Abstract
More than a hundred proteins comprise the RAS superfamily of small GTPases. This family can be divided into RAS, RHO, RAB, RAN, ARF, and RAD subfamilies, with each shown to play distinct roles in human cells in both health and disease. The RAS subfamily has a well-established role in human cancer with the three genes, HRAS, KRAS, and NRAS being the commonly mutated in tumors. These RAS mutations, most often functionally activating, are especially common in pancreatic, lung, and colorectal cancers. Efforts to inhibit RAS and related GTPases have produced inhibitors targeting the downstream effectors of RAS signaling, including inhibitors of the RAF-mitogen-activated protein kinase/extracellular signal-related kinase (ERK)-ERK kinase pathway and the phosphoinositide-3-kinase-AKT-mTOR kinase pathway. A third effector arm of RAS signaling, mediated by RAL (RAS like) has emerged in recent years as a critical driver of RAS oncogenic signaling and has not been targeted until recently. RAL belongs to the RAS branch of the RAS superfamily and shares a high structural similarity with RAS. In human cells, there are two genes, RALA and RALB, both of which have been shown to play roles in the proliferation, survival, and metastasis of a variety of human cancers, including lung, colon, pancreatic, prostate, skin, and bladder cancers. In this review, we summarize the latest knowledge of RAL in the context of human cancer and the recent advancements in the development of cancer therapeutics targeting RAL small GTPases.
I. Introduction
Two closely related G-proteins, RALA and RALB, constitute the RAL branch of the RAS superfamily of small GTPases (Chardin and Tavitian, 1986). Activating oncogenic RAS mutations have been shown to drive many different types of human cancer (Cox et al., 2014). Since its discovery in 1982, numerous efforts have been made to target RAS in human cancer with little success (Cox et al., 2014). Many different approaches have been tried, including direct targeting of the active site, targeting RAS cell membrane localization, targeting the interaction with its upstream activators, and targeting its downstream effector signaling pathway. To date, the most effective strategy to target RAS oncogenic signaling has been inhibiting the downstream RAF-MEK-ERK and PI3K-AKT-mTOR effector pathways (Roberts and Der, 2007; Yap et al., 2008). In the last decade, however, the RALGEF-RAL signaling pathway has emerged as a third important effector signaling axis downstream of RAS (Gentry et al., 2014). This has prompted the search for ways to manipulate RAL signaling as a potential therapeutic strategy for targeting RAS-driven human cancer.
II. Evolutionary Biology and Structure of RAL
More than 150 small GTPases have been identified in the RAS superfamily (Cox and Der, 2010), which can be divided into five main families (RAS, RHO, RAN, RAB, and ARF) based on differences in sequence, structure, and function (Goitre et al., 2014). The RAS family itself is further divided into six subfamilies: RAS, RAL, RIT, RAP, RHEB, and RAD. RAL is the closest relative of RAS on the phylogenetic tree, sharing a high degree of sequence similarity with RAS (>50%), hence the name RAL (RAS like). RAL gene was initially identified in 1986 during a search for RAS-related genes in a simian cDNA library (Chardin and Tavitian, 1986). The human RALA and RALB genes were identified 3 years later using the simian RAL cDNA as a probe in a human pheochromocytoma cDNA library (Chardin and Tavitian, 1989). The two protein isoforms, RALA and RALB, despite sharing 82% similarity to each other at amino acid level, exhibit distinctive biologic functions especially in human cancer, which will be discussed in section V. Although RAL genes and proteins are highly conserved across species, invertebrates only harbor single RAL gene. Studies in Caenorhabditis elegans and Drosophila have confirmed the interaction between RAL and its known effectors, like TANK-binding kinase 1 (TBK1), SEC5, and RALBP1 (Mirey et al., 2003; Chien et al., 2006), and shed light on new effector signaling pathways such as Msn MAP4 kinase and mTOR signaling (Balakireva et al., 2006; Martin et al., 2014).
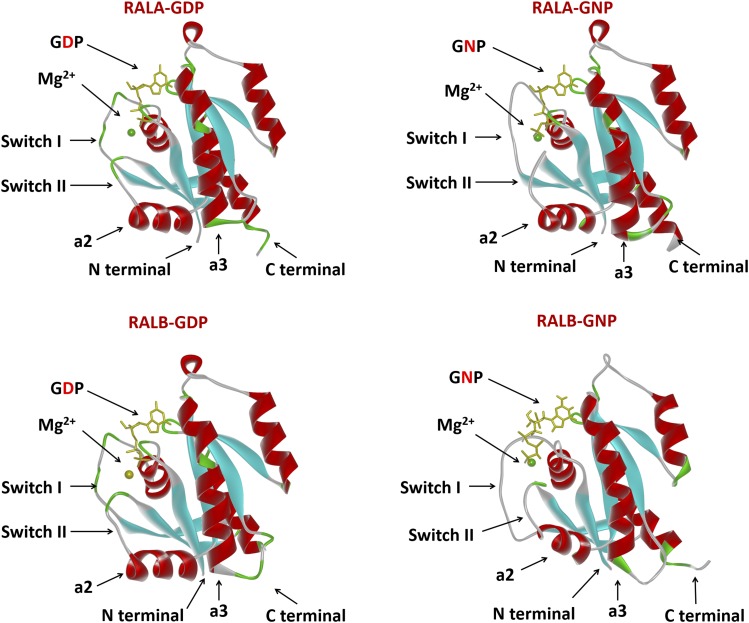
The protein structure for both RALA and RALB has been resolved, either alone or together with their binding partners (Nicely et al., 2004; Fenwick et al., 2009; Popovic et al., 2016). The tertiary protein structure of RALA and RALB is also very similar; both proteins contain a free-floating N-terminal 11-amino acid sequence, followed by the G-domain, involved in GDP/GTP binding, and the C-terminal membrane targeting sequence. RAL proteins share a similar G-domain architecture with RAS, consisting of six β-sheets connected by loops and five α-helices, which form the highly flexible switch I and switch II regions (Fig. 1). Structural comparison between the GDP- and GTP-bound form of RAL revealed that guanine nucleotide exchange mainly induces conformational changes in the two flexible switch regions, which are responsible for the selective binding to both regulators and effectors of RAL. RALA and RALB share almost identical sequence in the switch regions but differ considerably in the C-terminal hypervariable region. The essential role of the C-terminal hypervariable region in the correct intracellular trafficking and membrane localization of RAS proteins has been extensively studied (Cox et al., 2015). Like RAS, RAL proteins also terminate with the CAAX tetrapeptide motif sequence (C = cysteine, A = aliphatic amino acids, X = any amino acid), which signals for a series of posttranslational modification events that are critical for the membrane association of RAL.
Fig. 1.
Structure comparison between human RALA and RALB proteins. Structure models of RALA-GDP, RALA-GNP, RALB-GDP, RALB-GNP are shown in ribbon representation. GDP and GNP (nonhydrolyzable form of GTP) is shown in yellow, Mg is shown as a green sphere, α-helices are shown in red, and β-sheets are shown in cyan. Models were generated with Accelrys Discovery Studio software (San Diego, CA) using published structures.
III. RAL Biochemistry and Enzymology
Small GTPases are small (20–25 kDa) G-proteins that can bind to and hydrolyze GTP. A typical G-protein is active when bound to GTP and inactive when bound to GDP and therefore can function as a molecular on and off switch in signaling pathways. RAL activity can be regulated by altering the cellular concentration of GTP and GDP bound states, by posttranslational modifications of the C-terminal end or phosphorylation by Aurora A kinase and protein kinase C (Fig. 2).
Fig. 2.
(A) The three most characterized effector signaling pathways downstream of RAS. (B) The GDP-GTP cycle of RAL small GTPases. Upstream signals stimulate binding of RALGEF that triggers GDP-GTP exchange, resulting in the activation of RAL. RAL-GTP can then bind to its downstream effector proteins such as RALBP1. Finally, RALGAPs bind to RAL-GTP and hydrolyze GTP to GDP, cycling RAL back to its inactive GDP-bound form.
