Abstract
Erb-B2 receptor tyrosine kinase 4 (ErbB4) is a kinase that can signal via a proteolytically released intracellular domain (ICD) in addition to classical receptor tyrosine kinase–activated signaling cascades. Previously, we have demonstrated that ErbB4 ICD is posttranslationally modified by the small ubiquitin-like modifier (SUMO) and functionally interacts with the PIAS3 SUMO E3 ligase. However, direct evidence of SUMO modification in ErbB4 signaling has remained elusive. Here, we report that the conserved lysine residue 714 in the ErbB4 ICD undergoes SUMO modification, which was reversed by sentrin-specific proteases (SENPs) 1, 2, and 5. Although ErbB4 kinase activity was not necessary for the SUMOylation, the SUMOylated ErbB4 ICD was tyrosine phosphorylated to a higher extent than unmodified ErbB4 ICD. Mutation of the SUMOylation site compromised neither ErbB4-induced phosphorylation of the canonical signaling pathway effectors Erk1/2, Akt, or STAT5 nor ErbB4 stability. In contrast, SUMOylation was required for nuclear accumulation of the ErbB4 ICD. We also found that Lys-714 was located within a leucine-rich stretch, which resembles a nuclear export signal, and could be inactivated by site-directed mutagenesis. Furthermore, SUMOylation modulated the interaction of ErbB4 with chromosomal region maintenance 1 (CRM1), the major nuclear export receptor for proteins. Finally, the SUMO acceptor lysine was functionally required for ErbB4 ICD-mediated inhibition of mammary epithelial cell differentiation in a three-dimensional cell culture model. Our findings indicate that a SUMOylation-mediated mechanism regulates nuclear localization and function of the ICD of ErbB4 receptor tyrosine kinase.
Keywords: breast cancer, nuclear transport, posttranslational modification (PTM), protein export, receptor tyrosine kinase, small ubiquitin-like modifier (SUMO), ErbB4, HER4, regulated intramembrane proteolysis (RIP), nuclear transport, RTK, breast cancer
Introduction
ErbB4 is a member of the epidermal growth factor receptor (EGFR)/ErbB4 subfamily of receptor tyrosine kinases (RTK) that includes EGFR, ErbB2, ErbB3, and ErbB4. The signaling of ErbB receptors is activated by binding of EGF-like ligands, such as neuregulins (NRG). The activity of ErbB receptors is tightly controlled during normal embryonic development and homeostasis of adult tissues, whereas aberrant ErbB signaling is frequently observed in cancer. Several cancer drugs specifically targeting ErbB receptors are currently in clinical use (1).
A unique feature of ErbB4 among the ErbB receptors is the presence of four structurally and functionally different isoforms that are generated by alternative mRNA splicing. Two of the isoforms differ in the extracellular juxtamembrane region (JM-a and JM-b) and two in the intracellular cytoplasmic domain (CYT-1 and CYT-2) (2, 3). ErbB4 JM-a isoform was the first RTK demonstrated to signal via regulated intramembrane proteolysis (RIP). Upon ligand binding, ErbB4 JM-a ectodomain is shed by tumor necrosis factor α–converting enzyme (TACE), which triggers intramembrane cleavage by γ-secretase complex, releasing a soluble intracellular domain (ICD) (4–6). The released ErbB4 ICD is an active tyrosine kinase (7, 8) that can translocate into the nucleus, where it interacts with transcriptional regulators (5, 9–16), or to the mitochondria, where it regulates apoptosis (17). ErbB4 signaling via the ICD has been implicated in mammary gland development (18, 19), but also in breast cancer, where nuclear localization of an intracellular ErbB4 epitope has been detected (18, 20, 21).
Posttranslational modification by the small ubiquitin-like modifier (SUMO) is a sensitive, rapid, and reversible way of modifying protein function. Mammalian cells express five SUMO proteins, SUMO1–5. Although the SUMO2 and SUMO3 are nearly identical, SUMO1 shares only ∼50% amino acid sequence identity with SUMO2/3. The expression of SUMO4 and SUMO5 is limited to specific tissues, and at present little is known about their functions (22). SUMO proteins are covalently conjugated to lysine residues of target proteins via a three-step enzymatic reaction involving E1 activating enzyme, E2 conjugating enzyme, and E3 ligating enzyme, which is often a member of the protein inhibitor of activated STAT (PIAS) protein family (23, 24). The removal of SUMO is catalyzed by SUMO isopeptidases such as the sentrin-specific protease (SENP) family, making SUMOylation a reversible and highly dynamic process (25). The consequences of SUMOylation are diverse and depend on the target protein. Mechanistically, SUMOylation may promote or interfere with protein-protein interactions, or induce conformational changes. Although SUMOylation has been shown to regulate various cellular processes, most SUMO targets are nuclear proteins. For example, activity or stability of many transcription factors, as well as subcellular distribution of proteins that shuttle between the cytoplasm and the nucleus, is often regulated by SUMOylation (24, 26).
In our previous study, we identified ErbB4 ICD as an interaction partner of PIAS3 SUMO E3 ligase, and showed that ErbB4 ICD was modified by SUMO (27). Although PIAS3 promoted the nuclear accumulation of functional ErbB4 ICD, direct evidence of SUMO modification in ErbB4 signaling remained elusive. Here, we demonstrate that even though the ErbB4 amino acid sequence contains SUMOylation consensus motifs, ErbB4 was SUMOylated at a non-consensus lysine. The SUMO acceptor lysine was located adjacent to a functional nuclear export sequence, and SUMOylation regulated the nuclear accumulation of ErbB4. ErbB4 SUMOylation was reversed by SENP1, SENP2, and SENP5. Although ErbB4 kinase activity was not necessary for SUMOylation, SUMOylated ErbB4 ICD was highly tyrosine phosphorylated. We also show that the SUMO acceptor lysine regulates RIP-mediated nuclear ErbB4 function in mammary epithelial cells, but not phosphorylation-dependent signaling cascades activated by full-length ErbB4 receptor at the cell surface.
Results
Identification of the SUMO modification site
Regulated intramembrane proteolysis of cleavable ErbB4 isoforms releases a soluble intracellular domain that can localize in the cytoplasm, mitochondria, or nuclei, and that contains tyrosine kinase activity (18). We have previously shown that ErbB4 ICD is posttranslationally modified by SUMO (27). SUMOylation frequently occurs on a consensus motif ΨKXE, where Ψ is a large hydrophobic residue, and X is any amino acid (24). Mass spectrometric screens have also identified an inverted consensus motif, where the acidic residue resides two positions upstream (E/DXKΨ) of the SUMOylated lysine, as well as frequent modification of a shorter KXE motif (26, 28).
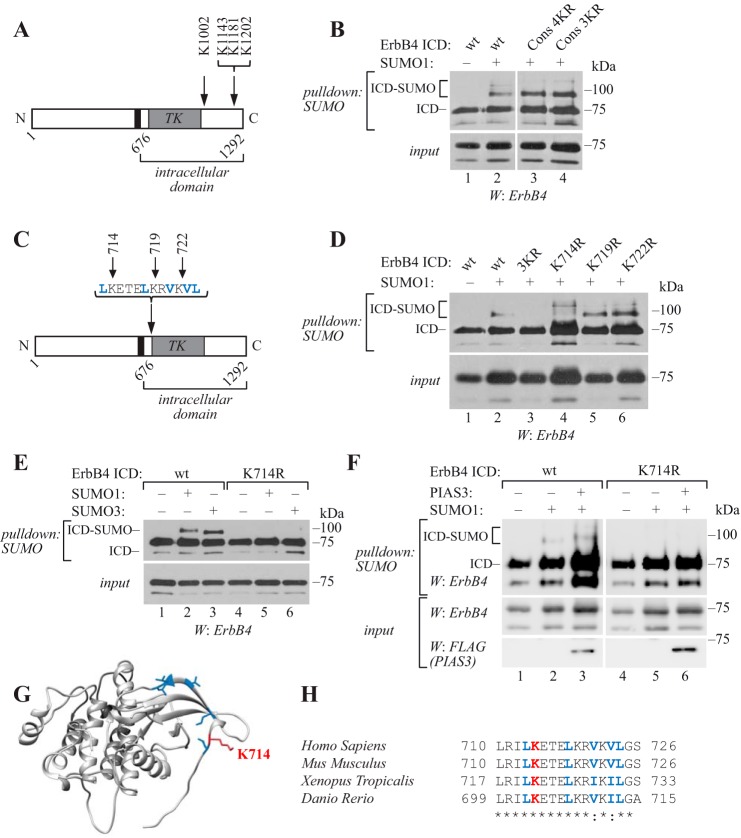
Amino acid sequence analysis of the ErbB4 ICD revealed two ΨKXE consensus motifs (PK1143QE and PK1181AE), a shorter KXE motif (GK1202AE) near the C terminus, and an inverted consensus motif DSK1002F adjacent to the kinase domain (Fig. 1A). To examine whether these lysine residues could serve as SUMO acceptor sites, we replaced them with arginines, cotransfected wild-type or lysine to arginine mutant soluble ErbB4 ICDs of CYT-2 type together with His6-tagged SUMO1 to COS-7 cells, and purified SUMOylated proteins using Ni2+-NTA agarose under denaturing conditions to inactivate SUMO proteases (24). Although some ErbB4 nonspecifically bound to Ni2+-NTA agarose, SUMOylated ErbB4 ICD was clearly detected as a slower migrating band of ∼90 kDa (Fig. 1B), corresponding to a single His6-SUMO residue conjugated to the ∼75-kDa ErbB4 ICD. A weaker, ∼100-kDa SUMOylated form of ErbB4 ICD was also detected. However, the replacement of the three C-terminal lysines (K1143R+K1181R+K1202R; Cons 3KR) and the inverted motif lysine (K1002R+K1143R+K1181R+K1202R; Cons 4KR) with arginines had no effect on content of SUMO-modified ErbB4 ICD (Fig. 1B), indicating that the lysine residue/residues targeted by SUMOylation are non-consensus sites. Similar results were obtained in MCF-7 breast cancer cells expressing wild-type or mutant ErbB4 ICD together with His6-tagged SUMO3 (supplemental Fig. S1).
Figure 1.
Lys-714 is the major SUMOylation site in ErbB4. A, schematic structure of ErbB4. Arrows indicate the mutated amino acids analyzed in B. Black color indicates transmembrane domain and TK indicates tyrosine kinase domain. B, COS-7 cells were transfected with wild-type or mutant HA-tagged ErbB4 ICD constructs and His6-tagged SUMO1. Cells were lysed in denaturing buffer, and lysates were incubated with Ni2+-NTA agarose to pull down His6-SUMO conjugates. Whole cell extracts or pulldown samples were analyzed by Western blotting with anti-ErbB4. Cons 4KR includes K1002R+K1143R+K1181R+K1202R; Cons 3KR includes K1143R+K1181R+K1202R. C, schematic structure of ErbB4 as in A. Blue color indicates the predicted NES and arrows indicate the mutated amino acids analyzed in D. D, COS-7 cells expressing wild-type or mutant HA-tagged ErbB4 ICD constructs and His6-SUMO1 were analyzed as in B. 3KR includes K714R+K719R+K722R. E, COS-7 cells expressing wild-type or K714R HA-tagged ErbB4 ICD and His6-SUMO1 or His6-SUMO3 were analyzed as in B. F, COS-7 cells expressing wild-type or K714R HA-tagged ErbB4 ICD and His6-SUMO1 with or without FLAG-tagged PIAS3 were analyzed as in B. G, crystal structure of the active ErbB4 kinase domain monomer (PDB ID: 3BCE). Lys-714 side chain is indicated in red, and side chains of the predicted NES amino acids are indicated in blue. H, sequence alignment of ErbB4 orthologues from the indicated species using Clustal Omega. The numbers refer to beginning and ending residues of each peptide. Red indicates Lys-714 and blue indicates the predicted NES. *, identical; :, conserved. ErbB4 ICD lysine-to-arginine mutant constructs were analyzed for SUMOylation in 4–11 independent experiments. The effect of PIAS3 on SUMOylation of wild-type and K714R ErbB4 ICD was analyzed in five independent experiments.
