Abstract
Ultraviolet light in the UVC range is a commonly used disinfectant to control viruses in clinical settings and water treatment. However, it is currently unknown whether human viral pathogens may develop resistance to such stressor. Here, we investigate the adaptation of an enteric pathogen, human echovirus 11, to disinfection by UVC, and characterized the underlying phenotypic and genotypic changes. Repeated exposure to UVC lead to a reduction in the UVC inactivation rate of approximately 15 per cent compared to that of the wild-type and the control populations. Time-series next-generation sequencing data revealed that this adaptation to UVC was accompanied by a decrease in the virus mutation rate. The inactivation efficiency of UVC was additionally compromised by a shift from first-order to biphasic inactivation kinetics, a form of ‘viral persistence’ present in the UVC resistant and control populations. Importantly, populations with biphasic inactivation kinetics also exhibited resistance to ribavirin, an antiviral drug that, as UVC, interferes with the viral replication. Overall, the ability of echovirus 11 to adapt to UVC is limited, but it may have relevant consequences for disinfection in clinical settings and water treatment plants.
Keywords: virus, experimental evolution, ultraviolet radiation, disinfection, ribavirin, resistance
1. Introduction
Human-hosted viruses, especially those with single-stranded RNA genomes, may exhibit high mutation rates, short generation times, and large census population sizes compared to other organisms. These traits contribute to a remarkable genetic and functional diversity and allow viruses to adapt to various challenges, such as environmental stressors or antiviral drugs (Duffy et al. 2008; Holmes 2009; Irwin et al. 2016). In recent years, the resistance of major viral pathogens such as influenza viruses or HIV to antiviral drugs has become one of the most serious concerns for public health (Hué et al. 2009; Kuritzkes 2011; Foll et al. 2014; Hurt 2014).
Disinfectants such as chemical oxidants (e.g. chlorine) or UVC radiation are of major importance to controlling the dissemination and transmission of viral pathogens via food, water or in clinical settings. Yet, the long-term effects of disinfectant use on the evolution of virus populations have not been investigated and may counteract the intended positive effects of disinfection. To date, only few studies explored the potential emergence of viral resistance to disinfectants. Notably, viruses isolated from chlorinated drinking water or sewage have been found to be less susceptible to disinfection than their corresponding laboratory strains (Shaffer et al. 1980; Payment et al. 1985). In laboratory studies, repeated exposure of poliovirus and the bacteriophage F116 to free chlorine led to the development of resistance (Bates et al. 1978; Maillard et al. 1998). Similarly, MS2 bacteriophages and human echovirus 11 (E11) became increasingly resistant to disinfection by chlorine dioxide upon repeated exposure in vitro (Zhong et al. 2016, 2017).
Combined, these studies suggest that human-hosted viruses with resistance to disinfection can emerge and indeed be sampled in the environment. However, infective viruses are not routinely monitored after disinfection because the costs and time demands associated with such a practice are currently prohibitive. As a result, there are no available data on the circulation of disinfection resistant viruses, and no information exists regarding their genetic composition. In order to investigate disinfection resistance mechanisms and the underlying genetic changes, we can therefore not rely on previous observations of genetic adaptation, but must instead produce resistant viruses and monitor the associated changes in vitro.
The UVC portion (100–280 nm) of UV radiation is one of the most frequently used physical disinfectants. Its most common applications include clinical sterilization of tools and surfaces in healthcare facilities, as well as drinking and wastewater treatment (Moeller et al. 2010). In water treatment, UVC owes its recent popularity to its efficiency against protozoan oocysts that are naturally resistant to chemical disinfection by chlorine (Hijnen et al. 2006), and to a lack of toxic disinfection byproducts. Besides engineered applications, genome damage mediated by UV light in the solar spectrum largely contributes to the natural inactivation of both autochthonous and pathogenic microorganisms in aquatic ecosystems (Carratalà et al. 2013; Mojica and Brussaard 2014). The antimicrobial activity of UVC arises from the random formation of photoproducts such as cyclobutane pyrimidine dimers (CPDs) in DNA and RNA chains upon exposure to radiation. These photoproducts act in two ways: they can block viral replication by interfering with the polymerase and can promote the random introduction of deleterious mutations across the entire viral genomes (Ikehata and Ono 2011). Thus, the emergence of resistant viruses to UVC may require a greater extent of genetic and phenotypic changes as compared to the mutations needed for the development of resistance against drugs or other stressors with specific genetic targets (Bank et al. 2016).
We here investigate the response of virus populations to continued exposure to UVC radiation using human E11, an enteric human pathogen. E11 is a representative of the Enterovirus genus of the Picornaviridae family, which also includes Coxsackievirus, Rhinovirus and Poliovirus. Enteroviruses are small (20–32 nm diameter), single-stranded (ss) RNA icosahedral viruses. Host entry is usually via the gastrointestinal tract, and thus the major mode of transmission is fecal-oral contact. Infection is associated with a wide variety of clinical symptoms, from asymptomatic to severe, which include poliomyelitis, common respiratory symptoms, and gastrointestinal disease as well as other non-specific symptoms (Tapparel et al. 2013). Since enterovirus virions are excreted by their hosts at high concentrations (up to 1010 viruses/g of feces) over long periods of time, their presence in raw wastewater is of concern (Gantzer et al. 1998; Sedmak et al. 2003; Lodder and de Roda Husman 2005; Okoh et al. 2010). Some studies have shown that enteroviruses can withstand wastewater treatments. Consequently, they are usually detectable in drinking water sources and surface waters that receive wastewater effluents (Shaffer et al. 1980; Lodder et al. 2010; Simmons and Xagoraraki 2011; Zhou et al. 2015), where they can persist for extended time periods and present a public health risk (Rzezutka and Cook 2004; Carter 2005).
Human enteroviruses are readily cultivated and easily handled in laboratory conditions, and, consequently, many recent studies of experimental evolution employ these viruses as a model. These studies have shown that human enteroviruses easily adapt to stressors, such as antiviral drugs targeting viral replication (e.g. ribavirin and guanidine hydrochloride), harmful environmental conditions (e.g. high temperatures), or even disinfectants (e.g. chlorine) (Bates et al. 1978; Shiomi et al. 2004; Sadeghipour et al. 2012, 2013). Furthermore, due to the typically high mutation rates of these viruses and to their high recombination rates, novel enterovirus genotypes emerge or re-emerge on a regular basis (Palacios and Oberste 2005; Modlin 2007).
The goal of the present study was to assess whether UVC resistant enteroviruses emerge under directed experimental adaptation and to characterize these populations with respect to both genotype and phenotype. To this end, we have combined experimental evolution, disinfection experiments and population genetics analyses to illuminate the mechanisms of virus adaptation to an infrequently studied causal agent (UVC radiation).
2. Materials and methods
2.1 Virus preparation and quantification
Human E11 stocks (Gregory strain, ATCC® VR737™) were produced by infecting sub-confluent monolayers of Buffalo green monkey kidney (BGMK) cells. The cells were maintained at 37 °C in 5 per cent CO2 with modified Eagle medium (MEM) (Gibco, Frederick, MD), supplemented with 1 per cent Penicillin-Streptomycin (Gibco, Frederick, MD) and 10 per cent (growth medium) or 2 per cent (maintenance medium) heat-inactivated fetal bovine serum (FBS; Gibco, Frederick, MD). The viruses were released from infected cells by freezing and thawing the culture flasks three times. To eliminate cell debris, the suspensions were centrifuged at 3,500 rpm for 5 minutes. To quantify the virus samples, we used a most probable number (MPN) infectivity assay. Briefly, BGMK cells were grown to 95 per cent confluence in flat-bottom ninety-six-well plates (Greiner CELLSTAR® ninety-six-well plates; Sigma Aldrich). Then, virus samples were diluted over a tenfold dilution series in MEM supplemented with 2 per cent FBS. The medium in each well was discarded and replaced with 150 µl of a diluted virus sample. Infected plates were then incubated for 5 days, after which inverted microscopy was used to differentiate infected from non-infected wells. E11 concentrations in the samples were then determined as the MPN of cytopathic units per milliliter (MPNCU/ml). The resulting stock solutions (≈109 MPNCU/ml) were stored at 4 °C until use.
