Abstract
Background
Infections and graft-vs-host disease (GvHD) still represent major, not easily predictable complications in allogeneic hematopoietic stem cell transplant (allo-HSCT). Both conditions have been correlated to altered enteric microbiome profiles during the peritransplant period. The main objective of this study was to identify possible early microbiome-based markers useful in pretransplant risk stratification.
Methods
Stool samples were collected from 96 consecutive patients at the beginning of the pretransplant conditioning regimen (T0) and at 10 (T1) and 30 (T2) days following transplant. When significant in univariate analysis, the identified microbiome markers were used in multivariate regression analyses, together with other significant clinical variables for allo-HSCT-related risk stratification. Four main outcomes were addressed: (1) septic complications, (2) GvHD, (3) relapse of the underlying disease, and (4) mortality.
Results
The presence of >5% proinflammatory Enterobacteriaceae at T0 was the only significant marker for the risk of microbiologically confirmed sepsis. Moreover, ≤10% Lachnospiraceae at T0 was the only significant factor for increased risk of overall mortality, including death from both infectious and noninfectious causes.
Finally, a low bacterial alpha-diversity (Shannon index ≤ 1.3) at T1 was the only variable significantly correlating with an increased risk of GvHD within 30 days.
Conclusions
Microbiome markers can be useful in the very early identification of patients at risk for major transplant-related complications, offering new tools for individualized preemptive or therapeutic strategies to improve allo-HSCT outcomes.
Keywords: allogeneic hematopoietic stem cell transplant (allo-HSCT), enteric microbiome, graft-vs-host disease (GvHD), microbiologically confirmed sepsis, severe sepsis and septic shock
The multifaceted role of the enteric human microbiome has emerged with the identification of its immunological and metabolic key functions. The establishment of stable enteric microbial communities early in life is associated with the correct development of the immune system [1, 2]. The infant enteric flora undergoes sequential developmental patterns and stabilizes in its core composition at approximately 3 years of age [3]. The dynamic stability (resilience) increases with age, and early alterations of the enteric flora may not be easily reversed in adult life [4]. These alterations (dysbioses) have been associated with several subsequent pathological conditions, and their role in the early risk stratification is currently under investigation [5–11].
Analogously, the role of the enteric microbiome is under investigation also in the case of allogeneic hematopoietic stem cell transplantation (allo-HSCT), the only cure for several hematological malignancies. Even before the next-generation sequencing era, it was clinically evident that broad changes in the enteric microbial flora could impact the outcomes of transplantation [12]. However, the clinical failure of non-fine-tuned “gut decontamination” approaches [13] clearly suggests that a deeper understanding of the enteric flora dynamics before and during the peri-allo-HSCT period is necessary for improved risk stratification.
The few predominantly US-based studies that have elaborated upon this point have suggested a possible predictive role of the enteric microbiome profile [14]. Overall, a low diversity of the intestinal microbiota at engraftment has been shown to be an independent predictor of mortality from both infectious and noninfectious causes [15–17]. As an example, the different overall survival rates at 3 years were 36%, 60%, and 67% in transplanted patients featuring low, intermediate, and high microbiome diversity at engraftment, respectively [15]. This was even more evident in cases of enteric domination (≥30%) by single pathogens (ie, Enterococcus spp.; Proteobacteria), which often preceded bacteremic episodes after allo-HSCT [15, 18]. On the other hand, high levels of 3-indoxyl sulfate, a tryptophan derivative produced by beneficial commensal bacteria (ie, Lachnospiraceae and Ruminococcaceae), have been correlated to lower transplant-related mortality [19]. Modifications of the enteric microbiome have also been correlated to different risk of GvHD in both mouse models and transplanted patients [20]. High relative amounts of proinflammatory enteric bacteria (ie, Enterococcus spp.), paralleled by a decrease of anaerobic commensals belonging to Lachnospiraceae and Ruminococcaceae (ie, Blautia spp. and Faecalibacterium spp.), have been associated to higher risk of GvHD both in children and adults [16, 17, 21]. Importantly, different antibiotic protocols for the treatment of fever in the setting of neutropenia have been shown to influence the risk of GvHD-related mortality by inducing differential shifts in the enteric flora [22]. However, all these pioneering studies mostly focused their attention on the post-transplant setting, not investigating the possible correlation of enteric microbial composition and allo-HSCT at earlier time points.
In this study, we longitudinally investigated the enteric microbiome profiles of patients undergoing allo-HSCT in an effort to identify possible early pretransplant microbiome-based markers, starting from the beginning of the conditioning regimen.
METHODS
Study Setting and Design
We conducted a prospective observational study of the enteric microbiome by next-generation sequencing (NGS) in 96 consecutive patients receiving an allo-HSCT at the Hematology and Bone Marrow Transplant Unit of Ospedale San Raffaele, Milan, Italy, from October 2014 to April 2016. All details of the cohort are reported in Table 1. All patients were closely monitored and treated for fever in the setting of neutropenia according to institutional guidelines; in particular, all patients were administered levofloxacin and trimethoprim-sulfamethoxazole as prophylaxis, and piperacillin-tazobactam as firstline empirical treatment.
Table 1.
