Abstract
T cell expression of TIM-3 following Ag encounter has been associated with a continuum of functional states ranging from effector memory T cells to exhaustion. We have designed an in vitro culture system to specifically address the impact of anti–TIM-3/TIM-3 engagement on human Ag-specific CD8 T cells during a normal response to Ag and found that anti–TIM-3 treatment enhances T cell function. In our in vitro T cell culture system, MART1-specific CD8 T cells were expanded from healthy donors using artificial APCs. To ensure that the T cells were the only source of TIM-3, cells were rechallenged with peptide-loaded artificial APCs in the presence of anti–TIM-3 Ab. In these conditions, anti–TIM-3 treatment promotes generation of effector T cells as shown by acquisition of an activated phenotype, increased cytokine production, enhanced proliferation, and a transcription program associated with T cell differentiation. Activation of mTORC1 has been previously demonstrated to enhance CD8 T cell effector function and differentiation. Anti–TIM-3 drives CD8 T cell differentiation through activation of the mTORC1 as evidenced by increased levels of phosphorylated S6 protein and rhebl1 transcript. Altogether these findings suggest that anti–TIM-3, together with Ag, drives differentiation in favor of effector T cells via the activation of mTOR pathway. To our knowledge, this is the first report demonstrating that TIM-3 engagement during Ag stimulation directly influences T cell differentiation through mTORC1.
Introduction
Functional CD8 T cell response requires recognition of peptide-loaded MHC class I complexes by TCR with appropriate costimulation. Such responses drive effective antiviral and antitumor responses, and are mediated by downstream signaling pathways that drive T cell differentiation and effector function. During activation, T cells upregulate inhibitory receptors to control the immune response, including T cell Ig and mucin domain containing molecule-3 (TIM-3).
The function of TIM-3 on CD8 T cells has been difficult to define because TIM-3 expression is associated with both T cell exhaustion (1–4) and T cell activation (5–7). Studies on TIM-3 signaling have reported that engagement of TIM-3 on T cells yields induction of tyrosine-phosphorylated proteins unique to T cells (8). Expression of TIM-3 in Jurkat T cells enhances TCR signaling under weak stimulation but not during stronger TCR signaling (9, 10). The functional outcome of TIM-3 engagement may depend on the strength of TCR activation such that optimal signaling results in a negative event, whereas TIM-3 engagement coincident with weaker TCR activation enhances T cell responses (11). This dichotomy could explain reports implicating TIM-3 in both T cell activation and exhaustion. Furthermore, prior experiments evaluating TIM-3 engagement on T cells have involved cross-linking of the CD3/TCR complex via anti-CD3 mAb and/or mitogen-induced activation. Although informative, these studies do not address immune regulation of Ag-specific effector mechanisms and T cell differentiation. Evaluating TIM-3 function through a more physiologically relevant activation signal via TCR-MHC recognition could illuminate pathways that were otherwise masked by artificial T cell activation. Moreover, most TIM-3 studies on Ag-specific T cells were conducted in the context of disease. Little is known about TIM-3 in a healthy Ag-specific T cell response.
In this study we explore how TIM-3 impacts in vitro–expanded Ag-specific CD8 T cells during stimulation via TCR-MHC engagement. We determined that engagement of TIM-3 with an Ab increases T cell effector function and drives changes in transcription factors and downstream genes associated with terminal differentiation (12–14). Under short-term stimulation, TIM-3 improves T cell activation by enhancing TCR signaling through PI3K (9). Mammalian target of rapamycin (mTOR) kinase is a highly conserved serine threonine kinase regulated by PI3K, and exists as a part of two distinct signaling complexes: mTORC1 and mTORC2. The mTORC1 complex is identified as a protein complex containing the scaffolding protein Raptor and is activated by the small GTPase Rheb (15, 16). Previous studies have shown mTORC1 as a crucial regulator of CD8 T cell effector function and memory (17–19). We show that with Ag-specific stimulation, engagement of TIM-3 on CD8 T cells promotes effector function through mTORC1 signaling correlating with increased expression of rhebl1, whose gene product is an isoform of mTOR activator Rheb (15, 16, 20).
Materials and Methods
Primary cells
Purified human negatively isolated CD8 T cells from healthy HLA-A2+ donors were purchased from Biological Specialty (Colmar, PA). T cells were >95% CD3+CD8+ by flow cytometry.
Artificial APCs
Artificial APCs (aAPC) derived from Drosophila Schneider 2 cells were cultured in Express V media (Life Technologies, Carlsbad, CA) transfected with pRMHa-3–derived vector encoding HLA-A2.1 Class I, B7.1, ICAM-1, LFA-3, and CD70 cultured under 200 μg/ml Geneticin (Life Technologies) selection. Gene expression was induced by addition of 1 mM CuSO4 (Sigma, St. Louis, MO). Protein expression was confirmed by flow cytometry using the following Abs: anti-CD54 Alexa Fluor 488 (BioLegend, San Diego, CA), anti–HLA-A,B,C FITC, CD70 PE, CD80 PE, CD58 PE (BD, San Jose, CA). Following induction, the aAPC were resuspended in Express V media and cross-linked for 10 min at 7.7 Joules per cm2 in media +5 μg/ml UVADEX (Johnson & Johnson, Skillman, NJ) in a VueLife bag (American Fluoroseal, Gaithersburg, MD) using an ILT72 UVA Radiometer (Life Technologies).
In vitro expansion of Ag-specific T cells
Following our standard protocol for expanding Ag-specific CD8 T cells, aAPC were loaded with a modified HLA-A2–restricted MART1 (ELAGIGILTV) peptide (CS Bio, Menlo Park, CA). Briefly, cells were incubated with 0.1 μg/ml MART1 peptide in the presence of 5 μg/ml β-2M (Janssen, in house) for 4 h at room temperature. CD8 T cells were cultured with MART1-loaded aAPC (1:10 ratio of aAPC/T cells) and 25 ng/ml IL-21 (PeproTech, Rocky Hill, NJ) weekly for 3 wk in RPMI 1640 (Life Technologies), supplemented with 10% heat-inactivated FBS (Life Technologies) at 37°C, 5% CO2. On days 6, 10, and 17, fresh media containing 20 U/ml of IL-2 and 30 U/ml IL-7 (PeproTech) was added to the cells. Following expansion, the Ag-specific T cells were frozen in 90% FBS and 10% DMSO (Sigma-Aldrich) for future in vitro restimulation experiments. For the terminal differentiation protocol, the expanded Ag-specific T cells were restimulated weekly with aAPC+MART1, as described above, for an additional three stimulations to drive terminal differentiation of the T cells.
Characterization and immune checkpoint blockade of Ag-specific T cells
T cells were phenotyped by flow cytometry with MART1 tetramer PE (MBL, Woburn, MA), anti-CD3 APC, anti-CD95 PerCP-Cy5.5, anti-CD27 APC (all BD), anti-CD8 BV570, anti-CD62L FITC, anti-CCR7 PE-Cy7, anti-CD45RA AF700, anti-CD45RO PE-Cy7, anti-CD127 PerCP-Cy5.5, anti-CD25 BV650, anti–PD-1 PerCP-Cy5.5 (all BioLegend), anti–LAG-3 FITC (Novus, St. Charles, MO), and anti–TIM-3 APC (R&D Systems, Minneapolis, MN). Proliferation was determined by labeling MART1 T cells with proliferation dye V450 (BD) per the manufacturer’s instructions and analyzed for dye dilution by flow cytometry. Cytokines from supernatants were measured by multiplex analyses (Meso Scale Discovery, Rockville, MD). Cytotoxicity assays were performed using Malme-3M and H1650 cells labeled with BATDA reagent (PerkinElmer, Waltham, MA) per the manufacturer’s instructions. Labeled targets were incubated with T cells at indicated E:T ratios and measured for BATDA release by tumor cells with EnVision reader (PerkinElmer). For ELISPOT analysis, restimulated T cells were incubated with Malme-3M or H1650 tumor cells overnight and analyzed for spot production using precoated plates (MabTech, Cincinnati, OH). For analysis of CD107a degranulation and FasL expression, restimulated T cells were incubated with Malme-3M or H1650 tumor cells for 6 h with GolgiStop (BD) and anti-CD107a PE-Cy7 (BioLegend) followed by anti-FasL APC (BD), and analyzed by flow cytometry.
