Abstract
The His-Me finger endonucleases, also known as HNH or ββα-metal endonucleases, form a large and diverse protein superfamily. The His-Me finger domain can be found in proteins that play an essential role in cells, including genome maintenance, intron homing, host defense and target offense. Its overall structural compactness and non-specificity make it a perfectly-tailored pathogenic module that participates on both sides of inter- and intra-organismal competition. An extremely low sequence similarity across the superfamily makes it difficult to identify and classify new His-Me fingers. Using state-of-the-art distant homology detection methods, we provide an updated and systematic classification of His-Me finger proteins. In this work, we identified over 100 000 proteins and clustered them into 38 groups, of which three groups are new and cannot be found in any existing public domain database of protein families. Based on an analysis of sequences, structures, domain architectures, and genomic contexts, we provide a careful functional annotation of the poorly characterized members of this superfamily. Our results may inspire further experimental investigations that should address the predicted activity and clarify the potential substrates, to provide more detailed insights into the fundamental biological roles of these proteins.
INTRODUCTION
The His-Me finger domain occurs in a large and diverse superfamily of endonuclease proteins present in all forms of life and involved in various cellular processes. In the literature, it is also dubbed the ‘HNH’ endonuclease (after its conserved sequence motif) or the ββα-metal endonuclease (denoting the conserved secondary structure of the fold and bound catalytic metal ion) (1).
Its function is associated with the catalytic cleavage of nucleic acids in multiple contexts, such as intron homing, phage DNA packaging, recombination or apoptotic DNA degradation. Moreover, His-Me finger-containing proteins are often a key part of the defence and stress response systems of bacteria and archaea, e.g. restriction–modification, toxin–antitoxin and CRISPR/Cas9 systems.
The His-Me finger is defined by a common structural core consisting of a β-hairpin followed by an α-helix, forming a binding site for a single catalytic metal ion (Figure 1). The finger might be considered as a part of the treble clef structural motif, which is formed by a zinc knuckle, loop, β-hairpin and α-helix (1). However, since many His-Me finger representatives lack the zinc knuckle (2,3), they escape the orthodox treble clef classification.
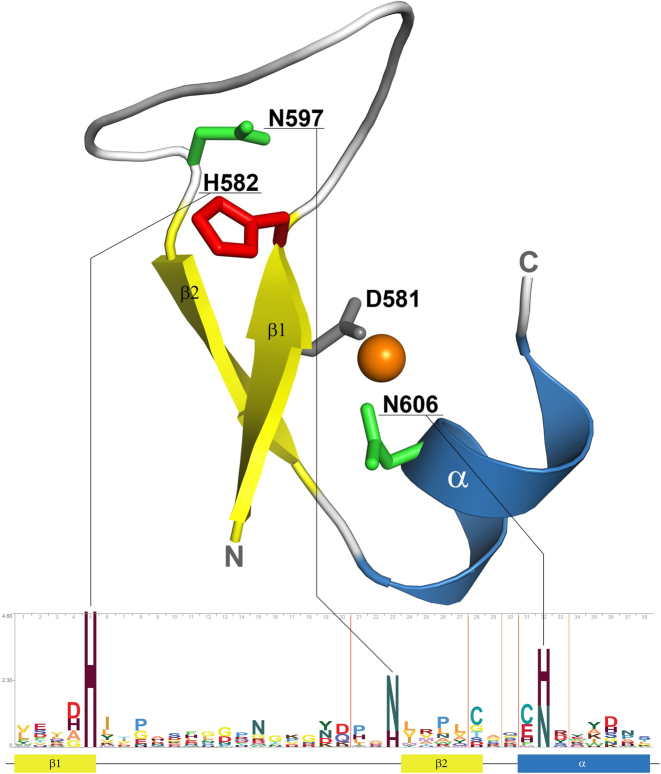
Figure 1.
The conserved structural core of His-Me finger fold of Cas9 (PDB ID: 4ogc). The core elements are coloured in yellow and blue for β-strands and α-helix, respectively. The catalytic histidine is shown as red sticks. The remaining HNH sequence motif residues are shown as green sticks. The residues from outside the motif, chelating catalytic zinc ion (orange), are denoted as grey sticks. The sequence logo was generated based on the structure-guided multiple sequence alignment of all superfamily sequences clustered at 70% sequence identity using the Skylign server (106). The total height of the letters depicts the information content of the position in bits.
The His-Me finger is thought to be suited for efficient, nonspecific DNA cleavage (4,5). The α-helix fits into the DNA minor groove, which in consequence precisely aligns the remaining β-hairpin against the DNA backbone for nucleolytic reaction (6–8). In structure-specific enzymes, such as the Holliday junction resolvases, the core α-helix contacts the minor groove side of the branch point (9). The finger alone displays little substrate specificity, but may be easily tailored to a particular function by the presence of additional domains or structural elements tethered to the core of the ββα-metal fold (3,10). The single metal ion-based catalytic site keeps the domain compact; however, its small size means it cannot easily achieve structural stability. Indeed, its three-element structural motif is unable to provide enough of a hydrophobic core for stability. Thus, many proteins containing the His-Me finger domain employ a variety of additional structural elements or domains for stabilization and/or specificity. These include embedded zinc binding motifs, like the zinc knuckle of the treble clef mentioned above (e.g. in restriction or CRISPR nucleases), or additional decoration ranging from single α-helices to extensive β-sheets—e.g. in DNA degrading nucleases like endonuclease G (EndoG) (11) and CPS-6 (12), or virulence factors like nuclease A (NucA) (13), Serratia marcescens nuclease (Sm) (14), and streptodornase (Spd1) (15).
The term ‘His-Me’ refers to a nearly invariant catalytic histidine (His) residue and a bound metal ion (Me). His-Me finger-containing proteins are commonly called HNH nucleases, due to the sequence motif present in the founding superfamily members. The HNH sequence motif consists of the central, nearly invariant, catalytic histidine at the C-terminus of the first β-strand (HNH), an asparagine (HNH) in the extensive loop connecting the two β-strands—known as the Ω-loop (16)—and a histidine (HNH) in the core α-helix. This second histidine is often substituted by an asparagine, which, in most cases, is the only residue that directly binds the metal ion (Figure 1).
The catalytic site of the His-Me finger consists of the catalytic histidine and a divalent metal ion ligand, coordinated mainly by the core α-helix residues through either direct or water-mediated interactions (Figure 1). During the nucleolytic reaction, the ion (predominantly Mg2+ (17,18)) destabilizes the scissile phosphodiester bond and neutralizes the negatively charged transition state. The catalytic histidine serves as a general base to deprotonate and activate a nucleophilic water molecule used for direct hydrolysis of the nucleic acid (19). In some cases, as in PacI restriction endonuclease (20), the histidine is absent, and its role is taken by a tyrosine, located in the Ω-loop (20). In other cases, such as RecA-dependent nuclease (2), Vibrio vulnificus nuclease (Vvn) (21) or endonuclease I (22), the active site is supplemented by an arginine, located in the Ω-loop or in the N-terminal α-helix, which stabilizes the cleaved phosphate.
Despite their common structural core and sequence motifs, His-Me fingers display extreme sequence diversity, which makes identification of new superfamily members difficult. Moreover, even for known 3D structures, it is difficult to annotate the HNH nuclease domain as it may be distorted and/or buried within a broader structural context. Nonetheless, due to the high biological importance and widespread occurrence of the His-Me proteins, there is a need for their systematic detection and classification. A previously published classification (23) was based on a sparse set of example proteins. However, since its publication in 2003 there has been an explosion of sequence, structural and biochemical data, rendering the classification somewhat outdated. Here, we apply state-of-the-art distant homology detection, combined with extensive sequence and literature searches, to collect the most complete set of proteins containing the His-Me finger domain. Detailed analysis of their sequence-structure-function relationships provides an updated, comprehensive classification of the His-Me finger superfamily.
MATERIALS AND METHODS
Detection of His-Me finger proteins
Firstly, all known His-Me finger families were obtained from the His-Me_finger ‘clan’, as defined by the Pfam database (24). This initial set of His-Me finger domains was used for further searches using our Gene Relational DataBase (GRDB) system based on Meta-BASIC (25), a highly sensitive method for distant homology detection, which compares sequence profiles, enriched by predicted secondary structure (meta-profiles). GRDB includes pre-calculated Meta-BASIC connections between PFAM, KOG and COG families, and proteins of known structure from PDB90, which contains representatives from the Protein Data Bank (PDB), filtered at 90% sequence identity. Each family and structure in the system was represented by its: (i) sequence (for PDB90 structures), or consensus sequence (for PFAM, COG and KOG families), (ii) sequence profile generated with PSI-BLAST (26) (3 iterations, with an inclusion threshold of 0.001) using the NCBI non-redundant protein sequence database derivative (NR70) and (iii) secondary structure, predicted using PSI-PRED (27). The search strategy was based on the concept of transitivity, where each newly identified PFAM, KOG, and COG family or PDB structure was used in further Meta-BASIC searches until no new additional His-Me fingers were found. In addition to the high-reliability Meta-BASIC predictions—with scores >40, corresponding to an E-value of 0.05—all hits scoring <40 were included to identify potentially correct predictions that may have been placed among the unreliable or incorrect ones. These potentially correct predictions were selected based on manual assessment of the conservation of the core secondary structure elements and sequence motifs deemed critical for the His-Me finger fold and nuclease function.
The resulting sequences of His-Me finger representatives retrieved from GRDB and literature searches served as a starting point for exhaustive transitive PSI-BLAST (26) searches against the NCBI non-redundant (NR) protein sequence database (as available in January 2016), with six iterations and an E-value threshold of 0.001. Specifically, at each step, the collected sequences were clustered at a 40% identity threshold with CD-HIT (28), filtered with regard to false positive hits, and used as an input for the subsequent PSI-BLAST runs until convergence.
Clustering of sequence space
The sequences of all the His-Me finger domains were compared, all-against-all, using protein BLAST to detect clusters of closely related proteins. Given that sequences within a superfamily can have different levels of similarity, we tested various thresholds, spanning from 10−5 to 10−11. As a result of our benchmarks, the E-value cut-off for clustering was set to 10−8. The sequence space was visualized and clustered using the Cytoscape software and prefuse force-directed layout (Supplementary Figures S1 and S2) (29).
HMM connectivity network
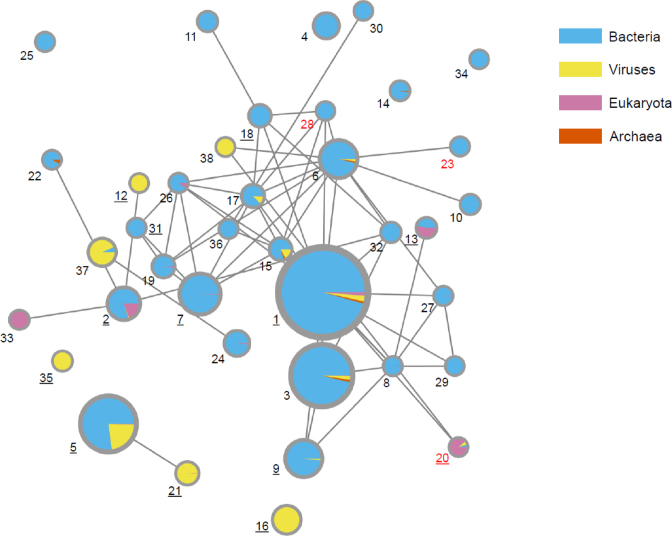
For each of 56 identified groups (clustered at 70% sequence identity, using CD-HIT), a multiple sequence alignment was generated using Mafft (with the –linsi option) (30). The alignments were used for building HMM profiles using hmmbuild (with default parameters) from the HMMER3 suite (31). The HMM profiles were then compared using hhsearch (with default parameters). Groups sharing high similarity (E-value < 10−9 between their representatives) were combined. The resulting network of 38 groups was visualized via Cytoscape (Figure 2).
