The crystal structure of SAM-dependent methyltransferase from P. horikoshii in complex with the cofactor SAM was solved by X-ray diffraction. The monomeric structure consists of a Rossmann-like fold (domain I) and a substrate-binding domain (domain II). The cofactor binds at the interface between adjacent subunits.
Keywords: methyltransferases, Rossmann-like fold, SAM-binding residues, Pyrococcus horikoshii
Abstract
Methyltransferases (MTs) are enzymes involved in methylation that are needed to perform cellular processes such as biosynthesis, metabolism, gene expression, protein trafficking and signal transduction. The cofactor S-adenosyl-l-methionine (SAM) is used for catalysis by SAM-dependent methyltransferases (SAM-MTs). The crystal structure of Pyrococcus horikoshii SAM-MT was determined to a resolution of 2.1 Å using X-ray diffraction. The monomeric structure consists of a Rossmann-like fold (domain I) and a substrate-binding domain (domain II). The cofactor (SAM) molecule binds at the interface between adjacent subunits, presumably near to the active site(s) of the enzyme. The observed dimeric state might be important for the catalytic function of the enzyme.
1. Introduction
Methyltransferases (MTs) are widely distributed in nature and are involved in methylation in various cellular processes such as biosynthesis, metabolism, gene expression, protein trafficking and signal transduction (Schubert et al., 2003 ▸). MTs act using an SN2-like nucleophilic substitution reaction mechanism for catalysis. During catalysis, the methyl group is transferred to an acceptor molecule, resulting in a methylated product and byproduct. The cofactor S-adenosyl-l-methionine (SAM) is used for catalysis by SAM-dependent methyltransferases (SAM-MTs), which are found in prokaryotic and eukaryotic organisms (Martin & McMillan, 2002 ▸; Liscombe et al., 2012 ▸; Kozbial & Mushegian, 2005 ▸). MTs are categorized depending on the methyl-accepting atom on the substrates: the categories are O, N, C or S. Based on sequence homology, SAM-MT from Pyrococcus horikoshii belongs to the UbiE/COQ5 family, which contains Caulobacter crescentus methyltransferase, which acts in a similar manner in ubiqinone synthesis. The major role of this methyltransferase is the regulation of the cellular concentration ratio of SAM to S-adenosyl-l-homocysteine, which plays a key role in SAM-dependent methyltransfer reactions. Methylation along with phosphorylation and acetylation comprise the integral components of the ‘histone code’, which combines with the genetic code as a critical determinant of chromosomal inheritance (Jenuwein & Allis, 2001 ▸). Furthermore, it is used in an enzyme-coupled calorimetric assay for salicylic acid carboxyl methyltransferase, which utilizes AdoMet as the methyl donor (Hendricks et al., 2004 ▸).
SAM-MTs share limited sequence identity, while structurally proteins of this family have common regions of conserved residues and correspond to Rossmann-like fold MTs (Fauman et al., 1999 ▸). The Rossmann fold is constituted of an α–β–α sandwich structure consisting of seven β-strands flanked by two layers of α-helices. The cores for interaction with SAM and the substrate are formed by the C-terminal regions of the β-strands and the adjoining loops from the catalytic site core. The cofactor-binding residues are poorly conserved (Martin & McMillan, 2002 ▸). In addition, the N-terminus plays an important role in substrate specificity and oligomerization (Kozbial & Mushegian, 2005 ▸).
Here, we report the structure of SAM-MT from P. horikoshii (PhSAM-MT) in complex with SAM obtained by X-ray diffraction at a resolution of 2.10 Å. The monomeric structure has two distinct domains: a Rossmann-fold domain and a nucleotide-binding domain. The structure of the complex provides a path to recognizing the substrate-binding residues, which may lead to an understanding of the structural basis of enzymatic catalysis.