2.2 Directed experimental evolution passages
Prior to the UVC-adaptation, an E11 population (hereafter referred to as WT) was adapted to cell culturing in BGMK cells by conducting three passages in the presence of MEM supplemented with 2 per cent FBS and Ribavirin (400 µM). The resulting population (henceforth named WT Rib+) served as the starting population for the following experimental evolution passages. Ribavirin was used here to increase the level of genetic diversity of the prototype laboratory strain used in the study, as higher diversity promotes adaptation (Vignuzzi et al. 2006). While the UVC inactivation kinetics of the WT and the WT Rib + were equivalent (P = 0.79, data not shown), the WT Rib + population grew to higher titers in BGMK cells as compared to the original WT (P < 0.05, data not shown), confirming that the WT Rib+ population is indeed better adapted to grow in the cell culture system used herein.
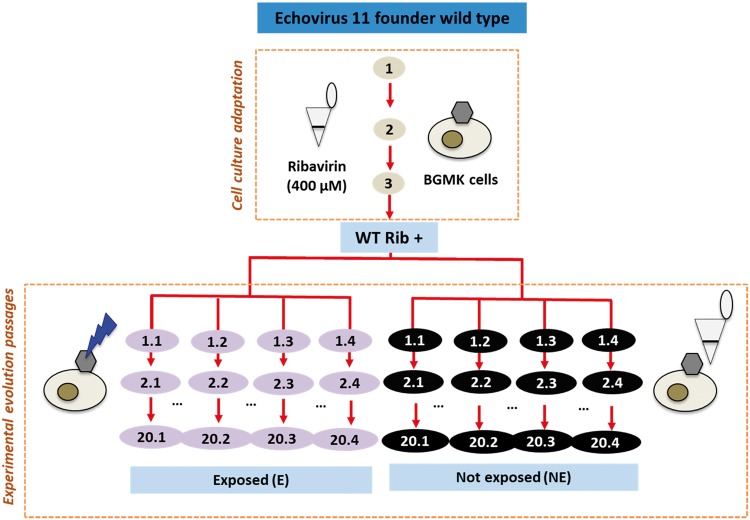
The cell culture pre-adapted WT populations (WT Rib+) were experimentally evolved in BGMK cells over twenty cycles. Each evolution passage included two steps: first the virus populations were exposed to UVC radiation, and then the survivors were regrown in BGMK cells for 3 days to increase the population size (Fig. 1). After regrowth, the viruses were released as detailed above and the experiment proceeded with a new round of UVC inactivation and regrowth. UVC radiation was produced from an 18 W low-pressure UVC lamp (model TUV T8; Philips), which emitted 253.7 nm light as quasi‐collimated beam. The fluence rate was determined by actinometry (Rahn 1997) and corresponded to 10.43 mJ/cm2 · min. The UVC fluence was assigned to achieve a 4 log10 reduction in infective virus titers, simulating a commonly used target threshold for virus inactivation in water treatment. Up to Passage 14, this fluence corresponded to 31.28 mJ/cm2. At Passage 15, due to the emergence of resistant viruses, the UVC fluence was increased to 52.14 mJ/cm2 until the end of the experiment. The 4 log10 reduction in virus titer resulted in a low multiplicity of infection (MOI) for regrowth of approximately 0.0001.
Figure 1.
Experimental set-up. Echovirus 11 was propagated infecting Buffalo green monkey kidney (BGMK) cells in monolayers. The viruses were adapted to cell culture for three passages in the presence of ribavirin and then serially passaged after exposure to UVC or after an equivalent dilution between cell culture infections. The multiplicity of infection (MOI) in the experimental evolution passages was 0.0001.
The typical replication time of enteroviruses is known to range between 5 and 10 hours (Knipe and Howley 2013). In the experimental evolution approach used herein, the regrowth phase in each passage lasted 72 hours, which corresponds to a maximum of 14.4 generations per passage. Assuming three to ten generations per passage, the twenty experimental evolution passages conducted thus corresponded to approximately 60–200 viral generations.
Control experiments with non-exposed populations were conducted in parallel, in reactors protected from UVC radiation with a cover of aluminum foil. Instead of inactivation by UVC, the concentration of these viruses was diluted with phosphate buffer by 4 log10 before being regrown, purified, and re-diluted. Hereafter, the final evolved populations (at Passage 20) will be referred to with the abbreviations E01 (exposed, Replicate 1) or NE01 (non-exposed, Replicate 1). Three biological replicates for both exposed (E02, E03, and E04) and non-exposed (NE02, NE03, and NE04) treatment groups were conducted using the same WT Rib+ as a founder population, in order to investigate the repeatability of our experimental evolution assays using one specific lineage.
2.3 UVC inactivation experiments
To determine the degree of UVC adaptation in the previously UVC exposed and control populations as compared to the WT Rib+, inactivation experiments were conducted every five passages. To this end, virus aliquots were spiked into 20 ml black glass beakers containing 2 ml of virus dilution buffer (VDB; 5 mM Na2HPO4, 10 mM NaCl, 1 mM NaOH; pH 7.2) to yield a concentration of approximately 107 MPNCU/mL, and were exposed to UVC light. The UV absorbance at 254 nm was measured for all samples (UV-2550; Shimadzu) to verify that samples did not differ in their extent of light screening. Aliquots were withdrawn periodically over 11 minutes, corresponding to a fluence of 114.70 mJ/cm2. The infective viruses in each sample were enumerated on the same day by MPN infectivity assays as detailed above. Inactivation rate constants k were determined by least-square fit of the data to a first-order model according to the following equation:
where N0 and N are the infective virus concentrations (MPNCU/mL) at times 0 and t, I is the fluence rate (mJ/cm2 · min), and t is the time (min).
2.4 Replicative fitness assays
To characterize the replicative fitness of the evolved populations, we compared the growth kinetics of E01 to that of the WT Rib+ in BGMK cells. Specifically, sub-confluent monolayers were infected at an MOI of 0.001 and incubated at 37 °C and 5 per cent of CO2. Aliquots of the culturing media were collected periodically over 96 hours post infection and the increase in virus titer was quantified by MPN assay.
2.5 Competition assays
We conducted competition experiments to determine whether under our experimental conditions the evolved populations could outcompete the WT Rib+ when co-cultured in BGMK cells. Specifically, clonal virus populations were established from the WT Rib+ and E01 populations by scission of isolated plaques using two successive plaque assays. The isolated plaques were transferred to 100 μl of VDB and were incubated overnight at 4 °C. Then, the liquid phase was transferred to a new tube and quantified as described above. Finally, the populations were mixed at 1:1 and 1:0.01 ratio of WT Rib+:E01. The mixtures were then spiked into cell culture flasks with sub-confluent BGMK monolayers and were passaged four times. Samples were retained at passages 0, 2, and 4, and a fragment in the 3Dpol region was then sequenced using Sanger sequencing as described below. In the sequenced fragment, the evolved viruses contain a known mutation (T347A) while the WT Rib+ viruses do not. This mutation thus allowed us to identify the dominant virus population after passaging.
2.6 Resistance to ribavirin
To determine if the evolved viruses exhibited cross-resistance to other agents that inflict random mutagenesis across viral genomes, we exposed different viral populations to an antiviral drug, ribavirin, and compared the viral titer in the stationary growth phase (48–72 hours post infection). This assay was conducted for populations WT, WT Rib+, E01, and NE01. In addition, several Rib- control populations (WT Rib-, E01, and NE01) were included in these analyses to ensure that resistance to ribavirin was not a result of the initial exposure the founder population to this drug. Rib-populations were obtained by passaging the WT three times in BGMK cells in the presence of MEM supplemented with 2 per cent of FBS and in the absence of ribavirin, and later evolved experimentally as previously detailed for the Rib+ lineage.
