Abstract
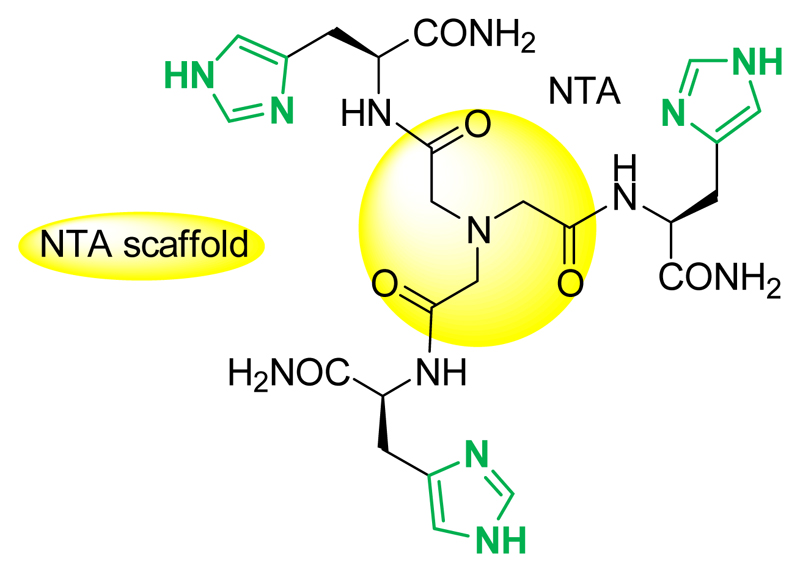
The pseudopeptide L, derived from a nitrilotriacetic acid scaffold and functionalized with three histidine moieties, is reminiscent of the amino acid side chains encountered in the Alzheimer’s peptide (Aβ). Its synthesis and coordination properties for Cui and Cuii are described. L efficiently complex Cuii in a square-planar geometry involving three imidazole nitrogen atoms and an amidate-Cu bond. By contrast, Cui is coordinated in a tetrahedral environment. The redox behavior is irreversible and follows an ECEC mechanism in accordance with the very different environments of the two redox states of the Cu center. This is in line with the observed resistance of the Cui complex to oxidation by oxygen and the Cuii complex reduction by ascorbate. The affinities of L for Cuii and Cui at physiological pH are larger than that reported for the Aβ peptide. Therefore, due to its peculiar Cu coordination properties, the ligand L is able to target both redox states of Cu, redox silence them and prevent reactive oxygen species production by the CuAβ complex. Because reactive oxygen species contribute to the oxidative stress, a key issue in Alzheimer’s disease, this ligand thus represents a new strategy in the long route of finding molecular concepts for fighting Alzheimer’s disease.
Keywords: Bioinorganic Chemistry, peptides, copper, Ligand design, Alzheimer’s disease
Introduction
Copper enzymes play crucial role in biology, where copper ions exert structural and/or catalytic function(s).[1] However the copper ions can also be detrimental as well illustrated by two genetic disorders, namely Menkes and Wilson’s disease, linked to a dysfunction of Cu homeostasis, which induces a depletion of Cu and an overload of Cu, respectively.[2] In Alzheimer’s disease (AD), copper ions have been proposed to play a harmful role as well (for recent reviews, see refs.[3]). Indeed, high level of copper ions is found in the senile plaques, one of the neuropathological hallmark of AD brains.[4] Due to its redox ability, copper is involved in the production of Reactive Oxygen Species (ROS) taking part in the oxidative stress linked to the etiology of the disease.[3b, 5] It has indeed been shown that when bound to the amyloid-β (Aβ) peptide, the amyloidogenic peptide encountered in AD, copper ions are able to cycle between the +I and +II oxidation states.[6] The resulting CuAβ complex can catalyze the formation of ROS in the presence of dioxygen and of a physiological reductant such as ascorbate,[7] via a complex mechanism.[8] While AD is a multi-factorial disease, involving many biological actors and complex interactions between them (Aβ and Tau protein, secretases responsible of the production of the (non-)amyloidogenic forms of the Aβ, acetylcholine esterase….),[9] CuAβ associated ROS over-production is recognized as a key event.[10] Copper ions thus remain a pertinent target for a therapeutic approach,[3d, 5a, 10a, 11] although the first clinical trials along this approach failed to benefit patients.[3d, 12] Targeting Cu ions requires well-defined coordination based approaches and ligand design that is, in general, difficult to include in multi-targeted drug in a first-line strategy.[9a] Hence, we designed the chelating moiety of a drug candidate that could be further implemented towards a multi-targeted purpose.
Up to now, most of the synthetic ligands designed to remove copper ions from Aβ have targeted Cuii (see refs.[3d, 9a, 11, 13] for recent reviews), including with peptide-based ligands.[14] Only few studies have focused on Cui synthetic chelators,[15] based on previous results using naturally occurring Cui proteins.[16] The reason for such a preference is not clear and has no real biological basis since the extracellular space of brain where the senile plaques are observed mainly represents a reducing environment.[17]
In the present study, another, more “pragmatic” approach is pursued: the ligand L (Scheme 1) was designed such as being able to remove either Cuii or Cui from Aβ. Indeed, in contrast to other diseases for which the redox state of the targeted copper is quite well-defined – Cuii for blood circulating Cu or Cui for the intracellular hepatic pool in Wilson's disease for instance[2a] – for neurodegenerative diseases, the situation is less clear although Cuii has been considered as the therapeutic target of choice. The synaptic cleft where the CuAβ interaction occurs[18] is an ill-defined space[19] with a unknown redox potential. In addition, the redox potential might be subject to spacio and/or temporal changes. It has for instance been shown that the level of Aβ can induce a reductive shift of the extracellular potential.[20] In addition, although the redox cycle between Cui and Cuii is the pre-requisite for ROS production implying that both redox states may be present, one redox state may be largely predominant and it is not known yet which one it is. Thus targeting either Cuii or Cui might be a uncertain approach ; chelators are, in general, specific for one given redox state,[3d, 9a, 11, 13, 15a] because Cuii and Cui coordination requirements are very different[21] (distorted square-planar, with N/O ligands for Cuii and tetrahedral with N,S ligands for Cui). Hence, the risk is to miss the good target since it has for instance been shown that although being able to remove Cuii from Aβ and being redox inert, tetraazamacrocycle ligands failed in stopping CuAβ induced ROS production.[22] Therefore, as the redox state of the Cu center in the brain regions has not been identified yet, targeting both Cu oxidation states may be considered as the safest approach.
Scheme 1.
NTA(HisNH2)3, noted L. The protonation state and charge of the ligand is given only when necessary.
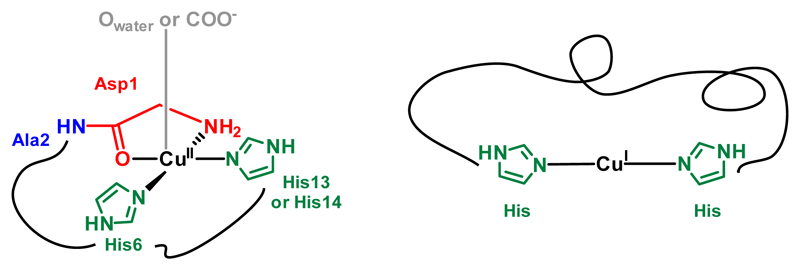
The coordination sites of Cui and Cuii to Aβ near physiological pH are reminded in Scheme 2. The Cui center lies in a digonal environment made by two imidazole groups from the Histidine (His) residues at position 6, 13 and 14, with no strong preference for one His couple among the three possible ones.[3b, 23] In the form mainly present at physiological pH, the Cuii center is surrounded by the N-terminal amine, the adjacent carbonyl group from the peptide backbone, and two imidazole rings from the His in a square-planar geometry.[3b, 24] To achieve removal of Cui or Cuii from Aβ, the ligand was based on functional group reminiscent of the amino-acid side-chains encountered in the Aβ peptide, i.e. three His.
Scheme 2.
Main coordination sites of Cuii and Cui in Aβ at physiological pH.
To benefit from preorganization of metal-binding groups, a tripodal pseudopeptide based on a chemical scaffold was chosen to introduce the three His moieties. The nitrilotriacetic (NTA) scaffold is a perfect candidate to anchor three amino acids that can be grafted to the three acidic functions with peptide amide bonds. Moreover, bioinspired pseudopeptides built up on the NTA template and extended with three sulfur amino acids such as cysteines[25] d-penicillamine[26] or methionine,[27] have proven their capacity to tightly bind Cui in trigonal planar coordination sites, CuiS3, with three sulfur donors in the first sphere. Therefore the novel ligand L (Scheme 1), based on a similar design with His is expected to make the N-donors of the three amino acids converge to the metal center. Regarding Cui, the preorganization of the chelating site due to the central amine anchor is expected to increase its affinity compared to that of Aβ by entropic effect. The entropic contribution may also help the ligand having a higher affinity for Cuii compared to Aβ, although changes in the nature of the coordinating moieties are expected.
In the present paper, the structural, thermodynamic and redox characterizations of the Cuii and Cui complexes with L are described. The ability of L to remove both Cuii and Cui ions from Aβ1-16 at physiological pH is probed by Electronic Paramagnetic Resonance (EPR) and X-ray Absorption Near-Edge Structure (XANES) spectroscopies, respectively. Last, the ability of L to abolish the ROS produced by the CuAβ complex is also demonstrated, which makes the ligand L a good candidate for further therapeutic purposes.
Results and Discussion
Synthesis and characterization of L
Synthesis
The histidine moiety to be grafted to the nitrilotriacetic acid scaffold may be chosen among the free acid[28] or C-terminus protected His derivatives. The latter are preferred here over the free carboxylate, which can coordinate to metal ions and have been demonstrated to induce harmful electrostatic repulsions for the formation of the preorganized mononuclear metal complexes with cysteine tripods.[25a] Therefore, the histidine moiety was chosen as the C-terminus primary amide derivative of His (H-His-NH2), which is neutral, highly soluble in water and stable in physiological conditions.
The synthetic procedure of the ligand L (Scheme S1) is similar to previously published procedures for thioether-based pseudopeptides.[27] The coupling reaction of the activated ester NTA(NHS)3 with commercially available H-His-NH2·2HCl was carried out in presence of DIEA as a base, leading to the desired pure compound in 11 % yield after RP-HPLC purification.
Protonation of the ligand
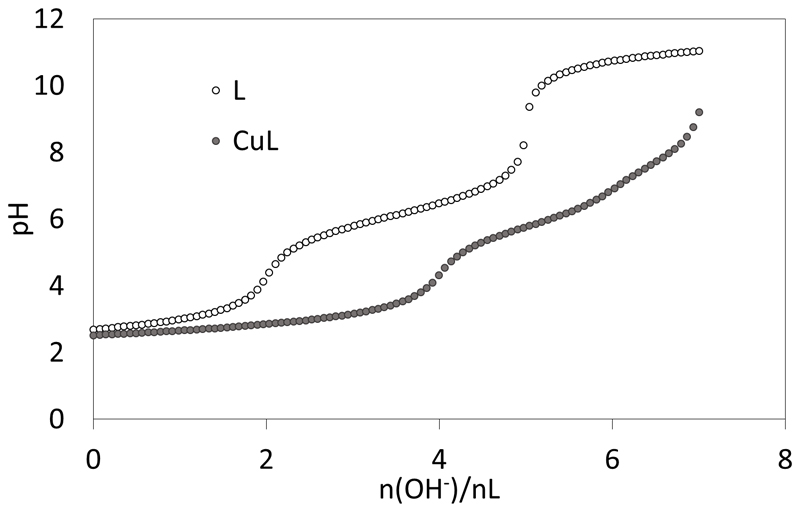
The potentiometric studies (Figure 1) have been performed in KCl 0.1 M at 298 K. The protonation constants of L could be obtained from the titrations of the free ligand with KOH and HCl and are listed in Table 1.