Figure 2.
Profile hidden Markov model (HMM) connectivity network for His-Me finger proteins. Nodes represent HMM profiles built with hmmbuild (using default parameters) from the HMMER3 suite, based on the alignments of sequences from respective groups clustered at 70% identity. Edges depict E-value scores (<0.001) between the groups calculated with hhsearch with default parameters. The size of the nodes is proportional to the number of all sequences in the NCBI NR database; the taxonomy distribution is also provided. All groups are numbered as in Table 1, those with available 3D structures are denoted with underlined numbers, whereas new groups are marked with red font. The network was visualized with the Cytoscape software (29).
Domain architecture and genomic context
The domain architectures of all the sequences were analyzed using hmmscan from the HMMER3 suite (31), run against the Pfam database, with an E-value threshold of 10−5. If two consecutive domains overlapped by >30% of the length of the shorter domain, the domain with lower E-value was retained. Transmembrane regions were detected using the TMHMM package (32). Gene neighbors were determined using the KEGG GENOME database, and their protein products were analyzed with hmmscan, run against the Pfam and CDD databases (E-value < 10−5).
Structure-based multiple sequence alignment
All identified His-Me finger domain structures were clustered at 90% sequence identity and manually superposed on their core structural elements (ββα) using Swiss PDB Viewer (33). The residue correspondence was assessed manually based on visual inspection of the superposition, with respect to their locations in secondary structure elements and contribution to the active site formation.
Representative sequences from structurally uncharacterized families were added to the structural alignment based on Meta-BASIC mappings as well as secondary structure predictions and conservation of key residues predicted to form the catalytic site.
RESULTS
The current release of PfamA (version 31.0) consists of 23 families belonging to the His-Me_finger clan (CL0263), whereas the SCOP database divides His-Me finger structures (SCOP ID: 54060) into six families. Nevertheless, to date, many of these families are poorly characterized, and their role in the cell remains elusive.
In this work, we conducted exhaustive distant homology detection searches, combining a previously established and successful methodology for studying large and diverse protein superfamilies (34,35) with a series of PSI-BLAST searches. We identified over 111 000 proteins in the NCBI NR database containing the His-Me finger fold, which we classified into 38 distinct groups (summarized in Table 1) based on their sequence similarity.
Table 1. 38 groups of proteins retaining the His-Me finger fold.
| Group | PDB90 | Pfam | COG/KOG | Functions of defining family members | Taxonomy | |||
|---|---|---|---|---|---|---|---|---|
| Viruses | Bacteria | Archaea | Eukaryota | |||||
| 1 | 4h9d | PF01844 (HNH) | COG3183 | GVE2 thermostable nicking nuclease (107) | 588 | 44036 | 404 | 1020 |
| 4ogc | PF13391 (HNH_2) | COG1403 | ATP-dependent nuclease EA31 | |||||
| 4cmp | PF13395 (HNH_4) | COG3440 | phiRv1 integrase (108) | |||||
| 5axw | PF14279 (HNH_5) | COG3513 | CRISPR RNA-guided endonuclease Cas9 | |||||
| 5h0o | McrA restriction endonuclease (109) | |||||||
| MnlI restriction endonuclease (51) | ||||||||
| Homing endonucleases F-LimI, F-LimIV | ||||||||
| Bacteriophage T4 homing endonucleases MobA, MobB, MobC, MobD, MobE | ||||||||
| F-TflIV nuclease (110) | ||||||||
| DNA annealing helicase and endonuclease ZRANB3 (52,53) | ||||||||
| PLC-like phosphodiesterase | ||||||||
| 2 | 1u3e | PF05551 (zf-His_Me_endon) | I-HmuI homing endonuclease (10) | 795 | 3288 | 13 | 28 | |
| PF13392 (HNH_3) | SPBc2 prophage-derived putative HNH homing endonuclease YosQ | |||||||
| Pathogenesis-related transcriptional factor and ERF protein (111) | ||||||||
| 3 | PF07510 (DUF1524) | COG3513 | GmrD from GmrSD restriction system (59) | 28 | 12124 | 174 | 263 | |
| COG1217 | ||||||||
| 4 | PF14414 (WHH) | COG5585 | WHH toxin domain in toxin-antitoxin systems (63) | 0 | 2098 | 2 (Methanosarcina barkeri) | 0 | |
| PF12639 (Colicin-DNase) | Cell wall assembly Smi1/Knr4 family proteins | |||||||
| Type VII secretion proteins | ||||||||
| 5 | 1g8t | PF01223 (Endonuclease_NS) | KOG3721 | YxiD toxin component of the YxiD-YxxD toxin-antitoxin system (66) | 22 | 8333 | 10 | 2456 |
| 1zm8 | PF13930 (Endonuclea_NS_2) | COG1864 | FhaB proteins | |||||
| 3ism | COG5444 | RhsA proteins (67) | ||||||
| 3s5b | Eukaryotic DNA/RNA mitochondrial endonuclease G (10) | |||||||
| 4a1n | Bacterial virulence factor NucA (12) | |||||||
| 5gkc | NET-degrading DNA-entry nuclease (EndA) (14) | |||||||
| 2xgr | Streptodornase (36) | |||||||
| 5fgw | ||||||||
| 6 | PF13391 (HNH_2) | COG3440 | Restriction endonucleases AlwI, BbrI (68), HphI (69) | 3 | 5351 | 88 | 120 (Fungi) | |
| 7 | 3g27 | PF06147 (DUF968) | Prophage-derived proteins YbcO, YdfU | 30 (Bacteriophages) | 6423 | 0 | 3 | |
| PF07102 (DUF1364) | ||||||||
| 8 | PF14279 (HNH_5) | 0 | 158 | 1 (Methanobrevibacter smithii) | 0 | |||
| 9 | 1oup | PF04231 (Endonuclease_1) | COG2356 | Bacterial periplasmic or secreted endonuclease I, Dns, DnsH, Vvn (21) | 0 | 5245 | 0 | 79 |
| 2pu3 | Mg2+-activated RNase Bsn (76) | |||||||
| 2vnd | ||||||||
| 10 | PF14412 (AHH) | AHH toxin domain in bacterial polymorphic toxin system (63) | 0 | 474 | 0 | 1 (Rhizophagus irregularis) | ||
| 11 | PF14411 (LHH) | LHH toxin domain in bacterial polymorphic toxin system (63) | 0 | 522 | 0 | 1 (Diachasma alloeum) | ||
| 12 | 1a73 | PF05551 (zf-His_Me_endon) | Homing endonucleases I-PpoI, I-CpiI, I-PchI (6,112) | 0 | 0 | 0 | 62 (Fungi) | |
| 13 | 3fc3 | PF02945 (Endonuclease_7) | Type II restriction endonuclease VII Hpy99I (7) | 330 | 289 | 3 | 0 | |
| 1en7 | Phage T4 recombination endonuclease VII (9) | |||||||
| 14 | PF14410 (GH-E) | GH-E toxin domain in bacterial polymorphic toxin system proteins (48) | 0 | 366 | 8 (Halogeometricum borinquense, Halovivax ruber) | 0 | ||
| 15 | PF01844 (HNH) | Putative mismatch repair endonuclease YisB (81) | 0 | 948 | 35 | 205 | ||
| 16 | 1ozj | PF03165 (MH1) | KOG3701 | MH1 domain of Smad proteins (43) | 0 | 0 | 0 | 2850 |
| 4zkg | ||||||||
| 17 | PF01844 (HNH) | COG3183 | Group II intron-encoded protein LtrA with reverse transcriptase and RNA maturase activities (82) | 16 | 1119 | 1 (Euryarchaeote) | 148 | |
| KOG4768 | Eukaryotic putative COX1/OXI3 intron 1 protein | |||||||
| Mitochondrial DNA mismatch repair protein MutS | ||||||||
| 18 | 3u43 | PF01844 (HNH) | Colicins, pyocins (4,113,114) | 0 | 1097 | 0 | 0 | |
| 7cei | ||||||||
| 1bxi | ||||||||
| 4uhp | ||||||||
| 4qko | ||||||||
| 19 | PF16784 (HNHc_6) | Phage Orf family recombinases (83) | 32 | 1163 | 0 | 1 (Homo sapiens) | ||
| 20* | 3ldy | Restriction endonuclease PacI (20,115) | 78 (Giant viruses) | 4 | 0 | 6 | ||
| 21 | 4gtw | PF01223 (Endonuclease_NS) | Ectonucleotide pyrophosphatase/phosphodiesterase (Enpp1, Enpp2, Enpp3) (84,86) | 5 (Birds viruses) | 0 | 0 | 1309 (Fish and marine species, i.e., tunicates) | |
| 4zg9 | Venom phosphodiesterase (87) | |||||||
| 22 | PF13392 (HNH_3) | Gyrase subunit B intein | 0 | 84 | 7 | 0 | ||
| Deoxycytidine triphosphate deaminase intein | ||||||||