BGMK cells were seeded in six-well plates, and sub-confluent monolayers were incubated with MEM 2 per cent supplemented with 1.6 mM ribavirin for 30 minutes before adding the viral suspensions at an initial concentration of approximately 103 MPNCU/ml. After 48 hours of incubation, the plates were subjected to three freeze–thaw cycles, and the samples were collected and centrifuged at 3,500 × g for 5 minutes to eliminate cell debris. The virus-containing supernatant was then collected, and the samples were quantified by MPN assay.
2.7 Sanger sequencing
Nucleic acid extractions were completed using the PureLinkTM Viral RNA/DNA kit (Invitrogen) following manufacturer’s instructions. The extracted RNA was reverse transcribed and amplified with a OneStep RT-PCR Kit (Qiagen) in a Trio thermal cycler (Biometra) using twenty-eight sets of primers, which cover almost the entire genome of E11, and were designed based on the reference strain (NCBI accession number X80059.1) (Supplementary Table S1). The obtained Polymerase chain reaction (PCR) amplicons were then purified using the MSB Spin PCRapace Kit (Stratec). The sequencing PCR reactions were conducted for twenty cycles as follows: 96 °C for 10 seconds, 50 °C for 10 seconds, 60 °C for 4 minutes with a final hold at 12 °C using the BigDye Terminator v3.1 Cycle Sequencing Kit (Applied Biosystems). Lastly, the nucleic acids were sequenced in an ABI 3130x Genetic Analyzer (Applied Biosystems).
2.8 Amplicon-based deep sequencing
Amplicon-based deep sequencing was conducted for the WT Rib + population, for Passages 5, 7, 10, 15, and 18 of E01 and NE01, and for Passage 20 of all biological replicates of E and NE. Sequencing libraries of 100 bp double-stranded cDNA were built using the TruSeq Stranded mRNA Library Prep kit (Illumina) from 300 bp PCR amplicons obtained and purified as described in the previous section. The PCR amplicons were pooled together to obtain 100 ng of nucleic acids. The concentration of nucleic acids and quality of the obtained libraries were determined by Nanodrop 2000 (Thermo Fisher Scientific Inc.) and Fragment analyzer™ (Advanced Analytical). Up to six independent libraries were barcoded and pooled for sequencing using the Illumina HiSeq 2500 platform at the Lausanne Genomics Technologies Core Facility. Single-read reads of 100 bp were generated, trimmed, and cleaned for further analyses in the Bioinformatics and Biostatistics Core Facility at EPFL. The reads were mapped to the reference strain of E11 (NCBI accession number X80059.1). The sequencing error threshold was experimentally determined as described elsewhere and corresponded to 1 per cent.
2.9 Accession numbers
Genome sequence obtained by next-generation sequencing (NGS) have been deposited in NCBI’s BioSample database with the accession numbers SAMN07654485 to SAMN07654503.
2.10 Effective population size analyses
Effective population size (Ne) can be estimated by the amount of fluctuation in allele frequencies through time (Jorde and Ryman 2007). The unbiased estimator Fs′ of Ne is given by Jorde and Ryman (2007) as:
where fm and fm+1 are the minor allele frequencies at two consecutive sampling time points (m and m + 1), fi = (fm + fm+1)/2, and ñ is the harmonic mean of the sample sizes nm and nm+1 at two consecutive sampling time points. From the pool-sequenced time-serial samples, only single nucleotide polymorphisms (SNPs) above our ascertainment frequency of 0.5 per cent in one of the sampling time points and with coverage depth of at least 100 were considered. For each site in the genome, a major allele was defined as the nucleotide with the highest frequency at the first sampling time point, and a minor allele as the SNP with the highest frequency at the last sampling time point. The minor allele frequencies from populations NE01 and E01 were collated into input files for the software WFABC (Foll et al. 2014, 2015), which was used to calculate the Ne of each population. The following command line was used to execute the estimation of Ne from these input allele frequency trajectories:
./wfabc_1 FILE –min_freq 0.005 –ploidy 1
We assumed 10 viral generations per passage and a sample size of 100 per sampling time point. The posterior distribution of Ne was drawn from 10,000 samples generated using a Bayesian bootstrap approach (Rubin 1981; Hall 1985). The results for whole-genome estimates of Ne for each population are given in Supplementary Fig. S3. Furthermore, the time-serial Ne of NE01 and E01 populations was calculated with WFABC software by considering allele frequency trajectories between consecutive sampling time points (Fig. 5). In addition, the protein-specific Ne for each protein in NE01 and E01 populations was calculated using the same method (Supplementary Fig. S4).
Figure 3.
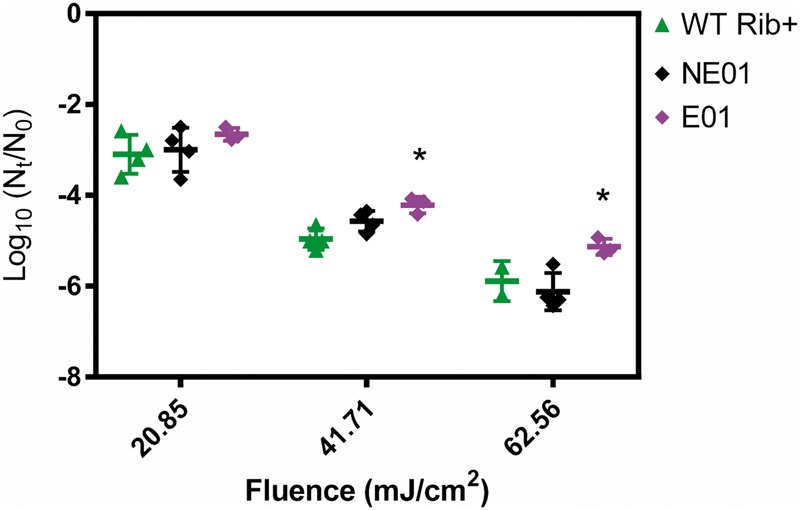
Virus log inactivation obtained for the WT Rib+, NE01 and E01 after exposure to different UVC fluences. Asterisks indicate statistically significant differences compared to the wild-type (WT) Rib+. Different data points represent replicate experiments.
2.11 Ratio of non-synonymous to synonymous segregating sites by protein
The ratio of non-synonymous to synonymous segregating sites () was calculated for each protein-coding gene. First, the WT Rib+ was used as the reference sequence to define segregating SNPs of each protein-coding gene, which were defined as those with minor alleles segregating at or above 1 per cent (the sequencing error threshold) at the last sampling time point. Secondly, the expected number of non-synonymous and synonymous sites in the reference sequence was calculated for any particular codon by denoting as the fraction of non-synonymous segregating sites at the ith codon position (i = 1, 2, 3). The number of expected non-synonymous (N) and synonymous (S) sites for a codon in the reference sequence (Morelli et al. 2013), was calculated as:
Lastly, the segregating SNPs in each protein-coding gene of the NE01 and E01 populations were categorized as either non-synonymous or synonymous mutations using Bioconductor software (Version 3.4), and compared to the expected number of non-synonymous/synonymous sites in the reference sequence respectively. The ratio of non-synonymous (Nsample) to synonymous (Ssample) segregating SNPs in the NE01 and E01 samples per expected non-synonymous (N) to synonymous (S) segregating sites in the reference was summarized for each protein in Table 1, where the ratio () is given as:
Table 1.