Figure 1.
Alkalimetric titrations of solutions containing 10-3 M L.4TFA + 10-3 M HCl with 0 and 1 equiv. of CuSO4 in water KCl 0.1 M at 298 K.
Table 1.
Protonation and complexation constants from potentiometric measurements in water KCl 0.1 M at 298 K.[a] The numbers m, l and h represent the numbers of metal, ligand and proton in the species, respectively.
| m | l | h | logβmlh | pKa | |
|---|---|---|---|---|---|
| LH+ | 0 | 1 | 1 | 6.75(3) | 6.75(3), LH ⇄ L + H |
| LH22+ | 0 | 1 | 2 | 12.87(3) | 6.12(6), LH2 ⇄ LH + H |
| LH33+ | 0 | 1 | 3 | 18.42(7) | 5.5(1), LH3 ⇄ LH2 + H |
| CuLH3+ | 1 | 1 | 1 | 16.65(5) | 5.5(1), CuLH ⇄ CuL + H |
| CuL2+ | 1 | 1 | 0 | 11.15(5) | |
| CuLH-1+ | 1 | 1 | -1 | 5.1(1) | 6.0(1), CuL ⇄ CuLH-1 + H |
| CuLH-2 | 1 | 1 | -2 | -2.6(2) | 7.7(3), CuLH-1 ⇄ CuLH-2 + H |
The figures in brackets correspond to the standard deviations of the last figure in three independent titrations.
The titration shown in Figure 1 is indicative of three protonation sites with pKa values characteristic of His nitrogen protonations.[29] The difference between two successive pKa values is expected to be 0.48 (= log3) if the three His were non interacting and therefore independently deprotonated.[30] Here these differences are 0.62 (pKa2-pKa1) and 0.63 (pKa3-pKa2) and are significantly larger than 0.48, due to the charge repulsion between the protonated arms, which is expected considering their proximity in the tripodal architecture of ligand L. The apical nitrogen protonation is not detected because the corresponding pKa value is too low, in accordance with previous studies with other pseudopeptides (pKa ≤ 2.8)[25a] or with the tripodal amide ligand N(CH2CONH2)3 (pKa = 2.6).[31] The three pKa values of the imidazole nitrogen atoms (pKa = 5.5 – 6.75) of the grafted histidine side-chains are consistent with values reported for trishistidyl peptides (pKa = 5.4 – 6.9).[32]
Cuii complexes
ESI-MS
The formation of the Cuii complex is clearly detected by electrospray-ionization mass spectrometry (ESI-MS). The spectrum recorded in the positive mode with 1 Cuii equiv. is shown in Figure S1. The complex CuiiL is the unique species detected as the monocation [L+Cuii-H]+ (m/z = 661.1) and its sodium and potassium adducts, with only traces of the free ligand.
Potentiometric titrations
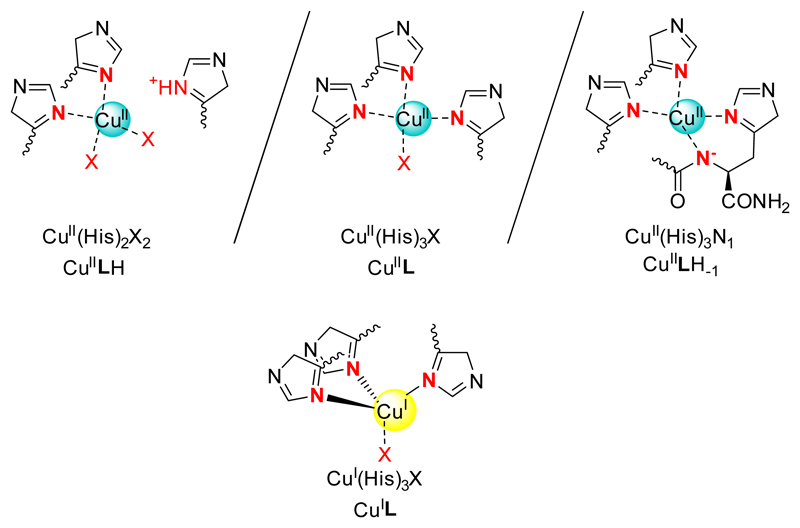
Potentiometric titrations also evidence the formation of Cuii complexes. A typical titration curve is shown in Figure 1. Titrations with 0.5 and 1 equiv. of Cuii could be fitted over the pH range 2.5-8 according to the formation of CuL with various protonation states: CuLH, CuL, CuLH-1 and CuLH-2. Titrations performed in excess of Cuii showed some precipitation and therefore they were not included in the fitting process. The corresponding stability constants are given in Table 1. The comparison of the stability constants with literature data,[32] in particular those obtained with polyhistidine peptides leads us to propose the structures drawn in Scheme 3 for CuLH, CuL and CuLH-1.
Scheme 3.
Proposed coordination in the Cuii and Cui complexes. X is the solvent or an O atom of an amide function of the ligand.
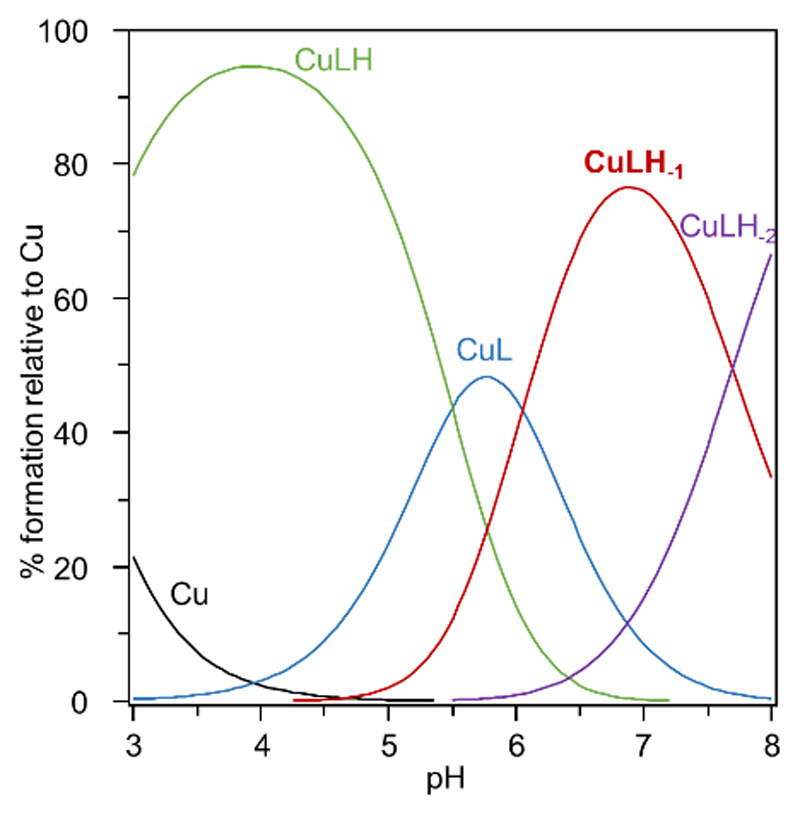
The stability constant calculated for the CuL complex (logβ11 = 11.15) is significantly larger than values reported for short peptides with three His: Ac-HxHxH-NH2 or Ac-GHHPHG-NH2 (logβ11 = 6.7-8.4),[32a, b] which evidences the greater preorganization of the tripodal pseudopeptide L in comparison to linear peptide sequences. The affinity of L for Cuii is similar to that reported for a cyclodecapeptide bearing three His residues (logβ11 = 11.44).[32c] This type of peptide scaffold is indeed known to be preorganized in a β–sheet structure that orients the side-chains of several amino acids in the same half-space for metal coordination.[33] The speciation in an equimolar solution of the ligand L and Cuii is presented in Figure 2 and shows that the CuL complex protonates below pH 6 to give the CuLH species with only two histidine coordinated to the metal center. The pKa value of CuLH is 5.5 and is similar to literature data.[32a, b] Interestingly the major species at physiological pH is CuLH-1 with a pKa value of the CuL complex of 6.0 in accordance with amide deprotonation to afford a metallacycle involving an amidate and the proximal imidazole nitrogen atom. The coordination sites of Cuii bound to L are summarized in Scheme 3 and are consistent with the potentiometric experiments and previously quoted literature data.
Figure 2.
Speciation diagram of a solution containing 1 mM L and Cuii. The stability constants tabulated in Table 1 were used to generate this diagram with the speciation program Hyss.[34]
Electron Paramagnetic Resonance
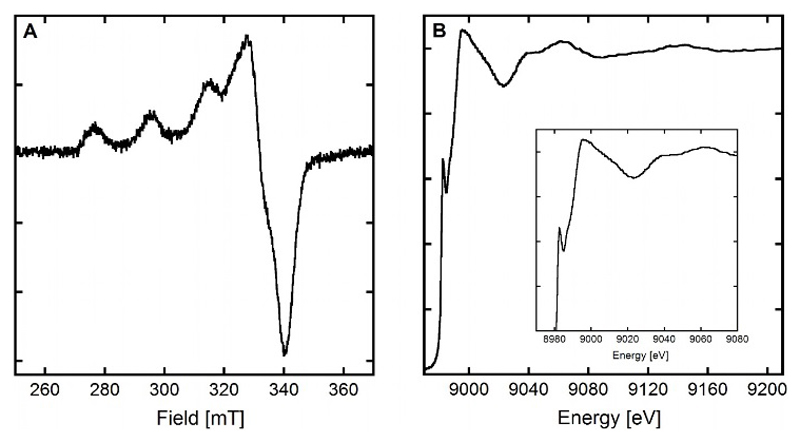
The EPR spectrum of the CuLH-1 species shown in Figure 3, panel A indicates that the Cuii center lies in a distorted square-planar geometry with an elongated Jahn-Teller effect.
Figure 3.
Panel A. EPR spectrum of CuiiLH-1, [L] = [65Cuii = 200 µM, [HEPES] = 50 mM, pH 7.1. 10% of glycerol was used as cryoprotectant. T = 110 K. Panel B. Normalized XANES spectrum of CuiL ; [L] = 1.0 mM, [Cuii] = 0.95 mM, [dithionite] = 10 mM, [HEPES] = 100 mM, pH 7.1. 10% of glycerol was used as cryoprotectant. T = 20 K.
The EPR parameters deduced from the spectrum (g// = 2.23, A//(65Cu) = 198 ± 5 10-4cm-1) best fit with a [2N2O] equatorial environment of the Cuii according to the Peisach and Blumberg correlation.[35] However divergences from this phenomenological correlation have previously been observed for imidazole containing ligands,[35–36] especially with constrained geometry.[36a] Such effect may be linked to an orientation of the imidazole rings that doesn’t permit the best delocalization of the unpaired density on the Cu center, leading to a weaker covalent character of the N-Cu bond (thus resembling a “O-Cu” bond). In the present case where a [4N] environment is proposed, this agrees well with the pre-organized and constrained Cuii environment when bound to LH-1 (Scheme 3).
