Abstract
We present the rapid-prototyping of type I collagen micropatterns on poly-dimethylsiloxane substrates for the biomimetic confinement of cells using the combination of a surface oxidation treatment and 3-aminopropyl triethoxysilane silanisation followed by glutaraldehyde crosslinking. The aim of surface treatment is to stabilise microcontact printing transfer of this natural extracellular matrix protein that usually wears out easily from poly-dimethylsiloxane, which is not suitable for biomimetic cell culture platforms and lab-on-chip applications. A low-cost CD-DVD laser was used to etch biomimetic micropatterns into acrylic sheets that were in turn replicated to poly-dimethylsiloxane slabs with the desired features. These stamps were finally inked with type I collagen for microcontact printing transfer on the culture substrates in a simple manner. Human hepatoma cells (HepG2) and rat primary hepatocytes, which do not adhere to bare poly-dimethylsiloxane, were successfully seeded and showed optimal adhesion and survival on simple protein micropatterns with a hepatic cord geometry in order to validate our technique. HepG2 cells also proliferated on the stamps. Soft and stiff poly-dimethylsiloxane layers were also tested to demonstrate that our cost-effective process is compatible with biomimetic organ-on-chip technology integrating tunable stiffness with a potential application to drug testing probes development where such cells are commonly used.
Keywords: Hepatocytes, microcontact printing, poly-dimethylsiloxane, cell patterning, cell proliferation
Introduction
Nowadays, biomimetic cell culture platforms are enabling a better manipulation of biological cell behaviour under in vitro or in silico study. Thanks to a finer technological control of increasingly complex synthetic microenvironments; it is now possible to imitate the native physicochemical properties that biological tissues undergo in vivo and hence guarantee a phenotype, organisation and function resembling that encountered in the native conditions. That makes possible to study the desired cells with greater fidelity on a biomimetic chip.1
Indeed, in addition to well-studied molecular signalling, more recent considerations of how superficial mechanics, fluid dynamics and static three-dimensional (3D) microfeatures of host substrates are perceived by the cells have shown that the culture microenvironment has a direct correlation with phenotypic cues that are important to preserve when developing organ-on-chip devices or cell culture platforms for testing probes of drug metabolism or cytotoxicity for instance. The adhesion and viability of cells,2 response to external stimuli,3,4 metabolism,5 growth6 and fate7 are some of the most critical examples of traits that are deeply affected by environmental parameters that need to and can be controlled by technology to progress in the development of biomimetic devices. One particular drawback of traditional static Petri dish culture is that it has a tendency to induce relatively rapid cell transdifferentiation of cells in primary culture that in turn limits its use in long-term studies as the cells evolve and diverge from their natural phenotype which is under study.8
One of the preferred materials to fabricate biomimetic cell culture platforms for studies of cell physiology and mechanobiology is poly-dimethylsiloxane (PDMS). Some of its well-known advantages for cell studies are its optical transparency, tunable mechanical properties,9,10 gas permeability, flexibility and non-toxicity when polymerisation is complete.11 Thanks to its ease of use in microfabrication, and it is also an excellent candidate for cell culture inside microstructured lab-on-chip and organ-on-chip platforms.12,13 However, this silicone is a hydrophobic material and this represents a challenge for its application in long-term cell culture due to a poor cell adhesion leading to detachment or transdifferentiation.14 This lack of affinity for cells such as HepG2 hepatic cells (human hepatocellular carcinoma) is a limiting factor for PDMS as it affects the growth and organised confluence of the cells required for the obtention of the desired phenotype, for example, for the formation of organoids or drug testing platforms.15 In this case, the organisation and polarisation of cells in PDMS biomimetic microstructures is impossible without an adhesion promoter such as a protein present in the native surroundings of the cells (extracellular matrix) in vivo. Many efforts have thus been made in order to achieve a stable PDMS hydrophilic surface to improve cell adhesion. For example, a method in which PDMS is functionalised with aminosilane 3-aminopropyl triethoxysilane (APTES) and then crosslinked with glutaraldehyde (GA) has been reported,16 showing a reduction in contact angle with water characteristic of a hydrophilic surface (~70°). Furthermore, this method allows to covalently immobilise extracellular matrix proteins, stabilising them for a longer period of time and thus enhancing biomimetic design.
In addition to adhesion promoter and in order to allow for cell patterning on a chip, the protein coating has to be transferred in the form of a micropattern, as a means to guarantee a better-controlled arrangement of confluent regions where cells organise themselves.17 One of the most common techniques to achieve a simple, rapid and cost-effective biomimetic cell patterning is microcontact printing (µCP), useful for transferring structured protein features from a microstructured stamp onto a host substrate.18 Although the feasibility of µCP on PDMS has been proven in previous reports,19–21 the obtention of the micropatterned stamps is not an easy task as it typically requires costly multi-step photolithographic methods, and the host substrates usually require preliminary chemical treatment, hence preventing the wide integration of µCP patterns in polymeric lab-on-chip devices. Moreover, PDMS is not a very suitable material to undergo microcontact printing of extracellular matrix due to its high hydrophobicity. Although a recent work has demonstrated the use of fibronectin and laminin on this particular polymer for nerve and muscle cells,22 we have been unable to reproduce a complete, stable transfer of the most common extracellular matrix, type I collagen (COL I), on PDMS using this procedure without further treatment of the surface as COL I structure is more complex and hence less stable on PDMS for such applications.
