SUMMARY
Aneuploidy is a hallmark of cancer, although its effects on tumorigenesis are unclear. Here, we investigated the relationship between aneuploidy and cancer development using cells engineered to harbor single extra chromosomes. We found that nearly all trisomic cell lines grew poorly in vitro and as xenografts, relative to genetically matched euploid cells. Moreover, the activation of several oncogenic pathways failed to alleviate the fitness defect induced by aneuploidy. However, following prolonged growth, trisomic cells acquired additional chromosomal alterations that were largely absent from their euploid counterparts and that correlated with improved fitness. Thus, while single-chromosome gains can suppress transformation, the genome-destabilizing effects of aneuploidy confer an evolutionary flexibility that may contribute to the aggressive growth of advanced malignancies with complex karyotypes.
INTRODUCTION
In the early 20th century, Theodor Boveri proposed that abnormal karyotypes altered the equilibrium between pro- and anti-proliferative cellular signals and were therefore capable of transforming primary cells into cancer cells (Boveri, 2008). Boveri’s hypothesis was one of the first genetic explanations of cancer development, and it helped motivate a century of research into the origins and consequences of chromosome segregation errors. Since Boveri’s time, it has been established that approximately 90% of solid tumors and 75% of hematopoietic cancers display whole-chromosome aneuploidy (Weaver and Cleveland, 2006). However, the precise relationship between aneuploidy and tumorigenesis remains unclear.
A preponderance of current evidence supports Boveri’s hypothesis (Gordon et al., 2012; Holland and Cleveland, 2009). First, individuals with Down syndrome (trisomy 21) frequently develop pediatric leukemia, suggesting a clear link between the gain of chromosome 21 and leukemogenesis (Seewald et al., 2012). Second, many human cancers exhibit recurrent aneuploidies (Ozery-Flato et al., 2011; Zack et al., 2013), and computational modeling has suggested that these patterns of chromosomal alterations reflect an evolutionary process in which cancer cells increase the copy number of loci encoding oncogenes and decrease the copy number of loci encoding tumor suppressors (Davoli et al., 2013). Finally, genetically engineered mice that harbor alleles that cause chromosomal instability (CIN) typically develop tumors at accelerated rates (Li et al., 2009; Michel et al., 2001; Park et al., 2013; Sotillo et al., 2007, 2010), particularly when combined with mutations in the p53 tumor suppressor (Li et al., 2010). Low levels of CIN have been reported to be particularly tumorigenic (Silk et al., 2013). Nonetheless, several observations suggest that the relationship between aneuploidy and cancer may be more complex than previously believed. While individuals with Down syndrome are at an increased risk of developing leukemia and germ cell tumors, they are at a significantly decreased risk of developing many other common solid tumors (Nižetić and Groet, 2012). Moreover, although mouse models of CIN are generally tumor prone, in certain organs or when combined with certain oncogenic mutations, CIN mice exhibit reduced tumor burden (Silk et al., 2013; Weaver et al., 2007). Thus, aneuploidy may have tumor-protective as well as tumor-promoting effects, which could differ depending on the genetic and environmental milieu.
In order to further our understanding of the effects of aneuploidy on cell and organismal physiology, systems have been developed to generate cells with a range of aneuploid karyotypes (Pavelka et al., 2010; Stingele et al., 2012; Torres et al., 2007; Williams et al., 2008). These cells have been constructed without CIN-promoting mutations, thereby allowing the study of aneuploidy in the absence of other genetic perturbations. This research has demonstrated the existence of a set of phenotypes that are shared among many different aneuploid cells and are largely independent of the specific chromosomal alteration: aneuploid cells display reduced fitness (Stingele et al., 2012; Torres et al., 2007; Williams et al., 2008), are deficient at maintaining proteostasis (Donnelly et al., 2014; Oromendia et al., 2012; Tang et al., 2011), and exhibit a specific set of gene expression changes that include the downregulation of cell-cycle transcripts and the upregulation of a stress-response program (Dürrbaum et al., 2014; Sheltzer, 2013; Sheltzer et al., 2012). A crucial question, however, is in what way(s) the cellular changes induced by aneuploidy affect (and possibly drive) tumorigenesis. Aneuploid cells may be poised to undergo transformation due to their increased dosage of oncogenes and decreased dosage of tumor suppressors (Davoli et al., 2013), the inherent instability of aneuploid genomes (Duesberg et al., 1999; Passerini et al., 2016), or a general misregulation of cell metabolism and other biological processes (Rasnick and Duesberg, 1999). However, the oncogenic potential of aneuploid cells has not been systematically tested.
RESULTS
Single-Chromosome Gains Are Insufficient to Induce Neoplastic Phenotypes
We have compared the tumorigenicity of genetically matched euploid and trisomic primary cells. To accomplish this, we took advantage of naturally occurring Robertsonian translocations to generate mouse embryonic fibroblasts (MEFs) trisomic for chromosome 1, 13, 16, or 19, as well as sibling-matched euploid controls (Williams et al., 2008). While advanced malignancies frequently harbor complex karyotypes that include multiple chromosome gains and/or losses, early-stage cancers typically exhibit one or a few arm-length or whole-chromosome aneuploidies (Balaban et al., 1986; Di Capua Sacoto et al., 2011; Lai et al., 2007; Magnani et al., 1994; El-Rifai et al., 2000). Thus, these trisomies likely recapitulate the karyotypic state of pre-malignant or early-stage cancer lesions, and their study can shed light on the role of aneuploidy in cancer development and evolution.
Read depth analysis from low-pass whole-genome sequencing of each MEF line demonstrated that these cells harbored complete trisomies without other chromosomal alterations and that the extra chromosomes were present clonally within the cell population (Figure 1A). Various oncogenes are encoded on these chromosomes, including BCL2 (mChr1), FGFR4 (mChr13), Jak2 (mChr19), and many others (Table S1). Gain of these oncogenes, or some other consequence of aneuploidy, could drive malignant growth or otherwise generate cancer-like phenotypes in primary cells. We therefore set out to discover whether single-chromosome aneuploidy in MEFs was sufficient to induce neoplastic or pre-neoplastic behavior in untransformed cells.
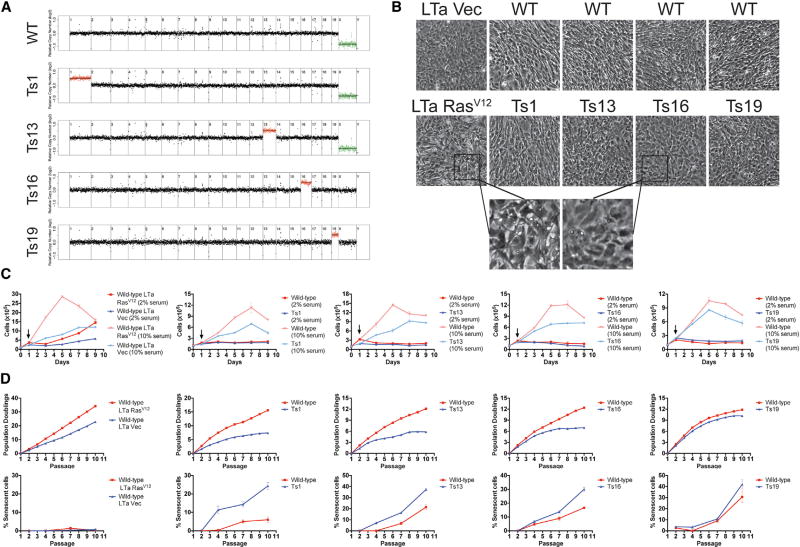
Figure 1. Single-Chromosome Aneuploidy Is Insufficient to Induce Neoplastic Phenotypes.
(A) MEF lines that were euploid or trisomic for chromosomes 1, 13, 16, or 19 were subjected to low-pass whole-genome sequencing. Normalized read depths across 500 kb bins are displayed. Note that only one euploid cell line is shown, although each trisomic MEF line had a separate euploid line that was derived from a euploid littermate.
(B) Photomicrographs of monolayers of the indicated cell lines. LTa + RasV12 MEFs, but not trisomic MEFs, lose contact inhibition when grown to confluence.
(C) Growth curves of the indicated cell lines are displayed. Cells were first plated in normal (10% serum) medium, then 24 hr after plating, the cells were re-fed or switched to reduced (2% serum) medium (indicated by an arrow). LTa + RasV12 MEFs, but not trisomic MEFs, continue to divide in low-serum medium.
(D) The indicated cells were passaged, counted, and plated in triplicate every third day for 10 passages (top row). On passages 2, 4, 7, and 10, β-galactosidase levels were measured (bottom row). LTa-transduced MEFs exhibit negligible levels of senescence, but trisomic cell lines senesce at an early passage. Error bars indicate the SEM.
See also Figures S1 and S2, and Table S1.
As a positive control for cancer-like growth, we generated MEF lines that had been stably transduced with the Large T antigen (LTa), which inhibits the Rb and p53 tumor suppressor pathways (Ahuja et al., 2005), and with either an empty vector or an activated allele of H-Ras (RasV12). As expected, the LTa + RasV12 MEFs exhibited several neoplastic phenotypes: the cells were not contact inhibited, and instead piled on top of each other when grown to confluence (Figure 1B), they formed colonies from single cells when plated at low density (Figure S1A), they grew independently of pro-proliferative signals, as demonstrated by their increase in cell number when plated in low-serum medium (Figure 1C), and they grew robustly without senescing over 10 passages in culture (Figure 1D). The LTa + vector MEFs displayed an intermediate, pre-neoplastic phenotype: the cells maintained contact inhibition and grew very poorly following serum withdrawal but were mildly clonogenic and doubled without noticeable senescence over 10 passages in culture. In contrast, both euploid and trisomic MEFs failed to display any cancer-like phenotypes: they exhibited appropriate contact inhibition, failed to proliferate in low-serum medium, were non-clonogenic, and senesced after 7–10 passages in culture (Figures 1 and S1A). Interestingly, trisomies 1, 13, and 16 senesced at earlier passages and to a significantly higher degree than their matched euploid lines, as judged by β-galactosidase staining (Figure 1D). The proliferation defect and increased senescence of the aneuploid cell lines was approximately proportional to the degree of aneuploidy: cells trisomic for mChr1 (the largest mouse autosome) displayed the most severe phenotypes, while trisomy of mChr19 (the smallest mouse autosome) induced more subtle effects. We conclude that single-chromosome aneuploidy is insufficient to generate neoplastic phenotypes, and many aneuploidies in fact induce a premature growth arrest.