The EPR spectra of the Cuii complexes recorded as a function of pH are shown in Figure S2. For the CuLH species, a g// = 2.25 and a A//(65Cu) = 202 ± 5 10-4cm-1 values are deduced in line with a [2N2O] Cuii environment according to the Peisach and Blumberg correlation.[35] The CuL species is not predominant enough (See the speciation diagram in Figure 2) to evaluate undoubtedly its EPR parameters. The close values observed for CuLH and CuLH-1 complexes mirror the flexibility of the ligand when only two His are bound to the metal center and the rigid structure imposed by the same ligand when the three His and a deprotonated amide are folded around the Cuii. In the former case, the imidazole rings may be positioned such as to maximize the covalent character of the Cu-N bond while in the latter case, this is not possible anymore.
Cui complex
ESI-MS
The ESI-MS spectrum of L with 1 Cui equiv. (Figure S3 in the SI) shows the formation of the CuiL complex as a unique metal species. The detailed observation of the isotopic pattern of the CuiL complex reveals the presence of a small amount of the CuiiL complex which may be formed when the solution is taken out the glovebox to perform the experiment. This is in accordance with the ability of this trishistidine ligand to also efficiently chelate Cuii as shown above.
Cu K-edge X-ray Absorption Spectroscopy (XAS)
The XANES spectrum of the CuiL complex is shown in Figure 3, panel B. It is characteristic of a Cui species and the intensity of the 1s → 4p transition (approx. 0.65) agrees with a tri- or tetra-coordination of the Cui center.[1, 37] Extended X-Ray Absorption Fine Structure data (Figure S4 and Table 2) are consistent with a coordination of Cui by the nitrogen atoms of the three histidine residues with Cu-N distances of 2.05 Å together with an extra O atom from the solvent/buffer at 1.92 Å, as proposed in Scheme 3. The relatively weak Debye-Waller values (< 0.003 Å2) are consistent with a relatively low structural disorder.
Table 2.
First coordination shell structural data obtained from R space fits of EXAFS spectra: N is the number of neighbors, R is the absorber-neighbor distance, σ is the Debye-Waller factor.
| Scattered-backscattered | N (± 20%) |
R (Å) (± 0.02 Å) |
σ2 (Å2) (± 0.0005 Å2) |
R factor (%)[a] |
|
|---|---|---|---|---|---|
| CuiL | Cu-N | 2.57 | 2.05 | 0.0021 | 0.70 |
| Cu-O | 1.24 | 1.92 | 0.0027 |
R factor represents the overall goodness-of-fit.
Affinity of L for Cui
The conditional stability constant of the CuiL complex was measured at pH 7.4 using ferrozine (Fz) as a competitor according to the reaction given in Equation 1. Indeed, bathocuproine disulfonate (BCS) or bicinchoninate anion (BCA) fully displace the Cui cation from the ligand even at low competitor concentration. Therefore Fz, which has a lower affinity for Cui was chosen as a competitor of appropriate affinity to conduct these experiments.[38]
| Equation 1 |
The conditional stability constant of the CuiL complex was calculated using two models found in the literature for the Cui(Fz)2 complex stability (β21pH 7.4).[38] This gives logβ11pH 7.4 values for the CuiL complex of 11.1 according to the model of Xiao et al[38a] and 7.6 according to the model of Alies et al.[38b] Importantly, regardless the model chosen for Cui(Fz)2, L exhibits an affinity for Cui slightly larger than that found for the Aβ1-16 peptide, corresponding to the C-terminally truncated part of the Aβ1-40/42 peptides.[38] The difference in the stability constants is Δlog β11pH 7.4 = 0.7 in favor of L. As expected, the preorganization of three His in the tripodal pseudopeptide L induces a larger affinity for the soft Cui cation than linear His–containing peptide sequences such as found in the Aβ peptides, which binds Cui with two His in a dynamic complex with digonal geometry. It is therefore expected that L is able to remove Cui from the Aβ1-16 peptide.
In the pseudopeptide series derived from the nitrilotriacetic acid scaffold, the trishistidine ligand L displays an affinity for Cui in between that of the triscysteine[25a] and the tristhioether[27] derivatives, as predicted for peptide sequences with these amino acids at physiological pH.[39]
Redox properties of the Cu complex at physiological pH
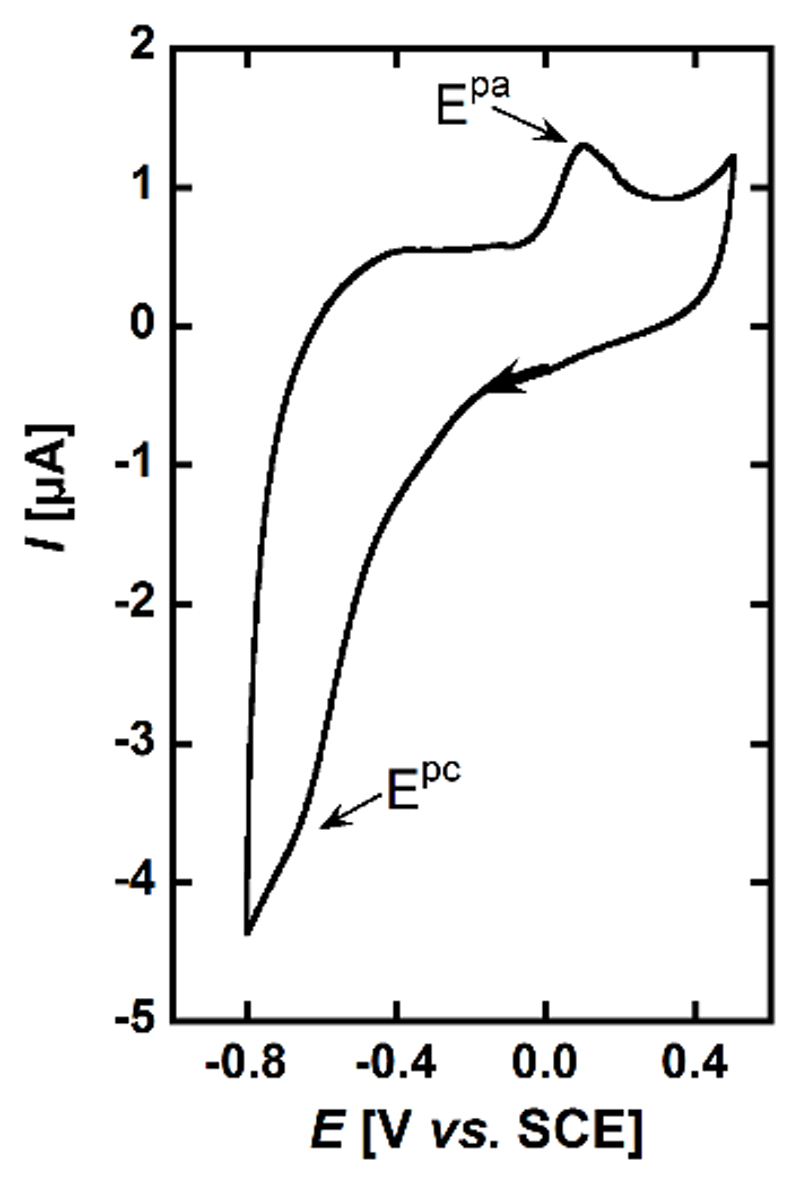
The cyclic voltammogram of the CuiiLH-1 complex predominant at pH 7.1 is shown in Figure 4. It shows one irreversible cathodic peak at Epc = -0.65 ± 0.02 V vs. SCE and an anodic peak on the reverse scan at Epa= 0.10 V vs. SCE. The cathodic peak is attributed to reduction of CuiiLH-1 complex followed by a structural rearrangement thus explaining the absence of reversibility.
Figure 4.
Cyclic voltamogram of CuiiLH-1. [L] = 0.2 mM, [Cuii] = 0.18 mM in [phosphate buffer] = 100 mM at pH 7.1 under Ar. Scan rate = 100 mV.s-1. WE = Glassy carbon, Ref = SCE, CE = Pt wire.
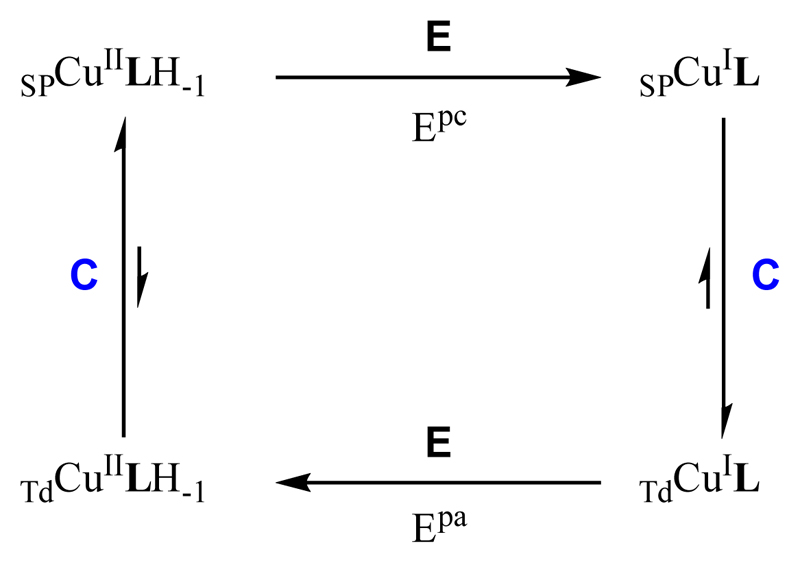
The origins of such rearrangement may be the change in the copper center geometry from square-planar (SP) to tetrahedral (Td) and the protonation of the amide bond upon reduction (See Scheme 3 and previous paragraphs). The anodic peak then corresponds to the oxidation of the tetrahedral CuiL species, and the irreversibility is due to the inverse changes: tetrahedral to square-planar and deprotonation of the amide bond. The mechanism is thus a classical ECEC (Electrochemical – Chemical - Electrochemical – Chemical) square scheme as shown in Scheme 4. Actually, the chemical reactions are double, i.e. structural rearrangement plus protonation/deprotonation processes. Determining whether this protonation / deprotonation steps are concerted with the structural rearrangement is beyond the scope of the present paper. It is worth noting that the cyclic voltammetry features is strongly reminiscent of what has been previously reported in case of calixarene-based Cu species.[40] However, in the present case, neither the TdCuii nor the SPCui has been observed.[41]
Scheme 4.
Proposed ECEC square scheme to explain the electrochemical data in Figure 4. E = Electrochemical process with the corresponding Ep of Figure 4; C = Chemical process, structural rearrangement and/or protonation/deprotonation events. SP indicates that the geometry of the complex is square-planar and Td that the geometry of the complex is tetrahedral.
It is important to note that (i) the reduction potential of the Cuii species is well beyond the oxidation potential of ascorbate and (ii) the oxidation of the Cui complex well above the reduction potential of dioxygen (recorded under the very same conditions, see supplementary Figure S5). Hence, the Cuii complex is expected to resist to reduction by ascorbate and the Cui complex to oxidation by dioxygen.
