Abstract
Legionellaceae are intracellular bacteria known as important human pathogens. In the environment, they are mainly found in biofilms associated with amoebas. In contrast to the gammaproteobacterial family Enterobacteriaceae, which established a broad spectrum of symbioses with many insect taxa, the only instance of legionella-like symbiont has been reported from lice of the genus Polyplax. Here, we sequenced the complete genome of this symbiont and compared its main characteristics to other Legionella species and insect symbionts. Based on rigorous multigene phylogenetic analyses, we confirm this bacterium as a member of the genus Legionella and propose the name Candidatus Legionella polyplacis, sp.n. We show that the genome of Ca. Legionella polyplacis underwent massive degeneration, including considerable size reduction (529.746 bp, 484 protein coding genes) and a severe decrease in GC content (23%). We identify several possible constraints underlying the evolution of this bacterium. On one hand, Ca. Legionella polyplacis and the louse symbionts Riesia and Puchtella experienced convergent evolution, perhaps due to adaptation to similar hosts. On the other hand, some metabolic differences are likely to reflect different phylogenetic positions of the symbionts and hence availability of particular metabolic function in the ancestor. This is exemplified by different arrangements of thiamine metabolism in Ca. Legionella polyplacis and Riesia. Finally, horizontal gene transfer is shown to play a significant role in the adaptive and diversification process. Particularly, we show that Ca. L. polyplacis horizontally acquired a complete biotin operon (bioADCHFB) that likely assisted this bacterium when becoming an obligate mutualist.
Keywords: symbiosis, horizontal gene transfer, genome evolution
Introduction
Legionellaceae are mainly known as important human bacterial pathogens (Diederen 2008), although their life strategy is generally bound to biofilms where they live in intracellular symbiotic associations with amoebas and other protists (Fields 1996). Consequently, all known species have therefore been described either from water sources or clinical materials. The only known exception to this rule is the obligate symbiont of the rodent lice Polyplax serrata and P. spinulosa originally characterized by light and electron microscopy (Ries 1931; Volf 1991). Based on the 16S rDNA sequence, this bacterium was later on suggested to be a member of the genus Legionella (Hypsa and Krizek 2007). In this work, the transition from a typical legionella to an obligate symbiont was inferred from the presence of the bacterium in all tested louse individuals, suggesting their transovarial transmission, and from the typical shift in GC content. These traits are common for many obligatory symbionts living intracellularly in various insects. Typically, such bacteria reside in specialized organs, usually called bacteriomes, and are presumed to supply the host with some essential compounds, mainly vitamins and amino acids, missing in their diet (Douglas 1989). Their evolution/adaptation toward this role is usually accompanied by extensive modifications of their genomes, consisting mainly of gene (or complete function) losses and decrease of GC content (Woolfit and Bromham 2003). Although the latter is considered a sign of relaxed selection, the reduction of metabolic capacities results mostly from adaptive processes. In the course of evolution, these bacteria lose metabolic functions fulfilled by the host and retain (sometimes even acquire by horizontal transfer; Husnik et al. 2013; Nakabachi et al. 2013; Nikoh et al. 2014) functions essential for the host’s development and/or reproduction. Due to these processes, the bacteria can evolve an array of different life strategies, from parasites/pathogens to obligate symbionts. It was for example demonstrated that the notorious insect parasite Wolbachia turned into a mutualistic bacterium in the bedbug Cimex lectularius upon horizontal acquisition of an operon for biotin biosynthesis (Nikoh et al. 2014; Gerth and Bleidorn 2017).
Several decades lasting research on insect-bacteria symbiosis has revealed a broad variety of these associations and has resulted in many fundamental discoveries (Moran 1996; Ochman and Moran 2001; McCutcheon and Keeling 2014; Moran and Bennett 2014). One of the interesting outcomes is the broad diversity of bacteria which are generally capable to establish obligate symbiosis with insects, including such diverse taxa as Enterobacteriales, Rickettsiales, Blattabacteriaceae, and Flavobacteriales. However, within this broad bacterial diversity, several groups show a particular tendency to this behavior (e.g., Enterobacteriaceae, more specifically the genera Arsenophonus and Sodalis), whereas others are found less frequently. Legionellaceae is one of these rare groups, with the Polyplax-associated bacterium being the only case of such intracellular obligate insect symbiont. This raises an interesting question on how this bacterium established its unique symbiotic relationships with lice and what changes it underwent during its adaption to obligate symbiosis. In this respect, it is important that there is a solid body of genomic information for both the genus Legionella and many other bacterial symbionts from various blood sucking insects. In a recent genomic work, comparing 38 Legionella species, Burstein et al. (2016) revealed a surprising diversity of their genomic traits, including major differences in the most general characteristics such as genome size or GC content. The main focus of this genomic comparison, particularly important in respect to the evolution of legionellae virulence, were effectors, especially those responsible for entering the host cell. The computational analysis inferred several thousands of candidate effectors and indicated that many of these factors have been recently acquired by horizontal transfer. This shows the genus Legionella as a highly dynamic system with ongoing genome rearrangements and adaptations toward the intracellular lifestyle. The newly characterized legionella-like louse symbiont thus provides us with a unique opportunity to study the processes of the genomic changes and compare them with “free living” Legionella on one hand and to the unrelated enterobacterial symbionts on the other hand.
In this study, we approach this issue by exploring genomic features of the symbiont from P. serrata, a louse associated with several species of the genus Apodemus (wood mice) across Europe (Stefka and Hypsa 2008). Among the 78 described species of the cosmopolitan genus Polyplax, associated exclusively with rodents and insectivores (Durden and Musser 1994), P. serrata and closely related species P. spinulosa, are the only two species for which the presence of symbiotic bacteria (and their origin within the genus Legionella) has so far been determined (Hypsa and Krizek 2007). Since all sucking lice rely on the limited resources of the mammalian blood, the presence of similar obligatory symbionts in other Polyplax species is very likely, but their taxonomic/phylogenetic positions are difficult to anticipate. Generally, different groups of lice were shown to host different taxa of symbiotic bacteria (Allen et al. 2016) which provides evidence for several replacements or independent acquisitions of the symbionts during the lice evolution. Using complete genomic data, we now confirm the unique phylogenetic origin of the P. serrata symbiont within the genus Legionella. We also show that its genome followed an evolutionary route typical for obligatory symbionts, and underwent a surprisingly convergent evolution with the unrelated enterobacterial symbiont from the hominid lice. Finally, we show that its adaptation to the role of obligatory symbiont included horizontal acquisition of six coding genes in a biotin operon, in analogy to the mutualistic Wolbachia from bedbugs.
Materials and Methods
Sample Preparation
The specimens of Polyplax serrata were collected during autumn 2011 from Apodemus flavicollis mice trapped around Baiersbronn, Germany and stored in absolute ethanol at − 4 °C. Since the obligate bacterial symbionts are uncultivable outside the host cells, total DNA was obtained by extraction from whole abdomens of 25 louse individuals (QiaAmp DNA Micro Kit, Qiaqen). DNA concentration was assessed by the Qubit High Sensitivity Kit (Invitrogen) and 1% agarose gel electrophoresis.