Oncogene-Transduced Trisomic Cells Exhibit Reduced Proliferation and Fitness Relative to Oncogene-Transduced Euploid Cells
As aneuploidy alone was insufficient to generate cancer-like phenotypes, we next set out to determine whether aneuploidy could have growth-promoting interactions with oncogenic mutations. In particular, loss of the p53 tumor suppressor has been linked with heightened proliferation of aneuploid cells (Thompson and Compton, 2010). To screen for synergistic interactions between aneuploidy and common oncogenic mutations, we stably transduced trisomic and euploid MEFs with plasmids encoding various oncogenes or with matched empty vectors. Trisomies 1, 13, 16, and 19, as well as matched euploid cell lines, were transduced with a dominant-negative allele of TP53 (p53dd; Shaulian et al., 1992), the E1a oncogene (which inhibits the Rb tumor suppressor, among several other cellular pathways; Gallimore and Turnell, 2001), the Large T oncogene, or with the MYC oncogene. Following selection for transduced cells, we confirmed that p53dd and MYC were expressed at similar levels in euploid and trisomic MEFs (Figure S2A). Low-pass whole-genome sequencing of a subset of oncogene-expressing cell lines at early passage further revealed that these cell lines had maintained their initial karyotypes following transduction and selection (Figure S2B). In total, these results confirmed that we had successfully constructed congenic, oncogene-expressing cell lines that differed by a single-chromosome copy number alteration (CNA). We then subjected the transduced euploid and trisomic cell lines to a series of assays designed to test their tumorigenic capacity.
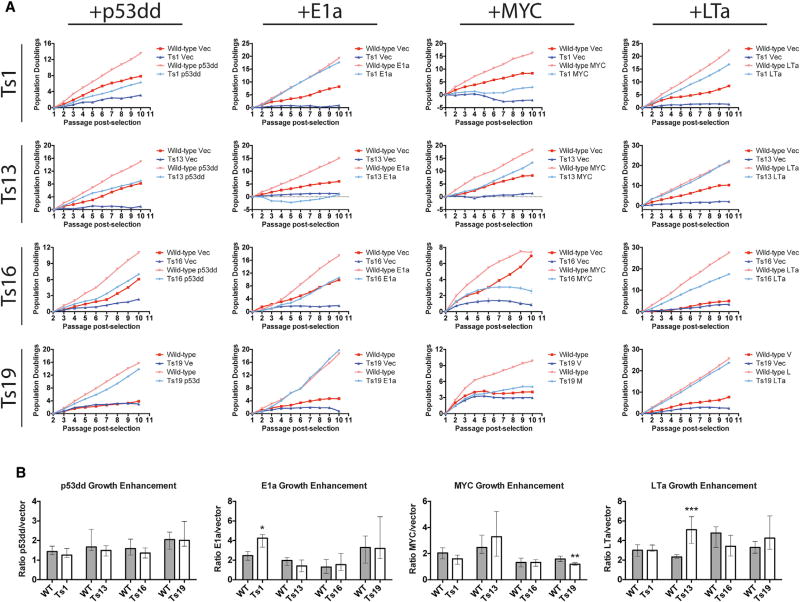
First, we counted and passaged oncogene-transduced euploid and trisomic MEFs in a modified 3T3 protocol up to 10 times over the course of 30 days. Empty vector-transduced trisomic MEFs generally underwent fewer population doublings than matched euploid lines (Figure 2A, compare dark red and dark blue lines). Transduction with oncogenes significantly enhanced the growth of trisomic cells, and the resultant lines doubled more frequently over the course of the experiment than the vector-matched controls. However, these oncogenes also enhanced the growth of euploid cell lines. In the majority of cases, the oncogene-transduced euploid cell line underwent more population doublings than the corresponding oncogene-transduced aneuploid line, and in only a few instances did we observe equivalent proliferation between euploid and trisomic MEFs (Figure 2A, compare light red and light blue lines). For instance, over 10 passages in culture, p53dd-transduced trisomy 13 cells doubled about 9 times, while a p53dd-transduced euploid line doubled 15 times. Cells trisomic for chromosome 1 (the largest mouse autosome) generally grew the slowest relative to wild-type (WT), while cells trisomic for chromosome 19 (the smallest mouse autosome) tended to proliferate at close to WT levels. Overall, expression of p53dd or MYC failed to suppress the growth differential between aneuploid and euploid cell lines, while the expression of E1a or LTa had more potent effects. Transduction with Large T, in particular, demonstrated the most significant suppression of the aneuploidy-induced proliferation defect, although LTa-expressing cells trisomic for chromosomes 1 and 16 still divided significantly more slowly than LTa-expressing euploid lines.
Figure 2. Trisomy Impedes the Proliferation of Oncogene-Transduced Cell Lines.
(A) Euploid and trisomic cell lines were stably transduced with plasmids harboring the indicated oncogene or a matched empty vector. Following selection, the cell lines were passaged every third day for up to 10 passages, and the cumulative population doublings over the course of each experiment are displayed. Note that the panel displaying Ts19 + LTa is reproduced in Figure S7. See also Figures S3 and S4.
(B) The number of cells recovered from oncogene-transduced MEFs was divided by the number of cells recovered from vector-transduced MEFs at every passage. Bar graphs display the median ratios and the interquartile ranges. *p < 0.05, **p < 0.005, ***p < 0.0005 (Wilcoxon rank-sum test). See also Figures S3 and S4.
We next tested several additional oncogenes in a subset of trisomies: euploid cell lines transduced with a stabilized allele of MYC (MYCT58A) outgrew matched cell lines trisomic for chromosomes 13 or 16, while expression of an activated allele of BRAF (BRAFV600E) or an allele of CDK4 resistant to p16 inhibition (CDK4R24C) resulted in senescence in both euploid and trisomic cell lines (Figure S3A).
As the experiments described above were conducted using initial populations of primary cells, we selected nine experiments to repeat on independently derived cell lines (Figure S3B). Replicate experiments displayed some line-to-line variability (e.g., Ts16 + LTa), but recapitulated the major features of our initial results: oncogene-transduced trisomic cells grew less rapidly than euploid cells, and the expression of Large T provided the most significant rescue of trisomic cell growth (Figure S3B).
The prolonged culture period also allowed us to follow the dynamics of aneuploid cell populations over time: rapidly dividing subpopulations could potentially arise during 30 days in culture that would enhance the apparent proliferative capacity of the aneuploid MEFs. Interestingly, in our replicate analysis on independently derived cell lines, we found that the proliferation rate of one Ts19 + LTa cell line increased during serial passaging, while an independent Ts19 + LTa line grew at approximately the same rate over the course of the experiment (compare Figures 2A and S3B). The reasons for this divergent behavior are explored below. In general, few trisomic cell lines doubled more rapidly over successive passages, while many trisomic cell lines grew more slowly over time instead. Consistent with their poor proliferation at high passage, p53-and MYC-transduced trisomic lines displayed elevated levels of senescence-associated β-galactosidase staining relative to euploid controls (Figure S4A). However, expression of E1a or LTa effectively blocked senescence in all cells, although a fitness differential between euploid and trisomic populations was still evident, suggesting that other factors also contribute to the growth delay in aneuploid cells. Across all of our experiments, we did not observe any instances in which transduction with an oncogene generated a trisomic cell line that consistently outgrew its matched euploid counterpart in multiple independent lines. We conclude that euploid lines harboring common oncogenic mutations generally proliferate more rapidly than trisomic lines harboring the same genetic alterations.
Although proliferative differences between aneuploid and euploid lines persisted following oncogene transduction, it remained possible that the oncogenes provided a relatively greater growth advantage to aneuploid cells than to euploid cells. To test this, we quantified the benefit conferred by each oncogene by comparing the number of cells recovered at every passage from oncogene-transduced and vector-transduced cell lines (Figure 2B). For most oncogene-trisomy combinations, the fold change in growth enhancement was equivalent between euploid and aneuploid lines. For instance, transduction with dominant-negative p53 resulted in an approximately 1.6-to 1.8-fold increase in doublings per passage, relative to the vector-transduced controls, in all euploid and trisomic cell lines. Thus, the abrogation of p53 signaling does not specifically enhance the growth of these trisomic cells. In total, across 16 oncogene-aneuploidy combinations, oncogene expression was found to confer a similar proliferative benefit to euploid and trisomic cells in 13 combinations, while in only two conditions (Ts1 + E1a and Ts13 + LTa) was oncogene expression found to have a stronger effect on the trisomic MEFs than on the euploid MEFs. These results suggest that oncogene-trisomy synergy is rarely observed and may be specific to certain chromosomes or oncogenes.
As an additional test of the proliferative capabilities of the oncogene-transduced lines, we assessed the focus-formation ability of cells that had been transduced with E1a or LTa (MYC and p53dd-transduced lines remained non-clonogenic). We found that transduced euploid lines exhibited uniformly superior colony-forming ability relative to the trisomic lines, even in instances when the euploid and aneuploid lines demonstrated approximately equal doubling times in culture (Figure S1B). For instance, while Ts19 + LTa and WT + LTa MEFs grew at nearly the same rate, the WT MEFs formed about 6-fold more colonies when plated as single cells than the trisomic line did. The differences between the colony-formation and population-doubling assays likely reflect the fact that forming a colony is a relatively greater challenge to a cell than doubling in a monolayer, and this challenge exacerbates the fitness differential between euploid and trisomic cells. In summary, our data indicate that single-chromosome gains cause a pervasive fitness defect, even in oncogene-transduced populations.
RasV12-Transformed Trisomic Cells Exhibit Reduced Tumorigenicity
Complete transformation of rodent cells requires transduction with two cooperating oncogenes (Land et al., 1983). We therefore took euploid and trisomic p53dd- or LTa-transduced cell lines and then stably transduced them with a vector harboring oncogenic H-RasV12 or a control empty vector. Whole-genome sequencing revealed that 14 out of 14 cell lines tested maintained their initial karyotype following two rounds of transduction and selection (Figure S2C).