In this work, we present a cost-effective technique to print stable, resistant micropatterns of COL I on PDMS previously treated with APTES-GA. After employing a low-cost, custom-made laser setup to fabricate microstructured poly-methyl methacrylate (PMMA) mould with the desired patterns in a single-step fashion,23 rigid PDMS stamps were fabricated by soft lithography and then inked with COL I before transferring the patterns onto PDMS substrates treated with APTES-GA adapting previously reported procedures16,24 to our process. Our technique offers the possibility to design and fabricate protein patterns on PDMS chips that can resemble the organisation of cells in an in vivo environment and are easily integrable in lab-on-chip and organ-on-chip platforms. Human hepatoma HepG2 cell line was selected to validate our process as such cells do not adhere on PDMS without COL I.25 Indeed, another important characteristic to pursue when biomimicry is sought to maintain a certain phenotype when cultured is a local planar26 or 3D organisation of the cells that ensures conditions similar to that of the desired tissue source. It is of particular importance for naturally polarised cells, such as epithelia, subject to an epithelial–mesenchymal transition when cultured in a foreign environment, as it occurs with hepatocytes.27 So far, hepatic spheroids have been used as 3D culture model for drug testing and toxicity studies.28 However, there are several disadvantages in spheroid cultures such as oxygen gradients within the aggregates, the absence of vascularisation, uniformity in cell proliferation and lack of acquisition of any biomimetic environment.29 Nakao and collaborators demonstrated that by confining primary hepatocytes in a thin linear space, they are arranged naturally in rows of one or two neighbouring cells similar to the hepatic cords present in hepatic lobules. They showed that hepatocytes present a proper organisation with a greater level of biomimicry: enclosed inside a microfluidics channel allowing nutrients exchange made them even capable of producing bile canaliculi on a chip.30 Unfortunately, the impact of cell attachment to extracellular matrix proteins or the influence of a substrate stiffness similar to liver tissue were not tested. The biomimetic geometry of the micropatterns was designed to organise a line of only a few cells alike the hepatic cord and study its impact on the cell viability, proliferation and organisation for further use in drug metabolism test probes, where a phenotype similar to that of an adult human hepatocyte is desired. Finally, after showing that our patterned scaffolds are useful to guarantee a cell behaviour more appropriate for lab-on-chip cell studies, as the biomimetic level increases, we report successful and very promising cell adhesion, viability and organisation of primary rat hepatocytes in an hepatic cord–type structure.
Materials and methods
PDMS host substrate preparation
As mentioned, PDMS is an ideal candidate to fabricate biomimetic cell culture platforms. In this work, it was used as a host substrate with a tunable stiffness for protein patterns transfer to validate our process aimed at organising biological cells on a microstructured biomimetic lab-on-chip platform. Sylgard 527 and Sylgard 184 (Dow Corning) were used for soft kPa-range and stiff MPa-range substrates, respectively, and glass was used as control. In case of the preparation of soft substrates, the prepolymer and the curing agent were mixed for 5 min in a 1:1 w/w ratio. Stiff substrates were prepared by mixing 10 parts of prepolymer with one part of the curing agent. Both mixtures were degassed under vacuum for 20 min, poured on the top of clean circular coverslips with a diameter of 18 mm and cured for 48 h at 60°C.
For protein immobilisation, PDMS substrates were silanised with APTES (440140; Sigma-Aldrich) followed by a treatment with GA (340855; Sigma-Aldrich), as can be seen in Figure 1. Some modifications were made to existing procedures16,24 as detailed in the following. A stock APTES solution was prepared by mixing 50% methanol, 47.5% APTES and 2.5% deionised water. It was stored for 1 h at 4ºC. The working solution was prepared at the moment diluting the stock solution 1:500 in methanol (the concentration of this solution was 0.095%).
Figure 1.
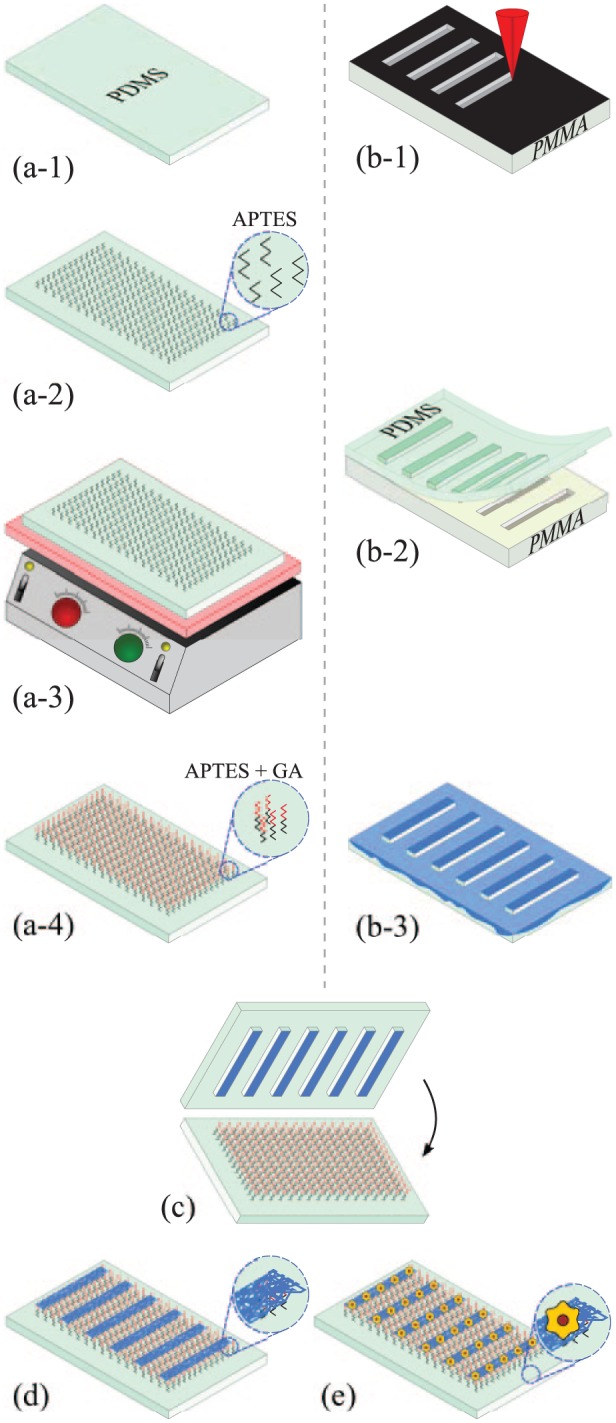
Fabrication process of micropatterned scaffolds: (a-1) soft and stiff PDMS host substrates were used; (a-2) surface chemical modification by silanisation with APTES; (a-3) thermal treatment on a hotplate at 110°C; (a-4) surface activation with GA; (b-1) laser-etching of a PMMA slide with the desired microstructures; (b-2) obtention of a stiff PDMS stamp by replica moulding; (b-3) incubation of the PDMS stamp with type I collagen; (c) microcontact printing onto the chemically modified PDMS host substrate; (d) generation of COL I micropatterns; and (e) culture and organisation of HepG2 cells or primary rat hepatocytes on COL I patterns.