| 23* | 0 | 180 | 0 | 0 | ||||
| 24 | PF05766 (NinG) | Rap (recombination adept with plasmid)/NinG proteins involved in recombination (73) | 27 | 1972 | 0 | 2 | ||
| 25 | PF15657 (Tox-HNH-EHHH) | Tox-HNH-EHHH toxin domain in bacterial polymorphic toxin system (48) | 0 | 197 | 0 | 0 | ||
| 26 | PF16786 (RecA_dep_nuc) | RecA-dependent nuclease involved in genome degradation (2) | 20 | 184 | 0 | 0 | ||
| 27 | PF05315 (ICEA) | Type II restriction endonucleases NlaIII and IceA1 (116,117) | 2 | 184 | 0 | 0 | ||
| 28* | SphI restriction endonuclease (48,118) | 0 | 34 | 0 | 0 | |||
| 29 | PF09665 (RE_Alw26IDE) | Restriction endonucleases RE_Alw26IDE, BsaI, BsmBI (119) | 0 | 57 | 0 | 0 | ||
| 30 | PF15652 (Tox-SHH) | Tox-SHH toxin domain in bacterial polymorphic toxin system (48) | 0 | 77 | 0 | 0 | ||
| 31 | 3plw | PF16786 (RecA_dep_nuc) | RecA-dependent nuclease involved in genome degradation (2) | 1 (Enterobacteria phage P7) | 203 | 0 | 0 | |
| 32 | PF01844 (HNH) | Hcp1 family polymorphic toxin system protein YhhZ (92) | 0 | 543 | 0 | 0 | ||
| 33 | PF13392 (HNH_3) | Putative early/late infection stage endonucleases from Chlorella viruses | 125 (Paramecium bursaria Chlorella virus, Acanthocystis turfacea Chlorella virus) | 0 | 0 | 0 | ||
| 34 | PF15637 (Tox-HNH-HHH) | Tox-HNH-HHH toxin domain in bacterial polymorphic toxin system (48) | 0 | 71 | 0 | 0 | ||
| 35 | 1v0d | PF09230 (DFF40) | Rep4, DffB, Drep-4 caspase-activated DNases (38) | 0 | 0 | 0 | 399 | |
| 36 | PF15635 (Tox-GHH2) | Tox-GHH2 toxin domain in bacterial polymorphic toxin system (48) | 0 | 166 | 0 | 0 | ||
| 37 | PF15636 (Tox-GHH) | KOG4659 | Tox-GHH toxin domain in bacterial polymorphic toxin system | 0 | 181 | 0 | 2476 | |
| Metazoan teneurin (48) | ||||||||
| 38 | PF13391 (HNH_2) | Eukaryotic pathogens effector CR-HNH (120) | 0 | 0 | 0 | 132 | ||
Structure analysis
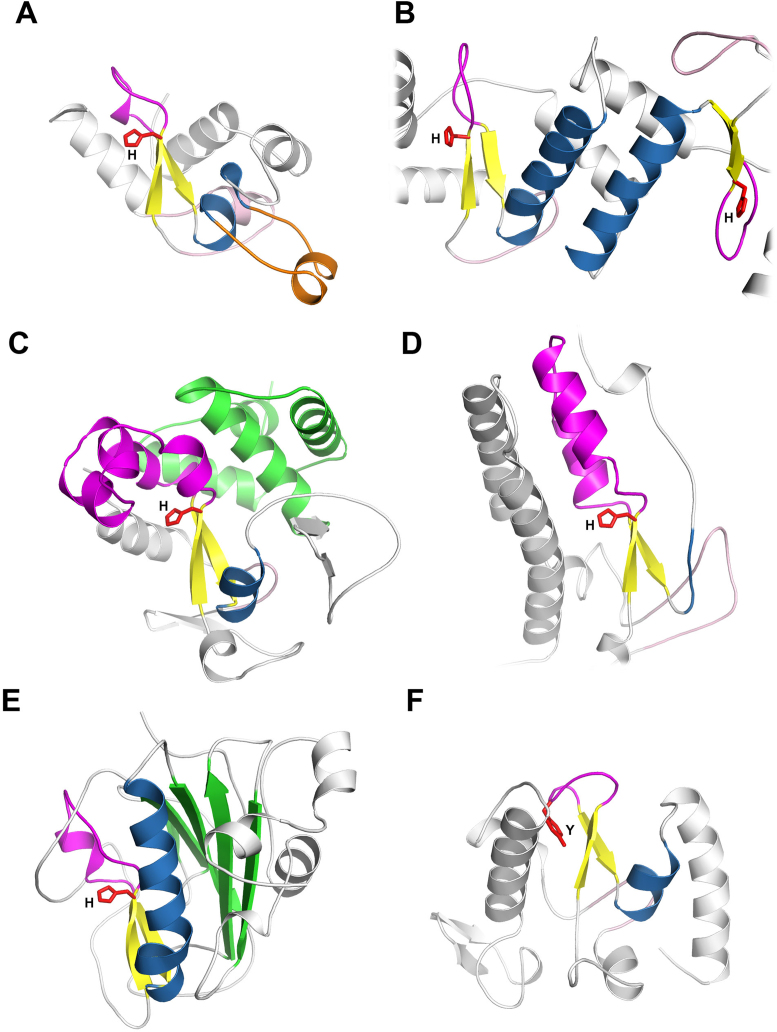
Proteins containing the His-Me finger domain display significant structural diversity, either within the His-Me finger itself, or in the additional structures supporting the catalytic domain. The core β-hairpin is always present, performing a strictly defined function. The hairpin along with the extensive Ω-loop, inserted between the consecutive β-strands, provides catalytic residues and stabilizes the overall structure. The core α-helix exhibits an extraordinarily broad repertoire of variants, given the overall small size of the His-Me finger. For instance, excepting obvious differences in length, this helix can be disrupted by a protruding ‘helical finger loop’ of unknown function, as observed in the NucA (13), Spd1 (15), Ref (2), EndA (36) and Sda1 (37) nucleases. An extreme example is provided by the viral RecA-dependent Ref nuclease, where the core α-helix hosts a helical finger almost twice as long as the helix itself (2). In all these cases, however, the α-helix retains its integrity and orientation relative to the β-hairpin (Figure 3A). It also preserves residues responsible for metal ion coordination. An additional role is assigned to the α-helix in the Holliday structure resolvase, ENDOVII (9), where it is essential for dimerization (Figure 3B), as well as for orienting the dimer towards the junction.
Figure 3.
Structural diversity of His-Me fingers. The core elements of the fold (ββα motif) are coloured in yellow and blue for β-strands and α-helix, respectively. The Ω-loop is coloured in magenta. The zinc knuckle preceding the core β-hairpin is shown in light pink. The catalytic site His/Tyr is presented as red sticks. (A) The structure of viral RecA-dependent Ref nuclease (PDB ID: 3plw) with finger loop insertion (orange). (B) Dimerized nuclease domains of Holliday Junction Resolvase ENDOVII (PDB ID: 2qnc). (C) The structure of Vibrio vulnificus nuclease (Vvn) (PDB ID: 1oup) with the α-helical domain (green) at the back of the HNH domain. (D) The structure of Caspase-Activated DNase (CAD) (PDB ID: 1v0d). The loop substituting for the core α-helix is coloured in blue. (E) The structure of Serratia marcescens nuclease (Sm) (PDB ID: 1g8t) with the β-sheet domain (green) at the back of HNH domain. (F) The structure of PacI (PDB ID: 3m7k) with tyrosine substituting for catalytic histidine.
The Ω-loop, being an insertion, exhibits a more obvious variation, from being strongly reduced as in ENDOVII resolvase (9) or Hpy99I restriction endonuclease (7), to becoming very elaborate, carrying additional secondary structure elements, as in e.g. CAD (38), Vvn (21) or endonuclease I (22) (Figure 3C). The Ω-loop may also host residues for coordination of a zinc ion, as observed in I-PpoI homing endonucleases (6) or inactive Smad (39), that additionally stabilizes the fold.
In general, zinc ions play an important role in stabilizing many His-Me finger structures. Apart from binding to the Ω-loop as described above, zinc may also be coordinated within a ‘zinc knuckle’ at the interface between the N-terminal part of the α-helix and an additional β-hairpin structure preceding the His-Me finger. This feature is present in a range of proteins bearing the His-Me finger domain, such as CRISPRs, homing endonucleases, CAD (Caspase-Activated DNase) DNA-degrading nuclease (38), ENDOVII HJC (9) and GmR87 nickase of unknown function (Figure 3D).
Stabilization of the His-Me finger core may also be provided by extensive structures. This is characteristic of certain virulence factors—such as Sm (14), NucA (13), Spd1 (15), Sda1 (37) – EndoG (11), EXOG or CPS-6 (12), that have an additional β-sheet at the ‘back’ of HNH structure (Figure 3E). In fact, in these cases, the His-Me finger domain is inserted into a larger β-sheet. Similarly, Vibrio vulnificus nuclease (Vvn) (21) and endonuclease I (22) retain a big α-helical domain located at the C-terminus sheltering the central His-Me finger nuclease (Figure 3C), whereas toxins (e.g. Colicin E9 (40), Pyocin S2 (41)) possess expanded structural decorations at the N-terminus.
Active site variation
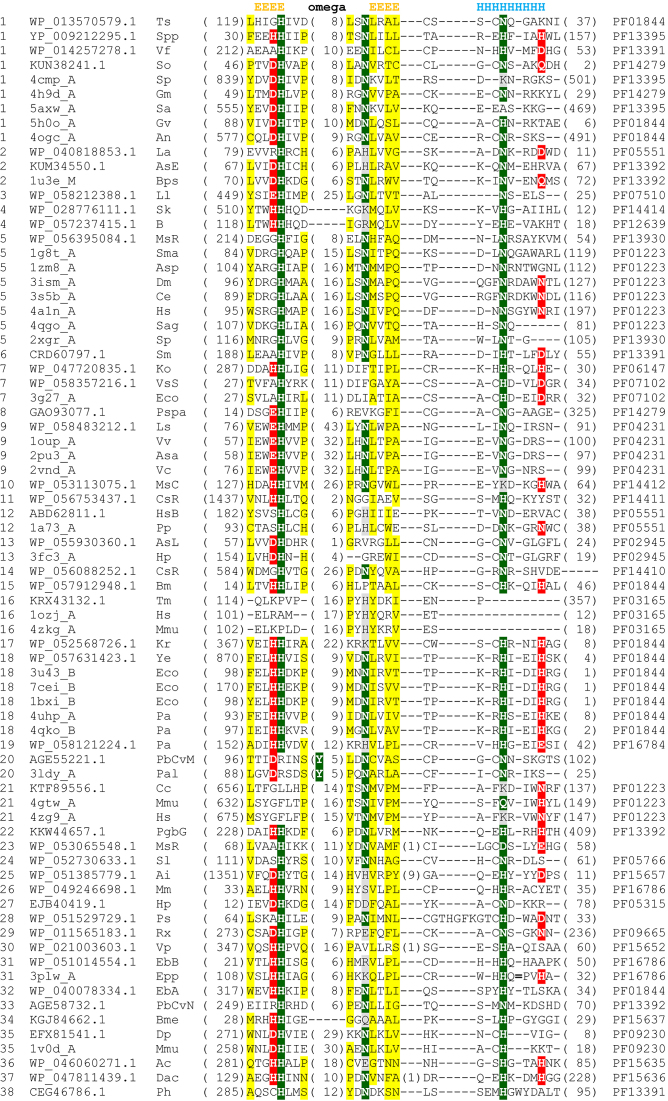
The His-Me catalytic site is set up around the axis connecting two key residues: the nearly invariant histidine, located in the first β-strand, and the asparagine/histidine in the N-terminal turn of the α-helix (Figure 3). The latter residue directly coordinates a divalent metal ion. In addition, the ion is often coordinated by D/E/H residues preceding the catalytic histidine in the first β-strand, and another polar residue along the α-helix, in the +4 or +8 position with respect to the asparagine/histidine mentioned above (Figure 4).
Figure 4.
Multiple sequence alignment of the conserved core regions of the His-Me finger superfamily. The numbers of excluded residues are specified in parentheses. The extensive finger loop in the sequence corresponding to PDB ID: 3plw is denoted with ‘ = ’. Sequences are labelled according to the group number followed by their NCBI accession number or PDB ID, and an abbreviation of the species name. Respective Pfam families are given in the last column. Residue conservation is denoted by the following scheme: uncharged, highlighted in yellow; polar, highlighted in grey; HNH motif residues, highlighted in green; remaining metal ion coordinating residues, highlighted in red. The secondary structure is shown above the corresponding alignment blocks.