Genome Sequencing and Assembly
The Polyplax sample was sequenced on one lane of Illumina HiSeq2000 (GeneCore, Heidelberg) using 2×100 paired-end reads (PE) library with an insert size of 150 bp. After quality checking and filtering in BBtools (https://jgi.doe.gov/data-and-tools/bbtools/; last accessed October 30, 2017), the resulting data set contained 309,892,186 reads in total. The reads were assembled using the SPAdes assembler v 3.10 (Bankevich et al. 2012), under default settings with the parameter careful, decreasing number of mismatches and indels. To check for possible presence of bacterial plasmid(s) in the data, we submitted complete assembly to the PlasmidFinder (Carattoli et al. 2014) with sensitivity set to three different thresholds (95%, 85%, and 60%). Phylogenetic affiliations of the contigs were determined by PhylaAMPHORA (Wu and Scott 2012). Of the total 124,985 contigs, 112 were assigned to the order Legionellales. Trimmed reads were mapped on these contigs and filtered using BWA v. 0.7.15 (Li and Durbin 2009) and retrieved by Samtools (Li et al. 2009). This set of reads was subsequently assembled by two alternative assemblers, SPAdes and A5 assembly pipeline (Coil, Jospin and Darling 2015). The latter software produced a closed 529,746-bp long genome. Its quality was checked and base calls were polished by Pilon v1.20 (Walker et al. 2014).
Genome Annotation
The genome was annotated using two tools services: RAST (Aziz et al. 2008), and PROKKA (Seemann 2014). The complete genome was deposited in GenBank with the accession number CP021497. To scan for potential horizontal gene transfer(s) (HGT), we retrieved the 50 most similar sequences for each protein using the BLASTp algorithm (Altschul et al. 1990) against the nr (nonredundant) protein database. Metabolic pathways for B vitamins and cofactors were reconstructed using KEGG Mapper (http://www.genome.jp/kegg/mapper.html; last accessed October 30, 2017) and EcoCyc database (https://ecocyc.org/; last accessed October 30, 2017). The absence of genes in important metabolic pathways was verified using BLAST searches. The number of proteins with transmembrane helices was predicted in TMHMM server (http://www.cbs.dtu.dk/services/TMHMM/; last accessed October 30, 2017) with the default parameters. The signal peptides were detected by SignalP online server (http://www.cbs.dtu.dk/services/SignalP/; last accessed October 30, 2017) with the cutoff values estimated by the program. The presence of CRIPSR repeats was checked using Geneious software (Katoh and Standley 2013). The origin and terminus of replication was determined in GenSkew online tool (http://genskew.csb.univie.ac.at/; last accessed October 30, 2017) according to the formula: GC skew = (G−C)/(G + C) and with the genome split into 9,000 windows.
Phylogenetic Analyses
Sixty-four proteins for a multigene analysis were selected based on the recent genomic study by (Burstein et al. 2016) and their orthologs in the L. polyplacis were determined using BLASTp search against the L. polyplacis genome. The sequences were aligned using MAFFT v. 1.3.5 (Katoh et al. 2002) implemented in the Geneious software. Equivocally aligned positions and divergent regions were eliminated by GBlocks (Castresana 2000) under the less stringent selection options. The alignments were created separately for each protein and multigene matrix was obtained by their concatenation. To minimize phylogenetic artifacts caused by rapid evolution and nucleotide bias of the symbiont sequences, we used PhyloBayes MPI v. 1.5a (Lartillot et al. 2013) with the CAT-GTR model and dayhoff6 amino acid recoding, and ran it for 32,000 generations. This approach has been previously shown to decrease phylogenetic artifacts affecting branching of symbiotic bacteria in bacterial phylogenies (Husnik et al. 2011).
For the candidate HGTs, that is, the biotin operon genes (see Results and Discussion), we prepared a representative set of orthologs covering several bacterial groups and used the alignment method described earlier to build amino acid matrices. The best evolutionary model for all matrices, determined in Prottest 3.2 (Darriba et al. 2011) by Akaike information criterion (AIC), was LG with a proportion of invariable sites and evolutionary rates separated in four categories of gamma distribution (LG + I+G). Phylogenetic reconstructions were done by maximum-likelihood analyses with 100 bootstrap replicates using PhyML v. 2.2.0 (Guindon et al. 2010) for each gene separately and also for a concatenated matrix of all six genes. Posterior probabilities for individual branches were determined by MrBayes v 3.2.6 (Huelsenbeck and Ronquist 2001) with the same evolutionary model (LG + I+G) and remaining parameters determined by the analysis. The analysis was run under the default four-chain setting for 10,000,000 generations. Convergence of all Bayesian analyses was checked in Tracer v1.6.0 (Rambaut et al. 2014), and for MrBayes also by the values of standard deviation of split (<0.01) and PSRF + (reached the value 1.0). Based on the availability for each analyzed gene, one of the following bacteria was used as an outgroup: Kurthia sp. (Firmicutes), Geobacter sulfurreducens (Deltaproteobacteria), or Cyanothece sp. (Cyanobacteria). Graphical representation of the trees was processed in FigTree (http://tree.bio.ed.ac.uk/software/figtree/; last accessed October 30, 2017) and Inkscape (http://www.inkscape.org/; last accessed October 30, 2017).
Comparative Genome Analyses
In order to reveal general similarity patterns and possible functional convergences between the symbiont of P. serrata and other symbiotic bacteria, we have treated the genomes and genes as communities and their components, respectively, and employed nonmetric multidimensional scaling (NMDS). This clustering analysis, routinely used in microbial ecology, produces an ordination based on a distance or dissimilarity matrix (Legendre and Legendre 2012). The dissimilarity matrix based on Bray–Curtis distances was calculated for 4,632 GOCs from 94 bacterial genomes (supplementary table S2 in supplementary file S1, Supplementary Material online). Draft genomes of poor quality have not been included. In particular we have omitted two genomes of high interest, that is, Riesia pediculischaeffi and Sodalis endosymbiont of Proechinophthirus fluctus (Boyd et al. 2016). All the genome data were retrieved as proteomes from GenBank and assigned to particular functional orthologs as defined by the COG database released December 2014 (Tatusov et al. 2000; Galperin et al. 2015) using rpsBLAST (Altschul et al. 1990) limited by a single hit and a maximal e-value set to 10−5. Although the data set includes “free living” Legionellaceae with large genomes, the matrix was transformed using a percentage proportion for each COG ortholog calculated from the COG sum of each genome. The matrix of 4,632 orthologs was converted into the biom format. The Bray Curtis dissimilarities were calculated and the NMDS analysis was performed using vegan package functions in R (Dixon 2003). In order to show the genome functional similarities among lice symbionts with distinct phylogenetic origin, a Neighbour Joining (NJ) tree was calculated in T-rex (Alix et al. 2012) using the same distance matrix.
Proposal for the Species Name of Legionella-like Endosymbiont
As we show that the symbiont clusters within the genus Legionella, we propose in accordance with the terms for species designation which have not been cultivated in a laboratory media the name “Candidatus Legionela polyplacis” sp. nov. (hereafter Legionella polyplacis for simplicity). The specific name “polyplacis” refers to the genus of its insect host, the louse Polyplax serrata. The bacterium is the only member of the genus for which endosymbiotic association with insects has been documented.