Treatment of Ag-specific T cells with anti–TIM-3 Ab
Expanded Ag-specific T cells were thawed and restimulated with aAPC that had been loaded with MART1 peptide, as described earlier. T cells were cultured with aAPC (1:10 ratio of aAPC/ T cells) and 25 ng/ml IL-21 for 4 d in RPMI 1640, supplemented with 10% heat-inactivated FBS at 37°C, 5% CO2. Rat anti-human TIM-3 mAb (rat IgG2a clone 344823) (R&D Systems) and/or purified mouse anti-human PD-1 mAb (mouse IgG1 clone J116) (eBioscience) were added to MART1 T cells at the start of the restimulation with aAPC+MART1 at the indicated concentrations. After 4 d in culture, cells were analyzed by flow cytometry and cell culture media supernatants were collected for cytokine analyses. For cytotoxicity measurements, expanded Ag-specific T cells were restimulated with MART1-loaded aAPC in the presence of anti–TIM-3 Ab at 10 μg/ml for 3 d prior to incubation with tumor targets. Target cell lysis, CD107a, and FasL expression were measured as described above. For gene expression analyses, expanded T cells were restimulated with MART1-loaded aAPC in the presence of anti–TIM-3 Ab at 10 μg/ml for 48 h prior to RNA extraction.
RNA extraction and sequencing library generation
Total RNA was extracted from cells using a QIASymphony RNA kit (Qiagen, Germantown, MD). Following extraction, the quantity and 260/280 ratio were determined on a NanoDrop (Thermo Fisher Scientific, Grand Island, NY), and the quality of RNA was confirmed on 2100 Agilent Bioanalyzer (Agilent, Santa Clara, CA). In total, 400 ng of RNA was used as input for generating sequencing libraries using TruSeq mRNA kit per the manufacturer’s protocol. Libraries were loaded on to a paired-end flow cell on cBot (Illumina, San Diego, CA), and sequenced on a HiSeq2500 (Illumina). For data analysis, transcripts with zero counts in more than two thirds of samples were discarded from downstream analysis to reduce noise. Filtered data were normalized using quantile normalization and differentially expressed transcripts were identified using Limma Voom. A p value cutoff of 0.05 and fold change ≥2 was used to classify transcripts as differentially expressed in treatment condition. Upstream regulator and pathway analyses on differentially expressed transcripts were performed using QIAGEN’s Ingenuity Pathway Analysis (IPA). For real-time PCR analyses, mRNA was extracted using RNeasy Mini kit (Qiagen), RNA quantity and 260/280 ratio were determined on a NanoDrop, and cDNA was generated using High Capacity cDNA kit (Thermo Fisher Scientific). Gene expression was detected using RT2 qPCR Primer Assays for Rheb and RhebL1 with the RT2 First Strand kit (Qiagen) and PCR reactions were run on the ViiA7 Real-Time PCR system (Thermo Fisher Scientific).
Quantification and statistical analysis
For all markers analyzed by flow cytometry, isotype controls were used to establish gates by setting gates between 0.5 and 1% positive events. Quantifications were made based on data generated from FlowJo and statistical analysis was performed using GraphPad Prism version 6. Statistical significance was determined using two-tailed paired t test (Holm–Sidak) and two-way ANOVA (Dunnett).
Results
In vitro expanded Ag-specific CD8 T cells exhibit effector memory phenotype and express immune regulatory receptor TIM-3
To study Ag-specific T cell responses in the presence of anti–TIM-3, ex vivo enrichment of T cells was performed to generate populations comprised of ≥80% Ag-specific T cells. Primary HLA-A2+ CD8 T cells were expanded by stimulating three times with Drosophila-derived aAPC loaded with HLA-A2-restricted melan-A–derived peptide, MART1. This protocol results in an enriched pool of MART1-specific CD8 T cells (Supplemental Fig. 1A). These T cells proliferated and produced cytokines in response to MART1 peptide stimulation but not when stimulated with HIV peptide as shown in Supplemental Fig. 1B, 1C. These MART1-specific T cells specifically lysed MART1-positive HLA-A2+ target cells but not MART1-negative HLA-A2+ targets (Supplemental Fig. 1D). Specific lysis of the MART1+ targets was mediated by the degranulation pathway as shown by CD107a staining and perforin deposition rather than the FAS/FASL pathway (Supplemental Fig. 1E, 1F).
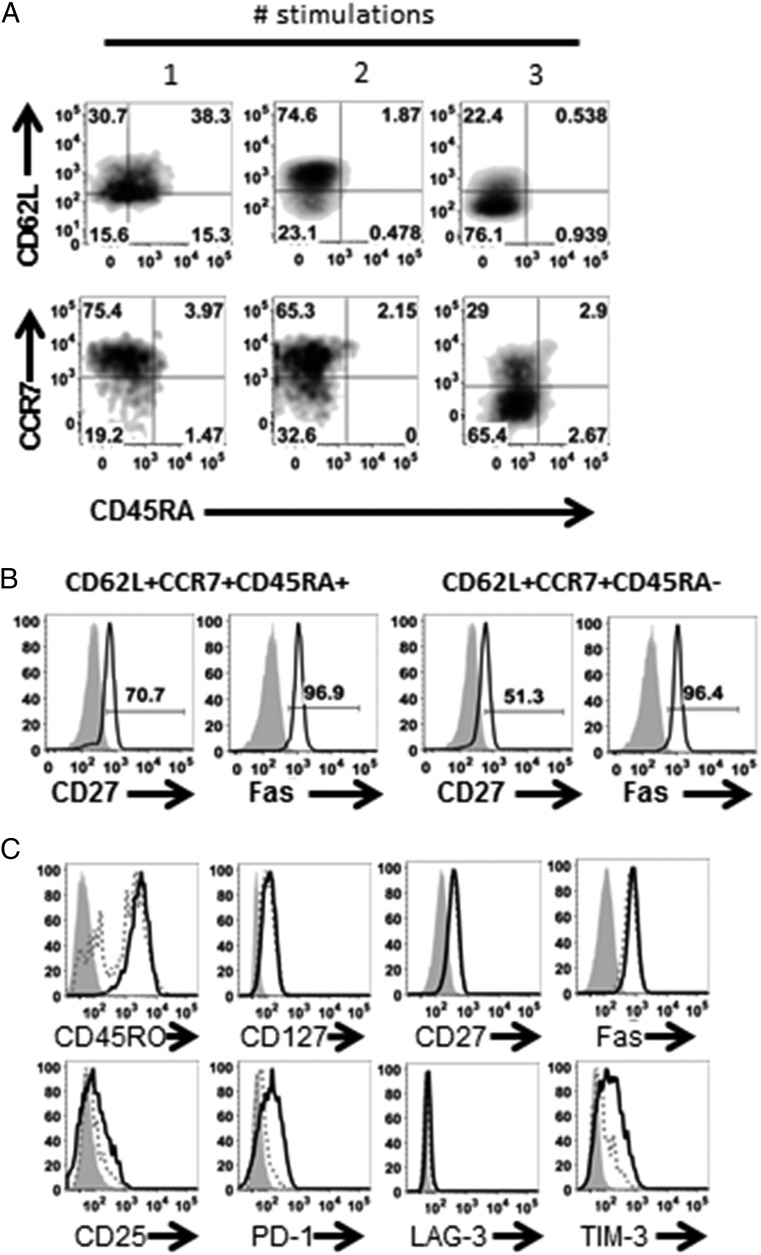
Increasing Ag exposure drives T cells toward terminal differentiation with a concomitant reduction in self-renewing potential (21, 22). Analysis of the T cells throughout the expansion process revealed T cells transitioning from a stem cell memory phenotype, in this study identified as CD62L+CCR7+CD45RA+, toward a T central memory (TCM) phenotype (CD62L+CCR7+CD45RA−) and effector memory T (TEM) or effector T (TEFF) phenotype (CD62L−CCR7−CD45RA−) with each subsequent stimulation (Fig. 1A). Gating on tetramer+ cells showed that the T stem cell memory and TCM populations were Ag-experienced because they expressed CD27 and Fas (Fig. 1B). In our protocol, T cells were stimulated three times for maximum cell expansion without terminal differentiation, resulting in cells that were predominantly TCM and TEM/TEFF. Furthermore, MART1-specific T cells expressed CD45RO, CD127 (IL-7Rα), CD27, and Fas, indicative of a more TEM phenotype rather than terminally differentiated TEFF (Fig. 1C) (23).
FIGURE 1.
