Abstract
Although a founder variant of RNF213 4810G>A is a major genetic risk factor for moyamoya disease (MMD) in East Asians, the frequency and disease susceptibility of RNF213 variants remain largely unknown. This study investigated the mutation analysis of RNF213 (4448, 4810, 4863, and 4950) between Korean MMD and healthy controls. We performed a polymerase chain reaction-restriction fragment length polymorphism analysis. To identify the association between RNF213 gene polymorphisms and MMD disease, we performed statistical analyses such as multivariable logistic regression and Fisher’s exact test. Genetic data from 117 MMD patients were analyzed and compared with 253 healthy controls. We assessed and compared single nucleotide polymorphisms of RNF213 (4448, 4810, 4863, and 4950) between MMD and control groups. We performed genome-wide association studies to investigate the genetic pathophysiology of MMD. Among the RNF213 variants (4448G>A, 4810G>A, 4863G>A, and 4950G>A), RNF213 4810G>A and 4950G>A variants were more frequent in MMD patients. In a subgroup analysis, the RNF213 4810G>A was more frequent in moyamoya disease, and the comparison with GG+AA genotype was also significantly different in moyamoya patients. These results confirm that RNF213 4810G>A and RNF213 4950G>A were more frequent in MMD patients. We have confirmed that RNF213 4810G>A and 4950G>A are strongly associated with Korean MMD in children and adults as well as for the ischemic and hemorrhagic types.
Keywords: moyamoya disease, single nucleotide polymorphism, genetic, stroke, cerebrovascular disease, RNF213
1. Introduction
Moyamoya disease (MMD) is a chronic cerebrovascular occlusive disorder that results in transient ischemia, cerebral infarcts, and hemorrhagic strokes [1,2,3,4,5]. MMD occurs worldwide, but its prevalence is highest in East Asian countries [5].
MMD has a bimodal age distribution for peak incidence, with peaks in children who are approximately five years of age and adults in their mid-forties [1,3,6,7,8]. Most children MMD patients develop transient ischemic attacks or cerebral infarctions [9], whereas adult MMD patients are more likely to have a hemorrhagic stroke [8,10,11]. This suggests some variant or impairment of the genetic sequence in the same disease. The majority of MMD cases are sporadic, even though familial MMD cases account for approximately 9–15% of all cases [12,13]. Genetic associations with loci on chromosome 3, 6, 8, 10, and 17 and a specific human leukocyte antigen (HLA) haplotype have been reported [14,15,16,17,18,19,20], but questions about genetic penetrations remain.
RNF213, which is located on chromosome 17q25, has been recognized as the major susceptibility gene for MMD in East Asians [21,22], as well as Caucasians and East/South Asians [22,23,24,25]. An association of the p.R4810K polymorphism with intracranial major artery stenosis/occlusion has been reported in Japanese and Korea populations [26,27,28,29]. However, this genetic variant associated with MMD was also observed in patients with non-MMD intracranial stenosis [26,28,29]. The association between RNF213 and clinical manifestations of MMD in young children and adults remain uncertain.
The RNF213 variant p.R4810K (c.14429G>A, rs112735431) was first reported to have a high level of association with MMD on a large scale [30]. The RNF213 variants R4859K [21] and R4810K [22] correspond to rs112735431, while R4859K is based on the computer predicted open-reading frame in the database [21].
In the present study, four single nucleotide polymorphisms (SNPs) in exon region were identified through previous studies [21,22,29] and a database search: RNF213 4448G>A, RNF213 4810G>A, RNF213 4863G>A, and RNF213 4950G>A. These four SNPs were selected on the basis of information in the HapMap database (http://hapmap.ncbi.nlm.nih.gov/). Currently, no study has addressed the frequency of RNF213 variants (4448, 4810, 4863, 4950) in a Korean population with a high prevalence of MMD. Therefore, we have investigated the frequency of MMD-related RNF213 variants in a cohort of Korean patients with MMD compared with healthy controls.
2. Results
2.1. Genetic Analysis
Table 1 shows the demographic characteristics for patients with MMD in the present study. A comparison of genotype frequencies between moyamoya patients and control subjects of the RNF213 4448G>A, RNF213 4810G>A, RNF213 4863G>A, and RNF213 4950G>A polymorphisms is shown in Table 2. There were statistically significant differences between moyamoya patients and controls in RNF213 4810G>A and RNF213 4950G>A (Table 2).
Table 1.
Demographic characteristics of moyamoya disease patients and subjects.
| Characteristics | Controls (n = 253) | Cases (n = 117) | p |
|---|---|---|---|
| Age (mean ± S.D.) | 25.60 ± 16.98 | 23.20 ± 17.75 | 0.213 |
| Age ≥ 18 | 37.69 ± 10.40 (151) | 37.70 ± 12.81 (60) | 0.995 |
| Age < 18 | 7.71 ± 4.05 (102) | 7.93 ± 4.11 (57) | 0.740 |
| Sex (F:M) | 145:108 | 77:40 | 0.151 |
| Clinical findings | - | ||
| Transient ischemic attack | - | 52 | |
| Cerebral infarction | - | 50 | |
| Cerebral hemorrhage | - | 17 | |
| Surgery | - | 65 | |
| Suzuki stage | - | ||
| Right | - | 3.17 ± 0.78 | |
| Left | - | 3.89 ± 1.27 |
p-Values were calculated by a two-sided t-test for continuous variables and a χ-square test for categorical data. Abbreviation: S.D., standard deviation. F, female; M, male.
Table 2.
Genotype frequencies of RNF213 in moyamoya disease (MMD) patients and control subjects.
| Genotype | Controls, n = 253 (%) | MMD, n = 117 (%) | COR (95% CI) | p | FDR-P | AOR (95% CI) * | p | FDR-P |
|---|---|---|---|---|---|---|---|---|
| RNF213 4448G>A | ||||||||
| GG | 219 (86.6) | 109 (93.2) | 1.000 (reference) | 1.000 (reference) | ||||
| GA | 33 (13.0) | 8 (6.8) | 0.487 (0.218–1.090) | 0.080 | 0.107 | 0.472 (0.210–1.061) | 0.069 | 0.092 |
| AA | 1 (0.4) | 0 | N/A | N/A | N/A | N/A | ||
| Dominant (GG vs. GA+AA) | 0.473 (0.212–1.056) | 0.068 | 0.091 | 0.463 (0.207–1.039) | 0.062 | 0.083 | ||
| HWE P | 0.837 | 0.702 | ||||||
| RNF213 4810G>A | ||||||||
| GG | 234 (92.5) | 14 (12.0) | 1.000 (reference) | 1.000 (reference) | ||||
| GA | 19 (7.5) | 102 (87.2) | 89.73 (43.31–185.9) | <0.001 | 0.004 | 94.43 (44.45–200.6) | <0.001 | 0.004 |
| AA | 0 | 1 (0.9) | N/A | N/A | N/A | N/A | ||
| Dominant (GG vs. GA+AA) | 90.61 (43.74–187.7) | <0.001 | 0.004 | 95.34 (45.45–192.8) | <0.001 | 0.004 | ||
| HWE P | 0.535 | 0 | ||||||
| RNF213 4863G>A | ||||||||
| GG | 223 (88.1) | 108 (92.3) | 1.000 (reference) | 1.000 (reference) | ||||
| GA | 30 (11.9) | 9 (7.7) | 0.619 (0.284–1.351) | 0.229 | 0.229 | 0.574 (0.258–1.278) | 0.174 | 0.174 |
| AA | 0 | 0 | N/A | N/A | N/A | N/A | ||
| Dominant (GG vs. GA+AA) | 0.619 (0.284–1.351) | 0.229 | 0.229 | 0.574 (0.258–1.278) | 0.174 | 0.174 | ||
| HWE P | 0.316 | 0.665 | ||||||
| RNF213 4950G>A | ||||||||
| GG | 224 (88.5) | 91 (77.8) | 1.000 (reference) | 1.000 (reference) | ||||
| GA | 29 (11.5) | 26 (22.2) | 2.207 (1.232–3.952) | 0.008 | 0.016 | 2.194 (1.216–3.958) | 0.009 | 0.018 |
| AA | 0 | 0 | N/A | N/A | N/A | N/A | ||
| Dominant (GG vs. GA+AA) | 2.207 (1.232–3.952) | 0.008 | 0.016 | 2.194 (1.216–3.958) | 0.009 | 0.018 | ||
| HWE P | 0.334 | 0.176 | ||||||
* Adjusted by age and gender. MMD, moyamoya disease. Abbreviation: MMD, moyamoya disease; COR, crude odd ratio; CI, confidence interval, FDR, false discovery rate; AOR, adjusted odd ratio; HWE, Hardy-Weinberg equilibrium; N/A, not application.