The surface of PDMS host substrates was exposed to ultraviolet light in the presence of ozone in a UV/Ozone ProCleaner (BioForce Nanosciences) for 15 min and then immersed in the APTES working solution for 2 h. At the end of incubation, the substrates were washed three times with methanol and thermally baked on a hotplate at 110°C for 30 min. The last step of the surface modification was the immersion of the substrates in a 2.5% GA aqueous solution for 1 h. After removing the solution, substrates were rinsed three times with deionised water.
Micromould fabrication
The micromoulds used in this work for microcontact-printing of collagen on PDMS were fabricated using a simple, direct laser ablation technique based on previous work that manages to avoid costly photolithographic steps and materials with a similar pixel resolution of 4 µm.23,31 Clean PMMA sheets were etched locally using this procedure to fabricate 17-µm-deep design-specific features with controlled width, pitch and length in a rapid-prototyping fashion, following the procedure reported in López-Aparicio et al.23 and as shown in Figure 1(b-1). The moulds were characterised after etching by profilometry (KLA Tencor D600) to verify the structural integrity and geometry of the microstructures before replicating on PDMS microstamps (see Supplementary Data). As said, although any desired pattern with different dimensions may be transferred using our low-cost technique, as reported in López-Aparicio et al.,23 only straight lines imitating hepatic cords were fabricated in this work to study how the confinement of hepatic cells in such COL I features promoting adhesion on PDMS induce a phenotype that is suitable for drug testing and organ-on-chip platforms.
Microcontact printing
Stamps for microcontact printing are routinely fabricated of PDMS Sylgard 184 in a 10:1 w/w proportion of prepolymer and curing agent. Indeed, rigid structures are desired for better pattern transfer of the ink in microcontact printing.32,33 After preparing the liquid polymer following the procedure used to construct the stiff substrates, the material was poured on the micromoulds and cured at 60°C for 48 h. Such temperature did not affect or deform the moulds and ensure a perfect replication of the features on the stamps. The PDMS stamps were peeled from the micromoulds and cut with a scalpel.
The stamps were placed in a UV/Ozone Procleaner for 15 min in order to oxidise its surface and increase its hydrophilicity. Then, a COL I solution (BD Biosciences) was prepared at a concentration of 0.1 mg/mL for HepG2 cells and 1 mg/mL for primary rat hepatocytes, using acetic acid 20 mM to dissolve collagen. The surface of the stamp was covered with a drop of 50 µL of this solution and incubated for 10 min at room temperature (RT). After the protein solution was aspirated, the stamps were washed three times with phosphate-buffered saline (PBS) and air-dried.
Finally, the stamps were placed on the substrates and pressed for 5 min with a 20 g weight. Stamps were then rinsed three times with PBS solution and sterilised by soaking in sterilised PBS and exposed to UV light for 30 min.
Characterisation of PDMS substrates
PDMS stiffness
PDMS micromechanical properties were characterised by a Micromechanical Testing and Assembly System FT-MTA-02 (FemtoTools). By indenting a 50 µm sphere into the PDMS slabs and measuring force against indentation distance, it was possible to evaluate Young’s moduli of the substrates used in this work using the Hertz model. Stiff PDMS presented Young’s modulus of 1–2 MPa, while soft PDMS layers showed a 12–13 kPa modulus. The measurements were very consistent at different positions on each sample and from run to run; more than five different runs were used to calculate the average values reported here.
Characterisation of microstamps
Microstamps were obtained by a conventional replica-moulding technique of the microstructured acrylic moulds using Sylgard 184. Moulds were characterised using a profilometer to measure an average depth of 17 µm for the microstructures. The stamps were characterised by optical microscopy (Figure 2) and with the same profilometry technique, confirming the correct and accurate replication of the patterns etched in PMMA onto the PDMS stamps. As said, Alexa594 fluorescence assays were used to finally verify the transfer of micropatterns on the substrates (Figure 2(c)).
Figure 2.
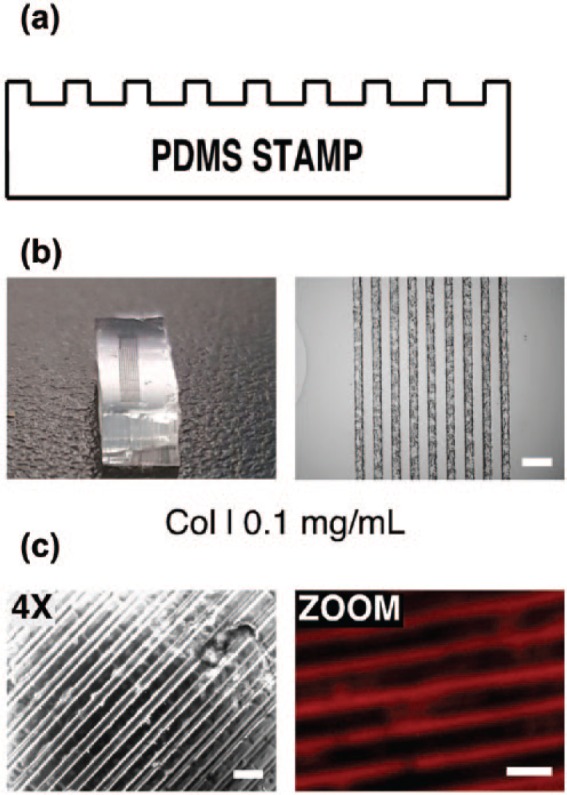
Details of microstructured PDMS stamps for microcontact printing: (a) diagram of the transversal section of a PDMS stamp showing the microstructure; (b) example of a stamp cutted in 0.5 cm × 1 cm and with an array of lines with a 80 width and with 80 μm spacing (scale = 200 μm); and (c) micropatterns of type I collagen (COL I) deposited onto soft PDMS surface. (Left: objective 4× micrograph (Nomarski); right: ZOOM micrograph of anti-collagen type I immunofluorescence (red). Scale = 200 μm.)