Heatmap of the frequency of the mutations detected by NGS in the NE and E populations after twenty experimental evolution passages. 
| Protein | NT | AA | WT | E20 |
NE20 |
||||||
|---|---|---|---|---|---|---|---|---|---|---|---|
| Replicate 1 | Replicate 2 | Replicate 3 | Replicate 4 | Replicate 1 | Replicate 2 | Replicate 3 | Replicate 4 | ||||
| VP4 | A849T | Y33F | 0 | 100 | 0 | 0 | 0 | 0 | 0 | 0 | 0 |
| VP2 | G1373C | G139R | 39 | 100 | 100 | 100 | 100 | 100 | 100 | 100 | 100 |
| VP3 | A2283G | / | 1 | 0 | 100 | 100 | 100 | 99 | 75 | 99 | 100 |
| VP1 | C2842T | / | 3 | 99 | 0 | 0 | 0 | 1 | 0 | 0 | 0 |
| G3164A | C236I | 0 | 0 | 70 | 0 | 75 | 19 | 81 | 27 | 40 | |
| A3170G | M238V | 6 | 100 | 99 | 99 | 100 | 100 | 98 | 100 | 100 | |
| A3233G | K259Q | 6 | 99 | 99 | 99 | 100 | 99 | 98 | 99 | 99 | |
| 2C | T4200C | V40A | 0 | 100 | 0 | 0 | 0 | 45 | 2 | 2 | 1 |
| G4262A | A61T | 1 | 1 | 100 | 99 | 100 | 95 | 90 | 51 | 100 | |
| C4263T | / | 1 | 0 | 100 | 100 | 100 | 4 | 88 | 38 | 98 | |
| A4552G | / | 4 | 100 | 100 | 100 | 100 | 96 | 75 | 99 | 100 | |
| T4990C | / | 1 | 100 | 0 | 0 | 0 | 1 | 0 | 0 | 0 | |
| 3A | G5170A | / | 0 | 100 | 0 | 0 | 0 | 0 | 0 | 0 | 0 |
| 3C | A5893T | / | 4 | 100 | 100 | 100 | 100 | 96 | 91 | 61 | 100 |
| 3D | C6061T | / | 5 | 100 | 100 | 100 | 100 | 96 | 91 | 62 | 100 |
| G6409A | / | 22 | 100 | 100 | 100 | 100 | 98 | 91 | 96 | 100 | |
| C6931T | / | 0 | 100 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | |
| A6989G | T347A | 3 | 99 | 100 | 99 | 100 | 99 | 99 | 99 | 99 | |
NT and AA stand for the nucleotide and amino acid position, respectively. The table includes all mutations that increased from minor to major allele compared to the starting population in at least one of the replicates. Heatmap of the frequency of the mutations detected by NGS in the NE and E populations after twenty experimental evolution passages. Yellow and blue colours stand for lower to higher frequencies, respectively.
2.12 Calculation of the mutation rate from time-series NGS data
In this mutation-accumulation experiment, identical lines of E11 were subjected to a series of consistent but extreme population bottlenecks such that mutations accumulate under effective neutrality (which does not apply in cases of complete lethality; Lynch et al. 2016). The control experiments (NE01) follow this scheme, while the UVC experiments (E01) also involve the selective pressure of UV damage. We do however use the same procedure to calculate the mutation rate in both experiments. The number of new mutations was taken as the number of newly arising mutations per generation per site between each pair of consecutive sampling time points as shown in Supplementary Table S2. Newly arising mutations are defined as nucleotides that appear de novo above the error threshold at a sampling time point. The error threshold was set at a conservative level of 1 per cent, and the protein-coding genome size (above the coverage of 100) was calculated as 6567. Finally, 10 generations were assumed to pass between each passaging. Using these parameters, the time-serial mutation rate was calculated by dividing the number of new mutations between a pair of passages by the time-serial effective population size. The results for the total mutation rate are summarized in Supplementary Table S2, and for the synonymous mutation rate in Supplementary Table S3, where the number of synonymous sites in the protein-coding genome was calculated to be 1696.3.
2.13 Data analyses
All statistical analyses were conducted using the Graph Pad Prism software (Prism 7 version 7.0a, 2016). Multiple comparison of inactivation rate constants and resistance against ribavirin between the WT and the evolved populations was performed by ordinary Analysis of variance (ANOVA) tests (Dunnett’s multiple test, with a single pool variance and α = 0.05).
3. Results
3.1 Phenotypic evolution of UVC-adapted populations
3.1.1 Adaptation to UVC radiation
Inactivation of the WT Rib+ and all biological replicates are shown in Fig. 2 as a function of UVC fluence. It is evident from these data that even after twenty rounds of UVC exposure and regrowth, UVC is still able to inactivate E11. Nevertheless, a significant enhancement in the resistance to UVC was observed in the UVC exposed (E) populations. This enhanced resistance to UVC was manifested in a smaller first-order inactivation rate constant k, which was determined from the first-order portion of the inactivation curve up to a UVC fluence of 41.71 mJ/cm2. Specifically, the k values associated with E01 at Passages 15 and 20 were reduced (P < 0.05) by 11 per cent (k = 0.24 cm2/mJ) and 15 per cent (k = 0.23 cm2/mJ), respectively, compared to the k value of the WT Rib+ (k = 0.27 cm2/mJ). Equivalent change in k was observed in all four biological replicates (Fig. 2) and was strictly associated with exposure to UVC radiation; the non-exposed populations did not exhibit a decreasing trend in k (Supplementary Fig. S1).
Figure 2.
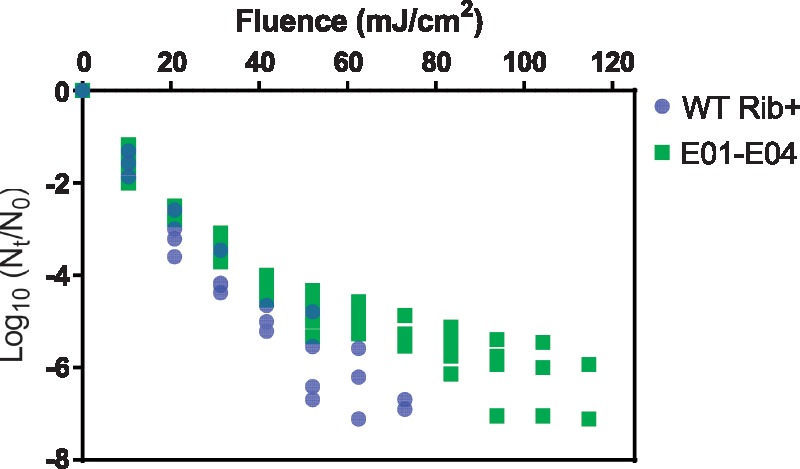
UVC inactivation of the wild-type (WT Rib+) and all the biological replicates of the population exposed to UVC (E01 to E04) after twenty passages of experimental adaptation. At minimum of three replicates were conducted for each population at initial concentrations of approximately 107 MPNCU/ml.
Figure 3 shows the measured log inactivation for populations WT Rib+, E01 and NE01 at the different UVC fluences tested. At fluences of 41.71 and 62.56 mJ/cm2, the extent of inactivation between E01 and WT Rib+ differed by 0.8 and 1.4 log units, respectively (P = 0.004 and P = 0.01), whereas no significant difference between NE01 and WT Rib+ was observed (P > 0.05). In summary, while the observed decrease in k is modest, it results in a significant reduction in the inactivation achieved at typical UVC fluences applied in water treatment.
3.1.2 Replicative fitness of UVC-resistant populations
In order to determine whether the increase in UVC resistance reduced the replicative fitness of the evolved populations, the growth of the E01 on BGMK cells was compared to that of the WT Rib+. If grown in independent cell cultures, the E01 population exhibited equivalent growth kinetics as WT Rib+ (data not shown). A different outcome was observed in competition assays, in which the E01 populations were mixed at 1:1 and 0.01:1 ratios with the WT Rib+, and were co-cultured on BGMK cells over four passages. Sequencing of the regrown populations revealed that—independent of the initial population ratio—the evolved populations (both exposed and non-exposed) rapidly outcompeted WT Rib+ (data not shown). Overall, these results indicate that, under our experimental conditions, UVC resistant viruses show no major replicative trade-offs as compared to the founding population.
