Short abstract
We performed whole transcriptome analysis of osteosarcoma bone samples. Initially, we sequenced total RNA from 36 fresh-frozen samples (18 tumoral bone samples and 18 non-tumoral paired samples) matching in pairs for each osteosarcoma patient. We also performed independent gene expression analysis of formalin-fixed paraffin-embedded samples to verify the RNAseq results. Formalin-fixed paraffin-embedded samples allowed us to analyze the effect of chemotherapy. Data were analyzed with DESeq2, edgeR and Reactome packages of R. We found 5365 genes expressed differentially between the normal bone and osteosarcoma tissues with an FDR below 0.05, of which 3399 genes were upregulated and 1966 were downregulated. Among those genes, BTNL9, MMP14, ABCA10, ACACB, COL11A1, and PKM2 were expressed differentially with the highest significance between tumor and normal bone. Functional annotation with the reactome identified significant changes in the pathways related to the extracellular matrix degradation and collagen biosynthesis. It was suggested that chemotherapy may induce the modification of ECM with important collagen biosynthesis. Taken together, our results indicate that changes in the degradation of extracellular matrix seem to be an important mechanism of osteosarcoma and efficient chemotherapy induces the genes related to bone formation.
Impact statement
Osteosarcoma is a rare disease but it is of interest to many scientists all over the world because the current standard treatment still has poor results. We sequenced total RNA from 36 fresh-frozen paired samples (18 tumoral bone samples and 18 non-tumoral paired samples) from osteosarcoma patients. We found that differences in the gene expressions between the normal and affected bones reflected the changes in the regulation of the degradation of collagen and extracellular matrix. We believe that these findings contribute to the understanding of OS and suggest ideas for further studies.
Keywords: Osteosarcoma, transcriptome, gene expression profiling, RNA sequencing, high-through put nucleotide sequencing, osteogenic sarcoma
Introduction
Osteosarcoma is a primary malignant tumor of the skeleton characterized by the direct formation of immature bone and osteoid tissue by tumor cells. Rarely, osteosarcoma may arise in the soft tissues.1 Osteosarcoma is the most common form of primary malignant bone tumors,2 accounting for 20–40% of all bone cancers.3 The incidence rate of osteosarcoma in young persons (0–24 years) averages 4.2 per million in males and 3.1 per million in females. The incidence rate in persons aged 25–59 is 1.7 and 1.4 in males and females, respectively. The incidence increases again among persons older than 60 years, about 4.6 and 3.3 per million in males and females, respectively.4 Therefore, there are two peaks in the incidence of disease. The first is around puberty, earlier in women (10–14 years) and a little bit later in men (15–19 years). The second incidence peak is in the elderly after 60 years.1–3 Osteosarcoma is more common in men than in women in most countries. The overall male-to-female ratio of osteosarcoma in the age ranges 0–24 years, 25–59 and over 60 years is 1.43:1; 1.28:1; 1.01: 1, respectively.4 Clinically, patients with osteosarcoma complain most often about pain and swelling. Systemic symptoms such as weight loss, pallor, fever, anorexia are very uncommon.1 Over half of all osteosarcomas arise in the long bones of the lower limb (56%), with the upper limb (10%) and pelvis (9%) being the next most affected sites.3
The exact cause of osteosarcoma is not yet determined but some risk factors have been collected. These include rapid bone growth, radiation, and some genetic predisposition such as Li-Fraumeni syndrome (LFS), hereditary retinoblastoma (RB), Rothmund-Thomson syndrome (RTS) type 2, Werner syndrome (WS), Bloom syndrome (BS), RAPADIL- INO syndrome, and Diamond Blackfan anemia (DBA).5 Osteosarcoma is a highly metastatic disease, as 80–90% patients are assumed to have micrometastatic disease at the time of diagnosis.6 This can explain the gain of outcome in treating this disease after introducing chemotherapy in the late 1970s and early 1980s.7
Several studies have been carried out to understand the molecular aspect and to define new therapeutic targets for osteosarcoma, which is a very heterogeneous disease.5 Mutations in the p53 pathway are found in about 95% of tumors, with a majority of the TP53 mutations being translocations or focal deletions. Other mutations in RB1, ATRX, and DLG2 genes have been identified.5,8 Some microRNAs have been found to be downregulated in osteosarcoma and are associated with poor prognosis of the disease, such as with miR-206, miR-195, miR-340, miR-503, while some others are upregulated, like miR-17.9–12
High-throughput technologies have been used in recent OS studies but there are still many limitations, including small sample sizes, variations in control groups, variations in laboratory protocols, and analysis techniques.13,14 In a transcriptome analysis of a single case of osteosarcoma in Estonia, 65 genes were found differentially expressed between a tumor sample and normal bone of the same patient. There were 7 upregulated genes in normal tissue and 58 upregulated genes in the sarcoma.15 In another study of 20 samples (14 osteosarcoma and 6 normal samples), three differentially expressed genes GJA1, COL1A2, and COL5A2 were identified. It was also observed that COL1A2 and COL5A2 interact with a number of genes of the matrix metalloprotease (MMP) family, including MMP1, MMP2, MMP3, and MMP14, TGFβ, and RUNX2.16
The aim of present study was to find alterations in the transcriptome of bone caused by the osteosarcoma. We analyzed 18 tumor-normal pairs of bone tissue samples, all collected during surgery after the chemotherapy. We also analyzed 15 FFPE samples from biopsies, of which 10 were collected before and 5 after chemotherapy.
Materials and methods
Patients
The Ethics Review Committee on Biomedical Research of Hue University of Medicine and Pharmacy approved the protocols and informed consent forms used in the study. All the participants or representatives of patients signed the informed consent.
Samples were collected from 18 patients in Vietnam who had histologically confirmed osteosarcoma and were allocated for surgery (limb sparing or amputation). Tumor and normal bone samples were collected from the removed bone right after the operation. The samples were transported with dry ice and then stored at −80°C until RNA extraction.
FFPE samples were all biopsy samples, 10 of them before and 5 after chemotherapy.
RNA sequencing
Bone samples (40–50 mg) were ground with nitrogen by pestle and mortar into powder and pretreated with trizol. Total RNA was extracted with an RNeasy Fibrous Tissue Mini Kit (Qiagen, Valencia CA, USA) according to the manufacturer’s protocol. Isolated RNA was dissolved in RNase free water and stored at −80°C. The quality of total RNA was evaluated with Agilent 2100 Bioanalyzer and the RNA 6000 Nano Kit (Agilent Technologies Inc., CA, USA).