HepG2 and rat hepatocyte primary cultures
HepG2 cell line was maintained in modified essential medium (MEM) supplemented with 10% fetal bovine serum (FBS) plus sodium pyruvate and antibiotics (penicillin/streptomycin). Hepatocytes were isolated from livers of male Wistar rats approximately 200–250 g of weight. Animals were maintained on an ad libitum diet and handled according to internal institutional guidelines and ethical agreements (Instituto de Fisiología Celular, Universidad Nacional Autónoma de México) for animal experimentation. Rat hepatocytes were isolated using the modified collagenase perfusion method from Berry and Friend as previously described in Caligaris et al.34 After digestion, hepatocytes were separated by centrifugation at 400 r/min for 2 min from non-parenchymal cells, and viable hepatocytes were isolated by iso-density percoll centrifugation (Amersham-GE Life Science). Cell viability was evaluated by trypan blue exclusion staining. For primary culture, hepatocytes were seeded on coverslips coated with PDMS and treated with 1 mg/mL rat tail collagen type 1 (BD Biosciences), and cells were cultured for 4 h at 37°C in attachment medium supplemented with 10% FBS and antibiotics. Then, media was changed for feeding medium (FBS-free) and hepatocytes were cultured for 24 h for further studies.
HepG2 cell transfection
HepG2 cell line, stably overexpressing mTurquoise2-coupled LifeAct, was generated by transfecting 5 µg of LifeAct-mTurquoise2 plasmid with 10 µL of Lipofectamine® 2000 as manufacturer describes (Invitrogen); the plasmid No. 36201 was obtained from Addgene (Goedhart et al., 2012). After 48 h post-transfection, cells were treated with 900 µg/mL of Geneticin® (G418) from GIBCO for stable cell selection. LifeAct-mTurquoise2 stable expression was analysed by confocal microscopy as previously reported.35
Fluorescence microscopy
HepG2 cells and rat hepatocytes were seeded on collagen-treated glass or PDMS-coated coverslips in 12-well tissue culture plates and cultured at different days as indicated. Glass was used as a GPa-range rigid control substrate commonly used in HepG2 culture and ideal for immunofluorescence assays. Those assays were performed as described in Vázquez-Victorio et al.35 Cells were fixed for 15 min with 4% paraformaldehyde in PBS at 37°C. Cells were permeabilised with 0.1% Triton X-100 in PBS for 10 min at RT and blocked with bovine albumin fraction V at 1% in PBS for 30 min. F-actin was stained with Alexa Fluor 594-coupled Phalloidin (Molecular probes, ThermoFisher), diluted 1:40. Actin-cytoskeleton dynamics was studied using two different approaches that stain actin filaments: (1) Phalloidin-coupled Alexa Fluor 594 and (2) LifeAct coupled to mTurquoise2 fluorescent protein. Cells were seeded on collagen-treated glass or PDMS-coated coverslips in 12-well tissue culture plates. Samples were mounted in glass coverslips (24 × 50 mm) using either VECTASHIELD mounting medium with 4′,6-diamidino-2-phenylindole (DAPI; Vector Laboratories) or staining nuclei with DAPI at 1:200 (Invitrogen) in PBS for 10 min and samples were mounted with Mowiol (Calbiochem) at RT. Cells were visualised in Zeiss LSM 800 Airyscan confocal microscope. Leica TCS SP8 confocal microscope and Olympus epifluorescence microscope IX71 were also used for image acquisition. ImageJ free software was used for imaging processing and analysis. For immunofluorescence, samples were incubated with 10% horse serum (HS) in PBS for 1 h at RT. Goat anti-COL 1 (1:100; SC-59772), goat anti-HNF4-α (SC-6556; 1:100) and goat anti-Albumin (SC-46293; 1:100) primary antibodies (Santa Cruz Biotechnology) were incubated in 10% HS PBS for 12 h at 4°C, and coverslips were incubated with anti-goat secondary antibody coupled to Alexa Fluor 594 (Jackson ImmunoResearch), diluted 1:500 for 1 h at RT. Anti-COL 1 was used to verify the proper transfer of COL I on the different substrates. Hepatocyte Nuclear Factor 4-α (HNF4-α) and albumin were used as specific biomarkers of hepatic phenotype and function in rat hepatocytes.
Cell adhesion, viability and proliferation
HepG2 cells and rat hepatocytes were seeded on collagen type I (#354236; Corning) treated glass or PDMS-coated coverslips in 12-well tissue culture plates. HepG2 cells were seeded at low density (1 × 105cells per well) and cultured for 4 or 48 h. After culture, cells were incubated with 1 µM Calcein AM (Molecular Probes, Thermo Fisher Scientific) and 5 µM propidium iodide (PI; P4170; Sigma) for 15 min in fresh medium at 37°C.36 Then, live and dead cells were visualised by epifluorescence or confocal microscopy, and images were processed and analysed in ImageJ software. Likewise, rat hepatocytes adhesion and viability were analysed after 24 h of culture.
Western blot
Confluent cells were lysed using radioimmunoprecipitation assay (RIPA) buffer (50 mM Tris-HCl (pH 7.5), 150 mM NaCl, 1 mM ethylenediaminetetraacetic acid (EDTA), 1% Nonidet-P40, 0.5% sodium deoxycholate and 0.1% sodium, dodecyl sulphate (SDS)) plus protease and phosphatase inhibitors and incubated 1 h at 4°C. Cell extracts were quantified by Bradford protein assay (#5000006; Bio-Rad) and normalised before loading in SDS-polyacrylamide gel electrophoresis (PAGE). After resolving, proteins were transferred to a polyvinylidene difluoride (PVDF) membrane, and it was incubated in blocking solution (5% of fat-free milk in Tris-buffered saline (TBS)-Tween 0.1%) for 1 h at RT, and primary antibodies were incubated overnight at 4°C. Later, membrane was washed and incubated by secondary antibodies in blocking solution for 1 h in RT. Proteins were detected by Immobilon Western chemiluminescent substrate (Millipore).
Statistical analysis
Student’s t-test was used to calculate statistical significance, and p < 0.05 was considered to be significant.
