The crystal structure of a fibronectin type III domain from human collagen α1 type XX (residues Pro386–Pro466) was solved at 2.5 Å resolution.
Keywords: crystal structure, fibronectin type III domain, FN3 domain, collagen α1 type XX
Abstract
Collagen α1 type XX, which contains fibronectin type III (FN3) repeats involving six FN3 domains (referred to as the FN#1–FN#6 domains), is an unusual member of the fibril-associated collagens with interrupted triple helices (FACIT) subfamily of collagens. The results of standard protein BLAST suggest that the FN3 repeats might contribute to collagen α1 type XX acting as a cytokine receptor. To date, solution NMR structures of the FN#3, FN#4 and FN#6 domains have been determined. To obtain further structural evidence to understand the relationship between the structure and function of the FN3 repeats from collagen α1 type XX, the crystal structure of the FN#2 domain from human collagen α1 type XX (residues Pro386–Pro466; referred to as FN2-HCXX) was solved at 2.5 Å resolution. The crystal structure of FN2-HCXX shows an immunoglobulin-like fold containing a β-sandwich structure, which is formed by a three-stranded β-sheet (β1, β2 and β5) packed onto a four-stranded β-sheet (β3, β4, β6 and β7). Two consensus domains, tencon and fibcon, are structural analogues of FN2-HCXX. Fn8, an FN3 domain from human oncofoetal fibronectin, is the closest structural analogue of FN2-HCXX derived from a naturally occurring sequence. Based solely on the structural similarity of FN2-HCXX to other FN3 domains, the detailed functions of FN2-HCXX and the FN3 repeats in collagen α1 type XX cannot be identified.
1. Introduction
Collagen is the most abundant structural protein in extracellular matrices (Gordon & Hahn, 2010 ▸). Studies of vertebrate extracellular matrices have identified a family of fibril-associated collagens with an interrupted triple helix (FACIT), which are found attached to the surfaces of fibrils and participate in the regulation of the sizes of banded collagen fibrils, and form connecting links between various components of the extracellular matrix and cells in different tissues (Gordon & Hahn, 2010 ▸; Ricard-Blum, 2011 ▸). The FACIT family currently consists of collagens α1 type IX, α1 type XII, α1 type XIV, α1 type XVI and α1 types XIX–XXII (Koch et al., 2001 ▸; Tuckwell, 2002 ▸; Gordon & Hahn, 2010 ▸; Ricard-Blum, 2011 ▸). Collagens α1 type XII, α1 type XIV and α1 type XX contain additional fibronectin type III (FN3) repeats; in contrast, collagens α1 type IX, α1 type XVI, α1 type XIX, α1 type XXIV and α1 type XXII do not contain FN3 repeats. Therefore, α1 type XII, α1 type XIV and α1 type XX should be divided into a particular class inside the FACIT family. The FN3 repeats from collagens α1 type XII, α1 type XIV and α1 type XX consist of 18, eight and six FN3 domains, respectively. The FN3 domain is a very common constituent of animal proteins, occurring in roughly 2% of known sequences (Bork & Doolittle, 1992 ▸). FN3 domains mediate a wide variety of cellular interactions in cell adhesion, migration, growth and differentiation (Lucena et al., 2007 ▸; Vakonakis et al., 2007 ▸). Current ideas on the structure and evolution of collagens have been reviewed by Ivanova & Krivchenko (2012 ▸, 2014 ▸). However, the functions of the FN3 repeats in collagen α1 type XX are still unknown.