For each FFPE sample, six pieces of 10 μm thick section were used. RNA was extracted with PureLink FFPE total RNA isolation kit from invitrogen (Invitrogen Corporation, Carlsbad, CA, USA). For all samples (fresh and FFPE), osteosarcoma was confirmed in the samples by two independent pathologists.
Total RNA (50 ng) was amplified by applying Ovation RNA-Seq System V2 (NuGen, Emeryville, CA, USA) after which the resulting cDNAs were pooled in equal amount and the pool was used to prepare the DNA fragment library with SOLiD System chemistry (Life Technologies Corp., Carlsbad, CA, USA). Sequencing was performed using the SOLiD 5500W platform and DNA sequencing chemistry (Life Technologies Corp., Carlsbad, CA, USA). Raw reads (75 bp) were color-space mapped to the human genome hg19 reference using the Maxmapper algorithm implemented in the Lifescope software (Life Technologies, Ltd). Mapping to multiple locations was permitted. The quality threshold was set to 10 giving the mapping confidence more than 90. Reads with a score less than 10 were filtered out. Average mapping quality was 30. Analysis of the RNA content and gene-based annotation was done within whole transcriptome workflow implemented in Lifescope.
Statistical analysis
For statistical analysis, DeSeq2 (for fresh samples) and edgeR (for FFPE samples) packages for R was used.17,18 DeSeq2 and edgeR are packages which are specifically developed for RNA-seq or other count data. The packages provide methods to test for differential expression by use of the negative binomial distribution and a shrinkage estimator for the distribution’s variance.19 The packages perform sample comparison and also adjust the P-value to overcome multiple testing problems. Both packages use the Benjamini–Hochberg procedure, which controls for false discovery rate (FDR).20
Functional annotation of differentially expressed genes
In order to find functional relations between differentially expressed genes, we applied pathway enrichment analysis. We used the R/Bioconductor package ReactomePA for reactome pathway analysis and visualization.21 Enrichment analysis is a widely used way to identify biological themes in the complex lists of differentially expressed genes. ReactomePA implements hypergeometric model to assess whether the number of selected genes associated with the reactome pathway is larger than expected. The P-values are calculated based on the hypergeometric model. After analysis, results were visualized using an enrichment map and category-gene-network tools.21–23 This method was used to analyse the pathways enrichment of differentially expressed genes between the affected bone and normal bone and to detect the pathways related to differential genes expression with chemotherapy.
Results
Clinical description of patients (Table 1)
Table 1.
Characteristics of osteosarcoma patients in the present study.
| Patient code | Age at diagnosis | Gender | Site of tumor | Chemotherapy | Sample type |
RIN |
|
|---|---|---|---|---|---|---|---|
| Cancer | Normal | ||||||
| OSVN001 | 16 | Female | Femur | Yes | Fresh | 4 | 4 |
| OSVN003 | 13 | Male | Femur | Yes | Fresh | 7.9 | 6.3 |
| OSVN004 | 16 | Female | Femur | Yes | Fresh | 6.3 | 6.4 |
| OSVN005 | 18 | Male | Femur | Yes | Fresh | 8.3 | 4.3 |
| OSVN006 | 18 | Male | Femur | Yes | Fresh | 6 | 2 |
| OSHN008 | 24 | Female | Tibia | No | Fresh | NA | 8.1 |
| OSVN008 | 52 | Male | Femur | Yes | Fresh | 5.9 | 6 |
| OSHN009 | 16 | Male | Femur | Yes | Fresh | 8.6 | 5.5 |
| OSHN010 | 20 | Female | Femur | Yes | Fresh | 6.8 | 7.4 |
| OSHN011 | 7 | Male | Tibia | Yes | Fresh | 8.7 | 7.7 |
| OSHN012 | 11 | Male | Humerus | No | Fresh | 8.3 | 6.3 |
| OSHN013 | 17 | Male | Femur | No | Fresh | 7.9 | 6.1 |
| OSHN014 | 16 | Female | Tibia | Yes | Fresh | 7.4 | 8.4 |
| OSVN015 | 15 | Male | Tibia | Yes | Fresh | 5.6 | 8.4 |
| OSHN015 | 8 | Female | Tibia | Yes | Fresh | NA | 8.4 |
| OSHN016 | 20 | Male | Femur | Yes | Fresh | 9.4 | 3.3 |
| OSHN017 | 16 | Male | Humerus | Yes | Fresh | 8.2 | 7.3 |
| OSDN001 | 23 | Male | Tibia | Yes | Fresh | 8.4 | 5.8 |
| EE4878 | 24 | Male | Femur | Yes | FFPE | NA | |
| EE6762 | 51 | Male | Tibia | Yes | FFPE | NA | |
| EE6065 | 80 | Female | Femur | Yes | FFPE | NA | |
| EE6311 | 9 | Male | Humerus | No | FFPE | NA | |
| EE1480 | 18 | Female | Tibia | Yes | FFPE | 1 | |
| EE9244 | 29 | Male | Pelvis | No | FFPE | 2.8 | |
| EE6921 | 31 | Female | Femur | No | FFPE | 2.4 | |
| EE3447 | 20 | Male | Femur | Yes | FFPE | 2.2 | |
| EE13536 | 22 | Female | Femur | No | FFPE | 2.5 | |
| EE648 | 32 | Male | Femur | No | FFPE | 2.4 | |
| EE8076 | 19 | Male | Femur | No | FFPE | NA | |
| VN26391 | 52 | Male | Femur | No | FFPE | 2.5 | |
| VN25065 | 20 | Male | Femur | No | FFPE | 2 | |
| VN23611 | 23 | Male | Fibula | No | FFPE | 2.4 | |
| VN21890 | 15 | Male | Femur | No | FFPE | 2.5 | |
We collected samples after surgical removal of affected bone from 18 Vietnamese osteosarcoma patients. Each surgically removed sample contained paired tumor and normal parts. Fifteen FFPE samples were retrieved from the pathology archives. A pathologist confirmed the diagnosis after histological analysis. Of the 33 patients, 10 (30%) were females and 23 (70%) were males. The mean age was 23.4, which ranged from 7 to 80 years.
General description about the analysed samples
Before sequencing, we evaluated the integrity of RNA. The quality of fresh samples was much higher than FFPE samples. Median RIN was 7.9 (4–9.4), 6.3 (2–8.4) and 2.4 (1–2.8) for fresh cancer samples, fresh control and FFPE, respectively. In some samples, RIN value was not readable but the electropherograms were acceptable to be further analyzed. RIN values were integrated in Table 1.