Detailed Clinical Description of 96 Patients Included in the Analysis
| Clinical Variables | No. (%) of Patients Analyzed |
|---|---|
| Age, y | |
| <35 | 22 (22.92) |
| 35–49 | 24 (25) |
| ≥50 | 50 (52.08) |
| Gender | |
| F | 38 (39.59) |
| M | 58 (60.41) |
| Underlying diseases | |
| Acute leukemia (AML, ALL) | 61 (63.54) |
| Lymphoma (HD, NHL) | 13 (13.54) |
| Multiple myeloma | 5 (5.20) |
| Myelodysplastic syndrome | 8 (8.33) |
| Others | 9 (9.4) |
| Time from diagnosis of the underlying disease, mo | |
| >12 | 47 (48.96) |
| <12 | 49 (51.04) |
| Number of transplant | |
| First HSCT | 82 (85.42) |
| Second or more HSCT | 14 (14.58) |
| HLA matching | |
| ≥9/10 | 47 (48.96) |
| <9/10 | 49 (51.04) |
| Stem cell source | |
| Sibling donors/matched unrelated donors | 48 (50) |
| Mismatched related donors/umbilical cord blood | 48 (50) |
| Donor gender | |
| F | 41 (42.70) |
| M | 55 (57.30) |
| Sorror scorea | |
| 0–2 | 54 (56.25) |
| >2 | 40 (41.66) |
| not applicable | 2 (2.08) |
| Disease status | |
| Complete remission | 43 (44.80) |
| Active disease | 51 (53.12) |
| Not applicable | 2 (2.08) |
| Disease Risk Index | |
| Low–intermediate | 34 (35.42) |
| High–very high | 50 (52.08) |
| Not applicable | 12 (12.5) |
| Conditioning intensity | |
| Reduced intensity | 21 (21.87) |
| Myeloablative | 75 (78.13) |
| GvHD prophylaxis (1) | |
| Rapamycin | 78 (81.25) |
| Cyclosporine | 18 (18.75) |
| GvHD prophylaxis (2) | |
| ATG | 25 (26.04) |
| CTX | 57 (59.38) |
| ATG plus CTX | 5 (5.20) |
| None | 9 (9.38) |
| Previous infectious episodes | |
| Yes | 52 (54.17) |
| No | 44 (45.83) |
| Rituximab administration | |
| Yes | 73 (76.04) |
| No | 23 (23.96) |
| Time to engraftment, d | |
| ≤15 | 24 (25) |
| >15 | 65 (67.70) |
| Missing value | 7 (7.30) |
Abbreviations: ALL, acute lymphoblastic leukemia; AML, acute myeloid leukemia; ATG, Antithymocyte globulin; CTX, Cyclophosphamide; GvHD, graft versus host disease; HD, Hodgkin disease; HLA, human leukocyte antigens; HSCT, hematopoietic stem cell transplantation; NHL, non-Hodgkin lymphoma.
aSorror score was calculated as in Elsawy and Sorror [38].
Sample Processing and Sequencing
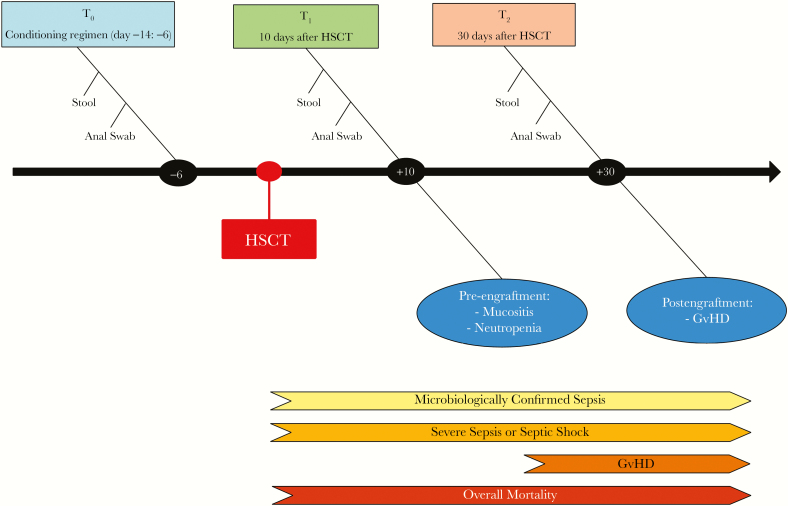
Stool samples were collected at 3 time points: at the beginning of the pretransplantation conditioning regimen, usually 6 days before transplant (T0); 10 days following transplant (T1), in correspondence with the period of full aplasia and the epidemiological peak of post-transplant sepsis [23]; and 30 days after transplant (T2) (Figure 1). The fecal microbiome was analyzed using the 454 GS Junior System (Roche Applied Science, Mannheim, Germany), using polymerase chain reaction (PCR) primers targeting the V3–V5 regions of the 16S ribosomal RNA gene. Full methods and details on the bioinformatic analysis are reported in the supplementary material.
Figure 1.
Timeline. We investigated the enteric microbiome in patients undergoing allogenic stem cell transplantation at 3 different time points: T0, the day of the initiation of the pretransplantation conditioning regimen (usually 6 days before the transplant), T1, 10 days following transplant, and T2, 30 days following transplant. We enrolled to protocol 100 patients, of which 4 were excluded, leaving 96 subjects for clinical and microbiome analysis. We investigated the role of the enteric bacterial microbiome in preventing or favoring microbiologically documented bloodstream infections, clinically suspected severe sepsis and septic shock, graft-vs-host disease, relapse, and overall mortality. Anal swabs were performed to detect colonization by multidrug-resistant bacterial isolates, as reported in the supplementary material. Abbreviations: GvHD, graft-vs-host disease; HSCT, hematopoietic stem cell transplantation.
Outcomes
The following outcomes were monitored from T0: septic complications, including microbiologically confirmed sepsis (ie, positive blood cultures) and clinically defined severe sepsis and septic shock (SSSS), relapse of the underlying disease, and mortality; GvHD was monitored from the day of HSCT (Supplementary Table 1). Further details on the outcomes definition are reported in the supplementary material.
Statistical Analysis
Receiver operating characteristic (ROC) curves of microbiome data were calculated at the family level for all outcomes. Curves with an area under the curve (AUC) >0.6 and a P value ≤.05 after Bonferroni family-wise correction were considered significant. From significant curves, the points with the best accuracy were chosen and used with all the other clinical variables to estimate their association with the different outcomes (Cox and binary logistic regression). In particular, Cox regression (hazard ratio) was used for each outcome, except GvHD within 30 and 100 days, for which binary logistic regression (odds ratio) was used. When multiple clinical or microbiome variables were significant for a given outcome in univariate analysis, a multivariate analysis with the same variables was performed using the same tests. A post hoc Bonferroni family-wise correction was performed both for univariate and multivariate analyses. More details on the statistical analysis are reported in the supplementary material.