In vertebrates, collagen α1 type XX is distributed mainly in the cornea, tendons, sternal cartilage and embryonic skin. The amino-acid sequences of collagen α1 type XX isolated from Gallus gallus and Homo sapiens have been reported. Chicken α1 type XX contains a polypeptide chain of 1472 amino-acid residues and human collagen contains a polypeptide chain of 1284 amino-acid residues (Koch et al., 2001 ▸; Strausberg et al., 2002 ▸). Collagen α1 type XX contains the following conserved structural domains: six FN3 domains (referred to as FN#1–FN#6) forming the FN3 repeats, one VWFA domain, one Tsp domain, one Col2 domain, one NC2 domain, one Col1 domain and one NC1 domain (Koch et al., 2001 ▸). In this study, we first carried out a standard protein BLAST search (http://www.ncbi.nlm.nih.gov/blast/) with the human collagen α1 type XX sequence. The results of the bioinformatics assay revealed that the three FN3 domains FN#1, FN#2 and FN#4 do not contain a cytokine-binding motif, while the three FN3 domains FN#3, FN#5 and FN#6 contain one individual cytokine-binding motif. The cytokine-binding motif sequences in the FN#3, FN#5 and FN#6 domains are as follows: EGSEA (Glu543–Ala547), RSDPV (Arg726–Val730) and RSEAV (Arg819–Val823). The same analysis revealed that the type 1 cytokine receptor common subunit gamma (γc) contains two FN3 domains. While the upstream FN3 domain does not contain a cytokine-binding motif, a cytokine-binding motif (amino-acid sequence HWSEWS) is present in the downstream FN3 domain (Ohbo et al., 1995 ▸; Junttila et al., 2012 ▸). Thus, we guess that the FN#2 domain uniting with the FN#3 domain, and the FN#4 domain uniting with the FN#5 domain, might form individual cytokine-binding sites and that each of the cytokine-binding site structures may show a resemblance to the γc structure. To date, there are no biochemical data to suggest that collagen α1 type XX acts as a cytokine receptor. Sequence alignment reveals no significant similarity between any of the FN3 domains from human collagen α1 type XX and those of γc. Solution NMR structures of the FN#3, FN#4 and FN#6 domains of human collagen α1 type XX have been determined. To obtain more structural evidence to understand the relationship between the structure and the function of collagen α1 type XX, crystallographic studies of the FN#2 domain from human collagen α1 type XX (residues Pro368–Pro466; referred to as FN2-HCXX) have been carried out in this study. The structural features of this FN3 domain are reported in this paper.
2. Materials and methods
2.1. Macromolecule production
The cDNA encoding FN2-HCXX (residues Pro368–Pro466; GenBank No. AAH43183.1) was amplified by PCR using full-length human complementary DNA, which was cloned by reverse transcription of the messenger RNA extracted from SH-SY5Y cells using TRIzol reagent (Invitrogen), as a template. This cDNA was cloned and inserted into the expression vector pEHISTEV (Invitrogen). The recombinant FN2-HCXX was synthesized using an Escherichia coli cell-free system (Kigawa, 2010 ▸; Kigawa et al., 2004 ▸) as a fusion with an N-terminal His tag and a TEV protease cleavage site. The E. coli cell-free protein synthesis was carried out using the protocol described in the Supporting Information. Macromolecule-production information is summarized in Supplementary Table S1. Prior to the purification of FN2-HCXX, the reaction solution was centrifuged at 16 000g and 277 K for 20 min. The supernatant was loaded onto a HisTrap column (5 ml) equilibrated with 20 mM Tris–HCl buffer pH 8.0 containing 500 mM NaCl and 20 mM imidazole. After washing the column with the buffer, the His-tagged protein was eluted with 20 mM Tris–HCl buffer pH 8.0 containing 500 mM NaCl and 200 mM imidazole. The sample buffer was exchanged to 20 mM Tris–HCl buffer pH 8.0 containing 500 mM NaCl and 20 mM imidazole using a HiPrep 26/10 desalting column. The His tag was cleaved with 100 µl TEV protease (4 mg ml−1) at 303 K for 1 h. To remove the His tag and the uncleaved His-tagged protein, the reaction solution was loaded onto a HisTrap column as described above. The flowthrough fractions were collected and concentrated to 1.0 ml using an Amicon Ultra-15 filter unit (5000 molecular-weight cutoff, Millipore). Finally, protein purification was carried out using a HiLoad 16/60 Superdex 75 column equilibrated with 20 mM Tris–HCl buffer pH 8.0 containing 150 mM NaCl and 2 mM dithiothreitol (DTT). All chromatography materials were purchased from GE Healthcare Biosciences. Nickel-affinity chromatography and gel-filtration chromatography were carried out at 277 K.