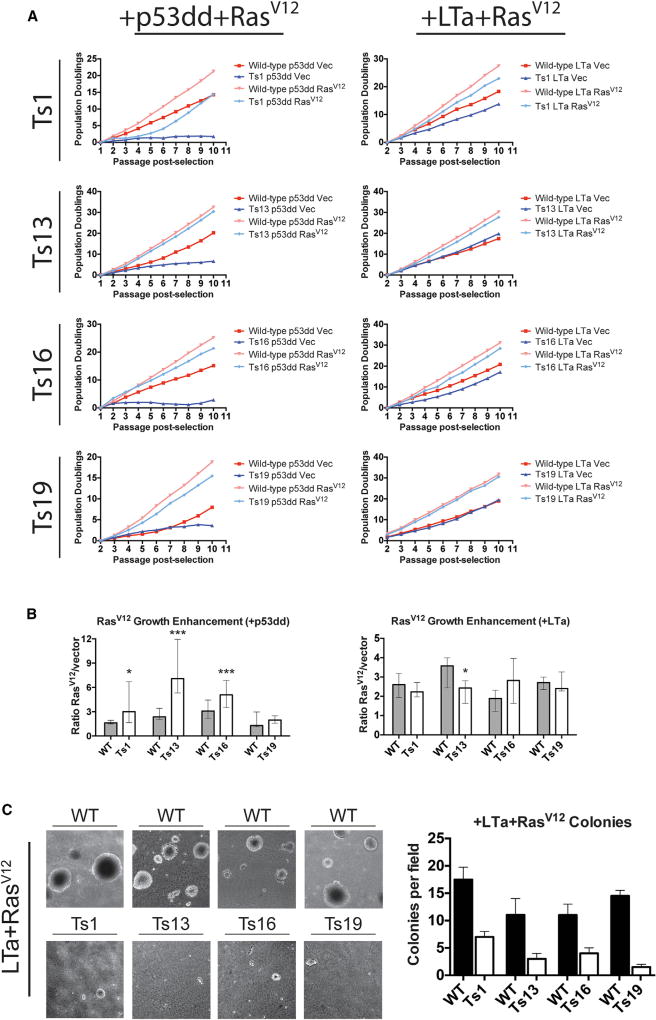
RasV12-transduced cell lines doubled significantly faster than vector-transduced lines over 10 passages in culture, and RasV12 narrowed or in some cases abolished the proliferative difference between euploid and trisomic MEFs (Figure 3A). For instance, while p53dd + vector-transduced WT and Ts13 MEFs doubled 20 and 6 times, respectively, p53dd + RasV12-transduced WT and Ts13 MEFs doubled 32 and 30 times, respectively. However, no RasV12-transduced trisomic line grew faster than its euploid counterpart, and in subsequent assays described below, a fitness benefit conferred by euploidy was clearly detected. RasV12-transduced trisomic cell lines were also found to display elevated levels of senescence-associated β-galactosidase, even when co-expressed with LTa (Figure S4A). In addition, we examined the effects of RasV12 expression in E1a- or MYC-transduced Ts16 cell lines, and we tested the expression of BRAFV600E, PIK3CAH1047R, or MYC in lieu of RasV12 as a driver oncogene (Figure S5). The latter oncogenes typically had little effect in this assay compared with experiments using RasV12 that were performed in parallel, and no other oncogene exhibited a strong rescue of the aneuploidy-induced growth delay.
Figure 3. Effects of RasV12 Expression on Immortalized Euploid and Trisomic MEFs.
(A) Euploid and trisomic cell lines were first stably transduced with p53dd or with LTa, and then transduced a second time with plasmids harboring RasV12 or a matched empty vector. The cell lines were passaged, counted, and plated in triplicate up to 10 passages following the second round of selection. Note that the panel displaying Ts13 + LTa + RasV12 is reproduced in part in Figure S5, the panel displaying Ts16 + p53dd + RasV12 is reproduced in Figure S6, and the panel displaying Ts19 + LTa + RasV12 is reproduced in part in Figures S5 and S7.
(B) The number of cells recovered from RasV12-transduced MEFs was divided by the number of cells recovered from vector-transduced MEFs at every passage. Bar graphs display the median ratios and the interquartile ranges. *p < 0.05, ***p < 0.0005 (Wilcoxon rank-sum test).
(C) 20,000 cells of the indicated cell lines were plated in soft agar and then grown for 20 days. For each comparison, the euploid MEFs formed more colonies than the trisomic MEFs (p < 0.01, Student’s t test). Bar graphs display the mean ± SEM colonies per field. See also Figures S5 and S6.
In order to assess the reproducibility of our results, we performed several replicate experiments on independently derived and immortalized cell lines (Figures S5 and S6). One line of Ts19 + LTa + PIK3CAH1047R grew slightly better than its euploid control, although an independent line of Ts19 + LTa + PIK3CAH1047R did not exhibit this phenotype (Figure S5, and see Figure S8 below). In other replicate experiments, we also observed some variability in the degree of growth rescue caused by oncogene expression. For instance, in an initial experiment, p53dd + RasV12 transduction resulted in an incomplete suppression of the proliferative difference between WT and Ts16 cells, while in a replicate experiment, strong suppression was induced by this oncogene combination. To address the origins and scope of this variability, we repeated this experiment a total of six times using five independent cell lines (Figure S7). When the same primary cell line was transduced with p53dd and RasV12, both sets of transformed lines exhibited comparable proliferation dynamics. When four other Ts16 cell lines were used, each transformed line grew at a moderately different rate, although no trisomic line was observed to divide more rapidly than its euploid counterpart (Figure S7). We conclude that variability within this assay results from genetic or epigenetic differences between primary cell lines, rather than from stochasticity in the experimental protocol. Moreover, while the growth penalty induced by aneuploidy varies among independently derived cell lines, across multiple replicate experiments, no oncogene cocktail tested resulted in reproducibly superior growth in a trisomic line compared with its euploid counterpart.
In order to determine the relative benefit conferred by RasV12 expression to euploid and aneuploid MEFs, we compared the number of cells recovered at each passage from RasV12-transduced and vector-transduced cell lines. RasV12 expression conferred a similar fold increase in proliferative capacity in euploid and trisomic MYC-, E1A-, and three LTa-transduced cell lines, while RasV12 expression had a proportionately greater effect on WT + LTa lines than on Ts13 + LTa lines (Figures 3B and S5B). Interestingly, when p53 signaling was compromised by the expression of p53dd, RasV12 accelerated proliferation to a greater degree in the trisomy 1, 13, and 16 lines than it did in the euploid cell lines (Figure 3B). This is likely due at least in part to increased senescence of the p53dd + vector doubly transduced trisomic MEFs (Figure S4A).
Despite the robust growth of RasV12-transduced trisomic MEFs, we found that RasV12-transduced euploid MEFs formed significantly more colonies from single cells than trisomic lines did (Figure S1C). Fully transformed cell lines are also capable of growing in soft agar, a phenotype that strongly correlates with in vivo tumorigenicity (Shin et al., 1975). We tested the ability of RasV12-transduced cell lines to form colonies in soft agar, and in each experiment the euploid control lines exhibited higher colony-forming ability than the equivalently transduced trisomic lines (Figure 3C).
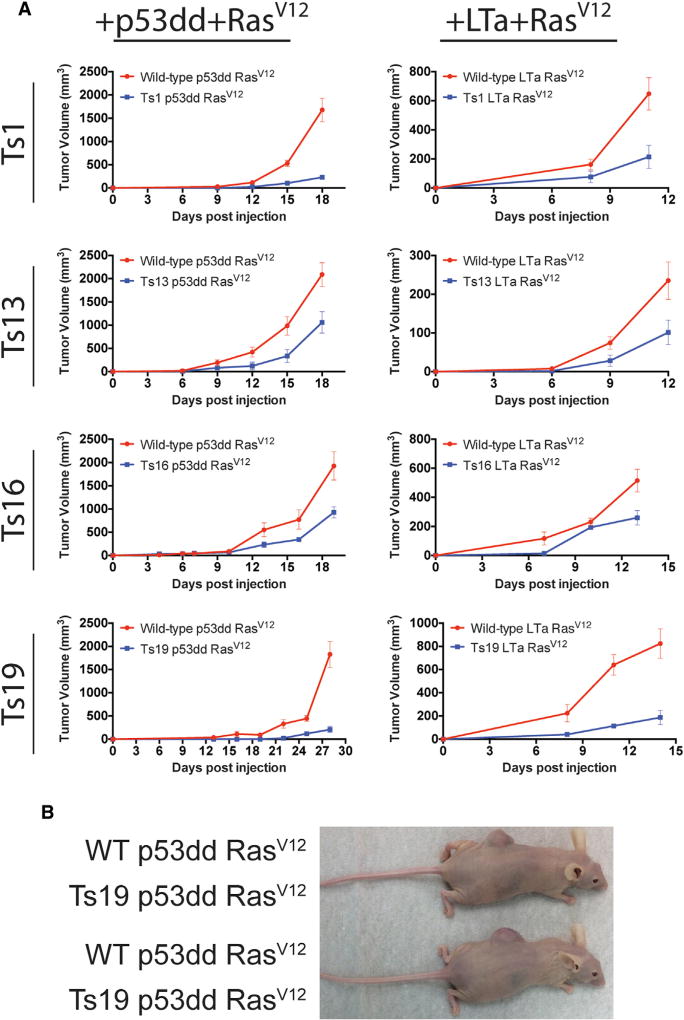
As a final test of the tumorigenicity of transformed trisomic MEFs, we examined the ability of RasV12-transduced euploid and trisomic cell lines to form tumors in xenograft assays. Euploid and trisomic p53dd + RasV12 cell lines were injected contralaterally into the flanks of nude mice, and tumor volume measurements were obtained every third day. While aneuploid and euploid cell lines grew at similar rates in vitro, the euploid lines invariably formed significantly larger tumors in vivo (Figure 4).
Figure 4. Trisomy Hampers Tumor Growth in Xenografts.
(A) 106 euploid or aneuploid cells transduced with either p53dd and RasV12 or with LTa and RasV12 were injected subcutaneously into the flanks of nude mice. Tumor volume was measured every 3 days. Note that mice injected with cells transduced with LTa + RasV12 had to be euthanized prematurely due to cachexia. Error bars indicate the SEM.
(B) Representative images of mice injected contralaterally with WT + p53dd + RasV12 cells or Ts19 + p53dd + RasV12 cells.
