Abstract
Genetic polymorphisms in human N-acetyltransferase 2 (NAT2) modify the metabolism of numerous drugs and carcinogens. These genetic polymorphisms modify both drug efficacy and toxicity and cancer risk associated with carcinogen exposure. Previous studies have suggested phenotypic heterogeneity among different NAT2 slow acetylator genotypes. NAT2 phenotype was investigated in vitro and in situ in samples of human hepatocytes obtained from various NAT2 slow and intermediate NAT2 acetylator genotypes. NAT2 gene dose response (NAT2*5B/*5B > NAT2*5B/*6A > NAT2*6A/*6A) was observed towards the N-acetylation of the NAT2-specific drug sulfamethazine by human hepatocytes both in vitro and in situ. N-acetylation of 4-aminobiphenyl, an arylamine carcinogen substrate for both N-acetyltransferase 1 and NAT2, showed the same trend both in vitro and in situ although the differences were not significant (p>0.05). The N-acetylation of the N-acetyltransferase 1-specific substrate p-aminobenzoic acid did not follow this trend. In comparisons of NAT2 intermediate acetylator genotypes, differences in N-acetylation between NAT2*4/*5B and NAT2*4/*6B hepatocytes were not observed in vitro or in situ towards any of these substrates. These results further support phenotypic heterogeneity among NAT2 slow acetylator genotypes, consistent with differential risks of drug failure or toxicity and cancer associated with carcinogen exposure.
Keywords: N-acetyltransferase 2, slow acetylator genotype, human hepatocytes, sulfamethazine, 4-aminobiphenyl
Introduction
Genetic polymorphism in arylamine N-acetyltransferase 2 (NAT2) modifies the therapeutic efficacy and toxicity of numerous drugs (Weber and Hein, 1985; McDonagh et al., 2014). Human NAT2 also catalyzes the N-acetylation and O-acetylation of xenobiotics and carcinogens (Hein 1988). Genetic polymorphisms in NAT2 have been shown to be associated with differential susceptibility to various cancers, particularly of the urinary bladder (Garcia-Closas et al., 2005; Moore et al., 2011). Associations with other cancers such as colon (Moslehi et al., 2006; Shin et al., 2008), and breast (Deitz et al., 2000; Ambrosone et al., 2008) also have been reported but the evidence is not as conclusive. As recently reviewed (Hein, in press), several studies have reported that associations of NAT2 genetic polymorphism with breast cancer are strongest in smokers who are “very slow” acetylators (van del Hel et al., 2003; Baumgartner et al., 2009; Conlon et al., 2010).
Common single nucleotide polymorphisms (SNPs) in the NAT2 coding exon modify N-acetylation capacity resulting in bimodal (rapid and slow) or trimodal (rapid, intermediate, and slow) acetylator phenotypes (McDonagh et al., 2014). The absence of SNPs defines the NAT2*4 haplotype associated with rapid acetylator phenotype. Within the slow acetylator phenotype(s), the most common NAT2 SNPs are rs1801280 T341C associated with the NAT2*5B haplotype and rs1799930 G590A associated with the NAT2*6A haplotype. Investigations of recombinant arylamine N-acetyltransferases (Hein, 2002) as well as recent in vivo measurements of N-acetylation capacity measured as urinary metabolite ratios of caffeine (Ruiz et al., 2012; Selinski et al., 2013) are consistent with genetic heterogeneity within the slow acetylator phenotype. A recent guest editorial (Selinski et al., 2015a) eloquently describes the importance of discriminating “ultra-slow” acetylators, particularly the NAT2*6A haplotype, to enable more accurate and robust assessments of the effects of NAT2 genetic polymorphism on not only drug toxicity and cancer risk, but in other broader health contexts such as cognitive function (Selinski et al., 2015b).
Published evidence for genetic heterogeneity within the slow NAT2 acetylator phenotype has been limited primarily to measurement of NAT2 expression following recombinant expression in (non-human) prokaryotic and eukaryotic expression systems or measurement of urinary metabolite ratios in vivo that also reflect other metabolic functions, including possible N-acetylation catalyzed by arylamine N-acetyltransferase 1 (NAT1). Previous studies have shown a very high degree of concordance between measurements of NAT2 catalytic activity of human liver in vitro with the human acetylator phenotype expressed in vivo (Meisel et al., 2001) as well as with human NAT2 genotype (Doll et al., 2010; Hein and Doll, 2012). Thus, we investigated N-acetylation capacity both in vitro and in situ in cryopreserved human hepatocytes possessing the most common slow acetylator haplotypes (NAT2*5B and NAT2*6A). Our investigations included substrates with specificity for NAT1 or NAT2, and a common arylamine carcinogen that is N-acetylated by both NAT1 and NAT2.