Ability of L to remove Cuii and Cui from the Aβ1-16 peptide proved by competition experiments
The removal of the Cuii and Cui ions from Aβ by the ligand L has been directly probed by EPR and XANES spectroscopy, respectively.
Figure 5 shows that 1 equiv. of L almost completely removes Cuii from the Aβ peptide. Indeed, the EPR signature in the presence of equimolar concentrations of the ligand L and the Aβ1-16 peptide is almost superimposable with the one of the CuiiLH-1 complex.
Figure 5.
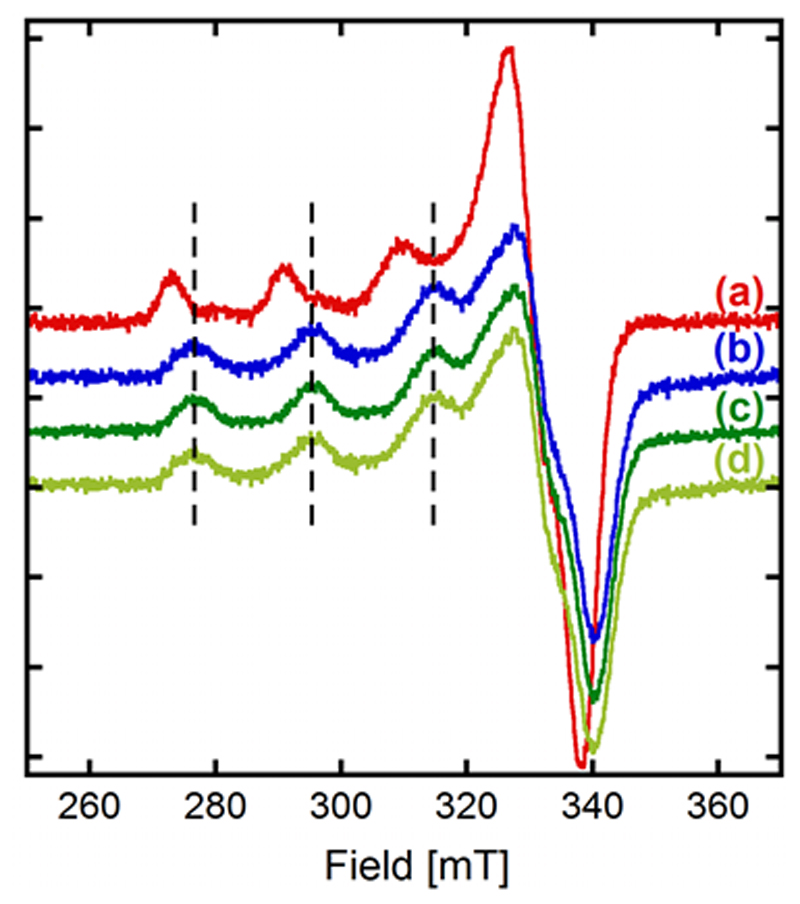
Competition experiments between the Aβ1-16 peptide and the ligand L. EPR experiments of (a) CuiiAβ1-16, (b) CuiiL, (c) Aβ1-16 + Cuii + L, (d) best linear combination representing (c): 5% (a) + 95% (b). [L] = [Aβ1-16] = [65Cuii] = 200 µM, [HEPES] = 50 mM, pH 7.1. 10% of glycerol was used as cryoprotectant. T = 110 K.
The proportion of Cuii remaining bound to Aβ was experimentally determined by linear combinations of the EPR spectra (see Figures 5 and S6) to be less than 5 %. The corresponding theoretical proportion was estimated with the speciation program Hyss.[34] The conditional stability constants used for this calculation were 9.2 (pH 7.1) and 9.7 (pH 7.4) for CuiiAβ1 16 reported in the literature,[42] and logβ11 = 12.1 (pH 7.1) or 12.6 (pH 7.4) for CuiiL, obtained from the global protonation and complexation constants given in Table 1. Given the possible modification in equilibria between room temperature and frozen solution studies, the calculated 3.4% value is in total accordance with the 5% experimental value from EPR measurements.
This 3 orders of magnitude between the affinity constants of the peptide and L for Cuii may be assigned to the preorganized tripodal chemical architecture of the pseudopeptide L and also to its ability to form an amidate bond with Cuii at physiological pH. These data demonstrate that Cuii can be removed from the N-terminally unmodified Aβ peptide, but not from the relatively abundant N-terminally truncated peptide at position 4 that forms a very high-affinity Cuii site.[43]
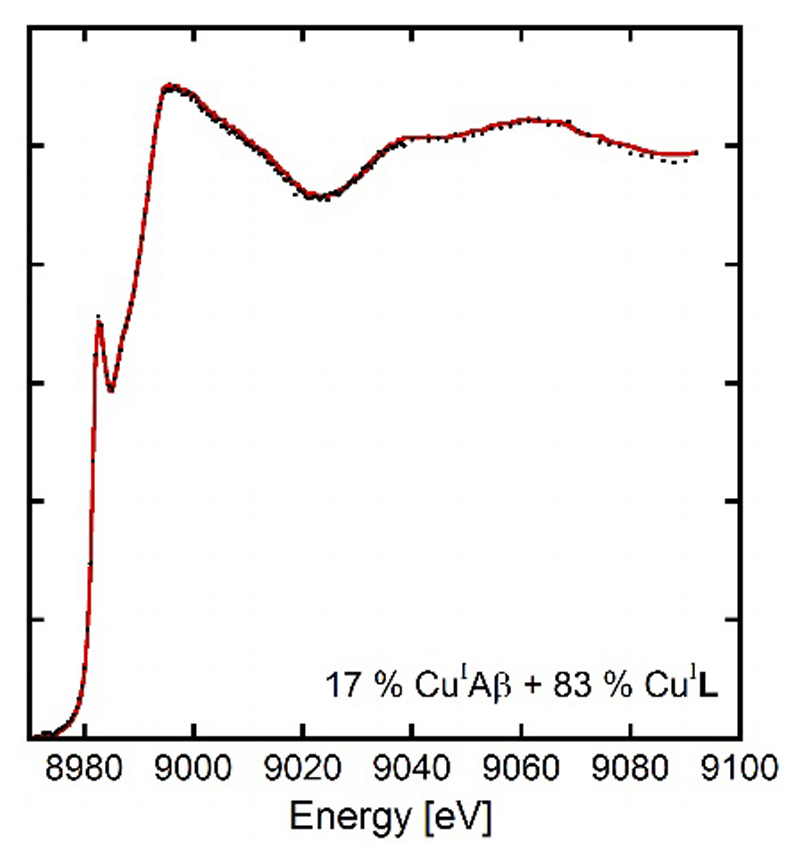
A similar competition followed by XANES for Cui is shown in Figure 6. In the presence of 1 equiv. of L, Cui is removed from the Aβ1-16 peptide but to a lesser extent than Cuii. Proportion of Cui remaining bound to Aβ is evaluated to 17% ± 5% by linear combinations (See Figure 6 and S7). This value agrees well with the log difference in the stability constants of 0.7 in favor of L obtained in a previous section. Indeed the speciation program Hyss[34] using the conditional stability constants measured for CuiL in this paper and previously published for CuiAβ[38] predicts 30% of Cui bound to Aβ in these conditions.
Figure 6.
Competition experiments between the Aβ1-16 peptide and the ligand L. XANES experiments for [L] = [Aβ1-16] = 1.0 mM, [Cuii] = 0.95 mM, [dithionite] = 10 mM, [HEPES] = 100 mM, pH 7.1. 10% of glycerol was used as cryoprotectant. The red line represents the experimental spectrum and black circles the best linear combination from Athena[44] (17%CuiAβ1-16 + 83% CuiL).
The trishistidine pseudopeptide L is thus able to remove Cui and/or Cuii from the Aβ peptide at physiological pH.
Reactive Oxygen Species Production
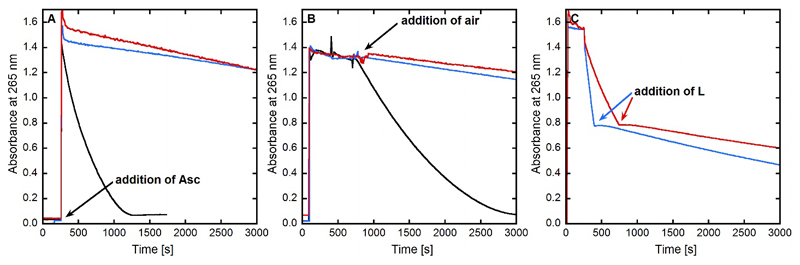
The impact of Cu removal from the monomeric CuAβ for both the Aβ1-16 model peptide (used for solubility issue in previous spectroscopic studies) and the Aβ1-40 peptide complex by the ligand L on ROS production was investigated by previously described methods in similar context.[45] We have focused our study on the monomeric peptidic complex, because it is the major form responsible for ROS production, fibrillary forms being less active by one order of magnitude.[46] ROS production corresponds to the incomplete reduction of dioxygen by ascorbate catalyzed by the CuI/IIAβ complex leading to O2°-, H2O2 and HO°. Thus, to probe ROS formation, either ascorbate consumption can be followed by UV-Vis at 265 nm or HO° formation can be monitored by the detection of the fluorescent 7-OH-CCA (7-hydroxy-coumarin-3-carboxylic acid) dye formed by reaction of HO° with the CCA (Coumarin-3-carboxylic acid) molecule.[7b] It has indeed been shown that the ascorbate consumption perfectly mirrors the H2O2 production.[7b, 47] The results of ascorbate consumption experiments are shown in Figure 7 and Figure S8 with Aβ1-40 and Aβ1-16 peptides, respectively. Those of the CCA experiments are described in the SI (Figure S9). Three different ascorbate consumption experiments were performed representing three starting conditions for ROS production: (i) starting from Cuii, (ii) starting from Cui and (iii) starting from a mixture of Cui and Cuii. In the first experiment, the ligand L is added to Cuii or CuiiAβ under aerobic conditions and then ascorbate is added. In the second one, the ligand L is added to Cui or CuiAβ under anaerobic conditions and then air is added. In the last one, ascorbate and air are mixed, then either Cuii or CuiiAβ is added and after a while (typically 8 minutes) the ligand L is added. Whatever the experiments, the ligand L is able to preclude the production of ROS, in the presence of Cu bound to the buffer only or to Aβ. This mirrors the double ability of the ligand to remove rapidly Cu from the peptide and to redox silence it. These experiments are fully consistent with affinity values as well as with the redox properties of the CuLH-1 previously described.
Figure 7.
Kinetics of ascorbate consumption, followed by UV-visible spectroscopy at 265 nm with subtraction of the background signal at 800 nm. Panel A. Aβ1-40 + Cuii + Asc (black curve), L + Cuii + Asc (blue curve), Aβ1-40 + Cuii + L +Asc (red curve). Panel B. Cuii + Asc + Aβ1-40 + air (black curve), Cuii + Asc + L + air (blue curve), Cuii + Asc + Aβ1-40 + L + air (red curve). Panel C. Asc + Cuii + L (blue curve), Asc + Aβ1-40 + Cuii + L (red curve). [L] = [Aβ1-40] = 12 µM, [Cuii] = 10 µM, [Asc] = 100 µM, [HEPES] = 100 mM, pH 7.1. For the experiments from Panel B, all the solutions were deoxygenated by bubbling Argon and were added under a little overpressure of Argon in order to keep Cu under its +I oxidation state.