We also examined the tumorigenicity of trisomic cell lines that had been transduced with LTa and RasV12. In preliminary experiments, a fraction of mice injected with these cell lines developed cachexia that resulted from metastatic disease. In order to more easily identify the cell line that the metastases were derived from, euploid and trisomic cell lines were injected into different mice during single experiments. In addition, mice were euthanized at 11–15 days post-injection, as cachexia began to develop. Consistent with our previous results, euploid LTa + RasV12 cell lines formed larger tumors than trisomic LTa + RasV12 cell lines (Figure 4). Following euthanasia, necropsies were performed on 29 mice: 3 of 14 mice injected with trisomic cells and 5 of 15 mice injected with euploid cells exhibited evidence of gross metastases (p = 0.68, Fisher’s exact test). Histological analysis identified the primary tumors and metastatic lesions as poorly differentiated fibrosarcomas, consistent with their embryonic fibroblast origins (data not shown). No gross differences in histology were apparent between euploid and trisomic tumors. In total, these results demonstrate that tumors derived from euploid and trisomic cells form histologically similar structures, but euploid cells typically outgrow genetically identical trisomic cells in xenografts.
Chromosome Gains Impede the Tumorigenicity of Human Colorectal Cancer Cells
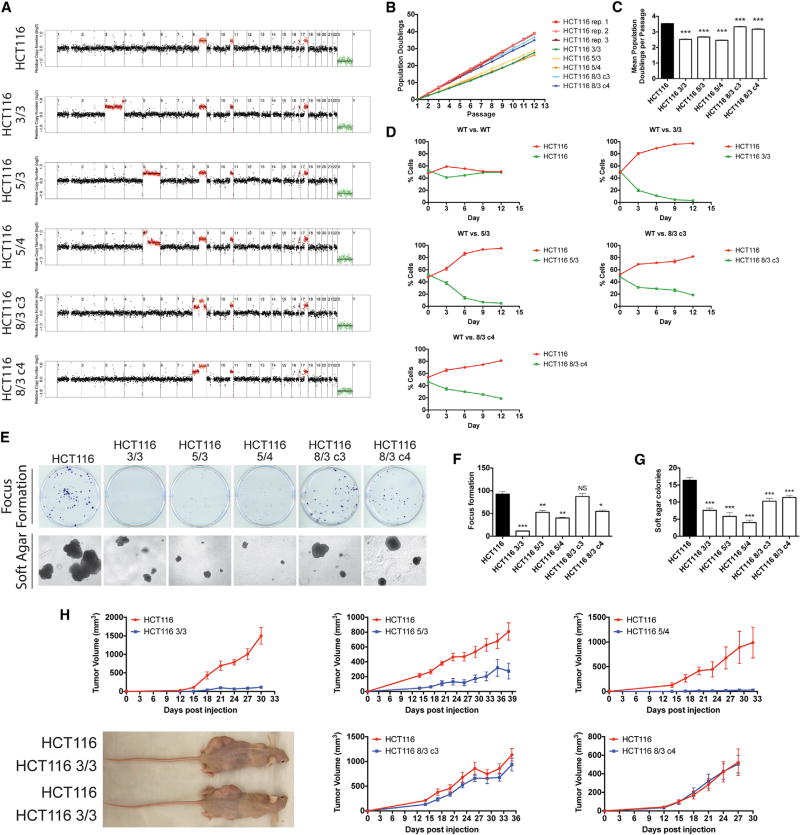
Our experiments in MEFs demonstrated that trisomies in primary cells commonly impede transformation. As aneuploidy is a nearly universal occurrence in cancer, we hypothesized that the acquisition of aneuploidy in previously transformed cells, rather than in primary cells, could have distinct, protumorigenic consequences. To test this, we utilized a series of chromosomally stable human colorectal cancer cells (HCT116) into which extra chromosomes had been introduced via microcell-mediated chromosome transfer (Donnelly et al., 2014; Stingele et al., 2012).
We first characterized each cell line by whole-genome sequencing (Figure 5A). Our panel of cell lines included a derivative of HCT116 that was trisomic for chromosome 5 (HCT116 5/3), a line that had regions of trisomy and tetrasomy on chromosome 5 (HCT115 5/4), a line that had regions of trisomy and tetrasomy on chromosome 3 (HCT116 3/3), and two lines that had regions of trisomy and tetrasomy on chromosome 8 (HCT116 8/3 c4, which had gained a complete extra copy of chromosome 8, and HCT116 8/3 c3, which had gained a partial copy of chromosome 8). Oncogenes encoded on these chromosomes include β-catenin (hChr3), PIK3CA (hChr3), TERT (hChr5), MYC (hChr8), and several others (Table S2). We tested the HCT116 cell lines in similar assays as described above for the trisomic MEFs in order to compare the relative fitness and tumorigenicity of the parental and the derived colon cancer cell lines. During serial passaging, the near-diploid parental line divided the most rapidly, while the HCT116 3/3, 5/3, and 5/4 lines underwent on average one fewer doubling per passage (Figure 5B). HCT116 8/3 c3 and c4 divided at nearly the same rate as the WT line, although over the course of the experiment a small but significant growth delay was evident in these lines (Figure 5C). As the overall difference in proliferation between these lines was small, we considered the possibility that a fitness benefit conferred by aneuploidy would be more readily apparent when the cells were put in direct competition. To test this, we transduced the parental HCT116 line with a vector expressing dsRed, and then co-cultured equal numbers of that line with either a dsRed-negative WT line or with the aneuploid derivatives. Over the course of 12 days in culture, the dsRed+ and dsRed− WT lines displayed equivalent proliferative capacity, as similar numbers of dsRed+ and dsRed− cells were recovered at each passage (Figure 5D). In contrast, when the parental cell line was co-cultured with an aneuploid cell line, the dsRed+ population inevitably expanded to dominate the culture. We conclude that, while the differences in doubling times between the parental and aneuploid HCT116 lines are small, aneuploidy has a profound and negative impact on cell fitness.
Figure 5. Single-Chromosome Gains Impede the Growth of Human Colorectal Cancer Cell Lines In Vitro and In Vivo.
(A) Normalized read depths from whole-genome sequencing of the HCT116 human colorectal cancer cell line as well as HCT116 derivatives that harbored extra chromosomes.
(B) Growth curves of colorectal cancer cell lines with different karyotypes. Cells were counted and passaged every third day.
(C) Quantification of the mean population doublings per passage of multiple replicates of the experiment shown in (B).
(D) Competition experiment between GFP+ aneuploid HCT116 cells and GFP + dsRed+ near-euploid HCT116 cells. Every third day, the cells were trypsinized, passaged, and analyzed for dsRed+ cell number.
(E) 200 cells of the indicated lines were grown for 14 days prior to staining with crystal violet (top), or 2,000 cells of the indicated lines were plated in soft agar and allowed to grow for 20 days before being imaged (bottom).
(F) Quantification of focus formation assayed in (E).
(G) Quantification of colony formation in soft agar in (E).
(H) 4 × 106 HCT116 cells or HCT116 cells with additional chromosome(s) were injected subcutaneously into the flanks of 5–10 nude mice. Tumor growth was measured every third day.
Error bars indicate the SEM. *p < 0.05, **p < 0.005, ***p < 0.0005 (Student’s t test). See also Table S2.
Consistent with these results, we observed that parental HCT116 cells exhibited the highest rates of focus formation and colony growth in soft agar (Figures 5E–5G). HCT116 3/3, 5/3, and 5/4 displayed significant impairments in both assays. HCT116 8/3 c4 exhibited a small reduction in both assays, while HCT116 8/3 c3 grew moderately worse in soft agar but was able to form foci on plastic dishes at WT levels. Several aneuploidies also caused an increase in the expression of senescence-associated β-galactosidase: while the parental line and both hChr8 trisomies displayed minimal senescence, the level of staining was greatly increased in HCT116 3/3 and slightly increased in HCT116 5/3 and 5/4 cells (Figure S4B). The different levels of senescence between HCT116 3/3, 5/3, and 5/4, all of which grow at similarly slow rates, further suggest that factors in addition to senescence contribute to the growth differential between the parental line and the derived aneuploid lines.
To determine how aneuploidy influences tumorigenesis in these cell lines in vivo, we performed contralateral subcutaneous injections of either the parental HCT116 line or the aneuploid derivative lines into flanks of nude mice. The parental HCT116 cell line formed large tumors in all animals into which it had been injected (Figure 5H). These tumors grew at a rapid rate, and each animal had to be euthanized 30–35 days after injection due to tumor burden. HCT116 3/3 and 5/ 4 formed small nodules at the site of injection that remained stable or increased in size only very slightly over the course of the experiment. Mice injected with HCT116 5/3 developed tumors that grew faster than either HCT116 3/3 or HCT116 5/4 but significantly less rapidly than the HCT116 tumors. Finally, consistent with our in vitro experiments, HCT116 8/3 c3 and c4 formed large tumors as xenografts, and there was no significant difference in tumor volume between these lines and the WT line. We conclude that gain of chromosomes in a cancer cell line can antagonize tumor formation.