Results and Discussion
Basic Genomic Properties
The complete genome of Legionella polyplacis is 529,746 bp long, has an extremely low CG content (23%) and a coding density 84.8%. According to the RAST annotation (supplementary table S3 in supplementary file S1, Supplementary Material online), it contains 484 protein encoding genes (pegs) with the average length of 939 bp, 3 genes coding for rRNAs and 36 genes for tRNAs. In two cases, pairs of adjacent pegs, annotated by identical names, corresponded clearly to a gene interrupted by a stop codon (DNA polymerase I [EC 2.7.7.7] and COG1565: Uncharacterized conserved protein). Since we confirmed the presence of the stop codons by mapping the reads on the assembled genome, we suppose that these cases represent either a functional split into two separate genes or an early stage of pseudogenization. To avoid an arbitrary determination of other possible pseudogenes, based on the sequence lengths, we provide in supplementary table S3 in supplementary file S1, Supplementary Material online, the comparison of length ratios between the L. polyplacis genes and their closest BLAST hits. In summary, of the 484 coding genes, only 48 are shorter than 90% of the BLAST-identified closest homologs, 23 of them shorter than 80%. The genome size and gene number place L. polyplacis among many other obligate symbionts of insects. In table 1, we show the comparison of these general genomic characteristics between typical legionellae, L. polyplacis and other hominid louse symbionts. The positions of the origin and terminus of DNA replication are shown on the GC-skew plot (supplementary file S4, Supplementary Material online), corresponding to the minimum and maximum values, respectively. The origin position coincides with the location of dnaA gene.
Table 1.
Comparison of the Main Genomic Characteristicsa
| Legionella spp. | Legionella polyplacis | Endosymbionts of Hominid Liceb | |
|---|---|---|---|
| Genome size (bp) | 2,367,087–4,818,052 | 529,746 | 528,700–580,415 |
| Number of proteins | 1,984–3,986 | 484 | 444–529 |
| CG content (%) | 36.60–51.10 | 23 | 23.75–31.80 |
| Number of COGs | 1578–2603 | 452 | 428–508 |
| ø coding density (%) | 85.43–93.10c | 86.50 | 79.17–85.96 |
| ø gene size (bp) | 744–1,113 | 931 | 849–896 |
The source data are provided in supplementary table S1 in supplementary file S1, Supplementary Material online.
The Riesia phthiripubis genome was not included into the comparison due to the presence of 444 psedogenes in the NCBI annotation (CP012846.1).
Coding densities were calculated only for the species with complete annotations available.
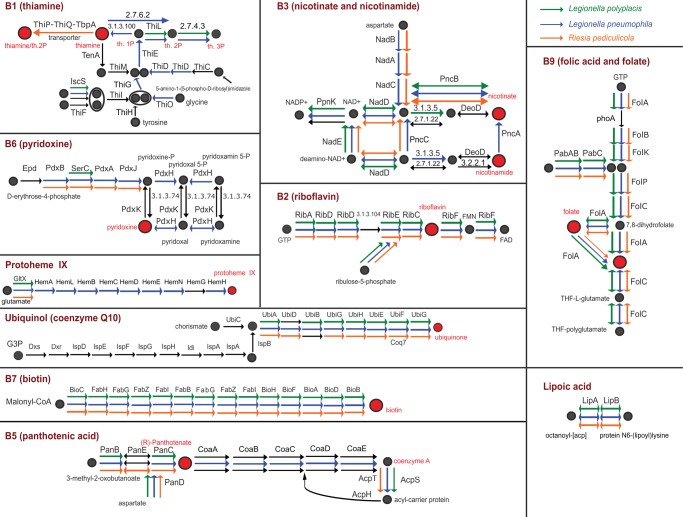
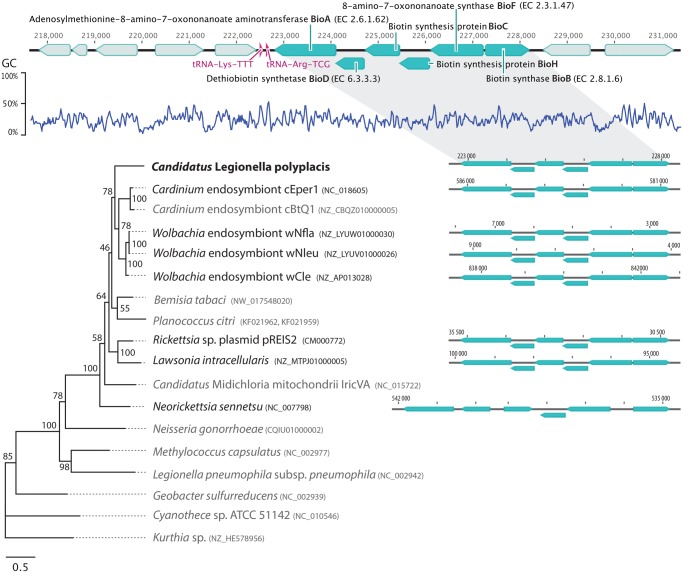
Of the coding genes, 105 contain transmembrane helices and 4 signal peptides. No CRIPSR repeats were identified. PlasmidFinder, run under the sensitivity range 95–60%, did not reveal any candidate plasmid sequence. Majority of the coding genes (445 genes) could be assigned by BLASTp unequivocally to the genus Legionella (i.e., 5 best hits corresponded to the genus Legionella), for 20 genes the search returned Legionella together with some other bacterial genera, and for 30 genes we either did not get any match or the hits corresponded to other bacteria than Legionella. Among the “non-legionella” genes, some were highly conserved (ribosomal proteins) or very short genes difficult to assign based on the BLAST algorithm. However, six of these “non-legionella” genes formed a complete biotin operon known from several nonrelated symbiotic bacteria, suggesting its horizontal acquisition in the L. polyplacis). A significance of this HGT in respect to the symbiotic nature of this legionella is discussed below. The genes with no BLAST hits were mostly annotated as hypothetical proteins, usually with very short sequences. Of the metabolic functions considered particularly significant in the endosymbiotic bacteria, that is, biosynthesis of the vitamins and cofactors, we detected several complete pathways (fig. 1) and the horizontally acquired capability of biotin synthesis (fig. 2).
Fig. 1.
—Comparison of B-vitamins and cofactors pathways for Ca. Legionella polyplacis, Riesia pediculicola, and Legionella pneumophila. Black arrows designate missing genes. Ellipses around two metabolites highlight the same following steps in the pathway.
Fig. 2.
—Structure of the horizontally acquired biotin operon in Ca. Legionella polyplacis and its putative evolutionary origin, based on ML analyses of all available genes. Blue arrowhead blocks represent genes for biotin synthesis organized in an intact operon. The adjacent genes code for the following proteins: for BioA from right to left: hypothetical protein, Isopentenyl-diphosphate delta-isomerase FMN-dependent, 4-hydroxy-tetrahydrodipicolinate synthase, 4Fe-4S ferredoxin, TsaB protein; for BioB from left: Aminomethyltransferase (glycine cleavage system T protein), and Glycine dehydrogenase [decarboxylating] (glycine cleavage system P2 protein). The position of the complete operon is visualized in the respective genomes (black font). The species with missing genes or disrupted operon structure (Legionella pneumophila) are in gray. The numbers at the tree nodes stand for the bootstrap values.