2. Materials and methods
2.1. Macromolecule production
P. horikoshii is a hyperthermophilic, anaerobic archaeon. The plasmid encoding the PhSAM-MT protein was digested with NdeI and the fragment was inserted into the expression vector pET-11a linearized with NdeI and BamHI. The recombinant plasmid was transformed into Escherichia coli BL21 (DE3) cells and grown at 310 K in Luria–Bertani medium containing 0.5 µg ml−1 ampicillin for 20 h. The cells were harvested by centrifugation at 6500 rev min−1 for 5 min at 277 K. The cell pellet was suspended in 20 mM Tris–HCl pH 8.0 containing 0.5 M sodium chloride and 5 mM β-mercaptoethanol and homogenized by ultrasonication. The supernatant was heated at 343 K for 12 min and cell debris and denatured proteins were removed by centrifugation at 14 000 rev min−1 for 30 min; the crude extract in the supernatant was subjected to purification. The crude extract was desalted using a HiPrep 26/10 desalting column and applied onto a Super Q Toyopearl 650M column equilibrated with 20 mM Tris–HCl pH 8.0. Fractions containing proteins were eluted with a linear gradient of 0–0.3 M sodium chloride. The proteins were then dialyzed against 20 mM Tris–HCl pH 8.0 and subjected to a Resource Q column (Amersham Biosciences) equilibrated with 20 mM Tris–HCl pH 8.0. Fractions containing proteins were again eluted with a linear gradient of 0–0.3 M sodium chloride. The proteins were desalted on a HiPrep 26/10 desalting column with 10 mM sodium phosphate pH 7.0 and applied onto a Bio-Scale CHT20-I column (Bio-Rad) equilibrated with 10 mM sodium phosphate pH 7.0. The proteins were again eluted with a linear gradient of 10–150 mM sodium phosphate. The proteins were desalted with a HiPrep 26/10 desalting column with 20 mM Tris–HCl pH 8.0 containing 0.05 M sodium chloride and applied onto a Mono Q column (Amersham Biosciences) equilibrated with 20 mM Tris–HCl pH 8.0 containing 0.05 M sodium chloride. The fractions containing proteins were eluted with a linear gradient of 0–0.5 M sodium chloride. The fractions containing proteins were pooled, concentrated by ultrafiltration (Vivaspin, 10 kDa cutoff) and loaded onto a HiLoad 16/60 Superdex 75 pg column (Amersham Biosciences) equilibrated with 20 mM Tris–HCl pH 8.0 containing 0.05 M sodium chloride.
For the preparation of selenomethionine-substituted protein, E. coli BL21 (DE3) Star cells (Invitrogen) were grown in M9 medium until they reached an absorbance at 600 nm (A 600) of 0.4. At this point, 100 mg l-lysine, 100 mg l-phenylalanine, 100 mg l-threonine, 50 mg l-isoleucine, 50 mg l-leucine and 60 mg selenomethionine (SeMet) were added to 1 l of culture and the cells were grown at 37°C for a further 1 h before inducing expression with 1 mM IPTG overnight at 25°C. The SeMet-substituted protein was purified similarly to the native protein. The homogeneity and identity of the purified sample were ascertained by SDS–PAGE and N-terminal sequence analysis. The protein concentrations were determined by the UV method and the Bio-Rad protein assay based on the Bradford dye-binding procedure, using bovine serum albumin as a standard. Finally, purified PhSAM-MT was concentrated to 20.13 mg ml−1 by ultrafiltration and stored at 203 K. The oligomeric state of purified PhSAM-MT was examined by dynamic light-scattering experiments performed using a DynaPro MS/X instrument (Protein Solutions). Macromolecule-production information is summarized in Table 1 ▸.