Materials and Methods
Human Hepatocytes
Cryopreserved human hepatocyte samples were obtained from Bioreclamation IVT (Baltimore, MD) and stored in liquid nitrogen until use. Upon removal from liquid nitrogen, hepatocytes were thawed according to the manufacturer’s instructions by warming a vial of the hepatocytes at 37°C for 90 s, and transferring the contents to a 50 mL conical tube containing 45 mL of InVitroGRO HT medium (Bioreclamation IVT). The cell suspension was centrifuged at 50 × g at room temperature for 5 min. The supernatant was discarded, and the cells washed once in ice-cold phosphate buffered saline (PBS) before lysing the cells in ice-cold 20 mM NaPO4, 1 mM dithiothreitol, 1 mM EDTA, 0.2% triton-X-100, 1 mM phenylmethylsulfonyl fluoride, 1 μM pepstatin A, and 1 μg/mL aprotinin. The lysate was centrifuged at 15,000 × g for 20 min and the supernatant was aliquoted and stored at 70°C. Protein concentrations in the lysates were determined using the Bio-Rad protein assay (Bio-Rad, Richmond, CA).
NAT2 Acetylator Genotyping Assay
Genomic DNA was isolated from pelleted cells prepared above using the QIAamp DNA mini Kit (Qiagen, Valencia, CA) according to manufacturer’s instruction. NAT2 genotype was assigned based on SNPs in the NAT2 coding region and their corresponding alleles and haplotypes determined with the seven-SNP panel assay as previously described (Doll and Hein, 2001). Briefly, SNP-specific PCR primers and fluorogenic probes were designed using Primer Express™ (Applied Biosystems, CA, USA). The fluorogenic probes are labeled with a reporter dye (either carboxyfluorescein or VIC®, Applied Biosystems) and are specific for one of the two possible bases identified at the seven SNPs rs1801279 (191G>A), rs1041983 (282C>T), rs1801280 (341T>C), rs1799929 (481C>T), rs1799930 (590G>A), rs1208 (803A>G) and rs1799931 (857G>A) in the NAT2 coding region. Assays were designed and optimized to work with TaqMan® Universal PCR Master Mix using thermal cycling conditions 50°C for 2 min, 95°C for 10 min followed by 40 cycles of 95°C for 15 s and 60°C for 1 min. Controls (no DNA template) were run to ensure no amplification of contaminating DNA. NAT2 genotype & inferred phenotype determination from the seven NAT2 coding region SNPs described above, defines most if not all of the reported human NAT2 alleles or haplotypes. NAT2 alleles possessing additional SNPs in the NAT2 coding region are quite rare.
Deduced NAT2 acetylator phenotype
Hepatocyte samples homozygous for NAT2 alleles associated with rapid acetylation activity (NAT2*4, NAT2*11, NAT2*12 and NAT2*13 clusters) were classified as rapid acetylator phenotype; samples heterozygous for one of these alleles and one NAT2 allele associated with slow acetylation (NAT2*5, NAT2*6, NAT2*7 and NAT2*14 clusters) were classified as intermediate acetylator phenotype, and samples homozygous for slow acetylation alleles were classified as slow acetylator phenotype.
In vitro N-acetyltransferase assay
N-acetyltransferase assays were conducted as described previously (Hein et al., 2006). Briefly, reactions containing hepatocyte lysate, arylamine substrate [300 μM for p-aminobenzoic acid (PABA) or sulfamethazine (SMZ)]; 1 mM for 4-aminobiphenyl (ABP), and 1 mM acetyl coenzyme A were incubated at 37°C for 10 min. Reactions were terminated by the addition of 1/10 volume of 1M acetic acid. The reaction tubes were centrifuged to precipitate protein. Reactants and products were separated and quantified by reverse-phase high-performance liquid chromatography.
In situ N-acetyltransferase assay
Hepatocytes were thawed as described above and contents of the vial were transferred into a 15 ml conical tube containing 12 ml of InVitroGRO CP (Bioreclamation IVT) media pre-warmed to 37°C. Cells (0.5 ml/well) were plated into Biocoat® collagen-coated 24-well plates (BD labware, Bedford, MA) and allowed to attach overnight. The next morning media was removed and attached cells washed with 1X PBS and replaced with fresh pre-warmed InVitroGRO CP media containing 10 or 200 μM SMZ, ABP or PABA. Hepatocytes were incubated with substrates for 24 h after which media was removed and protein precipitated by addition of 1/10 volume of 1 M acetic acid. Media was centrifuged at 15,000 × g for 10 min and supernatant used to quantitate acetylated reaction products as above.
