Abstract
The expression modulation of the immunosuppressive non-classical Human leukocyte antigen-G (HLA-G) molecule and its soluble isoforms is an immune evasion strategy being deployed by cytomegalovirus (CMV). The +3142 C>G single nucleotide polymorphism (SNP) located within the 3′ untranslated region (3′UTR) is of crucial importance for the regulation of HLA-G expression. Therefore, we analyzed the influence of the +3142 C>G HLA-G SNP on the occurrence of CMV infection in a cohort of 178 living-donor kidney recipients and their 178 corresponding donors. In addition, soluble HLA-G (sHLA-G) levels were quantified before and after transplantation. The presence of the HLA-G +3142 CC genotype in recipients, but not donors of our cohort as along with elevated sHLA-G levels (≥6.1 ng/mL) were associated with higher susceptibility to CMV infection after transplantation. Our results provided evidence that (i) HLA-G is implicated in the establishment of CMV after living-donor kidney transplantation and (ii) recipient HLA-G +3142 CC genotype and sHLA-G concentration levels could represent important predictive risk markers for CMV infection.
Keywords: Human leukocyte antigen-G, HLA-G 3′UTR Polymorphisms, Living Kidney Transplantation, Cytomegalovirus
1. Introduction
Human cytomegalovirus (CMV) is the prototypic member of the β-herpesvirus subfamily that causes widespread, life-long human infections, which are particularly life threatening for immunosuppressed patients after solid organ transplantation [1,2]. In order to escape immune responses, CMV has developed multiple strategies to counteract numerous aspects of the host immune system [3]. In this process, the immunological antagonism of the human leukocyte antigen (HLA) presentation represents an essential mechanism of immune evasion. CMV encodes several gene products (e.g., US2, US3, US6, US10, US11, pp71-UL82, miR-US4-1 etc.) which target classical HLA class I molecules and/or prevent antigen presentation on the surface of infected cells [4,5]. Some of these inhibitory proteins discriminate between classical and non-classical HLA molecules [6], while others affect both types of HLA molecules [7,8]. In this context, the accumulating line of evidences demonstrates that an up-regulated expression of the immune checkpoint molecule HLA-G facilitates viral immune escape [9,10]. HLA-G has the capacity to inhibit immune competent cells thereby modulating the innate and adaptive immune system [11]. It alters natural killer (NK) cell- and T lymphocyte-mediated cytotoxicity and B cell activation by interacting with the corresponding inhibitory receptors immunoglobulin-like transcript 2 (ILT-2) and ILT-4 as well as killer cell immunoglobulin-like receptor 2DL4 (KIR-2DL4) [12,13,14]. Remarkably, similar to the membrane bound forms, the soluble HLA-G facilitates these inhibitory effects [11,15,16,17]. HLA-G is characterized by a low number of allelic variations resulting in limited variability in the protein-encoding region. However, polymorphic sites within the non-coding region such as the 5′ upstream regulatory region (5′URR) and the 3′ untranslated region (3′UTR) are of crucial importance for the regulation of HLA-G expression levels [18,19]. The HLA-G +3142 C>G (rs1063320) single-nucleotide polymorphism (SNP) located within the 3′UTR, affecting the binding site for microRNAs and thereby influencing the expression levels is of particular relevance [20].
In kidney transplantation, CMV reactivation remains one of the most common infectious complications despite the availability of generally effective antiviral therapies [2]. Enhanced HLA-G expression has been associated with allograft tolerance after kidney transplantation [11]. Studies aiming to demonstrate the relevance of HLA-G on the occurrence of CMV infection are limited in transplantation setting and mostly exclusively focused on the recipient HLA-G genotype [21,22]. Considering the immune inhibitory features of HLA-G, differences in recipient and donor HLA-G +3142 C>G polymorphism induce altered protein expression levels, and therefore may possibly have implications on the occurrence of CMV replication during the first year of living-donor kidney transplantation. To address this question, the recipient and the donor HLA-G +3142 C>G SNP was genotyped, in addition to the quantification of pre- as well as post-transplant sHLA-G concentrations. The results were correlated with the clinical outcome in terms of allograft loss and the occurrence of CMV infection.
2. Results
We found that the incidence of CMV infection was significantly higher among living-donor kidney transplant recipients carrying at least one +3142 C allele and the homozygous +3142 CC genotype, compared to non-carriers (p < 0.0462 and 0.0394, respectively, Table 1A).
Table 1.
The genotype distribution and allele frequencies of the +3142 C>G (rs1063320) gene polymorphism in living-donor kidney transplant recipients; (A) and corresponding donors (B) with respect to cytomegalovirus (CMV) infection.