Ag-specific CD8 T cells differentiate in response to Ag stimulation. HLA-A2+ CD8 T cells were stimulated weekly with MART1 peptide-loaded aAPC for three stimulations in total and (A) analyzed for CD62L, CCR7, and CD45RA expression following each stimulation. (B) Representative histograms of CD27 and Fas expression on CD62L+CCR7+CD45RA+ and CD62L+CCR7+CD45RA− populations (gated on tetramer+; black lines) compared with isotype (shaded gray) on the expanded MART1-specific CD8 T cell population after three stimulations. (C) Representative histograms of CD45RO, CD127, CD27, CD25, Fas, CD25, PD-1, LAG-3, and TIM-3 on MART-1 tetramer+ (black solid lines), tetramer− (dotted lines) compared with isotype (shaded gray) on the expanded MART1-specific CD8 T cell population after three stimulations. Data representative of four individual donors.
T cells can upregulate immune checkpoint molecules during activation (7, 24). Ag-specific CD8 T cells upregulated activation marker CD25 and the immune regulatory molecules PD-1 and TIM-3, but not LAG-3 (Fig. 1C). To confirm that these findings were a result of Ag-specific activation and not an artifact of the aAPC, HLA-A2+ CD8 T cells were stimulated with autologous dendritic cells (DCs) loaded with the MART-1 peptide in parallel. We demonstrated that the output CD8 T cells had similar phenotype and expression of PD-1, LAG-3, and TIM-3 on Ag-specific cells comparable to that of T cells stimulated with aAPC (Supplemental Fig. 2).
TIM-3 engagement during TCR-MHC-mediated stimulation promotes CD8 T cell effector function
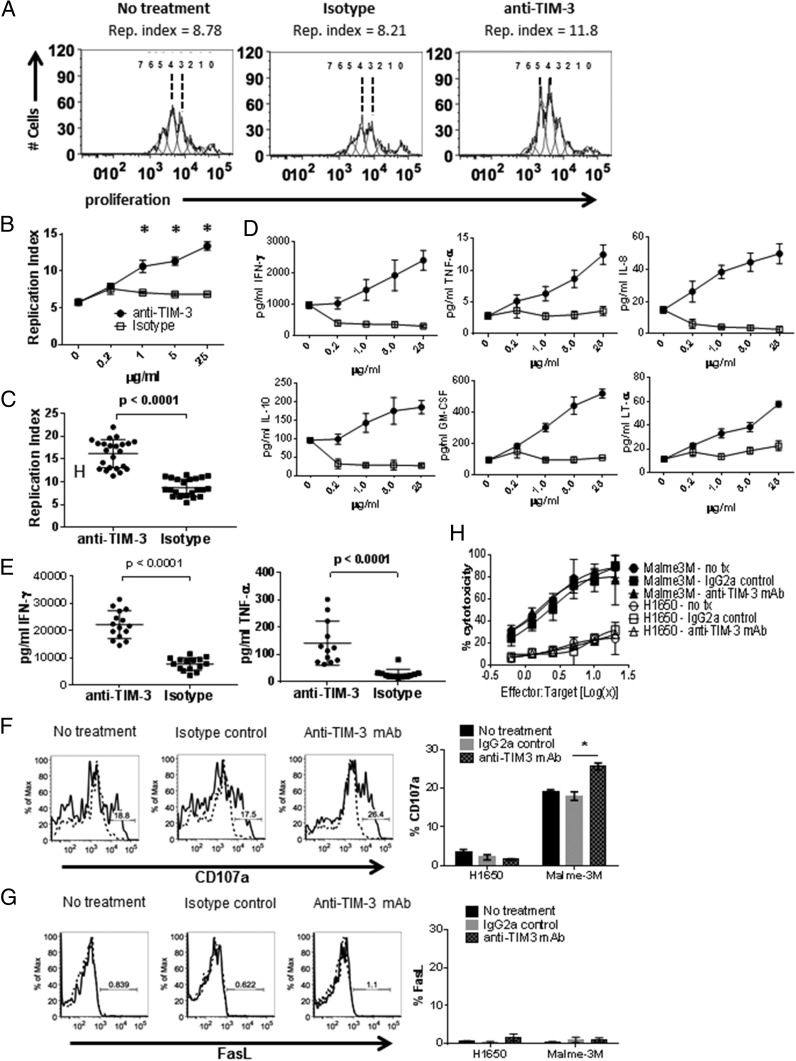
T cell activation through TCR-MHC provides a more physiologically relevant condition to evaluate immune-modulatory mechanisms as compared with T cell activation via CD3 cross-linking or mitogen treatment. This is particularly germane when considering how the strength of TCR signal impacts effector responses. We have shown that TIM-3 is expressed on healthy donor T cells during a polyclonal response to allogeneic DC (7). However, dissecting the biological impact of TIM-3 targeting remained a challenge due to expression of TIM-3 on both the T cells and DC. Using aAPC that lack TIM-3 expression allowed for specific targeting of TIM-3 on T cells with a mAb without interference from other cell types. To determine the effect of TIM-3 engagement on Ag-specific T cells, the expanded MART1-specific T cells were stimulated with aAPC loaded with cognate Ag in the presence of purified rat IgG2a (clone 344823) anti-human TIM-3 Ab, which was added at the start of the restimulation. Ab-treated T cells exhibited increased proliferation in response to Ag compared with isotype control treatment (Fig. 2A), in this study represented as the increase in Replication Index (25). The enhancement of proliferation directly correlated with anti–TIM-3 Ab concentration (Fig. 2B), which was consistent across multiple donors (Fig. 2C). Treatment with anti–TIM-3 Ab during stimulation also increased cytokine production by T cells (Fig. 2D, 2E). This was especially evident for IFN-γ, TNF-α, IL-8, IL-10, GM-CSF, and LT-α in agreement with results seen by others (3). The impact of anti–TIM-3 treatment on cytotoxic activity was assessed as it has been reported that antagonistic TIM-3 Ab improved CD8 T cell effector function (4, 26). Stimulated MART1-specific CD8 T cells treated with anti–TIM-3 Ab significantly increased CD107a deposition on the cell surface in the presence of MART1+ targets (Fig. 2F), affirming previously published results (4). Ag-specific T cell killing was not Fas/FasL mediated (Supplemental Fig. 1E), thus it was not surprising that anti–TIM-3 Ab treatment did not have any effect on that pathway (Fig. 2G). Despite the increase in degranulation, treatment with anti–TIM-3 Ab did not impact MART1+ tumor target lysis (Fig. 2H), likely due to the limited sensitivity of this method to measure subtle changes in T cell cytotoxicity. It is also possible that there are differences in kinetics of killing between anti–TIM-3 treated and isotype Ab-treated T cells, and these changes are not captured with the BATDA–release cytotoxicity assay because it is limited to detection in cytotoxicity for periods of 4 h, and incubation times beyond 4 h result in high nonspecific background cytotoxicity.
FIGURE 2.
Anti–TIM-3 Ab treatment increases T cell effector function. Ag-specific CD8 T cells were cultured as in Fig. 1 with addition of anti–TIM-3 mAb, isotype control, or left untreated, and analyzed by flow cytometry for proliferation dye dilution. Representative histograms of proliferation platform of individual cell generations and calculated replication indices are shown in (A). The increase in T cell proliferation upon anti–TIM-3 Ab treatment is dose dependent (B) and is observed in T cells from multiple donors, in this study shown at 25 μg/ml anti–TIM-3 Ab concentration (C) (*p < 0.0001, paired two-tailed t test). Anti–TIM-3 impacts cytokine production in a dose-dependent manner (D) in T cells from multiple donors, in this study shown at 25 μg/ml anti–TIM-3 Ab concentration for IFN-γ and TNF-α production (E) (*p < 0.0001, paired two-tailed t test). To measure CTL activity, Ag-specific CD8+ T cells were restimulated as described in Fig. 1 in the presence of anti–TIM-3 mAb, isotype control, or left untreated. The restimulated T cells were incubated with Malme3M (MART1+) or H1650 (MART1−) tumor targets. CD107a (F) and FasL (G) cell–surface expression was measured by flow cytometry (*p < 0.001, two-way ANOVA) and target cell killing was measured by BATDA release (H). Data representative of four to six biological replicates per treatment condition performed in three independent experiments.