Number of reads in fresh samples was 30–35 million reads with 65% mapping in average, while read counts for FFPE samples were much lower, 8–10 million reads with 60% mapping in average.
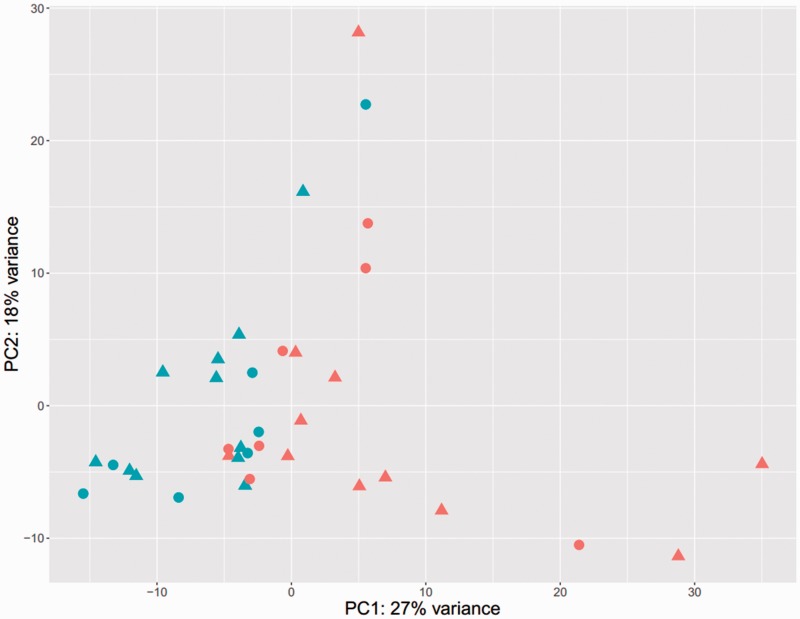
Distribution of gene expression among fresh samples (Figure 1)
Figure 1.
Visualization of sample-to-sample distances in a principal-components analysis (PCA). All 21,632 transcripts were used in the PCA analysis. The variations of gene expression among samples from 18 Vietnamese osteosarcoma patients (in paired samples, tumor vs normal) showed pretty clear difference between tumor samples and normal control samples. In this figure, the triangle dots were for males and round ones were for females; the red ones illustrated for normal adjacent bone, while blue ones were for tumor samples.
We used principal-components analysis (PCA) to visualize sample-to-sample distances. As PCA identified that the major component for sample distances was the presence of a tumor, we concluded that our sample collection quality was sufficient for the following analysis.
Differential expression analysis between paired fresh tumor and normal samples
We applied pairwise analysis of the signals from 21,632 genes. We identified 7645 genes differentially expressed with a P-value below 0.05. In order to eliminate the false positives, we used the Benjamini–Hochberg (BH) adjustment which is implemented in DESeq2. These values, called the BH-adjusted P-values (FDR), are given in the column "padj" in Tables 2 and 3. Hence, if we consider a proportion of 10% false positives to be acceptable, we can consider all genes with an adjusted P-value below 10% = 0.1 as significant. We consequently identified 6775 genes expressed differentially between the normal and osteosarcoma tissues with an FDR adjusted P-value below 0.1. With adjusted P-value below 0.05, we found 5365 genes differentially expressed between the two groups, of which 3399 genes were upregulated and 1966 were downregulated.
Table 2.
The most significantly down-regulated genes in osteosarcoma.
| Symbol | log2FoldChange | padj | chr | Genename | |
|---|---|---|---|---|---|
| BTNL9 | −1.54 | 1.20E-15 | 5 | Butyrophilin-like 9 | |
| DNASE1L3 | −1.45 | 2.90E-07 | 3 | Deoxyribonuclease I-like 3 | |
| CAMP | −1.45 | 4.47E-06 | 3 | Cathelicidin antimicrobial peptide | |
| LEPR | −1.43 | 1.21E-09 | 1 | Leptin receptor | |
| MIR223 | −1.43 | 6.84E-06 | X | microRNA 223 | |
| MS4A3 | −1.41 | 7.31E-06 | 11 | Membrane-spanning 4-domains, subfamily A, member 3 | |
| LTF | −1.40 | 1.00E-05 | 3 | Lectotransferrin | |
| LCN2 | −1.40 | 1.00E-05 | 9 | Lipocalin 2 | |
| MMP8 | −1.40 | 8.74E-06 | 11 | Matrix metallopeptidase 8 | |
| S100A12 | −1.39 | 1.17E-05 | 1 | S100 calcium binding protein A12 | |
| S100A8 | −1.39 | 1.17E-05 | 1 | S100 calcium binding protein A8 | |
| MPO | −1.39 | 1.17E-05 | 17 | Myeloperoxidase | |
| EPB42 | −1.39 | 8.52E-06 | 15 | Erythrocyte membrane protein band 4.2 | |
| HEMGN | −1.39 | 1.03E-05 | 9 | Hemogen | |
| AHSP | −1.39 | 1.30E-05 | 16 | Alpha hemoglobin stablizing protein | |
| ABCA10 | −1.38 | 3.43E-11 | 17 | ATP-binding cassette, sub-family A (ABC1), member 10 | |
| BPI | −1.38 | 1.16E-05 | 20 | Bactericidal/permeability-increasing protein | |
| CEACAM6 | −1.38 | 5.56E-06 | 19 | Carcinoembryonic antigen-related cell adhesion molecule 6 | |
| DEFA4 | −1.37 | 1.63E-05 | 8 | Definsin, alpha 4, corticostatin | |
Table 3.
The most significantly up-regulated genes in osteosarcoma.
