Abstract
Phytochemical examination of Convallaria majalis (Liliaceae) whole plants yielded 15 steroidal glycosides (1–15), including nine new compounds (4–6, 10–15) with a lycotetrose unit. The structures of the new compounds were determined using two-dimensional Nuclear magnetic resonance (NMR) analyses and chemical methods. The isolated compounds were evaluated for cytotoxicity against HL-60 human promyelocytic leukemia cells, A549 human lung adenocarcinoma cells, and HSC-4 and HSC-2 human oral squamous cell carcinoma cell lines. Of these, (25S)-spirost-5-en-3β-yl O-β-d-glucopyranosyl-(1→2)-O-[β-d-xylopyranosyl-(1→3)]-O-β-d-glucopyranosyl-(1→4)-β-d-galactopyranoside (1) exhibited cytotoxic activity against HL-60, A549, HSC-4, and HSC-2 cells with IC50 values ranging from 0.96 to 3.15 μM. The corresponding furostanol glycoside of 1, (25S)-26-[(β-d-glucopyranosyl)oxy]-22α-hydroxyfurost-5-en-3β-yl O-β-d-glucopyranosyl-(1→2)-O-[β-d-xylopyranosyl-(1→3)]-O-β-d-glucopyranosyl-(1→4)-β-d-galactopyranoside (8), was cytotoxic to the adherent cell lines of A549, HSC-4, and HSC-2 cells with IC50 values of 2.97, 11.04, and 8.25 μM, respectively. The spirostanol lycotetroside (1) caused necrotic cell death in A549 cells in a dose-dependent manner. Alternatively, the furostanol lycotetroside (8) induced apoptotic cell death in A549 cells in a time-dependent manner, as was evident by morphological observations and flow cytometry analyses.
Keywords: Convallaria majalis, Liliaceae, steroidal glycoside, apoptosis, necrosis, A549, HL-60, HSC-4, HSC-2
1. Introduction
Convallaria majalis L. (Liliaceae), commonly called lily of the valley, is a popular ornamental garden plant [1]. It has been reported to contain cardenolide glycosides, such as convallatoxin and convallatoxol, and is well-known to be a toxic plant [2]. Previously, we conducted phytochemical examinations of the rhizomes of C. majalis and isolated and characterized convallasaponin A, a new 5β-spirostanol triglycoside, along with two known cardenolide glycosides, a known cholestane glycoside, and polyhydroxylated steroidal saponins [3,4]. However, thin-layer chromatography (TLC) analysis of the MeOH extract of C. majalis indicates that it still contains numerous steroidal glycosides. Our continuing search for steroidal glycosides have revealed that some of them showed potent cytotoxic activity and induced apoptosis against cultured tumor cells. In this study, we conducted further phytochemical analysis of the methanol extract from C. majalis, paying particular attention to steroidal glycosides. As a result, nine new steroidal glycosides with an O-β-d-glucopyranosyl-(1→2)-O-[β-d-xylopyranosyl-(1→3)]-O-β-d-glucopyranosyl-(1→4)-β-d-galactopyranose (lycotetrose) unit (4–6, 10–15) and six known compounds (1–3, 7–9) were isolated. The structures of the new steroidal glycosides were determined based on spectroscopic analysis, including two-dimensional Nuclear magnetic resonance (NMR) data, and chemical methods. The isolated compounds were evaluated for their cytotoxicity against HL-60 human promyelocytic leukemia cells, A549 human lung adenocarcinoma cells, and HSC-4 and HSC-2 human oral squamous cell carcinoma cells. Apoptosis-inducing activity in A549 cells was also examined.
2. Results and Discussion
2.1. Structural Elucidation
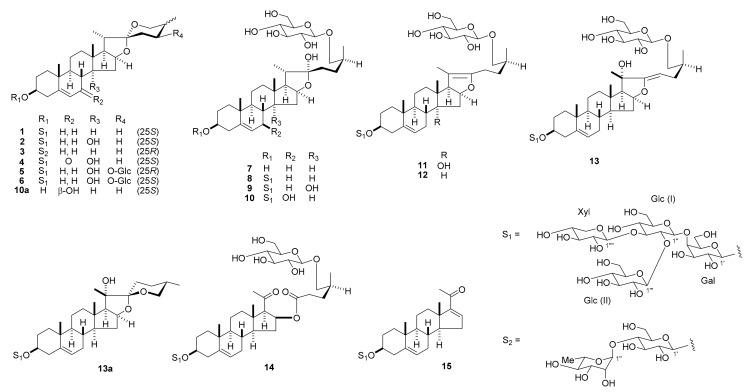
Convallaria majalis (3.0 kg, dry weight) whole plants were extracted with hot MeOH. The MeOH extract was passed through a porous-polymer polystyrene resin (Diaion HP-20, Mitsubishi-Chemical, Tokyo, Japan) column. The MeOH-eluted and MeOH-H2O (6:4)-eluted fractions were then subjected to silica gel and octadecylsilanized (ODS) silica gel column chromatography (CC) and reversed-phase preparative high-performance liquid chromatography (HPLC) to obtain compounds 1–15 (Figure 1). The structures of the known compounds 1–3 and 7–9 were identified as (25S)-spirost-5-en-3β-yl O-β-d-glucopyranosyl-(1→2)-O-[β-d-xylopyranosyl-(1→3)]-O-β-d-glucopyranosyl-(1→4)-β-d-galactopyranoside (1) [5], (25S)-14α-hydroxyspirost-5-en-3β-yl O-β-d-glucopyranosyl-(1→2)-O-[β-d-xylopyranosyl-(1→3)]-O-β-d-glucopyranosyl-(1→4)-β-d-galactopyranoside (2) [5], (25R)-spirost-5-en-3β-yl O-α-L-rhamnopyranosyl-(1→4)-β-d-glucopyranoside (3) [6], (25S)-26-[(β-d-glucopyranosyl)oxy]-22α-hydroxyfurost-5-en-3β-ol (7) [7], (25S)-26-[(β-d-glucopyranosyl)oxy]-22α-hydroxyfurost-5-en-3β-yl O-β-d-glucopyranosyl-(1→2)-O-[β-d-xylopyranosyl-(1→3)]-O-β-d-glucopyranosyl-(1→4)-β-d-galactopyranoside (8) [8], and (25S)-26-[(β-d-glucopyranosyl)oxy]-14α,22α-hydroxyfurost-5-en-3β-yl O-β-d-glucopyranosyl-(1→2)-O-[β-d-xylopyranosyl-(1→3)]-O-β-d-glucopyranosyl-(1→4)-β-d-galactopyranoside (9) [9], respectively.
Figure 1.
Steroidal glycosides from Convallaria majalis.