| (A) Recipient | CMV Infection N = 18 | No CMV Infection N = 160 | p Value | OR | CI (95%) |
| C/C | 8 (44.4%) | 34 (21.3%) | 0.0394 | 2.965 | 1.082–8.092 |
| C/G | 5 (27.8%) | 80 (50%) | 0.0855 | 0.384 | 0.131–1.129 |
| G/G | 5 (27.8%) | 46 (28.7%) | 1 | 1.037 | 0.35–3.07 |
| Allele | |||||
| C | 26 | 148 | 0.0462 | 2.01 | 1.028–3.947 |
| G | 15 | 172 | |||
| (B) Donor | CMV Infection N = 18 | No CMV Infection N = 160 | p Value | OR | CI (95%) |
| C/C | 4 (22.2%) | 41 (25.6%) | 1 | 0.82 | 0.26–2.66 |
| C/G | 8 (44.4%) | 85 (53.1%) | 0.62 | 0.7059 | 0.26–1.882 |
| G/G | 6 (33.3%) | 34 (21.3%) | 0.24 | 1.85 | 0.64–5.3 |
| Allele | |||||
| C | 16 | 167 | 0.386 | 0.732 | 0.366–1.46 |
| G | 20 | 153 | |||
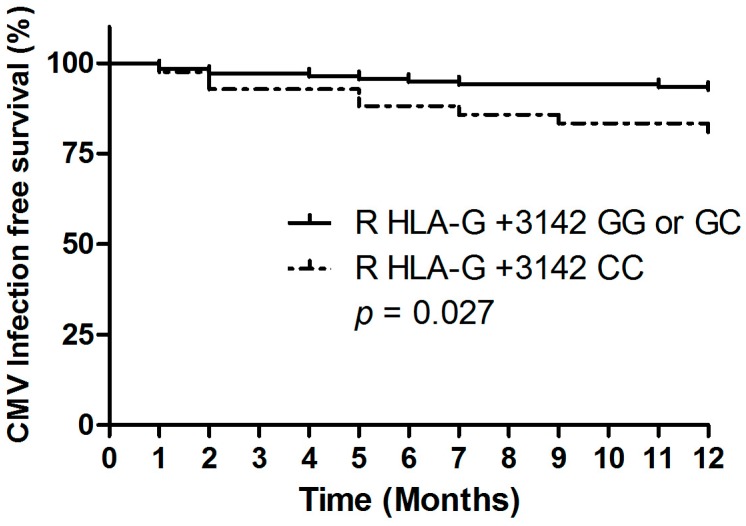
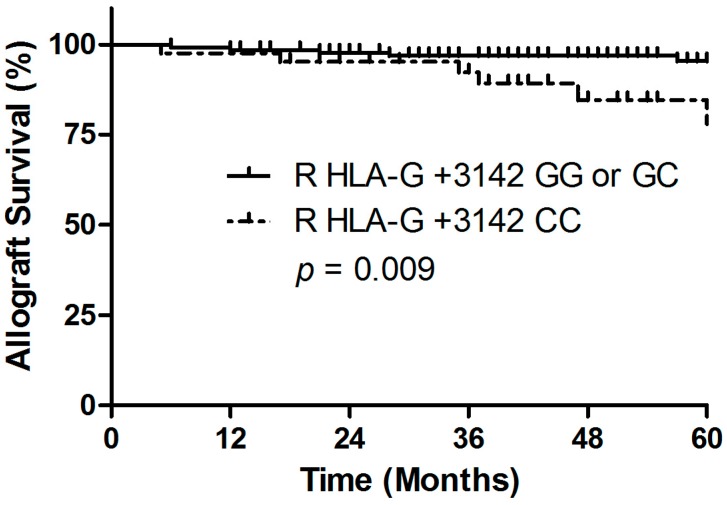
Taking into account the time course, the results of the Kaplan-Meier plot analysis combined with the log-rank test indicate that the development of CMV during the first year after living-donor kidney transplantation was significantly higher among recipients with a homozygous +3142 CC genotype (p = 0.027; Hazard Ratio (HR) 2.7; 95% confidence interval (95% CI): 1.0–6.9, Figure 1), compared to +3142 G allele carriers (i.e., homozygous +3142 GG and heterozygous +3142 CG). Interestingly, we found that the recipient homozygous +3142 CC genotype was associated with an increased risk of 5-year allograft loss (p = 0.009, HR 4.3; 95% CI: 1.3–14.2, Figure 2 and Table 2A). The recipient HLA-G +3142 C>G genotype was not associated with acute cellular rejection within the first year after living-donor kidney transplant (data not shown).
Figure 1.
Association between the transplant recipient HLA-G +3142 C>G polymorphism and CMV infection during first year of living-donor kidney transplantation. Recipients with a HLA-G +3142 CC genotype had a significantly increased likelihood of CMV infection.
Figure 2.
Association between the transplant recipient HLA-G +3142 C>G polymorphism and 5 year allograft survival. Recipients with a HLA-G +3142 CC genotype had a significantly reduced allograft survival.
Table 2.