| Symbol | log2FoldChange | padj | chr | genename |
|---|---|---|---|---|
| COL11A1 | 1.51 | 3.43E-11 | 1 | Collagen, type XI, alpha 1 |
| TGFBI | 1.40 | 1.46E-08 | 5 | Transforming growth factor, beta-induced, 68kDa |
| TREM2 | 1.39 | 7.91E-08 | 6 | Triggering receptor expressed on myeloid cells 2 |
| COL2A1 | 1.38 | 1.09E-05 | 12 | Collagen, type II, alpha 1 |
| COL10A1 | 1.35 | 5.05E-06 | 6 | Collagen, type II, alpha 2 |
| HAPLN1 | 1.26 | 1.60E-05 | 5 | Hyaluronan and proteoglycan link protein 1 |
| MMP14 | 1.24 | 2.86E-11 | 14 | Matrix metallopeptidase 14 (membrane-inserted) |
| PANX3 | 1.22 | 8.83E-05 | 11 | Pannexin 3 |
| CTHRC1 | 1.21 | 3.90E-08 | 8 | Collagen triple helix repeat containing 1 |
| STEAP1 | 1.20 | 1.40E-07 | 7 | Six transmembrane epithelial antigen of prostate 1 |
| COL3A1 | 1.19 | 2.94E-07 | 2 | Collagen, type III, alpha 1 |
| CA12 | 1.18 | 5.99E-08 | 15 | Carbonic anhydrase XII |
| GJB2 | 1.17 | 5.26E-05 | 13 | Gap junction protein, beta 2, 26kDa |
| PLOD1 | 1.15 | 3.61E-10 | 1 | Procollagen-lysine, 2-oxoglutarate 5-dioxygenase 1 |
| LEPRE1 | 1.15 | 2.78E-08 | 1 | Prolyl 3-hydroxylase 1 |
| PSAT1 | 1.13 | 2.78E-08 | 9 | Phosphoserine aminotransferase 1 |
| COL6A1 | 1.13 | 1.23E-06 | 21 | Collagen, type VI, alpha 1 |
| FGFBP2 | 1.12 | 0.000404 | 4 | Fibroblast growth factor binding protein 2 |
| CTSB | 1.11 | 1.27E-07 | 8 | Cathepsin B |
| UCHL1 | 1.10 | 7.90E-05 | 4 | Ubiquitin carboxyl-terminal esterase L1 (ubiquitin thiolesterase) |
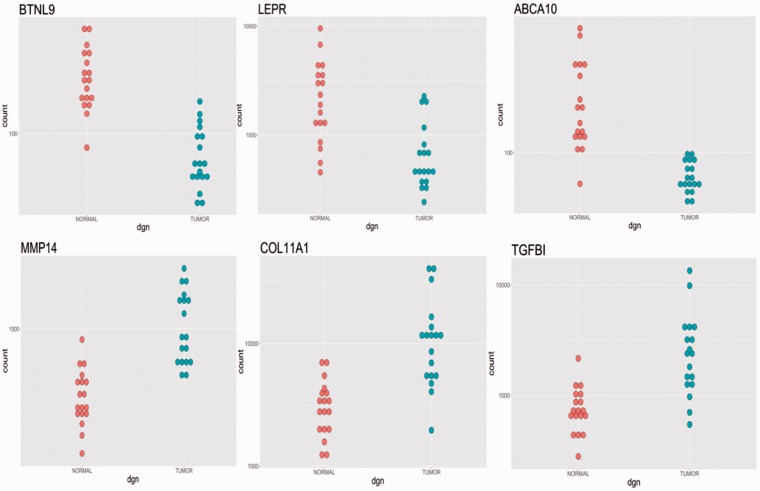
Based on the fold change (log2fold) differences between tumor and control samples, we generated the list of genes that were the most downregulated and the most upregulated in tumor tissues. It means that these genes got the lowest log2fold and largest log2fold, respectively, with significant P value. The most downregulated genes in osteosarcoma are the following: BTNL9, DNASE1L3, CAMP, LEPR, MIR223, MS4A3, LTF, LCN2, MMP8, S100A12, S100A8, MPO, EPB42, HEMGN, AHSP, ABCA10, BPI, CEACAM6, DEFA (Table 2). The most upregulated genes in osteosarcoma were COL11A1, TGFBI, TREM2, COL2A1, COL10A1, HAPLN1, MMP14, PANX3, CTHRC1, STEAP1, COL3A1 (Table 3). From ranking by FDR value, we got the list of genes with the lowest FDR which were considered as the most significantly differential expression between osteosarcoma and control samples. The most significant (by FDR value) differentially expressed genes were BTNL9, MMP14, ABCA10, ACACB, COL11A1, and PKM2 (PKM) showed in the supplementary Table 1. In order to visualize the differential expression between tumor and non-tumor samples, we plotted the individual values of some significantly changed genes. Figure 2 illustrates the three downregulated genes and the three upregulated genes in osteosarcoma tissue. It is evident that all these genes have very consistent patterns of expression. Moreover, we also used pairwise plotting of the gene expression values from the same patients. In supplementary Figure 1, BTNL9, LEPR, MMP14, and COL11A1 expression is compared in tumor-normal pairs, in which the data from same individuals are connected with the lines. This figure indicates that the expressional changes between individual data points are very consistent and our findings are reliable.
Figure 2.
We chose four genes (BTNL9, ABCA10, MMP14, and Col11A1) which had the lowest padj and we chose LEPR and TGFBI on the top lists of downregulated and upregulated genes for illustration in Figure 2. The expression of six genes (three upregulated and three downregulated) with the largest differences between tumor and control samples indicates very good consistency between different samples. We can easily see the BTNL9, LEPR, ABCA10 were downregulated (log2foldchange < −1 with padj much less than 0.001) and MMP14, COL11A1, TGFBI (log2foldchange > 1 with padj much less than 0.001) were upregulated in tumor samples. The blue dots were illustrated for tumor samples and the red ones were for normal samples. (A color version of this figure is available in the online journal.)
Reactome analysis of fresh samples
In addition to the list of differentially expressed genes, we aimed to characterize the disease-specific transcriptome signature in paired samples. We used the Reactome database which is a manually curated resource describing chemical reactions, biological processes, and pathways.21–23 Reactome analysis indicated significant enrichment of the pathways related to the collagen degradation, extracellular matrix organization and to the erythrocyte activation (Supplementary table 1). Supplementary Figure 2 indicates the eight most significantly activated pathways, enrichment P-values, and the number of genes involved in each particular pathway. All these eight pathways are related to degradation of the extracellular matrix or to the hematopoiesis. Supplementary Figure 3 describes the genes and their expression levels from the pathways illustrated in the Supplementary Figure 2.
Gene expression changes induced by chemotherapy
Next we decided to analyse the transcriptional changes after chemotherapy in FFPE samples using edgeR package. In our analysis set, we had 5 samples taken after and 10 samples taken before chemotherapy. By using padj < 0.1, 22 genes were found to be differentially expressed with chemotherapy. They all have log2foldchange more than 1 which means that they are all upregulated with chemotherapy. These genes are listed in Table 4 including POSTN, AMBN, SLC35F3, TAC3, TYROBP, STX7, HEATR1, OGN, USP6, SPRR1B, ASPN, SULF1, DNAJC14, NOB1, FAP, SP3, C8ORF59, CCNB2, KRTDAP, MRPS24, C2ORF89, and RPS6. To understand the pathways related to these genes, a gene-set enrichment analysis of these 22 genes was done which detected the following pathways with padj <0.1: Collagen biosynthesis and modifying enzymes, collagen formation, Influenza viral RNA transcription and replication, eukaryotic translation elongation, integrin cell surface interactions, SRP-dependent cotranslational protein targeting to membrane, collagen degradation (Table 5). Interestingly, a majority of the genes upregulated with chemotherapy was involved in the collagen formation and support osteogenesis.