2.2. Crystallization
The purified protein was concentrated to 600 µl (approximately 6 mg ml−1) using an Amicon Ultra-15 filter unit (3000 molecular-weight cutoff; Millipore). Crystallization was carried out using the sitting-drop vapour-diffusion method at 293 K. Each sitting drop consisted of 1 µl protein solution and 1 µl reservoir solution. Crystal Screen HT, Natrix and Index (Hampton Research) were used to establish the initial crystallization conditions. Single crystals were obtained from condition No. 25 [80 mM magnesium acetate, 50 mM sodium cacodylate pH 6.5, 30%(w/v) PEG 4000] of the Natrix kit within one week.
2.3. Data collection and processing
A single crystal with approximate dimensions of 0.2 × 0.1 × 0.1 mm was mounted on a nylon loop (Hampton Research) with reservoir solution and dipped into 5 µl of a cryoprotectant solution consisting of 66.6%(v/v) Paratone N, 28.6%(v/v) paraffin, 4.8%(v/v) glycerol. The crystal was swished about using the nylon loop to remove the reservoir solution. The crystal was mounted on the nylon loop in a liquid-nitrogen gas stream at 100 K. X-ray diffraction data from the crystal were collected on beamline BL17U at the Shanghai Synchrotron Radiation Facility (SSRF), Shanghai, People’s Republic of China. Diffraction intensity data were processed and scaled using the HKL-2000 program (Otwinowski & Minor, 1997 ▸). Data-collection and processing statistics are summarized in Table 1 ▸.
Table 1. Data collection and processing.
| Diffraction source | BL17U, SSRF |
| Wavelength (Å) | 1.000 |
| Temperature (K) | 100 |
| Detector | ADSC Quantum 315r |
| Crystal-to-detector distance (mm) | 200 |
| Rotation range per image (°) | 1 |
| Total rotation range (°) | 180 |
| Exposure time per image (s) | 10 |
| Space group | P212121 |
| a, b, c (Å) | 56.48, 78.60, 81.99 |
| α, β, γ (°) | 90, 90, 90 |
| Mosaicity (°) | 2.4 (2.9) |
| Resolution range (Å) | 48.3–2.50 (2.62–2.50) |
| Total No. of reflections | 114950 |
| No. of unique reflections | 12912 |
| Completeness (%) | 98.1 (99.9) |
| Multiplicity | 6.8 (6.3) |
| CC1/2 † | 0.981 (0.989) |
| 〈I/σ(I)〉 | 22.0 (11.3) |
| R r.i.m. ‡ | 0.053 (0.203) |
| Overall B factor from Wilson plot (Å2) | 26.99 |
CC1/2 is Pearson’s correlation coefficient between random half data sets.
R r.i.m. = R merge[N/(N − 1)]1/2, where N is the data multiplicity.
2.4. Structure solution and refinement
The crystal structure was solved using the molecular-replacement (MR) method. The solution was obtained with Phaser-MR in the PHENIX suite (Adams et al., 2010 ▸) using the structure of tencon, a consensus domain (PDB entry 3tes; Jacobs et al., 2012 ▸), as the search model. After MR, several cycles of manual model building and refinement were performed using Coot (Emsley et al., 2010 ▸) and phenix.refine in the PHENIX suite (Adams et al., 2010 ▸). The coordinates and the structure factors have been deposited in the Protein Data Bank (PDB entry 5kf4). The refinement parameters are summarized in Table 2 ▸.
Table 2. Structure solution and refinement.
| Resolution range (Å) | 45.86–2.50 |
| Completeness (%) | 92.2 |
| σ Cutoff | F > 4.450σ(F) |
| No. of reflections, working set | 12128 |
| No. of reflections, test set | 596 |
| R cryst † | 0.258 |
| R free ‡ | 0.284 |
| No. of non-H atoms | |
| Protein | 2917 |
| Water | 68 |
| R.m.s. deviations | |
| Bonds (Å) | 0.004 |
| Angles (°) | 1.13 |
| Average B factors (Å2) | |
| Protein | 32.9 |
| Water | 26.8 |
| Ramachandran plot | |
| Most favoured (%) | 93.73 |
| Allowed (%) | 6.27 |
R
cryst = 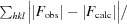
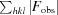 .