A significant proportion of SUMOylated lysines identified in mass spectrometric screens do not match the consensus motif (26, 28–31). The selection of a non-consensus SUMOylation site can be dependent on non-covalent interaction of the SUMO-carrying E2 conjugating enzyme with a SUMO interacting motif (SIM) present in a target protein (24). However, a possibility of SIM-directed SUMOylation was excluded, as the ErbB4 ICD did not interact non-covalently with SUMO1 in a GST pulldown assay, unlike PIASy, which is known to contain a SIM (23) (supplemental Fig. S2).
There are in total 39 lysine residues in the ErbB4 ICD sequence. Most of these lysine residues (35/39) are predicted to be solvent-exposed (supplemental Fig. S3; predicted using NetSurfP, www.cbs.dtu.dk/services/NetSurfP)5 and may thus be available for SUMOylation. Roughly half of them (18/39) are within the predicted kinase domain spanning the residues 718–985 (supplemental Fig. S3). The analysis of ErbB4 deletion constructs indicated that a construct containing the N-terminal region of ErbB4 ICD, which includes the kinase domain, was SUMOylated (supplemental Fig. S4). Interestingly, three lysines in this region (Lys-714, Lys-719, and Lys-722) were located within a leucine-rich sequence 713LKETELKRVKVL724, which is one of the three sequences suggested to resemble a nuclear export signal (NES) (5) (Fig. 1C). To analyze their role in ErbB4 SUMOylation, we constructed a series of lysine to arginine ErbB4 ICD mutants containing all three (K714R+K719R+K722R) point mutations or single mutations (Fig. 1C), transfected them in COS-7 (Fig. 1D) or MCF-7 (supplemental Fig. S1) cells together with His6-tagged SUMO1 or SUMO3, respectively, and analyzed ErbB4 SUMOylation as in Fig. 1B. Notably, the replacement of Lys-714, Lys-719, and Lys-722 with arginines disrupted the formation of the ∼90-kDa SUMO1- or SUMO3-modified ErbB4 ICD (Fig. 1D; supplemental Fig. S1). Furthermore, K714R mutation alone, but not K719R or K722R, was sufficient to abolish the formation of the ∼90-kDa SUMOylated ErbB4 (Fig. 1, D and E; supplemental Fig. S1). Coexpression of PIAS3, a SUMO E3 ligase previously shown (27) to promote ErbB4 ICD SUMOylation, had no effect on the level of modification of the K714R mutant, but strongly enhanced the SUMOylation of wild-type ErbB4 ICD (Fig. 1F).
Upon the replacement of lysine 714 or three lysines 714, 719, and 722 to arginines, higher molecular weight bands of ∼100 kDa were occasionally detected in longer exposure (Fig. 1D; supplemental Fig. S1B), suggesting that other sites could be used for SUMOylation when Lys-714 was not available. These SUMOylated species potentially represented ErbB4 ICD modified to consensus sites, as adding the four consensus site mutations to the same construct inhibited their formation (supplemental Fig. S1B). However, these SUMOylated species were present in smaller quantities compared with the Lys-714–modified ErbB4 ICD and were not always readily detectable.
The crystal structure of active ErbB4 kinase domain obtained from the Research Collaboratory for Structural Bioinformatics Protein Data Bank (RCSB PDB ID: 3BCE) (32) indicated that the region around Lys-714 was nonhelical, and that Lys-714 was exposed and thus accessible to the SUMOylation machinery (Fig. 1G). Furthermore, ErbB4 SUMO modification site is conserved in vertebrates, including mammals, Xenopus tropicalis and Danio rerio (Fig. 1H), suggesting functional importance. Taken together, these data demonstrate that Lys-714 is the major SUMO acceptor site in ErbB4 ICD.
SENP1, SENP2, and SENP5 function as deSUMOylating enzymes for ErbB4
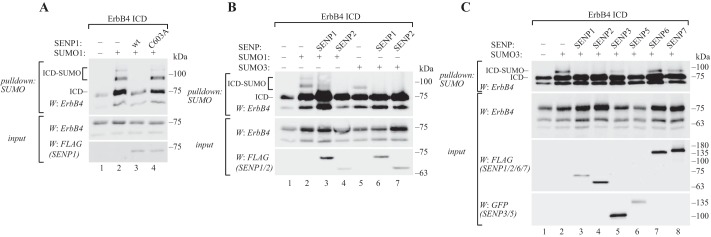
SUMOylation is a reversible modification (25). To characterize factors negatively regulating ErbB4 SUMOylation, activities of the SENP family of SUMO isopeptidases toward SUMOylated ErbB4 ICD were compared. COS-7 cells were transfected with ErbB4 ICD, together with SENP1 or SENP2, and His6-SUMO1 or His6-SUMO3, or with SENP3, SENP5, SENP6, or SENP7, and His6-SUMO3, according to the SUMO paralog preferences of SENP proteins (25). ErbB4 ICD SUMOylation was analyzed by Ni2+-NTA purification and Western blotting. Both SENP1 and SENP2 could efficiently deconjugate SUMO1 and SUMO3 from ErbB4 ICD (Fig. 2, A and B). DeSUMOylation was dependent on the catalytic activity of SENP1, as mutating the catalytic cysteine to alanine (C603A) abolished its deSUMOylating activity toward ErbB4 ICD (Fig. 2A). Unlike SENP1 and SENP2, which have specificity for all SUMO paralogs, the SUMO2/3-specific SENPs (SENP3, SENP5, SENP6, SENP7) were less potent in reducing the level of SUMO-modified ErbB4 ICD (Fig. 2C). Only SENP5 overexpression resulted in decreased SUMOylation of ErbB4 ICD. These results indicate that ErbB4 ICD SUMOylation is a reversible process and that the deSUMOylation is catalyzed by SENP1, SENP2, and SENP5. qRT-PCR analyses indicated that SENP1 and SENP2 are expressed in breast cancer cell lines representing estrogen receptor–positive breast cancer (supplemental Fig. S5A), a subtype in which ErbB4 is also expressed (33). SENP5 expression was not detected in our analysis (supplemental Fig. S5A).
Figure 2.
ErbB4 ICD is deSUMOylated by SENP1, SENP2, and SENP5. A, COS-7 cells were transfected with HA-tagged ErbB4 ICD, His6-tagged SUMO1, and FLAG-tagged wild-type or C603A mutant SENP1 as indicated. Cells were lysed in denaturing buffer, and lysates were incubated with Ni2+-NTA agarose to pull down His6-SUMO conjugates. Whole cell extracts or pulldown samples were analyzed by Western blotting with anti-ErbB4. SENP1 expression was analyzed by Western blotting with anti-FLAG. B, COS-7 cells were transfected with HA-tagged ErbB4 ICD, His6-SUMO1, or His6-SUMO3 and FLAG-tagged SENP1 or SENP2 as indicated, and analyzed as in A. C, COS-7 cells were transfected as indicated with HA-tagged ErbB4 ICD, His6-SUMO3, and FLAG-tagged SENP1, SENP2, SENP6, or SENP7 or GFP-tagged SENP3 or SENP5, and analyzed as in A. The effect of each SENP on deSUMOylation of ErbB4 ICD was analyzed in three to six independent experiments.
SUMOylation promotes the nuclear accumulation of ErbB4 ICD
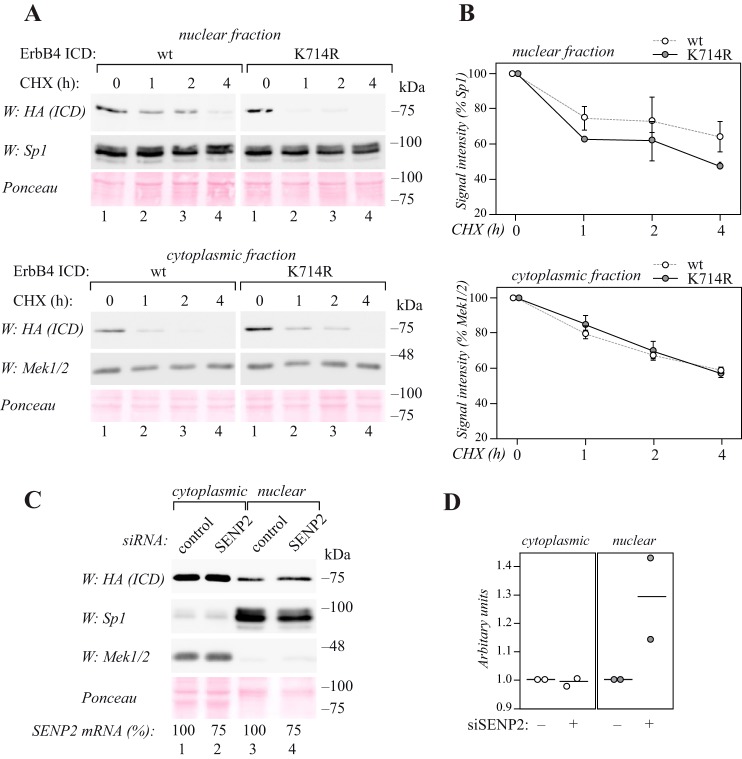
Our previous results have demonstrated that PIAS3, a SUMO E3 ligase that stimulates SUMOylation of ErbB4, also promotes the nuclear localization of the ICD (27). To analyze the role of the major SUMOylation site for nuclear ErbB4 localization and stability in a biologically relevant context, estrogen receptor–positive MCF-7 breast cancer cells, which naturally express PIAS3 (supplemental Fig. S5B), were chosen as a model. MCF-7 cells were transiently transfected with wild-type or K714R mutant HA-tagged ErbB4 ICD and treated with the protein synthesis inhibitor cycloheximide. The abundances of the ICDs were analyzed at different time points in nuclear and cytoplasmic fractions. Both wild-type and K714R mutant ErbB4 ICD were detected in the nuclear and cytoplasmic fractions (Fig. 3A). Compared with wild-type ErbB4 ICD, the abundance of K714R mutant in the nuclear fraction reduced faster over time (Fig. 3, A and B). However, in the cytoplasmic fractions or in the total cell lysates no differences were observed (Fig. 3, A and B; supplemental Fig. S6, A and B).
Figure 3.
SUMOylation regulates the nuclear accumulation of ErbB4 ICD. A, MCF-7 cells were transfected with HA-tagged wild-type or K714R ErbB4 ICD, treated with 100 μg/ml cycloheximide (CHX) for the indicated periods of time, and subjected to subcellular fractionation. Nuclear and cytoplasmic fractions were analyzed by Western blotting with anti-HA (H3663, Sigma-Aldrich). Equal loading of the nuclear fraction was controlled with anti-Sp1, and loading of the cytoplasmic fraction with anti-Mek1/2. Total protein loading in both fractions was controlled with Ponceau staining. Representative data of two independent experiments are shown. B, quantification of HA (ErbB4 ICD) signal intensities (shown in A) normalized to Sp1 or Mek1/2. Data are represented as mean ± range from two independent experiments. C, MCF-7 cells were transfected with HA-tagged ErbB4 ICD together with control or SENP2 siRNA and subjected to subcellular fractionation. Nuclear and cytoplasmic fractions were analyzed by Western blotting with anti-HA (H3663, Sigma-Aldrich). Fractionation was controlled with anti-Sp1 and anti-Mek1/2, and total protein loading with Ponceau staining. Efficiency of RNA interference analyzed by qRT-PCR is indicated as relative SENP2 mRNA levels in cells treated with SENP2-targeting siRNA compared with cells treated with control siRNA (SENP2 mRNA %). Representative data of two independent experiments are shown. D, quantification of HA (ErbB4 ICD) signal intensities (shown in C) normalized to Ponceau. The circles indicate individual data points from two independent experiments and the horizontal line indicates the mean.