Table 1. Macromolecule-production information.
| Source organism | P. horikoshii |
| DNA source | Genomic DNA |
| Cloning vector | pET-11a |
| Expression vector | pET-11a |
| Expression host | E. coli BL21 (DE3) |
| Complete amino-acid sequence of the construct produced | MGFKEYYRVFPTYTDINSQEYRSRIETLEPLLMKYMKKRGKVLDLACGVGGFSFLLEDYGFEVVGVDISEDMIRKAREYAKSRESNVEFIVGDARKLSFEDKTFDYVIFIDSIVHFEPLELNQVFKEVRRVLKPSGKFIMYFTDLRELLPRLKESLVVGQKYWISKVIPDQEERTVVIEFKSEQDSFRVRFNVWGKTGVELLAKLYFTKEAEEKVGNYSYLTVYNPK |
2.2. Crystallization
Cystallization screens from Hampton Research (Jancarik & Kim, 1991 ▸) were used to determine initial crystallization conditions. Crystals of the complex of PhSAM-MT with SAM were obtained using the microbatch sitting-drop method in NUNC HLA plates (Nalge Nunc International). Each crystallization drop was prepared by mixing 1.0 µl precipitant solution (8% PEG 4000, 0.1 M sodium acetate trihydrate pH 4.6) and 1.0 µl protein solution at 20.13 mg ml−1. The crystallization drop was then overlaid with a 1:1 mixture of silicon and paraffin oils, allowing the slow evaporation of the water in the drop. The crystals grew to full size in 6–8 d and were flash-cooled in liquid nitrogen at 100 K for data collection. Crystallization information is summarized in Table 2 ▸.
Table 2. Crystallization.
| Method | Sitting-drop vapour diffusion |
| Plate type | NUNC HLA plates |
| Temperature (K) | 295 |
| Protein concentration | 20.13 |
| Buffer composition of protein solution | 8% PEG 4000, 0.1 M sodium acetate trihydrate pH 4.6 |
| Volume and ratio of drop | 1 µl, 1:1 |
2.3. Data collection and processing
Complete data sets were collected on the BL26B1 beamline at SPring-8, Japan under cryogenic conditions. Crystals were flash-cooled with liquid nitrogen at 100 K in their mother liquor or in a soaking solution containing 26%(v/v) glycerol as a cryoprotectant. A Rigaku R-AXIS V image-plate detector was used for data collection. The data were processed and scaled using the HKL-2000 suite (Otwinowski & Minor, 1997 ▸). The data sets were completed by including all possible hkl and R free columns using UNIQUE from the CCP4 suite (Winn et al., 2011 ▸). The data-collection parameters and processing statistics are given in Table 3 ▸.
Table 3. Summary of data collection and phasing.
Values in parentheses are for the outer shell.
| SeMet peak | SeMet inflection | SeMet remote | |
|---|---|---|---|
| Resolution range (Å) | 30.0–2.10 | 30.0–2.10 | 30.0–2.10 |
| Wavelength (Å) | 0.97882 | 0.97945 | 1.0 |
| Space group | P3221 | ||
| Unit-cell parameters (Å) | |||
| a | 58.167 | ||
| b | 58.167 | ||
| c | 252.463 | ||
| Total No. of reflections | 155595 | 155449 | 154615 |
| No. of unique reflections | 29778 | 28942 | 29753 |
| Completeness (%) | 98.2 (98.5) | 97.8 (98.1) | 98.6 (98.0) |
| Multiplicity | 5.2 | ||
| 〈I/σ(I)〉 | 18.0 (5.1) | 16.9 (4.9) | 18.0 (5.1) |
| R r.i.m. | 0.049 | ||
| Overall B factor from Wilson plot (Å2) | 29.5 | ||
| Overall figure of merit | |||
| Before density modification | 0.61 | ||
| After density modification | 0.82 | ||
2.4. Structure solution and refinement
The structure of PhSAM-MT was determined by the multi-wavelength anomalous dispersion (MAD) method using selenomethionine-derivatized crystals (Hendrickson et al., 1990 ▸). Phases were obtained from MAD data using eight Se atoms (Table 3 ▸). The Se-atom coordinates and the initial electron-density map were obtained with SOLVE/RESOLVE (Terwilliger & Berendzen, 1999 ▸). The coordinates were refined using CNS (Brünger et al., 1998 ▸). Clear positive electron density was observed for the SAM molecule which was bound to the enzyme. The model was further improved using real-space fitting and interactive manual building. The topology and parameter files of the SAM molecules were obtained from the Hetero-compound Information Center of the Uppsala website (Kleywegt & Jones, 1998 ▸). The stereochemical quality of the model was checked with PROCHECK (Laskowski et al., 1993 ▸). The refinement statistics are summarized in Table 4 ▸. The final atomic coordinates have been deposited in the Protein Data Bank (http://www.rcsb.org/pdb) with accession code 1ve3.