Results
N-acetyltransferase catalytic activity in vitro
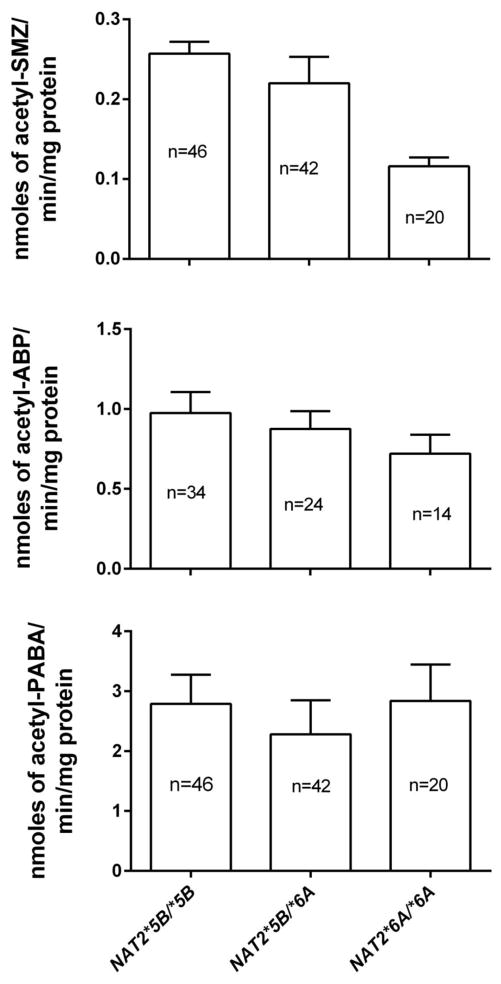
N-acetyltransferase catalytic activity in human hepatocyte lysates was measured from different slow acetylator NAT2 genotypes (NAT2*5B/*5B, NAT2*5B/*6A, NAT2*6A/*6A) using three different substrates. As shown in Fig 1, SMZ N-acetyltransferase catalytic activity in vitro differed significantly (p=0.0031) among these slow acetylator NAT2 genotypes. Samples with NAT2*5B/*5B had the highest catalytic activity followed by samples with NAT2*5B/*6A genotype and samples with NAT2*6A/*6A. N-acetyltransferase catalytic activity towards the arylamine carcinogen ABP showed the same trend as SMZ (NAT2*5B/*5B > NAT2*5B/*6A > NAT2*6A/*6A although the differences for ABP failed to meet statistical significance (p>0.05). N-acetyltransferase catalytic activity towards PABA did not reflect this same trend.
Fig. 1.
SMZ (top), ABP (middle) and PABA (bottom) N-acetyltransferase catalytic activities in vitro in human hepatocyte lysates of NAT2*5B/*5B, NAT2*5B/*6A or NAT2*6A/*6A genotypes. Differences in N-acetyltransferase catalytic activity differed significantly as NAT2*5B/*5B > NAT2*5B/*6A > NAT2*6A/*6A towards SMZ (p=0.0031), but not towards ABP (p=0.4402) or PABA (p=0.7321) following one way analysis of variance. Each bar illustrates mean ± SEM for the number of individual human hepatocyte samples (n) listed. Portions of this data were published previously (Hein and Doll, 2012) where the statistical analysis did not focus on these three NAT2 genotypes.
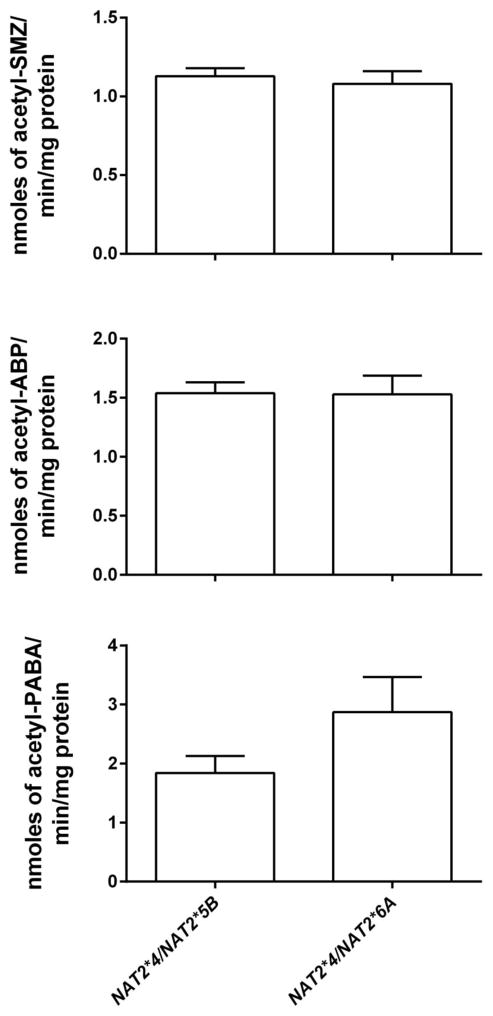
We also measured N-acetyltransferase activity in human hepatocyte lysates with heterozygous NAT2 genotypes NAT2*4/*5B and NAT2*4/*6A. N-acetyltransferase catalytic activities towards SMZ, ABP and PABA did not differ significantly (p>0.05) between these intermediate NAT2 acetylator genotypes (Fig 2).