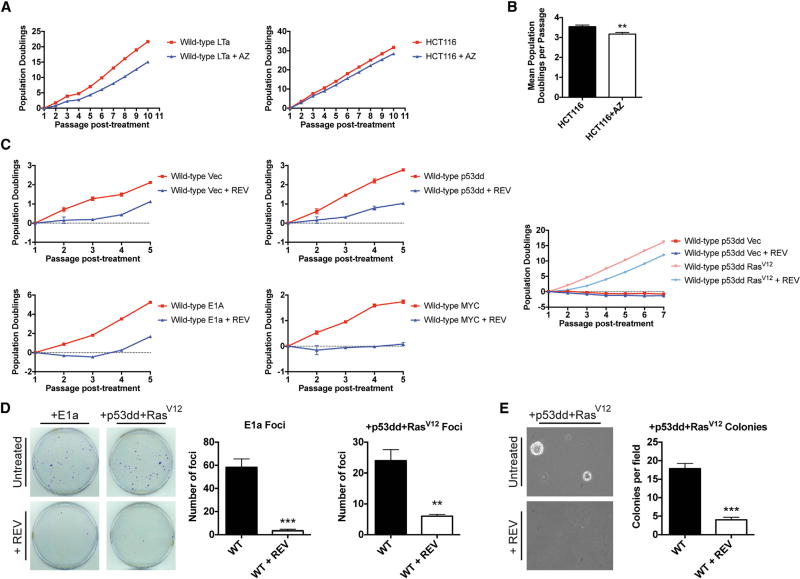
Treatment with Spindle Assembly Checkpoint Inhibitors Fails to Transform MEFs
As our experiments had revealed that single-chromosome gains tended to inhibit tumorigenesis, we sought to expand our analysis by analyzing cells with a wider range of whole-chromosome karyotypic abnormalities. To accomplish this, we treated cells with either reversine or AZ3146, which inhibit the essential spindle assembly checkpoint kinase Mps1 (Hewitt et al., 2010; Santaguida et al., 2010). Treatment with either drug allows cells to enter anaphase with misaligned chromosomes and leads to the generation of daughter cells that harbor various whole-chromosome gains and losses (Santaguida et al., 2015). In order to avoid the confounding effects of continuous CIN, we treated either HCT116 cells or MEFs expressing oncogenes with reversine or AZ3146 for a single passage and then assessed their proliferative capacities in the absence of the inhibitor. In every condition tested, cells that had previously been subjected to the Mps1 inhibitor grew more slowly than untreated control cells (Figure 6). In some cell lines, the poor proliferation remained apparent up to a month after the initial drug treatment (e.g., HCT116 in Figures 6A and 6B). In other cell lines, the proliferation rates between treated and untreated cells equalized over time, potentially due to negative selection against deleterious aneuploid karyotypes (e.g., WT + E1a, Figure 6C). In addition, we found that transient Mps1 inhibition significantly impaired focus formation and growth in soft agar in oncogene-expressing MEFs (Figures 6D and 6E). These results indicate that, like simple trisomies, many complex karyotypes fail to induce cellular transformation and instead inhibit the process.
Figure 6. Spindle Assembly Checkpoint Inhibitor Treatment Fails to Transform MEFs.
(A) MEFs expressing LTa or HCT116 cells were treated for 24 hr with 2 µM AZ3146, and then counted and passaged every third day following removal of the drug.
(B) Quantification of the HCT116 growth curve from (A). Although the slopes of these growth curves are similar, the untreated HCT116 cells undergo significantly more population doublings per passage than the AZ3146-treated HCT116 cells.
(C) MEFs expressing either an empty vector control, p53dd, MYC, E1A, or p53dd and RasV12 were treated with 500 nM reversine for 24 hr and then counted and passaged every third day following removal of the drug.
(D) 1,000 cells of the indicated cell lines that either had or had not been treated with 500 nM reversine for 24 hr were plated and then allowed to grow for 10 days before being stained with crystal violet. Representative plates are shown on the left, while average foci counts are displayed on the right.
(E) 20,000 MEFs expressing p53dd and RasV12 that either had or had not been treated with 500 nM reversine for 24 hr were plated in soft agar and then grown for 20 days. Representative images are shown on the left, while average colonies per field are displayed on the right.
**p < 0.005, ***p < 0.0005 (Student’s t test). Error bars indicate the SEM.
Karyotype Evolution and Improved Growth in Trisomic Cell Lines
The robust growth of aneuploid tumors suggests that cancer cells are able to adapt to the adverse effects of aneuploidy. Our results argue that one commonly hypothesized aneuploidy-tolerating mechanism, the activation of oncogenes and the inactivation of p53 or other tumor suppressors, is largely insufficient to equalize growth between euploid and aneuploid cells. We therefore sought to uncover other changes that could explain how cells are able to adapt to the aneuploid state.
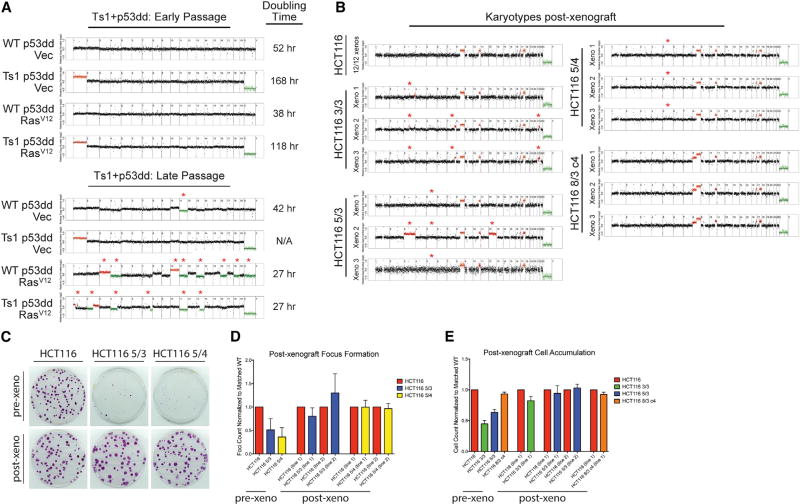
During the course of the oncogene transduction experiments, we observed that the proliferation rate of euploid and aneuploid cell lines occasionally increased following serial passaging. For instance, immediately following selection, Ts1+p53dd + RasV12 cells grew poorly, doubling once every 118 hr (Figures 3A and 7A). However, after several passages in culture, the doubling time had decreased to 27 hr, a rate indistinguishable from the WT + p53dd + RasV12 line. We hypothesized that, if the gain of mChr1 had an inhibitory effect on cell division, then the improved growth of these cells could potentially result from the loss of the trisomy. Indeed, while whole-genome sequencing at early passage revealed the expected karyotypes, a second round of sequencing at late passage revealed that the cells had lost most of the extra copy of mChr1 and displayed several other chromosome gains and losses (Figure 7A). In contrast, the Ts1+p53dd + Vec cell line proliferated at a slow rate over the course of the experiment and maintained an extra copy of mChr1 into late passage. Thus, in certain circumstances, cells can evolve in response to aneuploidy stress by losing a growth-inhibitory chromosome.
Figure 7. Karyotype Evolution Correlates with Enhanced Growth in Aneuploid Cell Lines.
(A) Karyotype analysis of the Ts1+p53dd + RasV12 experiment, initially presented in Figure 3A, is displayed. Whole-genome sequencing at passage 2 revealed that the slow-growing line maintained its initial karyotype, while whole-genome sequencing at passage 10 revealed that the rapidly growing line had lost the trisomy of mChr1 and displayed several further chromosomal gains and losses. Note that the early-passage karyotype analysis is also displayed in Figure S2B, and is presented here for reference.
(B) HCT116 xenografts, initially presented in Figure 5H, were extracted, digested with trypsin, and then plated on plastic. Low-pass whole-genome sequencing revealed that 12 HCT116 xenografts and 3 HCT116 8/3 c4 xenografts maintained their initial karyotypes. However, all HCT116 3/3, 5/3, and 5/4 xenografts lost their initial trisomies or tetrasomies during in vivo growth, and several lines displayed additional chromosomal CNAs. Deviations from each cell line’s initial karyotype are indicated with an asterisk in (A) and (B).
(C) Comparison of the focus-formation ability of HCT116 cells before or after growth as a xenograft.
(D) Quantification of the focus-formation assay in (C).
(E) Proliferation assay of HCT116 cells before or after growth as a xenograft. Error bars indicate the SEM.
See also Figures S7 and S8.
To further our understanding of how cells adapt to aneuploidy, we closely examined the behavior of Ts19 MEFs that had been transduced with LTa. In a series of experiments with different cell lines, we found that one Ts19 + LTa line doubled at a consistent rate over the course of 10 passages in culture, while two independent Ts19 + LTa lines grew more rapidly as the experiment progressed (Figures 2A and S7A). In addition, in a set of Ts19 cell lines transduced with two oncogenes, we noted that one line of Ts19 + LTa + PIK3CAH1047R MEFs grew more rapidly during serial passaging, while an independent Ts19 + LTa + PIK3CAH1047R line proliferated at a consistently slower rate (Figures S5A, S7B, and S7C). Karyotype analysis of all five Ts19 cell lines at late passage revealed that two of three Ts19 + LTa cell lines that exhibited the improved growth phenotype had maintained their mChr19 trisomy but had unexpectedly gained additional copies of mChr2. In contrast, none of the slow-growing Ts19 + LTa cell lines exhibited a +mChr2+mChr19 karyotype. These results imply that, in addition to simply losing a growth-inhibitory chromosome, cells may adapt to karyotype imbalances by acquiring additional aneuploidies that potentially compensate for or “balance” the growth defect induced by the initial trisomy. However, not every rapidly growing Ts19 line gained mChr2, suggesting that for any particular aneuploidy, there may exist multiple evolutionary routes to an improved fitness state.
We next set out to test whether aneuploid cells exhibit karyotype adaptations to growth in vivo. Toward this end, we re-derived cell lines from several MEF + p53dd + RasV12 and HCT116 xenografts and then subjected the post-xenograft lines to whole-genome sequencing. We found that one Ts13 + p53dd + RasV12 xenograft lost its extra copy of mChr13 in vivo, while one Ts19 + 53dd + RasV12 xenograft lost part of mChr19 and also gained an extra copy of mChr3 (Figure S8). In contrast, the Ts19-matched WT line remained euploid during xenograft growth. Interestingly, the WT line matched to Ts13 appeared to have lost copies of mChr11 and mChr13 prior to injection, but following growth in vivo, it reverted to a near-euploid state (Figure S8). The apparent selection for euploidy in this cell line underscores our finding that many single-chromosome aneuploidies hinder tumor growth and that certain aneuploidies can be actively selected against.
Among the parental HCT116 xenografts, 12 of 12 re-derived cell lines had near-euploid karyotypes that were indistinguishable from the karyotype of the pre-xenograft cell line (Figure 7B). However, every HCT116 3/3, 5/3, and 5/4 cell line was found to have lost its original trisomic or tetrasomic chromosome. Three of these nine aneuploid lines exhibited other chromosomal alterations: one cell line initially trisomic for chromosome 5 gained an extra copy of chromosomes 3 and 12, while two cell lines initially trisomic for chromosome 3 gained parts of hChr7 and hChr21. HCT116 8/3 c4, which proliferated at nearly the same rate as the parental line, maintained its trisomy when grown as a xenograft, and did not exhibit any further chromosomal alterations. These results indicate that growth-inhibitory trisomies in both mouse and human cells are selected against during in vivo tumor formation, while aneuploidies that are neutral can escape negative selection.