Table 4. Structure refinement.
Values in parentheses are for the outer shell.
| Resolution range (Å) | 30.0–2.10 |
| Completeness (%) | 98.6 (98.0) |
| No. of reflections, working set | 29753 |
| No. of reflections, test set | 1468 |
| Final R cryst (%) | 23.3 |
| Final R free (%) | 27.0 |
| No. of non-H atoms | |
| Protein | 3655 |
| Ligand | 54 |
| Water | 178 |
| Total | 3887 |
| R.m.s. deviations | |
| Bonds (Å) | 0.007 |
| Angles (°) | 1.27 |
| Average B factors (Å2) | |
| Protein | 31 |
| Ligand | 26 |
| Water | 34 |
| Ramachandran plot | |
| Favoured regions (%) | 95.12 |
| Additionally allowed (%) | 4.88 |
The DALI server (Holm & Sander, 1995 ▸; http://ekhinda.biocenter.helsinki.fi/dali_server/start) was utilized for a structural similarity search against all structures deposited in the PDB. All figures were prepared using PyMOL (http://www.pymol.org). Surface areas for the dimer and hydrogen bonds were calculated using PISA (Krissinel & Henrick, 2005 ▸). The ionic bridges were calculated using ESRBI (Kumar & Nussinov, 1999 ▸). Aromatic–aromatic interactions and cation–π interactions were calculated using PIC (Tina et al., 2007 ▸).
3. Results and discussion
3.1. Overall structure of PhSAM-MT
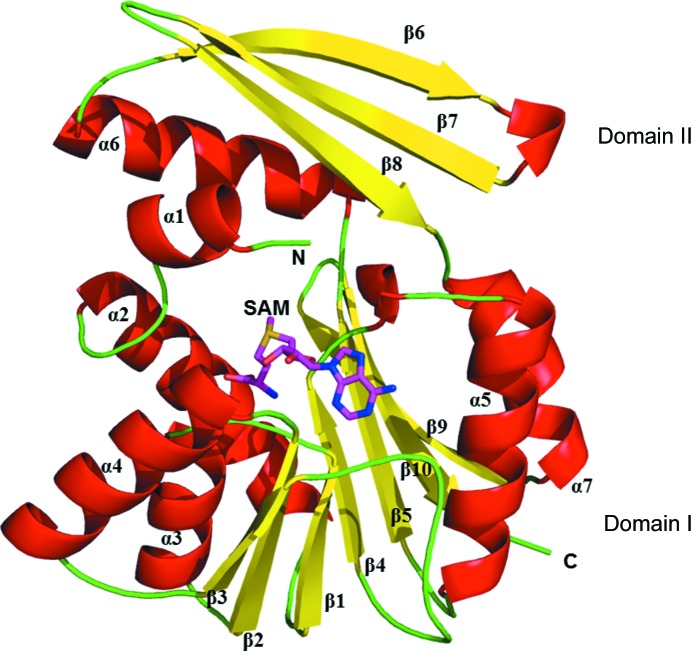
The structure of the SAM-bound complex of PhSAM-MT (Fig. 1 ▸) was determined to 2.1 Å resolution. The Matthews coefficient for PhSAM-MT is 2.3 Å3 Da−1 and the estimated solvent content is 46.1% (Matthews, 1968 ▸). The overall monomeric structure (Fig. 1 ▸) consists of a chain of 226 amino acids mainly assembled into two domains: a typical α/β Rossmann fold (domain I) and a substrate-binding domain (domain II). Protomer B has continuous density for all 226 residues, whereas protomer A shows no density for residues 155–163 and 181–185. These residues were not included. Domain I consists of a seven-stranded β-sheet sandwiched between two helical regions consisting of five α-helices, and domain II consist of an α-helix and three antiparallel β-strands. Compared with other SAM-MTs, the Rossmann fold of domain I adopts the strand topology β3–β2–β1–β4–β5–β10–β9. In this topology, the β10 strand folds antiparallel when compared with other strands in the sequential topology (Fig. 1 ▸).