Origin and Phylogeny
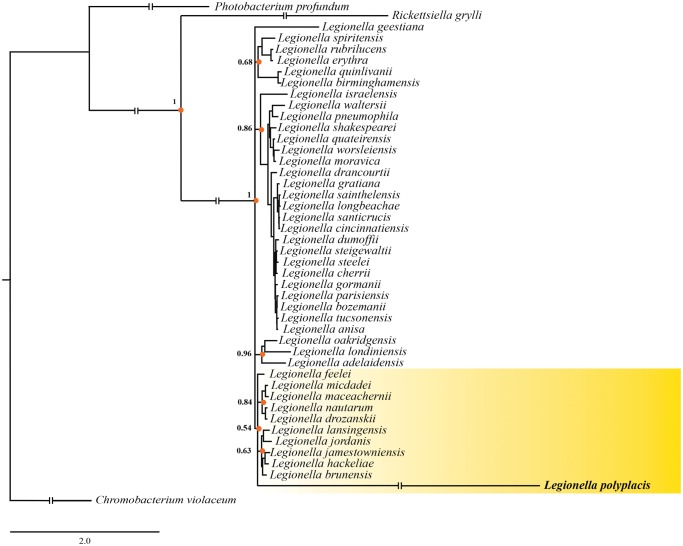
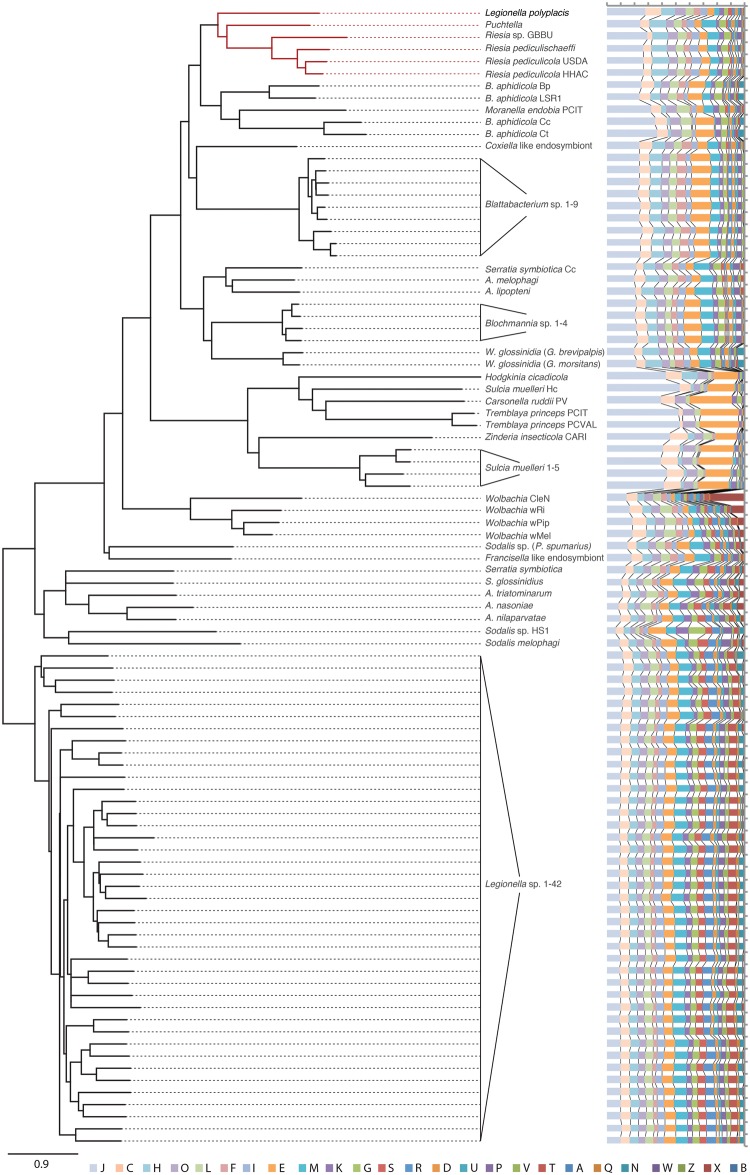
The multigene analyses confirm that L. polyplacis, the symbiotic bacterium from the lice of the genus Polyplax, have indeed originated within the genus Legionella (fig. 3 and supplementary file S2, Supplementary Material online), as previously suggested based on 16S rDNA (Hypša and Křížek 2007). Although in the PhyloBayes analysis the chain did not converge even after 32,000 generation, the maximum likelihood PhyML analysis yielded an identical result in respect to the L. polyplacis position. In both analyses, it clustered on an extremely long branch, but with a strong support, within the group corresponding to the L. micdadei clade of Burstein et al. (2016). This position was further supported by a BLAST analysis of the genes conserved in all species, including the symbiont, where the first eight hits belonged to the L. micdadei clade (i.e., L. feeleii, L. lansingensis, L. brunensis, L. hackeliae, L. jamestowniensis, L. maceachernii, L. jordanis, and L. nautarum). The basic structure of the whole Legionella cluster (i.e., monophyly of the main groups) corresponds to that reported by Burstein et al. (2016). The PhyML-derived topology retains closer similarity to their ML-based tree. This general agreement among results of our ML, Bayesian inference based on the recoded matrix, and the Burstein’s et al. (2016) topologies indicate that the data provide a reliable information for reconstructing the relationships within this group.
Fig. 3.
—Phylogenetic tree inferred by PhyloBayes analysis of concatenated 64-gene matrix. The orange dots (with posterior probability values) highlight arbitrary selected monophyletic clusters as main components of the tree structure. The interrupted branches were shortened by 50%.
The position of L. polyplacis deep within the Legionella phylogeny clearly indicates that it most likely evolved from a “free living” ancestor with similar ecology to other legionellae. Considering the capability of the legionellae species to switch between symbiotic forms living in amoebas and pathogenic forms infecting mammal cells, it is difficult to hypothesize which of these forms gave rise to the mutualistic L. polyplacis. Although the L. micdadei clade possess several distinctive features when compared with the L. pneumophila clade, for example, smaller genomes, lower number of effectors, general tendency to loosing genes (Burstein et al. 2016), their possible evolutionary significance is unclear. More generally, due to the considerable reduction of L. polyplacis genome and our current tools for exploring this association, it is impossible to deduce which of the inherited genetic factors and traits could play role in transition toward the nutritional insect symbionts and how much were these factors taxonomically specific. For the same reason, no meaningful differential comparison could be done for L. polyplacis and other Legionella species.