When evaluating the karyotypes of all evolved MEFs and HCT116 cell lines, we noted an intriguing difference between the copy number changes associated with aneuploid and euploid cell lines: CNAs found in cells that were initially euploid almost always encompassed whole chromosomes, while both whole-chromosome and segmental CNAs were observed in aneuploid cells (Table 1). For instance, 8/8 copy number changes in the high-passage WT + p53dd + RasV12 line affected whole chromosomes, while 5/6 copy number changes in the Ts1+p53dd + RasV12 affected chromosome segments. In total, across 30 euploid and 30 aneuploid cell lines that were sequenced after prolonged growth, only a single segmental CNA was observed in a line that was initially euploid, while 14 segmental CNAs were observed in lines that initially harbored chromosome gains. These results suggest that certain trisomies may trigger chromosome structure instability and are consistent with previous reports of defective DNA replication and repair in aneuploid cell lines (Blank et al., 2015; Passerini et al., 2016; Sheltzer et al., 2011). Although our experiments were not designed to directly measure the rates of chromosome missegregation or chromosome structure alterations, we speculate that underlying genomic instability in some aneuploid lines may speed adaptation.
Table 1.
Additional Copy Number Alterations Following the In Vitro and In Vivo Growth of Aneuploid Strains
| Experiment | Strain | Lines Sequenced |
Lines with Whole- Chromosome CNAs |
Chromosomes with Whole- Chromosome CNAs |
Lines with Segmental CNAs |
Chromosomes with Segmental CNAs |
|---|---|---|---|---|---|---|
| WT p53dd versus Ts1 p53dd | WT | 4 | 2 | 9 | 0 | 0 |
| Ts1 | 4 | 1 | 1 | 1 | 5 | |
| WT LTa versus Ts19 LTa | WT | 10 | 0 | 0 | 1 | 1 |
| Ts19 | 10 | 4 | 8 | 1 | 2 | |
| WT p53 versus Ts13 p53dd, xeno | WT | 2 | 2 | 4 | 0 | 0 |
| Ts13 | 2 | 1 | 2 | 0 | 0 | |
| WT p53dd versus Ts19 p53dd, xeno | WT | 2 | 0 | 0 | 0 | 0 |
| Ts19 | 2 | 1 | 1 | 1 | 1 | |
| HCT116 xenografts | HCT116 | 12 | 0 | 0 | 0 | 0 |
| HCT116 3/3 | 3 | 1 | 1 | 3 | 6 | |
| HCT116 5/3 | 3 | 3 | 5 | 0 | 0 | |
| HCT116 5/4 | 3 | 3 | 3 | 0 | 0 | |
| HCT116 8/3 c4 | 3 | 0 | 0 | 0 | 0 | |
| Euploid total | 30 | 4 | 13 | 1 | 1 | |
| Aneuploid total | 30 | 14 | 21 | 6 | 14 |
Finally, we set out to determine whether the karyotype changes that occurred during prolonged growth correlated with improved fitness. We tested the proliferation rates and colony-formation ability of several cell lines derived from the aneuploid and near-euploid HCT116 xenografts. Interestingly, the evolved aneuploid HCT116 cell lines exhibited proliferation and colony-formation rates that were significantly greater than the pre-xenograft cell lines and that were comparable with those of the WT line (Figures 7C–7E). These findings were true for both aneuploid cell lines that reverted to a WT karyotype (e.g., HCT116 5/3 line 1) and for a cell line that acquired additional trisomies during growth in vivo (HCT116 5/3 line 2). These results suggest that the karyotype evolution that occurs in aneuploid cells, both toward and away from a euploid state, can increase cellular fitness.
DISCUSSION
Aneuploidy is a nearly universal feature of human cancers. However, we have found that in carefully controlled experiments, cells harboring extra chromosomes display reduced tumorigenicity relative to genetically matched euploid cells. Gain of six different chromosomes tested so far (mChr1, mChr13, mChr16, mChr19, hChr3, and hChr5) impedes several measures of tumorigenic capacity, while gain of only one chromosome (hChr8) has nearly neutral consequences. Moreover, the activation of several oncogenic pathways (HRAS, BRAF, PIK3CA, and MYC) as well as the ablation of p53 and RB function are insufficient to overcome the fitness penalty induced by aneuploidy. While certain oncogene cocktails, particularly those that include LTa or RasV12, result in a partial and variable suppression of the aneuploidy-induced proliferation delay, trisomic cells transduced with multiple oncogenes exhibit consistent and severe defects in focus formation, anchorage-independent growth, and growth as xenografts. We failed to detect any conditions in which aneuploidy reproducibly triggered the transformation of primary cells or exhibited synergistic behavior with an oncogenic mutation. Instead, we found that trisomic cells underwent rapid karyotype evolution, often losing their extra chromosome but occasionally acquiring other aneuploidies as well. Although our assays were not designed to measure rates of genomic instability, both whole-chromosome and segmental CNAs were observed more frequently in aneuploid than in euploid cells, suggesting that one potential mechanism to enable the robust growth of aneuploid cells may be via evolution toward a more favorable karyotype.
While the tumor-suppressive effects of aneuploidy are unexpected, they are consistent with much of what has been learned about the effects of whole-chromosome aneuploidy on normal cell physiology. Aneuploid chromosomes are transcribed and translated proportional to their copy number (Dephoure et al., 2014; Stingele et al., 2012; Torres et al., 2007, 2010), which can lead to stoichiometric imbalances in endogenous proteins and protein complexes (Oromendia et al., 2012; Sheltzer and Amon, 2011). To compensate, cells rely on a set of protein quality control mechanisms, including the HSF1/HSP90 folding pathway (Donnelly et al., 2014; Oromendia et al., 2012), autophagy, and proteasomal degradation (Dephoure et al., 2014; McShane et al., 2016; Santaguida et al., 2015; Stingele et al., 2012; Torres et al., 2010). The energetic cost of expressing, folding, and turning over excess proteins, as well as the downstream consequences of unmitigated protein imbalances, impose a significant fitness cost on the cell. The severity of aneuploid phenotypes is generally proportional to the degree of aneuploidy, and indeed, we found that transformed cells trisomic for mouse chromosome 1 grew much more slowly than cells trisomic for mouse chromosome 19. Alternately, the number of anti-proliferative or tumor-suppressive genes on certain chromosomes may “outweigh” the number of proto-oncogenes on that chromosome, causing certain chromosome gains to result in a net inhibition of tumorigenesis.
The molecular pathway(s) that cause slow cell division and tumor suppression, in response either to trisomy or to the gain of specific anti-proliferative genes, remains an area of active investigation. We found that aneuploidy commonly induces senescence in both primary and transformed cells, as has previously been reported in cells that display chromosomal instability (Baker et al., 2004; Lentini et al., 2012). How aneuploidy induces senescence is an important question that remains to be addressed. However, our findings that HCT116 5/3 and E1a-transduced trisomic MEFs exhibit both minimal senescence and poor proliferation suggest that other factors contribute to the growth defect of aneuploid cells as well.
If aneuploidy can function as a tumor suppressor, then why is aneuploidy such a frequent occurrence in cancer? Several possibilities remain. First, our study has primarily focused upon the consequences of single-chromosome trisomies and tetrasomies. This degree of aneuploidy is frequently observed in early-stage lesions in a variety of tumor types (Balaban et al., 1986; Di Capua Sacoto et al., 2011; Lai et al., 2007; Magnani et al., 1994; El-Rifai et al., 2000). It could be the case that these low levels of aneuploidy are in fact tumor protective, while complex karyotypes or the multiple chromosome gains found in more advanced malignancies are tumor promoting. Monosomies could also have oncogenic consequences that trisomies and tetrasomies lack. We have begun to investigate the consequences of complex karyotypes generated via transient inhibition of the checkpoint kinase Mps1, although our initial experiments did not reveal evidence of cellular transformation or a fitness benefit conferred by this treatment. While further study into the phenotypic differences caused by various types of chromosome CNAs is warranted, our results suggest that the simple chromosome gains found in pre-malignant cells may not be oncogenic.
Alternately, aneuploidy could function predominantly as a tumor suppressor, yet still exhibit tumor-promoting effects under rare circumstances that involve specific cell types, chromosomes, or environments. For example, trisomy of chromosome 21 predisposes individuals to leukemia (Seewald et al., 2012), and gain of chromosome 21 is a common occurrence in sporadic leukemia (Loncarevic et al., 1999; Ozery-Flato et al., 2011), but trisomy of chromosome 21 appears to protect against the development of many other cancer types, including breast, lung, and prostate cancers (Nižetić and Groet, 2012). Similarly, a recent report suggests that, while aneuploidy is typically detrimental to cell fitness, under certain conditions, such as hypoxia or chemotherapy treatment, aneuploid cells may proliferate better than euploid cells do (Rutledge et al., 2016). Thus, while the nine aneuploid lines that we examined hinder or are neutral with regard to tumor growth in every assay that we performed, it is conceivable that a wider survey of aneuploidies, oncogenes, or growth conditions would reveal unusual cases in which aneuploidy provides a fitness advantage. Under natural selection in a tumor, rare growth-promoting aneuploidies could rise to clonal levels while a large number of growth-inhibitory aneuploidies are continually selected against. Finally, chromosome missegregation, rather than aneuploidy per se, could be a crucial driver of tumorigenesis. Lagging chromosomes can be damaged during anaphase (Janssen et al., 2011) or dramatically altered following encapsulation in a micronucleus (Crasta et al., 2012); missegregation-induced DNA damage could therefore promote transformation while any subsequent aneuploidy exists mainly as a “passenger” mutation. This mechanism could at least partially explain the observation that p53 mutations accelerate tumorigenesis in genetically engineered mouse models that lack spindle checkpoint proteins (Foijer et al., 2014; Li et al., 2010), as chromosome damage caused by faulty mitoses fails to trigger cell-cycle arrest or apoptosis in the absence of p53.