RESULTS
Subjects and Clinical Data
One hundred patients undergoing allo-HSCT were enrolled. Fecal samples were collected at all time points (T0, T1, and T2) in 54 (54%) patients, whereas adequate samples were available at T0 and T1 in 18 (18%) patients, at T0 and T2 in 14 (14%) patients, and only at T0 in 10 (10%) patients. Although enrolled, it was not possible to collect samples from 4 (4%) patients. Further details on transplantation procedure and other clinical data are reported in the supplementary material.
Effect of Previous Antibiotic Therapy on the Pretransplantation Enteric Microbiome
The effects of antibiotics administered in the 3 months before HSCT on the pretransplantation enteric microbiome composition are shown in Table 2. As expected, antibiotics significantly influenced the overall α-diversity (P = .001), with a marked decrease observed for antibiotics with anti-anaerobic activity (piperacillin-tazobactam, ticarcillin, meropenem, clindamycin, metronidazole, and intravenous vancomycin; P = .003). A significant decrease in families with the most dominant anaerobic features was observed, including Clostridiaceae (P = .038), Ruminococcaceae (P = .038), and Veillonellaceae (P = .016). However, no significant antibiotic-induced differences were detected in the 2 most important microbiome markers identified in the study, that is, Lachnospiraceae (P = .188) and Enterobacteriaceae (P = .896) (Supplementary Table 2).
Table 2.
Effect on Enteric Microbiome at T0 of Antibiotic Therapy in the 3 Months Before HSCT
| Any Antibiotic | β-Lactams | Fluoroquinolones | Anti-anaerobic Therapy | |||
|---|---|---|---|---|---|---|
| P | P | P | P | |||
| Shannon index | .001↓ | .02↓ | .05↓ | .003↓ | ||
| Phylum | Firmicutes | Clostridiaceae | .038↓ | .061 | .041↓ | .038↓ |
| Enterococcaceae | .138 | .045↑ | .219 | .055 | ||
| Peptostreptococcaceae | .072 | .011↓ | .099 | .019↓ | ||
| Ruminococcaceae | .038↓ | .016↓ | .525 | .012↓ | ||
| Veillonellaceae | .016↓ | .040↓ | .863 | .015↓ | ||
| Bacteroidetes | Rickinellaceae | .327 | .018↓ | .812 | .016↓ |
Only significant differences between patients taking a given class of antibiotics and the rest of the cohort are reported (↑ increase; ↓ decrease). All data, including further details on the statistical analysis, are reported in Supplementary Table 2.
Enteric Microbiome Shifts During the Peritransplant Period
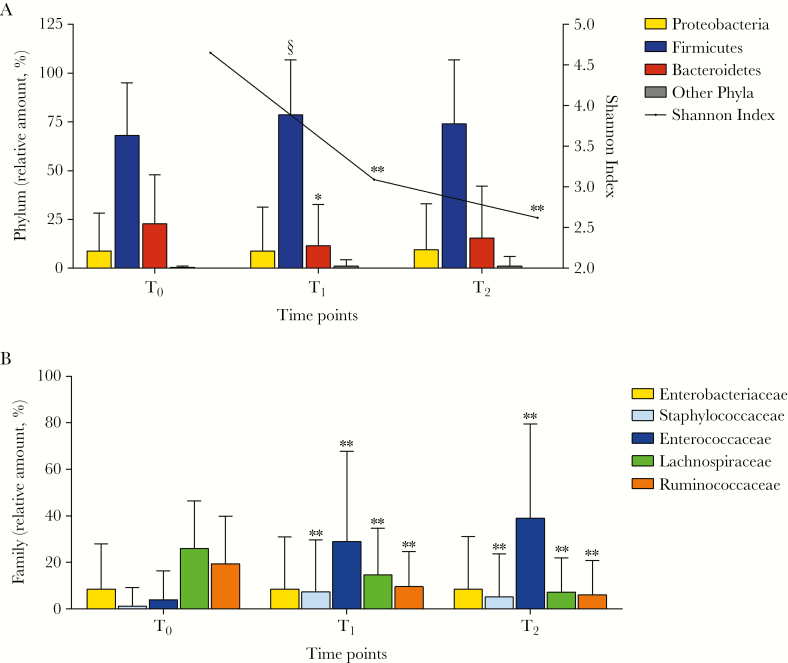
The transplant procedures impacted the enteric microbiome, with a dramatic decrease in bacterial α-diversity (Shannon index), particularly between T0 and T1 (P < .001) (Figure 2A). The loss of diversity was mainly due to the decrease in the relative amounts of strict anaerobic bacteria, as noted for the phylum Bacteroidetes (P = .002). An even more significant decrease of gram-positive strict anaerobes within the phylum Firmicutes was observed, in particular for the gut mucosa–protective Lachnospiraceae (P < .001) and Ruminococacceae (P < .001) families (Figure 2B). Conversely, within the Firmicutes, a significant increase was observed for the proinflammatory Enterococcaceae (P < .001) and Staphylococcaceae families (P < .001) (Figure 2B). No significant changes were observed in the facultative anaerobic gram-negative phylum Proteobacteria, almost completely represented by the Enterobacteriaceae (Figure 2B). The complete summary of different bacterial groups from the class to genus level is reported in Supplementary Table 4. Importantly, the observed shifts were more evident in patients undergoing myeloablative conditioning regimens (Supplementary Tables 5 and 6).
Figure 2.