.
R free was calculated using randomly selected reflections (5%).
3. Results and discussion
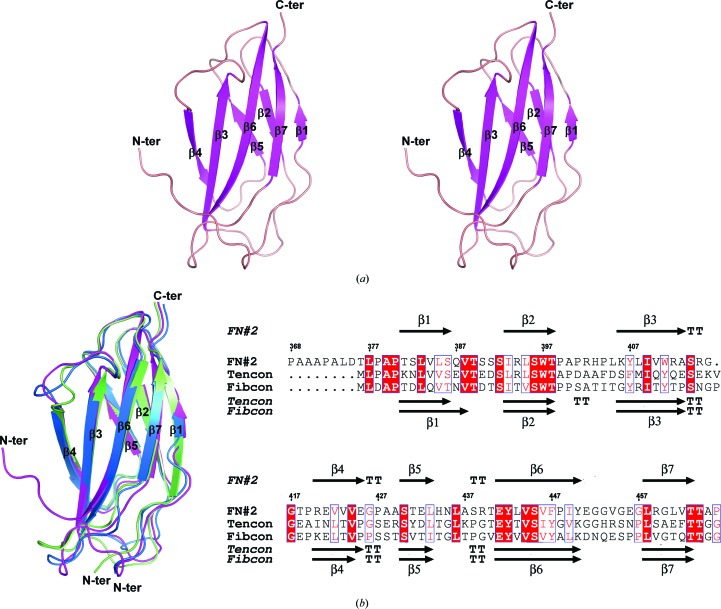
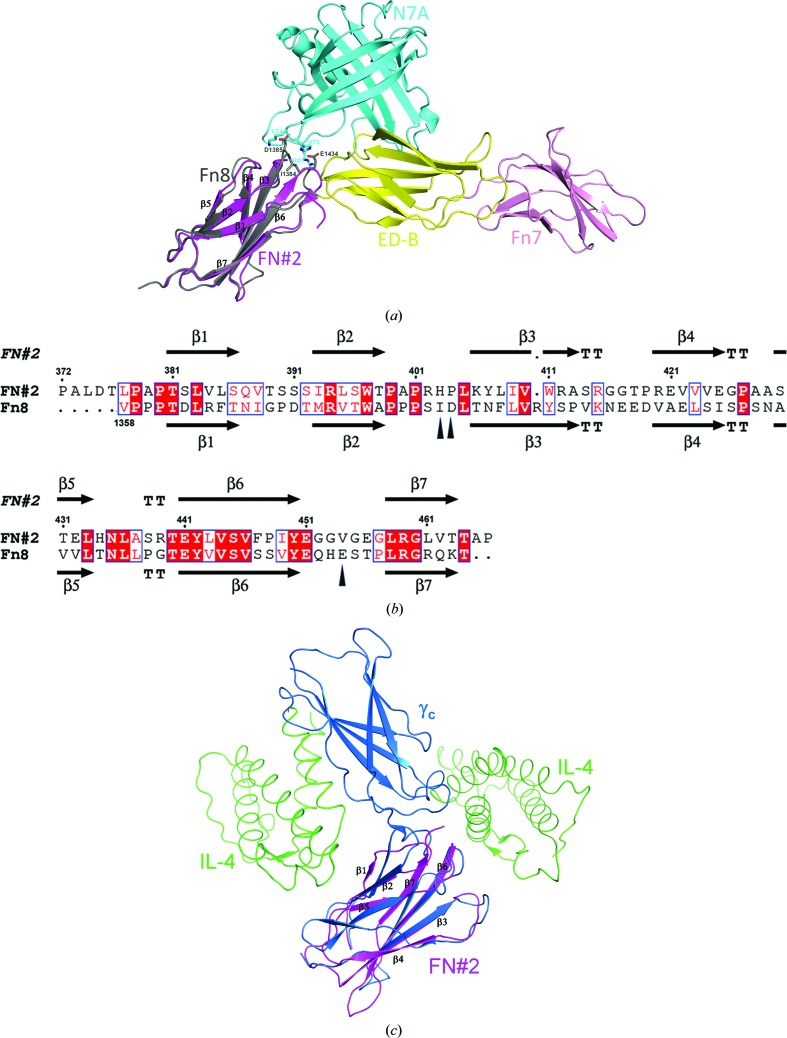
The crystals of FN2-HCXX belonged to the orthorhombic space group P212121, with unit-cell parameters a = 56.48, b = 78.60, c = 81.99 Å. Four FN2-HCXX molecules are present in the asymmetric unit, resulting in a Matthews coefficient (Matthews, 1968 ▸) of 2.66 Å3 Da−1 and an approximate solvent content of 54%. The crystal structure of FN2-HCXX shows an immunoglobulin-like fold (Koide et al., 2012 ▸) containing a β-sandwich structure, which is formed by a three-stranded β-sheet (β1, β2 and β5) packed onto a four-stranded β-sheet (β3, β4, β6 and β7) (Fig. 1 ▸ a). A structural homology search using the DALI server (Holm et al., 2006 ▸) revealed that two designed consensus domains, tencon (PDB entry 3tes) and fibcon (PDB entry 3teu), are structural homologues of the FN#2 domain (Jacobs et al., 2012 ▸). Superimposition of the FN2-HCXX structure with those of tencon and fibcon showed r.m.s. deviations of Cα atoms of 0.641 and 0.876 Å, respectively (Fig. 1 ▸ b). FN2-HCXX shares a sequence identity of 38% with tencon and 37% with fibcon (Fig. 1 ▸ c). The FN3 domain Fn8 from human oncofoetal fibronectin (Gebauer et al., 2013 ▸) is the closest structural analogue of FN2-HCXX derived from a naturally occurring sequence. The FN3 domain has been observed to form a binding surface in some cases. The FN3 domain can use the surface of a loop region between the β3 and β4 strands, the surface above the β2-strand terminus and the β6-strand terminus, as well as the surfaces of the β-sheet regions, to interact with other molecules (Wojcik et al., 2010 ▸; Junttila et al., 2012 ▸; Gebauer et al., 2013 ▸). These interaction surfaces of the FN3 domain are referred to as the top–base interface, the bottom–base interface and the side–base interface, respectively. Superimposition of the FN2-HCXX structure with the Fn8 domain from the crystal structure of the N7A–Fn7B8 complex (PDB entry 4gh7; Gebauer et al., 2013 ▸) yielded an r.m.s. deviation of Cα atoms of 0.88 Å (Fig. 2 ▸ a). Ile1384, Asp1385 and Glu1438 of Fn8 form a bottom–base interface to bind to N7A. The Fn8 domain shares a sequence identity of 28% with FN2-HCXX. These three residues are not