Tumor cells may also adapt to suppress certain adverse effects of aneuploidy, and our results demonstrate one potential mechanism by which this can occur. Following serial passaging or growth in vivo, aneuploid cells frequently exhibit further karyotypic alterations. Remarkably, these changes correlate with improved growth. Aneuploid cells can revert to euploidy by losing their extra chromosomes, and we have found that this is a common occurrence in vitro and in vivo when the extra chromosome(s) induces a significant growth disadvantage. Alternately, cells can acquire other chromosome copy number changes, including both chromosome gains and losses. We propose that aneuploidy + oncogene “sweet spots” exist in which the detrimental effects of aneuploidy are neutralized while a pro-proliferation phenotype is uncovered. In particular, the gain of mChr2 was found to correlate with enhanced growth in multiple independent experiments with Ts19. It may be the case that the gain of mChr2 and mChr19 in cells expressing LTa is one such sweet spot, and that these aneuploidy sweet spots represent frequently observed karyotypes in cancer. Consistent with this notion, many distinct chromosome CNAs are observed together in the same tumors more often than expected by chance (Ozery-Flato et al., 2011). For instance, tumors that have gained an extra copy of hChr7 are significantly more likely to have also gained an extra copy of hChr17, while loss of hChr7 is correlated with loss of hChr17 (Ozery-Flato et al., 2011). Such changes could potentially function to maintain stoichiometry in key protein complexes while also promoting tumorigenic growth through as yet undiscovered pathways. Moreover, we found that during prolonged growth, aneuploid cells acquired both segmental and whole-chromosome CNAs, while euploid cells tended to exhibit only whole-chromosome changes. Previous studies have reported that aneuploid cells display elevated rates of mitotic recombination and double-strand break formation (Blank et al., 2015; Sheltzer et al., 2011), which may result from the decreased expression of key replication proteins in these strains (Passerini et al., 2016). Thus, the inherent plasticity of aneuploid genomes that we have uncovered could drive the eventual development of cancers that harbor certain favorable karyotypic combinations.
EXPERIMENTAL PROCEDURES
A Robertsonian breeding scheme was utilized to generate sibling-matched euploid and trisomic MEFs as described in Williams et al. (2008). All animal studies and procedures were approved by the MIT Institutional Animal Care and Use Committee. Aneuploid cell lines derived from HCT116 cells were previously described in Stingele et al. (2012). Further experimental details are provided in the Supplemental Information.
Supplementary Material
Highlights.
Single-chromosome gains inhibit anchorage-independent growth and xenograft formation
P53 loss and Ras activation fail to normalize the fitness of trisomic cells
Trisomic cells acquire additional karyotype changes during prolonged growth
Trisomic cells can evolve over time to a higher-fitness state
Significance.
The prevalence of chromosome segregation errors in cancer was first noted more than 100 years ago and is commonly interpreted to imply that aneuploidy plays a crucial role in tumor development. Our current results complicate this hypothesis. We find that aneuploidy exerts a significant fitness penalty on the cell, and the introduction of oncogenic mutations into aneuploid cells is not sufficient to fully overcome its cost. Instead, aneuploid cells evolve over time, and occasionally give rise to rapidly proliferating descendants. Thus, some aneuploidies may directly suppress tumor growth while also enhancing the long-term evolutionary capacity of pre-malignant cells.
Acknowledgments
We thank the Koch Institute Flow Cytometry facility for assistance with cell sorting. We thank the MIT BioMicro Center for performing sequencing reactions, and Charlie Whittaker and Jie Wu in the Koch Institute Bioinformatics Core for assistance with data analysis (NIH grant P30-CA14051). This work was supported by the National Institute of Health GM056800 to A.A and the Kathy and the Curt Marble Cancer Research Fund. J.S. was supported by a Whitaker Health Sciences Fund Fellowship and an MIT School of Science Fellowship in Cancer Research. A.A. is an investigator of the Howard Hughes Medical Institute and the Glenn Foundation for Biomedical Research.
Footnotes
ACCESSION NUMBERS
Sequence reads from comparative genomic hybridization sequencing experiments were deposited under SRA project number SRP094642.
Supplemental Information includes Supplemental Experimental Procedures, eight figures, and two tables and can be found with this article online at http://dx.doi.org/10.1016/j.ccell.2016.12.004.
AUTHOR CONTRIBUTIONS
Conceptualization, J.M.S. and A.A. Investigation, J.M.S., J.H.K., J.M.R., N.C.H.B., E.S.C., C.M.M., and N.M.S. Resources, V.P. and Z.S. Writing, J.M.S. J.H.K. and J.M.R. contributed equally to this article.
References
- Ahuja D, Sáenz-Robles MT, Pipas JM. SV40 large T antigen targets multiple cellular pathways to elicit cellular transformation. Oncogene. 2005;24:7729–7745. doi: 10.1038/sj.onc.1209046. [DOI] [PubMed] [Google Scholar]
- Baker DJ, Jeganathan KB, Cameron JD, Thompson M, Juneja S, Kopecka A, Kumar R, Jenkins RB, de Groen PC, Roche P, et al. BubR1 insufficiency causes early onset of aging-associated phenotypes and infertility in mice. Nat. Genet. 2004;36:744–749. doi: 10.1038/ng1382. [DOI] [PubMed] [Google Scholar]
- Balaban GB, Herlyn M, Clark WH, Jr, Nowell PC. Karyotypic evolution in human malignant melanoma. Cancer Genet. Cytogenet. 1986;19:113–122. doi: 10.1016/0165-4608(86)90378-x. [DOI] [PubMed] [Google Scholar]
- Blank HM, Sheltzer JM, Meehl CM, Amon A. Mitotic entry in the presence of DNA damage is a widespread property of aneuploidy in yeast. Mol. Biol. Cell. 2015;26:1440–1451. doi: 10.1091/mbc.E14-10-1442. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Boveri T. Concerning the origin of malignant tumours by Theodor Boveri. Harris Henry., translator. J. Cell Sci. 2008;121:1–84. doi: 10.1242/jcs.025742. and annotated. [DOI] [PubMed] [Google Scholar]
- Di Capua Sacoto C, Budia Alba A, Alapont Alacreu JM, Ruiz Cerda JL, Jimenez Cruz JF. In vivo aneuploidization during the expansion of renal adenocarcinoma. Urol. Int. 2011;86:466–469. doi: 10.1159/000324101. [DOI] [PubMed] [Google Scholar]
- Crasta K, Ganem NJ, Dagher R, Lantermann AB, Ivanova EV, Pan Y, Nezi L, Protopopov A, Chowdhury D, Pellman D. DNA breaks and chromosome pulverization from errors in mitosis. Nature. 2012;482:53–58. doi: 10.1038/nature10802. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Davoli T, Xu AW, Mengwasser KE, Sack LM, Yoon JC, Park PJ, Elledge SJ. Cumulative haploinsufficiency and triplosensitivity drive aneuploidy patterns to shape the cancer genome. Cell. 2013;155:948–962. doi: 10.1016/j.cell.2013.10.011. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Dephoure N, Hwang S, O’Sullivan C, Dodgson SE, Gygi SP, Amon A, Torres EM. Quantitative proteomic analysis reveals posttranslational responses to aneuploidy in yeast. Elife. 2014;3:e03023. doi: 10.7554/eLife.03023. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Donnelly N, Passerini V, Dürrbaum M, Stingele S, Storchová Z. HSF1 deficiency and impaired HSP90-dependent protein folding are hallmarks of aneuploid human cells. EMBO J. 2014;33:2374–2387. doi: 10.15252/embj.201488648. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Duesberg P, Rasnick D, Li R, Winters L, Rausch C, Hehlmann R. How aneuploidy may cause cancer and genetic instability. Anticancer Res. 1999;19:4887–4906. [PubMed] [Google Scholar]
- Dürrbaum M, Kuznetsova AY, Passerini V, Stingele S, Stoehr G, Storchová Z. Unique features of the transcriptional response to model aneuploidy in human cells. BMC Genomics. 2014;15:139. doi: 10.1186/1471-2164-15-139. [DOI] [PMC free article] [PubMed] [Google Scholar]
- El-Rifai W, Sarlomo-Rikala M, Andersson LC, Knuutila S, Miettinen M. DNA sequence copy number changes in gastrointestinal stromal tumors: tumor progression and prognostic significance. Cancer Res. 2000;60:3899–3903. [PubMed] [Google Scholar]
- Foijer F, Xie SZ, Simon JE, Bakker PL, Conte N, Davis SH, Kregel E, Jonkers J, Bradley A, Sorger PK. Chromosome instability induced by Mps1 and p53 mutation generates aggressive lymphomas exhibiting aneuploidy-induced stress. Proc. Natl. Acad. Sci. USA. 2014;111:13427–13432. doi: 10.1073/pnas.1400892111. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gallimore PH, Turnell AS. Adenovirus E1A: remodelling the host cell, a life or death experience. Oncogene. 2001;20:7824–7835. doi: 10.1038/sj.onc.1204913. [DOI] [PubMed] [Google Scholar]
- Gordon DJ, Resio B, Pellman D. Causes and consequences of aneuploidy in cancer. Nat. Rev. Genet. 2012;13:189–203. doi: 10.1038/nrg3123. [DOI] [PubMed] [Google Scholar]
- Hewitt L, Tighe A, Santaguida S, White AM, Jones CD, Musacchio A, Green S, Taylor SS. Sustained Mps1 activity is required in mitosis to recruit O-Mad2 to the Mad1–C-Mad2 core complex. J. Cell Biol. 2010;190:25–34. doi: 10.1083/jcb.201002133. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Holland AJ, Cleveland DW. Boveri revisited: chromosomal instability, aneuploidy and tumorigenesis. Nat. Rev. Mol. Cell Biol. 2009;10:478–487. doi: 10.1038/nrm2718. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Janssen A, van der Burg M, Szuhai K, Kops GJPL, Medema RH. Chromosome segregation errors as a cause of DNA damage and structural chromosome aberrations. Science. 2011;333:1895–1898. doi: 10.1126/science.1210214. [DOI] [PubMed] [Google Scholar]