Phylogenetic changes and alpha diversity in allogeneic hematopoietic stem cell transplant patients across all time points using mean values and the Wilcoxon signed-rank test. (A) During the peritransplant period, patients show extreme shifts in the intestinal microbiota, including an overall loss of diversity and richness, as well as a significant average increase in phylum Firmicutes and a decrease in phylum Bacteroidetes between T0 and T1. (B) Enterobacteriaceae (phylum Proteobacteria) did not change significantly throughout the 3 time points. On the other hand, there were both a significant increase in Staphylococcaceae and Enterococcaceae (phylum Firmicutes) and a significant decrease in Lachnospiraceae and Ruminococcaceae (phylum Firmicutes). §P < .05; *P < .01; **P < .001. A full list of phylogenetic changes and alpha diversity is reported in Supplementary Table 4.
Identification of Clinically Significant Microbiome Cutoffs
ROC curves were plotted at all studied time points in order to identify possible early microbiome-based risk stratification markers (Supplementary Figure 1). The microbiome cutoffs chosen at T0 and T1 from significant ROC curves are reported in Table 3. No significant microbiome cutoffs were identified at T2 (data not shown), The microbiome cutoffs were then used in univariate and multivariate regression analyses, together with the other clinical variables for risk stratification of transplant-related infectious and noninfectious complications.
Table 3.
Microbiome Markers Included in the Univariate Analyses for Specific Clinical Outcomes
| Microbiome Markers | |
|---|---|
| At T0 | Shannon index ≤4 at T0 |
| Enterobacteriaceae >5% at T0 | |
| Lachnospiraceae ≤10% at T0 | |
| Ruminococcaceae ≤10% at T0 | |
| At T1 | Shannon index ≤1.3 at T1 |
| Staphylococcaceae >40% at T1 |
Abbreviations: T0, the day of the initiation of the pretransplantation conditioning regimen (approximately 6 days before the transplant); T1, 10 days following transplant.
The significant microbiome cutoffs and clinical variables associated with specific clinical outcomes emerging from the univariate analysis are reported in Table 4 for microbiologically confirmed sepsis, relapse of the underlying disease and GvHD, and in Supplementary Table 7 for death. As summarized in Figure 3, A–F, and in Figure 4, A–E, and as discussed below, the 2 most relevant microbiome-based markers were identified at T0: >5% relative amount of Enterobacteriaceae and ≤10% of Lachnospiraceae, representing >73th percentile and ≤24th percentile of the studied cohort, respectively.
Table 4.
Significant Predictors for Different Clinical Outcomes After Regression Analyses and Bonferroni Correction
| Positive Blood Cultures by GN | Relapse | Gvhd Within 30 Days | ||||||
|---|---|---|---|---|---|---|---|---|
| Univariate Analysis | Multivariate Analysis | |||||||
| Risk Factor | P | HR (95% CI) |
P | HR (95% CI) |
P | HR (95% CI) |
P | OR (95% CI) |
| Disease status | .019 | 5.633 (2.067–15.353) |
.006 | 6.214 (1.900–20.323) |
||||
| Disease Risk Index | .019 | 7.174 (2.170–23.719) |
.042 | 4.503 (1.255–16.166) |
||||
| Shannon index ≤4 at T0 | n.s. | |||||||
| Enterobacteriaceae >5% at T0 | .021 | 6.577 (2.246–19.255) |
||||||
| Lachnospiraceae ≤10% at T0 | n.s. | |||||||
| Ruminococcaceae ≤10% at T0 | n.s. | |||||||
| Shannon ≤1.3% at T1 | n.s. | .038 | 7.833 (2.141–28.658) |
|||||
| Staphylococcaceae >40% at T1 | n.s. | n.s. | ||||||
Analyses were performed with Cox regression for the following outcomes: positive blood cultures by all causes, by gram-negative and by gram-positive, severe sepsis or septic shock, relapse. Binary logistic regression was performed for GvHD within 30 or 100 days. Only outcomes with significant markers are listed in the table, and confidence intervals are reported only for significant variables after Bonferroni family-wise correction. For further details, see the supplementary material.
Abbreviations: CI, confidence interval; GN, gram-negative; GvHD, graft-vs-host disease; HR, hazard ratio; OR, odds ratio; n.s., not significant.
Figure 3.
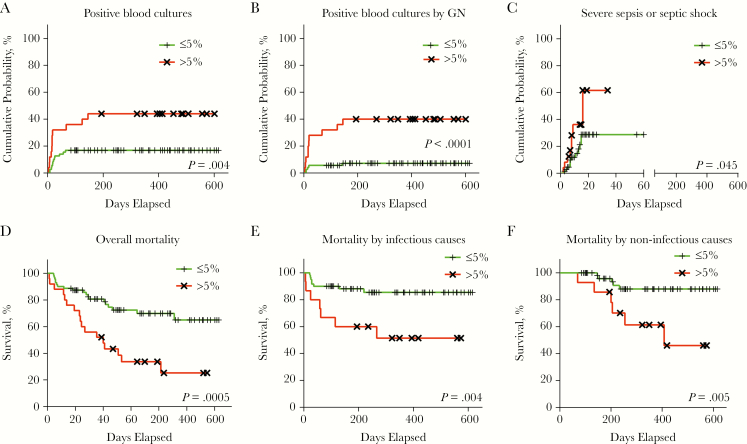
Enterobacteriaceae 5% cutoff at T0 for different clinical outcomes (log rank test). Within the studied cohort, 25 (26%) patients featured >5% Enterobacteriaceae at T0, with 8 (8.33%) of them also displaying ≤10% Lachnospiraceae. (A) Cumulative probability for all cases of microbiologically confirmed sepsis (ie, positive blood cultures by both gram-negative and gram-positive enteric pathogens). (B) Cumulative probability curves for microbiologically confirmed sepsis by gram-negative enteric pathogens. (C) Cumulative probability curves for severe sepsis or septic shock. (D) Kaplan-Meier overall mortality curves. (E) Kaplan-Meier curves for transplant-related mortality by infectious causes. (F) Kaplan-Meier curves for transplant-related mortality by noninfectious causes. Abbreviation: GN, gram-negative.