Genomic Evolution
Comparing the genome size of L. polyplacis (529,746 bp and 484 coding genes) to that determined for other legionellae shows that similarly to other insect symbionts, the genome of L. polyplacis experienced considerable reduction in course of its adaptation to the symbiotic lifestyle. Although the 38 Legionella spp. compared by Burstein et al., (2016) varied considerably in their genome sizes (2–5 Mbp; 2,000–4,500 genes), they do not approach the degree of reduction determined for the L. polyplacis genome. Due to this dramatic reduction, L. polyplacis lost many complete pathways and systems present in other Legionella spp. Since provisioning B-vitamins and cofactors is considered as fundamental function of symbionts in blood-sucking insects (Nogge 1981; Hosokawa et al. 2010; Snyder et al. 2010), we assessed this metabolic capacity of L. polyplacis in comparison to its phylogenetic relative Legionella pneumophila and phylogenetically distant louse symbiont Riesia pediculicola (fig. 1). All three bacteria possess complete pathways for riboflavin (B2), biotin (B7), lipoic acid, and folate (B9); the last one without a known enzyme responsible for removing the triphosphate motif from 7,8-dihydroneopterin 3′-triphosphate. The rest of the pathways are in various stages of incompleteness/degradation, as demonstrated in figure 1. Perhaps the most striking example of genomic degradation is a complete lack of virulence-related secretion systems in the L. polyplacis genome. The “free living”/pathogenic legionellae, like other intracellular bacteria, possess molecular machinery for entering the host cell, surviving, and reproducing. Major components of this machinery are the Dot/Icm type IVB secretion system, Lsp type II secretion system, and depending on the species, also various forms of the type IVA secretion systems (Joseph et al. 2016). None of these systems is encoded by the L. polyplacis genome, and its only remaining secretion machinery is thus the Sec-SRP system. This is in line with the suggested characteristic of L. polyplacis as an obligate mutualist. This type of symbionts are highly adapted to their host and in contrast to the facultative secondary symbionts (Masui et al. 2000; Dale et al. 2001; Wilkes et al. 2010), they often do not use secretion systems (Perez-Brocal 2006). Another capability often lost or compromised in the insect bacterial symbionts is the cell wall formation and shape determination (Moran et al. 2008). In supplementary table S4 in supplementary file S1, Supplementary Material online, we show a more detailed comparison of the L. polyplacis genome capacity to that of L. pneumophila, R. pediculicola, and Wigglesworthia glossinidia. To make the overview also comparable to other bacterial symbionts, we followed the pathways/genes list published by Moran et al. (2008) for a broader taxonomic range of insect symbionts. As shown in this table, L. polyplacis retains the capacity to synthesize peptidoglycan, but it lost the genes required for building the lipopolysacharide outer layer, particularly for the lipid A biosynthesis. However, compared with the “free living” legionellae, the peptidoglycan pathway in L. polyplacis lacks penicillin binding protein class A (aPBP). Interestingly, the same applies to the R. pediculicola genome. In addition, both L. polyplacis and R. pediculicola possess all four genes required for rod shape determination (ftsZ, ispA, mreB, rodA). Considering the high degree of completeness of the cell wall formation, it is likely that the function of missing aPBP is substituted by another enzyme. Such possibility is in line with the most recent discoveries showing that some of the cell wall formation functions can be fulfilled by the SEDS (shape, elongation, division, and sporulation) proteins (Meeske et al. 2016). More specifically, in Bacilus subtilis the function of the aPBP was shown to be replaced by the rodA enzyme, also present in the L. polyplacis and R. pediculicola genomes. Among the other major systems, a significant degradation occurred in the biosynthesis of amino acids with no complete pathway retained, and in the recombination/repair system with a high proportion of lost genes (supplementary table S4 in supplementary file S1, Supplementary Material online).
Both of the characteristics discussed earlier, that is, the strong compositional bias of the sequences toward GC, and considerable genome reduction, including loss of complete pathways and metabolic competencies, clearly place L. polyplacis among other obligatory mutualists of insects, despite the absence of a direct functional evidence.
Since nothing is known about the origin of bacterial symbionts in Polyplax species other than P. serrata and P. spinulosa, it is difficult to estimate the time frame of this reduction. In phylogenetic study published by Light et al., (2010), the family Polyplacidae are revealed as a paraphyletic taxon, with the genus Polyplax branching as sister group to a Pediculidae/Phthiridae/Pedicinidae cluster. The estimated diversification time for these two groups, ∼45 Ma, can thus be considered an upper limit for the origin of L. polyplacis. This would place L. polyplacis close in age to another obligatory symbiont associated with blood sucking insect, namely Wigglesworthia glossinidia, with the estimated origin of 40 Ma (Naito and Pawlowska 2016). However, since the Polyplax’s long branch in the Light et al. (2010) analysis indicates that other closer relatives of Polyplax might be missing in the taxa set, this time frame may be considerably overestimated. For example, another louse symbiont, Riesia pediculicola from the human louse, was shown to had reached similar genome state within ∼13–25 Ma (Boyd et al. 2014).
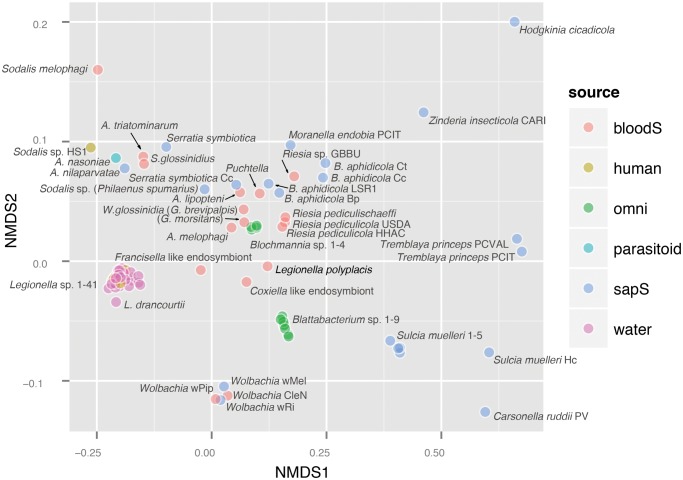
Regardless the uncertainty in time estimates, L. polyplacis, R. pediculicola, and the other recently described symbionts of lice (Boyd et al. 2017) reached similar basic genome characteristics (table 1). More interestingly, the NMDS analysis, depicturing the overall similarities among the genomes (fig. 4), and particularly the distance based NJ tree (fig. 5) placed L. polyplacis far from other legionellae but remarkably close to the phylogenetically unrelated lice symbionts from the genus Riesia (i.e., member of the Arsenophonus cluster). In the NJ tree, all of the louse symbionts, including Puchtella, even form a monophyletic lineage. It is important to notice that the clustering is not determined by the genome size as the other highly modified symbionts are scattered across the whole NMDS plot. Even more interestingly, the primary symbionts associated with blood feeding pupiparans, that is, Wigglesworthia spp., Arsenophonus melophagi, and A. lipopteni, cluster within a distant independent group relatively close to each other. In other words, the symbionts of blood feeding insects show clear tendency toward clustering according to their host phylogeny (and hence perhaps biology) rather than their own phylogeny. Although the sample of the symbionts from the blood-feeding insects is small for any decisive inference, the pattern suggests that the clustering reflects the convergence in the genome contents and consequently their functions rather than a general genome reduction and the host’s source of nutrients.
Fig. 4.
—NMDS analysis based on Bray–Curtis dissimilarities calculated for the genome content of selected bacteria analyzed across all COG orthologs. The position of each genome represents as closely as possible the pairwise dissimilarity between the genomes. The legend abbreviations for the bacterial source are as follows: bloodS: blood sucking host, human: clinical isolate, omni: omnivorous insect host, parasitoid: that is, Nasonia vitripennis host, sapS: sap sucking insect host, water: environmental water sample. List of the analyzed genomes is provided in supplementary table S2 in supplementary file S1, Supplementary Material online.