Results
HepG2 cell adhesion, viability and proliferation
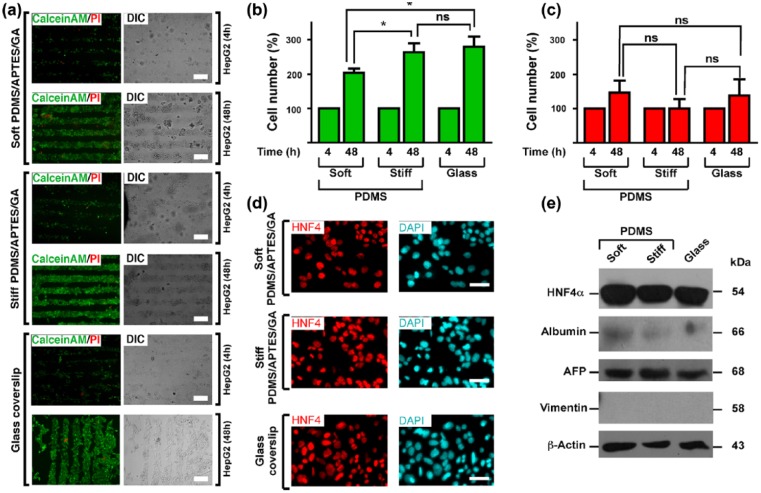
HepG2 cells were cultured on COL I micropatterns for 4 and 48 h. Cell viability was analysed by Calcein AM/propidium iodide assays. As we expected, HepG2 cells adhered exclusively on the COL I patterns and showed high cell viability after 4 and 48 h of culture (Figure 3(a)). Interestingly, HepG2 cells proliferated on the collagen micropatterns and were confined to the COL I stamps even when cells reached confluence (Figure 3(a)). Moreover, as can be observed from calcein levels in Figure 3(b), HepG2 cells seem to have a higher proliferation rate as the substrates grow stiffer and diverge from natural conditions (a few kPa). HepG2 cell monolayers cultured on collagen-treated glass or PDMS-coated coverslips showed nuclear HNF4-α staining (Figure 3(c)).
Figure 3.
High adhesion, viability and proliferation of HepG2 cells confined on stiff- and soft-PDMS/APTES/GA scaffolds patterned with COL 1 in lines: (a) HepG2 cell adhesion, viability and proliferation were evaluated by calcein AM/PI assay. HepG2 cells were seeded at low density (1 × 105 cells per well) on glass, stiff- and soft-PDMS/APTES scaffolds patterned by microcontact printing with collagen type I micropatterns. Live cells were stained with calcein AM (green) and dead cells were stained with propidium iodide (PI; red). Epifluorescence microscopy images were acquired 4 and 48 h after cell culture. Cells were visualised by differential interference contrast microscopy (DIC). Scale bars = 200 μm; (b and c) Viable and dead cells were presented as cell number percentage based on cell occupancy area for calcein (live cells/green bars) and PI (dead cells/red bars). Data are represented as mean ± SEM (n = 6). *p < 0.0318 (soft vs stiff, viable cells), * p < 0.0201 (soft vs glass, viable cells) and ns = not significant (stiff vs glass and in every condition on dead cells). (d) HepG2 cells were seeded at low density (1 × 105 cells per well) for 24 h on glass, stiff- and soft-PDMS/APTES scaffolds coated with 0.1 mg/mL COL I. Epifluorescence microscopy images show that HNF4-α marker exhibited a nuclear localisation. Nuclei were stained with DAPI. Scale bars = 50 μm. (e) Expression levels of protein makers of phenotype and function in primary hepatocytes and HepG2. Total extracts of freshly isolated hepatocytes and hepatocytes cultured for 24 h and 7 days were analysed, as well as HepG2 cells cultured for 24 h on soft PDMS, stiff PDMS and glass. Vimentin, albumin, AFP and HNF4 were detected by western blot. Molecular weights of each protein are indicated in kilodalton.
Fluorescence labelling of actin cytoskeleton
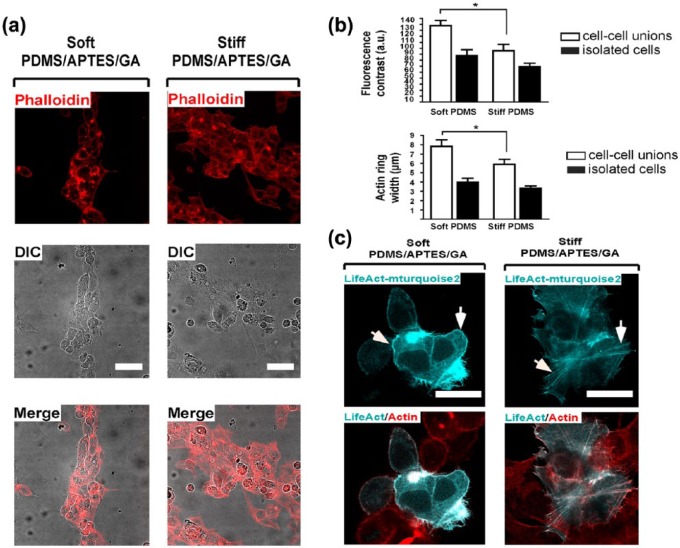
Confluent HepG2 cells were cultured on collagen stamps and the organisation of actin filaments was observed by Alexa Fluor 594-coupled phalloidin. Surprisingly, HepG2 cells cultured on collagen micropatterns on soft PDMS exhibited a greater cortical actin arrangement than cells cultured on stiff PDMS (as seen in Figure 4(a)). The width of the F-actin ring and its density, demonstrated in this case by fluorescence intensity contrast between the ring and the interior of the cells (Figure 4(b)), indeed shows a clear difference between soft and stiff PDMS for HepG2 culture on the microcontact-printed COL I stamps. A comparative analysis of F-actin staining with phalloidin and LifeAct showed that both methods detected a greater cortical actin layer on HepG2 cells cultured on soft PDMS (Figure 4(c)).
Figure 4.