Figure 1.
The overall monomeric structure (cartoon representation; red helices and yellow sheets) of PhSAM-MT is divided into two domains (domain I, Rossmann-like fold; domain II, substrate-binding domain). The SAM molecule is represented as a stick model. The N-terminus and C-terminus are marked.
3.2. SAM-binding interactions
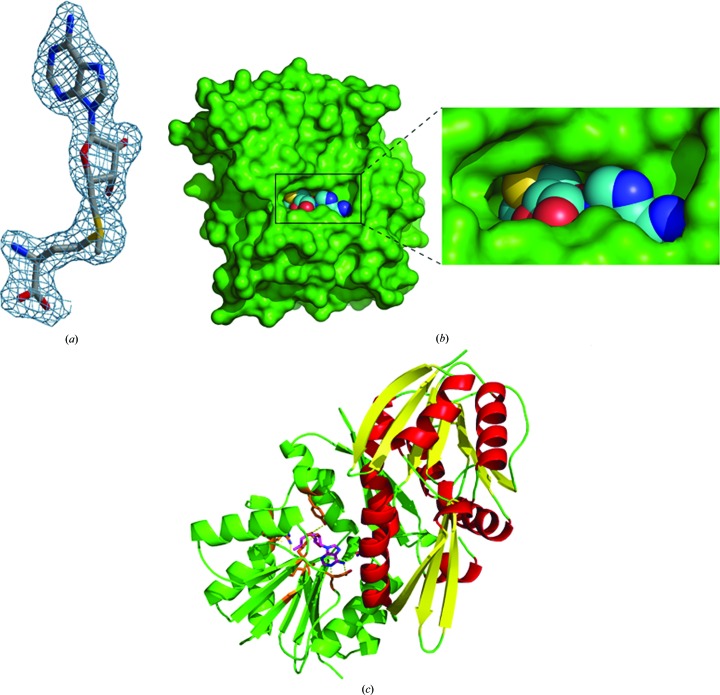
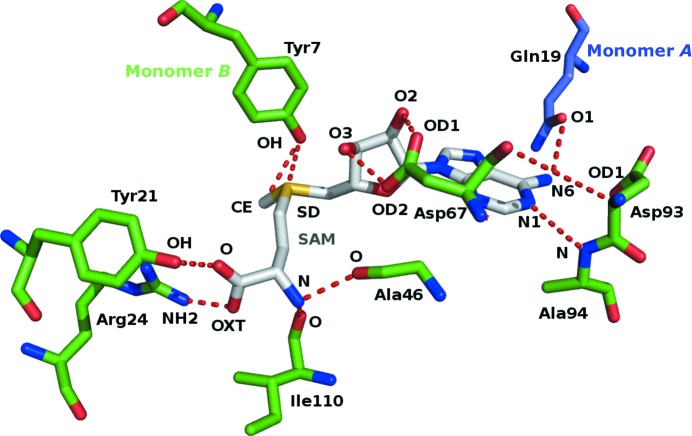
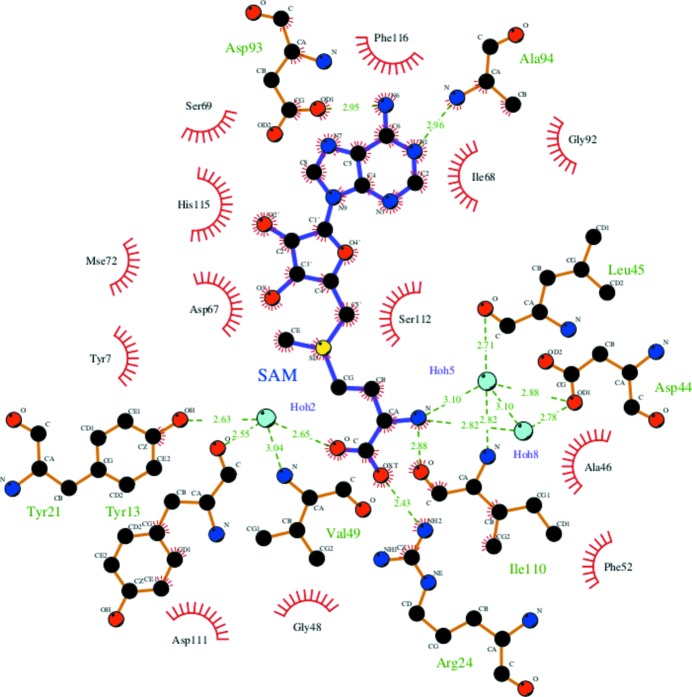
The SAM molecule has well defined electron density in both protomers (Fig. 2 ▸ a), and the surface of the enzyme with the binding pocket is shown in Fig. 2 ▸(b). The SAM molecule was not added to the protein solution deliberately during protein expression, purification and crystallization. The SAM molecule is mostly associated with one monomer, and a few residues from domain I of the other monomer help to define the binding site for the ADP portion of the cofactor. This suggests that formation of the dimer is essential for the function of this enzyme (Fig. 2 ▸ c). Binding-pocket residue interactions and hydrogen bonds are shown in Fig. 3 ▸. The adenine-base residues (N1 and N6) of SAM are in a narrow groove formed by the side chains of Asp93 (OD1) and Gln19 from the other monomer (O1) which form hydrogen-bond interactions. The hydroxyl group of the ribose moiety (O2 and O3 atoms) hydrogen-bonds to the side-chain O atoms (OD1 and OD2) of Asp67. The S atom (SD) of SAM and the methyl C