The genotype distribution and allele frequencies of the +3142 C>G (rs1063320) gene polymorphism in living-donor kidney transplant recipients (A) and the corresponding donors; (B) with respect to allograft loss.
| (A) Recipient | Graft Loss N = 11 | No Allograft Loss N = 167 | p Value | OR | CI (95%) |
| C/C | 6 (54.5%) | 36 (21.6%) | 0.022 | 4.37 | 1.26–15.14 |
| C/G | 4 (36.4%) | 81 (48.5%) | 0.54 | 0.61 | 0.17–2.15 |
| G/G | 1 (9.1%) | 50 (29.9%) | 0.29 | 0.25 | 0.03–2.03 |
| Allele | |||||
| C | 16 | 153 | 0.0158 | 3.155 | 1.204–8.263 |
| G | 6 | 181 | |||
| (B) Donor | Graft Loss N = 11 | No Allograft Loss N = 167 | p Value | OR | CI (95%) |
| C/C | 5 | 40 | 0.125 | 3.175 | 0.87–11.5 |
| C/G | 5 | 88 | 0.76 | 0.75 | 0.22–2.54 |
| G/G | 1 | 39 | 0.46 | 0.33 | 0.04–2.65 |
| Allele | |||||
| C | 15 | 168 | 0.125 | 2.11 | 0.84–5.33 |
| G | 7 | 166 | |||
With respect to the HLA-G +3142 C>G donor genotype, neither it was associated with occurrence of CMV infection (p = 0.77, HR 0.85; 95% CI: 0.28–2.59; Table 1B) or with acute cellular rejection (p = 0.39; HR 0.7; 95% CI: 0.3–1.6), while there was a borderline significance with allograft loss (p = 0.068; HR 2.9; 95% CI: 0.8–9.5; Table 2B). There was no association with outcome parameters and presence of the HLA-G +3142 C>G mismatch.
Soluble HLA-G molecules are of clinical relevance and exhibit equal immunosuppressive properties as their membrane bund counterparts [17]. In an attempt to identify a predictive marker for CMV infection based on sHLA-G levels, we quantified soluble molecule concentrations in patient sera. In contrast to previous studies that linked levels of sHLA-G to the HLA-G +3142 C>G SNP [19] there was no significant association in our cohort (data not shown).
The sHLA-G levels prior kidney transplantation were significantly elevated in recipients exhibiting productive CMV replication after transplantation compared to those with no viral replication (38.9 ± 13.0 ng/mL vs. 25.1 ± 1.74 ng/mL; p = 0.04). After transplantation, the sHLA-G levels dramatically decreased (sHLA-G before 26.8 ± 16.7 ng/mL vs. after transplantation 6.1 ± 4.8 ng/mL; p = 0.0002). Soluble HLA-G levels remained nearly twice as high in recipients with CMV reactivation without reaching the level of significance (10.32 ± 1.73 vs. 5.4 ± 0.9 ng/mL; p = 0.082). To evaluate whether post-transplant sHLA-G levels can help to identify patients with increased risk of CMV after transplantation, the ROC analysis was performed (Figure 3A). Using an optimal cut-off value of 6.1 ng/mL (AUC = 0.75, sensitivity: 80.0%, specificity: 75.8%), Kaplan-Meier curve analyses combined with the log-rank test revealed that sHLA-G concentrations were significantly associated with an increased occurrence of CMV within the first year after transplant (p = 0.010; HR 10.1, 95% CI: 1.1–90.8; Figure 3B).
Figure 3.
The correlation between plasma levels of soluble human leukocyte antigen-G (sHLA-G) and the occurrence of CMV viremia during the first year of living-donor kidney transplantation. (A) Receiver operating characteristic curve-based stratification was performed to predict CMV-infection free survival based on sHLA-G levels. The red open circle indicates the optimal cut-off value with a sensitivity of 80.0% and a specificity of 75.8%; black dots represent differing cut-off values (B) patients with higher sHLA-G levels (≥6.1 ng/mL) had a significantly increased likelihood of CMV infection compared to patients with low sHLA-G levels (<6.1 ng/mL).
3. Discussion
HLA-G is a naturally occurring immune suppressive molecule [23]. Its surface expression is physiologically restricted to the maternal-fetal interface and to immune privileged adult tissues. However, secreted soluble forms of HLA-G are detectable in a variety of body fluids [24] The clinical implications of the differential modulation of HLA-G gene expression by the regulatory polymorphism within the 3′UTR has been emphasized by a number of studies in a wide range of pathological conditions [11,13,25]. Many reports highlighted the importance of HLA-G either in viral infections or in solid allograft acceptance after transplantation [26,27,28,29], though only a minority focused on the role of HLA-G for CMV infection in an allogeneic setting after transplantation [21,22,30]. In a cohort of living-donor kidney transplant pairs, we were able to uncover that (i) the recipient HLA-G +3142 CC genotype is associated with CMV infection within the first year after transplantation and to the five year allograft loss; (ii) donor HLA-G +3142 C>G polymorphism had no impact on allograft outcome; and (iii) elevated sHLA-G serum concentrations might be useful to discriminate recipients with an occurrence of CMV, but do not predict acute rejection.
There are strong evidences that CMV infection after transplantation promotes inflammation, vasculopathy and cellular rejection of an allograft which impacts on allograft survival [2,31,32,33,34]. Considering the negative impact of CMV infection on allograft outcome the identification of predictive markers for detection of CMV prone transplant recipients benefiting from a more intense monitoring or alternative therapeutic approach is urgently needed. While in healthy individuals most CMV infections are asymptomatic, life-threatening diseases occur under the influence of immunosuppressive regimens in the context of allograft transplantation. Implementing prophylactic antiviral therapy has reduced the occurrence of early CMV disease, but the development of late-onset disease and drug resistance is increasingly recognized [2]. In this context, HLA-G +3142 CC recipient genotype constitutes a promising genetic candidate marker for the identification of CMV prone potential recipients even before kidney transplantation. The functional polymorphism +3142 C>G is of specific interest as it has been shown that the C allelic variation is associated with enhanced HLA-G protein expression [11]. Our results are strengthened by the few previous reports clearly outlining the influence of the HLA-G genotype to CMV-susceptibility [21,22,30]. Considering the various immune suppressive functions mediated by HLA-G, the HLA-G genotype has an impact on level of HLA-G expression and thus provides the background for facilitating the CMV escape mechanism [11].