A. GTP-GDP Cycling
Like other small GTPases, RAL cycles between the GDP-bound inactive state and GTP-bound active state in cells. The intrinsic GTPase activity of RAL GTPases is very weak, hence they rely on guanine nucleotide exchange factors (RALGEF) and GTPase activating proteins (RALGAP) to catalyze the GDP-GTP exchange. RALGEFs favor formation of RAL-GTP by stimulating exchange of GDP for GTP when intracellular concentrations of GTP are 10 times higher than GDP. RALGAPs catalyze the hydrolysis of GTP to GDP, returning RAL to an inactive state. RALA and RALB have the same enzymology, are regulated by the same set of RALGEFs and RALGAPs, and can bind to the same set of downstream effectors. However, as discussed below, they exhibit distinct roles in cellular function and human cancer.
The seven known RALGEFs, RALGDS, RGL1, RGL2, RGL3, RGL4, RALGPS1, and RALGPS2, can be divided into two groups. The first (RALGDS, RGL1, RGL2, RGL3) can directly bind to the effector binding region of activated RAS and hence relies on RAS activation, while the other group (RALGPS1, RALGPS2, RGL4) can be activated by RAS-independent mechanisms (Gentry et al., 2014). The most characterized RALGEF is RALGDS, which is also the first RALGEF discovered by yeast two-hybrid screening aimed at identifying RAS effectors (Albright et al., 1993). RALGDS was shown to be specific for RALA and RALB but not for other small GTPases. Subsequently, RGL1, RGL2, and RGL3 were also discovered using yeast two-hybrid screening (Kikuchi et al., 1994; Isomura et al., 1996; Wolthuis et al., 1996; Shao and Andres, 2000) and found to share a similar domain structure with RALGDS, consisting of the N-terminal REM (RAS exchanger motif) domain, the CDC25 catalytic domain, and the C-terminal RA (RAS association) domain (Ferro and Trabalzini, 2010). Activated RAS recruits RALGEFs onto the plasma membrane and delivers them to RALA and RALB. The other subset of RALGEFs, RALGPS1, RALGPS2, and RGL4, retain the CDC25 catalytic domain but lack the RAS-interacting domains REM and RA and are therefore activated independent of RAS (D’Adamo et al., 1997; de Bruyn et al., 2000; Rebhun et al., 2000; Ceriani et al., 2007). RALGPS1 and RALGPS2 contain a C-terminal PH (pleckstrin homology) domain that is believed to be important for its membrane targeting and activation of RAL. RGL4 has neither the RA domain nor the PH domain, and its activation mechanism is still poorly understood. Very recently, a chromosomal passenger complex protein, TD-60, was identified as an eighth RALGEF that activates RALA but not RALB during mitosis. However, little is known regarding the structural characteristics or the exact mechanism of Ral activation by TD-60 (Papini et al., 2015). Relatively little is known about RAL specific GAP proteins. Two closely related proteins, RALGAP1 and RALGAP2, were identified to have GAP activity for RAL (Shirakawa et al., 2009; Chen et al., 2011). RALGAPs are heterodimers containing one α-catalytic subunit and one β-regulatory subunit. Supporting an inhibitory role on RAL activity, knockdown of RALGAP2α promoted bladder cancer metastasis through enhancement of RAL activity (Saito et al., 2013). RALGAP-α subunit was also shown to be involved in insulin-stimulated RALA activation and GLUT4 trafficking (Chen et al., 2011, 2014). The regulatory subunit RALGAP-β was found to play an essential role in RAL-mediated regulation of spindle formation and chromatin separation during mitosis (Personnic et al., 2014).
B. Posttranslational Modifications
The requirement of posttranslational modifications for correct membrane association of RAS has been widely acknowledged (Gentry et al., 2015). Like RAS, RAL proteins also bear the C-terminal CAAX motif that are subject to similar posttranslational modification. The CAAX motif of RALA is CCIL and of RALB is CCLL (Gentry et al., 2015). The first step of the posttranslational modifications is the covalent addition of an isoprenoid lipid to the first cysteine residue by the geranylgeranyltransferase-I (GGTase-I) (Falsetti et al., 2007). The AAX residues were then cleaved by RAS-converting endopeptidase 1; and the terminal lipid-modified cysteine methylated by isoprenylcysteine carboxymethytransferase-1. This process was shown to be essential for RAL membrane association and subcellular localization.
In addition to prenylation of the CAAX motif, phosphorylation of RAL at the C-terminal region plays an important role in the regulation of RAL activity. Aurora A kinase and protein kinase C were shown to phosphorylate C-terminal serine residues of RALA (S194) and RALB (S198), respectively (Lim et al., 2010; Wang et al., 2010; Martin et al., 2012). Similar to RAS, protein kinase-mediated phosphorylation of the C-terminal of RAL has been shown to regulate the subcellular localization and function of RAL. Phosphorylation of RALA by Aurora A relocates RALA from plasma membrane to endomembranes where RALA interacts with RALBP1 and is required for RALA mediated anchorage-independent growth and tumorigenicity in human cancer cells (Lim et al., 2010). Phosphorylation of RALB by protein kinase C translocated RALB from plasma membrane to the perinuclear region of the cell and is also required for RALB-mediated migration and metastasis (Wang et al., 2010; Martin et al., 2012).
IV. Effectors of RAL
The RAS-RAL signaling axis is involved in various physiologic and pathologic processes, such as membrane trafficking, actin cytoskeletal reorganization, transcription regulation, and kinase cascade signaling. Like RAS, after activation, RAL exerts its biologic function via binding to various effector proteins, most of which were discovered by two-hybrid screening.
A. RAL Binding Protein 1/RLIP76
RAL binding protein 1 (RALBP1, also known as RLIP or RLIP76) was the first identified and most extensively studied effector of RAL. RALBP1 was originally discovered as RAL binding partner in two independent experiments using either a yeast two hybrid library or a cDNA library screen (Cantor et al., 1995; Jullien-Flores et al., 1995). RALBP1 is a 76 kDa protein with a RAL binding domain that binds to both switch I and II regions of RAL-GTP (Fenwick et al., 2010). RALBP1 binds a diverse group of proteins and mediates different cellular functions. RALBP1 bears GAP activity for CDC42/Rac1 small GTPases, which are involved in actin dynamics, filopodia formation, and membrane ruffling (Park and Weinberg, 1995). RALBP1 can also promote invadopodium formation upon activation by RALB (Neel et al., 2012). Furthermore, RALBP1 is necessary for migration of human bladder and prostate cancers (Wu et al., 2010). Collectively, these results suggest an important role of RALBP1 in cancer cell adhesion, invasion, and migration. RALBP1 is also important in endocytosis, especially receptor-mediated endocytosis of growth factor receptors through direct interaction with proteins known to be involved in receptor-mediated endocytosis such as REPS1, REPS2, and AP2 (Nakashima et al., 1999; Jullien-Flores et al., 2000). RALBP1 functions as a drug transporter in cancer cells and can render cells resistant to certain chemotherapeutic drugs (Awasthi et al., 2000). This function is independent from RAL binding and attributed to the two ATP binding motifs. This allows RALBP1 to function as an ATP-dependent transporter and increase the efflux of glutathione conjugates of electrophilic compounds and therefore protect cancer cells from cytotoxic drugs. RAL-dependent RALBP1 activation is required for mitochondrial fission at mitosis (Kashatus et al., 2011). Mitochondrial fission is prerequisite for the proper distribution of mitochondria to daughter cells during cell division. RALA-RALBP1 signaling plays an essential role in this process by inducing phosphorylation of the enzyme DRP1 GTPase and facilitating recruitment of DRP1 to mitochondrial membranes.
B. SEC5/EXOcyst Complex 84
SEC5/EXO84 are critical components of the exocyst complex, which plays an essential role in exosome generation and cell migration. The exocyst complex is a multiprotein complex which is responsible for the appropriate trafficking of various secretory vesicles to specific domains of plasma membrane (He and Guo, 2009). RAL proteins regulate exocyst-dependent vesicle trafficking and are required for exocyst complex assembly (Camonis and White, 2005). SEC5 was first identified as a direct target of RAL and was found to mediate the effects of RALA on both the basolateral delivery of membrane proteins in epithelial cells and the secretion of vesicle contents in neuronal cells (Moskalenko et al., 2002). EXO84 was then identified as a second subunit of the exocyst complex that interacts with RAL and is essential for RAL-mediated assembly of the full octameric exocyst complex (Moskalenko et al., 2003). SEC5 binds to the switch I region (Fukai et al., 2003) while EXO84 binds to both switch I and II regions (Jin et al., 2005) of activated RAL.