Fig. 5.
—NJ tree calculated for Bray–Curtis distance matrix based on COG orthologs. The bar chart represents COG category content for individual genomes. The categories abbreviations are as follows: A: RNA processing and modification, B: Chromatin Structure and dynamics, C: Energy production and conversion, D: Cell cycle control and mitosis, E: Amino Acid metabolism and transport, F: Nucleotide metabolism and transport, G: Carbohydrate metabolism and transport, H: Coenzyme metabolism, I: Lipid metabolism, J: Translation, K: Transcription, L: Replication and repair, M: Cell wall/membrane/envelop biogenesis, N: Cell motility, O: Posttranslational modification, protein turnover, chaperone functions, P: Inorganic ion transport and metabolism, Q: Secondary Structure, T: Signal Transduction, U: Intracellular trafficking and secretion, V: Defense mechanism, W: Extracellular structures, X: Mobilome: prophages, transposons, Y: Nuclear structure, Z: Cytoskeleton, R: General Functional Prediction only, S: Function Unknown.
Metabolism, Adaptive Processes, and HGT
To some degree, however, the convergent evolution/adaptation described earlier, is limited by phylogenetic constraints, that is, availability of different metabolic machineries inherited from unrelated bacterial ancestors. For example, both L. polyplacis and the Riesia species lost capacity to synthetize thiamin. In Riesia, this incapacity is compensated by a specific thiamin ABC transporter inherited from its ancestor (as inferred from the presence of ABC thiamin transporter in other Arsenophonus spp.). Boyd et al. (2017) hypothesized that the loss of thiamin synthesis by Riesia in hominid lice and its retention by Puchtella in colobus monkey lice may reflect diet differences of the two mammal hosts. They suggest that the complex diet of the hominids makes thiamin available for scavenging by Riesia. According to this view, L. polyplacis from the rodent associated lice could also scavenge thiamin from its host. However, the specific thiamin transporter is not present in the known Legionella genomes and L. polyplacis thus lacks both the synthetic pathway as well as specific transporter. Using the CoFactor database (Fischer et al. 2010), we determined thiamine as the essential compound required as a cofacor by at least two of the L. polyplacis enzymes (transketolase and pyruvate dehydrogenase). It is therefore likely that L. polyplacis utilizes some alternative system for its acquisition. As it is known that the ABC transporters can bound to more ligands (ter Beek et al. 2014), we suggest that among the possible candidates for this role are the other transporters which could be adapted for nonspecific transfer. For example, the ability to bind thiamin is known for the ABC putrescine transporter (ter Beek et al. 2014). This transporter is present in L. polyplacis and was obviously inherited from its legionellae ancestor. Apart from the ABC putrescine transporter subunits, only 11 additional sequences were identified as potential transporters using the TCDB (Saier et al. 2006). Four of these genes were associated with cytochrome c biogenesis, two with sec-SRP system, and the additional two were identified as NhaA antiporter and phospho-N-acetylmuramoyl-pentapeptide transferase. Of the remaining three sequences, one was directly annotated as Major facilitator family transporter, whereas the affiliation of the two unannotated proteins (designated in supplementary table S3 in supplementary file S1, Supplementary Material online, as FIG00758517: hypothetical protein and putative transport protein) to the same transporter family was confirmed by the BLAST search.
A further important difference between L. polyplacis and R. pediculicola introduces horizontal gene transfer (HGT) as yet another determinant of a symbiont’s evolutionary pathway. The significance of HGT for adaptation to a particular life style in bacteria is well recognized. For Legionella, Burstein et al. (2016) suggested that many of effectors, with possibly important functions in virulence and intracellular lifestyle, have been recently acquired by this mechanism. Within the system of thousands of predicted effectors, they were able identify only seven effectors shared by all tested Legionella species. This, together with the variability of main genome characteristics, shows Legionella as highly dynamic system capable of rapid adaptations. Unlike the “free living” legionellae, the HGT in L. polyplacis is bound to its symbiotic function known from other blood feeding insects, that is, provisioning the host with vitamins. In the case of B7 vitamin, this role in L. polyplacis is fulfilled by a horizontally acquired complete biotin operon with gene order conserved across the known homologues (fig. 2 and supplementary file S3, Supplementary Material online). Since only few instances of the complete homologous operon transfer are known from other bacteria, it would be difficult to establish a meaningful evolutionary scenario for this HGT and possible source of these genes in L. polyplacis. It is however interesting to note that the two most closely related operon homologs are also associated with insect symbionts, namely Wolbachia (strains wCle from bed bugs and wNfla, Wnleu from bees) and Cardinium (strain cEper1 from parasitic wasps and cBtQ1 from whiteflies). In addition to the biotin operon, for which a rickettsial origin was suggested by Penz et al. (2012), Cardinium was shown to harbor high number of other HGT-acquired genes (Penz et al. 2012). Regarding possible source(s) of these HGTs, the authors suggested two main possibilities, both highly relevant in respect to this study. First, for part of the genes the closest homologs are associated with other insect symbionts. The second possible source are bacteria associated with amoebas, including Legionella. In addition, a considerable portion of the putative bacterial donors are capable to infect both insects and amoebas (Penz et al. 2012). Considering its presumable ancestral life style, that is, association with amoebas, and current symbiotic relationships with insects, the acquisition of biotin operon by L. polyplacis from either of these sources seems plausible hypothesis. In Wolbachia from the bed bug Cimex lectularius, the acquisition of the operon by HGT has been explicitly claimed to had enabled ecological transition of otherwise parasitic Wolbachia to an obligate mutualist (Nikoh et al. 2014). However, in this case, the parasitic ancestor of the symbiotic Wolbachia was likely to lack the biotin synthesis capacity altogether. Since legionellae generally possess the biotin synthesis capacity, the acquisition of the biotin operon by L. polyplacis appears as a replacement rather than a de novo acquisition of this function. However, in contrast to this compact six-gene biotin operon, in the genomes of “free living” legionellae BioC is separated from the rest of the genes. As a remnant of this arrangement, L. polyplacis retains the original BioC (position 269231-269893) apart from the complete horizontally transferred biotin operon (222799-228219). Regarding the absence of any known intermediate form between the pathogenic legionellae and L. polyplacis, it is difficult to hypothesize on the absence/presence of the biotin synthesis capacity in the L. polyplacis ancestor, and therefore on the possible role of this HGT in triggering its transition to symbiosis. Various circumstances could possibly drive L. polyplacis toward adaptive acceptance of this operon. For example, within the highly economized L. polyplacis genome, an arrangement of biotin synthesis within a single operon could prove more efficient than the genes scattered around the genome as in other legionellae. In addition, the acquisition of this operon could follow preceding loss of the biotin synthesis in L. polyplacis parasitic ancestor (similar to the transition from parasitic to mutualistic Wolbachia described earlier). Altogether, the patterns discussed earlier show how the combination of different evolutionary forces (phylogenetic constraint, adaptive pressure, and HGT) resulted in emergence of a unique genome/phenotype in the genus Legionella.
Supplementary Material
Supplementary data are available at Genome Biology and Evolution online.