Conclusions
Pseudopeptides based on chemical scaffolds grafted with amino acids with a large affinity for Cui, such as cysteines, have proven their efficacy in chelating Cui with coordination properties similar to metallothioneins. Whereas these chelators are proposed as intracellular drugs to treat Cu overload in Wilson’s disease, they are not appropriate for Alzheimer’s disease (AD), which is not a classical metal overload disease. Such high affinity chelators could deplete the brain in essential metals, whereas AD requires chelating molecules with moderate affinities to not disturb Cu homeostasis. Therefore, the novel pseudopeptide L based on functional groups reminiscent of the amino-acid side-chains encountered in the Aβ peptide, i.e. three histidines, was designed to get a moderate affinity for Cui and Cuii and also to target both oxidation states of Cu.
The trishistidine ligand L promotes a tetrahedral (Td) geometry around Cui as suggested by EXAFS, with three nitrogen of the histidines and an extra O-donor from the solvent. Interestingly, the conditional stability constant of the CuiL complex at physiological pH is significantly lower than those measured with thiolate derivatives, but slightly larger than the one of the Aβ1-16 peptide that coordinates Cui with only two histidines. Potentiometric studies point to a CuiiLH-1 complex at physiological pH, with a metallacycle involving an amidate and the proximal imidazole nitrogen atom. According to the EPR parameters, L binds the Cuii ion in a square-planar geometry (SP). The conditional stability constant of the Cuii complex at physiological pH is three orders of magnitude larger than the one measured for the CuiiAβ1-16 complex. This might arise from the pre-organized structure of the ligand L and also from its ability to form an amidate bond with Cuii at low pH (pKa of CuiiL = 6.0). L can therefore remove Cuii from the N-terminally unmodified Aβ peptide. Other derivatives of the Aβ peptide, such as the abundant N-terminally truncated peptide at position 4 were not studied in this work.[43] The stability constants measured in the present paper indicate that the ligand L should not be able to remove Cuii from such truncated peptides that exhibit high-affinity Cuii sites. However, this is not a major issue since when Cuii is bound in this high-affinity site, it is redox silent and doesn't produce ROS.[43b]
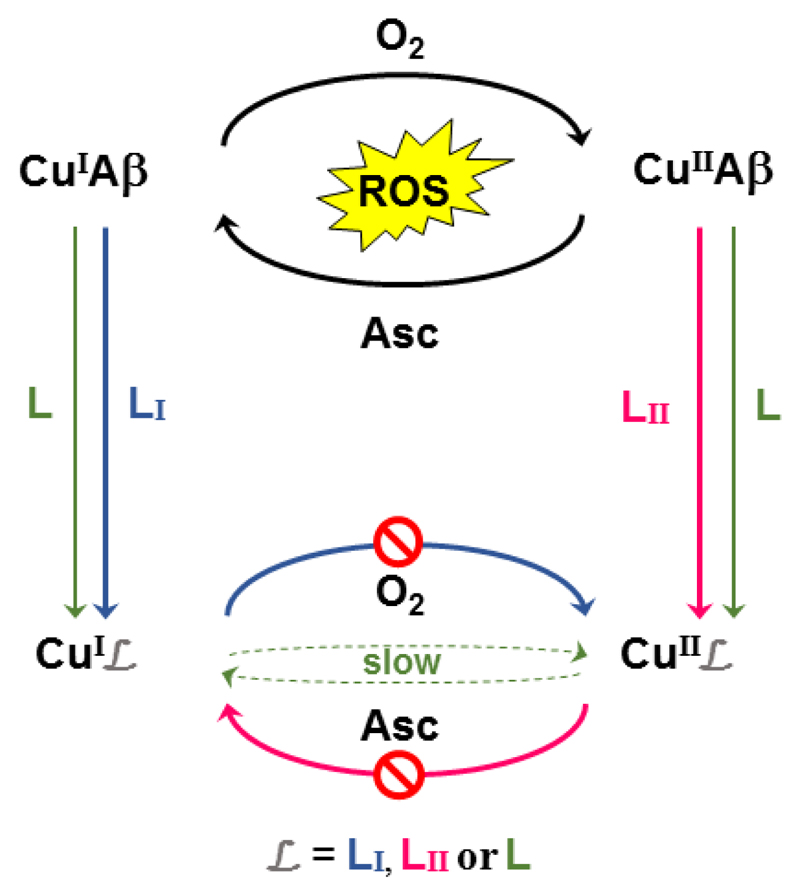
The pseudopeptide L has therefore a higher affinity than Aβ for both Cui and Cuii ions enabling it to remove Cu from Aβ regardless of the redox state of the metal center. The very different coordination of Cui and Cuii in complexes with ligand L account for the redox properties obtained by cyclic voltammetry where a ECEC mechanism is observed. It is indicative of a two steps (first the reduction, then a chemical reorganization) process going from SPCuii to TdCui and of a second two steps (first the oxidation, then a chemical reorganization) process going back from TdCui to SPCuii. Such redox properties are in line with the resistance of CuiiLH-1 to reduction by ascorbate and of CuiL to oxidation by oxygen and hence with an almost inactive CuL complex with respect to ROS production. They are due to the flexibility of the ligand and to its multi-site character. It actually reconciles what seems irreconcilable: (i) In the case of Cuii/Cui complexes exhibiting redox processes, the possibility to not produce ROS relies on either a very low redox potential or very high potential of the Cuii/Cui couple. This stabilizes only either the Cuii or the Cui state of the complex, respectively. Hence, in such a case, only one redox state can be targeted, which is as described in the introduction not the safest option ; (ii) in the case of ligands able to target both redox states, the formed complex is generally redox active. In our case, we can fulfil the two required properties (i.e. targeting both redox states and stopping ROS production) because the CuL complex undergoes sluggish redox process (Scheme 5).
Scheme 5.
Two possible approaches to stop CuAβ ROS production. On the one hand, Cui or Cuii is targeted and the system CuiLI or CuiiLII is inert toward O2 or Asc reaction, respectively. Note that this is a pre-requisite (see for instance [15a] and [45, 48] for ligands LI and LII ligands) but may be not enough (see ref.[22]). On the other hand, Cui and Cuii are targeted and the associated redox couple is slow. This is what is observed with L.
In conclusion, due to its unique Cu coordination properties, the ligand L is able to target both redox states of the Cu center and redox silence them. The delivery of L and in particular its crossing through the blood brain barrier (BBB) is beyond the scope of the present paper. However, the pseudopeptide L structure is versatile enough to be grafted with various functions either to target BBB receptors or for encapsulation in biological vectors. Some of these strategies have been previously developed to target hepatic cells for localized Cu detoxification in Wilson’s disease.[2a, 49] This ligand thus represents a new strategy in the long route of finding new coordination concepts for fighting AD.
Experimental Section
Synthesis
General information - Solvents and starting materials were purchased from ALDRICH, FLUKA, ACROS ORGANICS, ALFA AESAR and BACHEM and used without further purification. NTA(NHS)3 was obtained according to Jullien et al.[27] Water solutions were prepared from ultrapure laboratory grade water that has been filtered and purified by reverse osmosis using a Millipore MILLIQ reverse-osmosis cartridge system (resistivity - 18 MΩ cm). The 1H NMR and 13C NMR spectra were recorded on a Bruker Avance 400 MHz spectrometer. The chemical shifts (δ) are reported in ppm with the solvent as the internal reference. The NMR coupling constants (J) are reported in Hz. The mass spectra were acquired with a FINIGAN LXQ-linear ion trap (THERMO Scientific, San Jose, USA) equipped with an electrospray source. Analytical and preparative HPLC were performed with a VWR system fitted with Chromolith® RP-18e columns (L = 100 mm, Ø = 4.6 mm for analytical column; L = 100 mm, Ø = 25 mm for preparative column) with A = CH3CN/H2O/TFA (v/v/v = 90/10/0.1) and B - H2O/TFA (v/v = 99.925/0.075) as solvents. Flow rates of 1 mL/min and 15 mL/min were used for analytical and preparative column, respectively.
To a suspension of NTA(NHS)3 (250 mg, 0.518 mmol) in CH3CN/DMF mixture (15/15 mL), H-His-NH2·2HCl (502 mg, 2.21 mmol) and DIEA (0.59 mL, 3.39 mmol) were successively added. After stirring for 48h at 30 °C, the resulting mixture was concentrated in vacuo. The crude product was purified by preparative C18 reversed-phase HPLC (gradient from A/B: 0/100 to 50/50 in 15 min, Rt = 3.7 min) followed by lyophilisation to afford the desired compound L·4TFA (60 mg, Yield 11 %) as a white powder. The number of TFA per ligand in the solid was confirmed by potentiometric titrations. (+)ESI-MS calculated for C24H34N13O6: [M+H]+ m/z 600.27, Experimental [M+H]+m/z 600.3. 1H NMR (400 MHz, D2O, 300 K): 8.63 (s, 3H, H2), 7.32 (s, 3H, H5), 4.68 (dd, 3H, 3J=6.7 Hz, CHα), 3.52 (dAB, 3H, 2JAB=16.6 Hz, NCH2), 3.47 (dAB, 3H, 2JAB=16.6 Hz, NCH2), 3.24 (ABX syst., 6H, JAB=15.4, JAX=6.2, JBX=8.2, CH2β). 13C NMR (100 MHz, D2O, 300K): 174.1 (CONH2), 171.9 (CONH), 133.6 (C2), 128.4 (C4), 117.2 (C5), 57.3 (NCH2), 52.3 (CHα), 26.5 (CH2β) (for 1H NMR and 13C NMR spectra, see Figures S10 and S11 respectively).
Complexation studies
Chemicals
Reagents, except the ligand L, were commercially available and were used as received. All the solutions were prepared in milliQ water (resistance: 18.2 MΩ.cm). The Cuii ion source was CuSO4.5H2O bought from Sigma-Aldrich. The Cui source, except for XAS samples, was Cu(CH3CN)4PF6 bought from Sigma-Aldrich. The stock solution was prepared in acetonitrile and the exact concentration was determined by adding excess sodium bathocuproine disulfonate (BCS) and measuring the absorbance of Cu(BCS)23-.HEPES buffer (sodium salt of 2-[4-(2-hydroxyethyl)piperazin-1-yl]ethanesulfonic acid) was bought from Sigma-Aldrich. A stock solution was prepared at 500 mM, pH = 7.1. Phosphate buffer was bought from Sigma-Aldrich. Two stock solutions, K2HPO4 and KH2PO4, were prepared at 500 mM, and they were mixed until to reach a stock solution at pH = 7.1.Sodium ascorbate was bought from Sigma-Aldrich. A stock solution was prepared at 5 mM each day because of the quick degradation of the ascorbate. Coumarin-3-carboxilic acid (CCA) was bought from Acros Organics. A stock solution at 5 mM was prepared in phosphate buffer at 500 mM, pH = 7.1. The stock solution was stored at 4°C. Sodium dithionite was bought from Sigma-Aldrich. Before each experiment, a stock solution at 1 M was prepared. Ferrozine was bought from sigma Aldrich. A stock solution was prepared 20 mM phosphate buffer, pH = 7.4, and titrated with the Cui solution to determine the exact Fz concentration. Peptide. Aβ1-16 (DAEFRHDSGYEVHHQK) was bought from Genecust. A stock solution was prepared around 10 mM and stored at 4°C. Peptide concentration was determined by UV-visible absorption of Tyr10 considered as free tyrosine (at pH 2, (ε276-ε296) = 1410 M-1cm-1). Aβ1-40 (DAEFRHDSGYEVHHQKLVFFAEDVGSNKGAIIGLMVGGVV) was bought from Genecust. Stock solution of the Aβ40 peptide was prepared by dissolving the powder in 50mM NaOH at approx. 400 µM. Peptide concentration was then determined by UV-visible absorption of Tyr10 considered as free tyrosine (at pH 13, (ε296-ε360) = 2400 M-1cm-1). The solutions were diluted down to the appropriate concentration in peptide. All pH values are given with a ± 0.2 pH unit error.