Treatment with anti–TIM-3 Ab induces gene signature associated with TEFF differentiation
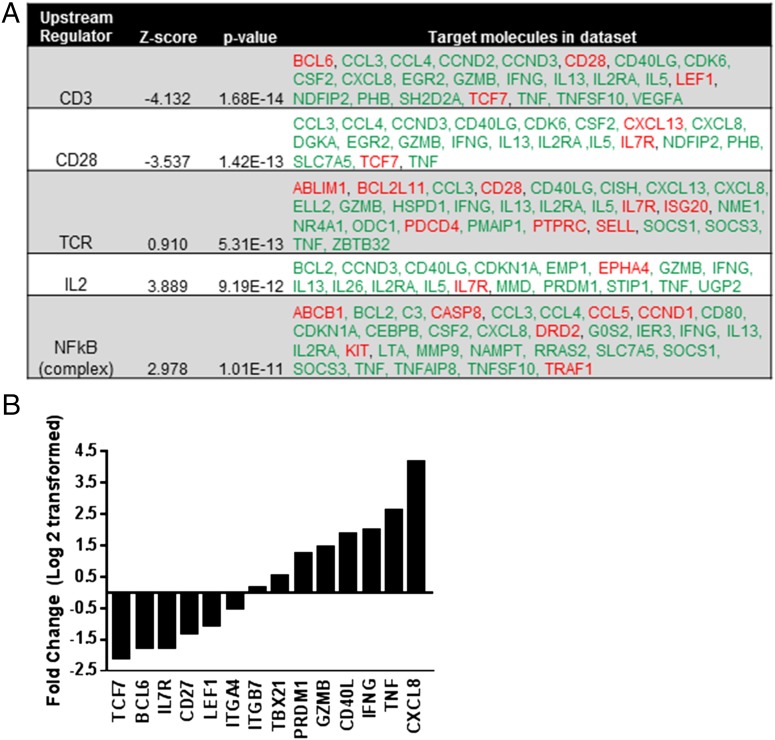
The observations of enhanced TEFF function in response to anti–TIM-3 Ab treatment prompted us to determine whether there were more global changes at the transcript level that could drive effector responses. Using IPA to compare gene expression of the stimulated Ag-specific CD8 T cells treated with either anti–TIM-3 Ab or isotype control revealed that TIM-3 engagement impacted signaling pathways associated with T cell activation (Fig. 3). TIM-3 expression in T cell lines can augment TCR- and CD28-dependent pathways leading to increased NF-κB–mediated transcription of downstream effector molecules (9). IPA showed that anti–TIM-3 Ab treatment of Ag-specific CD8 T cells increased expression of effector function genes including Cxcl8, Tnf, Ifng, Cd40l, and Gzmb and a decrease in Il7r, implicating TIM-3 in the regulation of CD3-, CD28-, TCR-, IL-2–, and NF-κB–dependent pathways (Fig. 3). Transcription factor Blimp1 promotes TEFF differentiation whereas Bcl-6 is essential for T cell memory (27, 28). Bcl6 gene expression decreased by 3.3-fold with anti–TIM-3 Ab treatment, coinciding with a 2.4-fold increase in transcript levels of prdm1, the gene that encodes Blimp-1 (Fig. 3B), indicative of TEM differentiation toward TEFF. CD27 is transiently upregulated during TCR stimulation and is expressed on memory T cells, but downregulated during TEFF cell differentiation (29). Indeed, we observe a 2.5-fold decrease in cd27 transcript in T cells treated with anti–TIM-3 Ab. Furthermore, gene transcript levels of Tcf7 and Lef1 decreased 5.3-fold and 2-fold respectively (Fig. 3B), suggesting a decrease in Wnt signaling, known to arrest T cell differentiation and promote T cell memory formation (30). Although a slight increase in the gene for T-bet (tbx21) was observed, the fold change was 1.43 and fell below our acceptance criteria (FC <2). Genes that encode tissue homing receptors or integrins, such as cxcr5, itga4 and itgb7, were either not detected or were minimally impacted by anti–TIM-3 treatment (Fig. 3B). However, this does not exclude the possibility of posttranslational regulation of these gene products (31). Taken together, these data show that treatment with anti–TIM-3 Ab increases transcription of genes involved in T cell effector function and differentiation with concomitant decrease in genes associated with T cell memory. This finding reveals a novel role of TIM-3 in the regulation of T cell effector function and TEM → TEFF differentiation in the context of Ag-specific stimulation.
FIGURE 3.
Engagement of TIM-3 by Ab increases transcription of genes involved in T cell effector function with concomitant decrease in genes associated with T cell memory. Ag-specific CD8 T cells were restimulated as described in Fig. 1. Total RNA was extracted from T cells treated with either anti–TIM-3 mAb or isotype control and analyzed by RNA sequencing. (A) Data were evaluated using IPA and the top upstream regulators are shown with predicted activation state, z-score, p value, and gene transcript levels of downstream targets that are increased (green) or decreased (red). (B) A p value cutoff of 0.05 and fold change ≥2 (up- and downregulation) was used to classify transcripts as differentially expressed in treatment condition. Data representative of two donors.
TIM-3 engagement augments re-expression of CCR7 and CD62L on Ag-specific CD8 T cells during activation
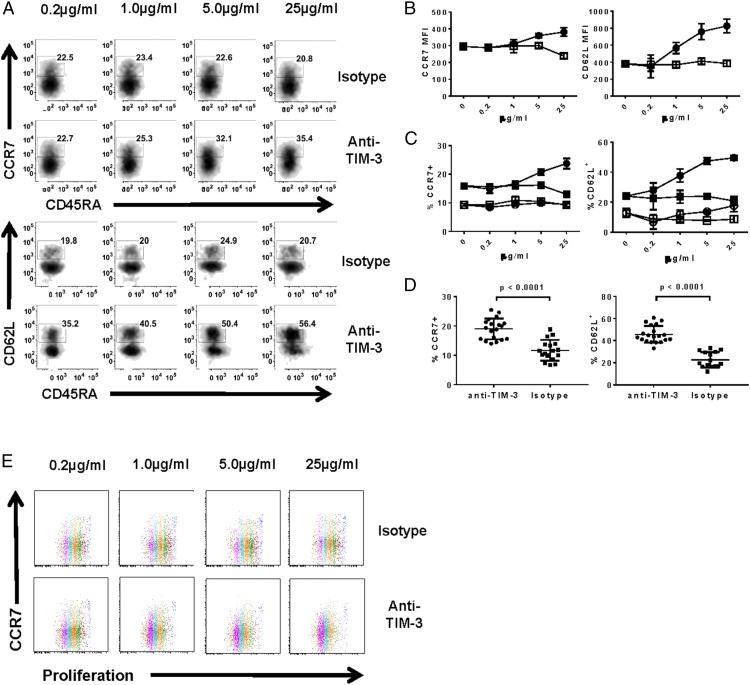
Considering the impact of an anti–TIM-3 Ab on T cell function, we wanted to investigate its role in T cell differentiation. Whereas the shift to TEFF phenotype in response to anti–TIM-3 treatment correlated with the enhancement in effector function, stimulation of Ag-specific CD8 T cells in the presence of an anti–TIM-3 Ab resulted in an increase in the frequency of T cells that phenotypically resemble TCM (CCR7+CD45RA− or CD62L+CD45RA− T cells) (Fig. 4) (32). The increase was observed both in the expression level (in this study determined by mean fluorescence intensity) of CCR7 and CD62L as well as apparent TCM frequency (Fig. 4B, 4C). This effect was only observed in the Ag-specific population (Fig. 4C) despite TIM-3 expression being present on Ag-specific and nonspecific cells (Fig. 1C). This finding was consistent across multiple donors (Fig. 4D). Although these data suggest an increase in TCM in response to anti–TIM-3 treatment, the apparent increase in CCR7 and CD62L expression may also represent a phase of transient activation as has been previously reported (33, 34). We expect bona fide TCM cells to have enhanced proliferative capacity (35); however, we found that the CCR7+ population underwent the same number of cell divisions as CCR7− cells (Fig. 4E). These results indicate that the CCR7+ population was similarly responding to Ag stimulation in the presence of anti–TIM-3 Ab, making it unlikely that these T cells are true TCM.
FIGURE 4.
Ag-specific CD8 T cells increase expression of CCR7 and CD62L in response to anti–TIM-3 Ab. Ag-specific CD8 T cells were cultured as in Fig. 1 in the presence of anti–TIM-3 mAb or isotype control, and analyzed by flow cytometry. (A) Representative density plots of CCR7 versus CD45RA and CD62L versus CD45RA on T cells treated with varying concentrations of anti–TIM-3 mAb/isotype control. (B) Graphical representation of CCR7 and CD62L geometric mean fluorescence intensities in response to anti–TIM-3 mAb (filled circles) or isotype control (open squares). (C) Frequency of CCR7+ and CD62L+ cells in tetramer+ (filled shapes) or tetramer− (open shapes) in response to anti–TIM-3 mAb (circles) or isotype control (squares). (D) CCR7 and CD62L expression on multiple donors when treated with 25 μg/ml anti–TIM-3 Ab (*p < 0.0001, paired two-tailed t test). (E) Representative dot plots of CCR7 versus proliferation dye dilution on T cells treated with varying concentrations of anti–TIM-3 mAb/isotype control. Cell generations are differentiated by color. Data shown are representative of four to six biological replicates per treatment condition performed in three independent experiments.