Based on an analysis of the conservation of the active site residues, most of the identified His-Me finger domains appear to be active nucleases with histidine serving as a general base (Figure 3). However, some groups display considerable variation within the predicted active site. Three groups defined in our sequence clustering (i.e. groups 16, 20, 21) lack the catalytic histidine. Structural and biochemical studies suggest that a tyrosine located in the extended Ω-loop in group 20 substitutes for histidine in the catalyzed reaction (Figure 3F) (20). As it was shown for PacI endonuclease, in the one-metal-ion mechanism, the tyrosine side-chain may act as a nucleophile, substituting for water utilized in histidine-dependent nucleases (19). A similar mode of action is typical for HUH endonucleases (42).
The remaining two groups, 16 and 21, lack both histidine and equivalent tyrosine residues, indicating a loss of nuclease function. Group 16 corresponds to the MH1 domain of Smad (dwarfin-A) proteins. According to previous studies (43,44), MH1 is a modified endonuclease that has been recruited as a transcriptional activator. Although it has lost the catalytic activity, it has retained the DNA-binding capability (43). Group 21 contains the predicted ectonucleotide pyrophosphatase/phosphodiesterase-1, -2 and -3 (Enpp1, Enpp2/autotaxin, Enpp3) and venom phosphodiesterases. The C-terminal regions of the proteins contain a nuclease-like His-Me finger domain, which is catalytically inactive and has been suggested to bind the substrate (45).
Newly detected His-Me finger superfamily members
To perform a systematic, context-independent classification of the His-Me finger superfamily, we clustered the collected sequences (as described in detail in Materials and Methods) and subsequently assigned them to 38 groups, corresponding to clusters of similar sequences (Figure 2).
Out of the 38 defined groups, three (designated here with numbers 20, 23 and 28) are novel, i.e., neither Pfam, CDD nor SCOP contains a corresponding entry. Sequence similarity to already described His-Me fingers and predicted secondary structure suggest that they possess the His-Me finger fold. For group 23 the molecular function remains unknown, whereas the remaining two (20 and 28) have been studied in the context of single proteins.
Additionally, we identified seven Pfam families: DUF1364, RE_Alw26IDE, Tox-HNH-HHH, Tox-HNH-EHHH, Tox-SHH, Tox-GHH and Tox-GHH2, which have not been assigned to the His-Me finger Pfam clan but are predicted to have both the His-Me finger fold and the active site (46–48). DUF1364 clusters with DUF968, together forming group 7, and includes the Escherichia coli prophage protein ybcO of known structure (PDB ID: 3g27). A manual structure superposition to other members of this superfamily unarguably classifies DUF1364 as a His-Me finger domain, which agrees with the hypothesis made previously (46).
Functional annotation of the defined groups
Table 1 gives a complete list of the defined groups, along with their taxonomic distribution, correspondence to solved structures, a short description of their founding members, and their Pfam, COG and KOG assignments. Below, we describe selected groups in more detail (including those that have been poorly characterized or functionally unannotated), proposing their function and highlighting distinctive properties. All the domain architectures (Supplementary Figure S3), genomic neighborhoods (Supplementary Figure S4), sequence logos (Supplementary Figure S5) together with accession numbers of superfamily members (Supplementary Dataset S1) are available as Supplementary Data.
Group 1 (HNH, HNH_2, HNH_4, HNH_5)
Group 1 embraces the RNA-guided Cas endonuclease from CRISR/Cas9 systems. Cas possesses two well-conserved nuclease domains: RuvC and His-Me finger (HNH). All known Cas9 enzymes contain an HNH domain that cleaves the DNA strand complementary to the guide RNA sequence (target strand), and a RuvC nuclease domain required for cleaving the non-complementary strand (non-target strand), yielding double-strand DNA breaks (49). Group 1 contains also characterized restriction endonucleases, e.g. McrA from E. coli (accession: NP_415677.1) and MnlI (accession: AAU87367.1) which are specific to 5-methylcytosines (50,51). This group also includes vertebrate (including Homo sapiens (accession: NP_115519.2)) ATP-dependent annealing helicase (AH2/ZRANB3). The structure-specific His-Me finger domain of this protein cleaves the D-loop structure and produces an accessible 3′-OH group, which enables DNA polymerase action (52,53). Another important subset of group 1 are the His-Me finger-containing endonucleases from bacteriophages—key components of phage DNA packaging machines (54). Among them is a recently biochemically and structurally studied thermostable nicking endonuclease from Geobacillus virus E2 (PDB ID: 5h0o). It was proposed that an additional α-helix preceding the core β-hairpin may contribute to the enzyme's thermostability by maintaining its structural conformation (55).
Group 2 (zf-His_Me_endon, HNH_3)
This group constitutes a large ensemble of homing endonucleases found in organisms from across the tree of life, including the structurally characterized endonuclease I-HmuI (PDB ID: 1u3e). Other proteins from this group show a domain architecture typical of homing endonucleases, i.e. a number of single or tandem repeats of nuclease-associated modular DNA-binding domains (NUMOD) and other helix-turn-helix-type (HTH-type) domains, along with an Apetala 2 (AP2) domain, which are known to serve as DNA-binding modules conferring substrate specificity (56,57).
Group 3 (DUF1524)
Proteins from this group belong to GmrSD restriction systems, which target and cleave DNA with glucosylated hydroxymethylcytosine (HMC) (58). GmrSD consists of GmrS and GmrD subunits, which both exhibit endonuclease activity towards DNA with sugar-modified HMCs (59). In addition, GmrS may also participate in NTP binding and hydrolysis. GmrS possesses a ParB/Srx fold, whereas GmrD has been shown to contain the HNH motif (59). These proteins usually possess additional C-terminal domains—DUF4357, DUF4268 and AlbA_2. The last of these, AlbA_2, belongs to the AlbA superfamily of DNA/RNA-binding domains (60). The molecular function of DUF4357 and DUF4268 remains unknown. According to our sequence-based predictions of their DNA-binding potential using DNABIND server (61), only DUF4268 is likely (with 98% probability) to bind DNA. Apart from REases, group 3 contains bacterial putative extracellular nucleases containing transmembrane helices and a C-terminal Excalibur domain (PF05901), which occurs in calcium-binding nucleases. The presence of close homologs in Agaricomycetes fungi and Agromyces bacteria may suggest a potential horizontal gene transfer between kingdoms.
Groups 4 (WHH, Colicin-DNase) and 11 (LHH)
Genes encoding bacterial proteins from these groups are located in direct genomic proximity to Smi1/Knr4 genes, whose products belong to the SUKH superfamily of immunity proteins. This suggests they might act as nucleic acid-degrading toxins in toxin-antitoxin systems, similarly to contact-dependent growth inhibition (CDI) operons (62). Proteins from groups 4 and 11 follow a widely conserved domain architecture. They contain pre-toxin secretion-related domains (PT-VENN, PT-HINT, RHS, filamentous hemagglutinin repeats), which facilitate export of a toxin from the host cell and eventually release the toxic nuclease domain upon uptake by the target cell. Additional transmembrane elements are presumably required for binding of the extruded toxin to the cell membrane (63). Interestingly, group 4 also contains single representatives from archaea and arthropods, e.g. two from an archaeon Methanosarcina barkeri and one from a wasp Diachasma alloeum, suggesting potential horizontal transfer.
Group 5 (Endonuclea_NS_2)
Non-specific endonucleases from this group constitute a broad set of bacterial virulence factors. They target and degrade both double-strand and linear DNA and RNA with no sequence specificity, making them well-tailored for successful competition. Well-studied examples include streptococcal NET-degrading DNA-entry nuclease (EndA) (accession: WP_001036779), streptodornase (Spd1, Sda1) (accession: NP_268944), and nuclease A (NucA) (accession: WP_010999911), which all contain a characteristic DRGH sequence motif preceding the catalytic histidine (35). They have been associated with invasive bacterial infections by degrading chromatin-rich neutrophil extracellular traps (NETs) and enabling Streptococci to overcome the mammalian immune response (15,37,64,65).
Some proteins from this group constitute polymorphic toxins, which are deployed to inhibit the growth of neighboring cells. The group contains Bacillus YxiD toxins from the YxiD-YxxD toxin-antitoxin system which show cytotoxic RNase activity (accession: P42296) (66) and RhsA toxins (accession: WP_013316542). Both proteins are thought to be secreted from the cell in a contact-dependent manner (66,67). However, this hypothesis still needs experimental confirmation.
Interestingly, the His-Me finger domain from this group is also present in some serine proteases, e.g., from Pseudomonas aeruginosa (accession: OKR41748) and other gram-negative species. These proteins are involved in virulence and secreted by the autotransporter pathway (70). Moreover, in the genomic neighborhood of the proteins from this group in the Campylobacter species, we identified genes encoding signal peptidases (accession: WP_002870274). They belong to the MEROPS S24 family (11), which embraces LexA and type I signal peptidases. These proteases cleave away the N-terminal signal peptide from the translocated protein precursor, thereby playing a crucial role in protein transport across membranes (71). Domains from this group are often coupled with peptidase-like domains, such as Trypsin_2 (PF13365), Peptidase_M8 (PF01457), and adhesin Mfa_like_1 (PF13149), which may participate in invading cell adhesion. This may imply that these His-Me finger nucleases are functionally linked to proteases and enhance virulent activity against the invaded host.
The group contains also a large number of eukaryotic members. These include mitochondrial DNA/RNA endonucleases G and endonuclease G-like 1 (EXOG), which participate in chromosomal DNA degradation during apoptosis (68). As an apoptosis-related factor, EXOG is heavily regulated. For instance, the multimerization induced by proteolysis has been shown to trigger a shift from endonuclease to 5′–3′ exonuclease activity in Caenorhabditis elegans EndoG (11).
Group 6 (HNH_2)
This group encompasses AlwI, BbrI and HphI restriction endonucleases (68–70). The exact mode of target sequence recognition for these restrictases is unknown. While the N-terminal domain of AlwI (accession: CBL24780) shows similarity to the BpuJI recognition domain (PF11564) (71), the recognition domain of BbrI and HphI could not be predicted reliably. As indicated by site-directed mutagenesis studies, the key residues responsible for substrate recognition lie outside the catalytic His-Me finger domain (70).
Uncharacterized members of this group appear to use various mechanisms to provide fidelity of sequence readout. A few of them have a SAD/SRA domain that recognizes hemi-methylated CpG dinucleotides and other 5mC containing dinucleotides, suggesting that some putative proteins may act as restriction enzymes that target methylated or hemi-methylated DNA rather than a specific nucleotide sequence (63). Interestingly, the DUF3427 domain (PF11907) tends to co-occur with the His-Me finger (e.g., in WP_052015257 and WP_052747245). Genes encoding proteins with DUF3427 together with the His-Me finger at the N- and C-terminus lie in close genomic proximity to genes that code for methyltransferases and ATPases (Supplementary Figure S4), respectively, suggesting a potential role in restriction for proteins containing HNH and DUF3427 domains.
Group 7 (DUF968, DUF1364)
This bacterial group contains multiple putative prophage-encoded proteins, including Qin prophage from Escherichia coli (YdfU) (accession: NP_416078), a phage transcriptional regulator from Arsenophonus nasoniae (accession: CBA76303), and DLP12 prophage (YbcO) of known structure (PDB ID: 3g27). The genomic neighbors of prophage-encoded members include, e.g., crossover junction endodeoxyribonuclease RusA, phage antitermination protein Q, and antirepressor protein, which are involved in transcription and recombination. Consistently, some members of this group have additional domains involved in single-strand DNA annealing during recombination, e.g. ERF (PF04404) and Rad52_Rad22 (PF04098), also of bacteriophage origin (72). Taken together, we hypothesize that the His-Me finger nuclease domain in these proteins acts as a recombinase that participates in DNA repair and replication, similarly to NinG and RecA-dependent endonucleases (73).