HepG2 cells confined on soft-PDMS/APTES/GA rather than on stiff scaffolds patterned with COL I in lines exhibit a greater cortical actin arrangement. (a) Actin filaments were observed with Alexa594-coupled phalloidin in HepG2 cells. Cells were seeded at high density on stiff- and soft-PDMS/APTES scaffolds patterned by microcontact-printing with type I collagen in micropatterns. Differential interference contrast (DIC) and confocal microscopy images were acquired 48 h after cell culture. Nuclei were stained with DAPI. Scale bars = 50 μm. (b) Relative F-actin levels were measured as fluorescence contrast (a.u. = arbitrary units) and cortical actin ring width (μm). Data are represented as mean ± SEM (n = 6). *p < 0.021 (cell–cell unions, contrast) and **p < 0.008 (cell–cell unions, width). (c) HepG2 cells were seeded at low density (1 × 105 cells per well) for 24 h on stiff- and soft-PDMS/APTES scaffolds coated with 0.1 mg/mL COL I. F-actin was stained with Alexa594-coupled phalloidin (red) and LifeAct-mTurquoise2 (cyan). Scale bars = 30 μm.
Primary rat hepatocytes cultured on micropatterned collagen
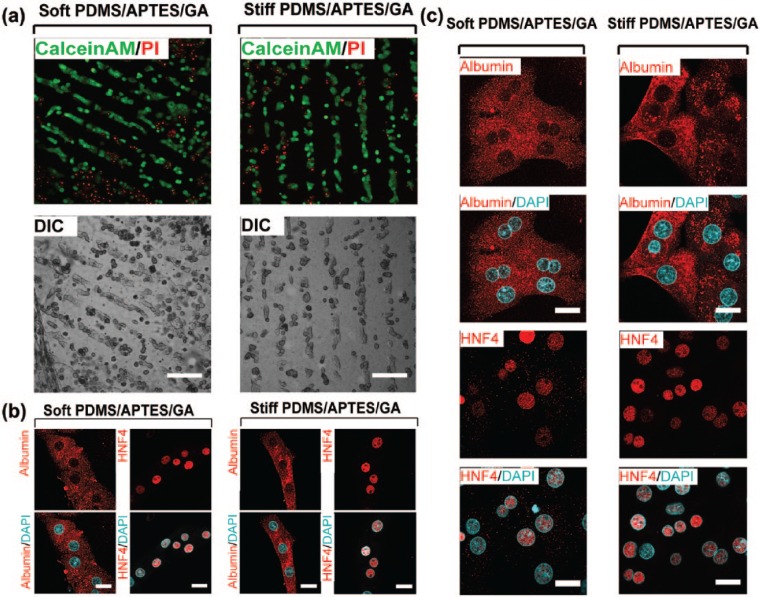
Here, primary rat hepatocytes were also cultured on similar platforms to those used for HepG2 cells in order to analyse cellular viability and organisation caused by confinement and stiffness. As can be seen in Figure 5(a) and (b), after 24 h of culture, hepatocytes showed high viability; intriguingly, hepatocytes were selectively attached to collagen micropatterns, promoting their confinement and organisation with an architecture that resemble a hepatic cord-like shape. Primary hepatocytes cultured on collagen-treated PDMS-coated coverslips showed cytoplasmic albumin and nuclear HNF4-α staining (Figure 5(b)). Figure 5(c) presents results of HNF4 and albumin on primary hepatocytes without any pattern as a control to validate that these cells present those hepatic markers.
Figure 5.
Confinement of primary hepatocytes cultured on stiff- and soft-PDMS/APTES/GA scaffolds patterned with COL I in lines. (a) Freshly isolated rat hepatocytes were seeded at high density (2 × 105 cells per well) on stiff- and soft-PDMS/APTES/GA scaffolds patterned by microcontact printing with type I collagen micropatterns. Primary rat hepatocytes viability was evaluated by calcein AM/PI assay. Live cells are shown in green and dead cells are shown in red. Differential interference contrast (DIC) and confocal microscopy images were acquired 24 h after cell culture. Scale bars = 100 μm. (b) Albumin (cytosol) and HNF4-α (nucleus) specific markers were expressed in primary rat hepatocytes cultured for 24 h at high density (2 × 105 cells per well) on stiff- and soft-PDMS/APTES/GA scaffolds patterned with 1 mg/mL COL I in lines. Scale bars = 20 μm. (c) Primary rat hepatocytes were cultured for 24 h at high density (2 × 105 cells per well) on stiff- and soft-PDMS/APTES/GA scaffolds coated with 1 mg/mL type I collagen. Confocal microscopy images show subcellular localisation of albumin (cytoplasm) and HNF4-α (nucleus) specific biomarkers of primary cultured hepatocytes. Nuclei were stained with DAPI. Scale bars = 20 μm.
Discussion
Characterisation of cell culture on collagen microstamps
Cell viability and proliferation
Because of many limitations in the use of isolated human hepatocytes, several hepatic cell lines are used as alternative models to study hepatocyte functions. Hepatoma cell lines such as HepG2 are used to study human hepatocyte functions as they still exhibit epithelial features; intriguingly, confluent HepG2 cells rather than spare cells enhance many of their hepatic metabolic functions.37 Our results suggest that it is possible to control hepatic cell proliferation on PDMS by tuning its stiffness, and it is very important to maintain this tendency on printed COL I micropatterns for organ-on-chip development.
Actin-cytoskeleton organisation in collagen stamps
Confluent HepG2 cells show cortical actin filaments along with cell–cell interactions when they reach confluence.37,38 We decided to study the influence of stiffness on actin-cytoskeleton organisation of HepG2 cells cultured on collagen micropatterns on PDMS in order to validate the compatibility of our process with organ-on-chip development. It is well known that substrate stiffness controls actin-cytoskeleton dynamics of hepatocytes, modifying their phenotype and functions.39 Actin-cytoskeleton arrangement observed in HepG2 cells cultured on soft and rigid PDMS-treated substrates supports that the substrates’ mechanical properties have differential effects on cell organisation and very likely on the rates of cell proliferation. Indeed, the assembly of cortical actin was promoted in HepG2 cells confined on soft substrates micropatterned with Col I, suggesting that the stiffness of soft PDMS seeks to resemble that of the native hepatic tissue, proving that biomimicry is critical for culturing cells.