Terminally differentiated TEFF exhibit decreased responsiveness to TIM-3 engagement compared with TEM
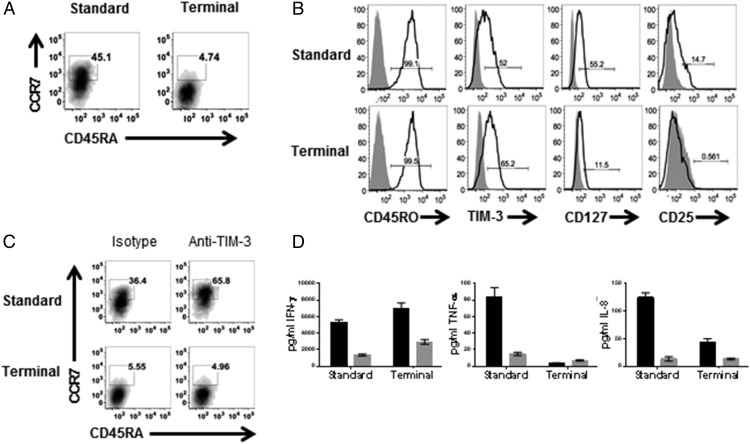
To further address whether the CD62L+CCR7+ phenotype was representative of a transient state during T cell differentiation or whether TIM-3 engagement was augmenting proliferation of existing TCM, the Ag-specific CD8 T cells were stimulated three times beyond the standard protocol to drive terminal differentiation. This shifted the population to a predominantly TEFF phenotype (CCR7−CD45RA−) with a smaller, but still present, TCM population (CCR7+CD45RA−) (Fig. 5A). These cells expressed CD45RO and TIM-3, but had decreased expression of CD127 and CD25, indicative of terminal differentiation (Fig. 5B) (36, 37). TIM-3 engagement enhanced the frequencies of T cells bearing the TCM phenotype when T cells were stimulated under standard conditions. However, treatment of terminally differentiated T cells with anti–TIM-3 Ab no longer induced an increase in CCR7+ T cells (Fig. 5C), indicating TIM-3 engagement does not increase proliferation of existing TCM. Although these cells increased IFN-γ production in response to anti–TIM-3 Ab, TNF-α and IL-8 production were reduced compared with cytokine production by cells that were stimulated with the standard protocol (Fig. 5D), indicating limited impact of TIM-3 engagement on effector T cells can re-express CCR7 in a transient activation state (33, 34). Rather than augmenting the TCM population, it is more likely that the increase in CCR7+CD45RA− and CD62L+CD45RA− T cells observed with anti–TIM-3 treatment represents a transient state during which TEM are differentiating toward TEFF. Together, these results further support our observations that TIM-3 engagement drives TEM → TEFF differentiation, with a transient re-expression of CCR7 and CD62L during TEM activation, and that terminally differentiated TEFF cells begin to lose responsiveness to anti–TIM-3 treatment.
FIGURE 5.
Ag-specific TEFF exhibit reduced responsiveness to anti–TIM-3 Ab compared with TEM. Ag-specific CD8 T cells were cultured as in Fig. 1 (standard) or stimulated an additional three times with aAPC loaded with MART1 peptide (terminal), as described in Materials and Methods, and analyzed by flow cytometry for expression of (A) CCR7 versus CD45RA and (B) CD45RO, TIM-3, CD127, and CD25. Cells stimulated with standard or terminal protocol were treated with anti–TIM-3 mAb (black bars) or isotype control (gray bars), and analyzed for (C) expression of CCR7 versus CD45RA by flow cytometry and (D) cytokine production. Data shown are representative of four to six biological replicates per treatment condition performed in three independent experiments.
PD-1 engagement does not impact or synergize with TIM-3 to enhance Ag-specific T cell effector function
Because coexpression of PD-1 and TIM-3 on T cells is associated with disease (3, 4, 38), coengagement of these two receptors on T cells during Ag stimulation was investigated. MART1-specific CD8 T cells were stimulated with peptide-loaded aAPC in the presence of either anti–TIM-3, anti–PD-1 Ab alone, or in combination. Treatment with anti–TIM-3 enhanced T cell proliferation, but inclusion of both Abs during Ag stimulation did not augment proliferation compared with treatment with anti–TIM-3 alone (Fig. 6A). Nor did anti–PD-1 alone induce any changes in T cell proliferation. Similarly, levels of IFN-γ and TNF-α were increased by treatment with anti–TIM-3 alone but not anti–PD-1, and the combination of Abs did not show any synergistic effect beyond what was observed with anti–TIM-3 treatment alone (Fig. 6B). Whereas treatment with anti–PD-1 did not induce changes in effector function, we wondered whether PD-1 engagement impacts expression of CCR7 as observed with TIM-3 engagement. Anti–PD-1 treatment did not increase the frequency of CCR7+ CD8 T cells nor did it enhance the ability of TIM-3 engagement to do so (Fig. 6C). Note that the aAPC do not express PD-1 ligands. Therefore, the only source of PD-L1 would be the T cells and it is unclear whether PD-L1 expression in cis, or trans interactions with other T cells, could trigger an immune regulatory mechanism. These data suggest that in the absence of trans expression of ligand on APC in the context of an immunological synapse, anti–PD-1 treatment has no impact, nor does it synergize with anti–TIM-3 in mediating T cell activation and driving TEM → TEFF differentiation.
FIGURE 6.
Combined engagement of TIM-3 and PD-1 does not impact T cell function. Ag-specific CD8 T cells were cultured as in Fig. 1 in the presence of anti–TIM-3 mAb, anti–PD-1 mAb, or anti–TIM-3 and anti–PD-1 combined, and analyzed for (A) proliferation by flow cytometry and (B) cytokine production. (C) Representative dot plots showing expression of CCR7 versus CD45RA expression and MART1 tetramer expression on Ag-specific CD8 T cells (*p < 0.0001, two-way ANOVA.) Data shown are representative of four to six biological replicates per treatment condition performed in three independent experiments.
TIM-3 functions in concert with TCR signaling through mTOR to drive effector differentiation in Ag-specific T cells
It has been reported that ectopic expression of TIM-3 in Jurkat cells leads to increased TCR signaling and downstream cytokine production (9). It remains unclear how TIM-3 and TCR signaling synergize during a TCR-MHC activation event. mTOR kinase, demonstrated to play a crucial role in T cell activation and differentiation, can signal via two distinct signaling complexes: mTORC1 and mTORC2. Signaling through mTORC1 has been linked to TEFF differentiation whereas inhibition of this pathway promotes TCM (17). We hypothesized that TIM-3 engagement enhances mTORC1 signaling (39–41). mTORC1 activation leads to phosphorylation of S6K1, pS6K1 phosphorylates ribosomal protein S6 at S240/244 and S235/236 (41). Therefore, mTORC1 activation can be quantitated by measuring pS6 levels in the presence or absence of rapamycin (42). We predicted that pS6 would increase in T cells following stimulation in the presence of anti–TIM-3 Ab if engagement of TIM-3 enhances TEM → TEFF differentiation (40, 43). Indeed, treatment with anti–TIM-3 increased and prolonged expression of pS6 and this event was reversible with rapamycin (Fig. 7A). Furthermore, RNA sequencing analysis indicated that anti–TIM-3 Ab treatment of Ag-specific CD8 T cells increased expression of rhebl1 whose gene product is an isoform of Rheb, the main activator protein of the mTOR pathway (15, 16), and this observation was confirmed by real-time PCR (Fig. 7B). Interestingly, no changes were observed in the main isoform of Rheb, which has reported function in T cell signaling (15, 16), thus showing, to our knowledge for the first time, the presence of RhebL1 in T cells and its role in the T cell differentiation cascade. Taken together, these data support the hypothesis that Ab-mediated engagement of TIM-3 drives T cell activation and differentiation of TEM toward TEFF, and that this occurs through increased rhebl1 transcription and enhanced mTORC1 activation (Fig. 8).
FIGURE 7.