3.1.3 Emergence of biphasic kinetics, a form of ‘viral persistence’
In addition to the reduction in k, a second mechanism leading to UVC resistance was observed. Specifically, the UVC inactivation of the WT Rib+ population reached the detection limit of the technique after being exposed to a fluence of 50 and 75 mJ/cm2, respectively (Supplementary Fig. S1). However, as passaging increased, we observed the onset of a tailing inactivation curve in all control (non-exposed) and UVC-exposed populations. Due to this tailing feature, the detection threshold increased to 100 mJ/cm2 or more for all NE and E populations (Supplementary Fig. S1). All E and NE populations were evolved under severe and serially repeated bottlenecks, even if the bottlenecks were due to different processes (UVC exposure versus dilution). The emergence of tailing may thus be linked to the process of frequent regrowth from low MOI.
3.1.4 Resistance to antiviral drugs
Like UVC, some antiviral drugs, such as ribavirin, lead to random mutagenesis during replication. Ribavirin is a mutagenic nucleotide analog that interferes with the replication of human enteroviruses by causing mutation accumulation in the viral progeny (Crotty et al. 2000; Graci and Cameron 2002; Palma et al. 2008). The genetic basis of ribavirin resistance in different enteroviruses, such as poliovirus or enterovirus 71, has been characterized in previous studies (Pfeiffer and Kirkegaard 2003; Sadeghipour et al. 2013). Here, we tested whether viruses capable of escaping UVC-induced mutagenesis can also escape ribavirin-induced mutagenesis. Specifically, we measured the inhibition of viral growth caused by ribavirin. Figure 4 shows that the NE01 and E01 populations became completely resistant to the effects of ribavirin (P = 0.0001 and P = 0.0002, respectively) and yielded equivalent final titers in both the presence and absence of the drug. Results obtained for control populations (Rib -) indicate that the observed drug resistance was not due to the initial cell culture adaptation of E11 in the presence of the drug (Supplementary Fig. S2), since resistance also emerged in these control populations.
Figure 4.
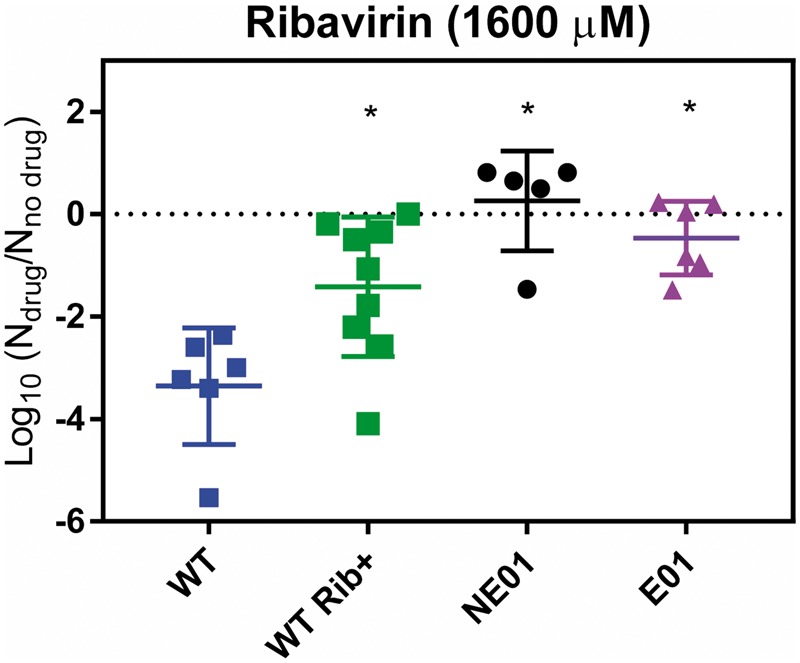
Viral titre of wild-type (WT), WT Rib+ and evolved populations (E01 and NE01) obtained in the stationary growth phase (48–72 hours) in the presence of ribavirin. Data are presented as the ratio of the titre in the presence versus absence of the drug. Asterisks indicate a statistically significant difference with respect to the WT.
3.2 Genome evolution
3.2.1 Mutations in the evolved populations
The heatmap in Table 1 shows the SNPs that increased from minor to major frequency or were fixed in one or more Rib+ populations, including the biological replicates. Both synonymous and non-synonymous mutations were fixed in all virus proteins except 3B. On the contrary, non-synonymous mutations were observed in the structural proteins VP4 (Y33F), VP2 (G139R) and VP1 (C236I, M238V, K259Q), and in the non-structural proteins 2 C (V40A, A61T) and 3Dpol (T347A). The mutations here identified in the VP4, VP2, and VP1 have been previously associated with changes in the thermal stability and alternative receptor use in EV11, respectively (Zhong et al. 2017).
3.2.2 Change in effective population size over time
To characterize the genetic evolution of our adapted viruses at the population-level, we estimated the overall and time-serial Ne (Fig. 5; Supplementary Fig. S3). The mode of Ne in the posterior distribution of the E01 is greater than that of the NE01 population (835 versus 538, respectively). This increase in diversity could be induced by the mutagenic effect of UVC in the E01 population. However, given that the number of segregating minor alleles (above the ascertainment frequency of 0.5%) was smaller in the E01 than in the NE population (439 versus 624, respectively), a larger census population size in the E01 population is the more likely explanation. The time-serial Ne in Fig. 5 shows that the Ne of the E01 population increases more than fourfold over the course of the experiment, whereas the Ne of the NE01 population remains relatively stable.
Figure 5.
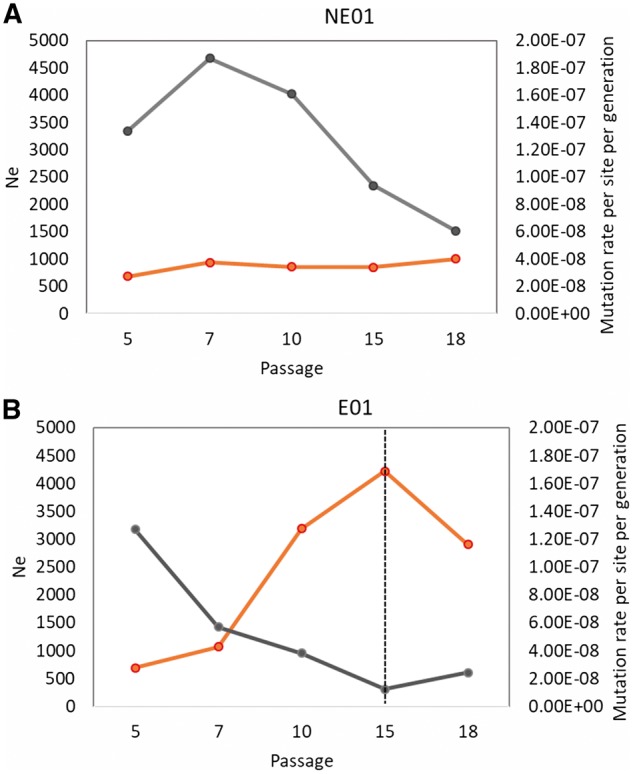
Time-serial effective population sizes (Ne) and mutation rate per site per generation of NE01 (A) and E01 (B) populations. The orange lines indicate the median of 1000 Ne estimates between consecutive sampling time points with the given passages. The grey line shows the mutation rate calculated for each population. The dashed line indicates an increase in the UVC fluence.