Primary hepatocyte alignment on collagen micropatterns
It is well known that primary hepatocytes modify their phenotype during long-term culture (5–7 days), and these changes can be stopped or delayed by culturing hepatocytes in a collagen sandwich or with chemical treatments.40 Dedifferentiation of primary hepatocytes is promoted by long-term culture, which initiates with changes on cell cytoskeleton rearrangements, mainly in actin cytoskeleton.26 To the best of our knowledge, the influence of micropatterns of this particular extracellular matrix proteins on PDMS has not been tested in primary hepatocytes, although it is a very useful feature for lab-on-chip and organ-on-chip platform development. The obtained results demonstrate that our low-cost collagen microstamp transfer technique allows for the culture of primary hepatocytes in a cord-like shape on stiff and soft PDMS. This is very important to control hepatocytes phenotype by modulating actin-cytoskeleton organisation not only by stiffness tuning.9 Further studies are needed to clarify the effect of both PDMS stiffness and micropattern on primary hepatocyte phenotype. The hepatocytes’ confinement to collagen lines also would allow coculturing with other hepatic cells that are able to adhere to bare PDMS, such as fibroblasts.
Conclusion
We have presented a low-cost custom-made laser method to fabricate microstructured PDMS stamps for microcontact printing and shown its efficiency to transfer COL 1, a natural extracellular matrix protein required for hepatic cell adhesion on PDMS with a precise geometric control. Thanks to the control offered by the laser technique,23 and the geometry and dimensions of patterns can be tuned to construct the microarchitecture of different tissues. Simple thin lines imitating hepatic cord patterns allowed to confine and organise HepG2 and primary rat hepatocytes. They were successfully deposited on different stiffness, enabling a greater level of biomimicry on chip, as required in modern cell culture platforms. It is a technique that is also compatible with 3D PDMS scaffolds and organ-on-chip devices integrating microfluidics as it offers a very resistant and stable union of COL I on PDMS.41 Moreover, as suggested by other works,10,16 this method can be naturally adapted to study other cell types besides hepatocytes.
Acknowledgments
G.V.-V., M.M.-S. and M.H. contributed equally as senior authors and correspondents to this work.
Footnotes
Declaration of conflicting interests: The author(s) declared no potential conflicts of interest with respect to the research, authorship and/or publication of this article.
Funding: This project has received financial support from CONACyT (projects 246988 and 280317), DGAPA-PAPIIT (TA100315 and IT102017). Authors would like to thank Aarón Cruz Ramírez for the fabrication of the moulds, Diego Zamarrón Hernández for profilometry, Jehú López-Aparicio for technical support, José Alfredo Jiménez Medina for his help with confocal microscopy and Fernando García Hernández (Unidad de Imagenología, IFC). This paper constitutes a partial fulfilment of the Graduate Program in Materials Science and Engineering at Universidad Nacional Autónoma de México (UNAM); Escutia-Guadarrama L. acknowledges the scholarship provided by Consejo Nacional de Ciencia y Tecnología (CONACyT No. 353349).
References
- 1. Sun W, Chen YQ, Luo GA, et al. Organs-on-chips and its applications. Chinese J Anal Chem 2016; 44: 533–541. [Google Scholar]
- 2. Wells R. The role of matrix stiffness in regulating cell behavior. Hepatology 2008; 47: 1394–1400. [DOI] [PubMed] [Google Scholar]
- 3. Kamble H, Barton MJ, Jun M, et al. Cell stretching devices as research tools: engineering and biological considerations. Lab Chip 2016; 16: 3193–3203. [DOI] [PubMed] [Google Scholar]
- 4. Ho CT, Lin RZ, Chen RJ, et al. Liver-cell patterning lab chip: mimicking the morphology of liver lobule tissue. Lab Chip 2013; 13: 3578–3587. [DOI] [PubMed] [Google Scholar]
- 5. Choe A, Ha SK, Choi I, et al. Microfluidic gut-liver chip for reproducing the first pass metabolism. Biomed Microdevices 2017; 19: 4. [DOI] [PubMed] [Google Scholar]
- 6. Jayaraj V, Wangikar PP, Jadhav SS. Microfluidic device optimization for cell growth. In: IEEE 5Th International Nanoelectronics Conference, Singapore, 2–4 June 2013 New York: IEEE. [Google Scholar]
- 7. Olsen AL, Bloomer SA, Chan EP, et al. Hepatic stellate cells require a stiff environment for myofibroblastic differentiation. Am J Physiol Gastrointest Liver Physiol 2011; 301: G110–G118. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8. Godoy P, Schmidt-Heck W, Natarajan K, et al. Gene networks and transcription factor motifs defining the differentiation of stem cells into hepatocyte-like cells. J Hepatology 2015; 63: 934–942. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9. Cozzolino AM, Noce V, Battistelli C, et al. Modulating the substrate stiffness to manipulate differentiation of resident liver stem cells and to improve the differentiation state of hepatocytes. Stem Cells Int 2016; 2016: 5481493. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10. Natarajan V, Berglund EJ, Chen DX, et al. Substrate stiffness regulates primary hepatocyte functions. RSC Adv 2015; 5: 80956–80966. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11. Regehr KJ, Domenech M, Koepsel JT, et al. Biological implications of polydimethylsiloxane-based microfluidic cell culture. Lab Chip 2009; 9: 2132–2139. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12. Huh D, Torisawa Y, Hamilton GA, et al. Microengineered physiological biomimicry: organs-on-chips. Lab Chip 2012; 12: 2156–2164. [DOI] [PubMed] [Google Scholar]
- 13. Bhatia SN, Ingber DE. Microfluidic organs-on-chips. Nat Biotechnol 2014; 32: 760–772. [DOI] [PubMed] [Google Scholar]