Engagement of TIM-3 increases mTORC1 activation. MART1-specific T cells were restimulated for either 48 or 72 h in the presence of anti–TIM-3 mAb or isotype control. (A) Representative histograms of pS6 240/S244 expression (black line) on T cells incubated with isotype, anti–TIM-3 mAb, or anti–TIM-3 mAb with rapamycin. (B) RNA from T cells treated with either anti–TIM-3 mAb or isotype control was isolated and gene expression of Rheb and RhebL1 was analyzed by RNA sequencing and real-time PCR. Flow cytometry data representative of four independent experiments. RNA analysis representative of two individual donors.
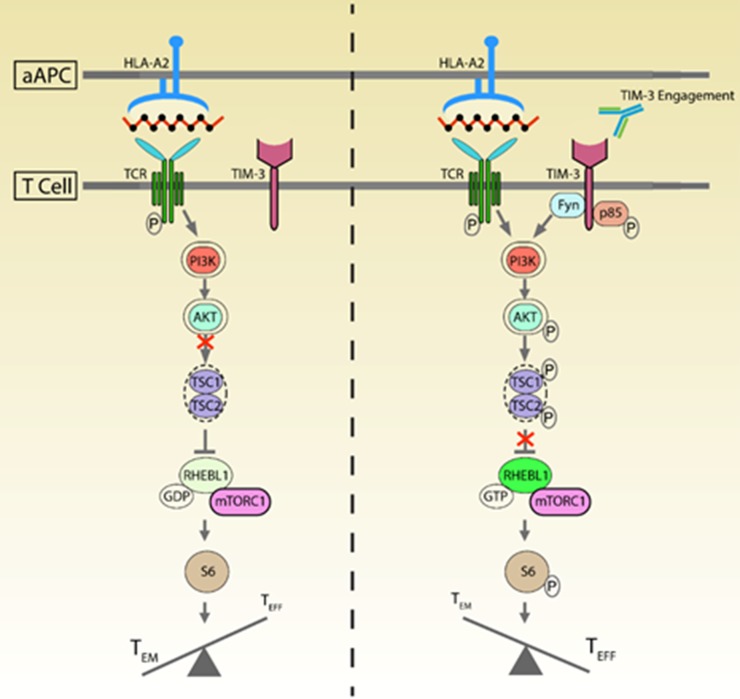
FIGURE 8.
Engagement of TIM-3 by Ab during Ag stimulation of CD8 T cells activates mTORC1 to drive differentiation into effector cells. CD8 T cells responding to Ag-specific stimulation in vitro act as predicted by proliferating and increasing cytokine production, and they exhibit target killing in a typical cytotoxicity assay. The known ligands, Gal9, HMGB1, and PtdS, are all likely present in that they are widely expressed by many different cell types including T cells, yet the mTOR pathway is not uniformly activated so there is a higher frequency of cells representing TEM compared with TEFF (left). Addition of anti–TIM-3 Ab during Ag stimulation results in specific activation of mTORC1 shown by increased pS6 and RhebL1 expression and ultimately the frequency of TEFF is increased (right).
Discussion
We demonstrated that TIM-3 is expressed on human primary Ag-specific CD8 T cells upon TCR-MHC–mediated stimulation of cells from healthy donors. Direct engagement of TIM-3 by a monoclonal anti–TIM-3 Ab enhanced effector function. Although TIM-3 has been associated with the negative regulation of T cell function (1, 3, 44), our data suggest that engagement of TIM-3 on Ag-specific CD8 T cells by mAb promotes the function of TEM/TEFF cells during Ag-stimulation. To date, there is no agreement on whether TIM-3 enhances or suppresses T cell function. The data in this study do not definitively show whether TIM-3 engagement with an Ab, in our studies rat clone 344823, is agonistic or antagonistic. We assume that known ligands, GAL-9, HMGB1, and phosphatidylserine, are present as the cultures contain dead cells. Although the rat clone 344823 anti–TIM-3 Ab inhibits GAL-9 binding to recombinant TIM-3 protein in competition studies (data not shown), we cannot conclude that this is the mechanism by which the Ab induces TIM-3 signaling. Furthermore, it is feasible that the Ab blocks interaction with one of the ligands, whereas other ligands could still engage TIM-3 (45–47). In addition, there is no consensus on the relevant ligand that drives TIM-3 function, thus confounding interpretation of the effect of anti–TIM-3 treatment as agonistic or antagonistic. To exclude the possibility of intrinsic agonism of TIM-3, future experiments could include treating Ag-specific cells with only the Fab portion of the rat anti-human TIM-3 Ab during restimulation, thus preventing bivalent cross-linking of TIM-3. In contrast, binding properties could change when the Ab is converted to only Fab fragments (i.e., affinity, stoichiometry), impacting TIM-3 ligand binding. Therefore, data from such experiments may be difficult to interpret and beyond the scope of this study. Instead, rather than defining the agonistic or antagonistic effect of the Ab, the findings reported in this study demonstrate how TIM-3 is involved in differentiation of Ag-specific CD8 TEM/TEFF cells, improving understanding of the function of this receptor, and the mechanism of action of Ab treatment in a therapeutic context.
We report that anti–TIM-3 treatment also increases expression of CCR7 and CD62L on Ag-specific CD8 T cells during restimulation. Our data suggest it is unlikely that CCR7+ T cells represent TCM because both CCR7− and CCR7+ populations proliferated similarly upon Ag stimulation. More likely, CCR7 and CD62L were re-expressed because of T cell activation (34, 48), and this transient re-expression is enhanced with TIM-3 engagement due to augmented T cell signaling. We also show diminished impact on effector function when anti–TIM-3 Ab is added to T cells that are terminally differentiated. If TIM-3 engagement solely impacted effector function in the absence of T cell differentiation, one would expect to see similar increases in effector responses on terminally differentiated cells as with less-differentiated cells. Our findings reveal how TIM-3 signaling drives T cell differentiation, which is coupled with effector function.
Although our results obtained from treatment with a combination of anti–TIM-3 and anti–PD-1 Abs in vitro seem to contradict results reported in vivo (2, 49), the Abs used in the referenced in vivo studies targeted PD-L1, a more broadly expressed Ag compared with PD-1 (50, 51). Abs targeting PD-L1 and TIM-3 in vivo may act on multiple cell populations, rendering the overall outcome more efficacious compared with treatment with either Ab individually. The culture system presented in this study addresses the impact of treatment specifically (and only) on Ag-specific CD8 T cells. PD-1 has been associated with inhibition of mTORC1 via PI3K and Akt resulting in loss of TEFF and increased memory T cells upon interaction with PD-L1 (52). Presumably, combined engagement of TIM-3 and PD-1 would have a synergistic effect on TEFF differentiation. However, the only (if any) source of PD-L1 in this system would be presented on T cells as there are no PD-1 ligands expressed on the aAPC. Therefore, any PD-1/PD-L1 interactions would occur beyond the immunological synapse and are likely less stable. In this context, anti–TIM-3 treatment is activating the mTOR pathway and any effect of the anti–PD-1 treatment is masked.
Our data reveal that RhebL1, but not Rheb, is involved in the T cell–differentiation signaling cascade via TIM-3. Although the differences in function between the two isoforms remain unclear, it has been suggested they impact mTOR signaling differently (53). To date there are no known reports about RhebL1 in T cells, however, it has been shown that overexpression of RhebL1 in mice enhances hematopoietic progenitor cell growth while impairing stem cell repopulation (54), implicating its involvement with cellular differentiation. Our data support the hypothesis that anti–TIM-3 treatment acts on CD8 T cells during Ag stimulation by activating mTORC1 signaling via RhebL1 and drives TEFF differentiation (Fig. 8). This hypothesis is supported by a similar model proposed by Ferris et al. (11) in that engagement of TIM-3 results in signaling through mTOR resulting in either exhaustion or activation depending on the duration of signaling.
We propose that TIM-3 signaling is necessary for TEFF differentiation. If the signaling is suboptimal, T cell differentiation shifts to memory T cell differentiation due to incomplete/weak activation of the mTORC1 (Fig. 8). However, robust TIM-3 signaling concomitant with Ag-stimulation results in sufficient mTORC1 activation to drive TEFF differentiation. Treatment of human Ag-specific CD8 T cells with anti–TIM-3 Ab during stimulation produces a population with a phenotype reminiscent of murine TSC2 KO T cells, which exhibit mTORC1 hyperactivity with heightened effector function as shown by increased proliferation and cytokine production but little difference in cytotoxic activity (17). Activation of the mTOR pathway by TIM-3 engagement has also been demonstrated in human leukocytes (55), although these studies were performed in the absence of Ag stimulation. Altogether these findings support our hypothesis that anti–TIM-3 Ab treatment enhances effector CD8 T cell differentiation via increased mTORC1 signaling.