Values of Ne estimated protein-by-protein (Supplementary Fig. S4) also exhibit a difference between the NE01 and E01 populations in their responses to serial bottlenecks. To investigate whether the differences in the Ne estimates of protein-coding genes was caused by differing selective pressures acting upon them, we investigated the role of each protein in the genome-wide response to the experimental evolution regime. Specifically, we examined , a partial indicator of selective pressure on each protein (i.e. with a ratio equal to one implying neutrality, and deviations implying the action of positive or negative selection). Although this ratio is not a monotonic function of the selection coefficient, as it is in the ratio of non-synonymous to synonymous substitutions () between species, it can be used as an indicator of selective pressure on protein-coding genes (Kryazhimskiy and Plotkin 2008). As shown in Table 2, as expected, all proteins in both the NE01 and E01 populations appear to be strongly constrained and relaxed constraint alone thus appears to be a poor explanation for the observed differences.
Table 2.
The ratio of non-synonymous to synonymous segregating sites () by protein-coding genes accumulated in the NE and E populations during the experiments.
| Protein | VP4 | VP2 | VP3 | VP1 | 2A | 2B | 2C | 3A | 3B | 3C | 3D |
|---|---|---|---|---|---|---|---|---|---|---|---|
| Coding sites | 752–958 | 959–1,744 | 1745–2,458 | 2,459–3,334 | 3,335–3,784 | 3,785–4,081 | 4,082–5,068 | 5,069–5,335 | 5,336–5,401 | 5,402–5,950 | 5,951–7,336 |
| Size | 207 | 786 | 714 | 876 | 450 | 297 | 987 | 267 | 66 | 549 | 1,386 |
| Rib+ NE pN/pS | 0 | 0.19 | 0.13 | 0.36 | 0.29 | 0.06 | 0.16 | 0.42 | 0 | 0.17 | 0.29 |
| Rib+ E pN/pS | 0.18 | 0.52 | 0.28 | 0.71 | 0.34 | 0.37 | 0.29 | 0.05 | NAa | 0.18 | 0.49 |
pN/pS values are averages of the four replicates (E01–E04).
aNA, no synonymous segregating sites.
3.2.3 Mutation rate estimated from time-series data
The mutation rates of NE01 and E01 estimated from time-series NGS data in the early passages of our directed evolution experiments (Fig. 5) corresponded to 1.20 × 10−7 and 1.40 × 10−7 site per cell infection, respectively). While the trend of the mutation rate in the NE01 population is more variable over the tested passages, the mutation rate in this population was consistently higher than that of the E01 population at each sequenced passage (Fig. 5), indicating that the E01 population has a higher replication fidelity than the NE01.
4. Discussion
In this study, we have demonstrated that the efficiency of UVC is compromised by the emergence of virus populations with increased resistance to UVC radiation. On the one hand, the virus populations that were repeatedly exposed to UVC radiation adapted to the stressor with an overall decrease (approximately 15%) in their sensitivity to UVC. However, the extent of UVC resistance observed in our work was relatively minor compared to that previously observed for chemical disinfectants (Bates et al. 1978; Maillard et al. 1998; Zhong et al. 2017). This implies that despite limited adaptation, UVC remains a useful tool to control viruses. Nevertheless, adaptation to UVC may be problematic given the current practices of UV disinfection for water treatment. For example, the most recognized and cited UV disinfection guidelines, developed by the German Gas and Water Association, require that a validated UVC system for water treatment deliver a fluence of 40 mJ/cm2, with the assumption that this fluence achieves a 4 log10 inactivation of pathogens (Wirtschafts und Verlagsgesellschaft Gas und Wasser 2006). However, this 4 log10 target is not met by populations with reduced UVC sensitivity, which show an approximate inactivation of 3.4 log10 at the fluence of 40 mJ/cm2. The current guidelines are thus not sufficiently protective for the UVC-resistant viruses produced herein.
The NGS data did not allow us to identify specific mutations uniquely responsible for the increased resistance to UVC (change in k), given that we did not identify any mutation that was present in all exposed replicates but absent from the non-exposed ones. However, putatively beneficial mutations were identified based on observed frequency changes (Table 1), which are thus candidates for acting epistatically, for underlying what may in fact be a highly polygenic trait, or indeed may simply represent a wide variety of mutational solutions for achieving a common phenotype.
Besides increased UVC resistance, an additional phenotypic trait observed in both non-exposed and UVC-exposed populations was the sequential onset of biphasic kinetics. This feature has also been observed in bacterial populations exposed to high concentrations of antibiotics or heavy metals (Harrison et al. 2005; Lewis 2010; Maisonneuve and Gerdes 2014). These so-called bacterial persisters may show multi-resistance to diverse antibiotics by entering a dormancy state and are therefore less affected by drug treatment. ‘Viral persistence’ as observed herein cannot be explained by a dormant state, and the molecular underpinnings of the observed biphasic kinetics are not understood.
Notably, all the evolved viruses with biphasic inactivation kinetics also showed resistance to ribavirin, a mutagenic antiviral drug that targets virus replication that is currently used in clinical therapies (e.g. against hepatitis C virus). Mutations in the 2 C region of enteroviruses have previously been shown to confer multi-resistance against ribavirin and other mutagenic drugs (Baltera and Tershak 1989; Sadeghipour et al. 2012; Agudo et al. 2016). In addition, poliovirus and human enterovirus 71 with mutations in the 3Dpol coding region were found to exhibit resistance to ribavirin (Sadeghipour et al. 2013). 2C is a multifunction protein that is involved in the binding of the polymerase to the nucleic acids and in the exit of virus particles from the infected cell. 3Dpol is a protein involved in the replication fidelity of viral polymerase. Here, we identified several mutations in 2 C (V40A, A61T) and 3Dpol (T347A), and these SNPs were shared by all evolved populations (E and NE). It is thus likely that these mutations are associated with the novel phenotypic characteristics (i.e. the biphasic UVC inactivation kinetics and the resistance to ribavirin) present in both E and NE populations.
The onset of biphasic kinetics are of importance for controlling the long-term effects of disinfection in clinical settings where complete sterilization may not be reached and drug resistant microorganisms may circulate (Moges et al. 2014). In this context, it is also important to consider that the UV-tolerant and drug-resistant viruses were capable of outcompeting the wild type in co-cultures of BGMK cells under our experimental conditions. It is not clear that this finding also applies to human infection by enteroviruses in clinical settings, which involves both person-to-person and environmental transmission (Lu et al. 2015). However, a subset of the mutations identified herein (Supplementary Table S4) have been previously described in clinical or environmental isolates of E11, indicating that these mutations may also be beneficial, or at least not detrimental, in the context of overall virus circulation. Notably, the 2C (V40A, A61T) and 3Dpol (T347A) mutations identified in this study were present in 6 per cent, 26 per cent, and 85 per cent of the analyzed sequences, respectively.
Viruses with resistance to mutagenic activity have previously been found to exhibit changes in their replication mechanism, specifically a lowering of their mutation rate (Pfeiffer and Kirkegaard 2003; Sadeghipour et al. 2013). While we do not have information on the specific mutation rate of the starting (WT Rib+) population, the evolved populations showed a lower mutation rate than those previously reported for other enteroviruses (e.g. 9 × 10−5 substitutions per site per cell infection; Sanjuan et al. 2010). Furthermore, the mutation rate of the E01 population was consistently lower across all passages compared to the NE01 population. This may be explained by the interplay of mutation rate and effective population size. Specifically, during the experimental evolution of NE, samples were consistently diluted at the start of each passage, resulting in a constant and low MOI in all passages. This, in turn, yielded a constant and low Ne across the entire experiment (Fig. 5). In contrast, the sample size in the E populations were reduced by UVC at the start of each passage. Thereby the applied UVC fluence was kept constant up to Passage 15 and was only then increased to correct for the emergence of resistant viruses (Fig. 5). It is possible that the resistance to UVC increased in the early passages and that consequently the UVC exposure was not sufficient to ensure the same MOI as in the NE populations. This would lead to a larger census population size in the E population and could explain the difference in the Ne values. After adjusting the UVC fluence at Passage 15, the MOI and hence Ne decreased. The larger Ne in the early passages of the E populations may have led to more efficient selection in favor of a lower mutation rate. The lower mutation rate of E at the nucleotide level compared to NE is further consistent with its more pronounced phenotypic changes, namely the decrease in the UVC inactivation constant (k), as well as a higher resistance to ribavirin.