In subgroup analyses (Table 3 and Table 4), the GA genotype of the RNF213 4810GA was more frequent in moyamoya disease (p < 0.001; GG vs. AA) and the comparison with the GG+AA genotype was also significantly different in moyamoya patients. The RNF213 4950G>A genotype was more frequent in moyamoya disease (p = 0.008, GG vs. AA), with high frequencies of the RNF213 4950G>A polymorphisms in MMD.
Table 3.
Genotype frequencies of RNF213 in moyamoya patient subtypes and control subjects.
| Genotype | Controls, n = 253 (%) | Ischemic Moyamoya, n = 100 (%) | AOR (95% CI) * | p | FDR-P | Hemorrhagic Moyamoya, n = 17 (%) | AOR (95% CI) * | p | FDR-P |
|---|---|---|---|---|---|---|---|---|---|
| RNF213 4448G>A | |||||||||
| GG | 219 (86.6) | 93 (93.0) | 1.000 (reference) | 16 (94.1) | 1.000 (reference) | ||||
| GA | 33 (13.0) | 7 (7.0) | 0.479 (0.204–1.129) | 0.093 | 0.124 | 1 (5.9) | 0.433 (0.055–3.429) | 0.428 | 0.428 |
| AA | 1 (0.4) | 0 | N/A | N/A | 0 | N/A | N/A | ||
| Dominant (GG vs. GA+AA) | 0.472 (0.201–1.110) | 0.085 | 0.113 | 0.415 (0.053–3.278) | 0.404 | 0.404 | |||
| RNF213 4810G>A | |||||||||
| GG | 234 (92.5) | 10 (10.0) | 1.000 (reference) | 4 (23.5) | 1.000 (reference) | ||||
| GA | 19 (7.5) | 89 (89.0) | 111.8 (49.07–254.8) | <0.001 | 0.004 | 13 (76.5) | 74.51 (16.57–335.0) | <0.001 | 0.003 |
| AA | 0 | 1 (1.0) | N/A | N/A | 0 | N/A | N/A | ||
| Dominant (GG vs. GA+AA) | 113.0 (49.60–257.4) | <0.001 | 0.004 | 74.51 (16.57–335.0) | <0.001 | 0.003 | |||
| RNF213 4863G>A | |||||||||
| GG | 223 (88.1) | 91 (91.0) | 1.000 (reference) | 17 (100.0) | 1.000 (reference) | ||||
| GA | 30 (11.9) | 9 (9.0) | 0.632 (0.282–1.414) | 0.264 | 0.264 | 0 | N/A | N/A | |
| AA | 0 | 0 | N/A | N/A | 0 | N/A | N/A | ||
| Dominant (GG vs. GA+AA) | 0.632 (0.282–1.414) | 0.264 | 0.264 | N/A | N/A | ||||
| RNF213 4950G>A | |||||||||
| GG | 224 (88.5) | 78 (78.0) | 1.000 (reference) | 13 (76.5) | 1.000 (reference) | ||||
| GA | 29 (11.5) | 22 (22.0) | 2.234 (1.199–4.163) | 0.011 | 0.022 | 4 (23.5) | 2.168 (0.649–7.244) | 0.209 | 0.314 |
| AA | 0 | 0 | N/A | N/A | 0 | N/A | N/A | ||
| Dominant (GG vs. GA+AA) | 2.234 (1.19–4.163) | 0.011 | 0.022 | 2.168 (0.649–7.244) | 0.209 | 0.314 | |||
* Adjusted by age and gender. MMD, moyamoya disease. Abbreviation: AOR, adjusted odd ratio; CI, confidence interval, FDR, false discovery rate; N/A, not application.
Table 4.
The frequencies of the RNF213 polymorphisms according to age.
| Genotype | Age < 18 Years | Age ≥ 18 Years | ||||||||
|---|---|---|---|---|---|---|---|---|---|---|
| Controls, n = 102 (%) | Moyamoya, n = 57 (%) | AOR (95% CI) * | p | FDR-P | Controls, n = 151 (%) | Moyamoya, n = 60 (%) | AOR (95% CI) * | p | FDR-P | |
| RNF213 4448G>A | ||||||||||
| GG | 89 (87.3) | 51 (89.5) | 1.000 (reference) | 130 (86.1) | 58 (96.7) | 1.000 (reference) | ||||
| GA | 13 (12.7) | 6 (10.5) | 0.800 (0.284–2.251) | 0.672 | 0.672 | 20 (13.2) | 2 (3.3) | 0.223 (0.050−0.986) | 0.048 | 0.064 |
| AA | 0 | 0 | N/A | N/A | 1 (0.7) | 0 | N/A | N/A | ||
| Dominant (GG vs. GA+AA) | 0.800 (0.284−2.251) | 0.672 | 0.672 | 0.214 (0.048−0.944) | 0.042 | 0.056 | ||||
| RNF213 4810G>A | ||||||||||
| GG | 89 (87.3) | 7 (12.3) | 1.000 (reference) | 1.000 (reference) | ||||||
| GA | 13 (12.7) | 49 (86.0) | 48.99 (18.00−133.3) | <0.001 | 0.004 | 145 (96.0) | 7 (11.7) | 199.6 (60.95−653.6) | <0.001 | 0.004 |
| AA | 0 | 1 (1.8) | N/A | N/A | 6 (4.0) | 53 (88.3) | N/A | N/A | ||
| Dominant (GG vs. GA+AA) | 50.06 (18.43−136.0) | <0.001 | 0.004 | 0 | 0 | 199.6 (60.95−653.6) | <0.001 | 0.004 | ||
| RNF213 4863G>A | ||||||||||
| GG | 78 (76.5) | 51 (89.5) | 1.000 (reference) | 145 (96.0) | 57 (95.0) | 1.000 (reference) | ||||
| GA | 24 (23.5) | 6 (10.5) | 0.395 (0.150−1.039) | 0.060 | 0.120 | 6 (4.0) | 3 (5.0) | 1.340 (0.321−5.592) | 0.688 | 0.688 |
| AA | 0 | 0 | N/A | N/A | 0 | 0 | N/A | N/A | ||
| Dominant (GG vs. GA+AA) | 0.395 (0.150−1.039) | 0.060 | 0.120 | 1.340 (0.321−−5.592) | 0.688 | 0.688 | ||||
| RNF213 4950G>A | ||||||||||
| GG | 91 (89.2) | 49 (86.0) | 1.000 (reference) | 133 (88.1) | 42 (70.0) | 1.000 (reference) | ||||
| GA | 11 (10.8) | 8 (14.0) | 1.260 (0.470−3.379) | 0.646 | 0.672 | 18 (11.9) | 18 (30.0) | 3.096 (1.473−6.506) | 0.003 | 0.006 |
| AA | 0 | 0 | N/A | N/A | 0 | 0 | N/A | N/A | ||
| Dominant (GG vs. GA+AA) | 1.260 (0.470−3.379) | 0.646 | 0.672 | 3.096 (1.473−6.506) | 0.003 | 0.006 | ||||
* Adjusted by age and gender. MMD, moyamoya disease. Abbreviation: AOR, adjusted odd ratio; CI, confidence interval, FDR, false discovery rate; N/A, not application.
We conducted subgroup analyses of the RNF213 polymorphisms (Table 3 and Table 4). RNF213 4950G>A was more frequent in the ischemic type than in the hemorrhagic type. RNF213 4810G>A was more frequent in both the ischemic and hemorrhagic types of MMD.
In the pediatric group (<18 years of age), RNF213 4810G>A was more frequent than in the control group (p < 0.001). In the adult MMD group, RNF213 4810G>A (p = 0.004) and RNF213 4950G>A were more frequent (p = 0.006).
2.2. Haplotype Analysis
Haplotype analysis was conducted, as shown in Table 5 and Table 6. The G-G-A-G (p = 0.009), G-A-G-G (p < 0.001), G-A-G-A (p < 0.001), and G-A-A-G (p < 0.001) (RNF213 4448/4810/4863/4950) haplotypes were significantly higher in moyamoya patients, while the G-G-A-G haplotype (RNF213 4448/4810/4863/4950) was lower in MMD patients compared to the group. The G-G-A and G-A-G haplotypes (RNF213 4448/4810/4950) were significantly higher in MMD patients, and the A-G-G (RNF213 4810/4863/4950) haplotype was more frequent in MMD patients. In addition, we performed haplotype analysis by dividing the MMD patients into two groups: the pediatric group (<18 years old) and the adult group (≥18 years old) (Tables S1–S4). Interestingly, the G-G-A-G (p = 0.005), G-A-G-G (p < 0.0001), and G-A-A-G (p = 0.033) (RNF213 4448/4810/4863/4950) haplotypes were associated with moyamoya disease risk in the pediatric group whereas G-A-G-A (p < 0.0001) elevated moyamoya disease risk in the adult group. The genotype combination frequency of RNF213 in MMD patients and control subjects is shown in Table 7. GG/GA (RNF213 4448G>A/4810G>A) was more frequent in the MMD group. GA/GG (RNF213 4810G>A/4863G>A) and GA/GA (RNF213 4810G>A/4863G>A) were more frequent in the MMD group. GA/GG and GA/GA (RNF213 4810G>A/4950G>A) were more frequent in the MMD group.