Translating these findings in vivo remains a challenge due to a microenvironment comprised of multiple cell populations contributing to a dynamic milieu of soluble factors. In this context, the Ab may function by targeting a subpopulation of functional cells present within the general T cell population. However, our conclusions reveal one aspect of a complicated role of TIM-3 in T cell differentiation during Ag encounter and its potential as a cancer immunotherapy.
Supplementary Material
Acknowledgments
We thank Helen Horton and Russell Lingham for helpful discussions and Jason Ekert and Jill Mooney for providing reagents to expand the aAPCs and MART-1–specific CD8 T cells.
S.S.-M. is the senior author.
The online version of this article contains supplemental material.
- aAPC
- artificial APC
- DC
- dendritic cell
- IPA
- Ingenuity Pathway Analysis
- mTOR
- mammalian target of rapamycin
- TCM
- T central memory
- TEFF
- effector T
- TEM
- effector memory T
- TIM-3
- T cell Ig and mucin domain containing molecule-3.
Disclosures
The authors have no financial conflicts of interest.
References
- 1.Jones R. B., Ndhlovu L. C., Barbour J. D., Sheth P. M., Jha A. R., Long B. R., Wong J. C., Satkunarajah M., Schweneker M., Chapman J. M., et al. 2008. Tim-3 expression defines a novel population of dysfunctional T cells with highly elevated frequencies in progressive HIV-1 infection. J. Exp. Med. 205: 2763–2779. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Jin H. T., Anderson A. C., Tan W. G., West E. E., Ha S. J., Araki K., Freeman G. J., Kuchroo V. K., Ahmed R. 2010. Cooperation of Tim-3 and PD-1 in CD8 T-cell exhaustion during chronic viral infection. Proc. Natl. Acad. Sci. USA 107: 14733–14738. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Fourcade J., Sun Z., Benallaoua M., Guillaume P., Luescher I. F., Sander C., Kirkwood J. M., Kuchroo V., Zarour H. M. 2010. Upregulation of Tim-3 and PD-1 expression is associated with tumor antigen-specific CD8+ T cell dysfunction in melanoma patients. J. Exp. Med. 207: 2175–2186. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.McMahan R. H., Golden-Mason L., Nishimura M. I., McMahon B. J., Kemper M., Allen T. M., Gretch D. R., Rosen H. R. 2010. Tim-3 expression on PD-1+ HCV-specific human CTLs is associated with viral persistence, and its blockade restores hepatocyte-directed in vitro cytotoxicity. J. Clin. Invest. 120: 4546–4557. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Qiu Y., Chen J., Liao H., Zhang Y., Wang H., Li S., Luo Y., Fang D., Li G., Zhou B., et al. 2012. Tim-3-expressing CD4+ and CD8+ T cells in human tuberculosis (TB) exhibit polarized effector memory phenotypes and stronger anti-TB effector functions. PLoS Pathog. 8: e1002984. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Gorman J. V., Starbeck-Miller G., Pham N. L., Traver G. L., Rothman P. B., Harty J. T., Colgan J. D. 2014. Tim-3 directly enhances CD8 T cell responses to acute Listeria monocytogenes infection. J. Immunol. 192: 3133–3142. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Sabins N. C., Harman B. C., Barone L. R., Shen S., Santulli-Marotto S. 2016. Differential expression of immune checkpoint modulators on in vitro primed CD4(+) and CD8(+) T cells. Front. Immunol. 7: 221. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Anderson A. C., Anderson D. E., Bregoli L., Hastings W. D., Kassam N., Lei C., Chandwaskar R., Karman J., Su E. W., Hirashima M., et al. 2007. Promotion of tissue inflammation by the immune receptor Tim-3 expressed on innate immune cells. Science 318: 1141–1143. [DOI] [PubMed] [Google Scholar]
- 9.Lee J., Su E. W., Zhu C., Hainline S., Phuah J., Moroco J. A., Smithgall T. E., Kuchroo V. K., Kane L. P. 2011. Phosphotyrosine-dependent coupling of Tim-3 to T-cell receptor signaling pathways. Mol. Cell. Biol. 31: 3963–3974. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Tomkowicz B., Walsh E., Cotty A., Verona R., Sabins N., Kaplan F., Santulli-Marotto S., Chin C. N., Mooney J., Lingham R. B., et al. 2015. TIM-3 suppresses anti-CD3/CD28-induced TCR activation and IL-2 expression through the NFAT signaling pathway. PLoS One 10: e0140694. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Ferris R. L., Lu B., Kane L. P. 2014. Too much of a good thing? Tim-3 and TCR signaling in T cell exhaustion. J. Immunol. 193: 1525–1530. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Johnston R. J., Poholek A. C., DiToro D., Yusuf I., Eto D., Barnett B., Dent A. L., Craft J., Crotty S. 2009. Bcl6 and Blimp-1 are reciprocal and antagonistic regulators of T follicular helper cell differentiation. Science 325: 1006–1010. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Crotty S., Johnston R. J., Schoenberger S. P. 2010. Effectors and memories: Bcl-6 and Blimp-1 in T and B lymphocyte differentiation. Nat. Immunol. 11: 114–120. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Xin A., Masson F., Liao Y., Preston S., Guan T., Gloury R., Olshansky M., Lin J. X., Li P., Speed T. P., et al. 2016. A molecular threshold for effector CD8(+) T cell differentiation controlled by transcription factors Blimp-1 and T-bet. Nat. Immunol. 17: 422–432. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Laplante M., Sabatini D. M. 2012. mTOR signaling in growth control and disease. Cell 149: 274–293. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Inoki K., Li Y., Xu T., Guan K. L. 2003. Rheb GTPase is a direct target of TSC2 GAP activity and regulates mTOR signaling. Genes Dev. 17: 1829–1834. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Pollizzi K. N., Patel C. H., Sun I. H., Oh M. H., Waickman A. T., Wen J., Delgoffe G. M., Powell J. D. 2015. mTORC1 and mTORC2 selectively regulate CD8+ T cell differentiation. J. Clin. Invest. 125: 2090–2108. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Pollizzi K. N., Sun I. H., Patel C. H., Lo Y. C., Oh M. H., Waickman A. T., Tam A. J., Blosser R. L., Wen J., Delgoffe G. M., Powell J. D. 2016. Asymmetric inheritance of mTORC1 kinase activity during division dictates CD8(+) T cell differentiation. Nat. Immunol. 17: 704–711. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Shrestha S., Yang K., Wei J., Karmaus P. W., Neale G., Chi H. 2014. Tsc1 promotes the differentiation of memory CD8+ T cells via orchestrating the transcriptional and metabolic programs. Proc. Natl. Acad. Sci. USA 111: 14858–14863. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Waickman A. T., Powell J. D. 2012. Mammalian target of rapamycin integrates diverse inputs to guide the outcome of antigen recognition in T cells. J. Immunol. 188: 4721–4729. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Gattinoni L., Lugli E., Ji Y., Pos Z., Paulos C. M., Quigley M. F., Almeida J. R., Gostick E., Yu Z., Carpenito C., et al. 2011. A human memory T cell subset with stem cell-like properties. Nat. Med. 17: 1290–1297. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Farber D. L., Yudanin N. A., Restifo N. P. 2014. Human memory T cells: generation, compartmentalization and homeostasis. Nat. Rev. Immunol. 14: 24–35. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Mahnke Y. D., Brodie T. M., Sallusto F., Roederer M., Lugli E. 2013. The who’s who of T-cell differentiation: human memory T-cell subsets. Eur. J. Immunol. 43: 2797–2809. [DOI] [PubMed] [Google Scholar]
- 24.Legat A., Speiser D. E., Pircher H., Zehn D., Fuertes Marraco S. A. 2013. Inhibitory receptor expression depends more dominantly on differentiation and activation than “Exhaustion” of human CD8 T cells. Front. Immunol. 4: 455. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Roederer M. 2011. Interpretation of cellular proliferation data: avoid the panglossian. Cytometry A 79: 95–101. [DOI] [PubMed] [Google Scholar]