C. Transcription and Other Factors
RAL signaling activates transcriptional activity of nuclear factor-κB through SEC5 and TBK1 (Chien et al., 2006). Active RALB causes the association of SEC5 with TBK1, which then directly phosphorylate and promote the nuclear localization and activation of nuclear factor-κB. Active RALA activates the ZONAB transcription factor to regulate the cell density of MDCK cells (Frankel et al., 2005). Other less studied examples include the activation of c-JUN through JNK (de Ruiter et al., 2000), ATF2 and STAT3 through SRC tyrosine kinase (Goi et al., 2000), and FOXO4 (De Ruiter et al., 2001) and NFAT (de Gorter et al., 2007). Filamin, an actin filament crosslinking protein interacts with RALA and is required for RALA-induced filopodia formation (Ohta et al., 1999). RAL proteins directly associate with phospholipase C delta 1 and phospholipase D1, important second messenger proteins of the G-protein coupled receptors (Luo et al., 1997; Sidhu et al., 2005).
V. RAL in Human Cancer
The role of RALGEF-RAL signaling in RAS-driven tumorigenesis was initially overlooked, because early studies in mouse fibroblast cells found the RAF effector pathway, but not PI3K or RALGEF-RAL pathway, to be sufficient to mediate RAS-driven tumor transformation (Urano et al., 1996). The Counter group (Hamad et al., 2002) was the first to report that the RALGEF-RAL pathway, but not RAF or PI3K, was sufficient for RAS transformation of immortalized human cells. In the following study, they further demonstrated that RALA but not RALB is the RALGEF effector protein that plays a critical role for RAS-mediated tumor transformation (Lim et al., 2005). The observation that RALA and RALB might play distinctive roles in tumorigenesis was also supported by a study from the White group (Chien and White, 2003), showing that RALA is required for anchorage-independent growth, whereas RALB is required for survival of human tumor cells. Subsequent studies in the past decade have established the critical role of RAL in both tumorigenesis and growth/metastasis of many types of human cancer.
A. Pancreatic Cancer
Pancreatic cancer is the most popular model for studying the role of RAL in human cancer because of its extremely high frequency of KRAS mutations (>90%, COSMIC database) and documented dependency of this tumor type on RAS. To determine which RAS effector signaling pathway mediates the oncogenic signaling, the Counter group examined the activation status of the three canonical effector pathways in panels of human pancreatic cancer cell lines (Lim et al., 2005) and tumor samples (Lim et al., 2006). Both studies showed consistent elevated levels of activated RALA and RALB across the cell lines or tumor samples. Importantly, both RALA and RALB were found to be more commonly activated compared with the other two RAS effector pathways PI3K and RAF, suggesting the critical importance of RAL signaling in KRAS driven pancreatic cancer. This central role of RALGEF-RAL signaling in pancreatic cancer was also confirmed by a more recent study showing that the RALGEF protein RGL2 is overexpressed in patient tumors and is necessary for both the anchorage-independent growth and invasion of tumor cells (Vigil et al., 2010). RALA and RALB were found to play distinct roles in pancreatic cancer. Stable shRNA suppression of RALA but not RALB in a panel of 10 genetically diverse human pancreatic cancer cell lines universally reduced their anchorage-independent growth in vitro and subcutaneous growth in vivo. In contrast, RALB but not RALA knockdown impaired in vitro invasion of these cell lines and metastatic colonization in vivo following tail-vein inoculation (Lim et al., 2006). The distinct roles of RAL in RAS-driven pancreatic cancers was confirmed by another study which showed RALB but not RALA plays a role in invadopodium formation in human pancreatic cancer cells, supporting a role for RALB in metastasis (Neel et al., 2012).
B. Colorectal Cancer
KRAS is the most frequently mutated (45%, COSMIC) oncogene in colorectal cancer. As in pancreatic cancer, upregulated RALA and RALB activation were found in both colorectal cancer cell lines and patient samples (Martin et al., 2011). RALA-responsive gene expression signature generated from three colorectal cancer cell lines was also found to correlate with progression-free survival of colorectal cancer patients in five independent data sets (Győrffy et al., 2015). However, RALA and RALB play antagonistic roles in colon cancer. Stable RNAi suppression of RALA reduced anchorage-independent growth of cancer cells, whereas stable suppression of RALB enhanced such growth. This discrepancy in isoform function was partially attributed to their differential usage of effector proteins. Both RALA and RALB required interaction with RALBP1 but with different components of the exocyst complex. RALA interacts with EXO84, whereas RALB interacts with SEC5 to mediate their effect on anchorage-independent growth in colorectal cancer cells (Martin et al., 2011). This antagonistic role of RALA and RALB in colorectal cancer demonstrated cancer cell type differences in RAL function and therefore calls for selective targeting of each RAL isoform in colorectal cancer.
C. Lung Cancer
In contrast to colorectal cancer, RALA and RALB were shown to have redundant effects on the growth of NSCLC tumor cells. The requirement of RAL in lung cancer tumorigenesis was also established in a genetic knockout mouse model (Peschard et al., 2012). Deletion of both RALA and RALB genes but not either one alone blocked KRAS-driven lung tumor development, suggesting that RALA and RALB have redundant functions in tumorigenesis in mouse model of lung cancer. By using the human NSCLC cell line A549, the Farassati group(Male et al., 2012) showed that transient knockdown of RALA reduced the proliferation and invasiveness of A549 cells in vitro and tumorigenesis in vivo. More recently, we analyzed RAL expression in a panel of 14 human NSCLC cell lines and found that, although both RALA and RALB had higher expression in KRAS mutant cell lines, only RALA activity was elevated (Guin et al., 2013), suggesting RALA drives tumor growth in NSCLC cell lines. A relationship between RAL dependence and RAS mutation type was established for NSCLC with cell lines carrying a G12C or G12V KRAS mutation found to be more sensitive to RAL depletion than those with other KRAS mutations or wild-type KRAS. KRAS mutations are found in about 30% of NSCLC, and G12C and G12V are the most common types of RAS mutations. This supported the importance of RAL signaling in driving tumor growth downstream of RAS in NSCLC. Immunohistochemical and expression analyses of 337 NSCLC patient tumors demonstrated high expression of both RALA and RALB proteins was associated with poor survival, supporting the simultaneously targeting of both RAL paralogs in NSCLC. In fact, BQU57 (6-amino-1,3-dimethyl-4-(4-(trifluoromethyl)phenyl)-1,4-dihydropyrano[2,3-c]pyrazole-5-carbonitrile), the first developed RAL inhibitor inhibited growth of NSCLC cell lines both in vitro and in vivo by inhibiting RALBP1 binding to both RAL paralogs (Yan et al., 2014).
D. Bladder Cancer
Overexpression and increased activation of RALA and RALB was detected in human bladder cancer cell lines with diverse genetic background and RAS mutation status (Smith et al., 2007; Saito et al., 2013). In the KRAS mutant UMUC3 human bladder cancer cell line, RALA and RALB have similar functions in tumor growth but opposite roles in motility (Oxford et al., 2005). Transient RNAi suppression of both RALA and RALB was required to reduce anchorage-independent growth, whereas RALA inhibits and RALB promotes the motility of UMUC3 cells. Importantly, this metastasis-promoting function of RALB was shown to be dependent on its phosphorylation by protein kinase C in UMUC3 cells (Wang et al., 2010). The importance of RAL signaling in patient bladder tumor samples was also revealed by us using a RAL transcriptional signature that was found to correlate with disease stage, progression to muscle invasion, and survival (Smith et al., 2007, 2012). In addition, overexpression of both the regulators and effectors of RAL was found in human bladder tumor samples. Some examples include RAL activators like RGL2 and Aurora A kinase, as well as RAL effectors like RALBP1 (Smith et al., 2007). Because RAS mutation is very infrequent in bladder cancer (∼10%), RAS-independent mechanisms have been proposed that mediate RAL activation in bladder cancer. Supporting this notion, work using both human bladder cancer cell lines and a genetic mouse model showed RAS-independent activation of RAL might be due to the reduced expression of RALGAP in RAS wild-type bladder cancer (Saito et al., 2013).