Group 9 (Endonuclease_1)
Escherichia coli endonuclease I (EndoI) is a bacterial sequence-independent periplasmic or secreted protein. Periplasmic nucleases, including Vibrio vulnificus nuclease (Vvn), EndoI, Dns, and DnsH protect the cell against the uptake of foreign DNA during transformation, which results in a decrease of transformation rate (21,74). These endonucleases are not active under reducing conditions in the cytoplasm, however the presence of disulfide bonds in their structures suggests that their activity is triggered by a transition to an oxidized form upon extrusion from the cell (21,75). Another known member of group 9 is an Mg2+-activated ribonuclease Bsn (accession: WP_009968119), whose role in the cell remains unknown (76).
In proteins from this group, the His-Me finger domain appears together with a wide range of cell-surface associated domains of an immunoglobulin-like fold, among which the most abundant are fibronectin type III (fn3, PF00041), lamin-tail domain (LTD, PF00932), and bacterial Ig-like domain (Big_2, PF02368). Big_2, along with the fibronectin domain, have been identified as virulence factors involved in adhesion of pathogenic bacterial strains to host cells (77). In the genomes of various Enterobacteria species, the genes encoding sprT peptidase-like proteins and RNA methyltransferases are located in direct proximity to the His-Me finger genes, suggesting potential involvement of the His-Me finger domain in the virulence machinery.
Group 13 (Endonuclease_7)
Group 16 contains endonuclease VII, i.e., a type II restriction enzyme Hpy99I from the gastric pathogen Helicobacter pylori (7), and phage T4 endonuclease VII, which is a Holliday junction resolvase that cleaves T4 DNA before packaging it into the phage head (9). In some uncharacterized members from Actinobacteria, genes encoding the His-Me finger-containing proteins are frequently found downstream to a prokaryotic ubiquitin-like gene whose protein product contains a Pup_ligase domain, or upstream to the proteasome beta subunit gene (Supplementary Figure S4), suggesting a role of these His-Me finger nucleases in the Pup-proteasome system (78).
Group 14 (GH-E)
This group comprises toxins, such as the Rhs family proteins from bacterial polymorphic toxin systems, whose sequences contain the characteristic GH-E sequence motif (48,63). In direct genomic proximity to genes coding for many members of group 14, from a wide variety of bacterial species, are genes that encode uncharacterized YjbI-like proteins. YjbI consists of multiple pentapeptide repeats of unknown function. According to the SecReT6 database (79), the genes from this group are frequently located in genomic regions occupied by genes encoding the type 6 secretion system (T6SS). The His-Me finger may therefore constitute an alternative toxin effector in these bacterial species. Intriguingly, we could not find any annotated adjacent gene whose product would provide immunity against self-detrimental nuclease activity. This may imply that these nucleases are benign, or have an additional, as yet uncharacterized, function as an antitoxin.
Group 15 (HNH)
This group contains restriction endonuclease-like proteins from various prokaryotic and eukaryotic organisms, including YisB protein from Bacillus subtilis (accession: NP_388947). In the MutS–MutL–MutH pathway of mismatch repair, in the absence of a MutH nuclease homologue, other nicking endonucleases might be recruited to serve as an analog of MutH. YisB has been proposed as a good candidate to fulfil this role in Bacillus species (80). Further experiments have confirmed the role of YisB in DNA repair (81). Our analysis of the genomic neighborhood of yisB in various Bacillus strains consistently finds adjacent genes involved in the DNA repair processes, such as ATP-dependent helicase/deoxyribonuclease subunits A and B with PDDEXK_1 nuclease domains, and ATP-dependent double-strand DNA exonucleases SbcC and SbcD. Although the role of fungal homologs (mostly from Rhizobiaceae) remains elusive, they may perform an equivalent function.
Group 17 (HNH)
This assembly contains a multitude of group II intron reverse transcriptases/maturases. They include LtrA protein from Lactococcus lactis (accession: WP_011835237), which exhibits three activities: DNA endonuclease for site-specific cleavage of the DNA target site and initiation of mobility, reverse transcriptase for intron duplication, and maturase that aids splicing (82). In some eukaryotes, mostly fungi and algae, proteins from this group are encoded by mitochondria or plastids.
Group 19 (HNHc_6)
Group 19 contains proteins from the phage Orf family of recombinases from Listeria phages (accession: CAC96489). Listeria phage members of the Orf family were indicated to be distantly related versions of a canonical Orf representatives and may constitute novel single-strand specific DNases that could have arisen from fusion of an Orf DNA binding domain to a His-Me finger nuclease domain (83). Our analyzes of genomic neighborhood support this hypothesis; the corresponding genes are potentially co-transcribed with genes encoding proteins from the SSB (PF00436) and DnaB_2 (PF07261) Pfam families. Products of these genes are involved in replication initiation, thus, bearing out the role of these His-Me finger proteins in replication.
Group 20
This predominantly viral group contains few bacterial and archaeal representatives, e.g. PacI rare-cutting restriction endonuclease from Pseudomonas alcaligenes of known structure (PDB ID: 3ldy). Interestingly, this REase appears to lack a coupled methyltransferase, so the protection from self-cleavage stems from the absence of recognized sequences in the P. alcaligenes genome (20). The His-Me finger domain is coupled with an HTH-like domain at the N-terminus which may contribute to DNA-binding specificity. Viral members of group 20 include proteins from giant viruses: Paramecium busaria and Pandoravirus species. It has been reported that the genome of Paramecium busaria chlorella virus 1 encodes multiple site-specific restriction endonucleases of other folds, capable of degrading host chromosome in early stages of infection (77,78). It is therefore tempting to speculate that PacI viral homologs might be other viral restrictases.
Group 21
This group embraces Enpp1, Enpp2 and Enpp3 transmembrane glycoproteins, which hydrolyze extracellular nucleotide triphosphates to produce pyrophosphates (Enpp1, Enpp3) (84,85) or lipid mediator lysophosphatidic acid (Enpp2/autotaxin) (86). These proteins typically contain four domains: catalytic phosphodiesterase, nuclease-like His-Me finger, and two somatomedin B (SMB)-like domains. Both the catalytic phosphodiesterase and His-Me finger domains contribute to structural stability (84), whereas the His-Me finger additionally participates in substrate binding (45). The nuclease-like domain retains neither the catalytic histidine nor its tyrosine counterpart and is presumed to be catalytically inactive. Interestingly, this cluster also includes proteins that function as venom phosphodiesterases in snakes and a variety of marine species (accession: J3SBP3) (87).
Group 23
This group contains unannotated bacterial proteins that are encoded in direct proximity to genes coding for MazG family proteins. In E. coli, MazG is a regulator of programmed cell death under conditions of nutritional stress. It possesses nucleotide pyrophosphohydrolase activity, inhibited by MazE–MazF toxin–antitoxin module. Within the module, the MazF toxin shows sequence- and single-strand-specific endoribonuclease activity and targets the cellular mRNA in response to nutrition stress (88). The presence of the His-Me finger-containing protein encoded closely to the MazG-coding gene suggests the involvement of an additional endonuclease domain in this complex regulatory system.
Group 24 (NinG)
This group comprises small bacterial and phage proteins. It contains Rap (recombination adept with plasmid) proteins (accession: NP_040639), which increase λ-by-plasmid recombination catalyzed in the native host bacteria recombination pathway (73). Rap is a structure-specific endonuclease that targets and nicks D-loops formed by recombination intermediates (89). It has been reported to function as a Holliday junction resolvase on the crossover of DNA substrates (90). Although its crystal structure is not known, our predictions suggest the Rap protein comprises a C-terminal His-Me finger within a full treble clef domain, with an additional zinc finger followed by a predicted HTH-like domain at the N-terminus. Therefore, its specificity towards branched DNA substrates is probably achieved via additional structural elements.
Group 28
This cluster includes SphI restriction endonuclease (accession: AAB40378), which belongs to the HNH-CIDE toxin family (48). It is hypothesized that the bacterial SphI nuclease domain arose in the milieu of bacterial competition, and has been horizontally transferred to animals (48). An animal CIDE (CAD/DFF40) protein (which belongs to group 35) is involved in DNA fragmentation during apoptosis. We propose that, similarly to PacI, the predicted HTH-like domain preceding the His-Me finger confers specificity to this restriction endonuclease.
Group 32 (HNH)
Group 32 contains Hcp1 family type VI secretion system effector proteins such as the hypothetical Yhhz from E. coli (accession: NP_417899). Type VI secretion system (T6SS) is used by Gram-negative bacteria to supply effector proteins to eukaryotic cells where they serve as virulence factors, and to neighboring prokaryotic cells to mediate competition (91). The C-terminal region of the effector proteins is usually occupied by domains responsible for effector activity—preferentially peptidoglycan hydrolases and phospholipases, but also endonucleases, including the His-Me finger. It has been suggested that HNH domains can mediate antibacterial toxicity in T6SS (92), indicating that the hypothetical Yhhz protein is, in fact, a His-Me finger nuclease.
DISCUSSION
The His-Me finger domain is a relatively small structural motif often embedded in the scaffolds formed by large domain architectures in proteins playing roles in various cellular activities. Predominantly, the His-Me finger exhibits endonuclease activity. However, the superfamily also includes a few examples of exonucleases and proteins that have undergone active site deterioration and, consequently, have lost their catalytic potential while retaining the ability to bind DNA. The origin and evolution of the His-Me finger domain remain elusive. Previous studies have suggested that the ancient treble clef fold might have been a common ancestor of HNH proteins (1,93). On the other hand, the presence of this domain in evolutionarily unrelated protein scaffolds, along with its overall low structural complexity, advocates the case for convergent evolution, in which independent evolutionary lines have resulted in a similar catalytic solution. However, it is tempting to speculate that a fold with such a narrow range of functions must have emerged from a single primordial structural motif.
In general, the rigid architecture of His-Me finger fold does not involve any significant conformational change upon binding to the substrate (21,94). Nonetheless, as suggested in (95), in RNA-guided DNA endonuclease Cas9 from Streptococcus pyogenes and Actinomyces naeslundii, the HNH active site in the apo state is disordered and might order upon nucleic acid binding (95,96). The α-helix of the ββα domain is parallel to the DNA minor groove and predominantly contacts the phosphate backbone, which results in a significant bending and widening of the minor groove downstream the cleavage site, as observed in the complex of homing endonuclease I-HmuI with cognate DNA substrate (10,18).