Table 5.
Haplotype analysis of RNF213 in MMD patients and control subjects.
| Haplotypes | Controls (2n = 506) | MMD (2n = 234) | OR (95% CI) | p | FDR-P |
|---|---|---|---|---|---|
| RNF213 4448G>A/4810G>A/4863G>A/4950G>A | |||||
| G-G-G-G | 0.7932 (401) | 0.4795 (112) | 1.000 (reference) | ||
| G-G-G-A | 0.0534 (27) | 0.0610 (14) | 1.856 (0.942–3.660) | 0.081 | 0.122 |
| G-G-A-G | 0.0488 (25) | 0 | 0.070 (0.004–1.159) | 0.004 | 0.009 |
| G-G-A-A | 0 | 0 | N/A | N/A | |
| G-A-G-G | 0.0226 (11) | 0.3421 (80) | 26.04 (13.40–50.60) | <0.001 | 0.003 |
| G-A-G-A | 0.0024 (1) | 0.0447 (10) | 35.80 (4.532–282.8) | <0.001 | 0.003 |
| G-A-A-G | 0.0105 (5) | 0.0385 (9) | 6.445 (2.117-19.62) | 0.001 | 0.003 |
| G-A-A-A | 0 | 0 | N/A | N/A | |
| A-G-G-G | 0.0656 (33) | 0.015 (4) | 0.434 (0.151–1.251) | 0.144 | 0.185 |
| A-G-G-A | 0.0015 (1) | 0 | 1.190 (0.048–29.43) | 1.000 | 1.000 |
| A-G-A-G | 0 | 0 | N/A | N/A | |
| A-G-A-A | 0 | 0 | N/A | N/A | |
| A-A-G-G | 0.0021 (1) | 0.0137 (3) | 10.74 (1.106–104.3) | 0.036 | 0.065 |
| A-A-G-A | 0 | 0.0054 (1) | 10.71 (0.433–264.8) | 0.220 | 0.248 |
| A-A-A-G | 0 | 0 | N/A | N/A | |
| A-A-A-A | 0 | 0 | N/A | N/A | |
| RNF213 4448G>A/4810G>A/4863G>A | |||||
| G-G-G | 0.8464 (428) | 0.5405 (126) | 1.000 (reference) | ||
| G-G-A | 0.0488 (25) | 0 | 0.066 (0.004–1.099) | 0.003 | 0.005 |
| G-A-G | 0.0252 (13) | 0.3863 (91) | 23.78 (12.86–43.95) | <0.001 | 0.003 |
| G-A-A | 0.0104 (5) | 0.0385 (9) | 6.114 (2.012–18.58) | 0.001 | 0.003 |
| A-G-G | 0.0671 (34) | 0.0150 (4) | 0.400 (0.139–1.148) | 0.103 | 0.103 |
| A-G-A | 0 | 0 | N/A | N/A | |
| A-A-G | 0.0020 (1) | 0.0192 (4) | 13.59 (1.504–122.7) | 0.012 | 0.015 |
| A-A-A | 0 | 0 | N/A | N/A | |
| RNF213 4448G>A/4810G>A/4950G>A | |||||
| G-G-G | 0.8406 (425) | 0.4333 (101) | 1.000 (reference) | ||
| G-G-A | 0.0539 (27) | 0.1054 (25) | 4.373 (2.468–7.748) | <0.001 | 0.004 |
| G-A-G | 0.0346 (18) | 0.4271 (100) | 27.62 (16.18–47.15) | <0.001 | 0.004 |
| G-A-A | 0.0017 (1) | 0 | 1.742 (0.070–43.10) | 1.000 | 1.000 |
| A-G-G | 0.0663 (34) | 0.0168 (4) | 0.552 (0.193–1.582) | 0.278 | 0.389 |
| A-G-A | 0.0017 (1) | 0 | 1.742 (0.070–43.10) | 1.000 | 1.000 |
| A-A-G | 0.0012 (1) | 0.0117 (3) | 15.74 (1.620–152.9) | 0.025 | 0.058 |
| A-A-A | 0 | 0.0057 (1) | 15.68 (0.634–387.9) | 0.194 | 0.400 |
| RNF213 4448G>A/4863G>A/4950G>A | |||||
| G-G-G | 0.8155 (413) | 0.8210 (192) | 1.000 (reference) | ||
| G-G-A | 0.0561 (28) | 0.1064 (25) | 1.921 (1.091–3.382) | 0.032 | 0.110 |
| G-A-G | 0.0593 (30) | 0.0385 (9) | 0.645 (0.300–1.386) | 0.290 | 0.387 |
| G-A-A | 0 | 0 | N/A | N/A | |
| A-G-G | 0.0679 (34) | 0.0295 (7) | 0.443 (0.193–1.017) | 0.055 | 0.110 |
| A-G-A | 0.0013 (1) | 0.0047 (1) | 2.151 (0.134–34.60) | 0.535 | 0.535 |
| A-A-G | 0 | 0 | N/A | N/A | |
| A-A-A | 0 | 0 | N/A | N/A | |
| RNF213 4810G>A/4863G>A/4950G>A | |||||
| G-G-G | 0.8587 (435) | 0.4947 (116) | 1.000 (reference) | ||
| G-G-A | 0.0549 (28) | 0.0608 (14) | 1.875 (0.956–3.677) | 0.080 | 0.080 |
| G-A-G | 0.0488 (25) | 0 | 0.073 (0.004–1.214) | 0.004 | 0.005 |
| G-A-A | 0 | 0 | N/A | N/A | |
| A-G-G | 0.0247 (12) | 0.3557 (83) | 25.94 (13.69–49.15) | <0.001 | 0.002 |
| A-G-A | 0.0024 (1) | 0.0503 (12) | 45.00 (5.789–349.8) | <0.001 | 0.002 |
| A-A-G | 0.0105 (5) | 0.0385 (9) | 6.750 (2.219–20.53) | 0.001 | 0.002 |
| A-A-A | 0 | 0 | N/A | N/A | |
Abbreviation: MMD, moyamoya disease; OR, odd ratio; CI, confidence interval, FDR, false discovery rate; N/A, not application.
Table 6.
Haplotype analysis of RNF213 in MMD patients and control subjects.
| Haplotype | Controls (2n = 506) | MMD (2n = 234) | OR (95% CI) | p | FDR-P |
|---|---|---|---|---|---|
| RNF213 4448G>A/4810G>A | |||||
| G-G | 0.8945 (453) | 0.5327 (125) | 1.000 (reference) | ||
| G-A | 0.0364 (18) | 0.4331 (101) | 19.53 (11.38–33.49) | <0.001 | 0.003 |
| A-G | 0.0680 (34) | 0.0229 (5) | 0.512 (0.196–1.336) | 0.227 | 0.227 |
| A-A | 0.0012 (1) | 0.0113 (3) | 10.44 (1.076–101.3) | 0.038 | 0.057 |
| RNF213 4448G>A/4863G>A | |||||
| G-G | 0.8715 (441) | 0.9274 (217) | 1.000 (reference) | ||
| G-A | 0.0593 (30) | 0.0385 (9) | 0.610 (0.284–1.307) | 0.222 | 0.222 |
| A-G | 0.0629 (35) | 0.0342 (8) | 0.465 (0.212–1.019) | 0.063 | 0.126 |
| A-A | 0 | 0 | N/A | N/A | |
| RNF213 4448G>A/4950G>A | |||||
| G-G | 0.8751 (443) | 0.8596 (201) | 1.000 (reference) | ||
| G-A | 0.0557 (28) | 0.1062 (25) | 1.968 (1.119–3.461) | 0.022 | 0.066 |
| A-G | 0.0675 (34) | 0.0293 (7) | 0.454 (0.198–1.041) | 0.078 | 0.117 |
| A-A | 0.0016 (1) | 0.0049 (1) | 2.204 (0.137–35.44) | 0.528 | 0.528 |
| RNF213 4810G>A/4863G>A | |||||
| G-G | 0.9136 (462) | 0.5556 (130) | 1.000 (reference) | ||
| G-A | 0.0489 (25) | 0 | 0.069 (0.004–1.150) | 0.004 | 0.004 |
| A-G | 0.0271 (14) | 0.4060 (95) | 24.12 (13.31–43.68) | <0.001 | 0.002 |
| A-A | 0.0104 (5) | 0.0385 (9) | 6.397 (2.107–19.42) | <0.001 | 0.002 |
| RNF213 4810G>A/4950G>A | |||||
| G-G | 0.9069 (459) | 0.4444 (104) | 1.000 (reference) | ||
| G-A | 0.0556 (28) | 0.1111 (26) | 4.098 (2.307–7.282 | <0.001 | 0.002 |
| A-G | 0.0358 (18) | 0.4444 (104) | 25.50 (14.80–43.93) | <0.001 | 0.002 |
| A-A | 0.0018 (1) | 0 | 1.466 (0.059–36.26) | 1.000 | 1.000 |
| RNF213 4863G>A/4950G>A | |||||
| G-G | 0.8834 (447) | 0.8504 (199) | 1.000 (reference) | ||
| G-A | 0.0573 (29) | 0.1111 (26) | 2.014 (1.156–3.509) | 0.016 | 0.032 |
| A-G | 0.0593 (30) | 0.0385 (9) | 0.674 (0.314–1.446) | 0.372 | 0.372 |
| A-A | 0 | 0 | N/A | N/A | |
Abbreviation: MMD, moyamoya disease; OR, odd ratio; CI, confidence interval, FDR, false discovery rate; N/A, not application.