E. Prostate Cancer
The contribution of RAL signaling to prostate cancer growth and progression has been implied in both androgen independence and metastasis (Yin et al., 2007). One study showed that RALA was activated upon androgen deprivation in human prostate cancer cells, which in turn leads to the transcriptional upregulation of vascular endothelial growth factor C (Rinaldo et al., 2007). On the other hand, RALA activation also leads to the interaction of RALBP1 with REPS2, which stimulates the endocytosis of the epidermal growth factor receptor (which has been shown to play a role in androgen independence (Oosterhoff et al., 2003). The RALGEF-RAL pathway was also shown to be both necessary and sufficient for the bone metastasis of prostate cancer. Activation of the RALGEF pathway enhanced bone metastasis of DU145, whereas depletion of either RALA or RALBP1 inhibited bone metastasis of the metastatic PC3 cell line (Yin et al., 2007; Wu et al., 2010). In human prostate tumor samples, RAL transcriptional signature was associated with seminal vesicle invasion, androgen-independent progression, and poor prognostics, supporting the central role of RAL in prostate cancer progression and metastasis (Smith et al., 2012).
F. Melanoma
Approximately one-third of melanomas harbor activating NRAS mutations and about 60% harbor BRAF mutations (COSMIC database). A role for RALGEF-RAL signaling in NRAS-driven melanoma tumorigenesis was first established in an immortalized mouse melanocyte model, where the transforming activities of the three NRAS downstream effectors, RAF, PI3K, and RALGEF were evaluated (Mishra et al., 2010). RALGEF signaling was found to have a major contribution to anchorage-independent growth in this model. RALA but not RALB activation was observed in a panel of human melanoma cancer cell lines regardless of their NRAS or BRAF mutational status (Zipfel et al., 2010). Stable knockdown of RALA, and to a lesser extent of RALB, inhibited the tumorigenic growth of melanoma cell lines both in vitro and in vivo (Zipfel et al., 2010).
G. Other Tumor Types
Elevated expression and activation of RAL is observed in various types of human cancer. In most models, RALA is the dominant isoform driving tumor progression and metastasis, including in hepatocellular carcinoma (Ezzeldin et al., 2014), malignant peripheral nerve sheath tumors (Bodempudi et al., 2009), and ovarian cancer (Wang et al., 2013). One exception, however, is squamous cell carcinoma (SCC), where RALA was found to suppress rather than promote tumor progression in SCC cancer cell lines (Sowalsky et al., 2010). This tumor suppressor function of RALA was validated by microarray analysis of SCC human tumor samples in which RALA gene expression signature was decreased in cancer tissues compared with normal mucosa (Smith et al., 2012).
In conclusion, in almost all cancer types examined, increased expression and activation of both RALA and RALB are observed in patient tumor samples compared with normal tissues, regardless of their RAS mutation status. Studies with genetic approaches in human cancer cell lines have established that, in general, RALA is important for anchorage-independent growth, whereas RALB is important for metastasis and invasion. However, in certain caner types such as lung cancer and melanoma, both isoforms seem to have overlapping effects. Therefore, targeting both RALA and RALB simultaneously seems the most appropriate anticancer approach.
VI. Therapeutic Targeting of RAL Signaling
A. Targeting RAL Guanine Exchange Factors
RALGEFs mediate RAS-dependent activation of RAL. Targeting RALGEFs should be as potent as inhibiting RAL itself if the predominant signal activating RAL is RAS mediated. Targeting RALGEFs or other GEFs of the small GTPase family is challenging, because targeting GEF means targeting protein-protein interactions. Nevertheless, inhibitors have been developed that specifically target GEF proteins of the RAS family small GTPases, including ARF, RHO, and RAC1 (Schmidt et al., 2002; Renault et al., 2003; Gao et al., 2004). For RALGEFs, limited structural information is available to understand their binding to RAS or RAL, thus structural studies will be needed before they can be exploited as feasible therapeutic targets of RAL signaling.
B. Targeting RAL Directly
As for RAS, several hurdles hamper the development of small molecule compounds that directly bind to and inhibit RAL activity: 1) the high affinity of RAL small GTPases for guanidine nucleotides (picomolar) and the high concentration of GTP/GDP in cells (millimolar) make competing with GTP/GDP binding at the active site of RAL extremely difficult (Cox et al., 2014), 2) the downstream signaling cascade of RAL is relayed through protein-protein interactions, which are difficult to target compared with the enzymatic activity of protein kinases (Arkin et al., 2014), and 3) the surface of activated RAL-GTP protein is very “smooth,” with no deep hydrophobic pockets suitable for tight binding of small molecules. Fortunately, unlike RAS, whose oncogenic activity is most commonly due to mutations that keep RAS in constitutively active state, RAL mutations in human cancer are extremely rare, allowing for more targeting options (Ostrem et al., 2013). In addition, recent advances in structure-based drug discovery have shed light on both RAS and RAL, allowing inhibitor development.
Structure-based drug discovery relies on the knowledge of the three-dimensional structure of the molecular target, usually determined by either X-ray crystallography or NMR, both of which have their intrinsic limitations for structure-based drug design (Chen et al., 2012). For example, X-ray structure of small GTPases only traps the protein in one low-energy confirmation, which might not be physiologically relevant, whereas NMR structures are usually too dynamic for computational modeling. Moreover, the original idea that RAS proteins exist in rigid inactive GDP-bound or active GTP-bound form has been altered when NMR studies of HRAS demonstrated existence of at least two conformation states for the GTP-bound form (Shima et al., 2010; Shima et al., 2013). This new consensus on the dynamic nature of conformational status of GTP-bound RAS and related GTPase, has resulted in several breakthroughs in targeting the RAS family small GTPases. In 2012, two small molecule compounds, DACI and VU0460009, were discovered as KRAS inhibitors using fragment based screening, both of which were shown to bind to the same pocket in the RAS-SOS1 binding surface resulting in the inhibition of RAS activation by the RASGEF SOS1 (Maurer et al., 2012; Sun et al., 2012). Importantly, this binding pocket did not exist on the apoprotein and was in fact formed by compound binding induced conformation changes, suggesting that the structure of RAS protein is dynamic. Fragment-based drug design is based on identifying small chemical fragments that can bind weakly to small pockets on the target, and then growing them or linking them together to produce a lead with a higher affinity (Zhao et al., 2017). This approach is particularly promising for structure-based drug design against RAS family small GTPase including RAL.
Another breakthrough in targeting small GTPases via structure-based drug design is the “stapled peptide” approach. The mimicking of α-helices in protein binding domains has been an emerging area in structure based inhibitor design (Walensky and Bird, 2014). Introduction of synthetic hydrocarbon braces (staples) helps to lock a small peptide in a specific conformation, thereby increasing stability, target affinity, and cell permeability of the peptide. Recently, this technique has been applied to target RAS (Patgiri et al., 2011; Leshchiner et al., 2015). Stapled peptides based on a helix from the RASGEF son of sevenless (SOS) has been used to target RAS. By using the RAL binding domain of RALBP1 as a template, the Mott group designed an α-helical stapled peptide that can bind selectively to active RALB-GTP and block downstream effector signaling of RAL (Thomas et al., 2016). These peptides bind to the switch I and II regions of RAL, which are the sites of interaction with RAL effectors like RALBP1 and regulators like RALGEF. However, because the peptides do not bind to inactive RAL-GDP, they could not inhibit RALGEF binding to RAL and hence could not inhibit RAL activation by RALGEF.
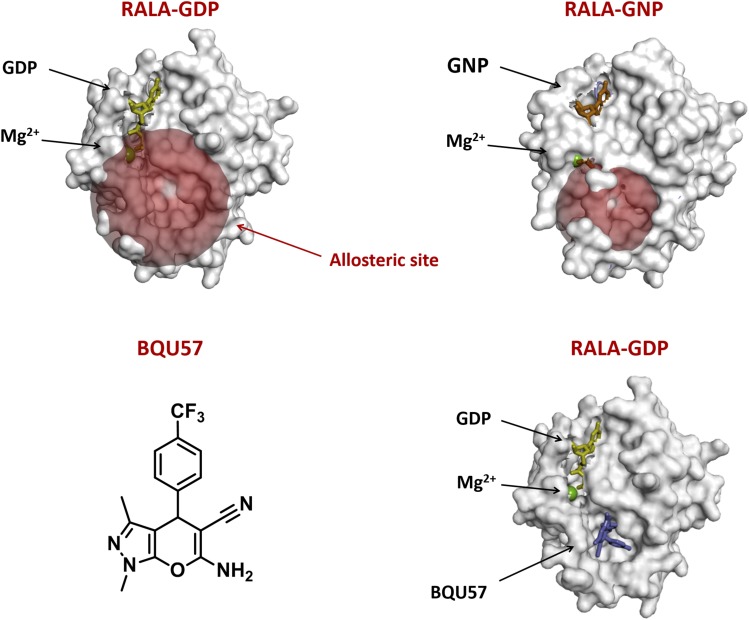
A third breakthrough in the targeting of small GTPases using structure-based approach is the development of allosteric inhibitors of the inactive GDP-bound form of RAL. Our group identified an allosteric binding site on the surface of RAL-GDP and searched for small molecule ligands that can bind to the allosteric site using a virtual chemical library screening (Yan et al., 2014) (Fig. 3). This generated drug candidates that bound to the allosteric site of RAL-GDP and lock RAL in the GDP-bound inactive state, thereby break up the GDP-GTP cycle and block RAL-mediated signaling. Importantly, these compounds demonstrated biologic activities in human NSCLC both in vitro and in vivo.