Table 4.
The most significantly up-regulated genes in FFPE OS samples after chemotherapy.
| Symbol | log2FoldChange | PValue | padj | Genename |
|---|---|---|---|---|
| POSTN | 3.16 | 2.59E-08 | 0.00 | Periostin. osteoblast specific factor |
| AMBN | 4.07 | 1.80E-07 | 0.00 | Ameloblastin (enamel matrix protein) |
| SLC35F3 | 3.84 | 1.93E-07 | 0.00 | Solute carrier family 35. member F3 |
| TAC3 | 3.90 | 1.71E-07 | 0.00 | Tachykinin 3 |
| TYROBP | 3.16 | 1.58E-07 | 0.00 | TYRO protein tyrosine kinase binding protein |
| STX7 | 2.99 | 5.36E-07 | 0.00 | Syntaxin 7 |
| HEATR1 | 2.12 | 5.18E-06 | 0.02 | HEAT repeat containing 1 |
| OGN | 1.97 | 7.49E-06 | 0.02 | Osteoglycin |
| USP6 | 2.49 | 7.34E-06 | 0.02 | Ubiquitin specific peptidase 6 |
| SPRR1B | 2.53 | 9.96E-06 | 0.02 | Small proline-rich protein 1B |
| ASPN | 1.94 | 1.54E-05 | 0.03 | Asporin |
| SULF1 | 1.07 | 1.52E-05 | 0.03 | Sulfatase 1 |
| DNAJC14 | 2.11 | 2.24E-05 | 0.04 | DnaJ (Hsp40) homolog. subfamily C. member 14 |
| NOB1 | 2.25 | 2.75E-05 | 0.04 | NIN1/RPN12 binding protein 1 homolog |
| FAP | 2.21 | 3.48E-05 | 0.05 | Fibroblast activation protein. Alpha |
| SP3 | 2.23 | 3.27E-05 | 0.05 | Sp3 transcription factor |
| C8orf59 | 2.79 | 4.50E-05 | 0.05 | Chromosome 8 open reading frame 59 |
| CCNB2 | 1.04 | 4.40E-05 | 0.05 | Cyclin B2 |
| KRTDAP | 3.24 | 4.54E-05 | 0.05 | Keratinocyte differentiation-associated protein |
| MRPS24 | 2.19 | 4.63E-05 | 0.05 | Mitrochondrial ribosomal protein S24 |
| C2orf89 | 2.34 | 6.99E-05 | 0.08 | TraB domain containing 2A |
| RPS6 | 2.38 | 7.47E-05 | 0.08 | Ribosomal protein S6 |
Table 5.
The result of gene set enrichment analysis of 22 genes differentially expressed after chemotherapy.
| ID | Description | setSize | enrichmentScore | P-adjust |
|---|---|---|---|---|
| 1650814 | Collagen biosynthesis and modifying enzymes | 9 | 0.61 | 0.09 |
| 1474290 | Collagen formation | 10 | 0.54 | 0.09 |
| 168273 | Influenza viral RNA transcription and replication | 22 | 0.52 | 0.09 |
| 156842 | Eukaryotic translation elongation | 21 | 0.56 | 0.09 |
| 216083 | Integrin cell surface interactions | 8 | 0.61 | 0.09 |
| 1799339 | SRP-dependent cotranslational protein targeting to membrane | 21 | 0.56 | 0.09 |
| 1442490 | Collagen degredation | 9 | 0.76 | 0.09 |
Note: We used hypergeometric model to assess whether the number of selected genes associated with the reactome pathway is larger than expected. We listed here the most significant pathways with P-adjusted <0.1 (after correction for multiple testing). The enrichment score reflects the degree to which a gene set is overrepresented at the extremes (top or bottom) of the entire ranked list.
Discussion
To our knowledge, the present study is the first to use whole transcriptome analysis (RNAseq) of osteosarcoma to reveal the differentially expressed genes by pairwise comparison of tumor and normal bone of each patient. Our study found 6775 and 5365 differentially expressed genes with an adjusted P-value below 0.1 and 0.05, respectively. This is a much higher number than in previous papers. In a meta-analysis composed of in vivo and in vitro studies by Yang et al.,13 979 genes were found to be differentially expressed. Our previous study using RNAseq for analyzing a single case of osteosarcoma in Estonia identified only 65 differentially expressed genes.15 One genechip study with 10 osteosarcoma and 2 normal tissue samples found 1608 genes as differentially expressed genes (DEGs) between samples.24 The large differences in the number of DEGs may come from the differences in how the control group was formed. In this last study, the authors used two normal samples from healthy persons as controls to compare with osteosarcoma samples. In contrast, we used paired tumor-normal samples that allow a direct comparison of the diseased and normal tissue from the same patient. This design reduces substantially the biological variability and increases statistical power.
The most significant differentially expressed genes were BTNL9, MMP14, ABCA10, ACACB, COL11A1, and PKM2. BTNL9 was not mentioned in previous osteosarcoma studies. In our study, BTNL9 was the most significantly differentially expressed. This gene is butyrophilin-like 9, also known as BTN3 or BTN8 or VDLS1900 and it is located in 5q35.3. It is one of the 13 members of human butyrophilin family which plays a role in immune homeostasis though the function of BTNL9 was not known.25 MMP14 belongs to the MMP family, which is a hallmark of invasive cancers, and was found to interact with COL1A2 and COL5A2 in osteosarcoma.16 It was found to be upregulated in breast cancer, mesothelioma and lung cancer26–28 and to be correlated to poor prognosis.28 Interestingly, inhibiting the MMP14- through targeting hemopexin domain of MMP14 showed promising approach in treating lung cancer.27 The ABCA10 is ATP binding cassette subfamily A member 10 and it is located in 17q24. ABCA10 was found to be downregulated in colorectal cancer and ovarian cancer.29,30 The relationship of ABCA10 with osteosarcoma has not been previously mentioned in the literature but we found it downregulated as in some others epithelial cancers mentioned above. It was shown significantly associated with PFS (progression-free survival) of ovarian cancer30 and it should be further evaluated for a prognostic marker in osteosarcoma. Similarly, in the present study, ACACB is here noted for the first time in relation to osteosarcoma. The COL11A1 gene codes for the α1 chain of procollagen and mature collagen of type XI, which is an extracellular minor fibrillar collagen.31 In our present study, COL11A1 was upregulated. It has not been discovered in recent studies of osteosarcoma, although COL1A2 and COL5A2 were found to be upregulated in osteosarcoma samples.16 COL11A1/(pro)collagen 11A1 is highly expressed by activated stromal cells of the desmoplastic reaction of human invasive carcinomas of the oral cavity, pharynx, head and neck, breast, lung, esophagus, stomach, pancreas, colon, and ovary.31 The expression of COL11A1/(pro)collagen 11A1 is correlated with carcinoma aggressiveness and progression, metastasis, and drug resistance. The suggested pathway model regulation of COL11A1 via TGF-β to regulates MMP3, MMP9, and finally it promotes cell proliferation, invasion, metastasis, and drug resistance may be promising in finding targeted therapies in the future for several cancers including osteosarcoma.31,32 PKM pyruvate kinase muscle known as PKM2 was found to be overexpressed in clear-cell renal carcinoma (ccRCC) and when it was knocked down in ccRCC cells, the rapid proliferation, high glucose consumption, and high lactate production were all clearly inhibited.33 This finding matched with ours and it is interesting for further evaluation on osteosarcoma.