Fig. 2.
SMZ (top), ABP (middle) and PABA (bottom) N-acetyltransferase catalytic activities in vitro in human hepatocyte lysates of heterozygous genotypes. Each bar illustrates mean ± SEM for NAT2*4/*5B (n=64) and NAT2*4/*6A (n=30) individual human hepatocyte samples. Differences in N-acetyltransferase catalytic activity between the heterozygous NAT2*4/*5B and NAT2*4/*6A genotypes were not significant towards SMZ (p=0.5851), ABP (p=0.9485) or PABA (p=0.0821) respectively for SMZ, ABP or PABA. Portions of this data were published previously (Hein and Doll, 2012) where the statistical analysis did not focus on these two NAT2 genotypes.
N-acetylation capacity in situ
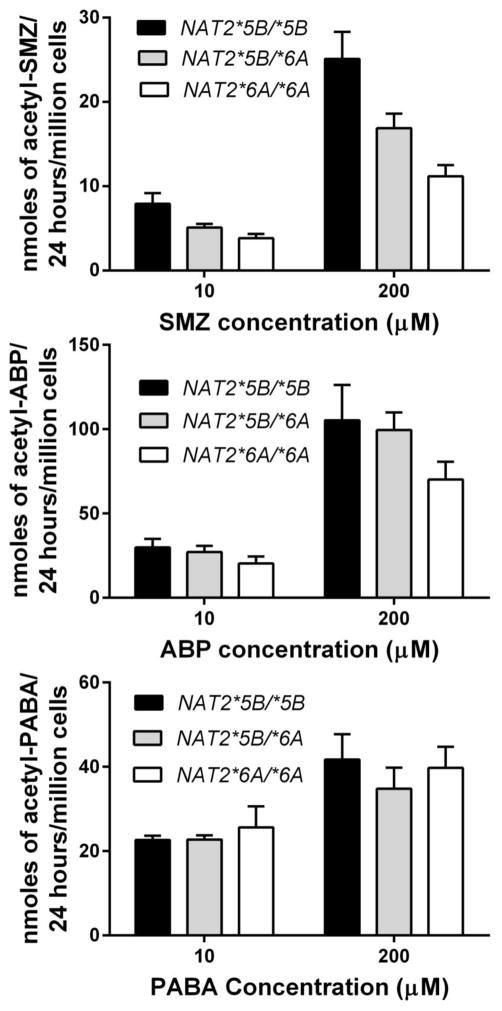
N-acetylation in human hepatocytes cultured in situ was determined towards SMZ, ABP and PABA at two concentrations. As shown in Fig. 3, N-acetylation of SMZ in situ differed significantly with respect to NAT2 genotype following incubations of 10 (p=0.0144) and 200 (p=0.0024) μM . Samples NAT2*5B/*5B had the highest capacity followed by samples with NAT2*5B/*6A genotype followed by samples with NAT2*6A/*6A. N-acetylation of the arylamine carcinogen ABP showed the same trend as SMZ (NAT2*5B/*5B > NAT2*5B/*6A > NAT2*6A/*6A although the differences for ABP failed to meet statistical significance (p>0.05). N-acetylation capacity towards PABA did not reflect this same trend.
Fig. 3.
N-acetylation capacity of cultured human hepatocytes in situ towards SMZ (top), ABP (middle) and PABA (bottom). Human hepatocyte samples with NAT2*5B/*5B, NAT2*5B/*6A or NAT2*6A/*6A genotype were incubated with SMZ, ABP or PABA at 10 or 200 μM for 24h. Each bar illustrates mean ± SEM for NAT2*5B/*5B (n=10), NAT2*5B/*6A (n=9), and NAT2*6A/*6A (n=7) individual human hepatocyte samples. Differences in N-acetylation differed significantly as NAT2*5B/*5B > NAT2*5B/*6A > NAT2*6A/*6A towards SMZ at both 10 and 200 μM (p=0.0144 and p= 0.0024) respectively, but not towards ABP (p=0.3548 and p=0.3163) and PABA (p= 0.6631 and p=0.6485) at 10 and 200 μM respectively.