In our cohort, the presence of the HLA-G CC recipient genotype was not only associated with occurrence of CMV infection but also with reduced allograft survival. It is noteworthy that chronic kidney allograft loss is attributed to multiple immune as well as non-immune injuries, against a background of various donor- and recipient-derived risk factors. In this setting, immune responses directed against donor antigens leading to allograft rejection play an eminent role. In our cohort the co-incidence of CMV infection occurred in 4 out of 35 acute rejection episodes. However, we did not find any association between recipient HLA-G +3142 CC genotype and occurrence of rejection. Acute rejection represents an immunological inflammation that occurs within the microenvironment of the allograft, while CMV infection may comprise a located as well as systemic disease. It is reasonable that the establishment of systemic CMV viremia is dominantly influenced by the recipient specific genetic characteristics whereas in cellular rejection both, recipient- and donor-derived factors influence alloimmune and inflammatory responses. The impact of the donor HLA-G genotype in solid organ transplantation has only been investigated in one liver transplantation study supporting our findings on the low influence of the donor HLA-G genotype on transplantation-outcome [30].
Of importance, there was a significant relationship between post-transplant elevated plasma sHLA-G levels and CMV occurrence. This result suggests that high levels of HLA-G promote susceptibility to viral infections. Nevertheless, no association between certain HLA-G +3142 C>G genotypes and soluble HLA-G levels was observed, which can be partly attributed to the low number of plasma samples available, but also to a variety of additional mechanistic factors affecting soluble molecule concentrations.
Taken together, in the context of transplantation the immunosuppressive checkpoint molecule HLA-G plays an eminent role and may discrepantly affect transplant outcome: while on the one hand HLA-G is involved in tolerogenic responses and high levels of HLA-G are associated with better allograft acceptance [11], on the other hand HLA-G promotes viral immune evasion and as showed in our study HLA-G expression is significantly increased during CMV infection [9,10]. Thus, the immunosuppressive properties of HLA-G create an anti-inflammatory environment and in doing so, contribute to increased susceptibility to CMV infection. Although the precise mechanisms contributing to the immunological hazards of CMV infection after transplantation have not been fully elucidated, the results of clinical studies clearly identify the presence of CMV infection as a risk factor for allograft rejection and loss [2,31,32,33,34]. Therefore, enhanced HLA-G expression upon CMV infection in the allogeneic setting of transplantation may indirectly contribute and predict a worse transplant outcome.
In summary, the results of our study suggest that the +3142 CC recipient genotype and enhanced soluble HLA-G levels are associated with a greater susceptibility to CMV infection after living-donor kidney transplantation. Thus, the HLA-G +3142 C>G polymorphism seems to represent a promising novel genetic biomarker for CMV infection after transplantation that is worth of being verified in future studies.
4. Materials and Methods
4.1. Study Population and Outcome Parameters
In total, 178 living-donor kidney transplant recipients and their 178 corresponding donors from the transplant program at the University Hospital Essen, Germany, were enrolled in this study. Occurrence of CMV infection was monitored during the first year after transplantation. The following clinical data were collected from electronic patient records: demographic and transplant-related characteristics of recipient and donor, underlying renal disease, CMV serostatus of recipient and donor, incidence of first episode of CMV viremia or disease within 12 months after transplant, and biopsy-proven acute cellular rejection within 12 months after transplant and 5 year allograft loss. CMV high risk patients (grafts derived from CMV-seropositive donor [D+] transplanted in CMV-seronegative recipients [R-] n = 38) and patients with a lymphocyte-depleting induction therapy received CMV prophylaxis for 100 days before the end of 2011. Because of emerging data suggesting that the incidence of CMV infection was lower among patients receiving a prolonged antiviral prophylaxis in 2012 the duration of the prophylactic CMV regimen was prolonged to 200 days for the high-risk population [35]. All other patients were pre-emptively monitored for CMV viremia weekly for the period of 3 months and afterwards monthly. CMV infection or disease was classified according to recent recommendations [36]: CMV infection was defined as CMV viremia (polymerase chain reaction >400 copies/mL or >1/100 pp65/pUL83 antigen positive cells); CMV disease was defined as CMV viremia in combination with attributable symptoms, such as fever, malaise, leukopenia, thrombocytopenia, or elevation of liver enzymes. CMV complications were analyzed within the first 12 months after transplantation. Incidence of first episode of clinically significant CMV viremia/disease within the 12 months follow-up was 10% (N = 18).
Additional clinical outcome parameters of this study were biopsy-proven rejection events within the first year and five year allograft loss. Allograft loss was defined as return to dialysis or estimated Glomerular filtration rate <15 mL/min/1.73 m². Detailed cohort characteristics are shown in Table 3.
Table 3.