The limited size of the HNH domain is a two-edged sword. On the one hand, its compactness allows for accommodation in multiple architectures and facilitates the secretion of the nuclease effector in various virulent and toxic activities. The small size is also required for homing endonucleases that are limited by mobility constraints (5). On the other hand, however, the domain alone is presumably not able to recognize the substrate specifically and thus requires additional features to gain specificity. The His-Me finger ββα catalytic core interacts with the target nucleic acid backbone to reach the scissile phosphate, therefore, it does not contribute significantly either to sequence-specificity or to the fidelity of these enzymes (10). Moreover, most sequence-specific interactions are thought to occur via the major groove of DNA, which is more accessible for direct readout by additional structural elements associated with the His-Me finger (97,98). In turn, the α-helix of the His-Me finger makes contacts with the DNA backbone from the side of the minor groove, which explains its explicit sequence-independence. Additionally, the domain shows rare contacts with DNA phosphate backbone, which might be another reason for its non-specificity (21,94). In order to achieve site-specific cleavage, His-Me finger enzymes incorporate a broad range of additional DNA-binding domains, such as various versions of HTH domains. Proteins that are highly specific, restriction endonucleases in particular, usually possess at least one tethered DNA recognition domain, in contrast to single-domain colicins or periplasmic nucleases, which cleave the target sequence with little, if any, specificity (21,99).
The His-Me finger specificity has many flavours. Among His-Me finger representatives, one may find both site-specific enzymes, such as I-Hmul, and proteins that show little sequence-specificity, such as colicin DNases. Colicins (e.g. ColE7) exhibit a universal mechanism of action—they cleave both single-strand and double-strand DNA and RNA without apparent base preference (100). On the other hand, some proteins show DNA-structure specificity, e.g. endonuclease VII prefers structurally perturbed branched and mismatched Holliday junctions, and related forms (101), while RecA-dependent nuclease Ref makes targeted double-strand breaks within a displacement loop formed by RecA (102,103).
The His-Me finger domain can be found on two sides of inter- and intra-organismal competition. It acts as a pathogenic module that, by degrading the infected organism's genetic material, leads to cell death. On the other hand, as an anti-pathogenic entity, it can damage the invading DNA and protect the host from the deleterious activity of the invader. Its overall structural compactness and intrinsic non-specificity make it a perfectly-tailored pathogenic module that successfully targets the competing organisms by various modes of action. We identified a number of groups that contain nucleic acid-targeting toxins in toxin–antitoxin systems and restriction endonucleases in restriction-modification systems. As reflected by this work, these systems exhibit rapid evolution and frequent lateral gene transfers. Nevertheless, since the His-Me finger domain itself exhibits low substrate and site specificity, its nuclease activity would be extremely toxic to the cell. To deal with the detrimental activity of His-Me finger endonucleases, various mechanisms have been utilized. One mode of action is enrichment of disulfide bonds in proteins containing the His-Me finger domain that renders the nuclease inactive (104), and allows for the activation of the enzyme upon extrusion from the reducing intracellular environment to the oxidizing extracellular one (105). Along with other secretion-related domains and immunity proteins, the His-Me finger is a part of a cellular secretion machinery able to kill competing strains. In groups that comprise bacterial toxins, we observed multiple pre-toxin domains coupled with the His-Me finger. The pre-toxin domains possess adhesive properties that help in displaying the C-terminal toxin domain on the cell surface and thus secreting the virulence factor.
Supplementary Material
ACKNOWLEDGEMENTS
The authors thank Roman Laskowski for proof-reading the manuscript.
SUPPLEMENTARY DATA
Supplementary Data are available at NAR Online.
FUNDING
Foundation for Polish Science [TEAM to K.G.]; Polish National Science Centre [2011/02/A/NZ2/00014 and 2014/15/B/NZ1/03357 to K.G.]. Funding for open access charge: Polish National Science Centre [2014/15/B/NZ1/03357].
Conflict of interest statement. None declared.
REFERENCES
- 1. Grishin N.V. Treble clef finger–a functionally diverse zinc-binding structural motif. Nucleic Acids Res. 2001; 29:1703–1714. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2. Gruenig M.C., Lu D., Won S.J., Dulberger C.L., Manlick A.J., Keck J.L., Cox M.M.. Creating directed double-strand breaks with the Ref protein: a novel RecA-dependent nuclease from bacteriophage P1. J. Biol. Chem. 2011; 286:8240–8251. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3. Zhuo W., Lai X., Zhang L., Chan S.H., Li F., Zhu Z., Yang M., Sun D.. Elimination of inter-domain interactions increases the cleavage fidelity of the restriction endonuclease DraIII. Protein Cell. 2014; 5:357–368. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4. Ko T.P., Liao C.C., Ku W.Y., Chak K.F., Yuan H.S.. The crystal structure of the DNase domain of colicin E7 in complex with its inhibitor Im7 protein. Structure. 1999; 7:91–102. [DOI] [PubMed] [Google Scholar]
- 5. Chevalier B.S., Stoddard B.L.. Homing endonucleases: structural and functional insight into the catalysts of intron/intein mobility. Nucleic Acids Res. 2001; 29:3757–3774. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6. Flick K.E., Jurica M.S., Monnat R.J. Jr, Stoddard B.L.. DNA binding and cleavage by the nuclear intron-encoded homing endonuclease I-PpoI. Nature. 1998; 394:96–101. [DOI] [PubMed] [Google Scholar]
- 7. Sokolowska M., Czapinska H., Bochtler M.. Crystal structure of the beta beta alpha-Me type II restriction endonuclease Hpy99I with target DNA. Nucleic Acids Res. 2009; 37:3799–3810. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8. Wang Y.T., Yang W.J., Li C.L., Doudeva L.G., Yuan H.S.. Structural basis for sequence-dependent DNA cleavage by nonspecific endonucleases. Nucleic Acids Res. 2007; 35:584–594. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9. Biertumpfel C., Yang W., Suck D.. Crystal structure of T4 endonuclease VII resolving a Holliday junction. Nature. 2007; 449:616–620. [DOI] [PubMed] [Google Scholar]
- 10. Shen B.W., Landthaler M., Shub D.A., Stoddard B.L.. DNA binding and cleavage by the HNH homing endonuclease I-HmuI. J. Mol. Biol. 2004; 342:43–56. [DOI] [PubMed] [Google Scholar]
- 11. Loll B., Gebhardt M., Wahle E., Meinhart A.. Crystal structure of the EndoG/EndoGI complex: mechanism of EndoG inhibition. Nucleic Acids Res. 2009; 37:7312–7320. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12. Rawlings N.D., Barrett A.J., Finn R.. Twenty years of the MEROPS database of proteolytic enzymes, their substrates and inhibitors. Nucleic Acids Res. 2016; 44:D343–D350. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13. Ghosh M., Meiss G., Pingoud A., London R.E., Pedersen L.C.. Structural insights into the mechanism of nuclease A, a betabeta alpha metal nuclease from Anabaena. J. Biol. Chem. 2005; 280:27990–27997. [DOI] [PubMed] [Google Scholar]
- 14. Miller M.D., Krause K.L.. Identification of the Serratia endonuclease dimer: structural basis and implications for catalysis. Protein Sci. 1996; 5:24–33. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15. Korczynska J.E., Turkenburg J.P., Taylor E.J.. The structural characterization of a prophage-encoded extracellular DNase from Streptococcus pyogenes. Nucleic Acids Res. 2012; 40:928–938. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16. Fetrow J.S. Omega loops: nonregular secondary structures significant in protein function and stability. FASEB J. 1995; 9:708–717. [PubMed] [Google Scholar]
- 17. Yang W. An equivalent metal ion in one- and two-metal-ion catalysis. Nat. Struct. Mol. Biol. 2008; 15:1228–1231. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18. Mate M.J., Kleanthous C.. Structure-based analysis of the metal-dependent mechanism of H-N-H endonucleases. J. Biol. Chem. 2004; 279:34763–34769. [DOI] [PubMed] [Google Scholar]
- 19. Yang W. Nucleases: diversity of structure, function and mechanism. Q. Rev. Biophys. 2011; 44:1–93. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20. Shen B.W., Heiter D.F., Chan S.H., Wang H., Xu S.Y., Morgan R.D., Wilson G.G., Stoddard B.L.. Unusual target site disruption by the rare-cutting HNH restriction endonuclease PacI. Structure. 2010; 18:734–743. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21. Li C.L., Hor L.I., Chang Z.F., Tsai L.C., Yang W.Z., Yuan H.S.. DNA binding and cleavage by the periplasmic nuclease Vvn: a novel structure with a known active site. EMBO J. 2003; 22:4014–4025. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22. Niiranen L., Altermark B., Brandsdal B.O., Leiros H.K., Helland R., Smalas A.O., Willassen N.P.. Effects of salt on the kinetics and thermodynamic stability of endonuclease I from Vibrio salmonicida and Vibrio cholerae. FEBS J. 2008; 275:1593–1605. [DOI] [PubMed] [Google Scholar]
- 23. Mehta P., Katta K., Krishnaswamy S.. HNH family subclassification leads to identification of commonality in the His-Me endonuclease superfamily. Protein Sci. 2004; 13:295–300. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24. Finn R.D., Coggill P., Eberhardt R.Y., Eddy S.R., Mistry J., Mitchell A.L., Potter S.C., Punta M., Qureshi M., Sangrador-Vegas A. et al. . The Pfam protein families database: towards a more sustainable future. Nucleic Acids Res. 2016; 44:D279–D285. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25. Ginalski K., von Grotthuss M., Grishin N.V., Rychlewski L.. Detecting distant homology with Meta-BASIC. Nucleic Acids Res. 2004; 32:W576–W581. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26. Camacho C., Coulouris G., Avagyan V., Ma N., Papadopoulos J., Bealer K., Madden T.L.. BLAST+: architecture and applications. BMC Bioinformatics. 2009; 10:421. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27. Jones D.T. Protein secondary structure prediction based on position-specific scoring matrices. J. Mol. Biol. 1999; 292:195–202. [DOI] [PubMed] [Google Scholar]
- 28. Li W., Godzik A.. Cd-hit: a fast program for clustering and comparing large sets of protein or nucleotide sequences. Bioinformatics. 2006; 22:1658–1659. [DOI] [PubMed] [Google Scholar]