The role of SUMOylation in the nuclear accumulation of ErbB4 ICD was also addressed by reducing SUMO isopeptidase expression using RNA interference. MCF-7 cells were transfected with nontargeting or SENP2 siRNA together with ErbB4 ICD, and subjected to subcellular fractionation. Knockdown of SENP2, which reversed SUMOylation of ErbB4 ICD (Fig. 2B), also promoted the accumulation of ErbB4 ICD specifically in the nuclear fraction (Fig. 3, C and D; supplemental Fig. S6, C and D).
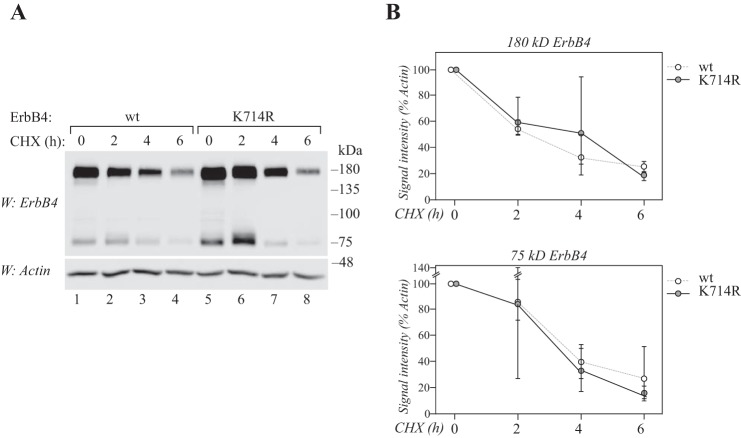
To assess the effect of the SUMO site mutation on the general stability of full-length ErbB4 receptor at the cell membrane, COS-7 cells were transfected with wild-type or K714R mutant ErbB4 and treated with cycloheximide for different time points. No differences were observed in the stability of wild-type and K714R full-length ErbB4, nor the 75-kDa cleavage product (Fig. 4, A and B). Moreover, the K714R mutant ErbB4 ICD was ubiquitinated to a similar level as wild-type ICD when transfected to MCF-7 cells (supplemental Fig. S7), indicating similar susceptibility to ubiquitin-mediated targeting to proteasomes (34). Taken together, these data suggest that SUMOylation at Lys-714 regulates accumulation of ErbB4 ICD in the nucleus, but not overall stability of the receptor in the cytoplasm or at the cell membrane.
Figure 4.
Lys-714 does not regulate the general stability of ErbB4. A, COS-7 cells expressing wild-type or K714R ErbB4 JM-a CYT-2 were starved without serum overnight and treated with 100 μg/ml cycloheximide (CHX) for the indicated periods of time. Lysates were analyzed by Western blotting with anti-ErbB4, and equal loading was controlled with anti-actin. B, quantification of 180 kDa or 75 kDa ErbB4 signal intensities (shown in A) normalized to actin. Data are represented as mean ± range from three independent experiments.
Characterization of ErbB4 nuclear export
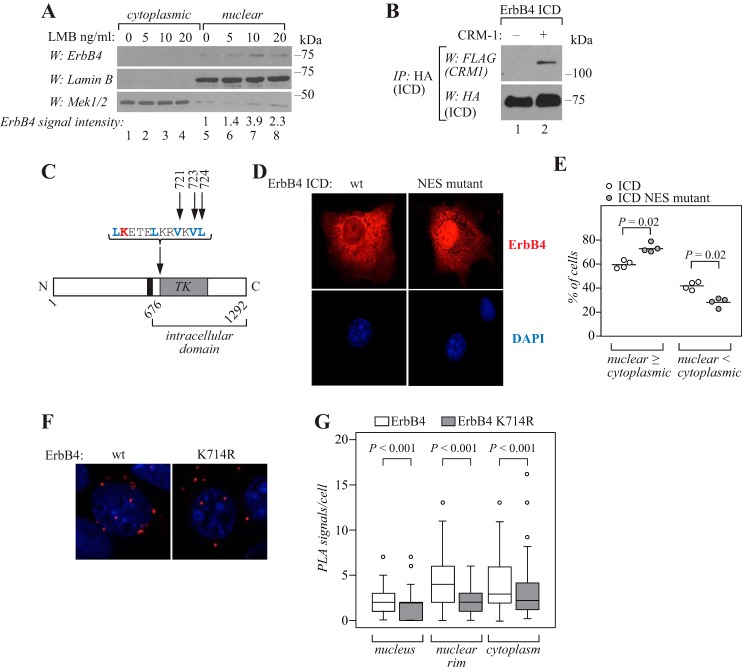
Because the identified SUMO modification site Lys-714 was located within a sequence resembling NES (5), the role of nuclear export in ErbB4 nuclear localization was further examined. In accordance with previous results produced with ectopically expressed ErbB4 (5, 6, 10), ICD from endogenously expressed ErbB4 receptor accumulated into the nuclear fraction of MCF-7 breast cancer cells upon treatment with leptomycin B (Fig. 5A). Leptomycin B is a chemical inhibitor of chromosomal region maintenance 1, the major nuclear export receptor for proteins, which recognizes the leucine-rich NES (35, 36). Indeed, reciprocal coimmunoprecipitation experiments demonstrated the interaction of ErbB4 ICD and FLAG-CRM1 (Fig. 5B; supplemental Fig. S8).
Figure 5.
Nuclear localization of ErbB4 ICD is regulated by CRM1-dependent nuclear export. A, MCF-7 cells were treated for 3 h without or with 5, 10, or 20 ng/ml leptomycin B (LMB) and subjected to subcellular fractionation. Nuclear and cytoplasmic fractions were analyzed by Western blotting with anti-ErbB4, anti-Lamin B, and anti-Mek1/2. Quantification of ErbB4 signal intensities normalized to Lamin B are presented as -fold change values relative to the nontreated control sample. The 75 kDa anti-ErbB4 reactive band represents the ICD. B, COS-7 cells were transfected with HA-tagged ErbB4 ICD2 with or without FLAG-tagged CRM1. Lysates were immunoprecipitated with anti-HA (2367, Cell Signaling Technology) followed by Western blotting with anti-FLAG. The membrane was reprobed with anti-HA (2367, Cell Signaling Technology). Representative data of four independent experiments are shown. C, schematic structure of ErbB4. Black color indicates the transmembrane domain and TK indicates tyrosine kinase domain. Blue color indicates the predicted NES amino acids and arrows indicate the mutated amino acids analyzed in D and E. Red color indicates the SUMOylation site analyzed in F and G. D, COS-7 cells expressing wild-type or NES-mutant (V721A+V723A+L724A) ErbB4 ICD2 were stained for anti-ErbB4 (red) and visualized by confocal microscopy using a 40× objective. Nuclei were stained with DAPI (blue). E, quantification of the nuclear staining intensity. Cells were scored for predominantly nuclear (equal signal in the nucleus and in the cytoplasm or more signal in the nucleus than in the cytoplasm; nuclear ≥ cytoplasmic) or cytoplasmic (more signal in the cytoplasmic than in the nucleus; nuclear < cytoplasmic) staining. Data are represented as a scatter plot, with circles indicating data points from four independent experiments and horizontal lines indicating the mean. 475 cells from four independent experiments were scored. F, MCF-7 cells were transfected with HA-tagged wild-type of K714R ErbB4, stimulated with 50 ng/ml NRG-1 for 15 min, and fixed. Complexes of ErbB4 and CRM1 were visualized with anti-HA and anti-CRM1 using in situ PLA. Red PLA foci represent ErbB4-CRM1 interactions. Nuclei were stained with DAPI (blue). G, quantification of PLA signals per cell. Signals were classified as nuclear, nuclear rim, or cytoplasmic using DAPI as a marker for cell nuclei. Data are presented as a box plot, with horizontal lines indicating the median, boxes indicating the second and third quartile, and error bars indicating the 1.5× interquartile range. Outliers are indicated as circles. n = 63 for cells expressing wild-type ErbB4; n = 53 for cells expressing ErbB4 K714R.
Like the SUMO acceptor site Lys-714, the hydrophobic, leucine-rich sequence 713LKETELKRVKVL724 is highly conserved in ErbB4 orthologues in different species (Fig. 1H). To experimentally test the role of this sequence in regulating the nuclear localization of ErbB4, point mutations converting hydrophobic residues Val-721, Val-723, and Leu-724 to alanine were introduced to ErbB4 ICD (Fig. 5C; supplemental S9A). COS-7 transfectants expressing wild-type ErbB4 ICD demonstrated both nuclear and cytoplasmic immunofluorescence (Fig. 5D), as previously shown (8). However, immunofluorescence analyses of the valine/leucine to alanine mutant construct revealed increased nuclear localization (Fig. 5, D and E), suggesting that amino acids Val-721, Val-723, and Leu-724 were critical for nuclear export of ErbB4. Mutation of Val-721, Val-723, and Leu-724 to alanine did not change the SUMOylation pattern of ErbB4 ICD, confirming that the nuclear accumulation of the mutant construct was not because of alterations in SUMO modification (supplemental Fig. S10).
To further study the role of SUMOylation in nuclear export of ErbB4, MCF-7 cells were transfected with HA-tagged wild-type or K714R mutant ErbB4, and analyzed for ErbB4-CRM1 interactions using the in situ proximity ligation assay (PLA). Fewer red PLA foci were detected in cells expressing K714R ErbB4 compared with cells expressing wild-type ErbB4 (Fig. 5, F and G; supplemental Fig. S9B), indicating that SUMOylation-deficient ErbB4 interacted with CRM1 less efficiently than the putatively SUMOylated wild-type ErbB4. Similar results were obtained in coimmunoprecipitation experiments (supplemental Fig. S8). Taken together, these data indicate that nuclear localization of ErbB4 ICD is regulated by CRM1-dependent nuclear export, mediated by hydrophobic amino acids of the leucine-rich NES. These results also suggest that ErbB4 SUMOylation at Lys-714 modulates the ErbB4-CRM1 interaction.
SUMOylation is independent of the ErbB4 kinase activity but increases its autophosphorylation
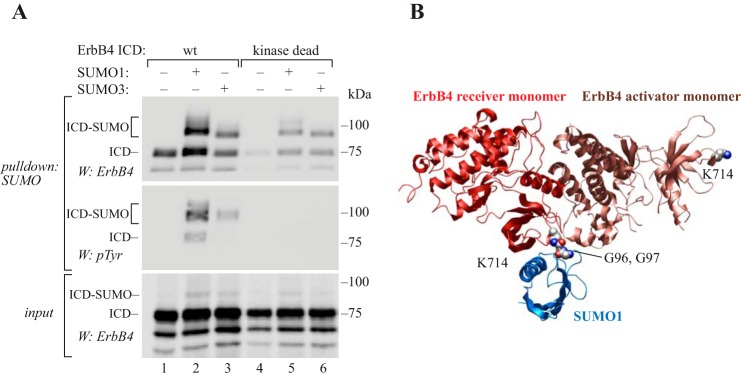
Because soluble ErbB4 ICD is a constitutively active tyrosine kinase (7, 8), we assessed whether catalytic activity was necessary for the SUMO modification. A previously characterized kinase-dead K751R mutant (7, 8) was expressed in COS-7 cells, and compared with wild-type ErbB4 ICD for its SUMOylation with His6-SUMO1 or His6-SUMO3. Both wild-type and K751R ErbB4 ICDs were efficiently SUMOylated, indicating that SUMOylation was independent of the catalytic activity of ErbB4 (Fig. 6A).