Table 7.
Genotype combination frequency of RNF213 in MMD patients and control subjects.
| Genotype | Controls, n = 253 (%) | MMD, n = 117 (%) | AOR (95% CI) * | p | FDR-P |
|---|---|---|---|---|---|
| RNF213 4448G>A/4810G>A | |||||
| GG/GG | 202 (79.8) | 13 (11.1) | 1.000 (reference) | ||
| GG/GA | 17 (6.7) | 95 (81.2) | 93.58 (42.23–207.4) | <0.001 | 0.002 |
| GG/AA | 0 | 1 (0.9) | N/A | N/A | |
| GA/GG | 31 (12.3) | 1 (0.9) | 0.483 (0.061–3.845) | 0.492 | 0.492 |
| GA/GA | 2 (0.8) | 7 (6.0) | 51.99 (9.165–294.9) | <0.001 | 0.002 |
| GA/AA | 0 | 0 | N/A | N/A | |
| AA/GG | 1 (0.4) | 0 | N/A | N/A | |
| AA/GA | 0 | 0 | N/A | N/A | |
| AA/AA | 0 | 0 | N/A | N/A | |
| RNF213 4448G>A/4863G>A | |||||
| GG/GG | 191 (75.5) | 100 (85.5) | 1.000 (reference) | ||
| GG/GA | 28 (11.1) | 9 (7.7) | 0.586 (0.260–1.318) | 0.196 | 0.196 |
| GG/AA | 0 | 0 | N/A | N/A | |
| GA/GG | 31 (12.3) | 8 (6.8) | 0.470 (0.207–1.068) | 0.072 | 0.144 |
| GA/GA | 2 (0.8) | 0 | N/A | N/A | |
| GA/AA | 0 | 0 | N/A | N/A | |
| AA/GG | 1 (0.4) | 0 | N/A | N/A | |
| AA/GA | 0 | 0 | N/A | N/A | |
| AA/AA | 0 | 0 | N/A | N/A | |
| RNF213 4448G>A/4950G>A | |||||
| GG/GG | 193 (76.3) | 85 (72.6) | 1.000 (reference) | ||
| GG/GA | 26 (10.3) | 24 (20.5) | 2.101 (1.131–3.903) | 0.019 | 0.057 |
| GG/AA | 0 | 0 | N/A | N/A | |
| GA/GG | 30 (11.9) | 6 (5.1) | 0.451 (0.180–1.127) | 0.088 | 0.132 |
| GA/GA | 3 (1.2) | 2 (1.7) | 1.201 (0.193–7.480) | 0.844 | 0.844 |
| GA/AA | 0 | 0 | N/A | N/A | |
| AA/GG | 1 (0.4) | 0 | N/A | N/A | |
| AA/GA | 0 | 0 | N/A | N/A | |
| AA/AA | 0 | 0 | N/A | N/A | |
| RNF213 4810G>A/4863G>A | |||||
| GG/GG | 210 (83.0) | 14 (12.0) | 1.000 (reference) | ||
| GG/GA | 24 (9.5) | 0 | N/A | N/A | |
| GG/AA | 0 | 0 | N/A | N/A | |
| GA/GG | 13 (5.1) | 94 (80.3) | 107.3 (48.21–238.9) | <0.001 | 0.001 |
| GA/GA | 6 (2.4) | 8 (6.8) | 18.08 (5.240–62.40) | <0.001 | 0.001 |
| GA/AA | 0 | 0 | N/A | N/A | |
| AA/GG | 0 | 0 | N/A | N/A | |
| AA/GA | 0 | 1 (0.9) | N/A | N/A | |
| AA/AA | 0 | 0 | N/A | N/A | |
| RNF213 4810G>A/4950G>A | |||||
| GG/GG | 207 (81.8) | 11 (9.4) | 1.000 (reference) | ||
| GG/GA | 27 (10.7) | 3 (2.6) | 1.961 (0.504–7.629) | 0.331 | 0.331 |
| GG/AA | 0 | 0 | N/A | N/A | |
| GA/GG | 17 (6.7) | 79 (67.5) | 89.25 (39.26–202.9) | <0.001 | 0.002 |
| GA/GA | 2 (0.8) | 23 (19.7) | 216.1 (44.62–1047) | <0.001 | 0.002 |
| GA/AA | 0 | 0 | N/A | N/A | |
| AA/GG | 0 | 1 (0.9) | N/A | N/A | |
| AA/GA | 0 | 0 | N/A | N/A | |
| AA/AA | 0 | 0 | N/A | N/A | |
| RNF213 4863G>A/4950G>A | |||||
| GG/GG | 195 (77.1) | 83 (70.9) | 1.000 (reference) | ||
| GG/GA | 28 (11.1) | 25 (21.4) | 2.060 (1.125–3.772) | 0.019 | 0.057 |
| GG/AA | 0 | 0 | N/A | N/A | |
| GA/GG | 29 (11.5) | 8 (6.8) | 0.557 (0.238–1.303) | 0.177 | 0.266 |
| GA/GA | 1 (0.4) | 1 (0.9) | 2.883 (0.174–47.67) | 0.460 | 0.460 |
| GA/AA | 0 | 0 | N/A | N/A | |
| AA/GG | 0 | 0 | N/A | N/A | |
| AA/GA | 0 | 0 | N/A | N/A | |
| AA/AA | 0 | 0 | N/A | N/A | |
* Adjusted by age and gender. Abbreviation: MMD, moyamoya disease; AOR, adjusted odd ratio; CI, confidence interval, FDR, false discovery rate; N/A, not application.
2.3. Screening with World Population for RNF213 Polymorphisms
Table 8 shows the major and minor allele frequencies of the RNF213 polymorphisms in different world populations according to the 1000 Genome Project database (http://www.internationalgenome.org/). Our study detected RNF213 4448G>A, 4810G>A, 4863G>A, and 4950G>A allele frequencies in control subjects and MMD patients.
Table 8.
Allele frequencies of RNF213 polymorphisms (4448G>A, 4810G>A, 4863G>A, and 4950G>A) in different world populations.
| Population | N | RNF213 4448G>A (rs148731719) | RNF213 4810G>A (rs112735431) | RNF213 4863G>A (rs760732823) | RNF213 4950G>A (rs371441113) | |||||
|---|---|---|---|---|---|---|---|---|---|---|
| G allele | A allele | G allele | A allele | G allele | A allele | G allele | A allele | Database | ||
| African | 661 | 0.9985 | 0.0015 | 1.0000 | 0.0000 | - | - | 1.0000 | 0.0000 | 1000 Genome a |
| Ad Mixed American | 347 | 0.9915 | 0.0085 | 1.0000 | 0.0000 | - | - | 1.0000 | 0.0000 | |
| East Asian | 504 | 0.9315 | 0.0685 | 0.9980 | 0.0020 | - | - | 0.9960 | 0.0040 | |
| Chinese Dai in Xishuangbanna, China | 93 | 0.8815 | 0.1185 | 1.0000 | 0.0000 | - | - | 0.9945 | 0.0055 | |
| Han Chinese in Beijing, China | 103 | 0.9370 | 0.0630 | 1.0000 | 0.0000 | - | - | 1.0000 | 0.0000 | |
| Southern Han Chinese | 105 | 0.9240 | 0.0760 | 1.0000 | 0.0000 | - | - | 0.9950 | 0.0050 | |
| Japanese in Tokyo, Japan | 104 | 0.9325 | 0.0675 | 0.9905 | 0.0095 | - | - | 1.0000 | 0.0000 | |
| Kinh in Ho Chi Minh City, Vietnam | 99 | 0.9800 | 0.0200 | 1.0000 | 0.0000 | - | - | 0.9900 | 0.0100 | |
| European | 503 | 0.9920 | 0.0080 | 1.0000 | 0.0000 | - | - | 1.0000 | 0.0000 | |
| South Asia | 489 | 0.9940 | 0.0060 | 0.9960 | 0.0040 | - | - | 1.0000 | 0.0000 | |
| African | 7650 | 0.9970 | 0.0030 | 1.0000 | 0.0000 | 1.0000 | 0.0000 | 1.0000 | 0.0000 | gnomAD b |
| Ad Mixed American | 16,791 | 0.9950 | 0.0050 | 1.0000 | 0.0000 | 1.0000 | 0.0000 | 1.0000 | 0.0000 | |
| Ashkenazi Jewish | 4925 | 0.9830 | 0.0170 | 1.0000 | 0.0000 | 1.0000 | 0.0000 | 1.0000 | 0.0000 | |
| East Asian | 8619 | 0.9470 | 0.0530 | 0.9970 | 0.0030 | 0.9990 | 0.0010 | 0.9970 | 0.0030 | |
| Finnish | 10,703 | 0.9960 | 0.0040 | 1.0000 | 0.0000 | 1.0000 | 0.0000 | 1.0000 | 0.0000 | |
| Non-Finnish European | 55,846 | 0.9920 | 0.0080 | 1.0000 | 0.0000 | 1.0000 | 0.0000 | 1.0000 | 0.0000 | |
| South Asia | 15,391 | 0.9960 | 0.0040 | 0.9996 | 0.0004 | 1.0000 | 0.0000 | 0.9996 | 0.0004 | |
| Other (population not assigned) | 2734 | 0.9890 | 0.0110 | 1.0000 | 0.0000 | 1.0000 | 0.0000 | 1.0000 | 0.0000 | |
| Korean (controls) | 253 | 0.9310 | 0.0690 | 0.9660 | 0.0340 | 0.9405 | 0.0595 | 0.9425 | 0.0575 | Present study |
| Korean (MMD) | 117 | 0.9660 | 0.0340 | 0.5560 | 0.4440 | 0.9615 | 0.0385 | 0.8890 | 0.1110 | Present study |
a The 1000 Genome Project website: https://www.internationalgenome.org/; we checked these polymorphisms frequencies based on GRCh38. b The Genome Aggregation Database website: http://gnomad.broadinstitute.org/; Official gnomAD release (version 2.0).