Figure 4.
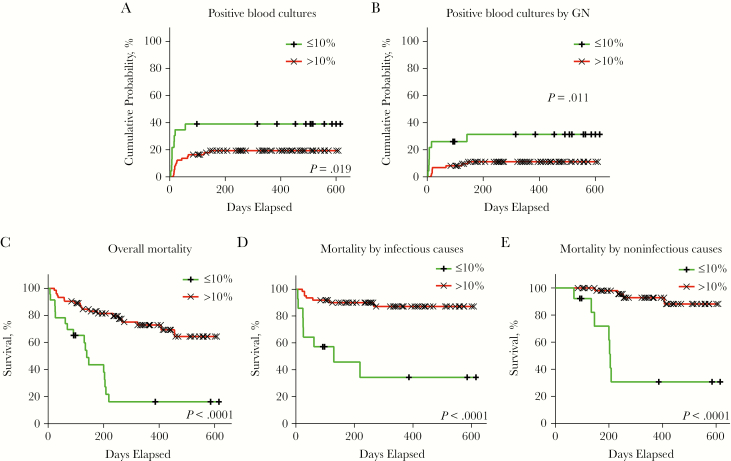
Lachnospiraceae 10% cutoff at T0 for different clinical outcomes (log rank test). Within the studied cohort, 23 (23.95%) patients featured ≤10% Lachnospiraceae, with 8 (8.33%) of them also displaying >5% Enterobacteriaceae at T0. (A) Cumulative probability for all cases of microbiologically confirmed sepsis (ie, positive blood cultures by both gram-negative and gram-positive enteric pathogens). (B) Cumulative probability curves for microbiologically confirmed sepsis by gram-negative enteric pathogens. (C) Kaplan-Meier overall mortality curves. (D) Kaplan-Meier curve for transplant-related mortality by infectious causes. (E) Kaplan-Meier curve for transplant-related mortality by noninfectious causes. Abbreviation: GN, gram-negative.
Risk of Sepsis
During hospitalization, 23 of the 96 patients (23.96%) had a microbiologically confirmed bloodstream infection from enteric bacteria. The presence of >5% Enterobacteriaceae was the only significant marker of sepsis by gram-negative (GN) pathogens in univariate analysis (hazard ratio [HR], 6.577; P = .021) (Table 4). Interestingly, the colonization by GN multidrug-resistant (MDR) pathogens was not correlated to an increased risk of microbiologically confirmed sepsis. No significant risk factors were identified for microbiologically confirmed enteric sepsis by gram-positive (GP) pathogens.
Analogously, no significant clinical or microbiome variables were identified in the 21 HSCT recipients (22.82%) who developed clinically defined severe sepsis or septic shock.
Risk of Graft-vs-Host Disease
Acute GvHD developed in 23 HSCT recipients (25.27%) within 30 days (early acute GvHD), and in 46 (50.55%) within 100 days (late acute GvHD). Further details regarding GvHD grade and organ site are reported in the Supplementary Methods.
In the risk stratification for early acute GvHD, no significant variables were identified at T0. However, microbiome shifts were observed between T0 and T1, with a more significant loss of Lachnospiraceae (P = .022) and an increase in Staphylococcaceae (P = .005) in patients with an early onset of acute GvHD (Supplementary Figure 2).
Microbiome markers were more informative at T1, still a clinically useful time point, with a low α-diversity (Shannon index ≤ 1.3) being significantly correlated to the risk of early acute GvHD (odds ratio, 7.833; P = .038) (Table 4). However, no significant associations were observed with late acute GvHD, GvHD severity, and GvHD organ localization.
Risk of Relapse
During the study period, relapse was observed in 30 (31.25%) patients. This was expected due to the high-risk patients admitted to our center and included in our cohort. Differently from a recent study [24], the risk of relapse was not significantly influenced by microbiome markers, with only disease-related variables emerging from the multivariate analyses (Table 4). This could be due to the different statistical approach followed and to the more limited and high-risk cohort investigated in our study.
Mortality Risk
A total of 36 (37.5%) deaths were registered, 15 (15.62%) of which were due to infectious causes (Supplementary Table 1). A relative amount of >5% Enterobacteriaceae (HR, 3.034; P = .021) and of ≤10% Lachnospiraceae (HR, 4.806; P < .001) at T0 were associated in univariate analysis to increased mortality, due to either infectious or noninfectious causes, and these were further investigated in multivariate analysis (Supplementary Table 7). The only other variable included in multivariate analysis was enteric colonization by GN-MDR pathogens (HR, 6.614; P < .001) (Supplementary Table 7).
Survival curves were plotted using the identified microbiome markers and highlighted an extremely significant rapid decline in survival in the first month, and even more so during the entire observation period (Figure 3D, Figure 4C).
Importantly, a relative amount of ≤10% Lachnospiraceae was the only independent risk factor in multivariate analysis for overall mortality (HR, 4.439; P < .001) (Table 5). Interestingly, ≤10% Lachnospiraceae was not only the unique independent variable for death due to infectious causes (HR, 7.051; P = .006), but also the most relevant for death due to noninfectious causes (HR, 21.211; P < .001) (Table 5).
Table 5.
Significant Predictors for Mortality After Multivariate Regression Analyses and Bonferroni Correction
| Deaths | Deaths due to Infection | Deaths due to Noninfectious Causes | ||||
|---|---|---|---|---|---|---|
| Risk Factor | P | HR (95% CI) |
P | HR (95% CI) |
P | HR (95% CI) |
| Enteric colonization by GN-MDR at T0 | n.s. | n.s. | .048 |
10.280
(1.536–68.795) |
||
| Enterobacteriaceae >5% at T0 | .060a | 2.488 (1.156–5.355) |
||||
| Lachnospiraceae ≤10% at T0 | <.001 |
4.439
(2.181–9.035) |
.006 |
7.051
(2.007–24.778) |
<.001 |
21.211
(4.949–90.915) |
The Cox regression model was used for multivariate analyses of mortality. Confidence intervals are reported only for significant variables after Bonferroni family-wise correction. For further details, see the supplementary material.
aData close to the limits of statistical significance after Bonferroni family-wise correction.