Figure 6.
SUMOylation increases autophosphorylation of ErbB4 ICD. A, COS-7 cells were transfected with wild-type or kinase dead K751R ErbB4 ICD2 with or without His6-tagged SUMO constructs as indicated. Cells were lysed in denaturing buffer, and lysates were incubated with Ni2+-NTA agarose to pull down His6-SUMO conjugates. Whole cell extracts were analyzed by Western blotting with anti-ErbB4, and pulldown samples with anti-ErbB4 or anti-phosphotyrosine (4G10, Upstate). Representative data of two independent experiments are shown. B, structural model (displayed as secondary structures) of the covalent complex between the ErbB4 kinase asymmetric dimer X-ray structure (RCSB PDB ID: 3BCE) and an NMR structure for SUMO1 (in blue) (RCSB PDB ID: 2ASQ). Lys-714 (CPK representations) in both the activator and receiver kinase domains are indicated but SUMO1 is shown ligated only to Lys-714 of the receiver kinase via an isopeptide bond represented between the lysine side chain and the C-terminal Gly-Gly sequence of SUMO (CPK representations).
Because autophosphorylation is a critical step of tyrosine kinase function, SUMOylated ErbB4 ICD was analyzed for its tyrosine phosphorylation. The SUMO1- or SUMO3-modified ErbB4 ICD was indeed tyrosine phosphorylated (Fig. 6A). Notably, the phosphotyrosine content of SUMOylated ErbB4 ICD was greater compared with the non-SUMOylated ErbB4 ICD, which may be an indication of increased autokinase activity of SUMOylated ErbB4.
The three-dimensional structure of the asymmetric ErbB4 ICD dimer (RCSB PDB ID: 3BCE) places Lys-714 at two very different locations, but both lysine side chains are exposed to solvent and should be available for SUMOylation. Lys-714 in the activator kinase is located far from the asymmetric dimer interface (Fig. 6B), whereas lysine Lys-714 in the receiver kinase is located right at the solvent-exposed junction of the receiver and activator kinase domains. Lys-714 of the receiver kinase is linked via residues spanning the interface to the juxtamembrane latch, which binds to the activator kinase domain and has an important role in receptor activation by docking the kinase monomers to each other (37). SUMOylation involves forming a covalent isopeptide bond between the SUMO C terminus and a lysine side chain. To assess the effect of SUMOylation at Lys-714 on the receiver kinase domain, a model was mocked up of the ErbB4 asymmetric kinase homodimer in complex with SUMO-1 (NMR structure; RCSB PDB ID: 2ASQ) (38). At Lys-714 of the receiver kinase domain, SUMO appears complementary to the asymmetric dimer at this location in terms of the size and shape and possibilities for non-covalent interactions among side chains. These complementary features would be consistent with SUMO-induced stabilization of the ternary complex leading to the observed increase in autophosphorylation.
SUMOylation is not required for signaling of full-length ErbB4 at the cell surface
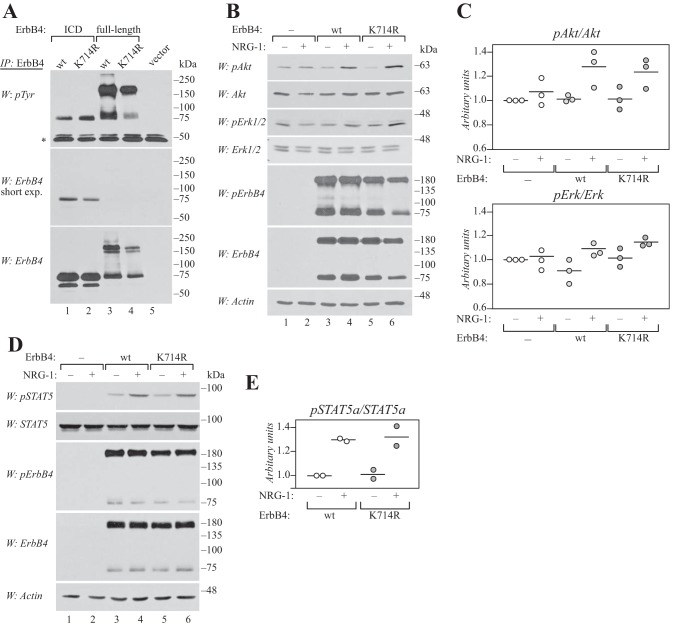
To study whether the major SUMOylation site is required for the basic functions of ErbB4, a SUMO site mutant and wild-type ErbB4 were compared for their phosphorylation and ability to activate signaling cascades. When expressed in COS-7 cells, wild-type and K714R ErbB4 demonstrated efficient constitutive tyrosine phosphorylation in the absence of ligand, both in the context of soluble ICD and full-length cleavable JM-a CYT-2 isoform (Fig. 7A). To address the ability of the K714R mutant to activate signaling pathways downstream of the full-length ErbB4 receptor, COS-7 transfectants were stimulated with the ErbB4 ligand NRG-1 and analyzed for phosphorylation of Akt and Erk kinases, as well as STAT5. Both SUMO site mutant and wild-type ErbB4 were equally efficient in activating Akt (Fig. 7, B and C) and STAT5a (Fig. 7, D and E) upon ligand stimulation, whereas phosphorylation of Erk1/2 was only weakly stimulated by wild-type and K714R ErbB4 (Fig. 7, B and C). These results indicate that the major SUMOylation site Lys-714 does not play a major role in regulating NRG-induced activation of canonical signaling pathways downstream of ErbB4 at the cell surface.
Figure 7.
Lys-714 is not required for signaling of full-length ErbB4 at the cell surface. A, COS-7 cells expressing wild-type or K714R ErbB4 ICD2 or JM-a CYT-2 were starved overnight without serum, lysed, and immunoprecipitated (IP) with anti-ErbB4. Phosphorylation was analyzed by Western blotting with anti-phosphotyrosine (4G10, Upstate), and the membrane was reprobed with anti-ErbB4. Representative data of five independent experiments are shown. *, IgG heavy chain. B, COS-7 cells expressing wild-type or K714R ErbB4 JM-a CYT-2 were starved without serum overnight, stimulated for 10 min with 50 ng/ml NRG-1, and lysed. Phosphorylation of Akt, Erk1/2, and ErbB4 was analyzed by Western blotting with phospho-specific antibodies. Loading was controlled using antibodies recognizing total Akt, Erk1/2, ErbB4, or actin. Representative data of three independent experiments are shown. C, quantification of pErk1/2 and pAkt signal intensities normalized to total Akt and Erk. Data are presented as scatter plots, with circles indicating data points from three independent experiments and horizontal lines indicating the mean. D, COS-7 cells expressing wild-type or K714R ErbB4 JM-a CYT-2 together with STAT5a were treated as in B. Phosphorylation of STAT5 and ErbB4 was analyzed by Western blotting with phospho-specific antibodies. Loading was controlled using antibodies recognizing total STAT5, ErbB4, or actin. Representative data of two independent experiments are shown. E, quantification of pSTAT5a signal intensity normalized to total STAT5a. Data are presented as a scatter plot, with circles indicating data points from two independent experiments and horizontal lines indicating the mean.
SUMOylation is required for the nuclear signaling of ErbB4 ICD
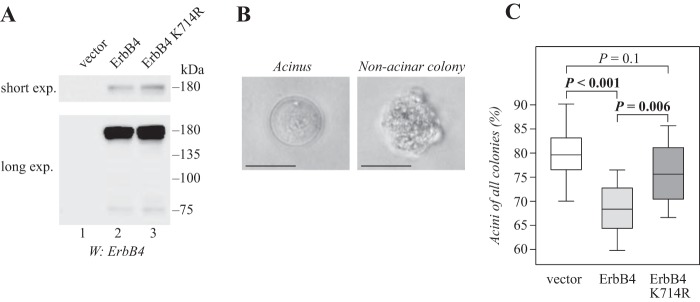
We and others have previously shown that ErbB4 inhibits the differentiation of normal and malignant mammary epithelial cells in three-dimensional cultures (19, 27). The inhibitory effect is limited to soluble ICD of ErbB4 CYT-2 isoform and requires PIAS3, suggesting that nuclear ErbB4 signaling is needed (19, 27). To address the role of SUMOylation in nuclear signaling of ErbB4 ICD, stable transfectants of HC11 mouse mammary epithelial cells expressing wild-type or K714R ErbB4 were generated by retroviral infection (Fig. 8A), and analyzed for three-dimensional growth in Matrigel. Whereas mammary epithelial cells are able to differentiate and form spherical or acinar structures in Matrigel, transformed cells grow in disorganized colonies (Fig. 8B) (39). Roughly 80% of HC11 vector control cells grew in spherical, organized colonies resembling mammary acini and were classified as differentiated (Fig. 8C). Reproducing our previous results (27), cells expressing ErbB4 demonstrated less differentiation than vector control cells (Fig. 8C). In contrast, HC11 cells expressing the SUMOylation-deficient K714R ErbB4 efficiently formed differentiated, acinar structures. Although these cells appeared slightly less potent than vector control cells in their ability to differentiate, no statistical significance was reached. These data indicate that SUMO modification to Lys-714 is required for the RIP-mediated nuclear signaling of ErbB4 ICD in mammary epithelial cells.
Figure 8.
Lys-714 is required for nuclear signaling of ErbB4 in mammary epithelial cells. A, HC11 cells stably expressing wild-type or K714R ErbB4 JM-a CYT-2 were analyzed for ErbB4 expression by Western blotting. B, representative images of control HC11 cells cultured for 15 days in growth factor reduced Matrigel in the presence of 50 ng/ml NRG-1. A photograph of a differentiated acinus (left) and an undifferentiated colony (right) are shown. Scale bar, 25 μm. C, quantification of the percentage of acini of all cell colonies of HC11 cells expressing the indicated constructs. Data of 12 wells from four independent experiments are presented as a box plot, with horizontal lines indicating the median, boxes indicating the second and third quartile, and error bars indicating the range of the data. n = 533 for vector cell colonies; n = 517 for ErbB4 wild-type colonies; n = 575 for ErbB4 K714R colonies.
Discussion
ErbB4 is a receptor tyrosine kinase that undergoes ligand-induced RIP, releasing a soluble ICD (5). Despite the accumulating evidence of nuclear localization and functions of ErbB4 ICD as a transcriptional coregulator, the factors determining its subcellular distribution and activity have remained poorly understood. Here, we characterized posttranslational SUMO modification of ErbB4 ICD, and showed that lysine residue Lys-714 in the kinase region is the major SUMO acceptor site. Although SUMOylation was not necessary for phosphorylation, or activity of phosphorylation-dependent signaling relayed by ErbB4, SUMOylation affects the nuclear accumulation and RIP-mediated signaling of ErbB4 ICD.
Although ErbB4 contains SUMOylation consensus motifs, the evolutionary conserved non-consensus lysine 714 located adjacent to the ErbB4 kinase domain was shown to be the major SUMO acceptor site. In addition to ErbB4, insulin-like growth factor-1 receptor (IGF-1R) and EGFR are currently among the few receptor tyrosine kinases that have been shown to be modified by SUMO. IGF-1R is SUMOylated at three non-consensus lysine residues within the kinase domain (40). One of these sites (Lys-1120) is conserved in the ErbB4 amino acid sequence (Lys-858). However, in ErbB4, this lysine residue was predicted to be buried (supplemental Fig. S3; by NetSurfP: www.cbs.dtu.dk/services/NetSurfP/)5 (41), and the K858R mutation did not change the content of ErbB4 SUMOylation (data not shown).6 More recently, EGFR has been reported to be SUMOylated at lysine 37 using mass spectrometry (42). However, the role of this extracellular lysine in EGFR SUMOylation remains to be elucidated, as the content of SUMO-interacting EGFR in coimmunoprecipitation experiments is not affected by K37R mutation (42). Although Lys-37 is conserved in ErbB4, the observations that EGFR translocates into the nucleus as a full-length receptor (43), whereas nuclear ErbB4 mainly represents the RIP-released ICD (5, 44), suggest differential regulation of the two modification processes.