- 29. Su G., Morris J.H., Demchak B., Bader G.D.. Biological network exploration with Cytoscape 3. Curr. Protoc. Bioinformatics. 2014; 47, doi:10.1002/0471250953.bi0813s47. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30. Katoh K., Standley D.M.. MAFFT multiple sequence alignment software version 7: improvements in performance and usability. Mol. Biol. Evol. 2013; 30:772–780. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31. Eddy S.R. Accelerated Profile HMM Searches. PLoS Comput. Biol. 2011; 7:e1002195. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32. Krogh A., Larsson B., von Heijne G., Sonnhammer E.L.. Predicting transmembrane protein topology with a hidden Markov model: application to complete genomes. J. Mol. Biol. 2001; 305:567–580. [DOI] [PubMed] [Google Scholar]
- 33. Guex N., Peitsch M.C.. SWISS-MODEL and the Swiss-PdbViewer: an environment for comparative protein modeling. Electrophoresis. 1997; 18:2714–2723. [DOI] [PubMed] [Google Scholar]
- 34. Steczkiewicz K., Muszewska A., Knizewski L., Rychlewski L., Ginalski K.. Sequence, structure and functional diversity of PD-(D/E)XK phosphodiesterase superfamily. Nucleic Acids Res. 2012; 40:7016–7045. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35. Kuchta K., Knizewski L., Wyrwicz L.S., Rychlewski L., Ginalski K.. Comprehensive classification of nucleotidyltransferase fold proteins: identification of novel families and their representatives in human. Nucleic Acids Res. 2009; 37:7701–7714. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36. Moon A.F., Midon M., Meiss G., Pingoud A., London R.E., Pedersen L.C.. Structural insights into catalytic and substrate binding mechanisms of the strategic EndA nuclease from Streptococcus pneumoniae. Nucleic Acids Res. 2011; 39:2943–2953. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37. Moon A.F., Krahn J.M., Lu X., Cuneo M.J., Pedersen L.C.. Structural characterization of the virulence factor Sda1 nuclease from Streptococcus pyogenes. Nucleic Acids Res. 2016; 44:3946–3957. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38. Woo E.J., Kim Y.G., Kim M.S., Han W.D., Shin S., Robinson H., Park S.Y., Oh B.H.. Structural mechanism for inactivation and activation of CAD/DFF40 in the apoptotic pathway. Mol. Cell. 2004; 14:531–539. [DOI] [PubMed] [Google Scholar]
- 39. Chai J., Wu J.W., Yan N., Massague J., Pavletich N.P., Shi Y.. Features of a Smad3 MH1-DNA complex. Roles of water and zinc in DNA binding. J. Biol. Chem. 2003; 278:20327–20331. [DOI] [PubMed] [Google Scholar]
- 40. Klein A., Wojdyla J.A., Joshi A., Josts I., McCaughey L.C., Housden N.G., Kaminska R., Byron O., Walker D., Kleanthous C.. Structural and biophysical analysis of nuclease protein antibiotics. Biochem. J. 2016; 473:2799–2812. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41. Joshi A., Grinter R., Josts I., Chen S., Wojdyla J.A., Lowe E.D., Kaminska R., Sharp C., McCaughey L., Roszak A.W. et al. . Structures of the ultra-high-affinity protein-protein complexes of pyocins S2 and AP41 and their cognate immunity proteins from Pseudomonas aeruginosa. J. Mol. Biol. 2015; 427:2852–2866. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42. Chandler M., de la Cruz F., Dyda F., Hickman A.B., Moncalian G., Ton-Hoang B.. Breaking and joining single-stranded DNA: the HUH endonuclease superfamily. Nat. Rev. Microbiol. 2013; 11:525–538. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43. Grishin N.V. Mh1 domain of Smad is a degraded homing endonuclease. J. Mol. Biol. 2001; 307:31–37. [DOI] [PubMed] [Google Scholar]
- 44. Makkar P., Metpally R.P., Sangadala S., Reddy B.V.. Modeling and analysis of MH1 domain of Smads and their interaction with promoter DNA sequence motif. J. Mol. Graph. Model. 2009; 27:803–812. [DOI] [PubMed] [Google Scholar]
- 45. Hausmann J., Kamtekar S., Christodoulou E., Day J.E., Wu T., Fulkerson Z., Albers H.M., van Meeteren L.A., Houben A.J., van Zeijl L. et al. . Structural basis of substrate discrimination and integrin binding by autotaxin. Nat. Struct. Mol. Biol. 2011; 18:198–204. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46. Kaur G., Subramanian S.. Classification of the treble clef zinc finger: noteworthy lessons for structure and function evolution. Sci. Rep. 2016; 6:32070. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47. Jakubauskas A., Giedriene J., Bujnicki J.M., Janulaitis A.. Identification of a single HNH active site in type IIS restriction endonuclease Eco31I. J. Mol. Biol. 2007; 370:157–169. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48. Zhang D., de Souza R.F., Anantharaman V., Iyer L.M., Aravind L.. Polymorphic toxin systems: Comprehensive characterization of trafficking modes, processing, mechanisms of action, immunity and ecology using comparative genomics. Biol. Direct. 2012; 7:18. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49. Chen H., Choi J., Bailey S.. Cut site selection by the two nuclease domains of the Cas9 RNA-guided endonuclease. J. Biol. Chem. 2014; 289:13284–13294. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50. Mulligan E.A., Hatchwell E., McCorkle S.R., Dunn J.J.. Differential binding of Escherichia coli McrA protein to DNA sequences that contain the dinucleotide m5CpG. Nucleic Acids Res. 2010; 38:1997–2005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 51. Kriukiene E., Lubiene J., Lagunavicius A., Lubys A.. MnlI–The member of H-N-H subtype of Type IIS restriction endonucleases. Biochim. Biophys. Acta. 2005; 1751:194–204. [DOI] [PubMed] [Google Scholar]
- 52. Yusufzai T., Kadonaga J.T.. Annealing helicase 2 (AH2), a DNA-rewinding motor with an HNH motif. Proc. Natl. Acad. Sci. U.S.A. 2010; 107:20970–20973. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 53. Weston R., Peeters H., Ahel D.. ZRANB3 is a structure-specific ATP-dependent endonuclease involved in replication stress response. Genes Dev. 2012; 26:1558–1572. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 54. Kala S., Cumby N., Sadowski P.D., Hyder B.Z., Kanelis V., Davidson A.R., Maxwell K.L.. HNH proteins are a widespread component of phage DNA packaging machines. Proc. Natl. Acad. Sci. U.S.A. 2014; 111:6022–6027. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 55. Zhang L., Xu D., Huang Y., Zhu X., Rui M., Wan T., Zheng X., Shen Y., Chen X., Ma K. et al. . Structural and functional characterization of deep-sea thermophilic bacteriophage GVE2 HNH endonuclease. Sci. Rep. 2017; 7:42542. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 56. Sitbon E., Pietrokovski S.. New types of conserved sequence domains in DNA-binding regions of homing endonucleases. Trends Biochem. Sci. 2003; 28:473–477. [DOI] [PubMed] [Google Scholar]
- 57. Wessler S.R. Homing into the origin of the AP2 DNA binding domain. Trends Plant Sci. 2005; 10:54–56. [DOI] [PubMed] [Google Scholar]
- 58. Bair C.L., Rifat D., Black L.W.. Exclusion of glucosyl-hydroxymethylcytosine DNA containing bacteriophages is overcome by the injected protein inhibitor IPI. J. Mol. Biol. 2007; 366:779–789. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 59. Machnicka M.A., Kaminska K.H., Dunin-Horkawicz S., Bujnicki J.M.. Phylogenomics and sequence-structure-function relationships in the GmrSD family of Type IV restriction enzymes. BMC Bioinformatics. 2015; 16:336. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 60. Aravind L., Iyer L.M., Anantharaman V.. The two faces of Alba: the evolutionary connection between proteins participating in chromatin structure and RNA metabolism. Genome Biol. 2003; 4:R64. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 61. Szilagyi A., Skolnick J.. Efficient prediction of nucleic acid binding function from low-resolution protein structures. J. Mol. Biol. 2006; 358:922–933. [DOI] [PubMed] [Google Scholar]
- 62. Aoki S.K., Pamma R., Hernday A.D., Bickham J.E., Braaten B.A., Low D.A.. Contact-dependent inhibition of growth in Escherichia coli. Science. 2005; 309:1245–1248. [DOI] [PubMed] [Google Scholar]
- 63. Zhang D., Iyer L.M., Aravind L.. A novel immunity system for bacterial nucleic acid degrading toxins and its recruitment in various eukaryotic and DNA viral systems. Nucleic Acids Res. 2011; 39:4532–4552. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 64. Midon M., Schafer P., Pingoud A., Ghosh M., Moon A.F., Cuneo M.J., London R.E., Meiss G.. Mutational and biochemical analysis of the DNA-entry nuclease EndA from Streptococcus pneumoniae. Nucleic Acids Res. 2011; 39:623–634. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 65. Derre-Bobillot A., Cortes-Perez N.G., Yamamoto Y., Kharrat P., Couve E., Da Cunha V., Decker P., Boissier M.C., Escartin F., Cesselin B. et al. . Nuclease A (Gbs0661), an extracellular nuclease of Streptococcus agalactiae, attacks the neutrophil extracellular traps and is needed for full virulence. Mol. Microbiol. 2013; 89:518–531. [DOI] [PubMed] [Google Scholar]
- 66. Holberger L.E., Garza-Sanchez F., Lamoureux J., Low D.A., Hayes C.S.. A novel family of toxin/antitoxin proteins in Bacillus species. FEBS Lett. 2012; 586:132–136. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 67. Koskiniemi S., Lamoureux J.G., Nikolakakis K.C., t’Kint de Roodenbeke C., Kaplan M.D., Low D.A., Hayes C.S.. Rhs proteins from diverse bacteria mediate intercellular competition. Proc. Natl. Acad. Sci. U.S.A. 2013; 110:7032–7037. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 68. O’Connell Motherway M., O’Driscoll J., Fitzgerald G.F., Van Sinderen D.. Overcoming the restriction barrier to plasmid transformation and targeted mutagenesis in Bifidobacterium breve UCC2003. Microb. Biotechnol. 2009; 2:321–332. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 69. Lubys A., Lubiene J., Kulakauskas S., Stankevicius K., Timinskas A., Janulaitis A.. Cloning and analysis of the genes encoding the type IIS restriction-modification system HphI from Haemophilus parahaemolyticus. Nucleic Acids Res. 1996; 24:2760–2766. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 70. Cymerman I.A., Obarska A., Skowronek K.J., Lubys A., Bujnicki J.M.. Identification of a new subfamily of HNH nucleases and experimental characterization of a representative member, HphI restriction endonuclease. Proteins. 2006; 65:867–876. [DOI] [PubMed] [Google Scholar]
- 71. Sukackaite R., Grazulis S., Bochtler M., Siksnys V.. The recognition domain of the BpuJI restriction endonuclease in complex with cognate DNA at 1.3-A resolution. J. Mol. Biol. 2008; 378:1084–1093. [DOI] [PubMed] [Google Scholar]
- 72. Iyer L.M., Koonin E.V., Aravind L.. Classification and evolutionary history of the single-strand annealing proteins, RecT, Redbeta, ERF and RAD52. BMC Genomics. 2002; 3:8. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 73. Tarkowski T.A., Mooney D., Thomason L.C., Stahl F.W.. Gene products encoded in the ninR region of phage lambda participate in Red-mediated recombination. Genes Cells. 2002; 7:351–363. [DOI] [PubMed] [Google Scholar]