Supplementary Material
Acknowledgments
We would like to thank Jana Martinu for providing the lice specimens. This work was supported by the Grant Agency of the Czech Republic (grant 14-07004S to V.H.). Access to computing and storage facilities owned by parties and projects contributing to the National Grid Infrastructure MetaCentrum provided under the programme “Projects of Large Research, Development, and Innovations Infrastructures” (CESNET LM2015042), is greatly appreciated.
Literature Cited
- Alix B, Boubacar D, Vladimir M.. 2012. T-REX: a web server for inferring, validating and visualizing phylogenetic trees and networks. Nucleic Acids Res. 40:W573–W579. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Allen J, Burleigh J, Light J, Reed D.. 2016. Effects of 16S rDNA sampling on estimates of the number of endosymbiont lineages in sucking lice. Peerj 4:e2187.. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Altschul SF, Gish W, Miller W, Myers EW, Lipman DJ.. 1990. Basic local alignment search tool. J Mol Biol. 215(3):403–410. [DOI] [PubMed] [Google Scholar]
- Aziz R, et al. 2008. The RAST server: rapid annotations using subsystems technology. BMC Genomics 9:75.. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bankevich A, et al. 2012. SPAdes: a new genome assembly algorithm and its applications to single-cell sequencing. J Comput Biol. 19(5):455–477.http://dx.doi.org/10.1089/cmb.2012.0021 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Boyd B, et al. 2016. Two bacterial genera, Sodalis and Rickettsia, associated with the seal louse Proechinophthirus fluctus (Phthiraptera: Anoplura). Appl Environ Microbiol. 82(11):3185–3197.http://dx.doi.org/10.1128/AEM.00282-16 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Boyd BM, Allen JM, de Crecy-Lagard V, Reed DL.. 2014. Genome sequence of Candidatus Riesia pediculischaeffi, endosymbiont of chimpanzee lice, and genomic comparison of recently acquired endosymbionts from human and chimpanzee Lice. G3 Genes Genomes Genet. 4(11):2189–2195. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Boyd BM, et al. 2017. Primates, lice and bacteria: speciation and genome evolution in the symbionts of hominid lice. Mol Biol Evol. 34(7):1743–1757.http://dx.doi.org/10.1093/molbev/msx117 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Burstein D, et al. 2016. Genomic analysis of 38 Legionella species identifies large and diverse effector repertoires. Nat Genet. 48(2):167–175.http://dx.doi.org/10.1038/ng.3481 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Carattoli A, et al. 2014. In silico detection and typing of plasmids using PlasmidFinder and plasmid multilocus sequence typing. Antimicrob Agents Chemother. 58(7):3895–3903.http://dx.doi.org/10.1128/AAC.02412-14 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Castresana J. 2000. Selection of conserved blocks from multiple alignments for their use in phylogenetic analysis. Mol Biol Evol. 17(4):540–552.http://dx.doi.org/10.1093/oxfordjournals.molbev.a026334 [DOI] [PubMed] [Google Scholar]
- Coil D, Jospin G, Darling A.. 2015. A5-miseq: an updated pipeline to assemble microbial genomes from Illumina MiSeq data. Bioinformatics 31(4):587–589.http://dx.doi.org/10.1093/bioinformatics/btu661 [DOI] [PubMed] [Google Scholar]
- Dale C, Young SA, Haydon DT, Welburn SC.. 2001. The insect endosymbiont Sodalis glossinidius utilizes a type III secretion system for cell invasion. Proc Natl Acad Sci U S A. 98(4):1883–1888.http://dx.doi.org/10.1073/pnas.98.4.1883 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Darriba D, Taboada G, Doallo R, Posada D.. 2011. ProtTest 3: fast selection of best-fit models of protein evolution. Bioinformatics 27(8):1164–1165.http://dx.doi.org/10.1093/bioinformatics/btr088 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Diederen B. 2008. Legionella spp. and Legionnaires’ disease. J Infect. 56(1):1–12. [DOI] [PubMed] [Google Scholar]
- Douglas A. 1989. Mycetocyte symbiosis in insects. Biol Rev Camb Philos Soc. 64(4):409–434.http://dx.doi.org/10.1111/j.1469-185X.1989.tb00682.x [DOI] [PubMed] [Google Scholar]
- Durden L, Musser G.. 1994. The sucking lice (Insecta, Anoplura) of the world – a taxonomic checklist with records of mammalian hosts and geographical distribution. Bull Am Museum Nat History 218:1–90. [Google Scholar]
- Fields B. 1996. The molecular ecology of legionellae. Trends Microbiol. 4(7):286–290.http://dx.doi.org/10.1016/0966-842X(96)10041-X [DOI] [PubMed] [Google Scholar]
- Fischer J, Holliday G, Thornton J.. 2010. The CoFactor database: organic cofactors in enzyme catalysis. Bioinformatics 26(19):2496–2497.http://dx.doi.org/10.1093/bioinformatics/btq442 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Galperin MY, Makarova KS, Wolf YI, Koonin EV.. 2015. Expanded microbial genome coverage and improved protein family annotation in the COG database. Nucleic Acids Res. 43(D1):D261–D269. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gerth M, Bleidorn C.. 2017. Comparative genomics provides a timeframe for Wolbachia evolution and exposes a recent biotin synthesis operon transfer. Nat Microbiol. 2:16241. [DOI] [PubMed] [Google Scholar]
- Guindon S, et al. 2010. New algorithms and methods to estimate maximum-likelihood phylogenies: assessing the performance of PhyML 3.0. Syst Biol. 59(3):307–321.http://dx.doi.org/10.1093/sysbio/syq010 [DOI] [PubMed] [Google Scholar]
- Hosokawa T, Koga R, Kikuchi Y, Meng X-Y, Fukatsu T.. 2010. Wolbachia as a bacteriocyte associated nutritional mutualist. Proc Natl Acad Sci U S A. 107(2):769–774. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Huelsenbeck J, Ronquist F.. 2001. MRBAYES: Bayesian inference of phylogenetic trees. Bioinformatics 17(8):754–755.http://dx.doi.org/10.1093/bioinformatics/17.8.754 [DOI] [PubMed] [Google Scholar]
- Husnik F, Chrudimsky T, Hypsa V.. 2011. Multiple origins of endosymbiosis within the Enterobacteriaceae (gamma-Proteobacteria): convergence of complex phylogenetic approaches. BMC Biol. 9:87. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Husnik F, et al. 2013. Horizontal gene transfer from diverse bacteria to an insect genome enables a tripartite nested mealybug symbiosis. Cell 153(7):1567–1578.http://dx.doi.org/10.1016/j.cell.2013.05.040 [DOI] [PubMed] [Google Scholar]
- Hypsa V, Krizek J.. 2007. Molecular evidence for polyphyletic origin of the primary symbionts of sucking lice (Phthiraptera, Anoplura). Microb Ecol. 54(2):242–251.http://dx.doi.org/10.1007/s00248-006-9194-x [DOI] [PubMed] [Google Scholar]