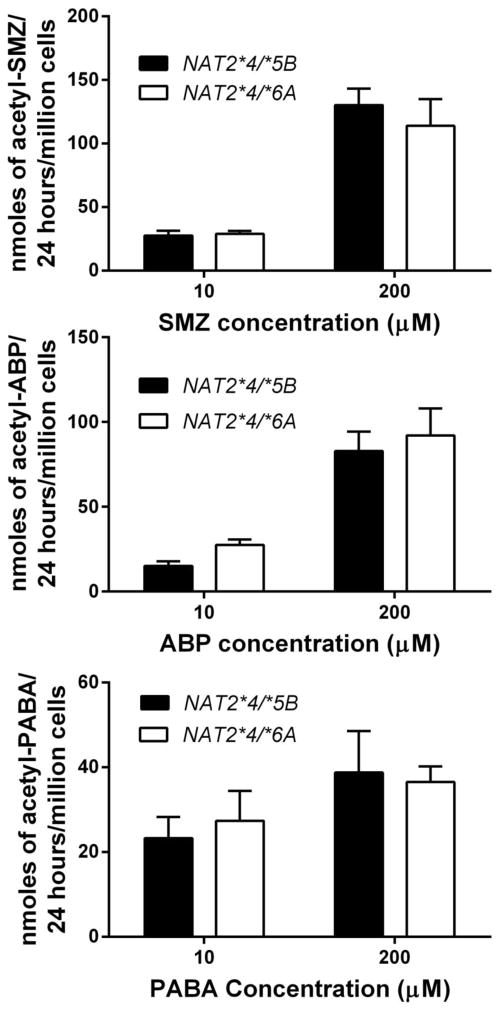
We also measured the level of N-acetylation in human hepatocytes with heterozygous NAT2 genotypes NAT2*4/*5B and NAT2*4/*6A. The N-acetylation of SMZ, ABP and PABA did not differ significantly (p>0.05) between these intermediate NAT2 acetylator genotypes following incubation at either 10 or 200 μM (Fig 4).
Fig. 4.
N-acetylation capacity of cultured human hepatocytes in situ towards SMZ (top), ABP (middle) and PABA (bottom). Human hepatocyte samples with NAT2*4/*5B or NAT2*4/*6A genotype were incubated with SMZ, ABP or PABA at 10 or 200 μM for 24h. Each bar illustrates mean ± SEM for NAT2*4/*5B (n=8) or NAT2*4/*6A (n=9) individual human hepatocyte samples. N-acetylation capacity did not differ significantly between samples with NAT2*4/*5B or NAT2*4/*6A genotype at 10 or 200 μM for SMZ (p=0.7547 and p= 0.5399) ABP (p=0.3096 and p=0.6534) or PABA (p= 0.6536 and p=0.8199) respectively.
Discussion
The N-acetylation of the NAT2-specific drug sulfamethazine was shown to differ in vitro and in situ among human hepatocytes among samples with different NAT2 genotypes associated with slow NAT2 acetylator phenotype. Samples with genotype of NAT2*5B/*5B had the highest activity followed by samples with NAT2*5B/*6A genotype followed by samples with NAT2*6A/*6A genotype. The results observed with the arylamine carcinogen ABP followed the same trend as that for SMZ although the differences did not reach statistical significance (p>0.05), most likely because ABP is N-acetylated by both human NAT1 and NAT2 (Hein et al., 1993). The N-acetylation of the NAT1-specific substrate PABA did not differ significantly among the various NAT2 genotypes and served as a useful control.
N-acetylation of SMZ in human hepatocytes with intermediate NAT2 genotypes NAT2*4/*5 and NAT2*4/*6 did not differ significantly between the two genotypes (p>0.05), probably due to the contribution of the NAT2*4 versus the NAT2*5B or NAT2*6A component of the heterozygous NAT2 genotype. The N-acetylation of SMZ was much higher, the N-acetylation of ABP was somewhat higher, and the N-acetylation of PABA was not higher in the intermediate NAT2 acetylator genotypes than in the slow NAT2 acetylator genotypes, which is consistent with the substrate specificity of SMZ for NAT2, PABA for NAT1, and ABP for both NAT1 and NAT2.