3. Discussion
At least 24 genetic changes in the RNF213 gene have been associated with moyamoya disease [31,32]. Three individual studies of MMD patients have revealed high frequencies of the same single base substitution (nonsynonymous mutation) as well as the c.14576G>A (p.R4859K) variant in RFP213 (a gene located in chromosome 17q) [21,22,23,24,25,26,27,28,29,30,31,32,33]. The c.14576G>A in RNF213 is present in ~2% of East Asian populations, which is a relatively higher rate compared with Caucasians [21,22,23,24,25,26,27,28,29,30,31,32,33]. The RNF213 gene can be used as a biomarker to predict prognosis, as it has been reported that the RNF213 gene correlates with the early-onset and severe forms of MMD [33]. The RNF213 p.Arg4810Lys variant is reportedly associated with ischemic-type MMD, while RNF213 non-p.Arg4810Lys variants are associated with hemorrhagic type MMD [23].
The single-nucleotide polymorphism of c.14576G>A, p.R4859K, in RNF-213 has been detected in 95% of familial cases and 79% of sporadic cases of MMD [21]. Nevertheless, some MMD patients do not carry the c.14576G>A variant and this portion is higher in western countries [34].
Miyawaki et al. found that RNF213 (14576G>A) was higher in intracranial major artery stenosis/occlusion (ICASO) patients, as in the MMD group [26], and they suggest ICASO without signs of MMD is a genetic variant associated with MMD. Bang et al. suggest that RNF213 is a susceptibility gene for both MMD and ICAS in East Asians. Further studies are needed on RNF213 variants in ICASO patients outside East Asian populations [27]. However, two variants of RNF213 (p.Arg2438Cys and p.Ala2826Thr) were found in intracranial aneurysm patients in a French-Canadian population [35]. Interestingly, our results differed with the results of a previous study [27] that suggested that 4810G>A is the only variant that is strongly associated with MMD in Korean patients. In this study, our results suggested that RNF213 4810 and 4950 were associated with MMD risk, furthermore our result found that frequencies of RNF213 4448G>A, 4863G>A, and 4950G>A hetero genotype. We think that this difference is due to the difference in how the genotypes were identified. We used the classical method of using polymerase chain reaction-restriction fragment length polymorphism (PCR-RFLP) and confirmed the genotypes of the individual samples, whereas the previous study performed the MALDI-TOF method, which is likely to cause errors because it confirms the genotype of many samples for many variants.
The RNF213 gene encodes a protein with 5256 amino acids harboring a RING (Really Interesting New Gene) finger motif and an AAA (ATPase associated with a variety of cellular activities) domain, indicating the presence of both E3 ubiquitin ligase activity and an energy-dependent E3 ubiquitin ligase (which has several subtypes), an enzyme that ubiquitinates specific target proteins, resulting in degradation by proteasomes [26]. The RNF213 variant associated with MMD prevails, but it is also found in other vascular diseases such as cerebrovascular stenosis [26], albeit not in the Caucasian MMD population [36]. In RNF213-deficient mice, an abnormal vascular network does not develop at the base of the brain [37]. The RNF213 variant is an important SNP that is not specific to MMD alone.
The c.14576G>A variant is mainly detected in Japanese, Korean, and Chinese populations [33,38,39]. However, the frequency in the latter population is much lower than those of the former two populations [34]. RNF213 exhibits strong and obvious ethnic diversity [32].
The biochemical function and pathological role of RNF213 have not been completely clarified. Disruption of the RNF213 first B motif disrupts ATP hydrolysis cyclicity, inhibiting angiogenesis, and this reduced anti-angiogenic activity of interferon beta 1 (IFNb) is partially mediated by RNF213, which acts as a mediator downstream of the IFNb signaling pathway [32]. The overexpression of RNF213 R4810K, but not wild type RNF213, suggests that RNF213 R4810K over expression mimics IFNb action. Koizumi et al. suggest that RNF213 R4810K is the major detrimental factor that elicits endothelial cell dysfunction. Pro-inflammatory signals such as IFNs can activate the transcription of RNF213 [32]. The model assumes that any of three independent abnormalities, such as endothelial dysfunction, smooth muscle cell dysfunction, and abnormal hemostasis, can exaggerate the proliferation of smooth muscle cells (SMCs) and that each abnormality can result in vascular stenosis [32].
Genome-wide association study (GWAS) approaches are now being applied to MMD with the hope of uncovering the underlying pathogenic mechanisms [40]. A GWAS was recently performed in Japanese MMD patients and demonstrated a strong association of MMD risk with chromosome 17q25-ter [21]. These GWAS studies will need further investigation to solidly replicate the results in modern genetic studies based on familial or non-familial MMD. Subgroup analysis was performed for adult versus pediatric and ischemic versus hemorrhagic groups. The former primarily presents with ischemia, while the latter presents with intracranial hemorrhage [6,41]. The progression of MMD has generally been considered to occur exclusively in childhood, with angiographic characteristics completed before adulthood [42,43]. Kuroda and colleagues reported in a multicenter observational study on adult onset MMD that the incidence of the disease progression in adult cases was not as rare as originally considered [44]. Miyatake et al. [33] reported that patients with childhood-onset MMD and the homozygous polymorphism of c.14576G>A in RNF213 were more likely to have an earlier onset and more severe mortality because of the rapid progression of vascular stenosis. Han et al. suggested that RNF213 R4810K is associated with the ischemic type , and A4399T is associated with the hemorrhagic type [23]. A genetic animal RNF213 knock-in model has been debated, however, the RNF213 genetic animal model failed to mimic the MMD model. Liu reported that RNF213 knock-down zebra fish have abnormal surrounding vessels [22]. Sonobe et al. [37] did not observe any modification of angiogenesis after they generated mice that lack RNF213. RN 213 knock out (KO) animal models have yielded conflicting results in the cerebrum and hind limbs [45]. Fujimura et al. speculated that RNF213 influences vascular remodeling in chronic ischemia [38]. Allele frequencies of RNF213 polymorphisms (4448G>A, 4810G>A, 4863G>A, and 4950G>A) in different world populations are presented in Table 8.
SNP studies have some limitations. First, a question remains about how the p.R4810K variant or nine other variants impair the physiological function of RNF213, resulting in moyamoya disease [22]. The second limitation is a lack of a pathologic process for MMD development from RNF213. Third, large population-based case-control analyses or analyses centered on family-based designs are needed. However, SNP studies have many advantages over other genetic studies, the benefits of which depend on how SNPs will be exploited in relevant study designs and what traits and diseases will be the focus of these studies [46].
We have considered some of the unique aspects of SNPs and their relative advantages and disadvantages in human population-based analyses [46]. We believe that wide-scale progress in genetically identifying MMD is needed because MMD appears to be a multifactorial, polygenic spectrum disorder.
4. Materials and Methods
4.1. Subjects
One hundred seventeen consecutive Korean patients with moyamoya disease (mean age, 23.20 ± 17.75 years; 77 females (65.8%), 40 males (34.2%)) were recruited for this study. MMD patients were diagnosed and enrolled based on the presence of clinical ischemic or hemorrhagic symptoms in combination with vascular lesions in magnetic resonance imaging (MRI) or magnetic resonance angiography (MRA) [47].