conserved in FN2-HCXX (Fig. 2 ▸ b). Collagen α1 type XX consists of conserved structural domains. The results of a standard protein BLAST (http://www.ncbi.nlm.nih.gov/blast/) search reveal that human collagen α1 type XX shares 50% sequence identity with chicken collagen type XX. The amino-acid sequence identities between the conversed structural domains of human collagen α1 type XX and those of chicken collagen α1 type XX are summarized in Supplementary Table S2. Human collagen α1 type XX does not contain the C-terminal NC1 domain, which contains a heparin-binding site (Montserret et al., 1999 ▸). Therefore, human collagen type XX is predicted not to act as a heparin interaction partner, in contrast to chicken collagen type XX. In addition, the amino-acid sequence identities between pairs of FN3 domains from human collagen type XX are summarized in Supplementary Table S3. To date, solution NMR structures of the FN#3 domain (PDB entry 2dkm), the FN#4 domain (PDB entry 2ee3) and the FN#6 domain (PDB entry 2ekj) from human collagen α1(XX) have been determined (RIKEN Structural Genomics/Proteomics Initiative, unpublished work). Superimposition of the structure of the FN#2 domain with solution NMR structures of the FN#3, FN#4 and FN#6 domains showed r.m.s. deviations of Cα atoms of 1.042, 1.021 and 1.049 Å, respectively. These results suggest that the structures of the FN#2, FN#3, FN#4 and FN#6 domains from human collagen α1 type XX are very similar. As posited in §1, FN2-HCXX together with the FN#3 domain might form an individual cytokine-binding site, and show resemblance to the γc structure. Superimposition of the FN2-HCXX structure with the upstream FN3 domain from the crystal structure of the γc–interleukin 4 (IL-4) complex (PDB entry 3qb7; Junttila et al., 2012 ▸) showed an r.m.s. deviation of Cα atoms of 1.339 Å, suggesting that the topological structure of FN2-HCXX is similar to that of the upstream Fn3 domain of γ c (Fig. 2 ▸ c). These results suggest that FN3 domains can use a top–base interface, bottom–base interface or side–base interface to bind to their interaction partner. To date, which proteins are interaction partners of FN2-HCXX remains unknown. To find the interaction partners of collagen α1 type XX, immunoprecipitation and GST-pulldown assays are presently being utilized.