Demographic and clinical characteristics of living-donor transplant recipients and corresponding donors at baseline. Abbreviations are as follows: y: years; SD: standard deviation, HLA: human leukocyte antigen, CMV: cytomegalovirus, R: recipient, D: donor, KTx: kidney transplant.
| Total | HLA-G +3142 GG or GC carrier | HLA-G +3142 CC carrier | p Value HLA-G +3142 GG/GC vs. CC | |
|---|---|---|---|---|
| Recipient | N = 178 | N = 136 | N = 42 | |
| Gender (men/women) | 106/72 | 85/51 | 21/21 | 0.15 |
| Age (y ± SD) | 41.9 ± 15.9 | 41.1 ± 15.6 | 44.6 ± 9.5 | 0.96 |
| CMV positive recipient (R+) | 91 | 73 | 18 | 0.22 |
| Donor | N = 178 | N = 133 | N = 45 | |
| Gender (men/women) | 71/107 | 56/77 | 15/30 | 0.29 |
| Age (y ± SD) | 51.4 ± 9.7 | 50.8 ± 9.6 | 53.0 ± 9.9 | 0.19 |
| CMV positive donor (D+) | 97 | 70 | 27 | 0.39 |
| Cause of end-stage renal disease | ||||
| Diabetes mellitus | 9 | 6 | 3 | 0.44 |
| Chronic glomerulonephritis | 56 | 42 | 14 | 0.85 |
| Polycystic kidney disease | 24 | 18 | 6 | 0.80 |
| Other or unknown | 89 | 70 | 19 | 0.60 |
| Transplant related characteristics | ||||
| Mean cold ischemia time (minutes ± SD) | 133.5 ± 49.4 | 131 ± 51.2 | 140.6 ± 42.9 | 0.67 |
| Lymphocyte-depleting induction therapy (yes/no) | 12/166 | 8/128 | 4/38 | 0.41 |
| AB0 incompatible transplant (yes/no) | 20/158 | 14/122 | 6/36 | 0.47 |
| HLA A, B mismatches (mean ± SD) | 2.0 ± 1.2 | 2.01 ± 1.15 | 2.0 ± 1.1 | 0.78 |
| HLA-DR mismatch (mean ± SD) | 1.1 ± 0.7 | 1.1 ± 0.7 | 1.2 ± 0.6 | 0.85 |
| Transplantation outcome parameters | ||||
| CMV infection first year after KTx total group (yes/no) | 21/157 | 12/124 | 9/33 | 0.027 |
| 5 year Allograft loss (yes/no) | 11/167 | 5/131 | 6/36 | 0.013 |
| Acute cellular rejection (yes/no) | 36/142 | 28/108 | 8/34 | 0.82 |
Signed informed consent was obtained from all patients in accordance with the Declaration of Helsinki, and the ethics committee of the University Hospital Essen approved the study (12-5312-BO; April 2013) .
4.2. HLA-G +3142 C>G SNP Typing and Soluble HLA-G Enzyme-Linked Immunosorbent Assay (ELISA)
Genotyping of the HLA-G +3142 C>G SNP was performed as previously described [37]. Briefly, for +3142 C>G polymorphism 50 ng of genomic DNA of genomic DNA was amplified in a reaction tube, with final concentrations as follows: PCR buffer 1.5 mM MgCl2; 1.8 mM of dNTP; Taq DNA polymerase and 100 pmol of each primer (GMIRNAF–5′-CATGCTGAACTGCATTCCTTCC-3′, GMIRNAR–5′-CTGGTGGGACAAGGTTCTACTG-3′). Thermocycling conditions were as follows: 94 °C for 5 min; 32 cycles of 94 °C for 30 s, 65.5 °C for 30 s and 72 °C for 60 s followed by a final extension step at 72 °C for 5 min. The amplified PCR products were cleaved with 3 U of the restriction enzyme BaeGI (New England Biolabs, Inc., Ipswich, MA, USA), according to the manufacturer’s instructions. RFLP products were analyzed by electrophoresis in a 2% (w/v) agarose gel stained with ethidium bromide, with amplicon sizes of 406 bp for the C allele and 316 and 90 bp for the G allele. The distribution of HLA-G +3142 C>G SNP genotypes and alleles for recipients and donors is summarized in Table 3. The HLA-G +3412 genotype and allele frequencies were similar among recipients and donor. The observed allelic distribution was in accordance with expectations indicated by the Hardy-Weinberg equilibrium (p > 0.05).
A sHLA-G-specific enzyme-linked immunosorbent assay (ELISA) was performed as previously described [38]. Briefly, plasma samples were used in a dilution of 1:2 in PBS. Purified HLA-G5 served as standard reagent. The sHLA-G levels were determined by four-parameter curve fitting. The ELISA detection limit of sHLA-G was 0.25 ng/mL. Plasma soluble HLA-G levels were determined in 57 patients before and consecutively in 34 recipients in a longitudinal follow-up after kidney transplantation.
4.3. Statistical Analysis
Baseline characteristics of donors and recipients were compared with two-sided Fisher’s exact test or the Wilcoxon rank-sum test, as appropriate. The occurrence of CMV infection was estimated by the method of Kaplan-Meier and survival curves were compared using the log-rank test. We used multivariate Cox proportional hazards modeling to assess the risk of CMV infection after transplantation. A two-sided p-value of 0.05 or lower was considered statistically significant.