Fig. 3.
Targeting the allosteric site of RAL. Structure of inactive (A) and active (B) RALA, red sphere represents the allosteric site that is only exposed in the GDP-bound form. GNP: nonhydrolyzable form of GTP used in X-ray crystallography. (C) Chemical structure of the first small molecule inhibitor of RAL, BQU57. (D) BQU57 binds to the allosteric site of RALA-GDP.
C. Targeting RAL Effectors
Another logical approach to interfere with RALGEF-RAL signaling is to inhibit downstream signaling of activated RAL. RAL proteins activate downstream signaling through direct binding with effector proteins. Although specific inhibitors for most RAL effectors do not exist, targeting the protein-protein interaction between RAL-GTP and effector proteins seems to be a reasonable approach that may lead to the discovery of novel inhibitors of RAL signaling.
D. Targeting RAL Localization
Similar to RAS, appropriate localization of RAL proteins to the inner surface of the plasma membrane is required for their biologic functions. As discussed earlier, all RAS family small GTPases have the CAAX C-terminal sequence motif that is subjected to prenylation by either farnesyltransferase (FTase) or geranylgeranyltransferase I (GGTase I) depending on the exact amino acid sequence of AAX. The initial discovery that RAS proteins undergo farnesylation by FTase prompted an intensive effort to develop farnesyltransferase inhibitors with two of them, lonafarnib and tipifarnib, progressing into phase III clinical trials (Berndt et al., 2011). However, both failed to show efficacy in human cancer. One basis for this failure was the alternative prenylation of RAS by GGTase I upon inhibition of FTase. Unlike RAS, which is primarily prenylated by FTase, RAL proteins are substrates for GGTase I (Kinsella et al., 1991) and do not undergo alternative prenylation in response to GGTI treatment (Falsetti et al., 2007). The effect of GGTase I inhibition on RAL signaling has been evaluated in the human Mia PaCa-2 pancreatic cancer cell line (Falsetti et al., 2007). RALA and RALB were shown to be involved in both mediating the inhibition of anchorage-independent growth and cell cycle progression in these cells. The importance of RALB in mediating the growth inhibition effect of GGTIs was also confirmed in other cancer types like glioma and squamous cell carcinoma (Hamada et al., 2011; Song et al., 2015). One potential problem with targeting GGTase is that many other RAS family members are also substrates for GGTase I, targeting of which might result in normal tissue toxicity. However, a recent phase I clinical trial with GGTI-2418 seemed to be well tolerated (Ullah et al., 2016). Therefore, GGTIs may still be a feasible approach for targeting RAL.
E. Combination Therapy
For a long time, combination therapy at various signaling knots of the RAS pathway has been confined to the RAF-MEK-ERK and PI3K-AKT-mTOR pathways, largely because of the lack of effective inhibitors of other components of RAS signaling. Recent breakthroughs in direct targeting of RAL have provided the conceptual possibility to block all three primary effector pathways downstream of RAS, namely the RAF-MEK-ERK kinase pathway, the PI3K-AKT-mTOR kinase pathway, and the RALGEF-RAL pathway. Potent synergistic activity between PI3K and RAF inhibitors has already been seen in human cell lines and mouse models (Cox et al., 2014). Concurrent inhibition of all three pathways should result in even more potent inhibition of RAS oncogenic signaling. Clinical useful inhibitors have been developed for almost every component of RAF-MEK-ERK and PI3K-AKT-mTOR signaling; for RALGEF-RAL pathway this is only the beginning.
VII. Summary
As the third primary signaling axis that mediates RAS oncogenic signaling, the RALGEF-RAL pathway has been extensively studied in human cancer and shown to play critical roles in the tumorigenesis, growth, progression, and metastasis of various types of human cancer. Although much remains to be understood about the detailed mechanism of RAL signaling and the distinct roles of RALA and RALB in human cancer, recent breakthroughs in targeting RAL and RAS have paved the way for the development of more potent and specific RAL-targeted therapies.
Abbreviations
- ERK
extracellular signal-regulated kinase
- EXO84
EXOcyst complex 84
- FTase
farnesyltransferase
- GGTase I
geranylgeranyltransferase I
- MEK
mitogen-activated protein kinase/ERK kinase
- mTOR
mechanistic target of rapamycin
- PI3K
phosphoinositide-3-kinase
- RAL
RAS like
- RALBP1
RAL binding protein 1
- RALGAP
RAL GTPase activating protein
- RALGEF
RAL gunaine exchange factor
- SCC
squamous cell carcinoma
- TBK1
TANK-binding kinase 1
Authorship Contributions
Wrote or contributed to the writing of the manuscript: Yan, Theodorescu.
Footnotes
This work was supported in part by National Institutes of Health National Cancer Institute [Grant CA075115].
References
- Albright CF, Giddings BW, Liu J, Vito M, Weinberg RA. (1993) Characterization of a guanine nucleotide dissociation stimulator for a ras-related GTPase. EMBO J 12:339–347. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Arkin MR, Tang Y, Wells JA. (2014) Small-molecule inhibitors of protein-protein interactions: progressing toward the reality. Chem Biol 21:1102–1114. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Awasthi S, Cheng J, Singhal SS, Saini MK, Pandya U, Pikula S, Bandorowicz-Pikula J, Singh SV, Zimniak P, Awasthi YC. (2000) Novel function of human RLIP76: ATP-dependent transport of glutathione conjugates and doxorubicin. Biochemistry 39:9327–9334. [DOI] [PubMed] [Google Scholar]
- Balakireva M, Rossé C, Langevin J, Chien YC, Gho M, Gonzy-Treboul G, Voegeling-Lemaire S, Aresta S, Lepesant JA, Bellaiche Y, et al. (2006) The Ral/exocyst effector complex counters c-Jun N-terminal kinase-dependent apoptosis in Drosophila melanogaster. Mol Cell Biol 26:8953–8963. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Berndt N, Hamilton AD, Sebti SM. (2011) Targeting protein prenylation for cancer therapy. Nat Rev Cancer 11:775–791. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bodempudi V, Yamoutpoor F, Pan W, Dudek AZ, Esfandyari T, Piedra M, Babovick-Vuksanovic D, Woo RA, Mautner VF, Kluwe L, et al. (2009) Ral overactivation in malignant peripheral nerve sheath tumors. Mol Cell Biol 29:3964–3974. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Camonis JH, White MA. (2005) Ral GTPases: corrupting the exocyst in cancer cells. Trends Cell Biol 15:327–332. [DOI] [PubMed] [Google Scholar]
- Cantor SB, Urano T, Feig LA. (1995) Identification and characterization of Ral-binding protein 1, a potential downstream target of Ral GTPases. Mol Cell Biol 15:4578–4584. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ceriani M, Scandiuzzi C, Amigoni L, Tisi R, Berruti G, Martegani E. (2007) Functional analysis of RalGPS2, a murine guanine nucleotide exchange factor for RalA GTPase. Exp Cell Res 313:2293–2307. [DOI] [PubMed] [Google Scholar]