Compound 4 was obtained as an amorphous solid, and its molecular formula was identified as C50H78O24, based on data from high-resolution electrospray ionization time-of-flight mass spectrometry (HR-ESI-TOF-MS; m/z 1085.4773 [M + Na]+, calcd. 1085.4781) and 13C NMR (50 carbon signals) spectrum. The 1H and 13C NMR spectral features of 4 were closely related to those of 2, showing the following signals: four steroid methyl groups at δH 1.21 (d, J = 7.0 Hz, Me-21), 1.07 (d, J = 7.0 Hz, Me-27), 1.04 (s, Me-18), and 0.99 (s, Me-19), in addition to δC 20.4 (C-18), 17.1 (C-19), 16.3 (C-27), and 15.2 (C-21); an olefinic group at δH 5.75 (s, H-6) and δC 166.6 (C-5) and 126.9 (C-6); oxygenated protons and carbons at δH 3.86 (m, H-3) and δC 78.1 (C-3), δH 5.06 (dd, J = 12.7, 9.0 Hz, H-16) and δC 82.2 (C-16), and δH 4.02 (dd, J = 10.9, 2.5 Hz, H-26ax) and 3.31 (br d, J = 10.9 Hz, H-26eq) and δC 65.0 (C-26); an oxygenated quaternary carbon at δC 110.0 (C-22); and four anomeric protons and carbons at δH 5.59 (d, J = 7.5 Hz), 5.25 (d, J = 7.6 Hz), 5.19 (d, J = 7.5 Hz), and 4.83 (d, J = 7.6 Hz) and δC 105.2, 105.0, 104.9, and 102.8 (Table 1). In addition, the IR and 13C NMR spectra of 4 suggest the presence of a conjugated carbonyl group (νmax 1650 cm−1; δC 200.6) (Table 2). In the heteronuclear multiple-bond correlation (HMBC) spectrum of 4, long-range correlations observed from H-6 at δH 5.75, H-9 at δH 2.41, and H-8 at δH 2.74 to C-7 at δC 200.6 indicated that the carbonyl group was located at C-7 of the aglycone. Nuclear Overhauser effect (NOE) correlations were found between the signals of H-8 at δH 2.74 and Me-18 at δH 1.04/Me-19 at δH 0.99, between the signals of H-12ax at δH 1.41 and H-9 at δH 2.41/H-17 at δH 2.72, between the signals of H-17 and Me-21, between the signals of H-20 at δH 2.02 and Me-18/H-23ax at δH 1.93, and between the signals of Me-27 at δH 1.07 and H-23ax at δH 1.93. These correlations in the NOE spectroscopy (NOESY) spectrum of 4 were consistent with the B/C trans, C/D trans, and D/E cis ring fusions, as well as the 14α, 20α, 22α, and 25S configurations. The linkage of lycotetrose, O-β-d-glucopyranosyl-(1→2)-O-[β-d-xylopyranosyl-(1→3)]-O-β-d-glucopyranosyl-(1→4)-β-d-galactopyranose, at C-3 of the aglycone, was ascertained by acid hydrolysis of 4 with 0.5 M HCl, obtaining d-galactose, d-glucose, and d-xylose. Additionally, long-range correlations from the anomeric proton (H-1′′′) of Glc (II) at δH 5.59 to C-2′′ of Glc (I) at δC 81.3, from H-1′′′′ of Xyl at δH 5.25 to C-3′′ of Glc (I) at δC 86.7, from H-1′′ of Glc (I) at δH 5.19 to C-4′ of Gal at δC 79.8, and from H-1′ of Gal at δH 4.83 to C-3 of the aglycone at δC 78.1 in the HMBC spectrum of 4 were also observed. Thus, 4 was labeled (25S)-3β-[(O-β-d-glucopyranosyl-(1→2)-O-[β-d-xylopyranosyl-(1→3)]-O-β-d-glucopyranosyl-(1→4) β-d-galactopyranosyl)oxy]-14α-hydroxyspirost-5-en-7-one.
Table 1.
1H and 13C Nuclear magnetic resonance (NMR) spectral data for the sugar moiety of 4 in C5D5N.
| Position | δH | J (Hz) | δC | Position | δH | J (Hz) | δC | ||||||
|---|---|---|---|---|---|---|---|---|---|---|---|---|---|
| Gal | 1′ | 4.83 | d | 7.6 | 102.8 | Glc (II) | 1′′′ | 5.59 | d | 7.5 | 105.0 | ||
| 2′ | 4.40 | dd | 8.5, 7.6 | 73.1 | 2′′′ | 4.09 | dd | 8.8, 7.5 | 76.2 | ||||
| 3′ | 4.10 | m | 75.5 | 3′′′ | 4.11 | dd | 8.8, 8.8 | 77.8 | |||||
| 4′ | 4.59 | br s | 79.8 | 4′′′ | 4.22 | dd | 8.8, 8.8 | 71.7 | |||||
| 5′ | 3.98 | m | 75.4 | 5′′′ | 3.96 | m | 78.7 | ||||||
| 6′ | a | 4.68 | dd | 15.1, 9.4 | 60.6 | 6′′′ | a | 4.60 | br d | 11.9 | 62.6 | ||
| b | 4.21 | br d | 15.1 | b | 4.38 | br d | 11.9 | ||||||
| Glc (I) | 1′′ | 5.19 | d | 7.5 | 105.2 | Xyl | 1′′′′ | 5.25 | d | 7.6 | 104.9 | ||
| 2′′ | 4.44 | dd | 8.8, 7.5 | 81.3 | 2′′′′ | 3.97 | dd | 8.0, 7.6 | 75.1 | ||||
| 3′′ | 4.12 | dd | 8.8, 8.8 | 86.7 | 3′′′′ | 4.08 | dd | 8.6, 8.0 | 78.8 | ||||
| 4′′ | 3.83 | dd | 9.5, 8.8 | 70.5 | 4′′′′ | 4.11 | m | 70.7 | |||||
| 5′′ | 3.88 | m | 9.5, 5.6, 2.5 | 77.6 | 5′′′′ | a | 4.24 | dd | 10.7, 4.9 | 67.4 | |||
| 6′′ | a | 4.53 | dd | 11.4, 5.6 | 63.0 | b | 3.68 | dd | 10.7, 10.4 | ||||
| b | 4.02 | dd | 11.4, 2.5 | ||||||||||
Table 2.
13C NMR spectral data for the aglycone moiety of 4–6 and 10–15 in C5D5N.
| Position | 4 | 5 | 6 | 10 | 11 | 12 | 13 | 14 | 15 |
|---|---|---|---|---|---|---|---|---|---|
| 1 | 36.3 | 37.6 | 37.6 | 37.1 | 37.7 | 37.5 | 37.4 | 37.4 | 36.8 |
| 2 | 30.1 | 30.1 | 30.1 | 30.1 | 30.1 | 30.1 | 30.0 | 30.1 | 30.1 |
| 3 | 78.1 | 78.2 | 78.0 | 78.0 | 78.1 | 78.1 | 78.1 | 78.0 | 77.8 |
| 4 | 38.9 | 39.2 | 39.2 | 38.7 | 39.2 | 39.2 | 39.2 | 39.2 | 39.0 |
| 5 | 166.6 | 140.5 | 140.4 | 141.6 | 140.5 | 141.0 | 141.0 | 141.2 | 141.2 |
| 6 | 126.9 | 122.2 | 122.1 | 128.4 | 122.1 | 121.6 | 121.5 | 121.4 | 121.1 |
| 7 | 200.6 | 26.6 | 26.5 | 72.6 | 26.7 | 31.6 | 31.9 | 31.9 | 31.5 |
| 8 | 49.5 | 35.5 | 35.4 | 40.8 | 35.0 | 31.3 | 31.0 | 30.9 | 30.0 |
| 9 | 44.4 | 43.5 | 43.5 | 48.6 | 43.5 | 50.2 | 50.0 | 50.3 | 50.4 |
| 10 | 38.5 | 37.3 | 37.3 | 36.9 | 37.3 | 37.0 | 36.9 | 36.9 | 37.0 |
| 11 | 20.5 | 20.3 | 20.3 | 21.1 | 20.5 | 21.2 | 20.5 | 20.6 | 21.0 |
| 12 | 31.4 | 31.8 | 31.8 | 39.9 | 31.7 | 39.6 | 39.2 | 38.1 | 34.9 |
| 13 | 45.9 | 44.9 | 44.9 | 41.2 | 47.8 | 43.4 | 40.3 | 42.3 | 46.0 |
| 14 | 84.6 | 86.3 | 86.3 | 56.2 | 84.7 | 54.9 | 56.8 | 54.0 | 56.2 |
| 15 | 41.9 | 39.6 | 39.6 | 35.4 | 42.4 | 34.4 | 33.4 | 35.5 | 32.2 |
| 16 | 82.2 | 82.3 | 82.3 | 81.6 | 85.1 | 84.4 | 84.2 | 74.7 | 144.5 |
| 17 | 58.3 | 59.2 | 59.4 | 63.3 | 61.5 | 64.5 | 67.8 | 66.6 | 155.0 |
| 18 | 20.4 | 19.9 | 20.0 | 16.4 | 17.7 | 14.1 | 13.5 | 13.8 | 15.7 |
| 19 | 17.1 | 19.2 | 19.3 | 18.8 | 19.3 | 19.3 | 19.3 | 19.4 | 19.0 |
| 20 | 42.5 | 42.4 | 42.1 | 40.7 | 103.9 | 103.5 | 76.7 | 205.5 | 196.1 |
| 21 | 15.2 | 14.9 | 15.1 | 16.4 | 11.9 | 11.8 | 21.8 | 30.5 | 26.9 |
| 22 | 110.0 | 111.5 | 111.9 | 110.6 | 152.2 | 152.4 | 163.0 | 173.2 | |
| 23 | 26.6 | 34.3 | 40.9 | 37.2 | 23.7 | 23.6 | 91.3 | 32.2 | |
| 24 | 26.2 | 72.8 | 81.5 | 28.3 | 31.4 | 32.3 | 29.6 | 29.0 | |
| 25 | 27.5 | 31.7 | 38.1 | 34.4 | 33.7 | 33.7 | 34.8 | 33.5 | |
| 26 | 65.0 | 64.2 | 65.0 | 75.4 | 75.2 | 75.4 | 75.3 | 74.7 | |
| 27 | 16.3 | 9.9 | 13.5 | 17.3 | 17.1 | 17.1 | 17.4 | 16.9 |
The 1H and 13C NMR spectral data of 5 (C56H90O29) suggest that 5 is a spirostanol glycoside resembling 2 in structure, with a lycotetrose unit at C-3 and a hydroxy group at C-14. However, the molecular formula of 5 was in excess of 2 by C6H10O6, which corresponded to a hexyloxy group. When the 1H and 13C NMR spectra of 5 were compared with those of 2, the signals for the C-24 methylene protons at δH 2.15 and 1.37 (each, m) and carbon at δC 26.2 were displaced by an oxymethine proton at δH 4.83 (ddd, J = 10.9, 5.5, 5.5 Hz) and carbon at δC 72.8. Acid hydrolysis of 5 gave d-galactose, d-glucose, and d-xylose as the sugar moieties. Analysis of the 1H-1H correlation spectroscopy (COSY) and heteronuclear multiple quantum coherence (HMQC) spectra allowed the hexosyl unit attached to C-24 to be assigned β-d-glucopyranosyl, and an HMBC correlation was observed from the anomeric proton at δH 5.04 (d, J = 7.7 Hz) to C-24 carbon at δC 72.8. In the NOESY spectrum of 5, NOE correlations between the signals of H-24 at δH 4.83 and H-25 at δH 2.24/H-26ax at δH 3.89, and J values of 3JH-24, H-23ax = 10.9 Hz, 3JH-24, H-23eq = 5.5 Hz, and 3JH-24, H-25 = 5.5 Hz provided evidence for the 24S and 25R configurations. Thus, the structure of 5 was determined to be (24S,25R)-24-[(β-d-glucopyranosyl)oxy]-14α-hydroxyspirost-5-en-3β-yl O-β-d-glucopyranosyl-(1→2)-O-[β-d-xylopyranosyl-(1→3)]-O-β-d-glucopyranosyl-(1→4)-β-d-galactopyranoside.