Early studies that reported lower levels of N-acetyltransferase (Hein et al., 1994a) and O-acetyltransferase (Hein et al., 1994b; 1995; Hickman et al., 1995) catalytic activities following recombinant expression of NAT2*5B compared to NAT2*6A in bacteria. More recent investigations reported similar N- and O-acetyltransferase catalytic activities following recombinant expression of NAT2*5B and NAT2*6A in mammalian COS-1 cells (Zang et al., 2007). Furthermore, recombinant expression of the G590A (R197Q) SNP present in NAT2*6A resulted in reduced NAT2 activity and protein levels secondary to reduced protein thermostability (Zang et al., 2007), whereas recombinant expression of T341C (I114T) SNP present in NAT2*5B did not change protein thermostability (Zang et al., 2007) but rather enhanced proteolytic degradation (Zang et al., 2004) in mammalian COS-1 cells. Protein degradation pathways vary from single cell organisms such as bacteria to mammalian/human cells lines (Wang et al, 2010) which likely accounts for the discrepancy between the early studies in bacteria and the more recent studies using mammalian cell expression systems.
Our findings are consistent with previous investigations of caffeine metabolism in which individuals with NAT2*5B/*5B genotype had a higher urinary metabolite ratio (surrogate for N-acetylation activity) than individuals with a NAT2*6A/*6A genotype following administration of caffeine (Cascorbi et al., 1995; Bolt et al., 2005; Selinski et al., 2013). Similar results also have been reported following administration of dapsone (Rothman et al., 1993) and isoniazid (Smith et al., 1997; Zabost et al., 2013). Notably, the results of the present study in human cryopreserved human hepatocytes are in complete agreement with a study by Ruiz et al. (2012) in which the rank order of NAT2 phenotypes as measured in vivo using the caffeine-based urinary metabolite ratio assay was: NAT2*4/*4 (rapid acetylator) > NAT2*4/5B and NAT2*4/6A (intermediate acetylators) > NAT2*5B/*5B > NAT2*5B/NAT2*6A > NAT2*6A/6A (slow acetylators) genotype.
In summary, we report a gene dose response (NAT2*5B/*5B > NAT2*5B/*6A > NAT2*6A/*6A) in N-acetylation capacity in vitro and in situ towards SMZ and the arylamine carcinogen ABP in cryopreserved human hepatocyte cultures that confirms the results previously obtained in vivo following administration of caffeine (Ruiz et al., 2012). Our results further support reviews (Hein, 2009) and editorials (Selinski et al., 2015a) which discuss the importance of genetic heterogeneity among the NAT2 slow acetylator phenotype(s) to enable more accurate and robust assessments of the effects of NAT2 genetic polymorphism on drug and carcinogen metabolism and toxicity.
Acknowledgments
We thank Timothy Moeller and Bioreclamation IVT (Baltimore, MD) for their valuable contributions towards this study.
Footnotes
Conflict of Interest None
References
- Ambrosone CB, Kropp S, Yang J, Yao S, Shields PG, Chang-Claude J. Cigarette smoking, N-acetyltransferase 2 genotypes, and breast cancer risk: pooled analysis and meta-analysis. Cancer Epidemiol Biomarkers Prev. 2008;17(1):15–26. doi: 10.1158/1055-9965.EPI-07-0598. [DOI] [PubMed] [Google Scholar]
- Baumgartner KB, Schlierf TJ, Yang D, Doll MA, Hein DW. N-acetyltransferase 2 genotype modification of active cigarette smoking on breast cancer risk among hispanic and non-hispanic white women. Toxicol Sci. 2009;112(1):211–20. doi: 10.1093/toxsci/kfp199. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bolt HM, Selinski S, Dannappel D, Blaszkewicz M, Golka K. Re-investigation of the concordance of human NAT2 phenotypes and genotypes. Arch Toxicol. 2005;79(4):196–200. doi: 10.1007/s00204-004-0622-8. [DOI] [PubMed] [Google Scholar]
- Cascorbi I, Drakoulis N, Brockmoller J, Maurer A, Sperling K, Roots I. Arylamine N-acetyltransferase (NAT2) mutations and their allelic linkage in unrelated Caucasian individuals: correlation with phenotypic activity. Am J Hum Genet. 1995;57(3):581–92. [PMC free article] [PubMed] [Google Scholar]
- Conlon MS, Johnson KC, Bewick MA, Lafrenie RM, Donner A. Smoking (active and passive), N-acetyltransferase 2, and risk of breast cancer. Cancer Epidemiol. 2010;34(2):142–9. doi: 10.1016/j.canep.2010.02.001. [DOI] [PubMed] [Google Scholar]