Acknowledgments
We especially thank the patients participating in this study. We are grateful to Sabine Schramm and Monika Collenburg for their technical support. Hana Guberina was supported by the “Programm zur internen Forschungsförderung Essen” (IFORES) Research fellowship program of the University Duisburg-Essen Medical School”. Rafael Tomoya Michita was supported by the “Coordenação de Aperfeicoamento de Pessoal de Nível Superior (CAPES) Foundation, Ministry of Education of Brazil, Brasília-DF Brazil (99999.000124/2016-08)”, the “Conselho Nacional de Desenvolvimento Científico e Tecnológico” (CNPq) Foundation, Ministry of Science and Technology of Brazil, Brasília-DF Brazil (142475/2015-7) and by the “Deutsche Akademische Austauschdienst” (DAAD) Scholarship of the German Federal Ministry of Education and Research.
Abbreviations
| HLA | Human leukocyte antigen |
| sHLA-G | (Soluble) Human leukocyte antigen-G |
| CMV | Cytomegalovirus |
| SNP | Single Nucleotid Polymorphism |
| 3′UTR | 3′ untranslated region |
| NK cell | Natural killer cell |
| ILT | Immunoglobulin-like transcript |
| KIR | Killer cell immunoglobulin-like receptor |
| 5′URR | 5′ upstream regulatory region |
| HR | Hazard Ratio |
| 95% CI | 95% Confidence interval |
| R | Recipient |
| D | Donor |
| US | Ultra short |
| eGFR | Estimated Glomerular Filtration Rate |
| ELISA | Enzyme-linked immunosorbent assay |
Author Contributions
Hana Guberina: Conceived and designed research, performed the experiments, interpreted data, performed statistical analysis, wrote the initial draft, and read and approved the final article. Rafael Tomoya Michita: Performed the experiments and read and approved the final article. Sebastian Dolff: Collected and provided clinical data, interpreted data and read and approved the final article. Anja Bienholz: Collected and provided clinical data, performed statistical analysis and read and approved the final article. Mirko Trilling: Interpreted data and read and approved the final article. Falko M. Heinemann: Contributed reagents and read and approved the final article. Peter A. Horn: Conceived and designed research and read and approved the final article. Andreas Kribben: Conceived and designed research and read and approved the final article. Oliver Witzke: Conceived and designed research and read and approved the final article. Vera Rebmann: Conceived and designed research, interpreted data, performed statistical analysis, wrote the initial draft, and read and approved the final article.
Conflicts of Interest
The authors declare no conflict of interest. The founding sponsors had no role in the design of the study; in the collection, analyses, or interpretation of data; in the writing of the manuscript, and in the decision to publish the results.
References
- 1.Asberg A., Jardine A.G., Bignamini A.A., Rollag H., Pescovitz M.D., Gahlemann C.C., Humar A., Hartmann A., VICTOR Study Group Effects of the intensity of immunosuppressive therapy on outcome of treatment for CMV disease in organ transplant recipients. Am. J. Transplant. 2010;10:1881–1888. doi: 10.1111/j.1600-6143.2010.03114.x. [DOI] [PubMed] [Google Scholar]
- 2.Fishman J.A. Infection in Organ Transplantation. Am. J. Transplant. 2017;17:856–879. doi: 10.1111/ajt.14208. [DOI] [PubMed] [Google Scholar]
- 3.Alcami A., Koszinowski U.H. Viral mechanisms of immune evasion. Trends Microbiol. 2000;8:410–418. doi: 10.1016/S0966-842X(00)01830-8. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Ploegh H.L. Viral strategies of immune evasion. Science. 1998;280:248–253. doi: 10.1126/science.280.5361.248. [DOI] [PubMed] [Google Scholar]
- 5.Halenius A., Gerke C., Hengel H. Classical and non-classical MHC I molecule manipulation by human cytomegalovirus: So many targets-but how many arrows in the quiver? Cell Mol. Immunol. 2015;12:139–153. doi: 10.1038/cmi.2014.105. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Schust D.J., Tortorella D., Seebach J., Phan C., Ploegh H.L. Trophoblast class I major histocompatibility complex (MHC) products are resistant to rapid degradation imposed by the human cytomegalovirus (HCMV) gene products US2 and US11. J. Exp. Med. 1998;188:497–503. doi: 10.1084/jem.188.3.497. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Park B., Spooner E., Houser B.L., Strominger J.L., Ploegh H.L. The HCMV membrane glycoprotein US10 selectively targets HLA-G for degradation. J. Exp. Med. 2010;207:2033–2041. doi: 10.1084/jem.20091793. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Jun Y., Kim E., Jin M., Sung H.C., Han H., Geraghty D.E., Ahn K. Human cytomegalovirus gene products US3 and US6 down-regulate trophoblast class I MHC molecules. J. Immunol. 2000;164:805–811. doi: 10.4049/jimmunol.164.2.805. [DOI] [PubMed] [Google Scholar]
