 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21. Gotoh B., Takeuchi K., Komatsu T., and Yokoo J. (2003) The STAT2 activation process is a crucial target of Sendai virus C protein for the blockade of α interferon signaling. J. Virol. 77, 3360–3370 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22. Komatsu T., Takeuchi K., Yokoo J., and Gotoh B. (2002) Sendai virus C protein impairs both phosphorylation and dephosphorylation processes of Stat1. FEBS Lett. 511, 139–144 [DOI] [PubMed] [Google Scholar]
- 23. Kato A., Cortese-Grogan C., Moyer S. A., Sugahara F., Sakaguchi T., Kubota T., Otsuki N., Kohase M., Tashiro M., and Nagai Y. (2004) Characterization of the amino acid residues of Sendai virus C protein that are critically involved in its interferon antagonism and RNA synthesis down-regulation. J. Virol. 78, 7443–7454 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24. Takeuchi K., Komatsu T., Yokoo J., Kato A., Shioda T., Nagai Y., and Gotoh B. (2001) Sendai virus C protein physically associates with Stat1. Genes Cells 6, 545–557 [DOI] [PubMed] [Google Scholar]
- 25. Lim C. P., and Cao X. (2006) Structure, function, and regulation of STAT proteins. Mol. Biosyst. 2, 536–550 [DOI] [PubMed] [Google Scholar]
- 26. Zhong M., Henriksen M. A., Takeuchi K., Schaefer O., Liu B., ten Hoeve J., Ren Z., Mao X., Chen X., Shuai K., and Darnell J. E. Jr. (2005) Implications of an antiparallel dimeric structure of nonphosphorylated STAT1 for the activation-inactivation cycle. Proc. Natl. Acad. Sci. U.S.A. 102, 3966–3971 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27. Mao X., Ren Z., Parker G. N., Sondermann H., Pastorello M. A., Wang W., McMurray J. S., Demeler B., Darnell J. E. Jr., and Chen X. (2005) Structural bases of unphosphorylated STAT1 association and receptor binding. Mol. Cell 17, 761–771 [DOI] [PubMed] [Google Scholar]
- 28. Mertens C., Zhong M., Krishnaraj R., Zou W., Chen X., and Darnell J. E. Jr. (2006) Dephosphorylation of phosphotyrosine on STAT1 dimers requires extensive spatial reorientation of the monomers facilitated by the N-terminal domain. Genes Dev. 20, 3372–3381 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29. Chen X., Vinkemeier U., Zhao Y., Jeruzalmi D., Darnell J. E. Jr., and Kuriyan J. (1998) Crystal structure of a tyrosine phosphorylated STAT-1 dimer bound to DNA. Cell 93, 827–839 [DOI] [PubMed] [Google Scholar]
- 30. Wenta N., Strauss H., Meyer S., and Vinkemeier U. (2008) Tyrosine phosphorylation regulates the partitioning of STAT1 between different dimer conformations. Proc. Natl. Acad. Sci. U.S.A. 105, 9238–9243 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31. Gotoh B., Takeuchi K., and Komatsu T. (2004) Inhibition of the γ interferon response by a Sendai virus C protein mutant with no STAT1-binding ability. FEBS Lett. 567, 291–296 [DOI] [PubMed] [Google Scholar]
- 32. Gotoh B., Komatsu T., Takeuchi K., and Yokoo J. (2003) The C-terminal half-fragment of the Sendai virus C protein prevents the γ-activated factor from binding to a gamma-activated sequence site. Virology 316, 29–40 [DOI] [PubMed] [Google Scholar]
- 33. Oda K., Matoba Y., Irie T., Kawabata R., Fukushi M., Sugiyama M., and Sakaguchi T. (2015) Structural basis of the inhibition of STAT1 activity by Sendai virus C protein. J. Virol. 89, 11487–11499 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34. Marq J. B., Brini A., Kolakofsky D., and Garcin D. (2007) Targeting of the Sendai virus C protein to the plasma membrane via a peptide-only membrane anchor. J. Virol. 81, 3187–3197 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35. Ota N., Brett T. J., Murphy T. L., Fremont D. H., and Murphy K. M. (2004) N-domain-dependent nonphosphorylated STAT4 dimers required for cytokine-driven activation. Nat. Immunol. 5, 208–215 [DOI] [PubMed] [Google Scholar]
- 36. Ho J., Pelzel C., Begitt A., Mee M., Elsheikha H. M., Scott D. J., and Vinkemeier U. (2016) STAT2 is a pervasive cytokine regulator due to its inhibition of STAT1 in multiple signaling pathways. PLos Biol. 14, e2000117. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37. Reddy Chichili V. P., Kumar V., and Sivaraman J. (2013) Linkers in the structural biology of protein-protein interactions. Protein Sci. 22, 153–167 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38. Kingston R. L., Hamel D. J., Gay L. S., Dahlquist F. W., and Matthews B. W. (2004) Structural basis for the attachment of a paramyxoviral polymerase to its template. Proc. Natl. Acad. Sci. U.S.A. 101, 8301–8306 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39. O'Sullivan L. A., Liongue C., Lewis R. S., Stephenson S. E., and Ward A. C. (2007) Cytokine receptor signaling through the Jak-Stat-Socs pathway in disease. Mol. Immunol. 44, 2497–2506 [DOI] [PubMed] [Google Scholar]