Abbreviations: CI, confidence interval; HR, hazard ratio; n.s., not significant.
DISCUSSION
Infections and GvHD still represent major allo-HSCT-related complications [25], stressing the need for noninvasive and reliable laboratory tests allowing a precision medicine–tailored prophylactic approach. The gastrointestinal tract is markedly impacted by allo-HSCT, being both the origin and target of infections and GvHD. The different enteric microbiome components can either elicit inflammation or promote immune tolerance [26, 27]. Current immune suppression strategies are only partially effective at preventing GvHD and increase the risk of infections and disease recurrence. Enteric microbiota-targeting strategies, reducing GvHD but leaving immune function intact, may thus potentially improve allo-HSCT outcomes.
This clinically oriented study represents the largest cohort of non-US-based allo-HSCT recipients characterized by their enteric microbiome consortia using NGS techniques. Weber et al. described a larger German cohort, but it included only 31 patients investigated by NGS, whereas all the other patients were analyzed by the detection of a urinary indole metabolite [19]. Interestingly, the amount of this metabolite correlated to the levels of Lachnospiraceae, the same beneficial bacterial families emerging from our study [19]. Overall, the representativeness of our cohort, a comprehensive picture of different causes leading to allo-HSCT in Europe [28], the different transplant strategies and the investigated time points, were of paramount importance to maximize the potential clinical utility of our study.
It is still debated whether patients undergo allo-HSCT with an already altered enteric microbiome [14]. Our study confirms that allo-HSCT-related treatments dramatically alter its composition, with a marked decrease of its richness and strict anaerobic components. Previous pioneering studies have evidenced that, during the peri-transplant period, severe dysbiosis, defined as bacterial domination (ie, relative amount >30%) by single proinflammatory taxon (ie, Enterococcus spp., Proteobacteria), significantly increases the risk of bacteremia from the same microorganisms [18, 29]. However, the same authors demonstrated that the presence of commensal bacterial genera somehow contrasts the gut domination and therefore decreases the infectious risk [29]. The importance of this balance was demonstrated also for noninfectious complications, evidencing the correlation between low gut microbiome diversity [15–17, 21], high levels of pro-inflammatory bacteria (ie, Enterococcus spp.) [17, 21] and low levels of beneficial commensals (ie, Faecalibacterium spp. [21], Blautia spp. [16]) with higher risk of GvHD [16, 17, 21] and higher mortality [15]. Under this perspective, the main goal of our study was to verify whether similar clinically important prognostic information could be derived from the enteric microbiome at the very beginning of the transplant procedure, that is, at the beginning of the conditioning regimen (T0).
Also following this study design, the dichotomy between noxious and protective taxa is evident, with high relative amounts (>5%) of proinflammatory Enterobacteriaceae and low levels (≤10%) of Lachnospiraceae at T0 emerging among the strongest biomarkers for both infectious and noninfectious complications. The Lachnospiraceae family is 1 of the major taxonomic groups of the enteric microbiome and includes notable commensal genera (ie, Blautia spp.), whose beneficial role has already been described above [16, 19, 20, 27]. Members of this family are barely absent in the first months of life, but rapidly increase, reaching and stabilizing at an average relative abundance of 12% to 20% at 3 years of age [8]. Conversely, Enterobacteriaceae (a family including well-known gram-negative pathogenic genera, such as Escherichia spp., Klebsiella spp., Proteus spp., and others) are significantly abundant in the first months (up to 25% at 2 months of life) but then steadily decrease, reaching the 1% to 3% relative abundance observed in healthy adults [8]. Of note, none of the above markers was significantly modified in our cohort by antibiotic therapy in the 3 months preceding HSCT. This finding is in contrast with studies evidencing the effect of antibiotic therapy on the enteric flora at baseline and in influencing HSCT outcomes [13, 22]. This is even more evident for GN-MDR colonization, whose role in predicting discrete HSCT outcomes, including risk of infection, is limited in our study. These results may be due to the limited size of our cohort, but may cast some doubt on the use of antibiotics in selective manipulation of enteric flora [13] or on the detection of multidrug resistance alone as a surrogate marker for dysbiosis and risk assessment [30].
As anticipated above, the most important point not fully evident in other studies [15, 16, 18, 31] is the potential predictive value of studying the enteric microbiome as early as at the beginning of the conditioning regimen. The advantages of such an early approach could be manifold, including both diagnostic and prophylactic/therapeutic benefits. Under the diagnostic point of view, the identification of early clinically significant cutoffs could allow the design of more high-throughput and easy-to-perform assays, such as, for example, the setup of quantitative real-time PCR, allowing the detection of discrete bacterial families of interest, or, as previously reported, the detection of significant amounts of bacterial metabolites [19]. Moreover, as pointed out later in this section, early time points could allow timely prophylactic and/or therapeutic management of infectious and GvHD risk even before the initiation of the allo-HSCT procedure. Other groups followed our same early approach, but their cohorts included only pediatric or non-Hodgkin lymphoma–affected adult patients [21, 32]. Overall, these prior studies and our more representative study highlight the relevance not only to reduce the presence of pathobionts, but also to increase the abundance of protective symbionts right from the beginning of the entire transplant procedure. Under this perspective, alternative preventive recolonizing strategies such as early fecal microbiota transplantation (FMT) [33, 34] ought to be explored. Such an approach could transfer high levels of diversity as well as components of the so-called “rare biosphere” to allo-HSCT recipients [9, 35]. A few reports on FMT in allo-HSCT-transplanted patients have already been performed, but only in GvHD cases not responding to conventional approaches [36, 37]. None of these studies, however, employed an early microbiota-modulating approach, as suggested by the results of our study. Larger multicenter studies are certainly needed to verify the feasibility of this fascinating hypothesis. Beyond FMT, other possible microbiota-modulating strategies could be postulated in order to durably impact the proinflammatory composition of the enteric microbiota, including the identification of immunomodulatory bacterial strains to be used as “probiotics,” of food components to be used as “prebiotics” conferring selective advantage to beneficial commensal bacteria, or of beneficial bacterial metabolites to be used as “postbiotics” [12].