Our data indicate that the SUMOylation-deficient K714R mutant was tyrosine phosphorylated at a level comparable with wild-type ErbB4, and equally potent to activate kinase-dependent downstream signaling via Akt, Erk, and STAT5. Moreover, SUMOylation did not affect ErbB4 stability. Similarly, compared with wild-type IGF-1R, SUMOylation-deficient mutant IGF-1R is phosphorylated, can activate phosphorylation-dependent downstream signaling pathways, and is internalized upon ligand stimulation equally efficiently (40). However, our results suggest that ErbB4 SUMOylation promotes the nuclear accumulation of ErbB4 ICD, as well as the ErbB4 RIP-mediated inhibition of mammary epithelial cell differentiation. Interestingly, the three SUMOylation sites of IGF-1R were required for the nuclear accumulation and function of IGF-1R (40, 45). As K714R did not disrupt the kinase activity or classical RTK-activated signaling pathways downstream of ErbB4, or influence its stability, it is likely that the loss of function phenotype in inhibition of differentiation is because of decreased nuclear accumulation. Notably, SUMOylation-deficient ErbB4 K714R functionally correlated with PIAS3 silencing, which similarly rescued the ErbB4 RIP-mediated inhibition of mammary epithelial differentiation in our previous study (27). This illustrates that although PIAS proteins sometimes regulate their interaction partners in a manner that is independent of their ability to promote SUMOylation (23), SUMO modification of ErbB4 ICD is mechanistically involved in the observed nuclear accumulation and altered nuclear function. SUMOylation is a dynamic reaction, and only a small fraction of a substrate is modified at a given time (24). Conditions that stimulate ErbB4 SUMOylation through increased PIAS3 activity or decreased SENP1 or SENP2 activity could thus promote nuclear signaling of the soluble ErbB4 ICD.
Both wild-type and SUMOylation-deficient ErbB4 were detected in the nuclear fraction. Nuclear import of ErbB4 ICD is, indeed, regulated by a nuclear localization signal (NLS) (10, 46). Moreover, PIAS3 promotes the nuclear accumulation of ErbB4 only when the NLS is intact (27). Together, these findings imply that SUMOylation is not required for the nuclear translocation of ErbB4. Here, we demonstrated that the ErbB4 SUMO modification site resides within one of the three sequences that resemble nuclear export signals (5). Inactivation of the putative NES by mutation of the critical amino acids resulted in nuclear sequestration of ErbB4 ICD, suggesting that this sequence could indeed function as an NES. Although the mechanisms of how SUMOylation regulates subcellular localization are not known for most target proteins, SUMOylation has been described to both stimulate and inhibit nuclear export (47–50). Like ErbB4, Krüppel-like factor 5 is SUMOylated to a lysine adjacent to its NES, and the SUMO modification facilitates nuclear localization of Krüppel-like factor 5 by inhibiting the NES function (48). SUMOylation of p53, unlike the SUMOylation of ErbB4, promotes cytoplasmic localization (50). Mechanistically, SUMOylation weakens the interaction of p53 and CRM1, which facilitates p53 export (50). The findings of this study suggest that SUMOylation promotes both the nuclear accumulation of ErbB4 and the formation of more stable ErbB4-CRM1 complexes. In both studies the more stable CRM1 interaction associates with more abundant nuclear localization. Indeed, low affinity of NES containing cargo proteins to CRM1 is critical for efficient export, as it enables complex dissociation at the cytoplasmic side of the nuclear pore complex (51, 52). Finally, SENP2, a SUMO isopeptidase that deSUMOylated ErbB4 and regulated the nuclear abundance, is localized to the nucleoplasmic side of the nuclear pore complex (53). SENP2 could thus regulate the level of ErbB4 SUMOylation at the nuclear pore, and hence the efficiency of nuclear export.
We also found that although ErbB4 kinase activity was not necessary for the efficient SUMOylation, SUMO modification enhanced the autophosphorylation of ErbB4, as illustrated by the increased tyrosine phosphorylation of SUMO-modified versus unmodified ErbB4 ICD. Intriguingly, SUMOylation has been shown to stimulate the autophosphorylation of focal adhesion kinase, a non-receptor cytoplasmic tyrosine kinase (54), as well as the catalytic activity of serine/threonine kinase Akt toward its substrate (55). Moreover, a mass spectrometry–based analysis has suggested that SUMOylation modulates the phosphorylation status of many target proteins (56). Notably, ErbB4 ICD kinase activity has been shown to promote its nuclear localization (8). Thus, SUMOylation could contribute to the nuclear accumulation of ErbB4 ICD through the increased tyrosine phosphorylation. It can also be speculated that the SUMOylated ErbB4 ICD could be more active toward its cytoplasmic or nuclear phosphorylation substrates.
Although it is not yet known how SUMOylation of kinases in general would regulate their functions, in the case of the ErbB4 ICD—the structural model—built from existing experimental three-dimensional structures, provides some clues in this case. SUMO, when attached to Lys-714 of the receiver kinase domain, is positioned to an important location, at the junction of the receiver and activator domains, with potential to interact with both domains simultaneously. The 12 residues preceding Lys-714 in the receiver kinase form part of the interface with the activator kinase, preceded by another 9 residues that form the juxtamembrane latch—critical for receptor activation—that binds to the activator kinase domain located on the opposite end of the asymmetric dimer interface. Thus, when Lys-714 is SUMOylated, these interface residues link both the juxtamembrane latch and SUMO on opposite ends of the interface.
The complementarity of both shape and polar groups on the surfaces of the ErbB4 ICD and SUMO are consistent with expectations if a ternary complex would be formed, providing additional stability. Indeed, it appears that SUMO functions regarding the ErbB4 ICD are more complex and nuanced than simply “tagging” the monomers, thus interactions formed in a ternary complex may have direct roles in functional regulation of the kinase activity, possibly even communicated along the dimer interface itself and to the juxtamembrane latch, and vice versa. Furthermore, SUMOylation may have direct effects on other processes related to nuclear transport, given that SUMO attaches to Lys-714 at the edge of the NES signal sequence and may even interact with polar residues along the NES signal sequence itself as seen in the structural model. (The nonpolar groups of the NES signal in the 3BCE structure are not exposed but packed against the rest of the kinase domain.)
The observed promotion of autophosphorylation of tyrosine on the ErbB4 kinase domains taking place on SUMOylation are likely the result of several effects. First, SUMOylation at Lys-714 could help shift the equilibrium toward the asymmetric complex by supporting the formation of the ternary complex in which the receiver and activator kinase domains have the appropriate conformations to support catalysis. Second, the increased interactions in the active, ternary complex may extend the lifetime of the complex and help ensure autophosphorylation takes place at multiple sites along the ICD C-terminal tail. Indeed, given a key function of the juxtamembrane latch on the receiver kinase is to “latch on” and stabilize the interactions with the activator kinase, SUMO may also increase these interactions at the opposite end of the dimer interface, increasing the stability and lifetime of the functional receptor assembly, and hence promote autophosphorylation. Resolving the role of these and perhaps other yet unrecognized contributions to the observed SUMO-promoted mechanism of autophosphorylation remains to be determined, but it seems clear that SUMOylation at Lys-714 on the receiver kinase places SUMO at a strategic location on the receptor, strongly suggesting SUMO does play an important functional role in ErbB4 functionality.
The expression of PIAS3 SUMO E3 ligase has been shown to be increased in breast cancer (57, 58). PIAS3 has also been reported to promote the proliferation of estrogen receptor–positive breast cancer cells, but conversely inhibit the proliferation of breast cancer cells that are estrogen receptor–negative (59). In addition, PIAS3 induces resistance to anti-estrogen hormone therapy in estrogen receptor–positive breast cancer cells (59). Together, these observations suggest an oncogenic role for PIAS3 in estrogen receptor–positive breast cancer. As ErbB4 is typically expressed in the same subtype of breast cancer (33), PIAS3-stimulated SUMOylation could regulate the activity of ErbB4 ICD in this context.
In conclusion, we have demonstrated that SUMOylation regulates RIP-mediated nuclear functions of ErbB4 ICD, but not classical RTK signaling pathways activated by full-length ErbB4 receptor. Furthermore, we provide evidence indicating that SUMO modification within an experimentally defined nuclear export sequence interferes with nuclear export, promoting the nuclear accumulation of ErbB4 ICD. These data provide a new understanding of the molecular mechanisms regulating the function of an ICD of a receptor tyrosine kinase. Finally, our findings describe novel regulatory networks of nuclear ErbB4 signaling that has been implicated in breast cancer.
Experimental procedures
Cell culture
COS-7 and Phoenix-AMPHO HEK 293T cells (a gift from Dr. Garry Nolan) were cultured in DMEM. MCF-7 human breast cancer cells and HC11 mouse mammary epithelial cells (a gift from Dr. Lars-Arne Haldosén) were cultured in RPMI 1640. Both media were supplemented with 10% fetal calf serum (Biowest), 2 mm L-glutamine (Lonza), and 50 units/ml penicillin-streptomycin solution (Lonza). The culture medium of MCF-7 cells was further supplemented with 10 μg/ml insulin (Sigma-Aldrich) and 1 nm 17-β-estradiol (Sigma-Aldrich), and the medium of HC11 cells with 5 μg/ml insulin (Sigma-Aldrich) and 10 ng/ml epidermal growth factor (Sigma-Aldrich).
Expression plasmids and transient transfection
The expression plasmids encoding the following inserts have been described: ErbB4 ICD2-HA, ErbB4 ICD2-K751R-HA, and ErbB4 JM-a CYT-2-HA (8); STAT5a (60); His6-SUMO1 (a gift from Dr. Lea Sistonen) (61); His6-SUMO3 (a gift from Dr. Erik Meulmeester) (62); FLAG-PIAS3, FLAG-SENP1, FLAG-SENP1-C603A, and FLAG-SENP6 (gifts from Dr. Jorma J. Palvimo) (63); FLAG-SENP2 and FLAG-SENP7 (gifts from Dr. Edward Yeh; Addgene plasmids 18047 and 42886) (64, 65); GFP-SENP3 and GFP-SENP5 (gifts from Dr. Mary Dasso; Addgene plasmids 34554 and 34555) (66); and FLAG-CRM1 (a gift from Dr. Xin Wang; Addgene plasmid 17647) (67). Point mutations K714R, K719R, K722R, K1002R, K1143R, K1181R, K1202R, V721A, V723A, and L724A (amino acid numbers refer to ErbB4 JM-a CYT-2 isoform) were introduced to pcDNA3.1hygro(+)–ErbB4ICD2-HA (8) by site-directed mutagenesis. K714R was also introduced to pcDNA3.1 hygro(+)–ErbB4JM-aCYT2-HA and pBABEpuro-ErbB4JM–aCYT-2-HA (68). All generated constructs were confirmed by sequencing. COS-7 and Phoenix-AMPHO HEK 293T cells were transiently transfected with expression plasmids with FuGENE6 transfection reagent (Promega), and MCF-7 cells with Lipofectamine (Thermo Fischer Scientific), according to the manufacturers' protocols.