atom (CE) are hydrogen-bonded to the side-chain O atom (OH) of Tyr7. The amide N atom of SAM (N) is hydrogen-bonded to the main-chain carbonyls of Ala46 (O) and Ile110 (O). The two hydroxyl groups (O and OXT) are hydrogen-bonded to the side chains of Tyr21 (OH) and Arg24 (NH2), respectively (Fig. 3 ▸ and Supplementary Table S1). In addition, water molecules are involved in forming hydrogen bonds between SAM and the enzyme. A water molecule (Hoh2) bridges between SAM (O) and Tyr13 (O), Tyr21 (OH) and Val49 (N). Asp44 (OD1) and Leu45 (O) interact with SAM through the environment created by water molecules Hoh5 and Hoh8, respectively (Fig. 4 ▸).
Figure 2.
(a) SAM (stick model) shown within a difference density map contoured at 1.3σ. (b) The binding pocket of the enzyme (green surface) with the SAM molecule shown as a sphere model. (c) The interaction of SAM (magenta stick model within the green monomer) with the other monomer (red α-helices and yellow β-sheets) leading to dimerization.
Figure 3.
SAM-binding residues: SAM (grey stick model) forms hydrogen bonds (red dashed lines) to Arg24, Tyr21, Ala46, Ile110, Ala94, Asp93, Tyr7, Asp67 and Gln19 from the other monomer (shown as stick models). The atoms involved in hydrogen bonding are labelled.
Figure 4.
SAM interaction: the residues binding through water molecules are depicted and the corresponding distances are labelled. This figure was drawn using LIGPLOT (Wallace et al., 1995 ▸).