ESI-MS spectrometry
100 µM pseudopeptide solutions were prepared in ammonium acetate buffer (20 mM, pH=6.9). Cu was added to the peptide solution from stock solutions of CuSO4 in water or Cu(CH3CN)4PF6 in acetonitrile, for Cuii and Cui samples, respectively. Mass spectra were recorded on a LXQ type THERMO SCIENTIFIC spectrometer equipped with an electrospray ionization source and a linear trap detector. Solutions were injected in the spectrometer at 10 µL/min flow rate. Ionization voltage and capillary temperature were about 2 kV and 250 °C, respectively.
Potentiometry
All titrant solutions were prepared using water purified by passing through a Millipore Milli-Q reverse-osmosis cartridge system (resistivity 18 MΩ cm). Carbonate-free 0.1 molL-1 KOH and 0.1 molL-1 HCl were prepared from Fisher Chemicals concentrates. Potentiometric titrations were performed in 0.1 molL-1 aqueous KCl under an argon atmosphere, the temperature was controlled to ±0.1 °C with a circulating water bath. The pH (pH = log[H+], concentration in molarity) was measured in each titration with a combined pH glass electrode (Metrohm) filled with 3 molL-1 KCl and the titrant addition was automated by use of a 751 GPD titrino (Metrohm). The electrode was calibrated in hydrogen ion concentration by titration of HCl with KOH in 0.1 molL-1 KCl.[50] A plot of meter reading versus pH allows the determination of the electrode standard potential (E°) and the slope factor (f). Ligand’s concentration was determined by potentiometric titrations and was in accordance with the formula NTA(HisNH2)3.4TFA. Continuous potentiometric titrations with KOH 0.1 molL-1 were conducted on 20 mL of aqueous solutions containing 10-3 molL-1 of the ligand and 0, 0.5, 1 and 2 equiv. of the Cuii cation. Back titrations with HCl 0.1 molL-1 were systematically performed after each experiment to check whether equilibration had been achieved. In a typical experiment, 100 points were measured with a 2 min delay between the measurements for the free ligand, and a 5 min delay for metallic complexes. Experimental data were refined using the computer program Hyperquad 2000.[34, 51] Some precipitation was detected in the experiments performed with 2 Cuii equiv. below pH 8. Therefore the latter titrations were not included in the fitting process. All equilibrium apparent constants are expressed as concentration ratio and not activities. The ionic product of water at 25 °C and 0.1 molL-1 ionic strength is pKw = 13.78.[29] The initial concentrations of ligand, metal and proton were fixed, as well as the ligand’s pKa values for the metallic complex stability constant determination. All values and errors (one standard deviation) reported represent the average of at least three independent experiments.
Affinity for Cui
The apparent affinity constants at pH 7.4 of the Cui complexes were measured by UV-visible titrations in presence of ferrozine (Fz) as a competitor. The spectra were recorded with a Varian Cary50 spectrophotometer equipped with optical fibers connected to an external cell holder in the glove box. A solution of the Cui complex with L in 20 mM phosphate buffer/ MeCN (9/1, v/v), pH = 7.4, in the UV cell was titrated with Fz. The spectra were then recorded and show the increase of the orange Cu(Fz)23- complex which absorbs at 470 nm with a molar extinction coefficient value ε = 4320 M-1 cm-1. The stability of the absorbance at 470 nm was controlled before the addition of any other aliquots of competitor. The stability constants were then determined using the two models described in the literature: binding constant of the Cu(Fz)23- complex (log β12 = 15.1[38a] or log β12 = 11.6[38b]).
Electron Paramagnetic Resonance
Electron Paramagnetic Resonance (EPR) data were recorded using an Elexsys E 500 Bruker spectrometer, operating at a microwave frequency of approximately 9.5 GHz. Spectra were recorded using a microwave power of 20 mW across a sweep width of 150 mT (centered at 310 mT) with modulation amplitude of 0.5 mT. Experiments were carried out at 110 K using a liquid nitrogen cryostat. EPR samples were prepared from stock solution of ligand diluted down to 0.2 mM in H2O. 0.9 eq. of 65Cuii was added from 25 mM 65Cu(NO3)2 stock solution home-made from a 65Cu foil. If necessary, pH was adjusted with H2SO4 and NaOH solutions. Samples were frozen in quartz tube after addition of 10% glycerol as a cryoprotectant and stored in liquid nitrogen until used.
X-ray Absorption Spectroscopy (XAS)
Cu K-edge XANES (X-ray absorption near edge structure) and EXAFS (Extended X-ray Absorption Fine Structure) spectra were recorded at the BM30B (FAME) beamline at the European Synchrotron Radiation Facility (ESRF, Grenoble, France).[52] The storage ring was operated in 7/8+1 mode at 6 GeV with a 200 mA current. The beam energy was selected using a Si(220) N2 cryo-cooled double-crystal monochromator with an experimental resolution close to that theoretically predicted (namely ~ 0.5 eV FWHM at the Cu energy).[53] The beam spot on the sample was approximately 300 x 100 µm2 (H x V, FWHM). Because of the low Cui and Cuii concentrations, spectra were recorded in fluorescence mode with a 30-element solid state Ge detector (Canberra) in frozen liquid cells in a He cryostat. The temperature was kept at 20 K during data collection. The energy was calibrated with a Cu metallic foil, such that the maximum of the first derivative was set at 8979 eV. EXAFS Cu data were collected from 8830 to 8970 eV using 2 eV step of 2 s, from 8970 to 9038.5 eV using 0.5 eV step of 3 s, and from 9038.5 to 9828.1 eV with a k-step of 0.05 Å-1 and an increasing time 3-10 s per step. At least five scans recorded on different spots were averaged. XANES Cu data were collected from 8830 to 8970 eV using 2 eV step of 2 s, from 8970 to 9038.5 eV using 0.5 eV step of 2 s, and from 9038.5 to 9320 eV with a k-step of 0.05 Å-1 and 2 s per step. At least three scans recorded on different spots were averaged. The data analysis was performed using the “Multi-Plateform Applications for X-ray Absorption” package, including Cherokee and Roundmidnight programs,[54] according to the standard and previously reported data analysis procedures.[55] Spectra were background-corrected by a linear regression through the pre-edge region and a polynomial through the post-edge region. The backscattering phase, Φi(k, Ri), and amplitude, Ai(k, Ri), functions were obtained using the ab initio FEFF7 code.[56] Since theoretical phase shifts were used, it is necessary to fit the energy threshold E0 by adding an extra fitting parameter, ΔE0. Moreover, the FEFF7 code was used to check if the multiple scattering of our reference compounds of known crystallographic structure is negligible in the 0-3 Å range. The estimated errors for distances and coordination numbers are ± 0.02 Å and ± 20%, respectively. XAS samples were prepared from stock solutions of ligand, peptide Aβ1-16 and Cuii diluted down to 1.0 mM in buffered solution (Phosphate buffer, pH 7.1). Dithionite was used to reduce Cuii in Cui. Samples were frozen in the sample holder after addition of 10% glycerol as a cryoprotectant and stored in liquid nitrogen until used. Cuii photoreduction was controlled by recording successive scans at the same spot. It was considered that during the first 20 minutes of recording the photoreduction is insignificant.
Electrochemistry
Cyclic voltamograms were recorded on a Autolab PGSTAT302N at 25°C. Saturated Calomel Electrode was used as a reference, Platine electrode was the counter electrode and the working electrode was a glassy carbon electrode. The working electrode was carefully polished before each measurement on a red disk NAP with 1 µm AP-A suspension under abundant distillate water flow during at least three minutes (Struers). The solution was deoxygenated by bubbling Argon before each measurement. Any support electrolyte was added because of the high concentration of phosphate buffer in the solution. The scanning speed was 0.1 V.s-1. The samples were prepared from stock solutions of ligand and Cuii down to approx. 1 mM and 0.9 mM respectively in a buffered solution. pH was adjusted with H2SO4 and NaOH solutions.
Competitions
EPR samples were prepared from stock solutions of peptide Aβ1-16 and ligand L in HEPES 50 mM pH 7.1 to reach 200 µM concentration of each compound. An aliquot of a 25 mM 65Cu(NO3)2 stock solution home-made from a 65Cu foil was then added to reach a 200 µM Cuii concentration. Samples were frozen in quartz tube after addition of 10% glycerol as a cryoprotectant and stored in liquid nitrogen until used. Remaining Cuii-bound to the Aβ1-16 peptide was evaluated by linear combinations of the spectra registered in the same conditions for the CuiiAβ1-16 and CuiiL complexes.
XAS samples were prepared from stock solutions of peptide Aβ1-16 and ligand L in HEPES 100 mM pH 7.1 to reach 1 mM concentration of each compound. An aliquot of a stock solution of Cu(NO3)2 was then added to reach a 0.95 mM Cu concentration. Excess dithionite was added (10 mM) to reduce Cuii in Cui. Samples were frozen in the sample holder after addition of 10% glycerol as a cryoprotectant and stored in liquid nitrogen until used. Remaining Cui-bound to the Aβ1-16 peptide was evaluated by linear combinations of the spectra registered in the same conditions for the CuiAβ1-16 and CuiLcomplexes with the program Athena.[44]
ROS formation
UV-Visible spectrophotometry. UV-vis kinetics were recorded on a spectrophotometer Agilent 8453 at 25°C in 1 cm path length quartz cuvette, with an 800 rpm stirring. The samples were prepared from stock solutions of ligand, peptide Aβ1-40 or Aβ1-16 and Cuii diluted down to 12, 12 and 10 µM respectively in HEPES solution, pH = 7.1. Ascorbate is diluted down to 100 µM.
Fluorimeter. CCA experiments were recorder on a FLUOstar OPTIMA BMG LABTECH at 25°C in a 96-well plate bought from Dutscher SAS. CCA was excited at 390 nm and the fluorescence was recorded at 450 nm. The gain used was 1350. The samples were prepared from stock solutions of ligand, peptide and Cuii diluted down to 12, 12 and 10 µM respectively in phosphate solution, pH = 7.1. CCA was added at a resulting concentration of 500 µM. Injector was used for the addition of ascorbate diluted down to 500 µM, 5 min after the beginning of the experiment.