- Chardin P, Tavitian A. (1986) The ral gene: a new ras related gene isolated by the use of a synthetic probe. EMBO J 5:2203–2208. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chardin P, Tavitian A. (1989) Coding sequences of human ralA and ralB cDNAs. Nucleic Acids Res 17:4380. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chen L, Morrow JK, Tran HT, Phatak SS, Du-Cuny L, Zhang S. (2012) From laptop to benchtop to bedside: structure-based drug design on protein targets. Curr Pharm Des 18:1217–1239. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chen Q, Quan C, Xie B, Chen L, Zhou S, Toth R, Campbell DG, Lu S, Shirakawa R, Horiuchi H, et al. (2014) GARNL1, a major RalGAP α subunit in skeletal muscle, regulates insulin-stimulated RalA activation and GLUT4 trafficking via interaction with 14-3-3 proteins. Cell Signal 26:1636–1648. [DOI] [PubMed] [Google Scholar]
- Chen XW, Leto D, Xiong T, Yu G, Cheng A, Decker S, Saltiel AR. (2011) A Ral GAP complex links PI 3-kinase/Akt signaling to RalA activation in insulin action. Mol Biol Cell 22:141–152. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chien Y, Kim S, Bumeister R, Loo YM, Kwon SW, Johnson CL, Balakireva MG, Romeo Y, Kopelovich L, Gale M, Jr, et al. (2006) RalB GTPase-mediated activation of the IkappaB family kinase TBK1 couples innate immune signaling to tumor cell survival. Cell 127:157–170. [DOI] [PubMed] [Google Scholar]
- Chien Y, White MA. (2003) RAL GTPases are linchpin modulators of human tumour-cell proliferation and survival. EMBO Rep 4:800–806. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Cox AD, Der CJ. (2010) Ras history: the saga continues. Small GTPases 1:2–27. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Cox AD, Der CJ, Philips MR. (2015) Targeting RAS Membrane association: back to the future for anti-RAS drug discovery? Clin Cancer Res 21:1819–1827. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Cox AD, Fesik SW, Kimmelman AC, Luo J, Der CJ. (2014) Drugging the undruggable RAS: mission possible? Nat Rev Drug Discov 13:828–851. [DOI] [PMC free article] [PubMed] [Google Scholar]
- D’Adamo DR, Novick S, Kahn JM, Leonardi P, Pellicer A. (1997) rsc: a novel oncogene with structural and functional homology with the gene family of exchange factors for Ral. Oncogene 14:1295–1305. [DOI] [PubMed] [Google Scholar]
- de Bruyn KM, de Rooij J, Wolthuis RM, Rehmann H, Wesenbeek J, Cool RH, Wittinghofer AH, Bos JL. (2000) RalGEF2, a pleckstrin homology domain containing guanine nucleotide exchange factor for Ral. J Biol Chem 275:29761–29766. [DOI] [PubMed] [Google Scholar]
- de Gorter DJ, Vos JC, Pals ST, Spaargaren M. (2007) The B cell antigen receptor controls AP-1 and NFAT activity through Ras-mediated activation of Ral. J Immunol 178:1405–1414. [DOI] [PubMed] [Google Scholar]
- De Ruiter ND, Burgering BM, Bos JL. (2001) Regulation of the Forkhead transcription factor AFX by Ral-dependent phosphorylation of threonines 447 and 451. Mol Cell Biol 21:8225–8235. [DOI] [PMC free article] [PubMed] [Google Scholar]
- de Ruiter ND, Wolthuis RM, van Dam H, Burgering BM, Bos JL. (2000) Ras-dependent regulation of c-Jun phosphorylation is mediated by the Ral guanine nucleotide exchange factor-Ral pathway. Mol Cell Biol 20:8480–8488. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ezzeldin M, Borrego-Diaz E, Taha M, Esfandyari T, Wise AL, Peng W, Rouyanian A, Asvadi Kermani A, Soleimani M, Patrad E, et al. (2014) RalA signaling pathway as a therapeutic target in hepatocellular carcinoma (HCC). Mol Oncol 8:1043–1053. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Falsetti SC, Wang DA, Peng H, Carrico D, Cox AD, Der CJ, Hamilton AD, Sebti SM. (2007) Geranylgeranyltransferase I inhibitors target RalB to inhibit anchorage-dependent growth and induce apoptosis and RalA to inhibit anchorage-independent growth. Mol Cell Biol 27:8003–8014. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Fenwick RB, Campbell LJ, Rajasekar K, Prasannan S, Nietlispach D, Camonis J, Owen D, Mott HR. (2010) The RalB-RLIP76 complex reveals a novel mode of ral-effector interaction. Structure 18:985–995. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Fenwick RB, Prasannan S, Campbell LJ, Nietlispach D, Evetts KA, Camonis J, Mott HR, Owen D. (2009) Solution structure and dynamics of the small GTPase RalB in its active conformation: significance for effector protein binding. Biochemistry 48:2192–2206. [DOI] [PubMed] [Google Scholar]
- Ferro E, Trabalzini L. (2010) RalGDS family members couple Ras to Ral signalling and that’s not all. Cell Signal 22:1804–1810. [DOI] [PubMed] [Google Scholar]
- Frankel P, Aronheim A, Kavanagh E, Balda MS, Matter K, Bunney TD, Marshall CJ. (2005) RalA interacts with ZONAB in a cell density-dependent manner and regulates its transcriptional activity. EMBO J 24:54–62. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Fukai S, Matern HT, Jagath JR, Scheller RH, Brunger AT. (2003) Structural basis of the interaction between RalA and Sec5, a subunit of the sec6/8 complex. EMBO J 22:3267–3278. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gao Y, Dickerson JB, Guo F, Zheng J, Zheng Y. (2004) Rational design and characterization of a Rac GTPase-specific small molecule inhibitor. Proc Natl Acad Sci USA 101:7618–7623. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gentry LR, Martin TD, Reiner DJ, Der CJ. (2014) Ral small GTPase signaling and oncogenesis: more than just 15minutes of fame. Biochim Biophys Acta 1843:2976–2988. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gentry LR, Nishimura A, Cox AD, Martin TD, Tsygankov D, Nishida M, Elston TC, Der CJ. (2015) Divergent roles of CAAX motif-signaled posttranslational modifications in the regulation and subcellular localization of Ral GTPases. J Biol Chem 290:22851–22861. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Goi T, Shipitsin M, Lu Z, Foster DA, Klinz SG, Feig LA. (2000) An EGF receptor/Ral-GTPase signaling cascade regulates c-Src activity and substrate specificity. EMBO J 19:623–630. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Goitre L, Trapani E, Trabalzini L, Retta SF. (2014) The Ras superfamily of small GTPases: the unlocked secrets. Methods Mol Biol 1120:1–18. [DOI] [PubMed] [Google Scholar]
- Guin S, Ru Y, Wynes MW, Mishra R, Lu X, Owens C, Barn AE, Vasu VT, Hirsch FR, Kern JA, et al. (2013) Contributions of KRAS and RAL in non-small-cell lung cancer growth and progression. J Thorac Oncol 8:1492–1501. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Győrffy B, Stelniec-Klotz I, Sigler C, Kasack K, Redmer T, Qian Y, Schäfer R. (2015) Effects of RAL signal transduction in KRAS- and BRAF-mutated cells and prognostic potential of the RAL signature in colorectal cancer. Oncotarget 6:13334–13346. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hamad NM, Elconin JH, Karnoub AE, Bai W, Rich JN, Abraham RT, Der CJ, Counter CM. (2002) Distinct requirements for Ras oncogenesis in human versus mouse cells. Genes Dev 16:2045–2057. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hamada M, Miki T, Iwai S, Shimizu H, Yura Y. (2011) Involvement of RhoA and RalB in geranylgeranyltransferase I inhibitor-mediated inhibition of proliferation and migration of human oral squamous cell carcinoma cells. Cancer Chemother Pharmacol 68:559–569. [DOI] [PubMed] [Google Scholar]
- He B, Guo W. (2009) The exocyst complex in polarized exocytosis. Curr Opin Cell Biol 21:537–542. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Isomura M, Okui K, Fujiwara T, Shin S, Nakamura Y. (1996) Isolation and mapping of RAB2L, a human cDNA that encodes a protein homologous to RalGDS. Cytogenet Cell Genet 74:263–265. [DOI] [PubMed] [Google Scholar]