The benefit of low mutation rates to withstanding UVC or other mutagenic agents lies in reducing the total mutational load of the viruses under consideration (Matuszewski et al. 2017). All viruses accumulate similar numbers of mutations during UVC or ribavirin treatment. In contrast, the number of additional mutations incurred during viral replication varies with mutation rate. The total mutational load, and hence also the number of deleterious mutations, will be lower in those viruses with a high replication fidelity. Thus, variants with the highest replication fidelity are thus the most likely to survive treatment with a mutagenic agents (such as UVC and ribavirin) followed by regrowth.
In conclusion, our results demonstrate that resistance of human E11 to UVC may arise and likely exhibits the same mechanism as previously observed for viruses with resistance to mutagenic antiviral drugs (i.e. a lowering of the mutation rate). As such, viruses resistant to UVC may be cross-resistant to antiviral drugs that act through analogous mechanisms, and vice versa. Hence, the practice of disinfection, while of course important, also may contribute to the emergence of multiresistant viruses. These findings are thus relevant for ensuring the long-term efficiency of UVC radiation in the context of disinfection in water treatment and in clinical settings.
Author contributions
AC, QZ, and TK contributed to the design of the work. AC and VB collected the experimental data. AC, HS, QZ, JJ, and TK were involved in the data analysis and interpretation. AC and TK drafted the article. All authors contributed to the critical revision of the manuscript and have agreed on the final version of this manuscript.
Supplementary Material
Acknowledgements
This work was funded by the Swiss National Foundation (project numbers 31003 A_138319 and 31003 A_163270), and by an ERC Starting Grant to JDJ. We thank the Lausanne Genomic Technologies Facility (GTF) for their technical support to this project. The authors declare no conflict of interests.
Conflict of interest: None declared.
Supplementary data
Supplementary data are available at Virus Evolution online.
References
- Agudo R. et al. (2016) ‘Involvement of a Joker Mutation in a Polymerase-Independent Lethal Mutagenesis Escape Mechanism’, Virology, 494: 257–66. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Baltera R. F., Tershak D. R. (1989) ‘Guanidine-Resistant Mutants of Poliovirus Have Distinct Mutations in Peptide 2C’, Journal of Virology, 63/10: 4441–4. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bank C. et al. (2016) ‘An Experimental Evaluation of Drug-Induced Mutational Meltdown as an Antiviral Treatment Strategy’, Evolution (N. Y), 70/11: 2470–84. [DOI] [PubMed] [Google Scholar]
- Bates R. C., Shaffer P. T. B., Sutherland S. M. (1978) ‘Development of Poliovirus Having Increased Resistance to Chlorine Inactivation’, Journal of Applied & Environmental Microbiology , 34: 849–53. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Carratalà A. et al. (2013) ‘Environmental Effectors on the Inactivation of Human Adenoviruses in Water’, Food and Environmental Virology, 5/4: 203–14. [DOI] [PubMed] [Google Scholar]
- Carter M. J. (2005) ‘Enterically Infecting Viruses: Pathogenicity, Transmission and Significance for Food and Waterborne Infection’, Journal of Applied Microbiology, 98/6: 1354–80. [DOI] [PubMed] [Google Scholar]
- Crotty S. et al. (2000) ‘The Broad-Spectrum Antiviral Ribonucleoside Ribavirin Is an RNA Virus Mutagen’, Nature Medicine, 6/12: 1375–9. [DOI] [PubMed] [Google Scholar]
- Duffy S., Shackelton L. A., Holmes E. C. (2008) ‘Rates of Evolutionary Change in Viruses: Patterns and Determinants’, Nature Review Genetics, 9/4: 267–76. [DOI] [PubMed] [Google Scholar]
- Foll M. et al. (2014) ‘Influenza Virus Drug Resistance: A Time-Sampled Population Genetics Perspective’, PLoS Genetics, 10/2: e1004185. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Foll M., Shim H., Jensen J. D. (2015) ‘WFABC: A Wright-Fisher ABC-Based Approach for Inferring Effective Population Sizes and Selection Coefficients From Time-Sampled Data’, Molecular Ecology Resources, 15/1: 87–98. [DOI] [PubMed] [Google Scholar]
- Gantzer C., Maul A., Audic J. M., Schwartzbrod L. (1998) ‘Detection of Infectious Enteroviruses, Enterovirus Genomes, Somatic Coliphages, and Bacteroides fragilis Phages in Treated Wastewater’, Applied and Environmental Microbiology, 64: 4307–12. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Graci J. D., Cameron C. E. (2002) ‘Quasispecies, Error Catastrophe, and the Antiviral Activity of Ribavirin’, Virology, 298: 175–80. [DOI] [PubMed] [Google Scholar]
- Hall P. (1985) ‘Resampling a Coverage Pattern’, Stochastic Processes and Their Applications, 20/2: 231–46. [Google Scholar]
- Harrison J. J. et al. (2005) ‘Persister Cells Mediate Tolerance to Metal Oxyanions in Escherichia coli’, Microbiology, 151/Pt 10: 3181–95. [DOI] [PubMed] [Google Scholar]
- Hijnen W. A. M., Beerendonk E. F., Medema G. J. (2006) ‘Inactivation Credit of UV Radiation for Viruses, Bacteria and Protozoan (oo)cysts in Water: A Review’, Water Research, 40/1: 3–22. [DOI] [PubMed] [Google Scholar]
- Holmes E. C. 2009. The evolution and emergence of RNA viruses. Oxford Series in Ecology and Evolution. Oxford University Press.
- Hué S. et al. (2009) ‘Demonstration of Sustained Drug-Resistant Human Immunodeficiency Virus Type 1 Lineages Circulating Among Treatment-Naïve Individuals’, Journal of Virology, 83/6: 2645–54. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hurt A. C. (2014) ‘The Epidemiology and Spread of Drug Resistant Human Influenza Viruses’, Current Opinion in Virology, 8: 22–9. [DOI] [PubMed] [Google Scholar]
- Ikehata H., Ono T. (2011) ‘The Mechanisms of UV Mutagenesis’, Journal of Radiation Research, 52/2: 115–25. [DOI] [PubMed] [Google Scholar]
- Irwin K. K., Renzette N., Kowalik T. F., Jensen J. D. (2016) ‘Antiviral Drug Resistance as an Adaptive Process’, Virus Evolution, 2/1: 1–10. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Jorde P. E., Ryman N. (2007) ‘Unbiased Estimator for Genetic Drift and Effective Population Size’, Genetics, 177/2: 927–35. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Knipe D. N., Howley P. M., 2013. Fields’ Virology, 6th edn. Philadelphia: Lippincott Williams and Wilkins, a Wolters Kluwer Business.