The molecular formula of 6 (C56H90O29) was the same as that of 5. The 1H and 13C NMR spectra of 6 showed close similarity with those of 5, suggestive of a stereoisomer with respect to the C-25 configuration. In the NOESY spectrum of 6, the NOE correlations between the signals of H-23ax at δH 2.00 and H-20 at δH 2.07/H-25 at δH 1.91, between the signals of H-26ax at δH 3.58 and H-16 at δH 5.05/H-24 at δH 4.06/Me-27 at δH 1.13, between the signals of Me-27 and H-24, and J values of 3JH-24, H-23ax = 12.4 Hz, 3JH-24, H-23eq = 5.3 Hz, and 3JH-25, H-26ax = 11.8 Hz confirmed the 24S and 25S configurations. The structure of 6 was characterized as (24S,25S)-24-[(β-d-glucopyranosyl)oxy]-14α-hydroxyspirost-5-en-3β-yl O-β-d-glucopyranosyl-(1→2)-O-[β-d-xylopyranosyl-(1→3)]-O-β-d-glucopyranosyl-(1→4)-β-d-galactopyranoside.
The hemi-acetal carbon signal at δC 110.6 and the positive color reaction in Ehrlich’s test suggest that 10 (C56H92O29) may be a 22-hydroxyfurostanol glycoside. The 1H and 13C NMR spectra of 10 were similar to those of 8; however, the molecular formula of 10 had one extra oxygen atom compared to that of 8, and a significant difference was observed in the 13C NMR signals from the B-ring portion. Enzymatic hydrolysis of 10 with β-d-glucosidase yielded the corresponding spirostanol saponin 10a. In the 1H-1H COSY spectrum of 10a, a proton signal of a hydroxy group at δH 5.73 (br d, J = 7.8 Hz), which disappeared following the addition of HCl vapor, showed a correlation peak with the C-7 oxymethine proton at δH 4.01 (m), indicating the presence of a hydroxy group at C-7. The NOE correlations between the signals of H-7 at δH 4.00 and H-9 at δH 1.05/H-12ax at δH 1.68/H-14 at δH 1.32 in the NOESY spectrum of 10a confirmed that the C-7 hydroxy group was β-oriented. Furthermore, the NOE correlations between the signals of Me-27 at δH 1.07 and H-23ax at δH 1.90 and between the signals of H-25 at δH 1.57 and H-24ax at δH 2.14 confirmed the 25S configuration. Thus, 10a was assigned as (25S)-7β-hydroxyspirost-5-en-3β-yl O-β-d-glucopyranosyl-(1→2)-O-[β-d-xylopyranosyl-(1→3)]-O-β-d-glucopyranosyl-(1→4)-β-d-galactopyranoside. The linkage of a β-d-glucopyranosyl group (Glc (III)) to the C-26 hydroxy group of the aglycone of 10 was ascertained by an HMBC correlation from H-1′′′′′ of Glc (III) at δH 4.80 (d, J = 7.7 Hz) to C-26 of the aglycone at δC 75.4. The NOE correlations between the signals of H-20 at δH 2.24 and H2-23 at δH 2.08 and 1.96 confirmed the C-22α configuration. Therefore, 10 was identified as (25S)-26-[(β-d-glucopyranosyl)oxy]-7β,22α-dihydroxyfurost-5-en-3β-yl O-β-d-glucopyranosyl-(1→2)-O-[β-d-xylopyranosyl-(1→3)]-O-β-d-glucopyranosyl-(1→4)-β-d-galactopyranoside.
The 1H and 13C NMR spectra of 11 (C56H90O28) showed similar features with those of 9; however, the molecular formula of 11 was smaller than that of 9 by H2O, and the signals assignable to H-17 and Me-21 were observed at δH 3.36 as a doublet (J = 9.7 Hz) and δH 1.67 as a singlet, respectively. Furthermore, signals for a pair of olefinic carbon were detected at δC 152.2 and 103.9, in addition to those attributable to C-5 and C-6. These data implied that 11 was the corresponding Δ20(22)-pseudo-furostanol glycoside of 9. This was confirmed by the fact that the peracetate (11a) of 11 was the same as the product obtained by treating 9 with Ac2O in pyridine at 110 °C for 3 h. Enzymatic hydrolysis of 11 with β-d-glucosidase gave the corresponding spirostanol saponin 2 and d-glucose. Accordingly, the structure of 11 was assigned (25S)-26-[(β-d-glucopyranosyl)oxy]-14α-hydroxyfurosta-5,20(22)-dien-3β-yl O-β-d-glucopyranosyl-(1→2)-O-[β-d-xylopyranosyl-(1→3)]-O-β-d-glucopyranosyl-(1→4)-β-d-galactopyranoside.
Compound 12 (C56H90O27) appeared to be the corresponding Δ20(22)-pseudo-furostanol glycoside of 8 (C56H92O28), based on the characteristic proton signals of the H-17 doublet at δH 2.44 (J = 9.9 Hz) and Me-21 singlet at δH 1.62, as well as the olefinic carbon signals at δC 152.4 and 103.5. Enzymatic hydrolysis of 12 with β-d-glucosidase gave the corresponding spirostanol saponin 1 and D-glucose. Thus, the structure of 12 was characterized as (25S)-26-[(β-d-glucopyranosyl)oxy]-furosta-5,20(22)-dien-3β-yl O-β-d-glucopyranosyl-(1→2)-O-[β-d-xylopyranosyl-(1→3)]-O-β-d-glucopyranosyl-(1→4)-β-d-galactopyranoside.