- Jin R, Junutula JR, Matern HT, Ervin KE, Scheller RH, Brunger AT. (2005) Exo84 and Sec5 are competitive regulatory Sec6/8 effectors to the RalA GTPase. EMBO J 24:2064–2074. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Jullien-Flores V, Dorseuil O, Romero F, Letourneur F, Saragosti S, Berger R, Tavitian A, Gacon G, Camonis JH. (1995) Bridging Ral GTPase to Rho pathways. RLIP76, a Ral effector with CDC42/Rac GTPase-activating protein activity. J Biol Chem 270:22473–22477. [DOI] [PubMed] [Google Scholar]
- Jullien-Flores V, Mahé Y, Mirey G, Leprince C, Meunier-Bisceuil B, Sorkin A, Camonis JH. (2000) RLIP76, an effector of the GTPase Ral, interacts with the AP2 complex: involvement of the Ral pathway in receptor endocytosis. J Cell Sci 113:2837–2844. [DOI] [PubMed] [Google Scholar]
- Kashatus DF, Lim KH, Brady DC, Pershing NL, Cox AD, Counter CM. (2011) RALA and RALBP1 regulate mitochondrial fission at mitosis. Nat Cell Biol 13:1108–1115. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kikuchi A, Demo SD, Ye ZH, Chen YW, Williams LT. (1994) ralGDS family members interact with the effector loop of ras p21. Mol Cell Biol 14:7483–7491. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kinsella BT, Erdman RA, Maltese WA. (1991) Carboxyl-terminal isoprenylation of ras-related GTP-binding proteins encoded by rac1, rac2, and ralA. J Biol Chem 266:9786–9794. [PubMed] [Google Scholar]
- Leshchiner ES, Parkhitko A, Bird GH, Luccarelli J, Bellairs JA, Escudero S, Opoku-Nsiah K, Godes M, Perrimon N, Walensky LD. (2015) Direct inhibition of oncogenic KRAS by hydrocarbon-stapled SOS1 helices. Proc Natl Acad Sci USA 112:1761–1766. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lim KH, Baines AT, Fiordalisi JJ, Shipitsin M, Feig LA, Cox AD, Der CJ, Counter CM. (2005) Activation of RalA is critical for Ras-induced tumorigenesis of human cells. Cancer Cell 7:533–545. [DOI] [PubMed] [Google Scholar]
- Lim KH, Brady DC, Kashatus DF, Ancrile BB, Der CJ, Cox AD, Counter CM. (2010) Aurora-A phosphorylates, activates, and relocalizes the small GTPase RalA. Mol Cell Biol 30:508–523. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lim KH, O’Hayer K, Adam SJ, Kendall SD, Campbell PM, Der CJ, Counter CM. (2006) Divergent roles for RalA and RalB in malignant growth of human pancreatic carcinoma cells. Curr Biol 16:2385–2394. [DOI] [PubMed] [Google Scholar]
- Luo JQ, Liu X, Hammond SM, Colley WC, Feig LA, Frohman MA, Morris AJ, Foster DA. (1997) RalA interacts directly with the Arf-responsive, PIP2-dependent phospholipase D1. Biochem Biophys Res Commun 235:854–859. [DOI] [PubMed] [Google Scholar]
- Male H, Patel V, Jacob MA, Borrego-Diaz E, Wang K, Young DA, Wise AL, Huang C, Van Veldhuizen P, O’Brien-Ladner A, et al. (2012) Inhibition of RalA signaling pathway in treatment of non-small cell lung cancer. Lung Cancer 77:252–259. [DOI] [PubMed] [Google Scholar]
- Martin TD, Chen XW, Kaplan RE, Saltiel AR, Walker CL, Reiner DJ, Der CJ. (2014) Ral and Rheb GTPase activating proteins integrate mTOR and GTPase signaling in aging, autophagy, and tumor cell invasion. Mol Cell 53:209–220. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Martin TD, Mitin N, Cox AD, Yeh JJ, Der CJ. (2012) Phosphorylation by protein kinase Cα regulates RalB small GTPase protein activation, subcellular localization, and effector utilization. J Biol Chem 287:14827–14836. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Martin TD, Samuel JC, Routh ED, Der CJ, Yeh JJ. (2011) Activation and involvement of Ral GTPases in colorectal cancer. Cancer Res 71:206–215. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Maurer T, Garrenton LS, Oh A, Pitts K, Anderson DJ, Skelton NJ, Fauber BP, Pan B, Malek S, Stokoe D, et al. (2012) Small-molecule ligands bind to a distinct pocket in Ras and inhibit SOS-mediated nucleotide exchange activity. Proc Natl Acad Sci USA 109:5299–5304. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mirey G, Balakireva M, L’Hoste S, Rossé C, Voegeling S, Camonis J. (2003) A Ral guanine exchange factor-Ral pathway is conserved in Drosophila melanogaster and sheds new light on the connectivity of the Ral, Ras, and Rap pathways. Mol Cell Biol 23:1112–1124. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mishra PJ, Ha L, Rieker J, Sviderskaya EV, Bennett DC, Oberst MD, Kelly K, Merlino G. (2010) Dissection of RAS downstream pathways in melanomagenesis: a role for Ral in transformation. Oncogene 29:2449–2456. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Moskalenko S, Henry DO, Rosse C, Mirey G, Camonis JH, White MA. (2002) The exocyst is a Ral effector complex. Nat Cell Biol 4:66–72. [DOI] [PubMed] [Google Scholar]
- Moskalenko S, Tong C, Rosse C, Mirey G, Formstecher E, Daviet L, Camonis J, White MA. (2003) Ral GTPases regulate exocyst assembly through dual subunit interactions. J Biol Chem 278:51743–51748. [DOI] [PubMed] [Google Scholar]
- Nakashima S, Morinaka K, Koyama S, Ikeda M, Kishida M, Okawa K, Iwamatsu A, Kishida S, Kikuchi A. (1999) Small G protein Ral and its downstream molecules regulate endocytosis of EGF and insulin receptors. EMBO J 18:3629–3642. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Neel NF, Rossman KL, Martin TD, Hayes TK, Yeh JJ, Der CJ. (2012) The RalB small GTPase mediates formation of invadopodia through a GTPase-activating protein-independent function of the RalBP1/RLIP76 effector. Mol Cell Biol 32:1374–1386. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Nicely NI, Kosak J, de Serrano V, Mattos C. (2004) Crystal structures of Ral-GppNHp and Ral-GDP reveal two binding sites that are also present in Ras and Rap. Structure 12:2025–2036. [DOI] [PubMed] [Google Scholar]
- Ohta Y, Suzuki N, Nakamura S, Hartwig JH, Stossel TP. (1999) The small GTPase RalA targets filamin to induce filopodia. Proc Natl Acad Sci USA 96:2122–2128. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Oosterhoff JK, Penninkhof F, Brinkmann AO, Anton Grootegoed J, Blok LJ. (2003) REPS2/POB1 is downregulated during human prostate cancer progression and inhibits growth factor signalling in prostate cancer cells. Oncogene 22:2920–2925. [DOI] [PubMed] [Google Scholar]
- Ostrem JM, Peters U, Sos ML, Wells JA, Shokat KM. (2013) K-Ras(G12C) inhibitors allosterically control GTP affinity and effector interactions. Nature 503:548–551. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Oxford G, Owens CR, Titus BJ, Foreman TL, Herlevsen MC, Smith SC, Theodorescu D. (2005) RalA and RalB: antagonistic relatives in cancer cell migration. Cancer Res 65:7111–7120. [DOI] [PubMed] [Google Scholar]
- Papini D, Langemeyer L, Abad MA, Kerr A, Samejima I, Eyers PA, Jeyaprakash AA, Higgins JM, Barr FA, Earnshaw WC. (2015) TD-60 links RalA GTPase function to the CPC in mitosis. Nat Commun 6:7678. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Park SH, Weinberg RA. (1995) A putative effector of Ral has homology to Rho/Rac GTPase activating proteins. Oncogene 11:2349–2355. [PubMed] [Google Scholar]
- Patgiri A, Yadav KK, Arora PS, Bar-Sagi D. (2011) An orthosteric inhibitor of the Ras-Sos interaction. Nat Chem Biol 7:585–587. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Personnic N, Lakisic G, Gouin E, Rousseau A, Gautreau A, Cossart P, Bierne H. (2014) A role for Ral GTPase-activating protein subunit β in mitotic regulation. FEBS J 281:2977–2989. [DOI] [PubMed] [Google Scholar]
- Peschard P, McCarthy A, Leblanc-Dominguez V, Yeo M, Guichard S, Stamp G, Marshall CJ. (2012) Genetic deletion of RALA and RALB small GTPases reveals redundant functions in development and tumorigenesis. Curr Biol 22:2063–2068. [DOI] [PubMed] [Google Scholar]