Figure 1.
(a) A stereoview of the overall structure of FN2-HCXX is shown using a ribbon presentation. (b) A superposition of the crystal structures of FN2-HCXX (magenta) with those of tencon (blue) and fibcon (green) is shown using a ribbon presentation (left) and an alignment of the amino-acid sequences of FN2-HCXX, tencon and fibcon is shown using ESPript (Robert & Gouet, 2014 ▸) (right). Identical residues are shown as white letters on a red background, while similar residues are shown as red letters. The secondary-structural elements of FN2-HCXX are shown at the top of the alignment. The secondary-structural elements of tencon and fibcon are shown below the alignment. Structural figures were rendered using PyMOL (http://www.pymol.org).
Figure 2.
(a) Superimposition of the FN2-HCXX structure (magenta) and the Fn8 domain structure (grey) from the crystal structure of the N7A–Fn7B8 complex. (b) Amino-acid sequence alignment between FN2-HCXX and the Fn8 domain is shown using ESPript (Robert & Gouet, 2014 ▸). The secondary-structural elements of FN2-HCXX and the Fn8 domain are shown above and below the alignment, respectively. The residues of the Fn8 domain interacting with N7A are marked using black triangles. (c) Superimposition of the FN2-HCXX structure (magenta) with the upstream Fn3 domain structure (sky blue) of the γc structure from the crystal structure of the γc–IL4 complex. Protein structures are shown using a ribbon presentation.
Supplementary Material
Large-scale cell-free protein-synthesis reaction protocol and Supplementary Tables.. DOI: 10.1107/S2053230X1701648X/dp5108sup1.pdf
PDB reference: FN3 domain (residues Pro368–Pro466) of human collagen XX, 5kf4
Acknowledgments
The authors are grateful to the staff for the use of beamline BL17U at the SSRF.
Funding Statement
This work was funded by National Natural Science Foundation of China grant 81473114. Program for New Century Excellent Talents in University grant to YX.
References
- Adams, P. D. et al. (2010). Acta Cryst. D66, 213–221.