- Lai LA, Paulson TG, Li X, Sanchez CA, Maley C, Odze RD, Reid BJ, Rabinovitch PS. Increasing genomic instability during premalignant neoplastic progression revealed through high resolution array-CGH. Genes. Chromosomes Cancer. 2007;46:532–542. doi: 10.1002/gcc.20435. [DOI] [PubMed] [Google Scholar]
- Land H, Parada LF, Weinberg RA. Tumorigenic conversion of primary embryo fibroblasts requires at least two cooperating oncogenes. Nature. 1983;304:596–602. doi: 10.1038/304596a0. [DOI] [PubMed] [Google Scholar]
- Lentini L, Barra V, Schillaci T, Di Leonardo A. MAD2 depletion triggers premature cellular senescence in human primary fibroblasts by activating a p53 pathway preventing aneuploid cells propagation. J. Cell Physiol. 2012;227:3324–3332. doi: 10.1002/jcp.24030. [DOI] [PubMed] [Google Scholar]
- Li M, Fang X, Wei Z, York JP, Zhang P. Loss of spindle assembly checkpoint-mediated inhibition of Cdc20 promotes tumorigenesis in mice. J. Cell Biol. 2009;185:983–994. doi: 10.1083/jcb.200904020. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Li M, Fang X, Baker DJ, Guo L, Gao X, Wei Z, Han S, van Deursen JM, Zhang P. The ATM-p53 pathway suppresses aneuploidy-induced tumorigenesis. Proc. Natl. Acad. Sci. USA. 2010;107:14188–14193. doi: 10.1073/pnas.1005960107. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Loncarevic IF, Roitzheim B, Ritterbach J, Viehmann S, Borkhardt A, Lampert F, Harbott J. Trisomy 21 is a recurrent secondary aberration in childhood acute lymphoblastic leukemia with TEL/AML1 gene fusion. Genes. Chromosomes Cancer. 1999;24:272–277. [PubMed] [Google Scholar]
- Magnani I, Guerneri S, Pollo B, Cirenei N, Colombo BM, Broggi G, Galli C, Bugiani O, DiDonato S, Finocchiaro G, et al. Increasing complexity of the karyotype in 50 human gliomas: progressive evolution and de novo occurrence of cytogenetic alterations. Cancer Genet. Cytogenet. 1994;75:77–89. doi: 10.1016/0165-4608(94)90157-0. [DOI] [PubMed] [Google Scholar]
- McShane E, Sin C, Zauber H, Wells JN, Donnelly N, Wang X, Hou J, Chen W, Storchova Z, Marsh JA, et al. Kinetic analysis of protein stability reveals age-dependent degradation. Cell. 2016;167:803–815. e21. doi: 10.1016/j.cell.2016.09.015. [DOI] [PubMed] [Google Scholar]
- Michel LS, Liberal V, Chatterjee A, Kirchwegger R, Pasche B, Gerald W, Dobles M, Sorger PK, Murty VVVS, Benezra R. MAD2 haplo-insufficiency causes premature anaphase and chromosome instability in mammalian cells. Nature. 2001;409:355–359. doi: 10.1038/35053094. [DOI] [PubMed] [Google Scholar]
- Nižetić D, Groet J. Tumorigenesis in Down’s syndrome: big lessons from a small chromosome. Nat. Rev. Cancer. 2012;12:721–732. doi: 10.1038/nrc3355. [DOI] [PubMed] [Google Scholar]
- Oromendia AB, Dodgson SE, Amon A. Aneuploidy causes proteotoxic stress in yeast. Genes Dev. 2012;26:2696–2708. doi: 10.1101/gad.207407.112. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ozery-Flato M, Linhart C, Trakhtenbrot L, Izraeli S, Shamir R. Large-scale analysis of chromosomal aberrations in cancer karyotypes reveals two distinct paths to aneuploidy. Genome Biol. 2011;12:R61. doi: 10.1186/gb-2011-12-6-r61. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Park I, Lee H, Choi E, Lee Y-K, Kwon M-S, Min J, Park P-G, Lee S, Kong Y-Y, Gong G, et al. Loss of BubR1 acetylation causes defects in spindle assembly checkpoint signaling and promotes tumor formation. J. Cell Biol. 2013;202:295–309. doi: 10.1083/jcb.201210099. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Passerini V, Ozeri-Galai E, de Pagter MS, Donnelly N, Schmalbrock S, Kloosterman WP, Kerem B, Storchová Z. The presence of extra chromosomes leads to genomic instability. Nat. Commun. 2016;7:10754. doi: 10.1038/ncomms10754. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Pavelka N, Rancati G, Zhu J, Bradford WD, Saraf A, Florens L, Sanderson BW, Hattem GL, Li R. Aneuploidy confers quantitative proteome changes and phenotypic variation in budding yeast. Nature. 2010;468:321–325. doi: 10.1038/nature09529. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Rasnick D, Duesberg PH. How aneuploidy affects metabolic control and causes cancer. Biochem. J. 1999;340:621–630. [PMC free article] [PubMed] [Google Scholar]
- Rutledge SD, Douglas TA, Nicholson JM, Vila-Casadesús M, Kantzler CL, Wangsa D, Barroso-Vilares M, Kale SD, Logarinho E, Cimini D. Selective advantage of trisomic human cells cultured in non-standard conditions. Sci. Rep. 2016;6:22828. doi: 10.1038/srep22828. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Santaguida S, Tighe A, D’Alise AM, Taylor SS, Musacchio A. Dissecting the role of MPS1 in chromosome biorientation and the spindle checkpoint through the small molecule inhibitor reversine. J. Cell Biol. 2010;190:73–87. doi: 10.1083/jcb.201001036. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Santaguida S, Vasile E, White E, Amon A. Aneuploidy-induced cellular stresses limit autophagic degradation. Genes Dev. 2015;29:2010–2021. doi: 10.1101/gad.269118.115. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Seewald L, Taub JW, Maloney KW, McCabe ERB. Acute leukemias in children with Down syndrome. Mol. Genet. Metab. 2012;107:25–30. doi: 10.1016/j.ymgme.2012.07.011. [DOI] [PubMed] [Google Scholar]
- Shaulian E, Zauberman A, Ginsberg D, Oren M. Identification of a minimal transforming domain of p53: negative dominance through abrogation of sequence-specific DNA binding. Mol. Cell. Biol. 1992;12:5581–5592. doi: 10.1128/mcb.12.12.5581. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sheltzer JM. A transcriptional and metabolic signature of primary aneuploidy is present in chromosomally unstable cancer cells and informs clinical prognosis. Cancer Res. 2013;73:6401–6412. doi: 10.1158/0008-5472.CAN-13-0749. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sheltzer JM, Amon A. The aneuploidy paradox: costs and benefits of an incorrect karyotype. Trends Genet. 2011;27:446–453. doi: 10.1016/j.tig.2011.07.003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sheltzer JM, Blank HM, Pfau SJ, Tange Y, George BM, Humpton TJ, Brito IL, Hiraoka Y, Niwa O, Amon A. Aneuploidy drives genomic instability in yeast. Science. 2011;333:1026–1030. doi: 10.1126/science.1206412. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sheltzer JM, Torres EM, Dunham MJ, Amon A. Transcriptional consequences of aneuploidy. Proc. Natl. Acad. Sci. USA. 2012;109:12644–12649. doi: 10.1073/pnas.1209227109. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Shin SI, Freedman VH, Risser R, Pollack R. Tumorigenicity of virus-transformed cells in nude mice is correlated specifically with anchorage independent growth in vitro. Proc. Natl. Acad. Sci. USA. 1975;72:4435–4439. doi: 10.1073/pnas.72.11.4435. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Silk AD, Zasadil LM, Holland AJ, Vitre B, Cleveland DW, Weaver BA. Chromosome missegregation rate predicts whether aneuploidy will promote or suppress tumors. Proc. Natl. Acad. Sci. USA. 2013;110:E4134–E4141. doi: 10.1073/pnas.1317042110. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sotillo R, Hernando E, Díaz-Rodríguez E, Teruya-Feldstein J, Cordón-Cardo C, Lowe SW, Benezra R. Mad2 overexpression promotes aneuploidy and tumorigenesis in mice. Cancer Cell. 2007;11:9–23. doi: 10.1016/j.ccr.2006.10.019. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sotillo R, Schvartzman J-M, Socci ND, Benezra R. Mad2-induced chromosome instability leads to lung tumour relapse after oncogene withdrawal. Nature. 2010;464:436–440. doi: 10.1038/nature08803. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Stingele S, Stoehr G, Peplowska K, Cox J, Mann M, Storchova Z. Global analysis of genome, transcriptome and proteome reveals the response to aneuploidy in human cells. Mol. Syst. Biol. 2012;8:608. doi: 10.1038/msb.2012.40. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tang Y-C, Williams BR, Siegel JJ, Amon A. Identification of aneuploidy-selective antiproliferation compounds. Cell. 2011;144:499–512. doi: 10.1016/j.cell.2011.01.017. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Thompson SL, Compton DA. Proliferation of aneuploid human cells is limited by a p53-dependent mechanism. J. Cell Biol. 2010;188:369–381. doi: 10.1083/jcb.200905057. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Torres EM, Sokolsky T, Tucker CM, Chan LY, Boselli M, Dunham MJ, Amon A. Effects of aneuploidy on cellular physiology and cell division in haploid yeast. Science. 2007;317:916–924. doi: 10.1126/science.1142210. [DOI] [PubMed] [Google Scholar]
- Torres EM, Dephoure N, Panneerselvam A, Tucker CM, Whittaker CA, Gygi SP, Dunham MJ, Amon A. Identification of aneuploidy-tolerating mutations. Cell. 2010;143:71–78. doi: 10.1016/j.cell.2010.08.038. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Weaver BA, Cleveland DW. Does aneuploidy cause cancer? Curr. Opin. Cell Biol. 2006;18:658–667. doi: 10.1016/j.ceb.2006.10.002. [DOI] [PubMed] [Google Scholar]
- Weaver BAA, Silk AD, Montagna C, Verdier-Pinard P, Cleveland DW. Aneuploidy acts both oncogenically and as a tumor suppressor. Cancer Cell. 2007;11:25–36. doi: 10.1016/j.ccr.2006.12.003. [DOI] [PubMed] [Google Scholar]
- Williams BR, Prabhu VR, Hunter KE, Glazier CM, Whittaker CA, Housman DE, Amon A. Aneuploidy affects proliferation and spontaneous immortalization in mammalian cells. Science. 2008;322:703–709. doi: 10.1126/science.1160058. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zack TI, Schumacher SE, Carter SL, Cherniack AD, Saksena G, Tabak B, Lawrence MS, Zhang C-Z, Wala J, Mermel CH, et al. Pan-cancer patterns of somatic copy number alteration. Nat. Genet. 2013;45:1134–1140. doi: 10.1038/ng.2760. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.