The Reactome analysis showed significant enrichment of specific pathways in case of osteosarcoma. The enrichment was focused on two main groups. One group is the hemoglobin interacting network which was also reported in a recent osteosarcoma study.24 In the present paper, we identified a hematopoiesis interacting network that is composed of HBA1, HBA2, HBB, CA1, and RHAG genes. These genes are responsible for the O2/CO2 exchange in erythrocytes. The second group of genes functions for collagen synthesis and degradation and this group is composed of related genes like COL11A1, COL10A1, COL2A1, MMP14, COL3A1, COL6A1, MMP11, CTSB, ACAN, COL6A3, CTSG, MMP8, ELANE. Therefore, although the list of genes is long, several genes indicate the enrichment of the cancer progression and extracellular matrix remodeling. COL was suggested a possible feature genes of OS.34 In a study on six samples (cell lines), they shared some common pathways with us including extracellular matrix organization, integrin cell surface interactions.35 Our findings herein on osteosarcoma –a very malignant, highly metastasizing tumor matched quite well with new findings on the role of extracellular matrix remodeling in sarcoma progression and metastasis. Remodeling of ECM by collagen degradation and re-deposition in interactions with matrix metalloproteinases (MMPs) promotes tumor infiltration, angiogenesis, invasion, and migration. Recent understanding about its mechanism leads to several clinical trials especially MMPs-inhibitor drugs and suggested for more targeted therapies in the future. MMPs were suggested to be further studied as a marker of invasiveness and risk of metastasis.36–38
In a previous meta-analysis, 979 genes were found to show altered expression in samples of OS compared with normal control.13 Among those genes, there were 472 upregulated genes and 507 downregulated genes. The most significantly upregulated genes were CPE, HEY1, PPT1, PTBP2, COLEC12, CPVL, FXYD6, RIN2, RPF1, EFNA1 and the most significantly downregulated genes were NPR3, LMOD1, GAS6, RDH5, AOX1, LIMS2, LTBP2, RGS4, PPAP2B, CLCF1.13 In this study, several different sources of data were combined (in vivo and in vitro), so the list of genes and gene networks is quite diverse. In our previous study with a single case of osteosarcoma in Estonia, we found 65 genes to be differentially expressed between the tumor sample and normal bone. There were 7 downregulated genes and 58 upregulated genes in the sarcoma. ADIPOQ was the most highly upregulated gene.15 Interestingly, in the present study, we found leptin receptor (LEPR) to be significantly downregulated in osteosarcoma. Leptin and adiponectin are functionally related hormones and in this respect our previous and present findings are coherent. In another study of 20 samples (14 osteosarcoma and 6 normal samples), three differentially expressed genes GJA1, COL1A2, and COL5A2 were identified.16 The authors observed that COL1A2 and COL5A2 interact with a number of genes of the matrix metalloprotease (MMP) family, including MMP1, MMP2, MMP3 and MMP14, TGFβ and RUNX2.16 Therefore, we can consider that results from present study fit very well with previously published results.
Additionally, FFPE samples were analysed to see if there is any changes due to chemotherapy. We compared gene expression before and after chemotherapy. Twenty-two genes were found to be upregulated significantly with chemotherapy (padj < 0.1 and log2foldchange >1). Of 22 upregulated genes with chemotherapy, POSTN and AMBN had the lowest P value. POSTN (periostin) is also known as PN; OSF2; OSF-2 or PDLPOSTN. It encodes a secreted extracellular matrix protein that functions in tissue development and regeneration.39 AMBN (ameloblastin) encodes the nonamelogenin enamel matrix protein ameloblastin. The encoded protein may be important in enamel matrix formation and mineralization. This gene is located in the calcium-binding phosphoprotein gene cluster on chromosome 4.39 The functional importance of the AMBN extracellular matrix protein in bone fracture prevention and rapid fracture healing was identified40 and interestingly, AMBN induced a tumor suppressive phenotype and chemosensitivity to doxorubicin (a main component of combination chemotherapy in OS) in osteosarcoma.41 By using gene-set enrichment analysis of these 22 genes, some significant pathways were identified with padj < 0.1 (Table 5). It is interesting that collagen biosynthesis and modifying enzymes, collagen formation, collagen degradation were among the most significant pathways. We can see that these pathways lead to the degradation and remodeling of the extracellular matrix (ECM). This finding suggests that chemotherapy may induce the remodeling of ECM. We emphasize that degradation and remodeling of ECM was found to be an important pathway in osteosarcoma samples compared with normal bone samples as we analyzed above. The activation of collagen biosynthesis-related pathways after chemotherapy supports the fact that degradation and remodeling of ECM may be an important mechanism of the disease . But we could not find a similar study to compare the role of chemotherapy in gene expression in OS.
In conclusion, we found 5365 genes to be differentially expressed between the normal and osteosarcoma tissues with FDR adjusted P-value below 0.05. Among them, BTNL9, MMP14, ABCA10, ACACB, COL11A1, and PKM2 were the most significantly differentially expressed genes. The degradation of collagen seems to be an important mechanism of osteosarcoma and should be further studied to serve as a biomarker of osteosarcoma. Robust activation of several COL family genes supports their involvement in malignancy and their potential role in tumorigenesis.