- Kryazhimskiy, S., and Plotkin, J. B. (2008) ‘The population genetics of dN/dS’, PLoS Genetics, 4/12: e1000304. [DOI] [PMC free article] [PubMed]
- Kuritzkes D. R. (2011) ‘Drug Resistance in HIV-1’, Current Opinion in Virology, 1/6: 582–9. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lewis K. (2010) ‘Persister Cells’, Annual Review of Microbiology, 64: 357–72. [DOI] [PubMed] [Google Scholar]
- Lodder W. J., de Roda Husman A. M. (2005) ‘Presence of Noroviruses and Other Enteric Viruses in Sewage and Surface Waters in The Netherlands’, Applied and Environmental Microbiology, 71/3: 1453–61. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lodder W. J., van den Berg H. H. J. L., Rutjes S. A., de Roda Husman A. M. (2010) ‘Presence of Enteric Viruses in Source Waters for Drinking Water Production in The Netherlands’, Applied and Environmental Microbiology, 76/17: 5965–71. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lu J. et al. (2015) ‘Enterovirus Contamination in Pediatric Hospitals: A Neglected Part of the Hand-Foot-Mouth Disease Transmission Chain in China?: Table 1’, Clinical Infectious Diseases, 20: civ940. [DOI] [PubMed] [Google Scholar]
- Lynch M. et al. (2016) ‘Genetic Drift, Selection and the Evolution of the Mutation Rate’, Nature Review Genetics, 17/11: 704–14 Nature Research. [DOI] [PubMed] [Google Scholar]
- Maillard J. Y., Hann A. C., Perrin R. (1998) ‘Resistance of Pseudomonas aeruginosa PAO1 Phage F116 to Sodium Hypochlorite’, Journal of Applied Microbiology, 85: 799–806. [DOI] [PubMed] [Google Scholar]
- Maisonneuve E., Gerdes K. (2014) ‘Molecular Mechanisms Underlying Bacterial Persisters’, Cell, 157/3: 539–48. [DOI] [PubMed] [Google Scholar]
- Matuszewski S., Ormond L., Bank C., Jensen J. D. (2017) ‘Two Sides of the Same Coin: A Population Genetics Perspective on Lethal Mutagenesis and Mutational Meltdown’, Virus Evolution, 3/1. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Modlin J. F. (2007) ‘Enterovirus Déjà vu’, The New England Journal of Medicine, 356/12: 1204–5. [DOI] [PubMed] [Google Scholar]
- Moeller R. et al. (2010) ‘Genomic Bipyrimidine Nucleotide Frequency and Microbial Reactions to Germicidal UV Radiation’, Archives of Microbiology, 192/7: 521–9. [DOI] [PubMed] [Google Scholar]
- Moges F., Endris M., Belyhun Y., Worku W. (2014) ‘Isolation and Characterization of Multiple Drug Resistance Bacterial Pathogens from Waste Water in Hospital and Non-Hospital Environments, Northwest Ethiopia’, BMC Research Notes, 7/1: 215 BioMed Central. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mojica K. D. A., Brussaard C. P. D. (2014) ‘Factors Affecting Virus Dynamics and Microbial Host-Virus Interactions in Marine Environments’, FEMS Microbiology Ecology, 89/3: 495–515. [DOI] [PubMed] [Google Scholar]
- Morelli M. J. et al. (2013) ‘Evolution of Foot-and-Mouth Disease Virus Intra-Sample Sequence Diversity During Serial Transmission in Bovine Hosts’, Veterinary Research, 44: 12 BioMed Central. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Okoh A. I., Sibanda T., Gusha S. S. (2010) ‘Inadequately Treated Wastewater as a Source of Human Enteric Viruses in the Environment’, International Journal of Environmental Research and Public Health, 7/6: 2620–37. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Palacios G., Oberste M. S. (2005) ‘Enteroviruses as Agents of Emerging Infectious Diseases’, Journal of NeuroVirology, 11/5: 424–33. [DOI] [PubMed] [Google Scholar]
- Palma A. M. D., Vliegen De Clercq I. E., Neyts J. (2008) ‘Selective Inhibitors of Picornavirus Replication’, Medicinal Research Reviews, 1–62. [DOI] [PubMed] [Google Scholar]
- Payment P., Tremblay M., Trudel M. (1985) ‘Relative Resistance to Chlorine of Poliovirus and Coxsackievirus Isolates from Environmental Sources and Drinking Water’, Applied and Environmental Microbiology, 49: 981–3. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Pfeiffer J. K., Kirkegaard K. (2003) ‘A Single Mutation in Poliovirus RNA-Dependent RNA Polymerase Confers Resistance to Mutagenic Nucleotide Analogs via Increased Fidelity’, Proceedings of the National Academy of Sciences of the United States of America, 100/12: 7289–94. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Rahn R. O. (1997) ‘Potassium Iodide as a Chemical Actinometer for 254 nm Radiation: Use of Iodate as an Electron Scavenger’, Photochemistry and Photobiology, 66/6: 885. [DOI] [PubMed] [Google Scholar]
- Rubin D. B. (1981) ‘The Bayesian Bootstrap’, Annals of Statistics, 9/1: 130–134. [Google Scholar]
- Rzezutka A., Cook N. (2004) ‘Survival of Human Enteric Viruses in the Environment and Food’, FEMS Microbiology Reviews, 28/4: 441–53. [DOI] [PubMed] [Google Scholar]
- Sadeghipour S., Bek E. J., McMinn P. C. (2012) ‘Selection and Characterisation of Guanidine-Resistant Mutants of Human Enterovirus 71’, Virus Research, 169: 72–9. [DOI] [PubMed] [Google Scholar]
- Sadeghipour S., Bek E. J., McMinn P. C. (2013) ‘Ribavirin-Resistant Mutants of Human Enterovirus 71 Express A High Replication Fidelity Phenotype during Growth in Cell Culture’, Journal of Virology, 87/3: 1759–69. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sanjua R. et al. (2010) ‘Viral Mutation Rates’, Journal of Virolology, 84/19: 9733–48. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sedmak G., Bina D., MacDonald J. (2003) ‘Assessment of an Enterovirus Sewage Surveillance System by Comparison of Clinical Isolates with Sewage Isolates from Milwaukee, Wisconsin, Collected August 1994 to December 2002’, Applied and Environmental Microbiology, 69/12: 7181–7. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Shaffer P. T. B., Metcalf T. G., Sproul O. J. (1980) ‘Chlorine Resistance of Poliovirus Isolants Recovered from Drinking Water’, Applied and Environmental Microbiology, 40/6: 1115–21. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Shiomi H. et al. (2004) ‘Isolation and Characterisation of Poliovirus Mutants Resistant to Heating at 50 Degrees Celsius for 30 Min’, Journal of Medical Virology, 74/3: 484–91. [DOI] [PubMed] [Google Scholar]
- Simmons F. J., Xagoraraki I. (2011) ‘Release of Infectious Human Enteric Viruses by Full-Scale Wastewater Utilities’, Water Research, 45/12: 3590–8. [DOI] [PubMed] [Google Scholar]
- Tapparel C., Siegrist F., Petty T. J., Kaiser L. (2013) ‘Picornavirus and Enterovirus Diversity with Associated Human Diseases’, Infection, Genetics and Evolution, 14: 282–93. [DOI] [PubMed] [Google Scholar]
- Vignuzzi M., Stone J. K., Arnold J. J., Cameron C. E., Andino R. (2006) ‘Quasispecies Diversity Determines Cooperative Interactions within a Viral Population’, Nature, 439/7074: 344–8. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wirtschafts und Verlagsgesellschaft Gas und Wasser. 2006. UV Disinfection Equipment for Water Supply Systems - Part 1: Design And Construction, Function and Operation.
- Zhong Q. et al. (2016) ‘Genetic, Structural, and Phenotypic Properties of MS2 Coliphage with Resistance to ClO 2 Disinfection’, Environmental Science & Technology, 50/24: 13520–528. [DOI] [PubMed] [Google Scholar]
- Zhong Q. et al. (2017) ‘Resistance of Echovirus 11 to ClO2 Is Associated with Enhanced Host Receptor Use, Altered Entry Routes, and High Fitness’, Environmental Science & Technology, 51/18: 10746–55. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhou J. et al. (2015) ‘Source Identification of Bacterial and Viral Pathogens and Their Survival/Fading in the Process of Wastewater Treatment, Reclamation, and Environmental Reuse’, World Journal of Microbiology and Biotechnology, 31/1: 109–20. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.