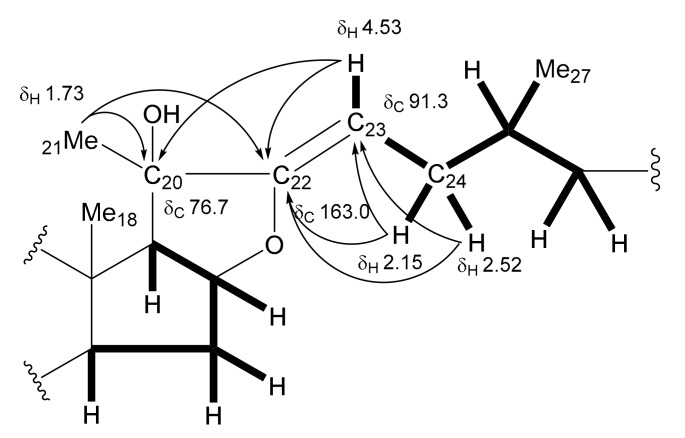
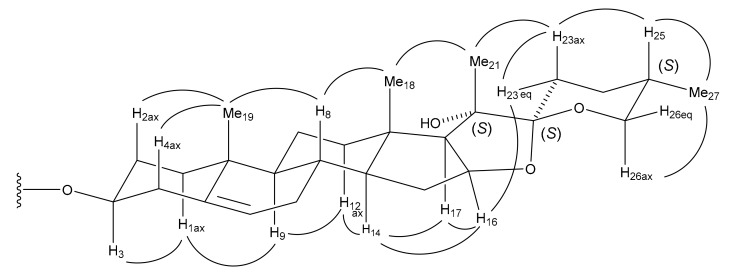
The 1H and 13C NMR spectral data for 13 (C56H90O28) suggest that it is a furostanol glycoside, having a structure similar to that of 8. However, significant differences were recognized in the signals from the ring E and the side chain moiety, where a tertiary hydroxy group [δC 76.7 (C)] and an oxygen-bearing trisubstituted olefinic group [δC 163.0 (-O-C(C)=CH-); δC 91.3 (-O-C(C)=CH-)/δH 4.53 (br d, J = 13.8 Hz)] were supposed to be located. The deshielded methyl singlet signal at δH 1.73 assignable to Me-21 and the olefinic proton at δH 4.53 exhibited long-range correlations with the quaternary carbon at δC 76.7 and the oxygen-bearing olefinic carbon at δC 163.0 in the HMBC spectrum of 13. The olefinic proton at δH 4.53 was shown to be coupled with the methylene protons at δH 2.52 and 2.15 (each, m) attributable to H2-24 in the 1H-1H COSY spectrum. These correlations allowed the tertiary hydroxy group and trisubstituted olefinic group to be placed at C-20 and between C-22 and C-23, respectively. The other correlations supporting the partial structure are depicted in Figure 2. The geometry of the olefinic group was determined to be Z by an NOE correlation between the signals of Me-21 at δH 1.73 and H-23 at δH 4.53. Enzymatic hydrolysis of 13 with β-d-glucosidase yielded the spirostanol saponin 13a and D-glucose. The NOE correlations between the signals of Me-21 at δH 1.73 and Me-18 at δH 1.19/H-23ax at δH 1.79, between the signals of H-23eq at δH 2.37 and H-16 at δH 5.06, and between the signals of H-25 at δH 1.69 and H-23ax/Me-27 at δH 0.75 in the NOESY spectrum of 13a, in addition to the large J value of the H-26ax proton (3JH-26ax, H-25 = 10.0 Hz), were indicative of the 20S, 22S, and 25S configurations (Figure 3). Thus, spirostanol saponin 13a was identified as (20S,22S,25S)-20-hydroxyspirost-5-en-3β-yl O-β-d-glucopyranosyl-(1→2)-O-[β-d-xylopyranosyl-(1→3)]-O-β-d-glucopyranosyl-(1→4)-β-d-galactopyranoside. An HMBC correlation from H-1′′′′′ of Glc (III) at δH 4.85 (d, J = 7.7 Hz) to C-26 of the aglycone at δC 75.3 in the HMBC spectrum of 13 confirmed that a β-d-glucopyranosyl group was attached to C-26. Therefore, the structure of 13 was elucidated to be (20S,22Z,25S)-26-[(β-d-glucopyranosyl)oxy]-20-hydroxyfurosta-5,22-dien-3β-yl O-β-d-glucopyranosyl-(1→2)-O-[β-d-xylopyranosyl-(1→3)]-O-β-d-glucopyranosyl-(1→4)-β-d-galactopyranoside.
Figure 2.
Heteronuclear multiple-bond correlation (HMBC) correlations of 13. Bold lines indicate the 1H-1H couplings and arrows indicate 1H/13C long-range correlations.
Figure 3.
Key Nuclear Overhauser effect (NOE) correlations of 13a.
The 1H and 13C NMR data for 14 (C56H90O29) were essentially analogous to those of 12, except for the lack of signals for the C-20(22)-tetrasubstituted olefinic group at δC 103.5 and 152.4. Instead, signals for a keto carbonyl carbon at δC 205.5 and an ester carbonyl carbon at δC 173.2 were newly observed in the 13C NMR spectrum of 14. All other signals appeared at almost the same positions between the two glycosides. In the HMBC spectrum of 14, the H-17 at δH 2.48 (d, J = 7.6 Hz) and Me-21 protons at δH 2.13 (s) showed long-range correlations with the keto carbonyl carbon at δC 205.5, which was assigned to C-20. Long-range correlations from the H-16 at δH 5.66 (m) and H2-23 protons at δH 1.84 (m) and 1.47 (m) to the ester carbonyl carbon at δC 173.2 resulted in the assignment of the ester carbonyl carbon to C-22. These data suggest that 14 was formed from 12 through the oxidative cleavage of the C-20(22) double bond. This was confirmed by the fact that the peracetate (14a) of 14 was identical to the product obtained by treating 12 with Ac2O in pyridine at room temperature for 12 h and then with CrO3 in AcOH. Accordingly, the structure of 14 was determined to be 16β-[[(4S)-5-(β-d-glucopyranosyloxy)-4-methyl-1-oxo-pentyl]oxy]-3β-[(O-β-d-glucopyranosyl-(1→2)-O-[β-d-xylopyranosyl-(1→3)]-O-β-d-glucopyranosyl-(1→4)-β-d-galactopyranosyl)oxy]-pregn-5-en-20-one.
The data suggest that 15 (C44H68O21) is a pregnane glycoside. Its 1H NMR spectrum showed signals for two angular methyl groups at δH 0.90 (s, Me-18) and 0.86 (s, Me-19), a methyl group of an acetyl moiety at δH 2.22 (s, Me-21), two olefinic protons at δH 6.57 (br s, H-16) and 5.29 (br d, J = 5.4 Hz, H-6), and four anomeric protons at δH 5.58 (d, J = 7.8 Hz, H-1′′′), 5.23 (d, J = 7.8 Hz, H-1′′′′), 5.18 (d, J = 7.8 Hz, H-1′′), and 4.88 (d, J = 7.8 Hz, H-1′). The existence of an α,β-unsaturated carbonyl group was verified by the IR (1660 cm−1), UV [239 nm (logε 3.81)], and 13C NMR [δC 196.1 (C=O), 155.0 (C), and 144.5 (CH)] spectra. These spectral data and comparison with those of previously reported compounds identified the aglycone of 15 as 3β-hydroxypregna-5,16-dien-20-one [10]. Acid hydrolysis of 15 and analysis of HMBC correlations provided evidence that a lycotetrose was attached to C-3 of the aglycone. Thus, the structure of 15 was assigned 3β-[(O-β-d-glucopyranosyl-(1→2)-O-[β-d-xylopyranosyl-(1→3)]-O-β-d-glucopyranosyl-(1→4)-β-d-galactopyranosyl)oxy]-pregna-5,16-dien-20-one.
2.2. Cytotoxic Activity
The isolated compounds 1–15 were evaluated for their cytotoxic activity against HL-60 human promyelocytic leukemia cells, A549 human lung adenocarcinoma cells, HSC-4, and HSC-2 human oral squamous cell carcinoma cells. Etoposide, cisplatin, and doxorubicin were used as positive controls. Compound 1 showed cytotoxic activity against the four tumor cell lines with IC50 values ranging from 0.96 ± 0.01 to 3.15 ± 0.43 μM. Compound 8, which is the corresponding furostanol glycoside of 1, was only cytotoxic to the adherent cell lines of A549, HSC-4, and HSC-2 cells, with IC50 values of 2.97 ± 0.06 μM, 11.04 ± 0.25 μM, and 8.25 ± 0.20 μM, respectively (Table 3). As we previously reported [11,12], the cytotoxicity of 1 compared to those of 2, 4, 5, and 6 and of 8 compared to those of 9 and 10 indicated that the introduction of polar substituents to the steroidal nuclei resulted in reduced the cytotoxicity. Compound 1 was cytotoxic to tumor cells, whereas 3, having the diglycoside did not show cytotoxic activity. These results implied that not only the structures of the aglycone moiety but also the sugar sequences in the steroidal glycosides considerably contributed to the appearance of cytotoxicity. In previous study, furostanol and pseudo-furostanol glycosides were generally inactive, and on the other hand, 3-O-lycotetroside showed significant cytotoxic activities [13]. In this study, correlation between the structure and cytotoxicity of steroidal glycosides have provided similar results. Isolated compounds were evaluated for their cytotoxic activity against TIG-3 normal human diploid fibroblasts. As a result, unfortunately, 1 and 8 gave IC50 values of 1.30 ± 0.72 and 3.27 ± 0.13 µM, respectively. However, cisplatin, a clinically used anticancer agent, was also toxic to healthy cells TIG-3 with IC50 value of 7.64 ± 0.08 μM in vitro evaluation.