Supplementary Material
Authors’ contributions
XDH contributed in study designing, samples collection, laboratory works, writing manuscript. PP, VQL, VHN, NTNL, LHT and HGN contributed in samples collection, data registry and follow up patients, manuscript review. ER, EP, GK contributed in RNA extraction, sequencing, data analysis. Associate professor, AM, Associate professor KM, professor SK contributed in study design, supervising, data analysis and manuscript writing.
Declaration of Conflicting Interests
The author(s) declared no potential conflicts of interest with respect to the research, authorship, and/or publication of this article.
Funding
This study was supported by institutional research funding (IUT20-46) from the Estonian Research Agency and by the European Union’s Seventh Framework Programme (FP7/2007-2013) under grant agreement no. 602398 (Hyporth).
Supplementary Materials
Our data GSE99671 can be accessed at GEO at NCBI. https://www.ncbi.nlm.nih.gov/geo/query/acc.cgi?acc=GSE99671
References
- 1.Picci P. Osteosarcoma (Osteogenic sarcoma). Orphanet J Rare Dis 2007; 2:6. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Mirabello L, Troisi RJ, Savage SA. Osteosarcoma incidence and survival rates from 1973 to 2004: data from the surveillance, epidemiology, and end results program. Cancer 2009. ;115:1531–43. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Whelan J, McTiernan A, Cooper N, Wong YK, Francis M, Vernon S, Strauss SJ. Incidence and survival of malignant bone sarcomas in England 1979-2007. Int J Cancer 2012; 131: E508–17 [DOI] [PubMed] [Google Scholar]
- 4.Mirabello L Troisi RJ andSavage SA.. International osteosarcoma incidence patterns in children and adolescents, middle ages and elderly persons. Int J Cancer 2009; 125:229–34. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Kansara M, Teng MW, Smyth MJ, et al. Translational biology of osteosarcoma. Nat Rev Cancer 2014; 14:722–35 [DOI] [PubMed] [Google Scholar]
- 6.Luetke A, Meyers PA, Lewis I, Juergens H. Osteosarcoma treatment – where do we stand? A state of the art review. Cancer Treat Rev 2014; 40:523–32 [DOI] [PubMed] [Google Scholar]
- 7.Chou A, S.Geller D, Gorlick R. Therapy for osteosarcoma: where do we go from here? Pediatr Drugs 2008; 10:315–27 [DOI] [PubMed] [Google Scholar]
- 8.Chen X, Bahrami A, Pappo A, Easton J, Dalton J, Hedlund E, Ellison D, Shurtleff S, Wu G, Wei L, Parker M, Rusch M, Nagahawatte P, Wu J, Mao S, Boggs K, Mulder H, Yergeau D, Lu C, Ding L, Edmonson M, Qu C, Wang J, Li Y, Navid F, Daw NC, Mardis ER, Wilson RK, Downing JR, Zhang J, Dyer MA. Recurrent somatic structural variations contribute to tumorigenesis in pediatric osteosarcoma. Cell Rep 2014; 7:104–12 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Gao Y, Luo L, Li S, Yang C. miR-17 inhibitor suppressed osteosarcoma tumor growth and metastasis via increasing PTEN expression. Biochem Biophys Res Commun 2014; 444:230–4. [DOI] [PubMed] [Google Scholar]
- 10.Chong Y, Zhang J, Guo X, Li G, Zhang S, Li C, Jiao Z, Shao M. MicroRNA-503 Acts as a Tumor Suppressor in Osteosarcoma by Targeting L1CAM. PLoS One 2014; 9:e114585. [DOI] [PMC free article] [PubMed] [Google Scholar] [Retracted]
- 11.Bao Y-P, Yi Y, Peng L-L, Fang J, Liu K-B, Li W-Z, Luo H-S. Roles of microRNA-206 in osteosarcoma pathogenesis and progression. Asian Pac J Cancer Prev 2013; 14:3751–5. [DOI] [PubMed] [Google Scholar]
- 12.Cai H, Zhao H, Tang J, Wu H. Serum miR-195 is a diagnostic and prognostic marker for osteosarcoma. J Surg Res 2015; 194:505–10. [DOI] [PubMed] [Google Scholar]
- 13.Yang Z, Chen Y, Fu Y, Yang Y, Zhang Y, Chen Y, Li D. Meta-analysis of differentially expressed genes in osteosarcoma based on gene expression data. BMC Med Genet 2014; 15:80. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Siddiqui AS, Delaney AD, Schnerch A, Griffith OL, Jones SJ, Marra MA. Sequence biases in large scale gene expression profiling data. Nucleic Acids Res 2006; 34:e83. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Märtson A, Kõks S, Reimann E, Prans E, Erm T, Maasalu K. Transcriptome analysis of osteosarcoma identifies suppression of WNT pathway and up-regulation of adiponectin as potential biomarker. Genom Discov 2013;[cited 2014 Sep 15]; 1(1):3. [Google Scholar]
- 16.Wu D, Chen K, Bai Y, Zhu X, Chen Z, Wang C, Zhao Y, Li M. Screening of diagnostic markers for osteosarcoma. Mol Med Rep 2014; 10:2415–20. [DOI] [PubMed] [Google Scholar]
- 17.Love MI, Huber W, Anders S. Moderated estimation of fold change and dispersion for RNA-seq data with DESeq2. Genome Biol 2014; 15:550. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Robinson MD, McCarthy DJ, Smyth GK. edgeR: a Bioconductor package for differential expression analysis of digital gene expression data. Bioinformatics 2010; 26:139–40. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Anders S, Huber W. Differential expression analysis for sequence count data. Genom Biol 2010. Jan [cited 2014 Jul 9]; 11:R106. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Benjamini Y, Hochberg Y. Controlling the False Discovery Rate : a Practical and Powerful Approach to Multiple Testing When researchers tend to select pursuing multiple the (statistically) and support of conclusions. An unguarded use in a greatly results of single-inference inc. JR Statist 1995; 57:289–300 [Google Scholar]
- 21.Yu G, He Q-Y. ReactomePA: an R/Bioconductor package for reactome pathway analysis and visualization. Mol BioSyst 2016; 12:477–9. [DOI] [PubMed] [Google Scholar]