- Bork, P. & Doolittle, R. F. (1992). Proc. Natl Acad. Sci. USA, 89, 8990–8994. [DOI] [PMC free article] [PubMed]
- Emsley, P., Lohkamp, B., Scott, W. G. & Cowtan, K. (2010). Acta Cryst. D66, 486–501. [DOI] [PMC free article] [PubMed]
- Gebauer, M., Schiefner, A., Matschiner, G. & Skerra, A. (2013). J. Mol. Biol. 425, 780–802. [DOI] [PubMed]
- Gordon, M. K. & Hahn, R. A. (2010). Cell Tissue Res. 339, 247–257. [DOI] [PMC free article] [PubMed]
- Holm, L., Kääriäinen, S., Wilton, C. & Plewczynski, D. (2006). Curr. Protoc. Bioinformatics, Unit 5.5. https://doi.org/10.1002/0471250953.bi0505s14. [DOI] [PubMed]
- Ivanova, V. P. & Krivchenko, A. I. (2012). Zh. Evol. Biokhim. Fiziol. 48, 118–128. [PubMed]
- Ivanova, V. P. & Krivchenko, A. I. (2014). Zh. Evol. Biokhim. Fiziol. 50, 245–254. [PubMed]
- Jacobs, S. A., Diem, M. D., Luo, J., Teplyakov, A., Obmolova, G., Malia, T., Gilliland, G. L. & O’Neil, K. T. (2012). Protein Eng. Des. Sel. 25, 107–117. [DOI] [PubMed]
- Junttila, I. S., Creusot, R. J., Moraga, I., Bates, D. L., Wong, M. T., Alonso, M. N., Suhoski, M. M., Lupardus, P., Meier-Schellersheim, M., Engleman, E. G., Utz, P. J., Fathman, C. G., Paul, W. E. & Garcia, K. C. (2012). Nature Chem. Biol. 8, 990–998. [DOI] [PMC free article] [PubMed]
- Kigawa, T. (2010). Methods Mol. Biol. 607, 101–111. [DOI] [PubMed]
- Kigawa, T., Yabuki, T., Matsuda, N., Matsuda, T., Nakajima, R., Tanaka, A. & Yokoyama, S. (2004). J. Struct. Funct. Genomics, 5, 63–68. [DOI] [PubMed]
- Koch, M., Foley, J. E., Hahn, R., Zhou, P., Burgeson, R. E., Gerecke, D. R. & Gordon, M. K. (2001). J. Biol. Chem. 276, 23120–23126. [DOI] [PubMed]
- Koide, A., Wojcik, J., Gilbreth, R. N., Hoey, R. J. & Koide, S. (2012). J. Mol. Biol. 13, 393–405. [DOI] [PMC free article] [PubMed]
- Lucena, S., Arocha Piñango, C. L. & Guerrero, B. (2007). Invest. Clin. 48, 249–262. [PubMed]
- Matthews, B. W. (1968). J. Mol. Biol. 33, 491–497. [DOI] [PubMed]
- Montserret, R., Aubert-Foucher, E., McLeish, M. J., Hill, J. M., Ficheux, D., Jaquinod, M., van der Rest, M., Deléage, G. & Penin, F. (1999). Biochemistry, 38, 6479–6488. [DOI] [PubMed]
- Ohbo, K., Takasawa, N., Ishii, N., Tanaka, N., Nakamura, M. & Sugamura, K. (1995). J. Biol. Chem. 270, 7479–7486. [DOI] [PubMed]
- Otwinowski, Z. & Minor, W. (1997). Methods Enzymol. 276, 307–326. [DOI] [PubMed]
- Ricard-Blum, S. (2011). Cold Spring Harb. Perspect. Biol. 3, a004978. [DOI] [PMC free article] [PubMed]
- Robert, X. & Gouet, P. (2014). Nucleic Acids Res. 42, W320–W324. [DOI] [PMC free article] [PubMed]
- Strausberg, R. L. et al. (2002). Proc. Natl Acad. Sci. USA, 99, 16899–16903.
- Tuckwell, D. (2002). Matrix Biol. 21, 63–66. [DOI] [PubMed]
- Vakonakis, I. D., Staunton, D., Rooney, L. M. & Campbell, I. D. (2007). EMBO J. 26, 2575–2583. [DOI] [PMC free article] [PubMed]
- Wojcik, J., Hantschel, O., Grebien, F., Kaupe, I., Bennett, K. L., Barkinge, J., Jones, R. B., Koide, A., Superti-Furga, G. & Koide, S. (2010). Nature Struct. Mol. Biol. 17, 519–527. [DOI] [PMC free article] [PubMed]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
Large-scale cell-free protein-synthesis reaction protocol and Supplementary Tables.. DOI: 10.1107/S2053230X1701648X/dp5108sup1.pdf
PDB reference: FN3 domain (residues Pro368–Pro466) of human collagen XX, 5kf4