Although an in-depth investigation on the mechanisms leading to the observed correlations was not the specific goal of this study, it is accepted that a general enteric dysbiosis, not necessarily represented by domination of a single definite bacterial group, could be correlated to local and systemic anomalies. This is salient in the case of the protective role played by Lachnospiraceae, whose low levels are associated in our study to various adverse outcomes. A role of Lachnospiraceae in exerting a beneficial effect on the GI tract mucosa and in epigenetically modifying immune cells through the production of highly beneficial short-chain fatty acids (SCFAs) in a time of immune re-education, could be a possible explanation of what has been observed. Some mechanistic studies have recently addressed this point, evidencing in animal models the selective decrease of SCFAs (in particular butyrate levels) in the intestinal tissues following allo-HSCT and its correlation to adverse outcomes [27]. An important observation of the same study was the lack of changes in stool levels of butyrate, despite a documented decrease in SCFA-producing bacterial species, positing a decreased uptake by intestinal epithelial cells. This observation casts some doubt on the clinical usefulness of using SCFAs levels in stool as a marker of allo-HSCT-related adverse outcomes evidencing the need for directly detecting the presence of discrete SCFA-producing microbiome groups. This is in line with what was observed in our clinical cohort for decreased levels of Lachnospiraceae, with the added value of the ≤10% cutoff highlighted in our study representing a markedly more inclusive attempt to determine clinically useful cutoffs than previous microbiome-based studies in this field [2, 14]. Unfortunately, it was not possible to evaluate the stool SCFA levels in our study and to correlate them with our identified microbiome cutoff.
We are aware of several limitations of our study, including its single-center design and the need to confirm its results in larger cohorts and to investigate the role of the enteric microbiome in predicting later transplant-related complications (ie, chronic GvHD). However, we believe that these data further highlight the great clinical potential of characterizing the microbiome in the earliest phases of HSCT and that this certainly deserves further scrutiny in larger, possibly interventional studies bringing novel microbiome-modulating strategies to the forefront of clinical practice. In this context, the results described identify multiple early microbiome-based biomarkers that could be useful in the risk stratification of allo-HSCT recipients, offering a new tool for individualized preemptive or therapeutic strategies to improve the allo-HSCT outcome.
Supplementary Data
Supplementary materials are available at Open Forum Infectious Diseases online. Consisting of data provided by the authors to benefit the reader, the posted materials are not copyedited and are the sole responsibility of the authors, so questions or comments should be addressed to the corresponding author.
Supplementary Material
Acknowledgements
Author contributions. NM, RG, GB, FC, and MCl designed the trial. RG, MCB, MM, LV, FG, MTLS, AF, SM, AA, MCa, MB, CC, and JP contributed to clinical data generation. RP, GP, OBM, and LI contributed to microbiome data generation. NM, RG, RP, GP, NC, RB, MPS, FC, and MCl analyzed the data and performed statistical analysis. NM, RG, RP, GP, FC, and MCl interpreted the data and wrote the final report.
Financial support. This work was supported by the Italian Ministry of Health.
Potential conflicts of interest. All authors: no reported conflicts of interest. All authors have submitted the ICMJE Form for Disclosure of Potential Conflicts of Interest. Conflicts that the editors consider relevant to the content of the manuscript have been disclosed.
References
- 1. Sommer F, Bäckhed F. The gut microbiota–masters of host development and physiology. Nat Rev Microbiol 2013; 11:227–38. [DOI] [PubMed] [Google Scholar]
- 2. Rooks MG, Garrett WS. Gut microbiota, metabolites and host immunity. Nat Rev Immunol 2016; 16:341–52. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3. Bokulich NA, Chung J, Battaglia T et al. . Antibiotics, birth mode, and diet shape microbiome maturation during early life. Sci Transl Med 2016; 8:343ra82. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4. Spor A, Koren O, Ley R. Unravelling the effects of the environment and host genotype on the gut microbiome. Nat Rev Microbiol 2011; 9:279–90. [DOI] [PubMed] [Google Scholar]
- 5. Becattini S, Taur Y, Pamer EG. Antibiotic-induced changes in the intestinal microbiota and disease. Trends Mol Med 2016; 22:458–78. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6. Korpela K, Salonen A, Virta LJ et al. . Intestinal microbiome is related to lifetime antibiotic use in Finnish pre-school children. Nat Commun 2016; 7:10410. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7. Marchesi JR, Adams DH, Fava F et al. . The gut microbiota and host health: a new clinical frontier. Gut 2016; 65:330–9. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8. Yassour M, Vatanen T, Siljander H et al. . Natural history of the infant gut microbiome and impact of antibiotic treatment on bacterial strain diversity and stability. Sci Transl Med 2016; 8:343ra81. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9. Clemente JC, Ursell LK, Parfrey LW, Knight R. The impact of the gut microbiota on human health: an integrative view. Cell 2012; 148:1258–70. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10. Dominguez-Bello MG, Costello EK, Contreras M et al. . Delivery mode shapes the acquisition and structure of the initial microbiota across multiple body habitats in newborns. Proc Natl Acad Sci U S A 2010; 107:11971–5. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11. Chu DM, Ma J, Prince AL et al. . Maturation of the infant microbiome community structure and function across multiple body sites and in relation to mode of delivery. Nat Med 2017; 23:314–26. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12. Peled JU, Jenq RR, Holler E, van den Brink MR. Role of gut flora after bone marrow transplantation. Nat Microbiol 2016; 1:16036. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13. Whangbo J, Ritz J, Bhatt A. Antibiotic-mediated modification of the intestinal microbiome in allogeneic hematopoietic stem cell transplantation. Bone Marrow Transplant 2017; 52:183–90. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14. Zama D, Biagi E, Masetti R et al. . Gut microbiota and hematopoietic stem cell transplantation: where do we stand? Bone Marrow Transplant 2017; 52:7–14. [DOI] [PubMed] [Google Scholar]