- 40. Yu H., and Jove R. (2004) The STATs of cancer–new molecular targets come of age. Nat Rev Cancer 4, 97–105 [DOI] [PubMed] [Google Scholar]
- 41. Begitt A., Droescher M., Meyer T., Schmid C. D., Baker M., Antunes F., Knobeloch K. P., Owen M. R., Naumann R., Decker T., and Vinkemeier U. (2014) STAT1-cooperative DNA binding distinguishes type 1 from type 2 interferon signaling. Nat. Immunol 15, 168–176 [DOI] [PubMed] [Google Scholar]
- 42. Domoszlai T., Martincuks A., Fahrenkamp D., Schmitz-Van de Leur H., Küster A., and Müller-Newen G. (2014) Consequences of the disease-related L78R mutation for dimerization and activity of STAT3. J. Cell Sci. 127, 1899–1910 [DOI] [PubMed] [Google Scholar]
- 43. Komatsu T., Takeuchi K., Yokoo J., Tanaka Y., and Gotoh B. (2000) Sendai virus blocks α interferon signaling to signal transducers and activators of transcription. J. Virol. 74, 2477–2480 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44. Kato A., Sakai Y., Shioda T., Kondo T., Nakanishi M., and Nagai Y. (1996) Initiation of Sendai virus multiplication from transfected cDNA or RNA with negative or positive sense. Genes Cells 1, 569–579 [DOI] [PubMed] [Google Scholar]
- 45. Kiyotani K., Takao S., Sakaguchi T., and Yoshida T. (1990) Immediate protection of mice from lethal wild-type Sendai virus (HVJ) infections by a temperature-sensitive mutant, HVJpi, possessing homologous interfering capacity. Virology 177, 65–74 [DOI] [PubMed] [Google Scholar]
- 46. Niwa H., Yamamura K., and Miyazaki J. (1991) Efficient selection for high-expression transfectants with a novel eukaryotic vector. Gene 108, 193–199 [DOI] [PubMed] [Google Scholar]
- 47. Saijo M., Qing T., Niikura M., Maeda A., Ikegami T., Sakai K., Prehaud C., Kurane I., and Morikawa S. (2002) Immunofluorescence technique using HeLa cells expressing recombinant nucleoprotein for detection of immunoglobulin G antibodies to Crimean-Congo hemorrhagic fever virus. J. Clin. Microbiol. 40, 372–375 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48. Irie T., Yoshida A., and Sakaguchi T. (2013) Clustered basic amino acids of the small Sendai virus C protein Y1 are critical to its RAN GTPase-mediated nuclear localization. PLoS ONE 8, e73740. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49. Shimizu N., Yatabe K., Nagatani Y., Saijyo S., Kosuge T., and Igarashi N. (2016) Software development for analysis of small-angle X-ray scattering data. AIP Conf. Proc. 1741, 050017 [Google Scholar]
- 50. Petoukhov M. V., Franke D., Shkumatov A. V., Tria G., Kikhney A. G., Gajda M., Gorba C., Mertens H. D., Konarev P. V., and Svergun D. I. (2012) New developments in the ATSAS program package for small-angle scattering data analysis. J. Appl. Crystallogr. 45, 342–350 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 51. Svergun D. I. (1992) Determination of the regularization parameter in indirect-transform methods using perceptual criteria. J. Appl. Crystallogr. 25, 495–503 [Google Scholar]
- 52. Glatter O., and Kratky O. (1982) Small-angle X-ray scattering, Academic Press, New York [Google Scholar]
- 53. Franke D., and Svergun D. I. (2009) DAMMIF, a program for rapid ab-initio shape determination in small-angle scattering. J. Appl. Crystallogr. 42, 342–346 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 54. Volkov V. V., and Svergun D. I. (2003) Uniqueness of ab initio shape determination in small-angle scattering. J. Appl. Crystallogr. 36, 860–864 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 55. Kozin M. B., and Svergun D. I. (2001) Automated matching of high- and low-resolution structural models. J. Appl. Crystallogr. 34, 33–41 [Google Scholar]
- 56. Bernadó P., Mylonas E., Petoukhov M. V., Blackledge M., and Svergun D. I. (2007) Structural characterization of flexible proteins using small-angle X-ray scattering. J. Am. Chem. Soc. 129, 5656–5664 [DOI] [PubMed] [Google Scholar]
- 57. Svergun D., Barberato C., and Koch M. H. J. (1995) CRYSOL: a program to evaluate x-ray solution scattering of biological macromolecules from atomic coordinates. J. Appl. Crystallogr. 28, 768–773 [Google Scholar]
- 58. DeLano W. L. (2002) The PyMOL Molecular Graphics System, DeLano Scientific, San Carlos, CA [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.