Table 3.
Cytotoxic activity of 1–15 against HL-60, A549, HSC-4, and HSC-2 cells a,b.
| Compound | IC50 (µM) | |||
|---|---|---|---|---|
| HL-60 | A549 | HSC-4 | HSC-2 | |
| 1 | 2.87 ± 0.15 | 2.54 ± 0.25 | 3.15 ± 0.43 | 0.96 ± 0.01 |
| 8 | >12 | 2.97 ± 0.06 | 11.04 ± 0.25 | 8.25 ± 0.20 |
| doxorubicin | ― | ― | 0.05 ± 0.01 | 0.11 ± 0.01 |
| etoposide | 0.40 ± 0.01 | 0.72 ± 0.03 | ― | ― |
| cisplatin | 1.33 ± 0.03 | 2.56 ± 0.05 | 1.88 ± 0.06 | 2.17 ± 0.06 |
a Data are represented as the mean value ± standard error of the mean (SEM) of three experiments performed in triplicate. b Compounds 2–7 and 9–15 were inactive against tumor cells (IC50 > 12 μM).
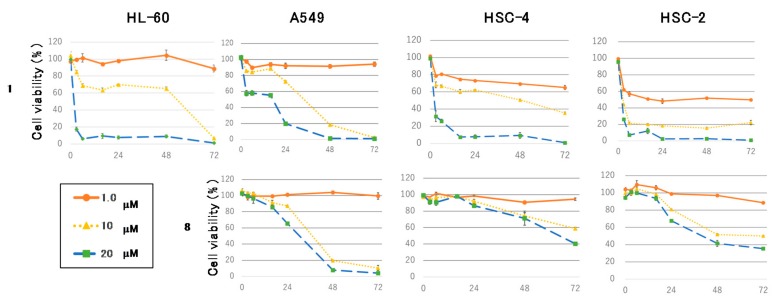
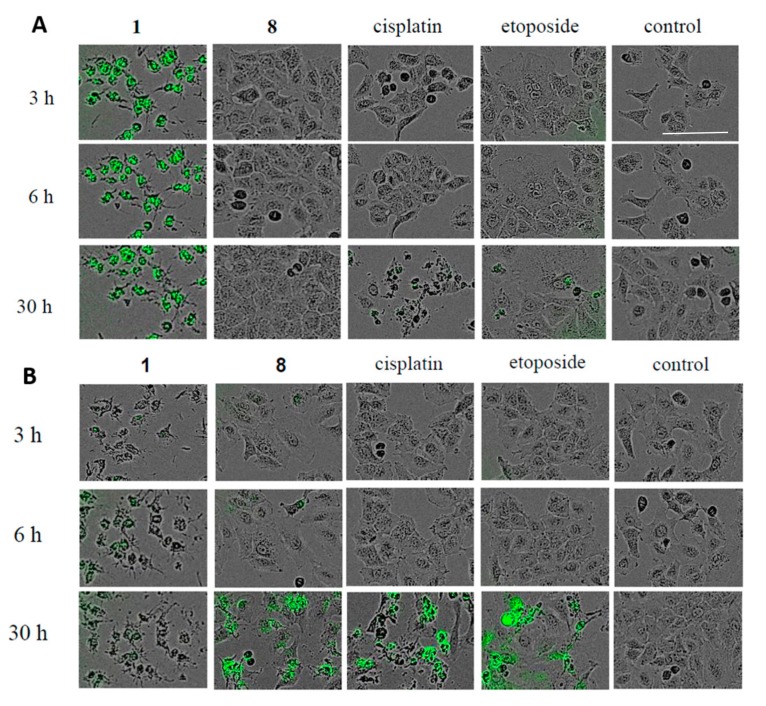
Figure 4 shows the time course of the antiproliferative effects of 1 or 8 at 1.0, 10, and 20 μM on HL-60, A549, HSC-4, and HSC-2 cells. Compound 1 decreased tumor cells viability in a dose-dependent manner within 16 h. In contrast, 8 reduced adherent cells viability in a time-dependent manner. HL-60, A549, HSC-4, and HSC-2 cells were exposed to 1 or 8 and then stained with 4′,6-diamidino-2-phenylindole dihydrochloride (DAPI). Treatment of the cells with 1 led to necrotic cell death in the four cell lines, whereas the cells exposed to 8 morphologically displayed nuclear chromatin condensation and nuclear disassembly, implying that 8 induced apoptosis in the adherent cells (Supplementary Material 1). Next, the activation of caspase-3/7 in A549 cells treated with 1 or 8 was evaluated using the Cell PlayerTM caspase-3/7 Apoptosis Assay kit. When A549 cells were treated with 20 μM of 8 for 30 h, a significant green fluorescence was detected (Figure 5A), indicating that 8 induced apoptotic cell death in A549 cells through the activation of caspase-3/7. Alternatively, phase-contrast and fluorescent images of the A549 cells treated with 20 μM of 1 followed by staining with oxazole yellow dimer (YOYO-1), which stains the nuclear DNA in permeabilized cells [14], showed that the cell membrane became permeable at an early stage of the necrotic cell death process (with green fluorescence) (Figure 5B). Finally, the cell cycle distribution of A549 cells treated with 8 for 30 h was analyzed using flow cytometry. Compound 8 increased the sub-G1 cell population from 4.07% to 14.10% in A549 cells (Table 4). These results suggest that 8 induced apoptotic death in A549 cells through caspase-3/7 activity in a time-dependent manner, whereas 1 induced necrotic cell death in A549 cells.
Figure 4.
Cell viability of HL-60, A549, HSC-4 and HSC-2 cells after treatment with 1.0, 10, and 20 μM of 1 and 8 for 0, 3, 6, 16, 24, 48, and 72 h. Data are represented as the mean value ± SEM of three experiments performed in triplicate.
Figure 5.
(A) Phase-contrast and fluorescent images of A549 cells stained with Cell PlayerTM 96-well caspase-3/7 Apoptosis Assay kit, showing morphological changes in response to 1 (20 μM), 8 (20 μM), cisplatin (33 μM), and etoposide (33 μM). (B) Phase-contrast and fluorescent images of A549 cells stained with YOYO-1, showing morphological changes in response to 1 (20 μM), 8 (20 μM), cisplatin (33 μM), and etoposide (33 μM). Scale bar = 100 μM.
Table 4.
Effects of 8 on cell cycle distribution of A549 cells a.
| % sub G0/G1 | % G0/G1 | % S | % G2-M | |
|---|---|---|---|---|
| control | 4.07 ± 2.00 | 80.43 ± 5.70 | 7.47 ± 2.02 | 6.17 ± 2.30 |
| 8 | 14.10 ± 1.33 | 71.63 ± 4.00 | 6.67 ± 2.17 | 5.30 ± 0.38 |
| etoposide | 31.15 ± 5.45 | 25.90 ± 2.40 | 24.10 ± 6.50 | 15.40 ± 1.20 |
| cisplatin | 40.90 ± 4.24 | 41.80 ± 4.72 | 11.90 ± 3.14 | 4.50 ± 0.61 |
The cell cycle distribution of A549 cells treated with 8 for 30 h was analyzed using flow cytometry. a Data are represented as the mean value ± SEM of three experiments performed in triplicate.