- Joseph S, et al. 2016. Dynamics of genome change among Legionella species. Sci Rep. 6:33442. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Katoh K, Misawa K, Kuma K, Miyata T.. 2002. MAFFT: a novel method for rapid multiple sequence alignment based on fast Fourier transform. Nucleic Acids Res. 30(14):3059–3066.http://dx.doi.org/10.1093/nar/gkf436 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Katoh K, Standley D.. 2013. MAFFT Multiple sequence alignment software version 7: improvements in performance and usability. Mol Biol Evol. 30(4):772–780.http://dx.doi.org/10.1093/molbev/mst010 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lartillot N, Rodrigue N, Stubbs D, Richer J.. 2013. PhyloBayes MPI: phylogenetic reconstruction with infinite mixtures of profiles in a parallel environment. Syst Biol. 62(4):611–615.http://dx.doi.org/10.1093/sysbio/syt022 [DOI] [PubMed] [Google Scholar]
- Legendre P, Legendre LFJ.. 2012. Numerical ecology. Amsterdam: Elsevier. Third English edition. ISBN 978-0-444-53868-0. [Google Scholar]
- Li H, Durbin R.. 2009. Fast and accurate short read alignment with Burrows-Wheeler transform. Bioinformatics 25(14):1754–1760.http://dx.doi.org/10.1093/bioinformatics/btp324 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Li H, Genome Project Data Processing S, et al. 2009. The Sequence Alignment/Map format and SAMtools. Bioinformatics 25(16):2078–2079., [DOI] [PMC free article] [PubMed] [Google Scholar]
- Light J, Smith V, Allen J, Durden L, Reed D.. 2010. Evolutionary history of mammalian sucking lice (Phthiraptera: Anoplura). BMC Evol Biol. 10:292.. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Masui S, Sasaki T, Ishikawa H.. 2000. Genes for the type IV secretion system in an intracellular symbiont, Wolbachia, a causative agent of various sexual alterations in arthropods. J Bacteriol. 182(22):6529–6531. [DOI] [PMC free article] [PubMed] [Google Scholar]
- McCutcheon JP, Keeling PJ.. 2014. Endosymbiosis: protein targeting further erodes the organelle/symbiont distinction. Curr Biol. 24(14):R654–R655. [DOI] [PubMed] [Google Scholar]
- Meeske A, et al. 2016. SEDS proteins are a widespread family of bacterial cell wall polymerases. Nature 537(7622):634..http://dx.doi.org/10.1038/nature19331 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Moran N, McCutcheon J, Nakabachi A.. 2008. Genomics and evolution of heritable bacterial symbionts. Annu Rev Genet. 42:165–190.http://dx.doi.org/10.1146/annurev.genet.41.110306.130119 [DOI] [PubMed] [Google Scholar]
- Moran NA. 1996. Accelerated evolution and Muller’s rachet in endosymbiotic bacteria. Proc Natl Acad Sci U S A. 93(7):2873–2878. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Moran NA, Bennett GM.. 2014. The tiniest tiny genomes. Annu Rev Microbiol. 68:195–215.http://dx.doi.org/10.1146/annurev-micro-091213-112901 [DOI] [PubMed] [Google Scholar]
- Naito M, Pawlowska TE.. 2016. The role of mobile genetic elements in evolutionary longevity of heritable endobacteria. Mobile Genet Elem. 6(1):7791–7796. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Nakabachi A, et al. 2013. Defensive bacteriome symbiont with a drastically reduced genome. Curr Biol. 23(15):1478–1484.http://dx.doi.org/10.1016/j.cub.2013.06.027 [DOI] [PubMed] [Google Scholar]
- Nikoh N, et al. 2014. Evolutionary origin of insect-Wolbachia nutritional mutualism. Proc Natl Acad Sci U S A. 111(28):10257–10262. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Nogge G. 1981. Significance of symbionts for the maintenance of an optimal nutritional state for successful reproduction in hematophagous arthropods. Parasitology 82:101–104. [Google Scholar]
- Ochman H, Moran NA.. 2001. Genes lost and genes found: evolution of bacterial pathogenesis and symbiosis. Science 292(5519):1096–1099.http://dx.doi.org/10.1126/science.1058543 [DOI] [PubMed] [Google Scholar]
- Penz T, et al. 2012. Comparative genomics suggests an independent origin of cytoplasmic incompatibility in Cardinium hertigii. PLoS Genet. 8(10):e1003012.. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Perez-Brocal V, et al. 2006. A small microbial genome: the end of a long symbiotic relationship? Science 314(5797):312–313. [DOI] [PubMed] [Google Scholar]
- Rambaut A, Suchard MA, Xie D, Drummond AJ.. 2014. Tracer v1.6. Available from: http://beast.community/tracer, last accessed October 30, 2017.
- Ries E. (co-authors). 1931. Die symbiose der läuse und federlinge. Z Morphol Ökologie Tiere 20(2–3):233–367. [Google Scholar]
- Saier MH Jr, Tran CV, Barabote RD.. 2006. TCDB: the Transporter Classification Database for membrane transport protein analyses and information. Nucleic Acids Res. 34(Database issue):D181–D186. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Seemann T. 2014. Prokka: rapid prokaryotic genome annotation. Bioinformatics 30(14):2068–2069.http://dx.doi.org/10.1093/bioinformatics/btu153 [DOI] [PubMed] [Google Scholar]
- Snyder AK, Deberry JW, Runyen-Janecky L, Rio RV.. 2010. Nutrient provisioning facilitates homeostasis between tsetse fly (Diptera: Glossinidae) symbionts. Proc R Soc. 277(1692):2389–2397. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Stefka J, Hypsa V.. 2008. Host specificity and genealogy of the louse Polyplax serrata on field mice, Apodemus species: a case of parasite duplication or colonisation? Int J Parasitol. 38(6):731–741. [DOI] [PubMed] [Google Scholar]
- Tatusov R, Galperin M, Natale D, Koonin E.. 2000. The COG database: a tool for genome-scale analysis of protein functions and evolution. Nucleic Acids Res. 28(1):33–36.http://dx.doi.org/10.1093/nar/28.1.33 [DOI] [PMC free article] [PubMed] [Google Scholar]
- ter Beek J, Guskov A, Slotboom D.. 2014. Structural diversity of ABC transporters. J Gen Physiol. 143(4):419–435. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Volf P. 1991. Postembryonal development of mycetocytes and symbionts of the spiny rat louse Polyplax spinulosa. J Invert Pathol. 58(1):143–146.http://dx.doi.org/10.1016/0022-2011(91)90172-M [Google Scholar]
- Walker B, et al. 2014. Pilon: an integrated tool for comprehensive microbial variant detection and genome assembly improvement. PLoS One 9(11):e112963. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wilkes TE, et al. 2010. The draft genome sequence of Arsenophonus nasoniae, son-killer bacterium of Nasonia vitripennis, reveals genes associated with virulence and symbiosis. Insect Mol Biol. 19(s1):59–73.http://dx.doi.org/10.1111/j.1365-2583.2009.00963.x [DOI] [PubMed] [Google Scholar]
- Woolfit M, Bromham L.. 2003. Increased rates of sequence evolution in endosymbiotic bacteria and fungi with small effective population sizes. Mol Biol Evol. 20(9):1545–1555.http://dx.doi.org/10.1093/molbev/msg167 [DOI] [PubMed] [Google Scholar]
- Wu M, Scott A.. 2012. Phylogenomic analysis of bacterial and archaeal sequences with AMPHORA2. Bioinformatics 28(7):1033–1034.http://dx.doi.org/10.1093/bioinformatics/bts079 [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.