SUMOylation assay
A protocol by Tatham et al. (69) was used to analyze the SUMOylation of ErbB4 ICD. COS-7 or MCF-7 cells growing on 10- or 6-cm plates, respectively, were transfected with expression plasmids as indicated in the figures. After 24 h, cells were washed with phosphate-buffered saline (PBS) and lysed in denaturing lysis buffer (8 m urea, 0.1 m Na2HPO4/NaH2PO4, 10 mm Tris-HCl pH 7.0, 10 mm imidazole, 10 mm β-mercaptoethanol). Lysates were centrifuged at 3000 × g for 5 min, and supernatants were incubated with 40 μl of Ni2+-NTA agarose (Qiagen) for 2 h to pull down His6-SUMO conjugates. After washing four times with 1 ml of denaturing lysis buffer, His6-SUMO conjugates were eluted with elution buffer (200 mm imidazole, 5% SDS, 150 mm Tris-HCl pH 6.8, 30% glycerol, 720 mm β-mercaptoethanol) and separated by SDS-PAGE. SUMOylation of ErbB4 ICD was analyzed by Western blotting with anti-ErbB4 (E200; Abcam), and tyrosine phosphorylation of SUMOylated ErbB4 ICD by Western blotting with anti-phosphotyrosine (4G10; Upstate). Expression of FLAG-tagged PIAS3 and FLAG- or GFP-tagged SENPs was analyzed by Western blotting with anti-FLAG (M2; Sigma-Aldrich) or anti-GFP (sc-9996; Santa Cruz Biotechnology).
Preparation of lysates and Western blotting
To prepare lysates for Western blotting and immunoprecipitation, cells were washed with PBS, lysed in lysis buffer (1% Triton X-100, 10 mm Tris-Cl pH 7.4, 150 mm NaCl, 1 mm EDTA, 5 mm NaF, 10 μg/ml of aprotinin, 10 μg/ml of leupeptin, 1 mm Na3VO4, 2 mm PMSF, and 10 mm Na4P2O7), and centrifuged at 16000 × g for 10 min. Protein concentration of the supernatants was measured by Bradford Protein Assay (Bio-Rad). Equal amounts of samples were denatured by heating at 95 °C for 5 min in SDS-PAGE loading buffer. Samples were separated by SDS-PAGE and transferred to nitrocellulose membranes. Membranes were incubated with indicated antibodies, and signals detected using enhanced chemiluminescence (Thermo Fischer Scientific).
Cell fractionation
MCF-7 cells growing on 6-cm plates were transfected with expression plasmids or siRNA oligonucleotides, or treated without or with 5, 10, or 20 ng/ml leptomycin B (Sigma-Aldrich) for 3 h as indicated in the figures. Cytoplasmic and nuclear fractions were prepared with an NE-PER kit (Thermo Fischer Scientific) and analyzed by Western blotting with anti-HA (H3663; Sigma-Aldrich) for the overexpression experiments, and anti-ErbB4 (E200; Abcam) for the leptomycin B experiment. Antibodies against Sp1 (sc-14027; Santa Cruz Biotechnology), Lamin B (sc-6217; Santa Cruz Biotechnology), and Mek1/2 (4694; Cell Signaling Technology) were used to control the fractionation, and Ponceau staining was used to control total protein loading when indicated. The signal intensities were quantified using ImageJ software. The ErbB4 signal intensities were normalized to the signal intensities of Mek1/2, Sp1, Lamin B, or Ponceau as described in figure legends.
RNA interference
siRNA oligonucleotides targeting human SENP2 (Hs_SENP2_1, 5′-ATGGTCGGAATCAGATTTGAA-3′) and AllStars Negative Control siRNA were purchased from Qiagen. MCF-7 cells growing on 6-cm plates were transfected with siRNA at final concentration of 20 nm using Lipofectamine (Thermo Fischer Scientific) according to the manufacturer's protocol. Knockdown efficacy was determined by qRT-PCR. Total RNA was extracted using TRIsure RNA isolation reagent (Bioline), and 1 μg of total RNA was subjected to cDNA synthesis using SensiFAST cDNA synthesis kit (Bioline). cDNA was analyzed with primers (Eurofins Scientific) and Universal ProbeLibrary probes (Roche) for SENP2 (left, 5′-TCACTGGCTCAATGATGAAGTC-3′; right, 5′-AGTGCTGGATAGCCTTGCTT-3′, probe 31) and TBP (used as a housekeeping gene) (left, 5′-GAACATCATGGATCAGAACAACA-3′; right, 5′-ATAGGGATTCCGGGAGTCAT-3′, probe 87) on a QuantStudioTM 12K Flex Real-Time PCR System (Thermo Fischer). Relative expression of SENP2 was estimated as previously described (70).
Analysis of ErbB4 half-life
MCF-7 or COS-7 cells growing on 6-well plates were transfected as indicated in the figures, starved overnight without serum, and treated with 100 μg/ml cycloheximide (Sigma-Aldrich) for the indicated time points to inhibit translation. Lysates were analyzed by Western blotting with anti-HA (H3663; Sigma-Aldrich) or anti-ErbB4 (E200; Abcam), and loading was controlled using anti-Mek1/2, anti-Sp1, or anti-actin (sc-1616; Santa Cruz Biotechnology). The signal intensities were quantified using ImageJ software. The ErbB4 signal intensities were normalized to the signal intensities of SP1, Mek1/2, or actin as described in figure legends.
Coimmunoprecipitation of ErbB4 and CRM1
MCF-7 cells growing on 10 cm plates were transiently transfected with expression plasmids and lysed 24 h after transfection. Lysates were precleared with 30 μl protein G-agarose (Santa Cruz Biotechnology) at 4 °C for 1 h, and subjected to immunoprecipitation with anti-HA (2367; Cell Signaling Technology) and 30 μl protein G-agarose at 4 °C overnight. Beads were washed four times with 1 ml of lysis buffer, and heated at 95 °C for 5 min in SDS-PAGE loading buffer. Precipitates were analyzed by Western blotting using anti-FLAG and anti-HA antibodies.
Immunofluorescence and confocal microscopy
COS-7 cells were cultured on coverslips, transfected with expression plasmids, and fixed 24 h after transfection with methanol at −20 °C for 10 min. After blocking in 3% bovine serum albumin in PBS, cells were stained with rabbit anti-ErbB4 (E200; Abcam) and Alexa Fluor 555 goat anti-rabbit (Thermo Fischer Scientific) antibodies. Nuclei were visualized with DAPI (Sigma-Aldrich). The images were acquired with a Zeiss LSM780 confocal microscope using a C-Apochromat 40× water objective (numerical aperture 1.20). For quantification, 475 cells from four independent experiments were scored for predominantly cytoplasmic (more signal in the cytoplasm than in the nucleus) or nuclear (equal signal in the nucleus and in the cytoplasm or more signal in the nucleus than in the cytoplasm) staining. Data were analyzed using the t test.
Proximity ligation assay
MCF-7 cells were cultured on coverslips, transfected with expression plasmids and starved without serum overnight. Twenty-four h after transfection, cells were treated with 50 ng/ml neuregulin-1 (NRG-1; R&D Systems) for 15 min, and fixed with methanol at −20 °C for 10 min. After blocking in 3% bovine serum albumin in PBS, cells were incubated with mouse anti-HA (H3663; Sigma-Aldrich) and rabbit anti-CRM1 (ab24189; Abcam) primary antibodies. In situ proximity ligation assay was performed using Duolink In Situ PLA Probe secondary antibodies (anti-mouse minus and anti-rabbit plus; Sigma-Aldrich) and Duolink In Situ Detection Reagents Orange (Sigma-Aldrich) according to the manufacturer's protocol. To visualize cells expressing HA-tagged ErbB4, cells were incubated with Alexa Fluor 488 goat anti-mouse (Thermo Fischer Scientific) secondary antibody after PLA reactions. Nuclei were visualized with DAPI. The images were acquired with Zeiss LSM780 confocal microscope using a C-Apochromat 63×water objective (numerical aperture 1.20). For quantification, Alexa Fluor 488 positive (i.e. ErbB4-HA positive) cells (n = 63 for cells expressing wild-type ErbB4 and n = 53 for cells expressing ErbB4 K714R) were quantified for PLA signals localized in cell nuclei, nuclear rim, or cytoplasm. Negative binomial regression analysis was used for statistical analysis of the data.
Analysis of ErbB4 tyrosine phosphorylation and signaling
To analyze ErbB4 tyrosine phosphorylation, COS-7 cells growing on 10-cm plates were transfected as indicated, serum starved overnight, and lysed. Lysates were subjected to immunoprecipitation with 1 μg of anti-ErbB4 (HFR-1; Abcam). ErbB4 tyrosine phosphorylation was analyzed by Western blotting with phosphotyrosine (4G10; Upstate) and ErbB4 (E200; Abcam) antibodies. For the analysis of ErbB4 signaling, COS-7 cells growing on 6-well plates were transfected as indicated, serum starved overnight, stimulated with 50 ng/ml NRG-1 for 10 min, and lysed. Lysates were analyzed by Western blotting with ErbB4 (E200; Abcam), phospho-ErbB4 (4757; Cell Signaling Technology), phospho-Akt (9271; Cell Signaling Technology), phospho-Erk1/2 (9101; Cell Signaling Technology), Akt (sc-1618; Santa Cruz Biotechnology), Erk (9102; Cell Signaling Technology), phospho-STAT5 (9351; Cell Signaling Technology), and STAT5 (sc-835; Santa Cruz Biotechnology) antibodies. Loading was controlled using anti-actin. The data from two to three independent experiments were quantified as signal intensities of phosphorylated Erk1/2, Akt, or STAT5a relative to respective total protein using ImageJ software.
Modeling and structural analysis
The asymmetric ErbB4 ICD was reconstructed from the 2.5 Å resolution X-ray structure of the human ErbB4 kinase domain (RCSB PDB ID: 3BCE) (32) and the NMR structure of SUMO1 (RCSB PDB ID: 2ASQ) (38), solved with bound SUMO binding motif peptide. BODIL (71) was used to visualize and build a complex between the asymmetric kinase homodimer and SUMO, by placing the terminal Gly-Gly adjacent to the Nζ side chain atom of Lys-714, to accurately represent the isopeptide bond formed on SUMOylation. At either of the Lys-714 lysines, SUMO is easily accommodated (including SUMO2 and SUMO3 structures that share high sequence and structural similarity; the C-terminal region, terminating in Gly-Gly-Oxt is flexible). To assess the role of SUMOylation with regard to the juxtamembrane latch, chain A of a second ErbB4 X-ray structure (2.4 Å resolution; RCSB PDB ID: 2R4B) (72) was superposed onto each subunit of the ErbB4 asymmetric dimer. The 2R4B structure includes an additional nine N-terminal residues that correspond to the juxtamembrane latch. Structural details of the model were examined using BODIL. Fig. 6B was produced in BODIL.
Generation of retroviral cell lines
For retrovirus production, pBABEpuro-ErbB4JM-aCYT-2-HA (68) and pBABEpuro-ErbB4JM-aCYT-2-K714R-HA (generated by site-directed mutagenesis and verified by sequencing) were transfected into Phoenix-AMPHO HEK 293T packaging cells. Retrovirus-containing media were harvested 36 h after transfection, and incubated on HC11 cells for 8 h in the presence of 8 μg/ml polybrene (Sigma-Aldrich). To generate stable cell lines, infected cells were selected with 2 μg/ml puromycin (Sigma-Aldrich). Pools of puromycin-resistant cells were used in experiments.
Differentiation of HC11 cells in three-dimensional culture
Three-dimensional culture of HC11 cells in Growth Factor Reduced Matrigel (Corning) has been described (27). Briefly, single cell suspensions of HC11 transfectants were suspended into Growth Factor Reduced Matrigel in triplicates on 96-well plates, supplemented with 50 ng/ml NRG-1, and maintained at 37 °C for 15–20 days. Colonies were counted using 200× magnification from four independent views through the whole thickness of the Matrigel, and classified as undifferentiated colonies of differentiated acini on the basis on their morphology (73). Data from four independent experiments were analyzed with one-way analysis of variance, followed by pairwise comparisons using the two-tailed t test. Correction for multiple testing was performed using the Benjamini-Hochberg procedure.