3. Experimental Section
3.1. General
Optical rotations were measured using a JASCO P-1030 (JASCO, Tokyo, Japan). UV spectra were recorded on a JASCO V-630. IR spectra were recorded on a JASCO FT-IR 410. 1H NMR spectra were recorded with a DRX-500 spectrometer (Bruker, Karlsruhe, Germany) using standard Bruker pulse programs at 300 K. Chemical shifts are given as δ values relative to tetramethylsilane (TMS), which was used as an internal standard. HR-ESI-TOF-MS data were recorded on the LCT mass spectrometer (Waters-Micromass, Manchester, UK). Diaion HP-20 (50 mesh; Mitsubishi-Chemical, Tokyo, Japan), BW-300 silica gel (200-300 mesh; Fuji Silysia Chemical, Kasugai, Japan), and ODS silica gel COSMOSIL 75C18-OPN (75 μM; Nacalai Tesque, Kyoto, Japan) were used for CC. TLC was carried out on precoated silica gel 60 F254 (0.25 mm thick; Merck, Darmstadt, Germany) and RP18 F254S plates (0.25 mm thick; Merck), and the spots were visualized by spraying the plates with 10% H2SO4 (aq) followed by heating. HPLC was performed using a system composed of a CCPM pump (Tosoh, Tokyo, Japan), a CCP PX-8010 controller (Tosoh), an RI-8010 detector (Tosoh), and a Rheodyne injection port (Rheodyne LLC, Rohnert Park, CA, USA). A TSK gel ODS-100Z column (10 mm i.d. × 250 mm, 5 μm; Tosoh) was employed for preparative HPLC. The purities of all the isolated compounds were confirmed by their 1H and 13C NMR spectra. The following materials and biochemical-grade reagents were used for the cell cultures and the cytotoxicity assays: a microplate reader (Spectra Classic, Tecan, Salzburg, Austria); a 96-well flat-bottom plate (Iwaki Glass, Funabashi, Japan); JCRB 0085 HL-60 and JCRB 0076 A549 cells (Human Science Research Resources Bank, Osaka, Japan); HSC-2 and HSC-4 cells (Riken Cell Bank, Tsukuba, Japan); fetal bovine serum (FBS; Nichirei Biosciences, Tokyo, Japan); 0.25% trypsin-ethylenediaminetetraacetic acid (EDTA) solution, RPMI-1640 medium, minimum essential medium (MEM), etoposide, Triton X-100, and 3-(4,5-dimethylthiazol-2-yl)-2,5-diphenyl-2H-tetrazolium bromide (MTT) (Sigma-Aldrich, St. Louis, MO, USA); penicillin G sodium salt and streptomycin sulfate (Gibco, Grand Island, NY, USA); paraformaldehyde and ribonuclease A (Wako Pure Chemical Industries, Osaka, Japan); and propidium iodide (PI) (Molecular Probes, Eugene, OR, USA).
3.2. Isolation and Structural Determination
3.2.1. Plant Material
C. majalis whole plants (dry weight, 3.0 kg) were obtained from Richters Co., Ltd. (Goodwood, ON, Canada). A voucher specimen was deposited at the Herbarium of the Tokyo University of Pharmacy and Life Sciences (KS-2011-004).
3.2.2. Extraction and Isolation
C. majalis whole plants (3.0 kg, dry weight) were extracted with MeOH under reflux for 4 h. After removing the solvent, the MeOH extract (460 g) was passed through a Diaion HP-20 column (2000 g, 8.5 cm i.d. × 60 cm) and successively eluted with MeOH-H2O (3:7, 6:4), MeOH, EtOH, and EtOAc (6 L of each eluent). CC of the MeOH-eluted fraction (100 g) on silica gel (2000 g, 8.0 cm i.d. × 40 cm), eluted with a stepwise gradient mixture of CHCl3-MeOH-H2O (9:1:0, 40:10:1, 20:10:1, 7:4:1) and finally with MeOH, provided 9 fractions (Frs. M-1 to M-9). Fr. M-3 was chromatographed on ODS silica gel (800 g, 6.0 cm i.d. × 25 cm) eluted with MeOH-H2O (1:1, 2:1, 3:1, 4:1) and finally with MeOH, providing 12 subfractions (Frs. M-3-1 to M-3-12). Fr. M-3-11 was separated by HPLC (1.0 cm i.d. × 25 cm) using MeCN-H2O (2:3) to isolate 3 (2.8 mg) and 7 (14.0 mg). Fr. M-6 was chromatographed on ODS silica gel (800 g, 6.0 cm i.d. × 30 cm) eluted with MeOH-H2O (2:1, 3:1, 4:1) to yield 1 (22.3 mg), 2 (68.7 mg), and 4 (5.1 mg). Fr. M-7 (94.0 mg) was separated by a silica gel column (1500 g, 6.0 cm i.d. × 30 cm) eluted with CHCl3-MeOH-H2O (7:4:1) and an ODS silica gel column (800 g, 6.0 cm i.d. × 28 cm) eluted with MeOH-H2O (3:2, 3:1) to yield 12 (13.7 mg), 13 (21.4 mg), 14 (26.4 mg), and 15 (14.5 mg). CC of the MeOH-H2O (6:4)-eluted fraction (40 g) on silica gel (1500 g, 7.5 cm i.d. × 40 cm), eluted with a stepwise gradient mixture of CHCl3-MeOH-H2O (7:4:1) and finally with MeOH, provided 8 fractions (Frs. MH-1 to MH-8). Fr. MH-7 (9.6 g) was chromatographed on ODS silica gel (800 g, 6.0 cm i.d. × 28 cm) eluted with MeOH-H2O (1:1, 2:1) and finally with MeOH, providing 9 subfractions (Frs. MH-7-1 to MH-7-9). Fr. MH-7-3 (487 mg) was separated by HPLC (1.0 cm i.d. × 25 cm) using MeCN-H2O (1:3) to isolate 5 (16.9 mg), 6 (11.9 mg), and 9 (98.3 mg). Fr. MH-7-6 (640 mg) was separated by ODS silica gel CC (500g, 4.0 cm i.d. × 30 cm) eluted with MeCN-H2O (1:4) to yield 8 (395 mg), 10 (20.9 mg), and 11 (20.3 mg).
3.2.3. Structural Characterization
Compound 4: An amorphous solid. [α]D25 –50.4 (c 0.25, MeOH). HR-ESI-TOF-MS m/z: 1085.4773 [M + Na]+ (calcd for C50H78NaO24: 1085.4781). IR νmax (film) cm−1: 3152 (OH), 2948 (CH), 1650 (C=O). UV λmax (MeOH) nm (log ε): 237 (3.86). For 1H and 13C NMR spectral data, see Supplementary Materials 1 and 2. For 1H and 13C NMR spectral data of the sugar moiety, see Table 1. For 13C-NMR spectral data of the aglycone moiety, see Table 2.
Compound 5: An amorphous solid. [α]D25 –64.6 (c 0.14, MeOH). HR-ESI-TOF-MS m/z: 1249.5472 [M + Na]+ (calcd for C56H90NaO29: 1249.5465). IR νmax (film) cm−1: 3420 (OH), 2930 (CH). For 1H and 13C NMR spectral data, see Supplementary Materials 1 and 2. For 13C-NMR spectral data of the aglycone moiety, see Table 2.
Compound 6: An amorphous solid. [α]D25 –38.2 (c 0.08, MeOH). HR-ESI-TOF-MS m/z: 1249.5477 [M + Na]+ (calcd for C56H90NaO29: 1249.5465). IR νmax (film) cm−1: 3365 (OH), 2929 (CH). For 1H and 13C NMR spectral data, see Supplementary Materials 1 and 2. For 13C-NMR spectral data of the aglycone moiety, see Table 2.
Compound 10: An amorphous solid. [α]D25 –49.4 (c 0.14, MeOH). HR-ESI-TOF-MS m/z: 1251.5625 [M + Na]+ (calcd for C56H92NaO29: 1251.5622). IR νmax (film) cm−1: 3398 (OH), 2874 (CH). For 1H and 13C NMR spectral data, see Supplementary Materials 1 and 2. For 13C-NMR spectral data of the aglycone moiety, see Table 2.
Compound 11: An amorphous solid. [α]D25 –29.1 (c 0.10, MeOH). HR-ESI-TOF-MS m/z: 1233.5516 [M + Na]+ (calcd for C56H90NaO28: 1233.5516). IR νmax (film) cm−1: 3397 (OH), 2924 (CH). For 1H and 13C NMR spectral data, see Supplementary Materials 1 and 2. For 13C-NMR spectral data of the aglycone moiety, see Table 2.
Compound 12: An amorphous solid. [α]D25 –33.3 (c 0.18, MeOH). HR-ESI-TOF-MS m/z: 1195.5739 [M + H]+ (calcd for C56H91O27: 1195.5748). IR νmax (film) cm−1: 3397 (OH), 2935 (CH). For 1H and 13C NMR spectral data, see Supplementary Materials 1 and 2. For 13C-NMR spectral data of the aglycone moiety, see Table 2.
Compound 13: An amorphous solid. [α]D25 –38.1 (c 0.09, MeOH). HR-ESI-TOF-MS m/z: 1233.5531 [M + Na]+ (calcd for C56H90NaO28: 1233.5516). IR νmax (film) cm−1: 3366 (OH), 2931 and 2872 (CH). For 1H and 13C NMR spectral data, see Supplementary Materials 1 and 2. For 13C-NMR spectral data of the aglycone moiety, see Table 2.