- 22.Fabregat A, Sidiropoulos K, Garapati P, Gillespie M, Hausmann K, Haw R, Jassal B, Jupe S, Korninger F, McKay S, Matthews L, May B, Milacic M, Rothfels K, Shamovsky V, Webber M, Weiser J, Williams M, Wu G, Stein L, Hermjakob H, D’Eustachio P. The reactome pathway knowledgebase. Nucleic Acids Res 2016; 44:D481–7. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Subramanian A, Tamayo P, Mootha VK, Mukherjee S, Ebert BL, Gillette MA, Paulovich A, Pomeroy SL, Golub TR, Lander ES, Mesirov JP. Gene set enrichment analysis: a knowledge-based approach for interpreting genome-wide expression profiles. Proc Natl Acad Sci USA 2005; 102:15545–50. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Zhao L, Zhang J, Tan H, Wang W, Liu Y, Song R, Wang L. Gene function analysis in osteosarcoma based on microarray gene expression profiling. Clin Exp Med 2015; 8:10401–10 [PMC free article] [PubMed] [Google Scholar]
- 25.Arnett HA, Viney JL. Immune modulation by butyrophilins. Nat Publ Gr 2014; 14:559–69 [DOI] [PubMed] [Google Scholar]
- 26.Crispi S, Calogero RA, Santini M, Mellone P, Vincenzi B, Vicidomini G, Fasano S, Meccariello R, Cobellis G, Menegozzo S, Pierantoni R, Facciolo F, Baldi A, Menegozzo M. Global gene expression profiling of human pleural mesotheliomas : identification of matrix metalloproteinase 14 (MMP-14) as potential tumour target. PLoS One 2009; 4: e7016. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Stawowczyk M, Wellenstein MD, Lee SB, Yomtoubian S, Durrans A, Choi H, Narula N, Altorki NK, Gao D, Mittal V. Matrix Metalloproteinase 14 promotes lung cancer by cleavage of Heparin-Binding. EGF-like Growth Factor. Neoplasia 2017; 19:55–64 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Têtu B, Brisson J, Wang CS, Lapointe H, Beaudry G, Blanchette C, Trudel D. The influence of MMP-14, TIMP-2 and MMP-2 expression on breast cancer prognosis. Breast Cancer Res 2006; 8:R28. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Hlavata I, Mohelnikova-Duchonova B, Vaclavikova R, Liska V, Pitule P, Novak P, Bruha J, Vycital O, Holubec L, Treska V, Vodicka P, Soucek P. The role of ABC transporters in progression and clinical outcome of colorectal cancer. Mutagenesis 2012; 27:187–96. [DOI] [PubMed] [Google Scholar]
- 30.Elsnerova K, Mohelnikova-Duchonova B, Cerovska E, Ehrlichova M, Gut I, Rob L, Skapa P, Hruda M, Bartakova A, Bouda J, Vodicka P, Soucek P, Vaclavikova R. Gene expression of membrane transporters: importance for prognosis and progression of ovarian carcinoma. Oncol Rep 2016; 35:2159–70 [DOI] [PubMed] [Google Scholar]
- 31.Vázquez-Villa F, García-Ocaña M, Galván JA, García-Martínez J, García-Pravia C, Menéndez-Rodríguez P, González-del Rey C, Barneo-Serra L, de Los Toyos JR, Rey CG, Barneo-Serra L, de Los Toyos JR. COL11A1/(pro)collagen 11A1 expression is a remarkable biomarker of human invasive carcinoma-associated stromal cells and carcinoma progression. Tumor Biol 2015; 36:2213–22. [DOI] [PubMed] [Google Scholar]
- 32.Raglow Z, Thomas SM. Tumor matrix protein collagen XIα1 in cancer. Cancer Lett 2015; 357:448–53. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Huang J, Kong W, Zhang J, Chen Y, Xue W, Liu D, Huang Y. c-Myc modulates glucose metabolism via regulation of miR-184/PKM2 pathway in clear-cell renal cell carcinoma. Int J Oncol 2016;1569–75. [DOI] [PubMed]
- 34.Zhang Y, Yan S, Lu D, Dong F, Lian Y. Screening feature genes of osteosarcoma with DNA microarray: a bioinformatic analysis. Int J Clin Exp Med 2015; 8:7134–42. [PMC free article] [PubMed] [Google Scholar]
- 35.Yang J, Wang N. Analysis of the molecular mechanism of osteosarcoma using a bioinformatics approach. Oncol Lett 2016; 12:3075–80 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Nerenberg PS, Salsas-Escat R, Stultz CM. Collagen – a necessary accomplice in the metastatic process. Cancer Genom Proteom 2007; 4:319–28 [PubMed] [Google Scholar]
- 37.Lu P, Takai K, Weaver VM, Werb Z. Extracellular matrix degradation and remodeling in development and disease. Cold Spring Harb Perspect Biol 2011; 3:1–24. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Jabłońska-Trypuć A, Matejczyk M, Rosochacki S. Matrix metalloproteinases (MMPs), the main extracellular matrix (ECM) enzymes in collagen degradation, as a target for anticancer drugs. J Enzyme Inhib Med Chem 2016; 31(sup1):177–83. [DOI] [PubMed] [Google Scholar]
- 39.O’Leary NA, Wright MW, Brister JR, Ciufo S, Haddad D, McVeigh R, Rajput B, Robbertse B, Smith-White B, Ako-Adjei D, Astashyn A, Badretdin A, Bao Y, Blinkova O, Brover V, Chetvernin V, Choi J, Cox E, Ermolaeva O, Farrell CM, Goldfarb T, Gupta T, Haft D, Hatcher E, Hlavina W, Joardar VS, Kodali VK, Li W, Maglott D, Masterson P, McGarvey KM, Murphy MR, O’Neill K, Pujar S, Rangwala SH, Rausch D, Riddick LD, Schoch C, Shkeda A, Storz SS, Sun H, Thibaud-Nissen F, Tolstoy I, Tully RE, Vatsan AR, Wallin C, Webb D, Wu W, Landrum MJ, Kimchi A, Tatusova T, DiCuccio M, Kitts P, Murphy TD, Pruitt KD. Reference sequence (RefSeq) database at NCBI: current status, taxonomic expansion, and functional annotation. Nucl Acids Res 2016; 44:D733–45 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Lu X, Li W, Fukumoto S, Yamada Y, Evans CA, Diekwisch T, Luan X. The ameloblastin extracellular matrix molecule enhances bone fracture resistance and promotes rapid bone fracture healing. Matrix Biol 2016. May; 52–54:113–26. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Ando T, Kudo Y, Iizuka S, Tsunematsu T, Umehara H, Shrestha M, Matsuo T, Kubo T, Shimose S, Arihiro K, Ogawa I, Ochi M, Takata T. Ameloblastin induces tumor suppressive phenotype and enhances chemosensitivity to doxorubicin via Src-Stat3 inactivation in osteosarcoma. Sci Rep 2017; 7:40187. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.