- Deitz AC, Zheng W, Leff MA, et al. N-Acetyltransferase-2 genetic polymorphism, well-done meat intake, and breast cancer risk among postmenopausal women. Cancer Epidemiol Biomarkers Prev. 2000;9(9):905–10. [PubMed] [Google Scholar]
- Doll MA, Hein DW. Comprehensive human NAT2 genotype method using single nucleotide polymorphism-specific polymerase chain reaction primers and fluorogenic probes. Anal Biochem. 2001;288(1):106–8. doi: 10.1006/abio.2000.4892. [DOI] [PubMed] [Google Scholar]
- Doll MA, Zang Y, Moeller T, Hein DW. Codominant expression of N-acetylation and O-acetylation activities catalyzed by N-acetyltransferase 2 in human hepatocytes. J Pharmacol Exp Ther. 2010;334(2):540–4. doi: 10.1124/jpet.110.168567. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Garcia-Closas M, Malats N, Silverman D, et al. NAT2 slow acetylation, GSTM1 null genotype, and risk of bladder cancer: results from the Spanish Bladder Cancer Study and meta-analyses. Lancet. 2005;366(9486):649–59. doi: 10.1016/S0140-6736(05)67137-1. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hein DW. Acetylator genotype and arylamine-induced carcinogenesis. Biochim Biophys Acta. 1988;948(1):37–66. doi: 10.1016/0304-419x(88)90004-2. [DOI] [PubMed] [Google Scholar]
- Hein DW. Molecular genetics and function of NAT1 and NAT2: role in aromatic amine metabolism and carcinogenesis. Mutat Res. 2002;506–507:65–77. doi: 10.1016/s0027-5107(02)00153-7. [DOI] [PubMed] [Google Scholar]
- Hein DW. N-acetyltransferase SNPs: emerging concepts serve as a paradigm for understanding complexities of personalized medicine. Expert Opin Drug Metab Toxicol. 2009;5(4):353–66. doi: 10.1517/17425250902877698. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hein DW. N-acetyltransferase 2 polymorphism and human urinary bladder and breast cancer risk. In: Sim E, Laurieri N, editors. Arylamine N-Acetyltransferases in Health and Disease. World Scientific Publishing; (in press) [Google Scholar]
- Hein DW, Doll MA. Accuracy of various human NAT2 SNP genotyping panels to infer rapid, intermediate and slow acetylator phenotypes. Pharmacogenomics. 2012;13(1):31–41. doi: 10.2217/pgs.11.122. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hein DW, Doll MA, Nerland DE, Fretland AJ. Tissue distribution of N-acetyltransferase 1 and 2 catalyzing the N-acetylation of 4-aminobiphenyl and O-acetylation of N-hydroxy-4-aminobiphenyl in the congenic rapid and slow acetylator Syrian hamster. Mol Carcinog. 2006;45(4):230–8. doi: 10.1002/mc.20164. [DOI] [PubMed] [Google Scholar]
- Hein DW, Doll MA, Rustan TD, Ferguson RJ. Metabolic activation of N-hydroxyarylamines and N-hydroxyarylamides by 16 recombinant human NAT2 allozymes: effects of 7 specific NAT2 nucleic acid substitutions. Cancer Res. 1995;55(16):3531–6. [PubMed] [Google Scholar]
- Hein DW, Doll MA, Rustan TD, et al. Metabolic activation and deactivation of arylamine carcinogens by recombinant human NAT1 and polymorphic NAT2 acetyltransferases. Carcinogenesis. 1993;14(8):1633–8. doi: 10.1093/carcin/14.8.1633. [DOI] [PubMed] [Google Scholar]
- Hein DW, Ferguson RJ, Doll MA, Rustan TD, Gray K. Molecular genetics of human polymorphic N-acetyltransferase: enzymatic analysis of 15 recombinant wild-type, mutant, and chimeric NAT2 allozymes. Hum Mol Genet. 1994a;3(5):729–34. doi: 10.1093/hmg/3.5.729. [DOI] [PubMed] [Google Scholar]
- Hein DW, Rustan TD, Ferguson RJ, Doll MA, Gray K. Metabolic activation of aromatic and heterocyclic N-hydroxyarylamines by wild-type and mutant recombinant human NAT1 and NAT2 acetyltransferases. Arch Toxicol. 1994b;68(2):129–33. doi: 10.1007/s002040050045. [DOI] [PubMed] [Google Scholar]
- Hickman D, Palamanda JR, Unadkat JD, Sim E. Enzyme kinetic properties of human recombinant arylamine N-acetyltransferase 2 allotypic variants expressed in Escherichia coli. Biochem Pharmacol. 1995;50(5):697–703. doi: 10.1016/0006-2952(95)00182-y. [DOI] [PubMed] [Google Scholar]