The control group was comprised of 253 healthy subjects (mean age 25.60 ± 16.98 years; 145 females (57.3%); 108 males (42.7%)) from the same regional background as the MMD patients. We recruited the age- and sex-matched subjects from outpatient clinics at Severance Hospital, CHA Bundang Medical Center, Chungbuk National University Hospital (Cheongju, Korea). Table 1 shows demographic characteristics of patients with the disease and control subjects. We divided the MMD patients into pediatric (<18 years) and adult (≥18 years) groups. We further divided the moyamoya patients into ischemic or hemorrhagic groups based on clinical and MRI findings.
All participants gave informed written consent prior to enrollment in the study. The institutional review boards of Severance Hospital (4-2008-0308), CHA Bundang Medical Center (PBC09-103), and Chungbuk National University Hospital (2014-08-010-005) approved this study.
4.2. RNF213 Genotyping
DNA was extracted from leukocytes using a G-DEX II Genomic DNA Extraction kit (Intron Biotechnology, Seongnam, Korea) according to the manufacturer’s instructions. To analyze RNF213 genotypes, we chose polymerase chain reaction-restriction fragment length polymorphism (PCR-RFLP).
RNF213 4448G>A was detected using a forward primer (5′-TTG CCA ACT AAG CCC TCG AAA CAA-3′) and a reverse primer (5′-CAA CAA TGG CAC AGA ATT GTC-3′). The 230-bp PCR product was then digested with 5U AluI. A digestion product of 230-bp represented the AA genotype; fragments of 230-bp, 135-bp, and 95-bp represented the AG genotype; 135-bp and 95-bp products represented the GG genotype.
The RNF213 4810G>A polymorphism was detected by PCR-RFLP analysis using forward (5′-AGC AGA GCT GAG GCT GGT AA-3′) and reverse (5′-CTG TCA GAG CAG AGC CAC AC-3′) primers. The 151-bp product was digested with 3U Hpy188I. A restriction fragment of 146-bp and 5-bp represented the AA genotype; fragments of 146-bp, 104-bp, 42-bp, and 5-bp represented the GA genotype; and 104-bp, 42-bp, and 5-bp products represented the GG genotype.
To detect the RNF213 4863G>A genotypes, PCR-RFLP analysis was performed with forward (5′-TGT GTG TGG AGC TGA TGG CT-3′) and reverse (5′-AGG GAG GAG ATA CAG ACC AGA CT-3′) primers. The length of the amplified fragment was 867-bp. PCR products were digested with 5U Hpy188I. For 4863G>A, restriction products of 867-bp identified the GG genotype; products of 867-bp, 719-bp, and 148-bp represented the GA genotype; and 719-bp and 148-bp products represented the AA genotype.
The RNF213 4950G>A polymorphism was detected by PCR-RFLP analysis using forward (5′-GGT GGA GGA GGG CAG AGA GAC CGT GCA CGA-3′) and reverse (5′- CTT CCC TCT CTC GAG AAA CAC ACC AA-3′) primers. The 188-bp product was digested with 3U BssSI. A restriction fragment of 162-bp and 26-bp represented the GG genotype; fragments of 188-bp, 162-bp, and 26-bp represented the GA genotype; and 188-bp products represented the AA genotype. The RNF213 4448G>A, 4810G>A, 4863G>A, and 4950G>A polymorphisms were digested by AluI, Hpy188I, Hpy188I, and BssSI, respectively, for 16 h at 37 °C (New England BioLabs, Beverly, MA, USA). The PCR annealing temperature was 64 °C for all polymorphisms, with 35 amplification cycles for RNF213 polymorphisms. The reaction product (12 μL) was run on a 3.0% ethidium bromide-stained agarose gel and confirmed under ultraviolet illumination.
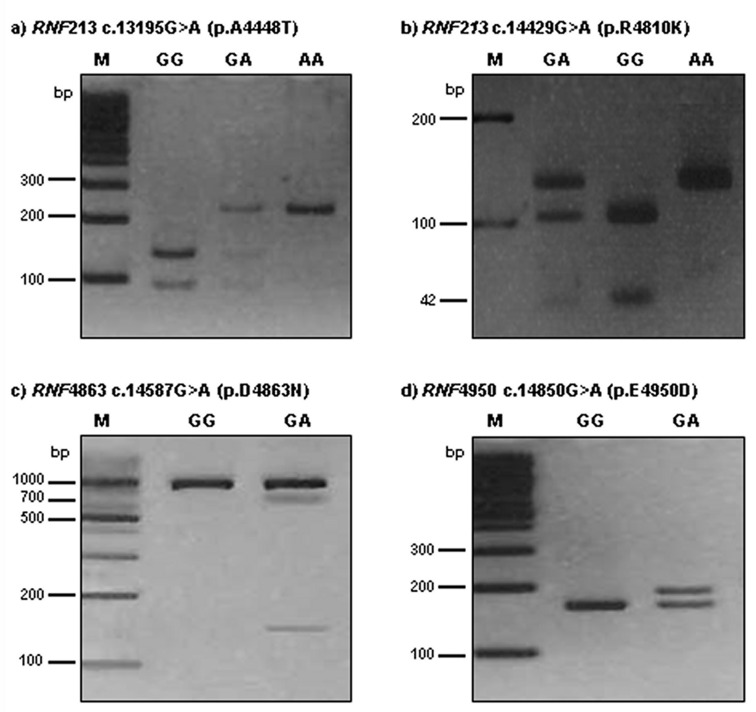
We randomly repeated approximately 10% of the PCR assays for each of the miRNA polymorphisms and checked the results for concordance by DNA sequencing using an automatic sequencer (ABI3730x1 DNA analyzer; Applied Biosystems, Foster City, CA, USA). The concordance of the quality control samples was 100%. Polymorphism analysis was performed for RNF213 4448, 4810, 4863, and 4950 in moyamoya disease patients. Polymorphism analysis of RNF213 gene amplicons was performed by agarose gel electrophoresis after restriction endonuclease digestion (Figure 1).
Figure 1.
Polymorphism analysis of RNF213 4448, 4810, 4863, and 4950 in moyamoya disease patients. Polymorphisms analysis of RNF213 genes amplicon by agarose gel electrophoresis after restriction endonuclease digestion. (a) RNF213 4448 c.13195G>A site was digested by AluI resulting in the appearance of the GG (wild type, 135/95 bp), GA (heterozygous type, 230/135/95 bp), and AA (mutant type, 230 bp) genotypes in moyamoya disease patients; (b) RNF213 4810 c.14429G>A site was digested by Hpy188I resulting in the appearance of the GG (wild type, 104/42/5 bp), GA (heterozygous type, 146/104/42/5bp), and AA (mutant type, 146 bp) genotypes in moyamoya disease patients. DNA fragments that were 5 bp or less were too small to be seen on 3% agarose gel; (c) RNF4863 c.14587G>A site was digested by Hpy188I resulting in the appearance of the GG (wild type, 867 bp) and GA (hetero type, 867/719/148 bp) genotypes in moyamoya disease patients. No mutant type (AA) was found in this study. (d) RNF4950 c.14850G>A site was digested by BssSαI resulting in the appearance of the GG (wild type, 162/26 bp) and GA (heterozygous type, 188/162/26 bp) genotypes. The mutant type (AA) was not found in results of restriction fragment length polymorphism (RFLP). DNA fragments that were 26 bp or less were too small to be seen on 3% agarose gel.
In this study, we investigated exome analysis and identified ring finger protein 213 (RNF213; DDBJ/EMBL/GenBank accession number AB537889) (National Center for Biotechnology Information (NCBI) in Table S5). Questions remain regarding how the p.R4810K variant or nine other variants impair RNF213 protein, thereby resulting in MMD.
4.3. Statistical Analyses
The genotype frequencies for RNF213 polymorphism were compared by Hardy-Weinberg equilibrium (HWE) test [48]. To analyze the demographic characteristics of moyamoya disease, we used the Mann–Whitney and chi-square (χ2) tests for continuous and categorical data, respectively. The relationships between RNF213 and MMD patients (pediatric or adult) were calculated according to the odds ratios (ORs) and 95% confidence intervals (CIs) using Fisher’s exact test. The adjusted odds ratios (AORs) for four polymorphisms of the RNF213 gene were calculated using multiple logistic regression analyses with gender and age. We considered Hardy-Weinberg equilibrium in the genotype distribution. We used GraphPad Prism 4.0 (GraphPad Software, Inc., San Diego, CA, USA) and StatsDirect software (version 2.4.4; StatsDirect Ltd., Altrincham, UK) to perform statistical analyses. We used HAPSTAT (version 3.0; University of North Carolina, Chapel Hill, NC, USA) and Haploview 4.2 (Broad Institute, Cambridge, MA, USA) to perform Haplotype analyses. The adjusted ORs and 95% CIs were calculated by statistical software (version 2.4.4; StatsDirect Ltd., Altrincham, UK).