Supplementary Material
Acknowledgements
The authors thank Drs. Clémence Cheignon and Fabrice Collin for their contribution in the recording of XAS data and Prof. P. Faller for fruitful discussions. This research was supported by the ERC StG-638712 (C.H.) and Labex ARCANE (Grant ANR-11-LABX-0003-01, P.D.) and the “Fondation pour la Recherche Médicale” (grant DCM20111223043, P.D.). ACM and CG thank FAPESP (Bepe 2012/06754-4). The ESRF is acknowledged for providing access to the beamline BM30B (experiments 30-02-1100).
References
- [1].Solomon EI, Heppner DE, Johnston EM, Ginsbach JW, Cirera J, Qayyum M, Kieber-Emmons MT, Kjaergaard CH, Hadt RG, Tian L. Chem Rev. 2014;114:3659–3653. doi: 10.1021/cr400327t. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [2].a) Delangle P, Mintz E. Dalton Trans. 2012;41 doi: 10.1039/c2dt12188c. [DOI] [PubMed] [Google Scholar]; b) Gateau C, Mintz E, Delangle P. In: Rational design of Cu and Fe chelators to treat Wilson’s disease and hemochromatosis. Storr T, editor. WILEY-BLACKWELL; 2014. pp. 287–319. [Google Scholar]
- [3].a) Pal A, Siotto M, Prasad R, Squitti R. J Alzheimers dis. 2015;44:343–354. doi: 10.3233/JAD-141194. [DOI] [PubMed] [Google Scholar]; b) Hureau C. Coord Chem Rev. 2012;256:2164–2174. [Google Scholar]; c) Faller P, Hureau C, La Penna G. Acc Chem Res. 2014;47:2252–2259. doi: 10.1021/ar400293h. [DOI] [PubMed] [Google Scholar]; d) Ayton S, Lei P, Bush AI. Neurotherapeutics. 2015;12:109–120. doi: 10.1007/s13311-014-0312-z. [DOI] [PMC free article] [PubMed] [Google Scholar]; e) Avan A, Hoogenraad TU. J Alzheimers dis. 2015;4:89–92. doi: 10.3233/JAD-150186. [DOI] [PubMed] [Google Scholar]; f) Kozlowski H, Luczkowski M, Remelli M, Valensin D. Coord Chem Rev. 2012;256:2129–2141. [Google Scholar]
- [4].Miller LM, Wang Q, Telivala TP, Smith RJ, Lanzirotti A, Miklossy J. J Struct Biol. 2006;155:30–37. doi: 10.1016/j.jsb.2005.09.004. [DOI] [PubMed] [Google Scholar]
- [5].a) Chiurchiù V, Orlacchio A, Maccarrone M. Oxid Med Cell Longev. 2016;2016 doi: 10.1155/2016/7909380. 7909380. [DOI] [PMC free article] [PubMed] [Google Scholar]; b) Jellinger KA. Int Rev Neurobiol. 2013;110:1–47. doi: 10.1016/B978-0-12-410502-7.00002-8. [DOI] [PubMed] [Google Scholar]
- [6].a) Balland V, Hureau C, Savéant J-M. P Natl Acad Sci USA. 2010;107:17113–17118. doi: 10.1073/pnas.1011315107. [DOI] [PMC free article] [PubMed] [Google Scholar]; b) Trujano-Ortiz LG, González FJ, Quintanar L. Inorg Chem. 2014;54:4–6. doi: 10.1021/ic501941a. [DOI] [PubMed] [Google Scholar]
- [7].a) Hureau C, Faller P. Biochimie. 2009;91:1212–1217. doi: 10.1016/j.biochi.2009.03.013. [DOI] [PubMed] [Google Scholar]; b) Chassaing S, Collin F, Dorlet P, Gout J, Hureau C, Faller P. Curr Top Med Chem. 2012;12:2573–2595. doi: 10.2174/1568026611212220011. [DOI] [PubMed] [Google Scholar]; c) Smith DG, Cappai R, Barnham KJ. Biochim Biophys Acta. 2007;1768:1976–1990. doi: 10.1016/j.bbamem.2007.02.002. [DOI] [PubMed] [Google Scholar]
- [8].a) Cassagnes L-E, Hervé V, Nepveu F, Hureau C, Faller P, Collin F. Angew Chem Int Ed. 2013;52:11110–11113. doi: 10.1002/anie.201305372. [DOI] [PubMed] [Google Scholar]; b) Cheignon C, Jones M, Atrián-Blasco E, Kieffer I, Faller P, Collin F, Hureau C. Chem Sci. doi: 10.1039/c7sc00809k. in press. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [9].a) Santos MA, Chand K, Chaves S. Coord Chem Rev. 2016:327–328. 287-303. [Google Scholar]; b) Bachurin SO, Bovina EV, Ustyugov AA. Med Res Rev. 2017 doi: 10.1002/med.21434. [DOI] [PubMed] [Google Scholar]; c) Wild K, August A, Pietrzik CU, Kins S. Front Mol Neurosci. 2017;10 doi: 10.3389/fnmol.2017.00021. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [10].a) Faller P, Hureau C. Chem Eur J. 2012;18:15910–15920. doi: 10.1002/chem.201202697. [DOI] [PubMed] [Google Scholar]; b) Dumont M, Beal MF. Free Radic Biol Med. 2011;51:1014–1026. doi: 10.1016/j.freeradbiomed.2010.11.026. [DOI] [PMC free article] [PubMed] [Google Scholar]; c) Jomova K, Vondrakova D, Lawson M, Valko M. Mol Cell Biochem. 2010;345:91–104. doi: 10.1007/s11010-010-0563-x. [DOI] [PubMed] [Google Scholar]
- [11].a) Xia N, Liu L. Mini Rev Med Chem. 2014;3 doi: 10.2174/1389557514666140123124841. [DOI] [PubMed] [Google Scholar]; b) Crouch PJ, Barnham KJ. Acc Chem Res. 2012;45:1604–1611. doi: 10.1021/ar300074t. [DOI] [PubMed] [Google Scholar]; c) Barnham KJ, Bush AI. Chem Soc Rev. 2014;43:6727–6749. doi: 10.1039/c4cs00138a. [DOI] [PubMed] [Google Scholar]; d) Robert A, Liu Y, Nguyen M, Meunier B. Acc Chem Res. 2015;48:1332–1339. doi: 10.1021/acs.accounts.5b00119. [DOI] [PubMed] [Google Scholar]
- [12].Drew SC. Front Neurosci. 2017;11:317. doi: 10.3389/fnins.2017.00317. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [13].a) Wang Y, Wang H, Chen HZ. Curr Neuropharmacol. 2016;14:364–375. doi: 10.2174/1570159X14666160119094820. [DOI] [PMC free article] [PubMed] [Google Scholar]; b) Rodriguez-Rodriguez C, Telpoukhovskaia M, Orvig C. Coord Chem Rev. 2012;256:2308–2332. [Google Scholar]; c) Guzior N, Wieckowska A, Panek D, Malawska B. Curr Med Chem. 2015;22:373–404. doi: 10.2174/0929867321666141106122628. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [14].a) Jensen M, Canning A, Chiha S, Bouquerel P, Pedersen JT, Østergaard J, Cuvillier O, Sasaki I, Hureau C, Faller P. Chem Eur J. 2012;18:4836–4839. doi: 10.1002/chem.201103546. [DOI] [PubMed] [Google Scholar]; b) Caballero AB, Terol-Ordaz L, Espargaró A, Vázquez G, Nicolás E, Sabaté R, Gamez P. Chem Eur J. 2016;22:7268–7280. doi: 10.1002/chem.201600286. [DOI] [PubMed] [Google Scholar]; c) Hu X, Zhang Q, Wang W, Yuan Z, Zhu X, Chen B, Chen X. ACS Chem Neurosc. 2016;7:1255–1263. doi: 10.1021/acschemneuro.6b00145. [DOI] [PubMed] [Google Scholar]
- [15].a) Atrian-Blasco E, Cerrada E, Conte-Daban A, Testemale D, Faller P, Laguna M, Hureau C. Metallomics. 2015;7:1229–1232. doi: 10.1039/c5mt00077g. [DOI] [PubMed] [Google Scholar]; b) Walke GR, Ranade DS, Ramteke SN, Rapole S, Satriano C, Rizzarelli E, Tomaselli GA, Trusso Sfrazzetto G, Kulkarni PP. Inorg Chem. 2017;56:3729–3732. doi: 10.1021/acs.inorgchem.6b02915. [DOI] [PubMed] [Google Scholar]
- [16].a) Meloni G, Sonois V, Delaine T, Guilloreau L, Gillet A, Teissié J, Faller P, Vasak M. Nat Chem Biol. 2008;4:366–372. doi: 10.1038/nchembio.89. [DOI] [PubMed] [Google Scholar]; b) Perrone L, Mothes E, Vignes M, Mockel A, Figueroa C, Miquel MC, Maddelein ML, Faller P. ChemBioChem. 2010;11:110–118. doi: 10.1002/cbic.200900474. [DOI] [PubMed] [Google Scholar]
- [17].Rice ME. Trends Neurosci. 2000;23:209–216. doi: 10.1016/s0166-2236(99)01543-x. [DOI] [PubMed] [Google Scholar]
- [18].D'Ambrosi N, Rossi L. Neurochem Int. 2015;90:36–45. doi: 10.1016/j.neuint.2015.07.006. [DOI] [PubMed] [Google Scholar]
- [19].Goch W, Bal W. PLoS One. 2017;12 doi: 10.1371/journal.pone.0170749. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [20].Garg SK, Vitvitsky V, Albin R, Banerjee R. Antioxidants & Redox Signaling. 2011;14:2385–2397. doi: 10.1089/ars.2010.3681. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [21].a) Telpoukhovskaia MA, Orvig C. Chemical Society Reviews. 2013;42:1836–1846. doi: 10.1039/c2cs35236b. [DOI] [PubMed] [Google Scholar]; b) Nguyen M, Rechignat L, Robert A, Meunier B. ChemistryOpen. 2015;4:27–31. doi: 10.1002/open.201402075. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [22].Chen TT, Wang XY, He YF, Zhang CL, Wu ZY, Liao K, Wang JJ, Guo ZJ. Inorg Chem. 2009;48:5801–5809. doi: 10.1021/ic900025x. [DOI] [PubMed] [Google Scholar]
- [23].a) Hureau C, Balland V, Coppel Y, Solari PL, Fonda E, Faller P. J Biol Inorg Chem. 2009:995–1000. doi: 10.1007/s00775-009-0570-0. [DOI] [PubMed] [Google Scholar]; b) Shearer J, Szalai VA. J Am Chem Soc. 2008;130:17826–17835. doi: 10.1021/ja805940m. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [24].Cherny RA, Legg JT, McLean CA, Fairlie DP, Huang X, Atwood CS, Beyreuther K, Tanzi RE, Masters CL, Bush AI. J Biol Chem. 1999;274:23223–23228. doi: 10.1074/jbc.274.33.23223. [DOI] [PubMed] [Google Scholar]