- McDonagh EM, Boukouvala S, Aklillu E, Hein DW, Altman RB, Klein TE. PharmGKB summary: very important pharmacogene information for N-acetyltransferase 2. Pharmacogenet Genomics. 2014;24(8):409–25. doi: 10.1097/FPC.0000000000000062. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Meisel P, Arndt D, Scheuch E, Klebingat KJ, Siegmund W. Prediction of metabolic activity from genotype: the gene-dose effect of N-acetyltransferase. Ther Drug Monit. 2001;23(1):9–14. doi: 10.1097/00007691-200102000-00003. [DOI] [PubMed] [Google Scholar]
- Moore LE, Baris DR, Figueroa JD, et al. GSTM1 null and NAT2 slow acetylation genotypes, smoking intensity and bladder cancer risk: results from the New England bladder cancer study and NAT2 meta-analysis. Carcinogenesis. 2011;32(2):182–9. doi: 10.1093/carcin/bgq223. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Moslehi R, Chatterjee N, Church TR, et al. Cigarette smoking, N-acetyltransferase genes and the risk of advanced colorectal adenoma. Pharmacogenomics. 2006;7(6):819–29. doi: 10.2217/14622416.7.6.819. [DOI] [PubMed] [Google Scholar]
- Rothman N, Hayes RB, Bi W, et al. Correlation between N-acetyltransferase activity and NAT2 genotype in Chinese males. Pharmacogenetics. 1993;3(5):250–5. doi: 10.1097/00008571-199310000-00004. [DOI] [PubMed] [Google Scholar]
- Ruiz JD, Martinez C, Anderson K, et al. The differential effect of NAT2 variant alleles permits refinement in phenotype inference and identifies a very slow acetylation genotype. PLoS One. 2012;7(9):e44629. doi: 10.1371/journal.pone.0044629. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Selinski S, Blaszkewicz M, Getzmann S, Golka K. N-Acetyltransferase 2: ultra-slow acetylators enter the stage. Arch Toxicol. 2015a;89(12):2445–7. doi: 10.1007/s00204-015-1650-2. [DOI] [PubMed] [Google Scholar]
- Selinski S, Blaszkewicz M, Ickstadt K, Hengstler JG, Golka K. Refinement of the prediction of N-acetyltransferase 2 (NAT2) phenotypes with respect to enzyme activity and urinary bladder cancer risk. Arch Toxicol. 2013;87(12):2129–39. doi: 10.1007/s00204-013-1157-7. [DOI] [PubMed] [Google Scholar]
- Selinski S, Getzmann S, Gajewski PD, et al. The ultra-slow NAT2*6A haplotype is associated with reduced higher cognitive functions in an elderly study group. Arch Toxicol. 2015b;89(12):2291–303. doi: 10.1007/s00204-015-1635-1. [DOI] [PubMed] [Google Scholar]
- Shin A, Shrubsole MJ, Rice JM, et al. Meat intake, heterocyclic amine exposure, and metabolizing enzyme polymorphisms in relation to colorectal polyp risk. Cancer Epidemiol Biomarkers Prev. 2008;17(2):320–9. doi: 10.1158/1055-9965.EPI-07-0615. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Smith CA, Wadelius M, Gough AC, Harrison DJ, Wolf CR, Rane A. A simplified assay for the arylamine N-acetyltransferase 2 polymorphism validated by phenotyping with isoniazid. J Med Genet. 1997;34(9):758–60. doi: 10.1136/jmg.34.9.758. [DOI] [PMC free article] [PubMed] [Google Scholar]
- van der Hel OL, Peeters PHM, Hein DW, et al. NAT2 slow acetylation and GSTM1 null genotypes may increase postmenopausal breast cancer risk in long-term smoking women. Pharmacogenetics. 2003;13(7):399–407. doi: 10.1097/00008571-200307000-00005. [DOI] [PubMed] [Google Scholar]
- Wang T, Darwin KH, Li H. Binding-induced folding of prokaryotic ubiquitin-like protein on the Mycobacterium proteasomal ATPase targets substrates for degradation. Nature structural & molecular biology. 2010;17(11):1352–7. doi: 10.1038/nsmb.1918. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zabost A, Brzezinska S, Kozinska M, et al. Correlation of N-acetyltransferase 2 genotype with isoniazid acetylation in Polish tuberculosis patients. BioMed Res Int. 2013;2013:853602. doi: 10.1155/2013/853602. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zang Y, Doll MA, Zhao S, States JC, Hein DW. Functional characterization of single-nucleotide polymorphisms and haplotypes of human N-acetyltransferase 2. Carcinogenesis. 2007;28(8):1665–71. doi: 10.1093/carcin/bgm085. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zang Y, Zhao S, Doll MA, States JC, Hein DW. The T341C (Ile114Thr) polymorphism of N-acetyltransferase 2 yields slow acetylator phenotype by enhanced protein degradation. Pharmacogenetics. 2004;14(11):717–23. doi: 10.1097/00008571-200411000-00002. [DOI] [PubMed] [Google Scholar]