3.3. Oligomerization and structural comparison
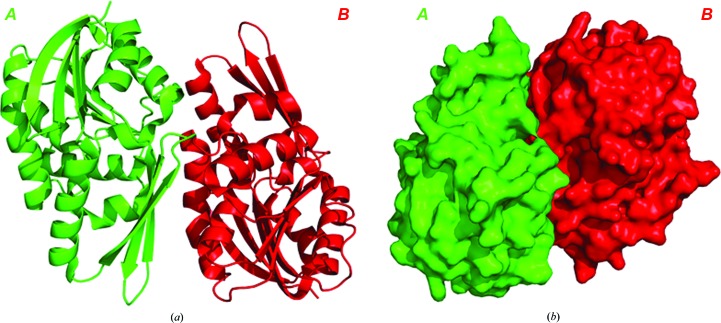
The observed dimers are consistent with the results of dynamic light-scattering studies showing the dimeric state of PhSAM-MT in solution. The dimeric interface between protomers contains 24 hydrogen bonds and has a dimeric surface area of 806 Å2 (Fig. 5 ▸). In addition, the SAM molecule forms hydrogen bonds to Gln19 (α1) of the other monomer (Fig. 2 ▸ c), providing additional interactions in the dimerization. The structural features responsible for thermostability were calculated (Supplementary Table S2) and consist of 19 ion pairs, 15 aromatic pairs, one aromatic–sulfur interaction (Phe89 and Cys47) and 12 cation–π interactions. Overall, the above parameters help to provide the enzyme with the thermal stability to perform catalysis at high temperature.
Figure 5.
The dimeric structure shown in (a) ribbon and (b) surface representation. The cofactor is removed for clarity.
A three-dimensional structural similarity search was performed using the DALI server for the domains of the PhSAM-MT complex structure (domain I and domain II) against structures available from the PDB. The results from the DALI server are listed in Supplementary Table S3. The two most similar structures to domain I are those of the SAM-MTs from P. horikoshii (PDB entry 1wzn; H. Mizutani & N. Kunishima, unpublished work) and Methanosarcina mazei (PDB entry 3sm3; Northeast Structural Genomics Consortium, unpublished work). The 1wzn coordinates show a Z-score of 22.8, an r.m.s.d. of 2.2 Å, 245 residues fitted and a sequence identity of 33%. The other structure (PDB entry 3sm3) shows a Z-score of 22.7, an r.m.s.d. of 1.5 Å, 212 residues fitted and a sequence identity of 28%. The remaining structures are from the SAM-MT family and the non-SAM-dependent methyltransferase (non-SAM-MT) family. The most similar structure to domain II is that of a protein of unknown function from Aquifex aeolicus (PDB entry 2arh; Midwest Center for Structural Genomics, unpublished work). The 2arh coordinates show a Z-score of 4.6, an r.m.s.d. of 2.5 Å, 198 residues fitted and a sequence identity of 8%. Other structures from the non-SAM-MT family show low Z-scores (4.3–3.9), r.m.s.d. values of 1.7–2.3 Å and very low sequence identity.
In this study, we have determined the three-dimensional structure of PhSAM-MT with SAM bound. The monomeric structure consists of two domains: a Rossmann-fold domain (domain I) and a substrate-binding domain (domain II). The cofactor SAM molecule is bound at the cleft between the two domains and interacts with residues from the other monomer, leading to oligomerization. This crystal structure may provide a structural platform for elucidating the mechanism of enzymatic catalysis. Further biochemical studies of the enzyme in substrate/cofactor and product/cofactor complexes will reveal further features of this enzyme.
Supplementary Material
Tables S1, S2 and S3. DOI: 10.1107/S2053230X17016648/ft5085sup1.pdf
PDB reference: SAM-dependent methyltransferase, 1ve3
Acknowledgments
The authors would like to thank the University of Mysore, Mysuru. The authors would also like to thank the staff of RIKEN Genomic Sciences Center for providing the plasmid and the beamline staff for assistance during data collection at beamline BL26B1 of SPring-8, Japan.