5. Conclusions
In summary, RNF213 4810G>A and RNF213 4950G>A were more frequent in MMD patient. Current RNF213 SNP studies suggest that RNF213 4810G>A and 4950G>A are strongly associated with Korean MMD, both in pediatric or adult patients as well as in the ischemic or hemorrhagic types. Our results show that polymorphism of RNF213 4810G>A and 4950G>A are associated with the occurrence of moyamoya disease, and that 4810G>A may affect the general moyamoya prevalence, while 4950G>A is particularly relevant to the occurrence of moyamoya in the adult group. It is therefore thought that it can serve as a potential biomarker of moyamoya disease in adult groups.
Acknowledgments
This work was supported by the National Research Foundation of Korea, Chungbuk National University Hospital (2013R1A2A2A01067990, NRF ICT 2016H1D5A1908909), and by the Korea Health Technology R&D Project through the Korea Health Industry Development Institute (KHIDI), funded by the Ministry of Health & Welfare, Republic of Korea (grant number: HI16C1559).
Supplementary Materials
Supplementary materials can be found at www.mdpi.com/1422-0067/18/11/2477/s1.
Author Contributions
Young Seok Park and Nam Keun Kim designed and directed the whole project. Won Seop Kim, In Bo Han, and Dong-Seok Kim collected the blood samples from osteoporosis patients and control subjects. Jung Oh Kim and Hui Jeong An performed the experiments, collected the results, and analyzed the data. Won Seop Kim, In Bo Han, Ok Joon Kim, Nam Keun Kim, and Dong-Seok Kim discussed and interpreted the data and results. Young Seok Park wrote the first draft of the manuscript. All authors contributed to and have approved the final manuscript.
Conflicts of Interest
The authors declare no conflict of interest.
References
- 1.Baba T., Houkin K., Kuroda S. Novel epidemiological features of moyamoya disease. J. Neurol. Neurosurg. Psychiatry. 2008;79:900–904. doi: 10.1136/jnnp.2007.130666. [DOI] [PubMed] [Google Scholar]
- 2.Starke R.M., Komotar R.J., Connolly E.S. Optimal surgical treatment for moyamoya disease in adults: Direct versus indirect bypass. Neurosurg. Focus. 2009;26:E8. doi: 10.3171/2009.01.FOCUS08309. [DOI] [PubMed] [Google Scholar]
- 3.Wakai K., Tamakoshi A., Ikezaki K., Fukui M., Kawamura T., Aoki R., Kojima M., Lin Y., Ohno Y. Epidemiological features of moyamoya disease in Japan: Findings from a nationwide survey. Clin. Neurol. Neurosurg. 1997;99:S1–S5. doi: 10.1016/S0303-8467(97)00031-0. [DOI] [PubMed] [Google Scholar]
- 4.Scott R.M. Arteriovenous malformation and moyamoya disease. Child’s Nerv. Syst. 1997;13:357. doi: 10.1007/s003810050097. [DOI] [PubMed] [Google Scholar]
- 5.Kuroda S., Houkin K. Moyamoya disease: Current concepts and future perspectives. Lancet Neurol. 2008;7:1056–1066. doi: 10.1016/S1474-4422(08)70240-0. [DOI] [PubMed] [Google Scholar]
- 6.Han D.H., Nam D.H., Oh C.W. Moyamoya disease in adults: Characteristics of clinical presentation and outcome after encephalo-duro-arterio-synangiosis. Clin. Neurol. Neurosurg. 1997;99:S151–S155. doi: 10.1016/S0303-8467(97)00058-9. [DOI] [PubMed] [Google Scholar]
- 7.Han D.H., Kwon O.K., Byun B.J., Choi B.Y., Choi C.W., Choi J.U., Choi S.G., Doh J.O., Han J.W., Jung S., et al. A co-operative study: Clinical characteristics of 334 Korean patients with moyamoya disease treated at neurosurgical institutes (1976–1994). The Korean Society for Cerebrovascular Disease. Acta Neurochir. 2000;142:1263–1274. doi: 10.1007/s007010070024. [DOI] [PubMed] [Google Scholar]
- 8.Kuroda S., Hashimoto N., Yoshimoto T., Iwasaki Y. Radiological findings, clinical course, and outcome in asymptomatic moyamoya disease: Results of multicenter survey in Japan. Stroke. 2007;38:1430–1435. doi: 10.1161/STROKEAHA.106.478297. [DOI] [PubMed] [Google Scholar]
- 9.Fukui M. Current state of study on moyamoya disease in Japan. Surg. Neurol. 1997;47:138–143. doi: 10.1016/S0090-3019(96)00358-8. [DOI] [PubMed] [Google Scholar]
- 10.Mineharu Y., Liu W., Inoue K., Matsuura N., Inoue S., Takenaka K., Ikeda H., Houkin K., Takagi Y., Kikuta K., et al. Autosomal dominant moyamoya disease maps to chromosome 17q25.3. Neurology. 2008;70:2357–2363. doi: 10.1212/01.wnl.0000291012.49986.f9. [DOI] [PubMed] [Google Scholar]
- 11.Nanba R., Kuroda S., Tada M., Ishikawa T., Houkin K., Iwasaki Y. Clinical features of familial moyamoya disease. Child’s Nerv. Syst. 2006;22:258–262. doi: 10.1007/s00381-005-1230-5. [DOI] [PubMed] [Google Scholar]
- 12.Burke G.M., Burke A.M., Sherma A.K., Hurley M.C., Batjer H.H., Bendok B.R. Moyamoya disease: A summary. Neurosurg. Focus. 2009;26:E11. doi: 10.3171/2009.1.FOCUS08310. [DOI] [PubMed] [Google Scholar]
- 13.Malek A.M., Connors S., Robertson R.L., Folkman J., Scott R.M. Elevation of cerebrospinal fluid levels of basic fibroblast growth factor in moyamoya and central nervous system disorders. Pediatr. Neurosurg. 1997;27:182–189. doi: 10.1159/000121249. [DOI] [PubMed] [Google Scholar]
- 14.Ikeda H., Sasaki T., Yoshimoto T., Fukui M., Arinami T. Mapping of a familial moyamoya disease gene to chromosome 3p24.2-p26. Am. J. Hum. Genet. 1999;64:533–537. doi: 10.1086/302243. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Nanba R., Tada M., Kuroda S., Houkin K., Iwasaki Y. Sequence analysis and bioinformatics analysis of chromosome 17q25 in familial moyamoya disease. Child’s Nerv. Syst. 2005;21:62–68. doi: 10.1007/s00381-004-1005-4. [DOI] [PubMed] [Google Scholar]
- 16.Inoue T.K., Ikezaki K., Sasazuki T., Matsushima T., Fukui M. Linkage analysis of moyamoya disease on chromosome 6. J. Child Neurol. 2000;15:179–182. doi: 10.1177/088307380001500307. [DOI] [PubMed] [Google Scholar]
- 17.Han H., Pyo C.W., Yoo D.S., Huh P.W., Cho K.S., Kim D.S. Associations of Moyamoya patients with HLA class I and class II alleles in the Korean population. J. Korean Med. Sci. 2003;18:876–880. doi: 10.3346/jkms.2003.18.6.876. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Sakurai K., Horiuchi Y., Ikeda H., Ikezaki K., Yoshimoto T., Fukui M., Arinami T. A novel susceptibility locus for moyamoya disease on chromosome 8q23. J. Hum. Genet. 2004;49:278–281. doi: 10.1007/s10038-004-0143-6. [DOI] [PubMed] [Google Scholar]
- 19.Kang H., Kim S., Cho B., Kim Y., Hwang Y., Wang K. Single nucleotide polymorphisms of tissue inhibitor of metalloproteinase genes in familial moyamoya disease. Neurosurgery. 2006;58:1074–1080. doi: 10.1227/01.NEU.0000215854.66011.4F. [DOI] [PubMed] [Google Scholar]
- 20.Guo D.C., Papke C.L., Tran-Fadulu V., Regalado E.S., Avidan N., Johnson R.J., Kim D.H., Pannu H., Willing M.C., Sparks E., et al. Mutations in smooth muscle α-actin (ACTA2) cause coronary artery disease, stroke, and moyamoya disease, along with thoracic aortic disease. Am. J. Hum. Genet. 2009;84:617–627. doi: 10.1016/j.ajhg.2009.04.007. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Kamada F., Aoki Y., Narisawa A., Abe Y., Komatsuzaki S., Kikuchi A., Kanno J., Niihori T., Ono M., Ishii N., et al. A genome-wide association study identifies RNF213 as the first moyamoya disease gene. J. Hum. Genet. 2011;56:34–40. doi: 10.1038/jhg.2010.132. [DOI] [PubMed] [Google Scholar]