- 9.Onno M., Pangault C., Le Friec G., Guilloux V., André P., Fauchet R. Modulation of HLA-G antigens expression by human cytomegalovirus: Specific induction in activated macrophages harboring human cytomegalovirus infection. J. Immunol. 2000;164:6426–6434. doi: 10.4049/jimmunol.164.12.6426. [DOI] [PubMed] [Google Scholar]
- 10.Rizzo R., Gabrielli L., Bortolotti D., Gentili V., Piccirilli G., Chiereghin A., Pavia C., Bolzani S., Guerra B., Simonazzi G. Study of Soluble HLA-G in Congenital Human Cytomegalovirus Infection. J. Immunol. Res. 2016;2016:3890306. doi: 10.1155/2016/3890306. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Rebmann V., da Silva Nardi F., Wagner B., Horn P.A. HLA-G as a tolerogenic molecule in transplantation and pregnancy. J. Immunol. Res. 2014;2014:297073. doi: 10.1155/2014/297073. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Carosella E.D., Moreau P., Le Maoult J., Le Discorde M., Dausset J., Rouas-Freiss N. HLA-G molecules: from maternal-fetal tolerance to tissue acceptance. Adv. Immunol. 2003;81:199–252. doi: 10.1016/s0065-2776(03)81006-4. [DOI] [PubMed] [Google Scholar]
- 13.Carosella E.D., Moreau P., Lemaoult J., Rouas-Freiss N. HLA-G: From biology to clinical benefits. Trends Immunol. 2008;29:125–132. doi: 10.1016/j.it.2007.11.005. [DOI] [PubMed] [Google Scholar]
- 14.Sullivan L.C., Hoare H.L., McCluskey J., Rossjohn J., Brooks A.G. A structural perspective on MHC class Ib molecules in adaptive immunity. Trends Immunol. 2006;27:413–420. doi: 10.1016/j.it.2006.07.006. [DOI] [PubMed] [Google Scholar]
- 15.Morandi F., Pistoia V. Soluble HLA-G modulates miRNA-210 and miRNA-451 expression in activated CD4+ T lymphocytes. Int. Immunol. 2013;25:279–285. doi: 10.1093/intimm/dxs108. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Shiroishi M., Kuroki K., Rasubala L., Tsumoto K., Kumagai I., Kurimoto E., Kato K., Kohda D., Maenaka K. Structural basis for recognition of the nonclassical MHC molecule HLA-G by the leukocyte Ig-like receptor B2 (LILRB2/LIR2/ILT4/CD85d) Proc. Natl. Acad. Sci. USA. 2006;103:16412–16417. doi: 10.1073/pnas.0605228103. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Pistoia V., Morandi F., Wang X., Ferrone S. Soluble HLA-G: Are they clinically relevant? Semin. Cancer Biol. 2007;17:469–479. doi: 10.1016/j.semcancer.2007.07.004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Castelli E.C., Veiga-Castelli L.C., Yaghi L., Moreau P., Donadi E.A. Transcriptional and posttranscriptional regulations of the HLA-G gene. J. Immunol. Res. 2014;2014:734068. doi: 10.1155/2014/734068. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Tan Z., Randall G., Fan J., Camoretti-Mercado B., Brockman-Schneider R., Pan L., Solway J., Gern J.E., Lemanske R.F., Nicolae D., et al. Allele-specific targeting of microRNAs to HLA-G and risk of asthma. Am. J. Hum. Genet. 2007;81:829–834. doi: 10.1086/521200. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Veit T.D., Chies J.A. Tolerance versus immune response—microRNAs as important elements in the regulation of the HLA-G gene expression. Transpl. Immunol. 2009;20:229–231. doi: 10.1016/j.trim.2008.11.001. [DOI] [PubMed] [Google Scholar]
- 21.Jin Z.K., Xu C.X., Tian P.X., Xue W.J., Ding X.M., Zheng J., Ding C.G., Ge G.Q., Mao T.C., Lin Y. Impact of HLA-G 14-bp polymorphism on acute rejection and cytomegalovirus infection in kidney transplant recipients from northwestern China. Transpl. Immunol. 2012;27:69–74. doi: 10.1016/j.trim.2012.06.008. [DOI] [PubMed] [Google Scholar]
- 22.Misra M.K., Prakash S., Kapoor R., Pandey S.K., Sharma R.K., Agrawal S. Association of HLA-G promoter and 14-bp insertion-deletion variants with acute allograft rejection and end-stage renal disease. Tissue Antigens. 2013;82:317–326. doi: 10.1111/tan.12210. [DOI] [PubMed] [Google Scholar]
- 23.Carosella E.D., Rouas-Freiss N., Tronik-Le Roux D., Moreau P., LeMaoult J. HLA-G: An Immune Checkpoint Molecule. Adv. Immunol. 2015;127:33–144. doi: 10.1016/bs.ai.2015.04.001. [DOI] [PubMed] [Google Scholar]