Author contributions
A. M. K., M. S., and K. E. designed the study. A. M. K., M. S., and M. H. carried out experimentation and A. M. K., M. H., M. S., and K. E. analyzed the results. M. S. J. performed structural modeling and analysis. A. M. K., M. S. J., M. S., and K. E. wrote the manuscript. All authors approved the final version of the manuscript.
Supplementary Material
Acknowledgments
We thank Deepankar Chakroborty, Johannes Merilahti, and Katri Vaparanta for their kind help with statistical analyses, and Minna Santanen, Mika Savisalo, and Maria Tuominen for their skillful technical assistance. Imaging was performed at the Cell Imaging Core in the Turku Centre for Biotechnology, Turku, Finland. Jukka Lehtonen, Biocenter Finland and CSC IT Center for Science are thanked for bioinformatics and computational infrastructure support.
This work was financially supported by the Academy of Finland; the Cancer Foundation of Finland; the Cancer Society of Southwest Finland; the Finnish Cultural Foundation; the Foundation of Åbo Akademi University; the Ida Montin Foundation; the Joe, Pentti, and Tor Borg Memorial Fund; the Orion-Farmos Research Foundation; the Sigrid Jusélius Foundation; the Turku Doctoral Programme of Biomedical Sciences; Turku University Central Hospital; and the Turku University Foundation. The authors declare that they have no conflicts of interest with the contents of this article.
This article contains supplemental Figs. S1–S10.
Please note that the JBC is not responsible for the long-term archiving and maintenance of this site or any other third party hosted site.
A. M. Knittle, M. Sundvall, and K. Elenius, unpublished observations.
- EGFR
- epidermal growth factor receptor
- ICD
- intracellular domain
- SUMO
- small ubiquitin-like modifier
- SENPs
- sentrin-specific proteases
- CRM1
- chromosomal region maintenance 1
- RTK
- receptor tyrosine kinase
- NRG
- neuregulin
- RIP
- regulated intramembrane proteolysis
- SIM
- SUMO interacting motif
- NES
- nuclear export signal
- PLA
- proximity ligation assay
- IGF-1R
- insulin-like growth factor-1 receptor
- qRT-PCR
- quantitative RT-PCR.
References
- 1. Yarden Y., and Pines G. (2012) The ERBB network: At last, cancer therapy meets systems biology. Nat. Rev. Cancer 12, 553–563 [DOI] [PubMed] [Google Scholar]
- 2. Elenius K., Corfas G., Paul S., Choi C. J., Rio C., Plowman G. D., and Klagsbrun M. (1997) A novel juxtamembrane domain isoform of HER4/ErbB4. Isoform-specific tissue distribution and differential processing in response to phorbol ester. J. Biol. Chem. 272, 26761–26768 [DOI] [PubMed] [Google Scholar]
- 3. Elenius K., Choi C. J., Paul S., Santiestevan E., Nishi E., and Klagsbrun M. (1999) Characterization of a naturally occurring ErbB4 isoform that does not bind or activate phosphatidyl inositol 3-kinase. Oncogene 18, 2607–2615 [DOI] [PubMed] [Google Scholar]
- 4. Rio C., Buxbaum J. D., Peschon J. J., and Corfas G. (2000) Tumor necrosis factor-α-converting enzyme is required for cleavage of erbB4/HER4. J. Biol. Chem. 275, 10379–10387 [DOI] [PubMed] [Google Scholar]
- 5. Ni C.-Y., Murphy M. P., Golde T. E., and Carpenter G. (2001) γ-Secretase cleavage and nuclear localization of ErbB-4 receptor tyrosine kinase. Science 294, 2179–2181 [DOI] [PubMed] [Google Scholar]
- 6. Määttä J. A., Sundvall M., Junttila T. T., Peri L., Laine V. J. O., Isola J., Egeblad M., and Elenius K. (2006) Proteolytic cleavage and phosphorylation of a tumor-associated ErbB4 isoform promote ligand-independent survival and cancer cell growth. Mol. Biol. Cell 17, 67–79 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7. Linggi B., Cheng Q. C., Rao A. R., and Carpenter G. (2006) The ErbB-4 s80 intracellular domain is a constitutively active tyrosine kinase. Oncogene 25, 160–163 [DOI] [PubMed] [Google Scholar]
- 8. Sundvall M., Peri L., Määttä J. A., Tvorogov D., Paatero I., Savisalo M., Silvennoinen O., Yarden Y., and Elenius K. (2007) Differential nuclear localization and kinase activity of alternative ErbB4 intracellular domains. Oncogene 26, 6905–6914 [DOI] [PubMed] [Google Scholar]
- 9. Komuro A., Nagai M., Navin N. E., and Sudol M. (2003) WW domain-containing protein YAP associates with ErbB-4 and acts as a co-transcriptional activator for the carboxyl-terminal fragment of ErbB-4 that translocates to the nucleus. J. Biol. Chem. 278, 33334–33341 [DOI] [PubMed] [Google Scholar]
- 10. Williams C. C., Allison J. G., Vidal G. A., Burow M. E., Beckman B. S., Marrero L., and Jones F. E. (2004) The ERBB4/HER4 receptor tyrosine kinase regulates gene expression by functioning as a STAT5A nuclear chaperone. J. Cell Biol. 167, 469–478 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11. Linggi B., and Carpenter G. (2006) ErbB-4 s80 intracellular domain abrogates ETO2-dependent transcriptional repression. J. Biol. Chem. 281, 25373–25380 [DOI] [PubMed] [Google Scholar]
- 12. Sardi S. P., Murtie J., Koirala S., Patten B. A., and Corfas G. (2006) Presenilin-dependent ErbB4 nuclear signaling regulates the timing of astrogenesis in the developing brain. Cell 127, 185–197 [DOI] [PubMed] [Google Scholar]
- 13. Zhu Y., Sullivan L. L., Nair S. S., Williams C. C., Pandey A. K., Marrero L., Vadlamudi R. K., and Jones F. E. (2006) Coregulation of estrogen receptor by ERBB4/HER4 establishes a growth-promoting autocrine signal in breast tumor cells. Cancer Res. 66, 7991–7998 [DOI] [PubMed] [Google Scholar]
- 14. Gilmore-Hebert M., Ramabhadran R., and Stern D. F. (2010) Interactions of ErbB4 and Kap1 connect the growth factor and DNA damage response pathways. Mol. Cancer Res. 8, 1388–1398 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15. Sundvall M., Veikkolainen V., Kurppa K., Salah Z., Tvorogov D., van Zoelen E. J., Aqeilan R., and Elenius K. (2010) Cell death or survival promoted by alternative isoforms of ErbB4. Mol. Biol. Cell 21, 4275–4286 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16. Paatero I., Jokilammi A., Heikkinen P. T., Iljin K., Kallioniemi O. P., Jones F. E., Jaakkola P. M., and Elenius K. (2012) Interaction with ErbB4 promotes hypoxia-inducible factor-1α signaling. J. Biol. Chem. 287, 9659–9671 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17. Naresh A., Long W., Vidal G. A., Wimley W. C., Marrero L., Sartor C. I., Tovey S., Cooke T. G., Bartlett J. M. S., and Jones F. E. (2006) The ERBB4/HER4 intracellular domain 4ICD is a BH3-only protein promoting apoptosis of breast cancer cells. Cancer Res. 66, 6412–6420 [DOI] [PubMed] [Google Scholar]
- 18. Jones F. E. (2008) HER4 Intracellular domain (4ICD) activity in the developing mammary gland and breast cancer. J. Mammary Gland Biol. Neoplasia 13, 247–258 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19. Muraoka-Cook R. S., Sandahl M. A., Strunk K. E., Miraglia L. C., Husted C., Hunter D. M., Elenius K., Chodosh L. A., and Earp H. S. 3rd (2009) ErbB4 splice variants Cyt1 and Cyt2 differ by 16 amino acids and exert opposing effects on the mammary epithelium in vivo. Mol. Cell. Biol. 29, 4935–4948 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20. Srinivasan R., Gillett C. E., Barnes D. M., and Gullick W. J. (2000) Nuclear expression of the c-erbB-4/HER-4 growth factor receptor in invasive breast cancers. Cancer Res. 60, 1483–1487 [PubMed] [Google Scholar]
- 21. Junttila T. T., Sundvall M., Lundin M., Lundin J., Tanner M., Härkönen P., Joensuu H., Isola J., and Elenius K. (2005) Cleavable ErbB4 isoform in estrogen receptor-regulated growth of breast cancer cells. Cancer Res. 65, 1384–1393 [DOI] [PubMed] [Google Scholar]
- 22. Pichler A., Fatouros C., Lee H., and Eisenhardt N. (2017) SUMO conjugation—a mechanistic view. Biomol. Concepts 8, 13–36 [DOI] [PubMed] [Google Scholar]
- 23. Rytinki M. M., Kaikkonen S., Pehkonen P., Jääskeläinen T., and Palvimo J. J. (2009) PIAS proteins: Pleiotropic interactors associated with SUMO. Cell. Mol. Life Sci. 66, 3029–3041 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24. Flotho A., and Melchior F. (2013) Sumoylation: A regulatory protein modification in health and disease. Annu. Rev. Biochem. 82, 357–385 [DOI] [PubMed] [Google Scholar]
- 25. Yeh E. T. (2009) SUMOylation and de-SUMOylation: Wrestling with life's processes. J. Biol. Chem. 284, 8223–8227 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26. Hendriks I. A., D'Souza R. C. J., Yang B., Verlaan-de Vries M., Mann M., and Vertegaal A. C. O. (2014) Uncovering global SUMOylation signaling networks in a site-specific manner. Nat. Struct. Mol. Biol. 21, 927–936 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27. Sundvall M., Korhonen A., Vaparanta K., Anckar J., Halkilahti K., Salah Z., Aqeilan R. I., Palvimo J. J., Sistonen L., and Elenius K. (2012) Protein inhibitor of activated STAT3 (PIAS3) protein promotes SUMOylation and nuclear sequestration of the intracellular domain of ErbB4 protein. J. Biol. Chem. 287, 23216–23226 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28. Matic I., Schimmel J., Hendriks I. A., van Santen M. A., van de Rijke F., van Dam H., Gnad F., Mann M., and Vertegaal A. C. O. (2010) Site-specific identification of SUMO-2 targets in cells reveals an inverted SUMOylation motif and a hydrophobic cluster SUMOylation motif. Mol. Cell 39, 641–652 [DOI] [PubMed] [Google Scholar]
- 29. Impens F., Radoshevich L., Cossart P., and Ribet D. (2014) Mapping of SUMO sites and analysis of SUMOylation changes induced by external stimuli. Proc. Natl. Acad. Sci. U.S.A. 111, 12432–12437 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30. Lamoliatte F., Caron D., Durette C., Mahrouche L., Maroui M. A., Caron-Lizotte O., Bonneil E., Chelbi-Alix M. K., and Thibault P. (2014) Large-scale analysis of lysine SUMOylation by SUMO remnant immunoaffinity profiling. Nat. Commun. 5, 5409. [DOI] [PubMed] [Google Scholar]
- 31. Tammsalu T., Matic I., Jaffray E. G., Ibrahim A. F. M., Tatham M. H., and Hay R. T. (2014) Proteome-wide identification of SUMO2 modification sites. Sci. Signal 7, rs2. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32. Qiu C., Tarrant M. K., Choi S. H., Sathyamurthy A., Bose R., Banjade S., Pal A., Bornmann W. G., Lemmon M. A., Cole P. A., and Leahy D. J. (2008) Mechanism of activation and inhibition of the HER4/ErbB4 kinase. Structure 16, 460–467 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33. Sundvall M., Iljin K., Kilpinen S., Sara H., Kallioniemi O.-P., and Elenius K. (2008) Role of ErbB4 in breast cancer. J Mammary Gland Biol. Neoplasia 13, 259–268 [DOI] [PubMed] [Google Scholar]