- 26.Golden-Mason L., Palmer B. E., Kassam N., Townshend-Bulson L., Livingston S., McMahon B. J., Castelblanco N., Kuchroo V., Gretch D. R., Rosen H. R. 2009. Negative immune regulator Tim-3 is overexpressed on T cells in hepatitis C virus infection and its blockade rescues dysfunctional CD4+ and CD8+ T cells. J. Virol. 83: 9122–9130. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Martins G., Calame K. 2008. Regulation and functions of Blimp-1 in T and B lymphocytes. Annu. Rev. Immunol. 26: 133–169. [DOI] [PubMed] [Google Scholar]
- 28.Ichii H., Sakamoto A., Arima M., Hatano M., Kuroda Y., Tokuhisa T. 2007. Bcl6 is essential for the generation of long-term memory CD4+ T cells. Int. Immunol. 19: 427–433. [DOI] [PubMed] [Google Scholar]
- 29.Tomiyama H., Matsuda T., Takiguchi M. 2002. Differentiation of human CD8(+) T cells from a memory to memory/effector phenotype. J. Immunol. 168: 5538–5550. [DOI] [PubMed] [Google Scholar]
- 30.Muralidharan S., Hanley P. J., Liu E., Chakraborty R., Bollard C., Shpall E., Rooney C., Savoldo B., Rodgers J., Dotti G. 2011. Activation of Wnt signaling arrests effector differentiation in human peripheral and cord blood-derived T lymphocytes. J. Immunol. 187: 5221–5232. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Jang E. J., Park H. R., Hong J. H., Hwang E. S. 2013. Lysine 313 of T-box is crucial for modulation of protein stability, DNA binding, and threonine phosphorylation of T-bet. J. Immunol. 190: 5764–5770. [DOI] [PubMed] [Google Scholar]
- 32.Unsoeld H., Pircher H. 2005. Complex memory T-cell phenotypes revealed by coexpression of CD62L and CCR7. J. Virol. 79: 4510–4513. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.van Leeuwen E. M., Gamadia L. E., Baars P. A., Remmerswaal E. B., ten Berge I. J., van Lier R. A. 2002. Proliferation requirements of cytomegalovirus-specific, effector-type human CD8+ T cells. J. Immunol. 169: 5838–5843. [DOI] [PubMed] [Google Scholar]
- 34.van Leeuwen E. M., van Buul J. D., Remmerswaal E. B., Hordijk P. L., ten Berge I. J., van Lier R. A. 2005. Functional re-expression of CCR7 on CMV-specific CD8+ T cells upon antigenic stimulation. Int. Immunol. 17: 713–719. [DOI] [PubMed] [Google Scholar]
- 35.Restifo N. P., Dudley M. E., Rosenberg S. A. 2012. Adoptive immunotherapy for cancer: harnessing the T cell response. Nat. Rev. Immunol. 12: 269–281. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Kaech S. M., Tan J. T., Wherry E. J., Konieczny B. T., Surh C. D., Ahmed R. 2003. Selective expression of the interleukin 7 receptor identifies effector CD8 T cells that give rise to long-lived memory cells. Nat. Immunol. 4: 1191–1198. [DOI] [PubMed] [Google Scholar]
- 37.Herndler-Brandstetter D., Schwaiger S., Veel E., Fehrer C., Cioca D. P., Almanzar G., Keller M., Pfister G., Parson W., Würzner R., et al. 2005. CD25-expressing CD8+ T cells are potent memory cells in old age. J. Immunol. 175: 1566–1574. [DOI] [PubMed] [Google Scholar]
- 38.Baitsch L., Legat A., Barba L., Fuertes Marraco S. A., Rivals J. P., Baumgaertner P., Christiansen-Jucht C., Bouzourene H., Rimoldi D., Pircher H., et al. 2012. Extended co-expression of inhibitory receptors by human CD8 T-cells depending on differentiation, antigen-specificity and anatomical localization. PLoS One 7: e30852. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Rao R. R., Li Q., Odunsi K., Shrikant P. A. 2010. The mTOR kinase determines effector versus memory CD8+ T cell fate by regulating the expression of transcription factors T-bet and Eomesodermin. Immunity 32: 67–78. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Li Q., Rao R. R., Araki K., Pollizzi K., Odunsi K., Powell J. D., Shrikant P. A. 2011. A central role for mTOR kinase in homeostatic proliferation induced CD8+ T cell memory and tumor immunity. Immunity 34: 541–553. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Salmond R. J., Emery J., Okkenhaug K., Zamoyska R. 2009. MAPK, phosphatidylinositol 3-kinase, and mammalian target of rapamycin pathways converge at the level of ribosomal protein S6 phosphorylation to control metabolic signaling in CD8 T cells. J. Immunol. 183: 7388–7397. [DOI] [PubMed] [Google Scholar]
- 42.Burnett P. E., Barrow R. K., Cohen N. A., Snyder S. H., Sabatini D. M. 1998. RAFT1 phosphorylation of the translational regulators p70 S6 kinase and 4E-BP1. Proc. Natl. Acad. Sci. USA 95: 1432–1437. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Sehgal S. N. 2003. Sirolimus: its discovery, biological properties, and mechanism of action. Transplant. Proc. 35(3 Suppl.): 7S–14S. [DOI] [PubMed] [Google Scholar]
- 44.Blackburn S. D., Shin H., Haining W. N., Zou T., Workman C. J., Polley A., Betts M. R., Freeman G. J., Vignali D. A., Wherry E. J. 2009. Coregulation of CD8+ T cell exhaustion by multiple inhibitory receptors during chronic viral infection. Nat. Immunol. 10: 29–37. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.DeKruyff R. H., Bu X., Ballesteros A., Santiago C., Chim Y. L., Lee H. H., Karisola P., Pichavant M., Kaplan G. G., Umetsu D. T., et al. 2010. T cell/transmembrane, Ig, and mucin-3 allelic variants differentially recognize phosphatidylserine and mediate phagocytosis of apoptotic cells. J. Immunol. 184: 1918–1930. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Cao E., Zang X., Ramagopal U. A., Mukhopadhaya A., Fedorov A., Fedorov E., Zencheck W. D., Lary J. W., Cole J. L., Deng H., et al. 2007. T cell immunoglobulin mucin-3 crystal structure reveals a galectin-9-independent ligand-binding surface. Immunity 26: 311–321. [DOI] [PubMed] [Google Scholar]
- 47.Freeman G. J., Casasnovas J. M., Umetsu D. T., DeKruyff R. H. 2010. TIM genes: a family of cell surface phosphatidylserine receptors that regulate innate and adaptive immunity. Immunol. Rev. 235: 172–189. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48.Champagne P., Ogg G. S., King A. S., Knabenhans C., Ellefsen K., Nobile M., Appay V., Rizzardi G. P., Fleury S., Lipp M., et al. 2001. Skewed maturation of memory HIV-specific CD8 T lymphocytes. Nature 410: 106–111. [DOI] [PubMed] [Google Scholar]
- 49.Sakuishi K., Apetoh L., Sullivan J. M., Blazar B. R., Kuchroo V. K., Anderson A. C. 2010. Targeting Tim-3 and PD-1 pathways to reverse T cell exhaustion and restore anti-tumor immunity [Published erratum appears in 2011 J. Exp. Med. 208: 1331.] J. Exp. Med. 207: 2187–2194. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50.Benson D. M., Jr., Bakan C. E., Mishra A., Hofmeister C. C., Efebera Y., Becknell B., Baiocchi R. A., Zhang J., Yu J., Smith M. K., et al. 2010. The PD-1/PD-L1 axis modulates the natural killer cell versus multiple myeloma effect: a therapeutic target for CT-011, a novel monoclonal anti-PD-1 antibody. Blood 116: 2286–2294. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 51.Graziotto R., Foresta C., Scannapieco P., Zeilante P., Russo A., Negro A., Salmaso R., Onisto M. 1999. cDNA cloning and characterization of PD1: a novel human testicular protein with different expressions in various testiculopathies. Exp. Cell Res. 248: 620–626. [DOI] [PubMed] [Google Scholar]
- 52.Staron M. M., Gray S. M., Marshall H. D., Parish I. A., Chen J. H., Perry C. J., Cui G., Li M. O., Kaech S. M. 2014. The transcription factor FoxO1 sustains expression of the inhibitory receptor PD-1 and survival of antiviral CD8(+) T cells during chronic infection. Immunity 41: 802–814. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 53.Bonneau A., Parmar N. 2012. Effects of RhebL1 silencing on the mTOR pathway. Mol. Biol. Rep. 39: 2129–2137. [DOI] [PubMed] [Google Scholar]
- 54.Campbell T. B., Basu S., Hangoc G., Tao W., Broxmeyer H. E. 2009. Overexpression of Rheb2 enhances mouse hematopoietic progenitor cell growth while impairing stem cell repopulation. Blood 114: 3392–3401. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 55.Gonçalves Silva I., Gibbs B. F., Bardelli M., Varani L., Sumbayev V. V. 2015. Differential expression and biochemical activity of the immune receptor Tim-3 in healthy and malignant human myeloid cells. Oncotarget 6: 33823–33833. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.