- Popovic M, Schouten A, Rensen-de Leeuw M, Rehmann H. (2016) The structure of the guanine nucleotide exchange factor Rlf in complex with the small G-protein Ral identifies conformational intermediates of the exchange reaction and the basis for the selectivity. J Struct Biol 193:106–114. [DOI] [PubMed] [Google Scholar]
- Rebhun JF, Chen H, Quilliam LA. (2000) Identification and characterization of a new family of guanine nucleotide exchange factors for the ras-related GTPase Ral. J Biol Chem 275:13406–13410. [DOI] [PubMed] [Google Scholar]
- Renault L, Guibert B, Cherfils J. (2003) Structural snapshots of the mechanism and inhibition of a guanine nucleotide exchange factor. Nature 426:525–530. [DOI] [PubMed] [Google Scholar]
- Rinaldo F, Li J, Wang E, Muders M, Datta K. (2007) RalA regulates vascular endothelial growth factor-C (VEGF-C) synthesis in prostate cancer cells during androgen ablation. Oncogene 26:1731–1738. [DOI] [PubMed] [Google Scholar]
- Roberts PJ, Der CJ. (2007) Targeting the Raf-MEK-ERK mitogen-activated protein kinase cascade for the treatment of cancer. Oncogene 26:3291–3310. [DOI] [PubMed] [Google Scholar]
- Saito R, Shirakawa R, Nishiyama H, Kobayashi T, Kawato M, Kanno T, Nishizawa K, Matsui Y, Ohbayashi T, Horiguchi M, et al. (2013) Downregulation of Ral GTPase-activating protein promotes tumor invasion and metastasis of bladder cancer. Oncogene 32:894–902. [DOI] [PubMed] [Google Scholar]
- Schmidt S, Diriong S, Méry J, Fabbrizio E, Debant A. (2002) Identification of the first Rho-GEF inhibitor, TRIPalpha, which targets the RhoA-specific GEF domain of Trio. FEBS Lett 523:35–42. [DOI] [PubMed] [Google Scholar]
- Shao H, Andres DA. (2000) A novel RalGEF-like protein, RGL3, as a candidate effector for rit and Ras. J Biol Chem 275:26914–26924. [DOI] [PubMed] [Google Scholar]
- Shima F, Ijiri Y, Muraoka S, Liao J, Ye M, Araki M, Matsumoto K, Yamamoto N, Sugimoto T, Yoshikawa Y, et al. (2010) Structural basis for conformational dynamics of GTP-bound Ras protein. J Biol Chem 285:22696–22705. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Shima F, Yoshikawa Y, Ye M, Araki M, Matsumoto S, Liao J, Hu L, Sugimoto T, Ijiri Y, Takeda A, et al. (2013) In silico discovery of small-molecule Ras inhibitors that display antitumor activity by blocking the Ras-effector interaction. Proc Natl Acad Sci USA 110:8182–8187. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Shirakawa R, Fukai S, Kawato M, Higashi T, Kondo H, Ikeda T, Nakayama E, Okawa K, Nureki O, Kimura T, et al. (2009) Tuberous sclerosis tumor suppressor complex-like complexes act as GTPase-activating proteins for Ral GTPases. J Biol Chem 284:21580–21588. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sidhu RS, Clough RR, Bhullar RP. (2005) Regulation of phospholipase C-delta1 through direct interactions with the small GTPase Ral and calmodulin. J Biol Chem 280:21933–21941. [DOI] [PubMed] [Google Scholar]
- Smith SC, Baras AS, Owens CR, Dancik G, Theodorescu D. (2012) Transcriptional signatures of Ral GTPase are associated with aggressive clinicopathologic characteristics in human cancer. Cancer Res 72:3480–3491. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Smith SC, Oxford G, Baras AS, Owens C, Havaleshko D, Brautigan DL, Safo MK, Theodorescu D. (2007) Expression of ral GTPases, their effectors, and activators in human bladder cancer. Clin Cancer Res 13:3803–3813. [DOI] [PubMed] [Google Scholar]
- Song X, Hua L, Xu Y, Fang Z, Wang Y, Gao J, Shi Q, Zhou X, Yu R. (2015) Involvement of RalB in the effect of geranylgeranyltransferase I on glioma cell migration and invasion. Clin Transl Oncol 17:477–485. [DOI] [PubMed] [Google Scholar]
- Sowalsky AG, Alt-Holland A, Shamis Y, Garlick JA, Feig LA. (2010) RalA suppresses early stages of Ras-induced squamous cell carcinoma progression. Oncogene 29:45–55. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sun Q, Burke JP, Phan J, Burns MC, Olejniczak ET, Waterson AG, Lee T, Rossanese OW, Fesik SW. (2012) Discovery of small molecules that bind to K-Ras and inhibit Sos-mediated activation. Angew Chem Int Ed Engl 51:6140–6143. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Thomas JC, Cooper JM, Clayton NS, Wang C, White MA, Abell C, Owen D, Mott HR. (2016) Inhibition of Ral GTPases using a stapled peptide approach. J Biol Chem 291:18310–18325. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ullah N, Mansha M, Casey PJ. (2016) Protein geranylgeranyltransferase type 1 as a target in cancer. Curr Cancer Drug Targets 16:563–571. [DOI] [PubMed] [Google Scholar]
- Urano T, Emkey R, Feig LA. (1996) Ral-GTPases mediate a distinct downstream signaling pathway from Ras that facilitates cellular transformation. EMBO J 15:810–816. [PMC free article] [PubMed] [Google Scholar]
- Vigil D, Martin TD, Williams F, Yeh JJ, Campbell SL, Der CJ. (2010) Aberrant overexpression of the Rgl2 Ral small GTPase-specific guanine nucleotide exchange factor promotes pancreatic cancer growth through Ral-dependent and Ral-independent mechanisms. J Biol Chem 285:34729–34740. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Walensky LD, Bird GH. (2014) Hydrocarbon-stapled peptides: principles, practice, and progress. J Med Chem 57:6275–6288. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wang H, Owens C, Chandra N, Conaway MR, Brautigan DL, Theodorescu D. (2010) Phosphorylation of RalB is important for bladder cancer cell growth and metastasis. Cancer Res 70:8760–8769. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wang K, Terai K, Peng W, Rouyanian A, Liu J, Roby KF, Wise AL, Ezzeldin M, Larson J, Woo RA, et al. (2013) The role of RalA in biology and therapy of ovarian cancer. Oncotarget DOI: 10.18632/oncotarget.1435.: [Google Scholar]
- Wolthuis RM, Bauer B, van ’t Veer LJ, de Vries-Smits AM, Cool RH, Spaargaren M, Wittinghofer A, Burgering BM, Bos JL. (1996) RalGDS-like factor (Rlf) is a novel Ras and Rap 1A-associating protein. Oncogene 13:353–362. [PubMed] [Google Scholar]
- Wu Z, Owens C, Chandra N, Popovic K, Conaway M, Theodorescu D. (2010) RalBP1 is necessary for metastasis of human cancer cell lines. Neoplasia 12:1003–1012. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yan C, Liu D, Li L, Wempe MF, Guin S, Khanna M, Meier J, Hoffman B, Owens C, Wysoczynski CL, et al. (2014) Discovery and characterization of small molecules that target the GTPase Ral. Nature 515:443–447. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yap TA, Garrett MD, Walton MI, Raynaud F, de Bono JS, Workman P. (2008) Targeting the PI3K-AKT-mTOR pathway: progress, pitfalls, and promises. Curr Opin Pharmacol 8:393–412. [DOI] [PubMed] [Google Scholar]
- Yin J, Pollock C, Tracy K, Chock M, Martin P, Oberst M, Kelly K. (2007) Activation of the RalGEF/Ral pathway promotes prostate cancer metastasis to bone. Mol Cell Biol 27:7538–7550. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhao W, Jamshidiha M, Lanyon-Hogg T, Recchi C, Cota E, Tate EW. (2017) Direct targeting of the Ras GTPase superfamily through structure- based design. Curr Top Med Chem 17:16–29. [DOI] [PubMed] [Google Scholar]
- Zipfel PA, Brady DC, Kashatus DF, Ancrile BD, Tyler DS, Counter CM. (2010) Ral activation promotes melanomagenesis. Oncogene 29:4859–4864. [DOI] [PMC free article] [PubMed] [Google Scholar]