- 34. Komander D., and Rape M. (2012) The ubiquitin code. Annu. Rev. Biochem. 81, 203–229 [DOI] [PubMed] [Google Scholar]
- 35. Fornerod M., Ohno M., Yoshida M., and Mattaj I. W. (1997) CRM1 is an export receptor for leucine-rich nuclear export signals. Cell 90, 1051–1060 [DOI] [PubMed] [Google Scholar]
- 36. Kudo N., Matsumori N., Taoka H., Fujiwara D., Schreiner E. P., Wolff B., Yoshida M., and Horinouchi S. (1999) Leptomycin B inactivates CRM1/exportin 1 by covalent modification at a cysteine residue in the central conserved region. Proc. Natl. Acad. Sci. U.S.A. 96, 9112–9117 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37. Jura N., Endres N. F., Engel K., Deindl S., Das R., Lamers M. H., Wemmer D. E., Zhang X., and Kuriyan J. (2009) Mechanism for activation of the EGF receptor catalytic domain by the juxtamembrane segment. Cell 137, 1293–1307 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38. Song J., Zhang Z., Hu W., and Chen Y. (2005) Small ubiquitin-like modifier (SUMO) recognition of a SUMO binding motif: a reversal of the bound orientation. J. Biol. Chem. 280, 40122–40129 [DOI] [PubMed] [Google Scholar]
- 39. Petersen O. W., Rønnov-Jessen L., Howlett A. R., and Bissell M. J. (1992) Interaction with basement membrane serves to rapidly distinguish growth and differentiation pattern of normal and malignant human breast epithelial cells. Proc. Natl. Acad. Sci. U.S.A. 89, 9064–9068 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40. Sehat B., Tofigh A., Lin Y., Trocmé E., Liljedahl U., Lagergren J., and Larsson O. (2010) SUMOylation mediates the nuclear translocation and signaling of the IGF-1 receptor. Sci. Signal. 3, ra10. [DOI] [PubMed] [Google Scholar]
- 41. Petersen B., Petersen T. N., Andersen P., Nielsen M., and Lundegaard C. (2009) A generic method for assignment of reliability scores applied to solvent accessibility predictions. BMC Struct. Biol. 9, 51. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42. Packham S., Lin Y., Zhao Z., Warsito D., Rutishauser D., and Larsson O. (2015) The nucleus-localized epidermal growth factor receptor is SUMOylated. Biochemistry 54, 5157–5166 [DOI] [PubMed] [Google Scholar]
- 43. Lin S.-Y., Makino K., Xia W., Matin A., Wen Y., Kwong K. Y., Bourguignon L., and Hung M.-C. (2001) Nuclear localization of EGF receptor and its potential new role as a transcription factor. Nat. Cell Biol. 3, 802–808 [DOI] [PubMed] [Google Scholar]
- 44. Vidal G. A., Naresh A., Marrero L., and Jones F. E. (2005) Presenilin-dependent gamma-secretase processing regulates multiple ERBB4/HER4 activities. J. Biol. Chem. 280, 19777–19783 [DOI] [PubMed] [Google Scholar]
- 45. Warsito D., Sjöström S., Andersson S., Larsson O., and Sehat B. (2012) Nuclear IGF1R is a transcriptional co-activator of LEF1/TCF. EMBO Rep. 13, 244–250 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46. Hsu S.-C., and Hung M.-C. (2007) Characterization of a novel tripartite nuclear localization sequence in the EGFR family. J. Biol. Chem. 282, 10432–10440 [DOI] [PubMed] [Google Scholar]
- 47. Wood L. D., Irvin B. J., Nucifora G., Luce K. S., and Hiebert S. W. (2003) Small ubiquitin-like modifier conjugation regulates nuclear export of TEL, a putative tumor suppressor. Proc. Natl. Acad. Sci. U.S.A. 100, 3257–3262 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48. Du J. X., Bialkowska A. B., McConnell B. B., and Yang V. W. (2008) SUMOylation regulates nuclear localization of Krüppel-like factor 5. J. Biol. Chem. 283, 31991–32002 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49. Bassi C., Ho J., Srikumar T., Dowling R. J. O., Gorrini C., Miller S. J., Mak T. W., Neel B. G., Raught B., and Stambolic V. (2013) Nuclear PTEN controls DNA repair and sensitivity to genotoxic stress. Science 341, 395–399 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50. Santiago A., Li D., Zhao L. Y., Godsey A., and Liao D. (2013) p53 SUMOylation promotes its nuclear export by facilitating its release from the nuclear export receptor CRM1. Mol. Biol. Cell 24, 2739–2752 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 51. Engelsma D., Bernad R., Calafat J., and Fornerod M. (2004) Supraphysiological nuclear export signals bind CRM1 independently of RanGTP and arrest at Nup358. EMBO J. 23, 3643–3652 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 52. Kutay U., and Güttinger S. (2005) Leucine-rich nuclear-export signals: Born to be weak. Trends Cell Biol. 15, 121–124 [DOI] [PubMed] [Google Scholar]
- 53. Hang J., and Dasso M. (2002) Association of the human SUMO-1 protease SENP2 with the nuclear pore. J. Biol. Chem. 277, 19961–19966 [DOI] [PubMed] [Google Scholar]
- 54. Kadaré G., Toutant M., Formstecher E., Corvol J.-C., Carnaud M., Boutterin M.-C., and Girault J.-A. (2003) PIAS1-mediated sumoylation of focal adhesion kinase activates its autophosphorylation. J. Biol. Chem. 278, 47434–47440 [DOI] [PubMed] [Google Scholar]
- 55. de la Cruz-Herrera C.F., Campagna M., Lang V., del Carmen González-Santamaría J., Marcos-Villar L., Rodríguez M. S., Vidal A., Collado M., and Rivas C. (2015) SUMOylation regulates AKT1 activity. Oncogene 34, 1442–1450 [DOI] [PubMed] [Google Scholar]
- 56. Yao Q., Li H., Liu B.-Q., Huang X.-Y., and Guo L. (2011) SUMOylation-regulated protein phosphorylation, evidence from quantitative phosphoproteomics analyses. J. Biol. Chem. 286, 27342–27349 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 57. Wang L., and Banerjee S. (2004) Differential PIAS3 expression in human malignancy. Oncol. Rep. 11, 1319–1324 [PubMed] [Google Scholar]
- 58. McHale K., Tomaszewski J. E., Puthiyaveettil R., LiVolsi V. A., and Clevenger C. V. (2008) Altered expression of prolactin receptor-associated signaling proteins in human breast carcinoma. Mod. Pathol. 21, 565–571 [DOI] [PubMed] [Google Scholar]
- 59. Yang S.-F., Hou M.-F., Chen F.-M., Ou-Yang F., Wu Y.-C., Chai C.-Y., and Yeh Y.-T. (2016) Prognostic value of protein inhibitor of activated STAT3 in breast cancer patients receiving hormone therapy. BMC Cancer 16, 20. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 60. Pircher T. J., Flores-Morales A., Mui A. L., Saltiel A. R., Norstedt G., Gustafsson J. A., and Haldosén L. A. (1997) Mitogen-activated protein kinase inhibition decreases growth hormone stimulated transcription mediated by STAT5. Mol. Cell. Endocrinol. 133, 169–176 [DOI] [PubMed] [Google Scholar]
- 61. Hietakangas V., Anckar J., Blomster H. A., Fujimoto M., Palvimo J. J., Nakai A., and Sistonen L. (2006) PDSM, a motif for phosphorylation-dependent SUMO modification. Proc. Natl. Acad. Sci. U.S.A. 103, 45–50 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 62. Meulmeester E., Kunze M., Hsiao H. H., Urlaub H., and Melchior F. (2008) Mechanism and consequences for paralog-specific sumoylation of ubiquitin-specific protease 25. Mol. Cell 30, 610–619 [DOI] [PubMed] [Google Scholar]
- 63. Rytinki M. M., and Palvimo J. J. (2009) SUMOylation attenuates the function of PGC-1α. J. Biol. Chem. 284, 26184–26193 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 64. Kang X., Qi Y., Zuo Y., Wang Q., Zou Y., Schwartz R. J., Cheng J., and Yeh E. T. H. (2010) SUMO-specific protease 2 is essential for suppression of polycomb group protein-mediated gene silencing during embryonic development. Mol. Cell 38, 191–201 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 65. Bawa-Khalfe T., Lu L.-S., Zuo Y., Huang C., Dere R., Lin F.-M., and Yeh E. T. H. (2012) Differential expression of SUMO-specific protease 7 variants regulates epithelial-mesenchymal transition. Proc. Natl. Acad. Sci. U.S.A. 109, 17466–17471 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 66. Yun C., Wang Y., Mukhopadhyay D., Backlund P., Kolli N., Yergey A., Wilkinson K. D., and Dasso M. (2008) Nucleolar protein B23/nucleophosmin regulates the vertebrate SUMO pathway through SENP3 and SENP5 proteases. J. Cell Biol. 183, 589–595 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 67. Wang W., Budhu A., Forgues M., and Wang X. W. (2005) Temporal and spatial control of nucleophosmin by the Ran-Crm1 complex in centrosome duplication. Nat. Cell Biol. 7, 823–830 [DOI] [PubMed] [Google Scholar]
- 68. Kurppa K. J., Denessiouk K., Johnson M. S., and Elenius K. (2016) Activating ERBB4 mutations in non-small cell lung cancer. Oncogene. 35, 1283–1291 [DOI] [PubMed] [Google Scholar]
- 69. Tatham M. H., Rodriguez M. S., Xirodimas D. P., and Hay R. T. (2009) Detection of protein SUMOylation in vivo. Nat. Protoc. 4, 1363–1371 [DOI] [PubMed] [Google Scholar]
- 70. Junttila T. T., Laato M., Vahlberg T., Söderström K.-O., Visakorpi T., Isola J., and Elenius K. (2003) Identification of patients with transitional cell carcinoma of the bladder overexpressing ErbB2, ErbB3, or specific ErbB4 isoforms: Real-time reverse transcription-PCR analysis in estimation of ErbB receptor status from cancer patients. Clin. Cancer Res. 9, 5346–5357 [PubMed] [Google Scholar]
- 71. Lehtonen J. V., Still D.-J., Rantanen V.-V., Ekholm J., Björklund D., Iftikhar Z., Huhtala M., Repo S., Jussila A., Jaakkola J., Pentikäinen O., Nyrönen T., Salminen T., Gyllenberg M., and Johnson M. S. (2004) BODIL: A molecular modeling environment for structure-function analysis and drug design. J. Comput. Aided Mol. Des. 18, 401–419 [DOI] [PubMed] [Google Scholar]
- 72. Wood E. R., Shewchuk L. M., Ellis B., Brignola P., Brashear R. L., Caferro T. R., Dickerson S. H., Dickson H. D., Donaldson K. H., Gaul M., Griffin R. J., Hassell A. M., Keith B., Mullin R., Petrov K. G., Reno M. J., Rusnak D. W., Tadepalli S. M., Ulrich J. C., Wagner C. D., Vanderwall D. E., Waterson A. G., Williams J. D., White W. L., and Uehling D. E. (2008) 6-Ethynylthieno[3,2-d]- and 6-ethynylthieno[2,3-d]pyrimidin-4-anilines as tunable covalent modifiers of ErbB kinases. Proc. Natl. Acad. Sci. U.S.A. 105, 2773–2778 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 73. Tvorogov D., Sundvall M., Kurppa K., Hollmén M., Repo S., Johnson M. S., and Elenius K. (2009) Somatic mutations of ErbB4: Selective loss-of-function phenotype affecting signal transduction pathways in cancer. J. Biol. Chem. 284, 5582–5591 [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.