- 15. Taur Y, Jenq RR, Perales MA et al. . The effects of intestinal tract bacterial diversity on mortality following allogeneic hematopoietic stem cell transplantation. Blood 2014; 124:1174–82. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16. Jenq RR, Taur Y, Devlin SM et al. . Intestinal blautia is associated with reduced death from graft-versus-host disease. Biol Blood Marrow Transplant 2015; 21:1373–83. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17. Holler E, Butzhammer P, Schmid K et al. . Metagenomic analysis of the stool microbiome in patients receiving allogeneic stem cell transplantation: loss of diversity is associated with use of systemic antibiotics and more pronounced in gastrointestinal graft-versus-host disease. Biol Blood Marrow Transplant 2014; 20:640–5. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18. Taur Y, Xavier JB, Lipuma L et al. . Intestinal domination and the risk of bacteremia in patients undergoing allogeneic hematopoietic stem cell transplantation. Clin Infect Dis 2012; 55:905–14. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19. Weber D, Oefner PJ, Hiergeist A et al. . Low urinary indoxyl sulfate levels early after transplantation reflect a disrupted microbiome and are associated with poor outcome. Blood 2015; 126:1723–8. [DOI] [PubMed] [Google Scholar]
- 20. Jenq RR, Ubeda C, Taur Y et al. . Regulation of intestinal inflammation by microbiota following allogeneic bone marrow transplantation. J Exp Med 2012; 209:903–11. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21. Biagi E, Zama D, Nastasi C et al. . Gut microbiota trajectory in pediatric patients undergoing hematopoietic SCT. Bone Marrow Transplant 2015; 50:992–8. [DOI] [PubMed] [Google Scholar]
- 22. Shono Y, Docampo MD, Peled JU et al. . Increased GVHD-related mortality with broad-spectrum antibiotic use after allogeneic hematopoietic stem cell transplantation in human patients and mice. Sci Transl Med 2016; 8:339ra71. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23. Mikulska M, Del Bono V, Bruzzi P et al. . Mortality after bloodstream infections in allogeneic haematopoietic stem cell transplant (HSCT) recipients. Infection 2012; 40:271–8. [DOI] [PubMed] [Google Scholar]
- 24. Peled JU, Devlin SM, Staffas A et al. . Intestinal microbiota and relapse after hematopoietic-cell transplantation. J Clin Oncol 2017; 35:1650–9. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25. Gooley TA, Chien JW, Pergam SA et al. . Reduced mortality after allogeneic hematopoietic-cell transplantation. N Engl J Med 2010; 363:2091–101. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26. Wang W, Xu S, Ren Z et al. . Gut microbiota and allogeneic transplantation. J Transl Med 2015; 13:275. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27. Mathewson ND, Jenq R, Mathew AV et al. . Gut microbiome-derived metabolites modulate intestinal epithelial cell damage and mitigate graft-versus-host disease. Nat Immunol 2016; 17:505–13. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28. Passweg JR, Baldomero H, Bader P et al. . Hematopoietic stem cell transplantation in Europe 2014: more than 40 000 transplants annually. Bone Marrow Transplant 2016; 51:786–92. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29. Ubeda C, Bucci V, Caballero S et al. . Intestinal microbiota containing Barnesiella species cures vancomycin-resistant Enterococcus faecium colonization. Infect Immun 2013; 81:965–73. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30. Bilinski J, Robak K, Peric Z et al. . Impact of gut colonization by antibiotic-resistant bacteria on the outcomes of allogeneic hematopoietic stem cell transplantation: a retrospective, single-center study. Biol Blood Marrow Transplant 2016; 22:1087–93. [DOI] [PubMed] [Google Scholar]
- 31. Taur Y, Jenq RR, Ubeda C et al. . Role of intestinal microbiota in transplantation outcomes. Best Pract Res Clin Haematol 2015; 28:155–61. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32. Montassier E, Al-Ghalith GA, Ward T et al. . Erratum to: Pretreatment gut microbiome predicts chemotherapy-related bloodstream infection. Genome Med 2016; 8:61. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33. Hamilton MJ, Weingarden AR, Unno T et al. . High-throughput DNA sequence analysis reveals stable engraftment of gut microbiota following transplantation of previously frozen fecal bacteria. Gut Microbes 2013; 4:125–35. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34. Choi HH, Cho YS. Fecal microbiota transplantation: current applications, effectiveness, and future perspectives. Clin Endosc 2016; 49:257–65. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35. Huffnagle GB, Noverr MC. The emerging world of the fungal microbiome. Trends Microbiol 2013; 21:334–41. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36. Kakihana K, Fujioka Y, Suda W et al. . Fecal microbiota transplantation for patients with steroid-resistant acute graft-versus-host disease of the gut. Blood 2016; 128:2083–8. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37. Staffas A, Burgos da Silva M, van den Brink MR. The intestinal microbiota in allogeneic hematopoietic cell transplant and graft-versus-host disease. Blood 2017; 129:927–33. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38. Elsawy M, Sorror ML. Up-to-date tools for risk assessment before allogeneic hematopoietic cell transplantation. Bone Marrow Transplant 2016; 51:1283–300. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.