- [25].a) Pujol AM, Gateau C, Lebrun C, Delangle P. Chem Eur J. 2011;17:4418–4428. doi: 10.1002/chem.201003613. [DOI] [PubMed] [Google Scholar]; b) Pujol AM, Gateau C, Lebrun C, Delangle P. J Am Chem Soc. 2009;131:6928–6929. doi: 10.1021/ja901700a. [DOI] [PubMed] [Google Scholar]; c) Jullien A-S, Gateau C, Kieffer I, Testemale D, Delangle P. Inorg Chem. 2013;52:9954–9961. doi: 10.1021/ic401206u. [DOI] [PubMed] [Google Scholar]; d) Pujol AM, Lebrun C, Gateau C, Manceau A, Delangle P. Eur J Inorg Chem. 2012:3835–3843. [Google Scholar]
- [26].a) Jullien AS, Gateau C, Lebrun C, Delangle P. Eur J Inorg Chem. 2015:3674–3680. doi: 10.1021/ic502962d. [DOI] [PubMed] [Google Scholar]; b) Jullien A-S, Gateau C, Lebrun C, Kieffer I, Testemale D, Delangle P. Inorg Chem. 2014;53:5229–5239. doi: 10.1021/ic5004319. [DOI] [PubMed] [Google Scholar]
- [27].Jullien AS, Gateau C, Lebrun C, Delangle P. Inorg Chem. 2015;54:2339–2344. doi: 10.1021/ic502962d. [DOI] [PubMed] [Google Scholar]
- [28].Dancs A, May NV, Selmeczi K, Darula Z, Szorcsik A, Matyuska F, Pali T, Gajda T. New J Chem. 2017;41:808–823. [Google Scholar]
- [29].Smith RM, Martell AE, Motekaitis RJ. NIST Critically Selected Stability Constants of Metal Complexes Database, NIST Standard Reference Database. 2001;46 [Google Scholar]
- [30].Adams EQ. J Am Chem Soc. 1916;38:1503–1510. [Google Scholar]
- [31].Siddons CJ, Hancock RD. Chem Commun. 2004;1632:1633. doi: 10.1039/b404316m. [DOI] [PubMed] [Google Scholar]
- [32].a) Kallay C, Varnagy K, Malandrinos G, Hadjiliadis N, Sanna D, Sovago I. Dalton Trans. 2006;45:45–4552. doi: 10.1039/b608190h. [DOI] [PubMed] [Google Scholar]; b) La Mendola D, Magri A, Santoro AM, Nicoletti VG, Rizzarelli E. J Inorg Biochem. 2012;111:59–69. doi: 10.1016/j.jinorgbio.2012.02.027. [DOI] [PubMed] [Google Scholar]; c) Fragoso A, Delgado R, Iranzo O. Dalton Trans. 2013;42:6182–6192. doi: 10.1039/c3dt32384f. [DOI] [PubMed] [Google Scholar]
- [33].a) Bonnet CS, Fries PH, Crouzy S, Sénèque O, Cisnetti F, Boturyn D, Dumy P, Delangle P. Chem Eur J. 2009;15:7083–7093. doi: 10.1002/chem.200900636. [DOI] [PubMed] [Google Scholar]; b) Fragoso A, Lamosa P, Delgado R, Iranzo O. Chem Eur J. 2013;19:2076–2088. doi: 10.1002/chem.201203545. [DOI] [PubMed] [Google Scholar]; c) Lebrun C, Starck M, Gathu V, Chenavier Y, Delangle P. Chem Eur J. 2014;20:16566–16573. doi: 10.1002/chem.201404546. [DOI] [PubMed] [Google Scholar]; d) Starck M, Sisommay N, Laporte FA, Oros S, Lebrun C, Delangle P. Inorg Chem. 2015;54:11557–11562. doi: 10.1021/acs.inorgchem.5b02249. [DOI] [PubMed] [Google Scholar]; e) Starck M, Laporte FA, Oros S, Sisommay N, Gathu V, Solari PL, Creff G, Roques J, Den Auwer C, Lebrun C, Delangle P. Chem Eur J. 2017;23:5281–5290. doi: 10.1002/chem.201605481. [DOI] [PubMed] [Google Scholar]
- [34].Alderighi L, Gans P, Ienco A, Peters D, Sabatini A, Vacca A. Coord Chem Rev. 1999;184:311–318. [Google Scholar]
- [35].Peisach J, Blumberg WE. Arch Biochem Biophys. 1974;165:691–708. doi: 10.1016/0003-9861(74)90298-7. [DOI] [PubMed] [Google Scholar]
- [36].a) Sakaguchi U, Addison AW. J Chem Soc Dalton Trans. 1979:600–608. [Google Scholar]; b) Jiang F, McCracken J, Peisach J. J Am Chem Soc. 1990;112:9035–9044. [Google Scholar]; c) Rasia RM, Bertoncini CW, Marsh D, Hoyer W, Cherny D, Zweckstetter M, Griesinger C, Jovin TM, Fernández CO. P Natl Acad Sci USA. 2005;102:4294–4299. doi: 10.1073/pnas.0407881102. [DOI] [PMC free article] [PubMed] [Google Scholar]; d) Manikandan P, Epel B, Goldfarb D. Inorg Chem. 2001;40:781–787. doi: 10.1021/ic0011361. [DOI] [PubMed] [Google Scholar]
- [37].Kau LS, Spira-Solomon DJ, Penner-Hahn JE, Hodgson KO, Solomon EI. J Am Chem Soc. 1987;109:6433–6442. [Google Scholar]
- [38].a) Xiao Z, Gottschlich L, van der Meulen R, Udagedara SR, Wedd AG. Metallomics. 2013;5:501–513. doi: 10.1039/c3mt00032j. [DOI] [PubMed] [Google Scholar]; b) Alies B, Badei B, Faller P, Hureau C. Chem Eur J. 2012;18:1161–1167. doi: 10.1002/chem.201102746. [DOI] [PubMed] [Google Scholar]
- [39].Rubino JT, Chenkin MP, Keller M, Riggs-Gelasco P, Franz KJ. Metallomics. 2011;3:61–73. doi: 10.1039/c0mt00044b. [DOI] [PubMed] [Google Scholar]
- [40].Le Poul N, Le Mest Y, Jabin I, Reinaud O. Acc Chem Res. 2015;48:2097–2106. doi: 10.1021/acs.accounts.5b00152. [DOI] [PubMed] [Google Scholar]
- [41].a) Le Poul N, Campion M, Douzieh B, Rondelez Y, Le Clainche L, Reinaud O, Le Mest Y. J Am Chem Soc. 2007;129:8801–8810. doi: 10.1021/ja071219h. [DOI] [PubMed] [Google Scholar]; b) Le Poul N, Campion M, Izzet G, Douzieh B, Reinaud O, Le Mest Y. J Am Chem Soc. 2005;127:5280–5281. doi: 10.1021/ja043073h. [DOI] [PubMed] [Google Scholar]
- [42].a) Conte-Daban A, Borghesani V, Sayen S, Guillon E, Journaux Y, Gontard G, Lisnard L, Hureau C. Anal Chem. 2017;89:2155–2162. doi: 10.1021/acs.analchem.6b04979. [DOI] [PMC free article] [PubMed] [Google Scholar]; b) Kowalik-Jankowska T, Ruta M, Wisniewska K, Lankiewicz L. J Inorg Biochem. 2003;95:270–282. doi: 10.1016/s0162-0134(03)00128-4. [DOI] [PubMed] [Google Scholar]; b) Alies B, Renaglia E, Rozga M, Bal W, Faller P, Hureau C. Anal Chem. 2013;85:1501–1508. doi: 10.1021/ac302629u. [DOI] [PubMed] [Google Scholar]
- [43].a) Wezynfeld NE, Stefaniak E, Stachucy K, Drozd A, Płonka D, Drew SC, Krężel A, Bal W. Angew Chem Int Ed. 2016;55:8235–8238. doi: 10.1002/anie.201511968. [DOI] [PubMed] [Google Scholar]; b) Mital M, Wezynfeld NE, Frączyk T, Wiloch MZ, Wawrzyniak UE, Bonna A, Tumpach C, Barnham KJ, Haigh CL, Bal W, Drew SC. Angew Chem Int Ed. 2015;54:10460–10464. doi: 10.1002/anie.201502644. [DOI] [PubMed] [Google Scholar]
- [44].Newville M. Data Processing with IFFEFIT, ATHENA & ARTEMIS, consortium for advanced radiation sources. University of Chicago; Jul 24, 2007. [Google Scholar]
- [45].Conte-Daban A, Day A, Faller P, Hureau C. Dalton Trans. 2016;45:15671–15678. doi: 10.1039/c6dt02308h. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [46].Pedersen JT, Chen SW, Borg CB, Ness S, Bahl JM, Heegaard NH, Dobson CM, Hemmingsen L, Cremades N, Teilum K. J Am Chem Soc. 2016;138:3966–3969. doi: 10.1021/jacs.5b13577. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [47].Alies B, Sasaki I, Proux O, Sayen S, Guillon E, Faller P, Hureau C. Chem Commun. 2013;49:1214–1216. doi: 10.1039/c2cc38236a. [DOI] [PubMed] [Google Scholar]
- [48].Noël S, Perez F, Pedersen JT, Alies B, Ladeira S, Sayen S, Guillon E, Gras E, Hureau C. Journal of Inorganic Biochemistry. 2012;117:322–325. doi: 10.1016/j.jinorgbio.2012.05.016. [DOI] [PubMed] [Google Scholar]
- [49].a) Pujol AM, Cuillel M, Jullien A-S, Lebrun C, Cassio D, Mintz E, Gateau C, Delangle P. Angew Chem Int Ed. 2012;51:7445–7448. doi: 10.1002/anie.201203255. [DOI] [PubMed] [Google Scholar]; b) Pujol AM, Cuillel M, Renaudet O, Lebrun C, Charbonnier P, Cassio D, Gateau C, Dumy P, Mintz E, Delangle P. J Am Chem Soc. 2011;133:286–296. doi: 10.1021/ja106206z. [DOI] [PubMed] [Google Scholar]
- [50].Martell AE, Motekaitis RJ. Determination and use of stability constants VCH New York. 1992 [Google Scholar]
- [51].Gans P, Sabatini A, Vacca A. Talanta. 1996;43:1739–1753. doi: 10.1016/0039-9140(96)01958-3. [DOI] [PubMed] [Google Scholar]
- [52].Proux O, Biquard X, Lahera E, Menthonnex JJ, Prat A, Ulrich O, Soldo Y, Trévisson P, Kapoujvan G, Perroux G, Taunier P, Grand D, Jeantet P, Deleglise M, Roux J-P, Hazemann J-L. Phys Scr. 2005;115:970–973. [Google Scholar]
- [53].Proux O, Nassif V, Prat A, Ulrich O, Lahera E, Biquard X, Menthonnex JJ, Hazemann J-L. J Synchrotron Radiat. 2006;13:59–68. doi: 10.1107/S0909049505037441. [DOI] [PubMed] [Google Scholar]
- [54].Michalowicz A, Moscovici J, Muller-Bouvet D, Provost K. J Phys: Conf Ser. 2009;190:012034–012035. [Google Scholar]
- [55].a) Lengeler B, Eisenberg P. Phys Rev B: Condens Matter Mater Phys. 1980;21:4507–4520. [Google Scholar]; b) Guillon, Merdy P, Aplincourt M. Chem Eur J. 2003:4479–4484. doi: 10.1002/chem.200304920. [DOI] [PubMed] [Google Scholar]
- [56].Zabinsky SI, Rehr JJ, Ankudinov AL, Albers RC, Eller MJ. Phys Rev B: Condens Matter Mater Phys. 1995;52:2995–3009. doi: 10.1103/physrevb.52.2995. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.