- 74. Dodd H.N., Pemberton J.M.. Cloning, sequencing, and characterization of the nucH gene encoding an extracellular nuclease from Aeromonas hydrophila JMP636. J. Bacteriol. 1996; 178:3926–3933. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 75. Wu S.I., Lo S.K., Shao C.P., Tsai H.W., Hor L.I.. Cloning and characterization of a periplasmic nuclease of Vibrio vulnificus and its role in preventing uptake of foreign DNA. Appl. Environ. Microbiol. 2001; 67:82–88. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 76. Nakamura A., Koide Y., Miyazaki H., Kitamura A., Masaki H., Beppu T., Uozumi T.. Gene cloning and characterization of a novel extracellular ribonuclease of Bacillus subtilis. Eur. J. Biochem. 1992; 209:121–127. [DOI] [PubMed] [Google Scholar]
- 77. Mei S., Zhang J., Zhang X., Tu X.. Solution structure of a bacterial immunoglobulin-like domain of the outer membrane protein (LigB) from Leptospira. Proteins. 2015; 83:195–200. [DOI] [PubMed] [Google Scholar]
- 78. Iyer L.M., Burroughs A.M., Aravind L.. Unraveling the biochemistry and provenance of pupylation: a prokaryotic analog of ubiquitination. Biol. Direct. 2008; 3:45. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 79. Li J., Yao Y., Xu H.H., Hao L., Deng Z., Rajakumar K., Ou H.Y.. SecReT6: a web-based resource for type VI secretion systems found in bacteria. Environ. Microbiol. 2015; 17:2196–2202. [DOI] [PubMed] [Google Scholar]
- 80. Malik H.S., Henikoff S.. Dual recognition-incision enzymes might be involved in mismatch repair and meiosis. Trends Biochem. Sci. 2000; 25:414–418. [DOI] [PubMed] [Google Scholar]
- 81. Pediaditakis M., Kaufenstein M., Graumann P.L.. Bacillus subtilis hlpB encodes a conserved stand-alone HNH nuclease-like protein that is essential for viability unless the hlpB deletion is accompanied by the deletion of genes encoding the AddAB DNA repair complex. J. Bacteriol. 2012; 194:6184–6194. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 82. Matsuura M., Saldanha R., Ma H., Wank H., Yang J., Mohr G., Cavanagh S., Dunny G.M., Belfort M., Lambowitz A.M.. A bacterial group II intron encoding reverse transcriptase, maturase, and DNA endonuclease activities: biochemical demonstration of maturase activity and insertion of new genetic information within the intron. Genes Dev. 1997; 11:2910–2924. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 83. Curtis F.A., Malay A.D., Trotter A.J., Wilson L.A., Barradell-Black M.M., Bowers L.Y., Reed P., Hillyar C.R., Yeo R.P., Sanderson J.M. et al. . Phage ORF family recombinases: conservation of activities and involvement of the central channel in DNA binding. PLoS One. 2014; 9:e102454. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 84. Kato K., Nishimasu H., Okudaira S., Mihara E., Ishitani R., Takagi J., Aoki J., Nureki O.. Crystal structure of Enpp1, an extracellular glycoprotein involved in bone mineralization and insulin signaling. Proc. Natl. Acad. Sci. U.S.A. 2012; 109:16876–16881. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 85. Zimmermann H., Zebisch M., Strater N.. Cellular function and molecular structure of ecto-nucleotidases. Purinergic Signal. 2012; 8:437–502. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 86. Stein A.J., Bain G., Prodanovich P., Santini A.M., Darlington J., Stelzer N.M., Sidhu R.S., Schaub J., Goulet L., Lonergan D. et al. . Structural basis for inhibition of human autotaxin by four potent compounds with distinct modes of binding. Mol. Pharmacol. 2015; 88:982–992. [DOI] [PubMed] [Google Scholar]
- 87. Stoynov S.S., Bakalova A.T., Dimov S.I., Mitkova A.V., Dolapchiev L.B.. Single-strand-specific DNase activity is an inherent property of the 140-kDa protein of the snake venom exonuclease. FEBS Lett. 1997; 409:151–154. [DOI] [PubMed] [Google Scholar]
- 88. Gross M., Marianovsky I., Glaser G.. MazG – a regulator of programmed cell death in Escherichia coli. Mol. Microbiol. 2006; 59:590–601. [DOI] [PubMed] [Google Scholar]
- 89. Sharples G.J., Corbett L.M., McGlynn P.. DNA structure specificity of Rap endonuclease. Nucleic Acids Res. 1999; 27:4121–4127. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 90. Hayes S., Asai K., Chu A.M., Hayes C.. NinR- and red-mediated phage-prophage marker rescue recombination in Escherichia coli: recovery of a nonhomologous immlambda DNA segment by infecting lambdaimm434 phages. Genetics. 2005; 170:1485–1499. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 91. Cascales E. The type VI secretion toolkit. EMBO Rep. 2008; 9:735–741. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 92. Salomon D., Kinch L.N., Trudgian D.C., Guo X., Klimko J.A., Grishin N.V., Mirzaei H., Orth K.. Marker for type VI secretion system effectors. Proc. Natl. Acad. Sci. U.S.A. 2014; 111:9271–9276. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 93. Belfort M. Homing Endonucleases and Inteins. 2005; Berlin; NY: Springer. [Google Scholar]
- 94. Hsia K.C., Chak K.F., Liang P.H., Cheng Y.S., Ku W.Y., Yuan H.S.. DNA binding and degradation by the HNH protein ColE7. Structure. 2004; 12:205–214. [DOI] [PubMed] [Google Scholar]
- 95. Jinek M., Jiang F., Taylor D.W., Sternberg S.H., Kaya E., Ma E., Anders C., Hauer M., Zhou K., Lin S. et al. . Structures of Cas9 endonucleases reveal RNA-mediated conformational activation. Science. 2014; 343:1247997. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 96. Nishimasu H., Ran F.A., Hsu P.D., Konermann S., Shehata S.I., Dohmae N., Ishitani R., Zhang F., Nureki O.. Crystal structure of Cas9 in complex with guide RNA and target DNA. Cell. 2014; 156:935–949. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 97. Luscombe N.M., Laskowski R.A., Thornton J.M.. Amino acid-base interactions: a three-dimensional analysis of protein-DNA interactions at an atomic level. Nucleic Acids Res. 2001; 29:2860–2874. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 98. Rohs R., West S.M., Sosinsky A., Liu P., Mann R.S., Honig B.. The role of DNA shape in protein-DNA recognition. Nature. 2009; 461:1248–1253. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 99. Eastberg J.H., Eklund J., Monnat R. Jr, Stoddard B.L.. Mutability of an HNH nuclease imidazole general base and exchange of a deprotonation mechanism. Biochemistry. 2007; 46:7215–7225. [DOI] [PubMed] [Google Scholar]
- 100. Lin Y.H., Liao C.C., Liang P.H., Yuan H.S., Chak K.F.. Involvement of colicin in the limited protection of the colicin producing cells against bacteriophage. Biochem. Biophys. Res. Commun. 2004; 318:81–87. [DOI] [PubMed] [Google Scholar]
- 101. Cheng Y.S., Hsia K.C., Doudeva L.G., Chak K.F., Yuan H.S.. The crystal structure of the nuclease domain of colicin E7 suggests a mechanism for binding to double-stranded DNA by the H-N-H endonucleases. J. Mol. Biol. 2002; 324:227–236. [DOI] [PubMed] [Google Scholar]
- 102. Gruber A.J., Olsen T.M., Dvorak R.H., Cox M.M.. Function of the N-terminal segment of the RecA-dependent nuclease Ref. Nucleic Acids Res. 2015; 43:1795–1803. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 103. Ronayne E.A., Wan Y.C., Boudreau B.A., Landick R., Cox M.M.. P1 Ref endonuclease: a molecular mechanism for phage-enhanced antibiotic lethality. PLoS Genet. 2016; 12:e1005797. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 104. Ball T.K., Suh Y., Benedik M.J.. Disulfide bonds are required for Serratia marcescens nuclease activity. Nucleic Acids Res. 1992; 20:4971–4974. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 105. Ball T.K., Saurugger P.N., Benedik M.J.. The extracellular nuclease gene of Serratia marcescens and its secretion from Escherichia coli. Gene. 1987; 57:183–192. [DOI] [PubMed] [Google Scholar]
- 106. Wheeler T.J., Clements J., Finn R.D.. Skylign: a tool for creating informative, interactive logos representing sequence alignments and profile hidden Markov models. BMC Bioinformatics. 2014; 15:7. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 107. Zhang L., Huang Y., Xu D., Yang L., Qian K., Chang G., Gong Y., Zhou X., Ma K.. Biochemical characterization of a thermostable HNH endonuclease from deep-sea thermophilic bacteriophage GVE2. Appl. Microbiol. Biotechnol. 2016; 100:8003–8012. [DOI] [PubMed] [Google Scholar]
- 108. Bibb L.A., Hancox M.I., Hatfull G.F.. Integration and excision by the large serine recombinase phiRv1 integrase. Mol. Microbiol. 2005; 55:1896–1910. [DOI] [PubMed] [Google Scholar]
- 109. Anton B.P., Raleigh E.A.. Transposon-mediated linker insertion scanning mutagenesis of the Escherichia coli McrA endonuclease. J. Bacteriol. 2004; 186:5699–5707. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 110. Akulenko N.V., Ivashina T.V., Shaloiko L.A., Shliapnikov M.G., Ksenzenko V.N.. Novel site-specific endonucleases F-TflI, F-TflII and F-TflIV encoded by the bacteriophage T5. Mol. Biol. (Mosk.). 2004; 38:632–641. [PubMed] [Google Scholar]
- 111. Magnani E., Sjolander K., Hake S.. From endonucleases to transcription factors: evolution of the AP2 DNA binding domain in plants. Plant Cell. 2004; 16:2265–2277. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 112. Haugen P., Reeb V., Lutzoni F., Bhattacharya D.. The evolution of homing endonuclease genes and group I introns in nuclear rDNA. Mol. Biol. Evol. 2004; 21:129–140. [DOI] [PubMed] [Google Scholar]
- 113. Wojdyla J.A., Fleishman S.J., Baker D., Kleanthous C.. Structure of the ultra-high-affinity colicin E2 DNase–Im2 complex. J. Mol. Biol. 2012; 417:79–94. [DOI] [PubMed] [Google Scholar]
- 114. Kushner V.D. Dispensary care of nephrological patients at a pediatric polyclinic. Pediatriia. 1982; 64–65. [PubMed] [Google Scholar]
- 115. Orlowski J., Bujnicki J.M.. Structural and evolutionary classification of Type II restriction enzymes based on theoretical and experimental analyses. Nucleic Acids Res. 2008; 36:3552–3569. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 116. Wong B.C., Yin Y., Berg D.E., Xia H.H., Zhang J.Z., Wang W.H., Wong W.M., Huang X.R., Tang V.S., Lam S.K.. Distribution of distinct vacA, cagA and iceA alleles in Helicobacter pylori in Hong Kong. Helicobacter. 2001; 6:317–324. [DOI] [PubMed] [Google Scholar]
- 117. Shiota S., Watada M., Matsunari O., Iwatani S., Suzuki R., Yamaoka Y.. Helicobacter pylori iceA, clinical outcomes, and correlation with cagA: a meta-analysis. PLoS One. 2012; 7:e30354. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 118. Guthrie E.P., Quinton-Jager T., Moran L.S., Slatko B.E., Kucera R.B., Benner J.S., Wilson G.G., Brooks J.E.. Cloning, expression and sequence analysis of the SphI restriction-modification system. Gene. 1996; 180:107–112. [DOI] [PubMed] [Google Scholar]
- 119. Bitinaite J., Mitkaite G., Dauksaite V., Jakubauskas A., Timinskas A., Vaisvila R., Lubys A., Janulaitis A.. Evolutionary relationship of Alw26I, Eco31I and Esp3I, restriction endonucleases that recognise overlapping sequences. Mol. Genet. Genomics. 2002; 267:664–672. [DOI] [PubMed] [Google Scholar]
- 120. Zhang D., Burroughs A.M., Vidal N.D., Iyer L.M., Aravind L.. Transposons to toxins: the provenance, architecture and diversification of a widespread class of eukaryotic effectors. Nucleic Acids Res. 2016; 44:3513–3533. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.