- 24.Rebmann V., Pfeiffer K., Pässler M., Ferrone S., Maier S., Weiss E., Grosse-Wilde H. Detection of soluble HLA-G molecules in plasma and amniotic fluid. Tissue Antigens. 1999;53:14–22. doi: 10.1034/j.1399-0039.1999.530102.x. [DOI] [PubMed] [Google Scholar]
- 25.Baricordi O.R., Stignani M., Melchiorri L., Rizzo R. HLA-G and inflammatory diseases. Inflamm. Allergy Drug Targets. 2008;7:67–74. doi: 10.2174/187152808785107615. [DOI] [PubMed] [Google Scholar]
- 26.Weng P.J., Fu Y.M., Ding S.X., Xu D.P., Lin A., Yan W.H. Elevation of plasma soluble human leukocyte antigen-G in patients with chronic hepatitis C virus infection. Hum. Immunol. 2011;72:406–411. doi: 10.1016/j.humimm.2011.02.008. [DOI] [PubMed] [Google Scholar]
- 27.Murdaca G., Contini P., Setti M., Cagnati P., Lantieri F., Indiveri F., Puppo F. Behavior of non-classical soluble HLA class G antigens in human immunodeficiency virus 1-infected patients before and after HAART: Comparison with classical soluble HLA-A, -B, -C antigens and potential role in immune-reconstitution. Clin. Immunol. 2009;133:238–244. doi: 10.1016/j.clim.2009.08.002. [DOI] [PubMed] [Google Scholar]
- 28.Cordero E.A., Veit T.D., da Silva M.A., Jacques S.M., Silla L.M., Chies J.A. HLA-G polymorphism influences the susceptibility to HCV infection in sickle cell disease patients. Tissue Antigens. 2009;74:308–313. doi: 10.1111/j.1399-0039.2009.01331.x. [DOI] [PubMed] [Google Scholar]
- 29.Rebmann V., Bartsch D., Wunsch A., Möllenbeck P., Golda T., Viebahn R., Grosse-Wilde H. Soluble total human leukocyte antigen class I and human leukocyte antigen-G molecules in kidney and kidney/pancreas transplantation. Hum. Immunol. 2009;70:995–999. doi: 10.1016/j.humimm.2009.07.016. [DOI] [PubMed] [Google Scholar]
- 30.Thude H., Janssen M., Sterneck M., Nashan B., Koch M. 14-bp ins/del polymorphism and +3142 C>G SNP of the HLA-G gene have a significant impact on acute rejection after liver transplantation. Hum. Immunol. 2016;77:1159–1165. doi: 10.1016/j.humimm.2016.09.009. [DOI] [PubMed] [Google Scholar]
- 31.Rubin R.H. The pathogenesis and clinical management of cytomegalovirus infection in the organ transplant recipient: The end of the “silo hypothesis”. Curr. Opin. Infect. Dis. 2007;20:399–407. doi: 10.1097/QCO.0b013e328285a358. [DOI] [PubMed] [Google Scholar]
- 32.Kas-Deelen A.M., de Maar E.F., Harmsen M.C., Driessen C., van Son W.J., The T.H. Uninfected and cytomegalic endothelial cells in blood during cytomegalovirus infection: Effect of acute rejection. J. Infect. Dis. 2000;181:721–724. doi: 10.1086/315266. [DOI] [PubMed] [Google Scholar]
- 33.Humar A., Gillingham K.J., Payne W.D., Dunn D.L., Sutherland D.E., Matas A.J. Association between cytomegalovirus disease and chronic rejection in kidney transplant recipients. Transplantation. 1999;68:1879–1883. doi: 10.1097/00007890-199912270-00011. [DOI] [PubMed] [Google Scholar]
- 34.Weill D. Role of cytomegalovirus in cardiac allograft vasculopathy. Transpl. Infect. Dis. 2001;3:44–48. doi: 10.1034/j.1399-3062.2001.00009.x. [DOI] [PubMed] [Google Scholar]
- 35.Humar A., Limaye A.P., Blumberg E.A., Hauser I.A., Vincenti F., Jardine A.G., Abramowicz D., Ives J.A., Farhan M., Peeters P. Extended valganciclovir prophylaxis in D+/R- kidney transplant recipients is associated with long-term reduction in cytomegalovirus disease: Two-year results of the IMPACT study. Transplantation. 2010;90:1427–1431. doi: 10.1097/TP.0b013e3181ff1493. [DOI] [PubMed] [Google Scholar]
- 36.Kotton C.N., Kumar D., Caliendo A.M., Asberg A., Chou S., Danziger-Isakov L., Humar A., Transplantation Society International CMV Consensus Group Updated international consensus guidelines on the management of cytomegalovirus in solid-organ transplantation. Transplantation. 2013;96:333–360. doi: 10.1097/TP.0b013e31829df29d. [DOI] [PubMed] [Google Scholar]
- 37.Castelli E.C., Mendes-Junior C.T., Deghaide N.H., de Albuquerque R.S., Muniz Y.C., Simões R.T., Carosella E.D., Moreau P., Donadi E.A. The genetic structure of 3′ untranslated region of the HLA-G gene: polymorphisms and haplotypes. Genes Immun. 2010;11:134–141. doi: 10.1038/gene.2009.74. [DOI] [PubMed] [Google Scholar]
- 38.Rebmann V., LeMaoult J., Rouas-Freiss N., Carosella E.D., Grosse-Wilde H. Quantification and identification of soluble HLA-G isoforms. Tissue Antigens. 2007;69:143–149. doi: 10.1111/j.1399-0039.2006.763_5.x. [DOI] [PubMed] [Google Scholar]