- 14. Chuah YJ, Koh YT, Lim K, et al. Simple surface engineering of polydimethylsiloxane with polydopamine for stabilized mesenchymal stem cell adhesion and multipotency. Sci Rep 2015; 5: 18162. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15. Murray LM, Nock V, Evans JJ, et al. The use of substrate materials and topography to modify growth patterns and rates of differentiation of muscle cells. J Biomed Mater Res A 2016; 104: 1638–1645. [DOI] [PubMed] [Google Scholar]
- 16. Kuddannaya S, Chuah YJ, Lee MHA, et al. Surface chemical modification of poly(dimethylsiloxane) for the enhanced adhesion and proliferation of mesenchymal stem cells. ACS Appl Mater Interfaces 2013; 5: 9777–9784. [DOI] [PubMed] [Google Scholar]
- 17. Tamura T, Sakai Y, Nakasawa K. Two-dimensional microarray of HepG2 spheroids using collagen/polyethylene glycol micropatterned chip. J Mater Sci Mater Med 2008; 19: 2071–2077. [DOI] [PubMed] [Google Scholar]
- 18. Ruiz SA, Chen CS. Microcontact printing: a tool to pattern. Soft Matter 2007; 3: 168–177. [DOI] [PubMed] [Google Scholar]
- 19. Bernard A, Delamarche E, Schmid H, et al. Printing patterns of proteins. Langmuir 1998; 14: 2225–2229. [Google Scholar]
- 20. Brock A, Chang E, Ho CC, et al. Geometric determinants of directional cell motility revealed using microcontact printing. Langmuir 2003; 19: 1611–1617. [DOI] [PubMed] [Google Scholar]
- 21. Ricoult SG, Nezhad AS, Knapp-Mohammady M, et al. Humidified microcontact printing of proteins: universal patterning of proteins on both low and high energy surfaces. Langmuir 2014; 30: 12002–12010. [DOI] [PubMed] [Google Scholar]
- 22. Palchesko RN, Zhang L, Sun Y, et al. Development of polydimethylsiloxane substrates with tunable elastic modulus to study cell mechanobiology in muscle and nerve. PLoS ONE 2012; 7: e51499. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23. López-Aparicio J, Hautefeuille M, Herrera-Domínguez S, et al. Use of a CD laser pickup head to fabricate microelectrodes in polymethylmethacrylate substrates for biosensing applications. Biomed Microdevices 2017; 19: 5. [DOI] [PubMed] [Google Scholar]
- 24. Yadav AR, Sriram R, Carter JA, et al. Comparative study of solution-phase and vapor-phase deposition of aminosilanes on silicon dioxide surfaces. Mater Sci Eng C Mater Biol Appl 2014; 35: 283–290. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25. Van Midwoud PM, Janse A, Merema MT, et al. Comparison of biocompatibility and adsorption properties of different plastics for advanced microfluidic cell and tissue culture models. Anal Chem 2012; 84: 3938–3944. [DOI] [PubMed] [Google Scholar]
- 26. Sung JH, Shuler ML. Microtechnology for mimicking in vivo tissue environment. Ann Biomed Eng 2012; 40: 1289–1300. [DOI] [PubMed] [Google Scholar]
- 27. Alépée N, Bahinski A, Daneshian M, et al. State-of-the-art of 3D cultures (organs-on-a-chip) in safety testing and pathophysiology. ALTEX 2014; 31: 441–447. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28. Ramaiahgari SC, den Braver MW, Herpers B, et al. A 3D in vitro model of differentiated HepG2 cell spheroids with improved liver-like properties for repeated dose high-throughput toxicity studies. Arch Toxicol 2014; 88: 1083–1095. [DOI] [PubMed] [Google Scholar]
- 29. Katt ME, Placone AL, Wong AD, et al. In vitro tumor models: advantages, disadvantages, variables, and selecting the right platform. Front Bioeng Biotechnol 2016; 4: 12. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30. Nakao Y, Kimura H, Sakai Y, et al. Bile canaliculi formation by aligning rat primary hepatocytes in a microfluidic device. Biomicrofluidics 2011; 5: 022212. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31. Hautefeuille M, Cabriales L, Pimentel-Domínguez R, et al. New perspectives for direct PDMS microfabrication using a CD-DVD laser. Lab Chip 2013; 13: 4848–4854. [DOI] [PubMed] [Google Scholar]
- 32. Kaufmann T, Ravoo B. Stamps, inks and substrates: polymers in microcontact printing. Polym Chem 2010; 1: 371–544. [Google Scholar]
- 33. Huang YY, Zhou W, Hsia KJ, et al. Stamp collapse in soft lithography. Langmuir 2005; 21: 8058–8068. [DOI] [PubMed] [Google Scholar]
- 34. Caligaris C, Vázquez-Victorio G, Sosa-Garrocho M, et al. Actin-cytoskeleton polymerization differentially controls the stability of Ski and SnoN co-repressors in normal but not in transformed hepatocytes. Biochim Biophys Acta 2015; 1850: 1832–1841. [DOI] [PubMed] [Google Scholar]
- 35. Vázquez-Victorio G, González-Espinosa C, Espinosa-Riquer ZP, et al. GPCRs and actin-cytoskeleton dynamics. Methods Cell Biol 2016; 132: 165–188. [DOI] [PubMed] [Google Scholar]
- 36. Haugland RP, MacCoubrey IC, Moore PL. Dual-fluorescence cell viability assay using ethidium homodimer and calcein AM. US5314805 Patent, 1994. [Google Scholar]
- 37. Chang TT, Hughes-Fulford M. Monolayer and spheroid culture of human liver hepatocellular carcinoma cell line cells demonstrate distinct global gene expression patterns and functional phenotypes. Tissue Eng Part A 2009; 15: 559–567. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38. Ong S-M, He L, Linh NTT, et al. Transient inter-cellular polymeric linker. Biomaterials 2007; 28: 3656–3667. [DOI] [PubMed] [Google Scholar]
- 39. Schrader J, Gordon-Walker TT, Aucott RL, et al. Matrix stiffness modulates proliferation, chemotherapeutic response and dormancy in hepatocellular carcinoma cells. Hepatology 2011; 53: 1192–1205. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40. Ezzell RM, Toner M, Hendricks K, et al. Effect of collagen gel configuration on the cytoskeleton in cultured rat hepatocytes. Exp Cell Res 1993; 208: 442–452. [DOI] [PubMed] [Google Scholar]
- 41. Chuah YJ, Kuddannaya S, Lee MHA, et al. The effects of poly(dimethylsiloxane) surface silanization on the mesenchymal stem cell fate. Biomater Sci 2015; 3: 383–390. [DOI] [PubMed] [Google Scholar]