Compound 14: An amorphous solid. [α]D25 –37.9 (c 0.18, MeOH). HR-ESI-TOF-MS m/z: 1249.5466 [M + Na]+ (calcd for C56H90NaO29: 1249.5465). IR νmax (film) cm−1: 3447, 3423, and 3400 (OH), 2923 and 2883 (CH), 1731 and 1707 (C=O). For 1H and 13C NMR spectral data, see Supplementary Materials 1 and 2. For 13C-NMR spectral data of the aglycone moiety, see Table 2.
Compound 15: An amorphous solid. [α]D25 –32.8 (c 0.10, MeOH). HR-ESI-TOF-MS m/z: 955.4127 [M + Na]+ (calcd for C44H68NaO21: 955.4151). IR νmax (film) cm−1: 3481 (OH), 2921 and 2889 (CH), 1660 (C=O). UV λmax (MeOH) nm (log ε): 239 (3.81). For 1H and 13C NMR spectral data, see Supplementary Materials 1 and 2. For 13C-NMR spectral data of the aglycone moiety, see Table 2.
Acid Hydrolysis of 4–6, 10a, 13a, and 15
A solution of 4 (3.0 mg) in 0.5 M HCl and dioxane-H2O (1:1; 2.0 mL) was heated at 95 °C for 2 h under an Ar atmosphere. The reaction mixture was neturalized by passing it through an Amberlite IRA-96SB column (16–50 mesh, 50 g, 1.5 cm i.d. × 15 cm; Organo, Tokyo, Japan). The mixture was then eluted through a Diaion HP-20 column (50 g, 1.5 cm i.d. × 15 cm) with MeOH-H2O (4:6) and EtOH-Me2CO (1:1). The sugar fraction was analyzed using HPLC under the following conditions: Capcell Pak NH2 UG80 column (4.6 mm i.d. × 25 cm, 5 μm; Shiseido, Tokyo, Japan); mobile phase of MeCN-H2O (85:15); detection by refractive index and optical rotation; and a flow rate of 1.0 mL/min. d-Glucose, d-galacose, and d-xylose were identified by comparing their retention times (tR) and optical rotation with those of authentic samples: d-galactose (12.66, positive optical rotation), d-glucose (14.34, positive optical rotation), and d-xylose (9.21, positive optical rotation). Compounds 5 (5.0 mg), 6 (3.5 mg), 10a (3.5 mg), 13a (3.0 mg), and 15 (2.0 mg) were independently subjected to acid hydrolysis as described for 4. HPLC analysis of the sugar fractions under the same conditions as in the case of 4 indicated the presence of d-galacose, d-glucose, and d-xylose.
Enzymatic Hydrolysis of 10, 11, 12, and 13
Compound 10 (10.0 mg) was treated with β-d-glucosidase (15.0 mg, EC 232-589-7; Sigma-Aldrich) in HOAc/NaOAc buffer (pH 5.0, 3.0 mL) at room temperature for 12 h. The reaction mixture was purified using CC on silica gel (100 g, 2 cm i.d. × 30 cm) eluted with CHCl3-MeOH-H2O (20:10:1) to obtain gitogenin (10a, 4.2 mg) and a sugar fraction. HPLC analysis of the sugar fraction under the same conditions as those used for 4 indicated the presence of d-glucose (14.28, positive optical rotation). Compounds 11 (5.0 mg), 12 (5.0 mg), and 13 (7.0 mg) were independently subjected to enzymatic hydrolysis as described for 10 to obtain 2 (2.0 mg; from 11), 1 (1.0 mg; from 12), 13a (3.3 mg; from 13), and sugar fractions. HPLC analysis of the sugar fractions under the same conditions as those used for 4 indicated the presence of d-glucose (11: 13.98, positive optical rotation; 12: 14.07, positive optical rotation; 13: 14.11, positive optical rotation).
Acetylation of 11 and 14
Compounds 11 (2.0 mg) and 14 (3.6 mg) were independently acetylated with Ac2O (1.0 mL) in pyridine (1.0 mL) at room temperature for 18 h. Each crude acetate was purified using preparative TLC with hexane-Me2CO (1:1) to yield 11a (1.9 mg) and 14a (2.8 mg).
Preparation of 11a from 9
A mixture of 9 (15.0 mg) and Ac2O (5.0 mL) in pyridine (3.0 mL) was stirred at 110 °C for 3 h. After the excess Ac2O was decomposed by H2O (10 mL), the reaction mixture was purified using preparative TLC with hexane-Me2CO (7:5) to yield 11a (4.5 mg).
Preparation of 14a from 12
Compound 12 (5.0 mg) was treated with Ac2O (2.0 mL) in pyridine (4.0 mL) at room temperature for 12 h. After the excess Ac2O was decomposed by H2O (10 mL), the reaction mixture was evaporated to dryness. The crude product was dissolved in 95% AcOH (aq.) (5.0 mL), to which the CrO3 (15.0 mg) solution in 95% AcOH (aq) (1.0 mL) was added, and the mixture was stirred at room temperature for 3 h. After the excess CrO3 was decomposed by MeOH (3.0 mL), the crude product was diluted with H2O (20.0 mL) and extracted with Et2O (20.0 mL × 3). The Et2O extract was purified using ODS Si CC (100 g, 3 cm i.d. × 25 cm) eluted with MeCN-H2O (3:1) to obtain 14a (8.2 mg).
Compound 10a: An amorphous solid. [α]D25 –21.7 (c 0.09, MeOH). HR-ESI-TOF-MS m/z: 1071.4994 [M + Na]+ (calcd for C50H80NaO23: 1071.4988). IR νmax (film) cm−1: 3335 (OH), 2928 (CH). For 1H and 13C NMR spectral data, see Supplementary Materials 1 and 2.
Compound 11a: An amorphous solid. [α]D25 –14.3 (c 0.05, MeOH). HR-ESI-TOF-MS m/z: 1925.7460 [M + H]+ (calcd for C90H125O45: 1925.7493). IR νmax (film) cm−1: 2924 and 2854 (CH), 1748 (C=O). 1H NMR (500 MHz, C5D5N): δH 5.65 (1H, br d, J = 1.9 Hz, H-6), 3.97 (1H, dd, J = 9.5, 5.3 Hz, H-26a), 3.70 (1H, m, W1/2 = 29.1 Hz, H-3), 3.45 (1H, dd, J = 9.5, 6.3 Hz, H-26b), 2.38, 2.34, 2.32, 2.16 × 2, 2.14, 2.13 × 2, 2.07, 2.04 × 2, 2.03, 2.01, 1.97 × 2, 1.96 (each 3H, s, Ac × 16), 1.69 (3H, s, Me-21), 1.15 (3H, s, Me-18 or Me-19), 0.98 (3H, d, J = 6.6 Hz, Me-27), 0.97 (3H, s, Me-18 or Me-19).
Compound 13a: An amorphous solid. [α]D25 –22.5 (c 0.15, MeOH). HR-ESI-TOF-MS m/z: 1071.4935 [M + Na]+ (calcd for C50H80NaO23: 1071.4988). IR νmax (film) cm−1: 3328 (OH), 2926 (CH). For 1H and 13C NMR spectral data, see Supplementary Materials 1 and 2.
Compound 14a: An amorphous solid. [α]D25 –21.2 (c 0.14, MeOH). HR-ESI-TOF-MS m/z: 1921.7156 [M + Na]+ (calcd for C88H122NaO45: 1921.7156). IR νmax (film) cm−1: 2936 (CH), 1750 (C=O). 1H NMR (500 MHz, C5D5N): δH 3.72 (1H, m, W1/2 = 28.0 Hz, H-3), 3.40 (1H, dd, J = 9.9, 6.4, H-26a), 2.39, 2.35, 2.33 (each 3H, s, Ac × 3), 2.17―2.13 (Ac × 5), 2.07 × 2, 2.05, 2.03, 2.02, 1.98, 1.97, 1.96 (each 3H, s, Ac × 8), 1.23 (3H, s, Me-18 or Me-19), 1.04 (3H, s, Me-18 or Me-19), 0.90 (3H, d, J = 6.5 Hz, Me-27).