Funding Statement
This work was funded by University Grants Commission grant MRP-Phys-2013-32718.
References
- Brünger, A. T., Adams, P. D., Clore, G. M., DeLano, W. L., Gros, P., Grosse-Kunstleve, R. W., Jiang, J.-S., Kuszewski, J., Nilges, M., Pannu, N. S., Read, R. J., Rice, L. M., Simonson, T. & Warren, G. L. (1998). Acta Cryst. D54, 905–921. [DOI] [PubMed]
- Fauman, E. B., Blumenthal, R. M. & Cheng, X. (1999). S-Adenosylmethionine-Dependent Methyltransferase: Structures and Functions, edited by X. Cheng & R. M. Blumenthal, pp. 1–38. Singapore: World Scientific. https://doi.org/10.1142/9789812813077_0001.
- Hendricks, C. L., Ross, J. R., Pichersky, E., Noel, J. P. & Zhou, Z. S. (2004). Anal. Biochem. 326, 100–105. [DOI] [PubMed]
- Hendrickson, W. A., Horton, J. R. & LeMaster, D. M. (1990). EMBO J. 9, 1665–1672. [DOI] [PMC free article] [PubMed]
- Holm, L. & Sander, C. (1995). Trends Biochem. Sci. 20, 478–480. [DOI] [PubMed]
- Jancarik, J. & Kim, S.-H. (1991). J. Appl. Cryst. 24, 409–411.
- Jenuwein, T. & Allis, C. D. (2001). Science, 293, 1074–1080. [DOI] [PubMed]
- Kleywegt, G. J. & Jones, T. A. (1998). Acta Cryst. D54, 1119–1131. [DOI] [PubMed]
- Kozbial, P. Z. & Mushegian, A. R. (2005). BMC Struct. Biol. 5, 19. [DOI] [PMC free article] [PubMed]
- Krissinel, E. & K. Henrick, K. (2005). Computational Life Sciences, edited by M. R. Berthold, R. Glen, K. Diederichs, O. Kohlbacher & I. Fischer, pp. 163–174. Berlin/Heidelberg: Springer-Verlag. https://doi.org/10.1007/11560500_15.
- Kumar, S. & Nussinov, R. (1999). J. Mol. Biol. 293, 1241–1255. [DOI] [PubMed]
- Laskowski, R. A., MacArthur, M. W., Moss, D. S. & Thornton, J. M. (1993). J. Appl. Cryst. 26, 283–291.
- Liscombe, D. K., Louie, G. V. & Noel, J. P. (2012). Nat. Prod. Rep. 29, 1238–1250. [DOI] [PubMed]
- Martin, J. L. & McMillan, F. M. (2002). Curr. Opin. Struct. Biol. 12, 783–793. [DOI] [PubMed]
- Matthews, B. W. (1968). J. Mol. Biol. 33, 491–497. [DOI] [PubMed]
- Otwinowski, Z. & Minor, M. (1997). Methods Enzymol. 276, 307–326. [DOI] [PubMed]
- Schubert, H. L., Blumenthal, R. M. & Cheng, X. (2003). Trends Biochem. Sci. 28, 329–335. [DOI] [PMC free article] [PubMed]
- Terwilliger, T. C. & Berendzen, J. (1999). Acta Cryst. D55, 849–861. [DOI] [PMC free article] [PubMed]
- Tina, K. G., Bhadra, R. & Srinivasan, R. (2007). Nucleic Acids Res. 35, W473–W476. [DOI] [PMC free article] [PubMed]
- Wallace, A. C., Laskowski, R. A. & Thornton, J. M. (1995). Protein Eng. Des. Sel. 8, 127–134. [DOI] [PubMed]
- Winn, M. D. et al. (2011). Acta Cryst. D67, 235–242.
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
Tables S1, S2 and S3. DOI: 10.1107/S2053230X17016648/ft5085sup1.pdf
PDB reference: SAM-dependent methyltransferase, 1ve3