- 22.Liu W., Morito D., Takashima S., Mineharu Y., Kobayashi H., Hitomi T., Hashikata H., Matsuura N., Yamazaki S., Toyoda A., et al. Identification of RNF213 as a susceptibility gene for moyamoya disease and its possible role in vascular development. PLoS ONE. 2011;6:e22542. doi: 10.1371/journal.pone.0022542. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Wu Z., Jiang H., Zhang L., Xu X., Zhang X., Kang Z., Song D., Zhang J., Guan M., Gu Y. Molecular analysis of RNF213 gene for moyamoya disease in the Chinese Han population. PLoS ONE. 2012;7:e48179. doi: 10.1371/journal.pone.0048179. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Cecchi A.C., Guo D., Ren Z., Flynn K., Santos-Cortez R.L., Leal S.M., Wang G.T., Regalado E.S., Steinberg G.K., Shendure J., et al. RNF213 rare variants in an ethnically diverse population with moyamoya disease. Stroke. 2014;45:3200–3207. doi: 10.1161/STROKEAHA.114.006244. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Ma J., Liu Y., Ma L., Huang S., Li H., You C. RNF213 polymorphism and moyamoya disease: A systematic review and meta-analysis. Neurol. India. 2013;61:35–39. doi: 10.4103/0028-3886.107927. [DOI] [PubMed] [Google Scholar]
- 26.Miyawaki S., Imai H., Takayanagi S., Mukasa A., Nakatomi H., Saito N. Identification of a genetic variant common to moyamoya disease and intracranial major artery stenosis/occlusion. Stroke. 2012;43:3371–3374. doi: 10.1161/STROKEAHA.112.663864. [DOI] [PubMed] [Google Scholar]
- 27.Bang O.Y., Chung J.W., Cha J., Lee M.J., Yeon J.Y., Ki C.S., Jeon P., Kim J.S., Hong S.C. A Polymorphism in RNF213 Is a Susceptibility Gene for Intracranial Atherosclerosis. PLoS ONE. 2016;11:e0156607. doi: 10.1371/journal.pone.0156607. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Bang O.Y., Ryoo S., Kim S.J., Yoon C.H., Cha J., Yeon J.Y., Kim K.H., Kim G.M., Chung C.S., Lee K.H., et al. Adult Moyamoya Disease: A Burden of Intracranial Stenosis in East Asians? PLoS ONE. 2015;10:e0130663. doi: 10.1371/journal.pone.0130663. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Miyawaki S., Imai H., Shimizu M., Yagi S., Ono H., Mukasa A., Nakatomi H., Shimizu T., Saito N. Genetic variant RNF213 c.14576G>A in various phenotypes of intracranial major artery stenosis/occlusion. Stroke. 2013;44:2894–2897. doi: 10.1161/STROKEAHA.113.002477. [DOI] [PubMed] [Google Scholar]
- 30.Koizumi A. Genetic Analysis of Familial Moyamoya. Labor and Welfare; Tokyo, Japan: 2010. pp. 25–26. [Google Scholar]
- 31.Park Y.S. Single Nucleotide Polymorphism in Patients with Moyamoya Disease. J. Korean Neurosurg. Soc. 2015;57:422–427. doi: 10.3340/jkns.2015.57.6.422. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Koizumi A., Kobayashi H., Hitomi T., Harada K.H., Habu T., Youssefian S. A new horizon of moyamoya disease and associated health risks explored through RNF213. Environ. Health Prev. Med. 2016;21:55–70. doi: 10.1007/s12199-015-0498-7. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Miyatake S., Miyake N., Touho H., Nishimura-Tadaki A., Kondo Y., Okada I., Tsurusaki Y., Doi H., Sakai H., et al. Homozygous c.14576G>A variant of RNF213 predicts early-onset and severe form of moyamoya disease. Neurology. 2012;78:803–810. doi: 10.1212/WNL.0b013e318249f71f. [DOI] [PubMed] [Google Scholar]
- 34.Ma Y.G., Zhang Q., Yu L.B., Zhao J.Z. Role of Ring Finger Protein 213 in Moyamoya Disease. Chin. Med. J. 2016;129:2497–2501. doi: 10.4103/0366-6999.191824. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Zhou S., Ambalavanan A., Rochefort D., Xie P., Bourassa C.V., Hince P., Dionne-Laporte A., Spiegelman D., Gan-Or Z., Mirarchi C., et al. RNF213 Is Associated with Intracranial Aneurysms in the French-Canadian Population. Am. J. Hum. Genet. 2016;99:1072–1085. doi: 10.1016/j.ajhg.2016.09.001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Liu W., Senevirathna S.T., Hitomi T., Kobayashi H., Roder C., Herzig R., Kraemer M., Voormolen M.H., Cahova P., Krischek B., et al. Genomewide association study identifies no major founder variant in Caucasian moyamoya disease. J. Genet. 2013;92:605–609. doi: 10.1007/s12041-013-0304-5. [DOI] [PubMed] [Google Scholar]
- 37.Sonobe S., Fujimura M., Niizuma K., Nishijima Y., Ito A., Shimizu H., Kikuchi A., Arai-Ichinoi N., Kure S., Tominaga T. Temporal profile of the vascular anatomy evaluated by 9.4-T magnetic resonance angiography and histopathological analysis in mice lacking RNF213: A susceptibility gene for moyamoya disease. Brain Res. 2014;1552:64–71. doi: 10.1016/j.brainres.2014.01.011. [DOI] [PubMed] [Google Scholar]
- 38.Fujimura M., Sonobe S., Nishijima Y., Niizuma K., Sakata H., Kure S., Tominaga T. Genetics and Biomarkers of Moyamoya Disease: Significance of RNF213 as a Susceptibility Gene. J. Stroke. 2014;16:65–72. doi: 10.5853/jos.2014.16.2.65. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Liu W., Hitomi T., Kobayashi H., Harada K.H., Koizumi A. Distribution of moyamoya disease susceptibility polymorphism p.R4810K in RNF213 in East and Southeast Asian populations. Neurol. Med. Chir. 2012;52:299–303. doi: 10.2176/nmc.52.299. [DOI] [PubMed] [Google Scholar]
- 40.Achrol A.S., Guzman R., Lee M., Steinberg G.K. Pathophysiology and genetic factors in moyamoya disease. Neurosurg. Focus. 2009;26:E4. doi: 10.3171/2009.1.FOCUS08302. [DOI] [PubMed] [Google Scholar]
- 41.Fukui M., Kono S., Sueishi K., Ikezaki K. Moyamoya disease. Neuropathology. 2000;20:S61–S64. doi: 10.1046/j.1440-1789.2000.00300.x. [DOI] [PubMed] [Google Scholar]
- 42.Ezura M., Yoshimoto T., Fujiwara S., Takahashi A., Shirane R., Mizoi K. Clinical and angiographic follow-up of childhood-onset moyamoya disease. Childs Nerv. Syst. 1995;11:591–594. doi: 10.1007/BF00300998. [DOI] [PubMed] [Google Scholar]
- 43.Houkin K., Yoshimoto T., Kuroda S., Ishikawa T., Takahashi A., Abe H. Angiographic analysis of moyamoya disease—How does moyamoya disease progress? Neurol. Med. Chir. 1996;36:783–787. doi: 10.2176/nmc.36.783. [DOI] [PubMed] [Google Scholar]
- 44.Kuroda S., Ishikawa T., Houkin K., Nanba R., Hokari M., Iwasaki Y. Incidence and clinical features of disease progression in adult moyamoya disease. Stroke. 2005;36:2148–2153. doi: 10.1161/01.STR.0000182256.32489.99. [DOI] [PubMed] [Google Scholar]
- 45.Ito A., Fujimura M., Niizuma K., Kanoke A., Sakata H., Morita-Fujimura Y., Kikuchi A., Kure S., Tominaga T. Enhanced post-ischemic angiogenesis in mice lacking RNF213; a susceptibility gene for moyamoya disease. Brain Res. 2015;1594:310–320. doi: 10.1016/j.brainres.2014.11.014. [DOI] [PubMed] [Google Scholar]
- 46.Schork N.J., Fallin D., Lanchbury J.S. Single nucleotide polymorphisms and the future of genetic epidemiology. Clin. Genet. 2000;58:250–264. doi: 10.1034/j.1399-0004.2000.580402.x. [DOI] [PubMed] [Google Scholar]
- 47.Park Y.S., Jeon Y.J., Kim H.S., Chae K.Y., Oh S.H., Han I.B., Kim W.C., Kim O.J., Kim T.G., Choi J.U., et al. The role of VEGF and KDR polymorphisms in moyamoya disease and collateral revascularization. PLoS ONE. 2012;7:e47158. doi: 10.1371/journal.pone.0047158. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48.Cho H., Kim W., Choi M.K., Le T.M., Choi H.J., Kim J.H., Kim K., Soundrarajan N., Park J.K., Lee Y.M., et al. Effects of natural resistance-associated macrophage protein 1 and toll-like receptor 2 gene polymorphisms on post-weaning piglet survivability. Genes Genom. 2016;38:171–178. doi: 10.1007/s13258-015-0355-5. [DOI] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.