3.3. Biological Activity
3.3.1. Cell culture and Cytotoxicity Assays
HL-60 cells were maintained in an RPMI-1640 medium, A549 and TIG-3 cells were maintained in MEM, and HSC-4 and HSC-2 cells were maintained in DMEM. The cell media contained heat-inactivated 10% (v/v) FBS supplemented with L-glutamine, penicillin G sodium salt (100 units/mL), and streptomycin sulfate (100 μg/mL). HL-60 (4 × 104 cells/mL), A549 (1 × 104 cells/mL), HSC-4 (2 × 104 cells/mL), HSC-2 (1 × 104 cells/mL), and TIG-3 (5 × 104 cells/mL) cells were continuously treated with each compound for 72 h, and cell growth was measured using an MTT reduction assay as previously described [15]. Data are represented as mean ± standard error of the mean (SEM) of three experiments performed in triplicate. The concentration, up to 12 μM, resulting in a 50% inhibition value (IC50) was calculated from the dose response curve. The time courses for 0, 3, 6, 16, 24, 48, and 72 h of the antiproliferative effects of 1 and 8 at 1.0, 10, and 20 μM on HL-60, A549, HSC-4, and HSC-2 cells were also examined using the MTT assay.
3.3.2. Cell Morphology Analysis
HL-60 (2 × 104 cells/well), A549 (1 × 104 cells/well), HSC-4 (1 × 104 cells/well) and HSC-2 (4 × 103 cells/well) were independently plated on coverslips in 96-well plates. After 24 h, the cells were treated with either 1 (20 μM) for 3 h, 8 (20 μM) for 24 h, etoposide (15 μM), or cisplatin (33 μM). The cells were fixed with 1% glutaraldehyde for 30 min at room temperature and before staining with DAPI (0.5 μg/mL). They were observed using a CKX41 fluorescence microscope (Olympus, Tokyo, Japan). A549 cells (1 × 105 cells/well) were seeded in 96-well plates. After 24 h, the A549 cells were treated with either 1 (20 μM) or 8 (20 μM) and stained with caspase-3/7 assay reagent (1 μM) (Essen BioScience, Hertfordshire, UK) or YOYO-1 (1 μM; Invitrogen, Carlsbad, CA, USA). Cells images were obtained after 3, 6, and 30 h, respectively, using the IncuCyte® ZOOM (Essen BioScience, Hertfordshire, UK) in 5% CO2/air at 37 °C.
3.3.3. Cell Cycle Analysis by Flow Cytometry
Cell cycle distribution was evaluated using a flow cytometer (FACSCanto II, BD Bioscience, San Jose, CA, USA). A549 cells (1 × 105 cells/ml) were treated with 8 (20 μM) for 30 h, in separate experiments, according to a previously reported method [11]. The DNA contents of the tumor cells were analyzed, and percentages of cells in each phase were calculated.
4. Conclusions
Chromatographic separation of the MeOH extract of Convallaria majalis (Liliaceae) whole plants yielded 15 steroidal glycosides (1–15), including nine new compounds (4–6, 10–15) with a lycotetrose unit. The structures of the new compounds were determined using two-dimensional NMR analyses and chemical methods. The isolated compounds were evaluated for their cytotoxicity against HL-60, A549, HSC-4, and HSC-2 cells. Of these, the spirostanol lycotetroside (1) exhibited cytotoxic activity against HL-60, A549, HSC-4, and HSC-2 cells with IC50 values ranging from 0.96 to 3.15 μM. Alternatively, the corresponding furostanol lycotetroside (8) was cytotoxic to the adherent cell lines of A549, HSC-4, and HSC-2 cells with IC50 values of 2.97, 11.04, and 8.25 μM, respectively. Compound 1 caused necrotic cell death in A549 cells in a dose-dependent manner. In contrast, the corresponding furostanol lycotetroside (8) induced apoptotic cell death in A549 cells in a time-dependent manner, as was evident by morphological observations and flow cytometry analyses. This study demonstrated that 8 has induced apoptosis against A549 cells through caspase-3/7 and had the potential for anti-lung cancer agent. Further investigation of the pathway of apoptosis induced by 8 should be addressed.
Supplementary Materials
Supplementary materials can be found at http://www.mdpi.com/1422-0067/18/11/2358/s1.
Author Contributions
Yukiko Matsuo, Hiroshi Sakagami, and Yoshihiro Mimaki conceived and designed the experiments; Yukiko Matsuo, Daisuke Shinoda, Aina Nakamaru, and Kuni Kamohara performed the experiments and analyzed the data; Yukiko Matsuo and Yoshihiro Mimaki wrote the paper. All authors have read and approved the manuscript.
Conflicts of Interest
The authors declare no conflict of interest.
References
- 1.Chwedorzewska K.J., Galera H., Kosiński I. Plantations of Convallaria majalis L. as a threat to the natural stands of the species: Genetic variability of the cultivated plants and natural populations. Biol. Conserv. 2008;141:2619–2624. [Google Scholar]
- 2.Erdogan O.I., Gokbulut A. Adonis sp., Convallaria sp., Strophanthus sp., Thevetia sp., and Leonurus sp.—Cardiotonic plants with known traditional use and a few preclinical and clinical studies. Curr. Pharm. Des. 2017;23:1051–1059. doi: 10.2174/1381612822666161010104548. [DOI] [PubMed] [Google Scholar]
- 3.Higano T., Kuroda M., Jitsuno M., Mimaki Y. Polyhydroxylated steroidal saponins from the rhizomes of Convallaria majalis. Nat. Prod. Commun. 2007;2:531–536. [Google Scholar]
- 4.Higano T., Kuroda M., Sakagami H., Mimaki Y. Convallasaponin A, a new 5β-spirostanol triglycoside from the rhizomes of Convallaria majalis. Chem. Pharm. Bull. 2007;55:337–339. doi: 10.1248/cpb.55.337. [DOI] [PubMed] [Google Scholar]
- 5.Son K.H., Do J.C., Kang S.S. Steroidal saponins from the rhizomes of Polygonatum sibiricum. J. Nat. Prod. 1990;53:333–339. doi: 10.1021/np50068a010. [DOI] [PubMed] [Google Scholar]
- 6.Espejo O., Llavot J.C., Jung H., Giral F. Spirostanic diosgenin precursors from Dioscorea composita tubers. Phytochemistry. 1982;21:413–416. doi: 10.1016/S0031-9422(00)95278-6. [DOI] [Google Scholar]
- 7.Bogacheva N.G., Sheichenko V.I., Kogan L.M. Structure of yamogenin tetroside from Trigonella foenum graecum seeds. Pharm. Chem. J. 1977;11:65–69. [Google Scholar]
- 8.Yang Q., Yang C. Steroidal constituents of Aspidistra elatior from Yongshan, Yunnan. Acta Bot. Yunnanica. 2000;22:109–115. [Google Scholar]
- 9.Sugiyama M., Nakano K., Tomimatsu T., Nohara T. Five steroidal components from the rhizomes of Polygonatum odoratum var. pluriflorum. Chem. Pharm. Bull. 1984;32:1365–1372. doi: 10.1248/cpb.32.1365. [DOI] [Google Scholar]
- 10.Szendi Z., Forgo P., Sweet F. Complete 1H and 13C NMR spectra of pregnenolone. Steroids. 1995;60:442–446. doi: 10.1016/0039-128X(94)00047-G. [DOI] [PubMed] [Google Scholar]
- 11.Matsuo Y., Akagi N., Hashimoto C., Tachikawa F., Mimaki Y. Steroidal glycosides from the bulbs of Bessera elegans and their cytotoxic activities. Phytochemistry. 2013;96:244–256. doi: 10.1016/j.phytochem.2013.09.023. [DOI] [PubMed] [Google Scholar]
- 12.Matsuo Y., Watanabe K., Mimaki Y. New steroidal glycosides from rhizomes of Clintonia udensis. Biosci. Biotechnol. Biochem. 2008;72:1714–1721. doi: 10.1271/bbb.80003. [DOI] [PubMed] [Google Scholar]
- 13.Lee K.T., Choi J.H., Kim D.H., Son K.H., Kim W.B. Constitutents and the antitumor principle of Allium victorialis var. platyphyllum. Arch. Pharm. Res. 2001;24:44–50. doi: 10.1007/BF02976492. [DOI] [PubMed] [Google Scholar]
- 14.ESSEN BIOSCIENCE. [(accessed on 6 November 2017)]; Available online: https://www.essenbioscience.com/media/uploads/files/8000-0210-A00_Supplemental_data_CellPlayer_96-Well_Kinetic_Cytotoxicity.pdf.
- 15.Matsuo Y., Mimaki Y. Lignans from Santalum album and their cytotoxic activities. Chem. Pharm. Bull. 2010;58:587–590. doi: 10.1248/cpb.58.587. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.